Abstract
Volatile anesthetics have been in clinical use for a long period of time and are considered to be promiscuous by presumably interacting with several ion channels in the central nervous system to produce anesthesia. Because ion channels and their existing evolutionary analogues, ion transporters, are very important in various organisms, it is possible that volatile anesthetics may affect some bacteria. In this study, we hypothesized that volatile anesthetics could affect bacterial behaviors. We evaluated the impact of anesthetics on bacterial growth, motility (swimming and gliding) and biofilm formation of four common bacterial pathogens in vitro. We found that commonly used volatile anesthetics isoflurane and sevoflurane affected bacterial motility and biofilm formation without any effect on growth of the common bacterial pathogens studied here. Using available Escherichia coli gene deletion mutants of ion transporters and in silico molecular docking, we suggested that these altered behaviors might be at least partly via the interaction of volatile anesthetics with ion transporters.
Introduction
Anesthesia is a critical component of surgical procedures, and volatile anesthetics have been the mainstay of drugs for more than a century. Although the anesthetic mechanism of volatile anesthetics has not been completely delineated yet, the general consensus is that they are promiscuous and work by directly interacting with several ion channels in the central nervous system [1]. Intravenous anesthetics also work via ion channels but have more specific targets, such as the anesthetic effect of propofol via gamma-aminobutyric acid type A (GABAA) receptors [2]. The primary goal of anesthesia providers is to make patients comfortable during surgery, and the effect of anesthetics on different organisms such as bacteria is not usually taken into consideration in the clinical setting. However, it is not rare to anesthetize patients with active bacterial infections. Changes in bacterial behavior after exposure to anesthetics at clinically relevant concentrations, if any, could have significant, clinical implications.
The interest in the effect of anesthetics on bacteria is not new, but studies are rather limited. Previous research solely studied the effect of anesthetics on bacterial growth, as the association between the severity of bacterial infection and higher bacterial load is intuitively clear, and has been supported by the literature [3–5]. Most of the previous studies suggest that volatile anesthetics at clinically relevant concentrations do not affect bacterial growth as summarized in Table 1[6–10]. In contrast, the effect of anesthetics on other bacterial behaviors including motility and biofilm formation has seldom been studied. Many bacteria are generally motile and some bacteria swim in liquid or swarm over solid surfaces using flagella [11]. While swarming is a form of bacterial movement in a group, swimming is an individual bacterial movement [12]. In addition, some bacteria can glide over solid surfaces without flagella. Bacteria can also accumulate and form non-motile multicellular aggregates. These aggregates of bacteria connected by extracellular polysaccharides are called biofilm. Biofilm can be more resistant to antibiotics than planktonic bacteria cells and contributes to persistent infection [13].
Table 1. The previous studies of effects of anesthetics on bacterial growth.
| Drug tested | Bacteria | Duration of exposure | Results | References |
|---|---|---|---|---|
| 3% Sevoflurane, 60% Nitrous oxide | Pseudomonas aeruginosa Acinetobacter lwoffii, Staphylococcus aureus | 3 hrs | No change in S.aureus, very small increase in A.lwoffii and P.aeruginosa under 3% sevoflurane, more than 2 fold increase in A.lwoffii and P. aeruginosa under 60% nitrous oxide | [10] |
| Isoflurane (2%) | Escherichia coli, Staphylococcus aurerus | 2 hrs | No chance | [9] |
| Isoflurane (21–25%) | Staphylococcus aureus, Streptococcus pneumoniae, coliform bacteria | 16 hrs | No effect | [8] |
| halothane (2–5%) | Staphylococcus aureus, Escherichia coli | 4 hrs | No effect | [7] |
| Isoflurane (1.1–2.3%), halothane (0.8–1.5%) | Psedumonas aeruginosa | 4 hrs | Reduction in growth under both isoflurane and halothane | [14] |
| Halothane (1–10%) | Escherichia coli, Bacillus licheniformis, Staphylococcus albus, Micrococcus lysodeikticus | 24 hrs | Effect seen only at > 5% of halothane | [6] |
Here we examined the effect of three commonly used anesthetics isoflurane, sevoflurane and propofol on four bacteria species; Gram-positive Staphylococcus aureus (S. aureus) and Enterococcus faecalis (E. faecalis) and Gram-negative Escherichia coli (E.coli) and Pseudomonas aeruginosa (P. aeruginosa). S. aureus and E. faecalis are non-flagellated bacteria and can form biofilm [15, 16]. S. aureus is known to glide [17], while E. faecalis is non-motile [18]. E. coli and P. aeruginosa are flagellated bacteria with swimming motility and also form biofilm [19–21]. First, we tested bacterial growth. Then, we tested the effect of anesthetics on swimming motility capability of E. coli and P. aeruginosa, on gliding motility of S. aureus and on biofilm formation in all four bacteria.
Materials and Methods
Bacterial species and plasmids
We used E. coli (K12 strain), S. aurerus (Newman strain), E. faecalis (strain 12030), and P. aeruginosa (strain PA14). E.coli K12 strains were purchased from E. coli Genetic Stock Center (CGSC; New Haven, CT, USA)[22]. E. faecalis and P. aeruginosa strains were kindly provided by Dr. Gregory Priebe (Boston Children’s Hospital, Boston, MA, USA). S. aureus Newman strain was kindly provided by Dr. Timothy Foster (Trinity College Dublin, Dublin, Ireland). In addition, plasmids used here (pKD3, pKD13, pKD46 and pCP20) [22] were purchased from E. coli Genetic Stock Center. All the bacteria used here were shown in Table 2.
Table 2. Bacterial strains used in the study.
| Bacteria strains or plasmids | Description of genotype | Reference |
|---|---|---|
| Bacteria | ||
| E. coli K12 (MG1655) | E. coli K12 Parent strain, wild-type | [22] |
| E. coli K12 (JW5012) | clcA deletion mutant, KanR | [22] |
| E. coli K12 (JW5263) | clcB deletion mutant, KanR | [22] |
| E. coli K12 (JW0018) | nhaA deletion mutant, KanR | [22] |
| E. coli K12 (JW1175) | nhaB deletion mutant, KanR | [22] |
| E. coli K12 (JW1207) | chaA deletion mutant, KanR | [22] |
| E. coli K12 (JW3313) | kefB deletion mutant, KanR | [22] |
| E. coli K12 (JW0046) | kefC deletion mutant, KanR | [22] |
| E. coli K12 (JW3251) | trkA deletion mutant, KanR | [22] |
| E. coli K12 (JW0454) | mscK deletion mutant, KanR | [22] |
| E. coli K12 (JW1908) | fliC deletion mutant, KanR | [22] |
| E. coli K12 (Triple mutant) | nhaA, nhaB, clcB deletion mutant, KanR, CatR | This study |
| E. coli K12 (double mutant 1) | nhaA, nhaB deletion mutant, KanR | This study |
| E. coli K12 (double mutant 2) | nhaA, clcB deletion mutant, KanR, CatR | This study |
| S. aureus Newman | CC8 clinical isolates | TF |
| E. faecalis 12030 | Clinical isolates | GP |
| P. aeruginosa PA14 | Multi host pathogen | GP |
| Plasmid | ||
| pCP20 | Fln recombinase gene (+) | [22] |
| pKD3 | Cat resistance gene flanked by FRT | [23] |
| pKD13 | Kan resistance gene flanked by FRT | [23] |
| pKD46 | Lambda red recombinase expression plasmid | [23] |
*GP = obtained from Dr. Gregory Priebe (Boston Children’s Hospital), TF = obtained from Dr. Timthy Foster (Ireland)
Bacterial growth
Overnight cultures of all four bacterial species were diluted with tryptic soy broth (TBS) to the optical density 600 (OD600) of 0.05. Bacteria were grown at 37°C for 4 or 6 hours with or without anesthetics. Volatile anesthetics were exposed to bacteria using an airtight chamber at clinically relevant concentrations as indicated in each experiment. Propofol was dissolved in dimethyl sulfoxide (DMSO). The final concentration of DMSO in propofol sample and its control was 0.1%. The concentration of propofol used in this study (50 μM) is significantly above the clinically relevant concentration [24]. After bacterial culture for the indicated time, samples were serially diluted, and plated on tryptic soy agar (TSA) plates and incubated at 37°C overnight for determination of colony forming units (CFU)/mL.
Swimming assay on agar plate
A 10-μL drop of overnight E. coli and P. aeruginosa cultures were added to plates consisting of lysogeny broth (LB) with 0.3% agar. E. coli was incubated for 12 hours and P. aeruginosa was incubated for 24 hours at 37°C with or without anesthetics. At the end of incubation, pictures of the swimming formation were taken and swimming area was measured using Image J (National Institutes of Health, Besthesda, MD, USA). Isoflurane and sevoflurane were exposed in an air-tight chamber. Control samples were also incubated in an airtight chamber to match the condition.
Swimming examination under microscope
E. coli K12 cultures were diluted into 2% of poly-ethylene glycol (PEG) aqueous solution as previously described [25]. A group of E. coli cells were exposed to isoflurane for 5 minutes. Movement was recorded and tracked using MetaMorph software (Molecular Devices, Sunnyvale, CA, USA).
Flagellin expression analysis using Western blot
We used E.coli K12 parent strains and mutants lysates to test flagellin expression. Some of E.coli parent strain cells were exposed to isoflurane for indicated durations. E. coli cells were suspended in 10mM Tris-HCl, pH 8.0, 100 mM NaCl, 1 mM MgCl2 as previously described [26]. After 10-second of sonication, the concentration of bacterial lysates was measured using BCA protein assay (Thermo Fisher Scientific, Waltham, MA, USA). Samples of the same weight were loaded on SDS-PAGE gel and blotted to nitrocellulose membrane. The membranes were probed with rabbit anti-flagellin antibody (abcam, Cambridge, UK) or goat anti-glyceraldehyde 3-phosphate dehydrogenase (GAPDH) antibody (Genscript, Piscataway, NJ, USA). Anti-rabbit IgG-horseradish peroxidase (HRP) conjugate (Cell Signaling, Danvers, MA, USA) or anti-goat IgG-HRP conjugate (Thermo Fischer Scientific) was used as the secondary antibody, respectively. Signal was detected with chemoluminescence (Thermo Scientific).
S. aureus gliding assay
A 10-μL drop of overnight S. aureus cultures were added to 0.3% LB agar plates with or without anesthetics for 19 hours. At the end of incubation, pictures of the gliding formations were taken and their areas were measured using Image J.
Biofilm assay
Following overnight growth of bacteria at 37°C, 100 μL of bacteria at OD600 of 0.05 was aliquoted to 96-well U-bottom plates and incubated at 37°C for 48 hours with or without anesthetics as previously described [27, 28]. Isoflurane and sevoflurane were exposed to samples as described in the swimming assay using an air-tight chamber. Control samples were also incubated in an air-tight chamber to match the condition. Wells were then washed and stained with 150 μL of 0.1% of crystal violet. Following incubation at room temperature for 10 minutes, plates were rinsed with water and dried for a few hours. Then, 30% of acetic acid was added to each well to solubilize the crystal violet, which was transferred to a new flat-bottomed plate. Absorbance was read at 590 nm using Versamax Tunable Microplate Reader (Molecular Devices, Sunnyvale, CA, USA).
Search of E. coli ion transporters
EcoCyc (http://ecocyc.org) [29] was used to search for ion transporters in E.coli K12 MG1655 strain.
Gene deletion
Gene deletion was performed as previously desribed [23]. clcB deletion mutant was transformed with pCP20 plasmid to remove kanamycin resistant cassette. Then, cells were transformed with pKD46. Red recombinase was induced with L-arabinose (Fischer scientific), and then competent cells were prepared and transformed with PCR products for nhaB gene deletion. PCR product was made using pCD13 as a template to have kanamycin resistant cassette. Similarly, PCR products made using pKD3 as a template to delete nhaA gene with chloramphenicol cassette were transformed to make nhaA(-)nhaB(-)clcB(-) mutant. Primer sets was refered to the paper by Baba et al [22].
Docking of isoflurane onto E.coli protein NhaA
Using previously reported structures in Protein Data Bank (PDB) (http://www.rcsb.org) of NhaA protein (PDB ID: 1ZCD), we performed the docking of isoflurane. Autodock software (The Scripps Research Institute; La Jolla CA, USA) was used as a docking software to identify the top ranking docked site of isoflurane.
Statistical analysis
All the statistical analyses were performed using Prism 6 software (GraphPad Software, Inc., La Jolla, CA, USA). Statistical analyses used were included in the corresponding figure legends. P < 0.05 was considered to be statistically significant.
Results
Isoflurane, sevoflurane and propofol did not affect bacterial growth
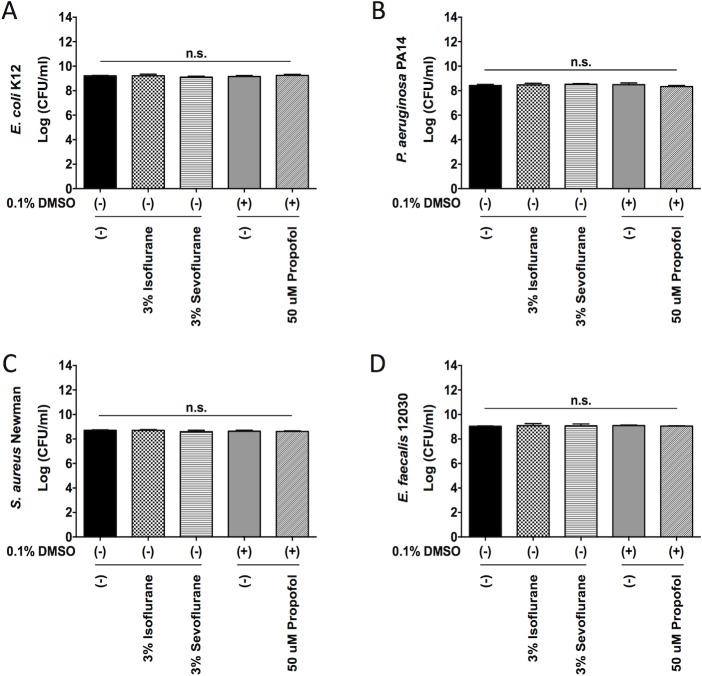
First, we tested the effect of isoflurane, sevoflurane at clinical relevant concentrations and propofol at a supraclinical concentration on the growth of four bacterial species. We used TSB as a growth media, which is considered as a medium rich in nutrition and protein content, similar to some human exudates [14]. We did not observe any significant difference in bacterial growth between the anesthetics-exposed group and the non-exposed group (Fig 1). This is in line with the majority of available literature demonstrating that volatile anesthetics at clinically relevant concentrations do not affect bacterial growth (Table 1).
Fig 1. The effect of anesthetics on bacterial growth.
The growth of E. coli (A), P. aeruginosa (B), S. aureus (C), and E. faecalis (D) under isoflurane, sevoflurane or propofol at various concentrations is shown. Data are shown as mean +/- S.D. of 4 replicates. Statistical analysis was performed using one-way analysis of variance with Bonferroni post hoc analysis. No statistical significance was noted. n.s. = not significant. CFU = colony forming unit.
Isoflurane and sevoflurane diminished bacterial swimming
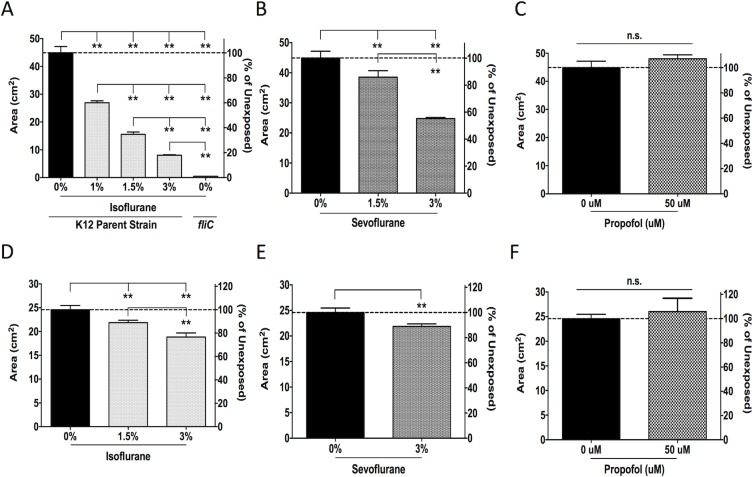
We tested the effect of anesthetics on swimming of flagellated bacteria E.coli and P. aeruginosa. Both isoflurane and sevoflurane significantly reduced E.coli swimming at all the concentrations tested. The reduction in swimming was dose-dependent and treatment with 3% isoflurane and sevoflurane resulted in 80% and 50% reduction compared to untreated E. coli, respectively (Figs 2A and 2B and S1). The fliC deletion mutant served as a non-motile control strain (Fig 2A). Propofol, on the other hand, did not affect E. coli swimming (Figs 2C and S1). The swimming of P. aeruginosa was also reduced by isoflurane and sevoflurane, but not by propofol (Figs 2D–2F and S1). The effect of isoflurane and sevoflurane on P. aeruginosa swimming was more modest than that on E. coli.
Fig 2. The effect of anesthetics on bacterial swimming.
Areas (cm2) of motility halos formed by E. coli K12 parent strain (A-C) and P. aeruginosa (D-F) exposed to isoflurane, sevoflurane or propofol at various concentrations are shown. E. coli K12 fliC deletion mutant, which lacks flagellin, was included as a control strain with no motility in (A). Data shown as mean ± SD; n = 4 for all conditions. Statistical analysis was performed using one-way analysis of variance with Bonferroni post hoc analysis for A, B and D. C, E, and F were analyzed using an unpaired t-test. * and ** denote p < 0.05 and p < 0.01, respectively. n.s. = not significant.
Isoflurane attenuated E.coli motility without the reduction of flagellin expression
We also evaluated the movement of E. coli under microscope. A brief exposure to isoflurane attenuated E. coli movement (Fig 3A), suggesting that the attenuation of E. coli motility by isoflurane occurred immediately and was unlikely to be caused by the change of protein expression such as flagellin expression. As expected, isoflurane exposure did not attenuate flagellin expression (Fig 3B).
Fig 3. The effect of isoflurane on E. coli motility and flagellin expression.
(A) The motility of E. coli K12 parent strain was observed in 2% PEG aqueous solution. A group of E. coli cells were exposed to isoflurane for 5 minutes before subjecting to microscope analysis. Movement over 10 second was tracked using MetaMorph software. (B) Flagellin expression was examined with or without isoflurane exposure for the indicated durations. GAPDH was used as a loading control [46]. fliC deletion mutant was used as a negative control of flagellin expression
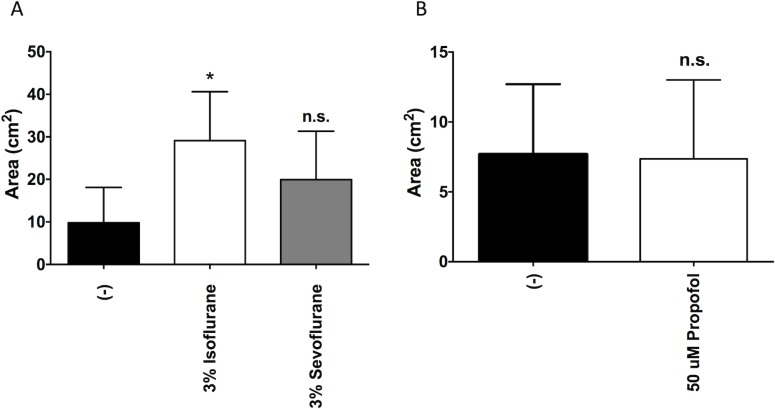
Isoflurane enhanced S. aureus motility
S. aureus is a non-flagellated bacteria but moves by gliding [17]. We found that isoflurane significantly increased motility of S. aureus approximately 3-fold, but sevoflurane and propofol did not (Fig 4).
Fig 4. The effect of anesthetics on S. aureus gliding.
Areas (cm2) of gliding formation of Newman S. aureus after exposure to isoflurane or sevoflurane (A) or propofol (B) for 19 hours are shown. Data shown as mean ± SD; n = 4 for A; n = 8 for B. Statistical analysis was performed using one-way analysis of variance with Bonferroni post hoc analysis for A. B was analyzed using an unpaired t-test. * denotes p<0.05. n.s. = not significant.
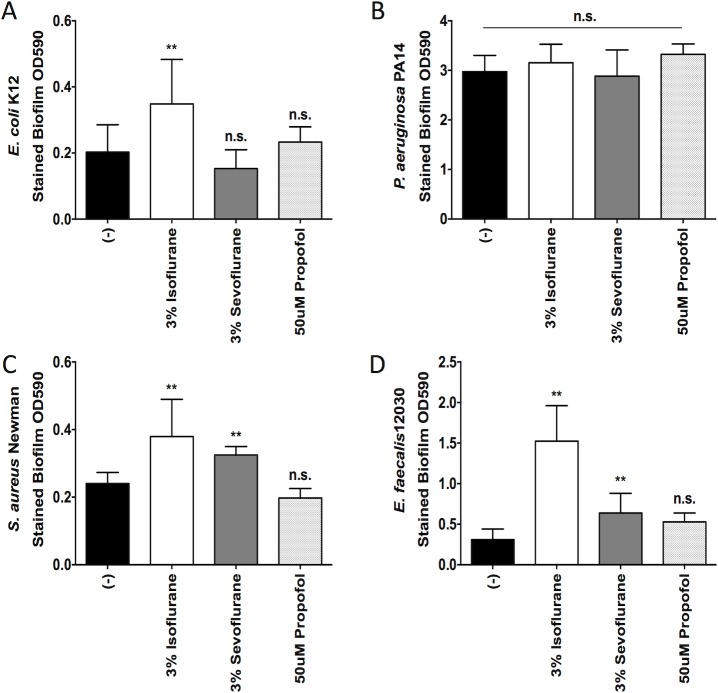
Isoflurane and sevoflurane exposure increased bacterial biofilm formation
We also evaluated the effect of anesthetics on biofilm formation by all four bacteria. P. aeruginosa, S. aureus and E. faecalis showed significantly more biofilm formation than E. coli (Fig 5). We found that isoflurane enhanced biofilm formation of S. aureus and E. faecalis by about 60% and 390% respectively, not P. aeruginosa (Fig 5). Isoflurane also enhanced biofilm formation by E. coli by 73%. In addition, sevoflurane enhanced biofilm formation by only S. aureus and E. faecalis by about 33% and 100% respectively. Propofol had no significant effect on biofilm formation.
Fig 5. The effect of anesthetics on bacterial biofilm formation.
The biofilm formation of E. coli (A), P. aeruginosa (B), S. aureus (C), and E. faecalis (D) after exposure to isoflurane, sevoflurane or propofol is shown. Data are shown as mean +/- S.D. of 8–12 replicates. Statistical analysis was performed using one-way analysis of variance with Bonferroni post hoc analysis. * and ** denote p < 0.05 and p < 0.01 of non-exposure vs exposure, respectively. n.s. = not significant.
The potential interaction of isoflurane with E. coli ion transporters
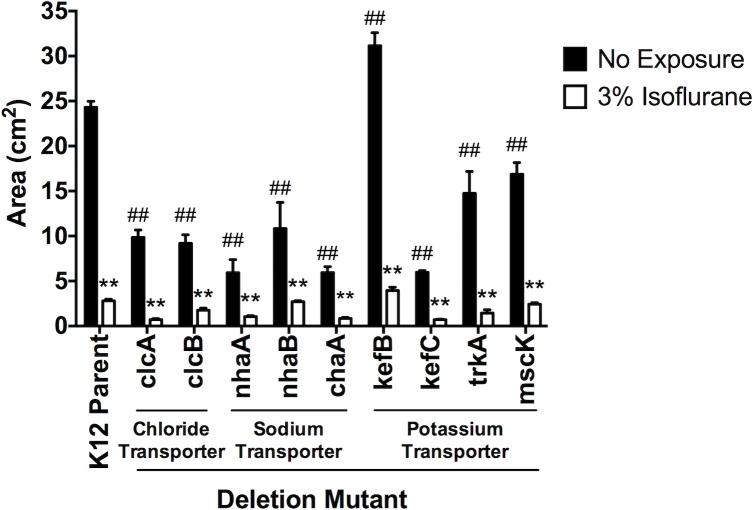
Mammalian ion channels and bacterial ion transporters are shown to be evolutionarily connected [30]. Because volatile anesthetics isoflurane and sevoflurane are considered to act via ion channels in the central nervous system, we hypothesized that they affected bacterial behaviors via their ion transporters. The genome of E. coli K12 strain has been completely sequenced [31], and a library of single gene deletion mutants has been created in this strain [22]. We identified nine ion transporters in E.coli K12 strain using EcoCyc database (Table 3) and we tested our hypothesis using nine ion transporter deletion mutants with or without isoflurane exposure. Because isoflurane reduced swimming of E. coli and we hypothesized that isoflurane would interact with ion transporters in E. coli, we predicted some of these ion transporter deletion mutants would show defect in swimming. As predicted, we found that the majority of ion transporter deletion mutants showed defect in swimming (Fig 6). In addition, these ion transporter mutants did not show any reduction in flagellin expression (S2 Fig). The majority of mutants had higher flagellin expression than that of parent strain. This result suggested that their swimming defect was not due to defect in flagellin expression. It is a general consensus that anesthetic effect by volatile anesthetics is through their interaction with multiple ion channels [32]. Therefore, we predicted that swimming impairment by isoflurane would derive from its interaction with multiple ion transporters. The addition of isoflurane to most of these mutants further attenuated swimming, supporting our prediction that multiple isoflurane target sites would exist in E. coli (Fig 6). When we evaluated the percentage of reduction in swimming under isoflurane, clcB, nhaA and nhaB deletion mutants showed less reduction by isoflurane than the parent strain (parent strain 88.5%, clcB(-) 80.8%, nhaA(-) 82.4%, and nhaB(-) 75.0%)(S3 Fig), suggesting that these three ion transporters could be isoflurane targets. We tested swimming of double and triple mutants. They swam significantly less than K12 parent strain (S4A Fig), suggesting that they were critical for swimming. When we compared the effect of isoflurane on clcB(-)nhaA(-)nhaB(-) mutant, swimming of this triple mutant was less reduced under isoflurane (parent strain 91.5%, triple deletion 6.4%), suggesting that they would be likely targets for isoflurane (S4B Fig). However, the reduction of the motility by isoflurane may be difficult to estimate because the triple mutant cells were already nearly non-motile without isoflurane.
Table 3. Known ion transporters in E.coli K12.
| Classification | Gene name |
|---|---|
| Chloride ion transporter | clcA |
| clcB | |
| Sodium ion transporter | nhaA |
| nhaB | |
| chaA | |
| Potassium ion transporter | MscK |
| KefB | |
| KefC | |
| trkA |
Fig 6. The effect of isoflurane on swimming of E. coli K12 parent strain and ion transporter deletion mutants.
The effect of isoflurane on the swimming of E. coli K12 strains is shown. Data are shown as mean +/- S.D. of 4 replicates. Statistical analysis was performed using two-way analysis of variance with Bonferroni post hoc analysis for differences between non-exposure and exposure groups. * and ** denote p<0.05 and p<0.01, respectively. For differences between deletion mutants and the K12 parent strain within the non-exposure group, statistical analysis was performed using one-way analysis of variance with Bonferroni post hoc analysis. # and ## denote p<0.05 and p<0.01, respectively.
Isoflurane increased biofilm formation in E.coli K12
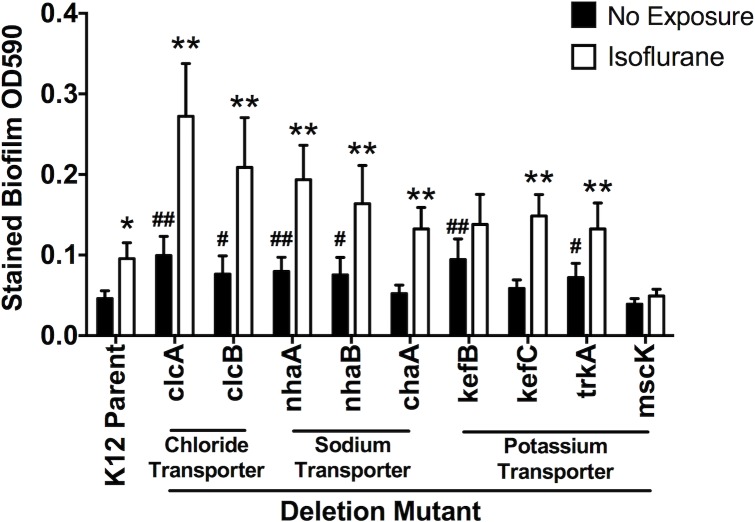
Because isoflurane increased biofilm formation by E. coli, we predicted that some of ion transporter deletion mutants would show increased biofilm formation than its parent strain (Fig 7). We found that clcA, clcB, nhaA, nhaB, kefB, and trkA deletion mutants had higher biofilm formation than its parent strain, and isoflurane further enhanced biofilm formation by these mutants (Fig 7). In addition, none of these mutants did not have any defect in growth (S5 Fig). Isoflurane targets are likely to be ones that are involved in both swimming and biofilm formation, not growth, and clcB, nhaA, and nhaB fitted for this category.
Fig 7. The effect of isoflurane on biofilm formation of E. coli K12 parent strain and ion transporter deletion mutants.
The effect of isoflurane on biofilm formed by E. coli K12 strains is shown. Data are shown as mean +/- S.D. of 8 replicates. Statistical analysis was performed using two-way analysis of variance with Bonferroni post hoc analysis for differences between non-exposure and exposure groups. * and ** denote p<0.05 and p<0.01, respectively. For differences between deletion mutants and the K12 parent strain within the non-exposure group, statistical analysis was performed using one-way analysis of variance with Bonferroni post hoc analysis. # and ## denote p<0.05 and p<0.01, respectively.
Isoflurane has predicted binding site in ion transporters in E. coli
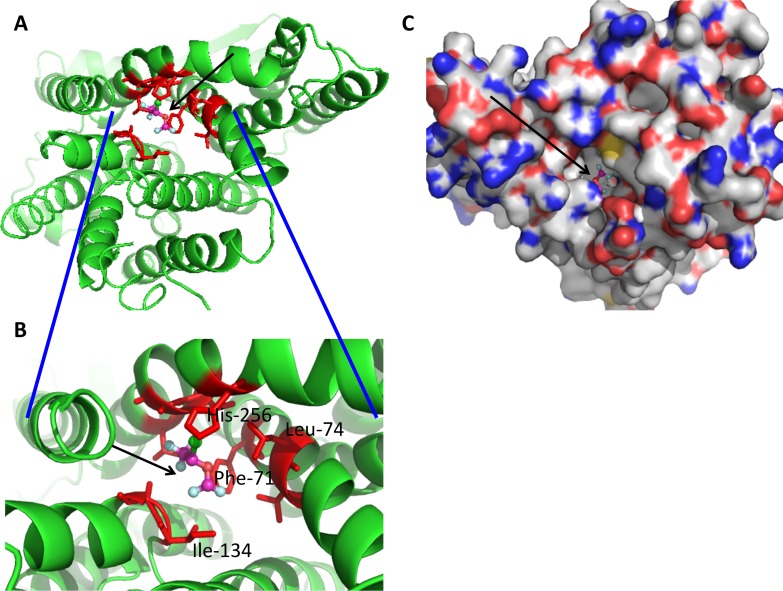
Using available structure of E.coli ion transporters, we sought for potential isoflurane binding site There were no reported structures of ClcB and NhaB. Protein structures for NhaA were reported [33–35]. Using NhaA structure, we performed docking simulation for the potential binding site (Fig 8). The trifluoromethyl head of isoflurane formed a hydrophobic interaction with Phe-71, Ala-135, Ala-260 and Leu-264, and the difluoromethyl head formed with Phe-71 and Ile-134. Amino acid residues within 4 angstrom from docked isoflurane were Phe-71, Asp- 133, Ile-134, Ala-135, Phe-136, Ala-137, Gly-139, His-256, Val-259, Ala-260 and Leu-264. The functional role of NhaA amino acid residues was reviewed in detail [36]. Asp-133 is at the iron binding site and His-256 is essential for pH signal transduction, suggesting that isoflurane docking site will be functionally critical for NhaA activity. This supports the idea that isoflurane affects ion transporter function.
Fig 8. Docking simulation of isoflurane onto NhaA protein (PDB ID: 1ZCD).
Docked image of isoflurane is shown (arrow). In isoflurane, light blue; floride, green; chloride and orange; oxygen. Red residues are within 4 angstroms of isoflurane. (A) Docked image in cartoon. (B) Blowout image of docked site. (C) Docked image in surface.
Discussion
Here we demonstrated that 1) volatile anesthetics tested at clinically relevant concentrations did not affect growth of four common bacterial pathogens, 2) isoflurane and sevoflurane significantly reduced swimming of E. coli and P. aeruginosa, and enhanced gliding of S. aureus, and 3) isoflurane enhanced biofilm formation by S. aureus, E. faecalis, E. coli, but not P. aeruginosa, with S. aureus and E. faecalis also being enhanced by sevoflurane. Our results suggest that exposure to volatile anesthetics isoflurane and sevoflurane significantly change the bacterial behaviors and their effects are different among bacteria species.
Swimming is a form of motility driven by flagella and allows bacteria to move toward a favorable environment [37]. From the host defense standpoint, however, flagella are potent stimulators of Toll-like receptor (TLR) 5 molecule and can also be a molecular marker of an invading pathogen for host immune cells. Thus impaired flagella motility could potentially act as protection for bacteria and would allow bacteria to evade host defenses. Several groups found that non-motile isolates of P. aeruginosa were resistant to macrophage phagocytosis and their presence was associated with poorer clinical scores in patients with cystic fibrosis than the normally motile isolates, which are often found in patients in the early stages of infection [38, 39]. Furthermore, the loss of flagella motility, not the loss of flagella expression was critical for the development of resistance against phagocytosis by immune cells via the activation of PI3K/Akt pathway [40, 41]. In our study, we found that isoflurane and sevoflurane significantly attenuated the swimming motility of E. coli and P. aeruginosa. Whether or not this attenuation by volatile anesthetics indicates impairment of phagocytosis by host immune cells and enhances bacterial virulence needs further investigation. In addition, we studied the role of anesthetics on S. aureus gliding. Our results showed that volatile anesthetic isoflurane increased the motility of S. aureus. The relationship between motility and virulence in S. aureus has not been studied and at this point, the clinical implication of our finding remains to be seen.
The formation of biofilm is observed in approximately 80% of bacterial infections and has been recognized to be a contributor in a number of diseases [42]. Biofilm is extremely resistant to clearance by immune cells and to antibiotics. Biofilm-protected cells are 10–1,000 times less susceptible to antibiotics than planktonic cells. Volatile anesthetics significantly enhanced biofilm formation by S. aureus and E. faecalis in our study. S. aureus is one of the most common bacterial pathogens [43]. Bacteremia and skin abscesses are usually caused by planktonic S. aureus, while osteomyelitis and endocarditis are caused by biofilm-forming strains. E. faecalis, on the other hand, is responsible for 80–90% of enterococcal infections. E. faecalis biofilm is important in periodontal infection as well as binding to various medical devices including urethral stents, intravascular catheters, and biliary stents [16]. Given that eradication of S. aureus and E. faecalis in biofilm are clinically important challenges to be solved, our data showing that volatile anesthetics facilitated biofilm formation needs to be evaluated further, including in vivo.
We also evaluated the growth under anesthetics and we did not find any difference among different anesthetics, which is line with the majority of literature. The study by Molliex et al. showed that the growth of Pseudomonas aeruginosa (P. aeruginosa) was attenuated under isoflurane and halothane exposure [14]. In contradiction, however, we did not observe any differences in P. aeruginosa as they did. Despite our study sharing similar methods, it was not clear if this difference in results is due to our strain differing from their strain, or culture conditions.
Because ion channels are considered to be prime volatile anesthetic targets and ion transporters are evolutionarily closely related to ion channels in humans [30], we hypothesized that bacterial behaviors were modulated via ion transporters. Taking advantage of multiple single gene deletion mutants that have been developed in E. coli K12 strains [22], we tested the hypothesis. Isoflurane reduced swimming, enhanced biofilm formation, and did not affect growth in E. coli parent strain. We identified that several ion transporter mutants showed reduction in swimming, increase in biofilm formation and no change in growth. Our result suggested that clcB, nhaA and nhaB would be isoflurane targets, although there may exist an additional target. We did not show the direct interaction of isoflurane with these molecules. Because specific agonists are not available, a docking simulation using NhaA suggested that isoflurane could bind within this ion transporter. Nearby residues from isoflurane are critical for pH signal transduction as well as serve as the iron binding site, further supporting the idea that isoflurane would affect ion transporter function by interacting its critical sites. The involvement of ion in biofilm formation has also been shown. Prindle et al showed that potassium channel regulates metabolic states for biofilm formation in Bacillus subtilis [44]. Therefore, it is possible that volatile anesthetics affected ion movement to modulate bacterial behaviors.
Our findings suggest the potential implication of anesthetic selection for patients with active bacterial infection. Choices of anesthetic drugs could potentially affect the course of infection. So far, the clinical impact of the type of anesthetics on infection has not been well reported. Von Dossow et al. reported their study of alcoholic patients who underwent surgical procedure either with propofol or isoflurane anesthesia and they found that postoperative infection rate was higher in isoflurane group [45]. Certainly the investigation of the in vivo impact of anesthetics on bacterial motility and biofilm formation is critical. In addition, identifying the underlying mechanism may allow practitioners to choose anesthetic drugs in a rationalized manner and also provide an opportunity to redesign better anesthetics.
In summary, we have demonstrated that volatile anesthetics modulated bacterial motility and biofilm formation in vitro and suggest the potential importance of anesthetic selection in patients with active infection. Future experiments will need to address in vivo significance.
Supporting Information
Swimming ring pictures for E. coli and P. aeruginosa were shown. Radius of each plate = 4.25 cm.
(TIFF)
Expression of flagellin in ion transporter deletion mutants was probed using anti-flagellin antibody. fliC deletion mutant was used as a negative control. GAPDH was used as a loading control.
(TIFF)
The percentage of swimming reduction in E. coli ion transporter deletion mutants by isoflurane exposure was calculated. Statistical analysis was performed using one-way analysis of variance with Bonferroni post hoc analysis. * denotes p < 0.05 versus parent strain.
(TIFF)
(A) The area of swimming holos from E. coli K12 parent strain, the double and triple ion transporter deletion mutants. Data are shown as mean +/- S.D. of 4–8 replicates. Statistical analysis was performed using one-way analysis of variance with Bonferroni post hoc analysis. ** denotes p < 0.01 versus parent strain. (B) The percentage reduction in E. coli triple deletion mutant by 3% isoflurane was shown. Data are shown as mean +/- S.D. of 8 replicates. Statistical analysis was performed using unpaired student t test. ** denotes p < 0.01 versus parent strain.
(TIFF)
The growth of the E. coli K12 parent strain and its ion transporter deletion mutants is shown. Data are shown as mean +/- S.D. of 4 replicates. Statistical analysis was performed using one-way analysis of variance with Bonferroni post hoc analysis. No statistical significance was observed. n.s. = not significant.
(TIFF)
Acknowledgments
We thanked Ms. Priya Shukla for her technical assistance and Dr. Kazumasa Tazawa, Dr. Erika Matsunami and Dr. Wei Wang (all Boston Children’s Hospital) for general help.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was supported by the Foundation of the National Institute of Health, NIH GM101345 and GM118277 (K.Y.), GM111421 (R.L.) and by the CHMC Anesthesia Foundation (K.Y.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Eckenhoff RG. Promiscuous ligands and attractive cavities: how do the inhaled anesthetics work? Mol Interv. 2001;1(5):258–68. [PubMed] [Google Scholar]
- 2.Jurd R, Arras M, Lambert S, Drexler B, Siegwart R, Crestani F, et al. General anesthetic actions in vivo strongly attenuated by a point mutation in the GABA(A) receptor beta3 subunit. FASEB J. 2003;17(2):250–2. 10.1096/fj.02-0611fje [DOI] [PubMed] [Google Scholar]
- 3.Rello J, Lisboa T, Lujan M, Gallego M, Kee C, Kay I, et al. Severity of pneumococcal pneumonia associated with genomic bacterial load. Chest. 2009;136(3):832–40. 10.1378/chest.09-0258 [DOI] [PubMed] [Google Scholar]
- 4.Nilsson AC, Bjorkman P, Welinder-Olsson C, Widell A, Persson K. Clinical severity of Mycoplasma pneumoniae (MP) infection is associated with bacterial load in oropharyngeal secretions but not with MP genotype. BMC Infect Dis. 2010;10:39 PubMed Central PMCID: PMCPMC2837002. 10.1186/1471-2334-10-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Darton T, Guiver M, Naylor S, Jack DL, Kaczmarski EB, Borrow R, et al. Severity of meningococcal disease associated with genomic bacterial load. Clin Infect Dis. 2009;48(5):587–94. 10.1086/596707 [DOI] [PubMed] [Google Scholar]
- 6.Wardley-Smith B, Nunn JF. The effect of halothane on bacterial growth rate. Br J Anaesth. 1971;43(10):919–25. [DOI] [PubMed] [Google Scholar]
- 7.Mehta S, Behr G, Kenyon D. The effect of volatile anaesthetics on bacterial growth. Can Anaesth Soc J. 1973;20(2):230–40. [DOI] [PubMed] [Google Scholar]
- 8.Slade JM. Bacterial growth in isoflurane vapour. Anaesthesia. 1993;48(12):1053–4. [DOI] [PubMed] [Google Scholar]
- 9.Asehnoune K, Cruaud P, Paries J, Gorce P, Pourriat JL. Effects of isoflurane on bacterial growth. Eur J Anaesthesiol. 2000;17(5):289–94. [DOI] [PubMed] [Google Scholar]
- 10.Karabiyik L, Turkan H, Ozisik T, Saracli MA, Haznedaroglu T. Effects of sevoflurane and/or nitrous oxide on bacterial growth in in vitro culture conditions. J Anesth. 2007;21(3):436–8. 10.1007/s00540-007-0520-3 [DOI] [PubMed] [Google Scholar]
- 11.Guttenplan SB, Kearns DB. Regulation of flagellar motility during biofilm formation. FEMS Microbiol Rev. 2013;37(6):849–71. PubMed Central PMCID: PMCPMC3718880. 10.1111/1574-6976.12018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kearns DB. A field guide to bacterial swarming motility. Nat Rev Microbiol. 2010;8(9):634–44. PubMed Central PMCID: PMCPMC3135019. 10.1038/nrmicro2405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stewart PS, Costerton JW. Antibiotic resistance of bacteria in biofilms. Lancet. 2001;358(9276):135–8. [DOI] [PubMed] [Google Scholar]
- 14.Molliex S, Montravers P, Dureuil B, Desmonts JM. Halogenated anesthetics inhibit Pseudomonas aeruginosa growth in culture conditions reproducing the alveolar environment. Anesth Analg. 1998;86(5):1075–8. [DOI] [PubMed] [Google Scholar]
- 15.Otto M. Staphylococcal biofilms. Curr Top Microbiol Immunol. 2008;322:207–28. PubMed Central PMCID: PMCPMC2777538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mohamed JA, Huang DB. Biofilm formation by enterococci. J Med Microbiol. 2007;56(Pt 12):1581–8. 10.1099/jmm.0.47331-0 [DOI] [PubMed] [Google Scholar]
- 17.Pollitt EJ, Crusz SA, Diggle SP. Staphylococcus aureus forms spreading dendrites that have characteristics of active motility. Sci Rep. 2015;5:17698 PubMed Central PMCID: PMCPMC4683532. 10.1038/srep17698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Horn K, Toth C, Kariyama R, Mitsuhata R, Kumon H. Evaluation of 15 motility media and a direct microscopic method for detection of motility in enterococci. J Clin Microbiol. 2002;40(7):2476–9. PubMed Central PMCID: PMCPMC120613. 10.1128/JCM.40.7.2476-2479.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beloin C, Roux A, Ghigo JM. Escherichia coli biofilms. Curr Top Microbiol Immunol. 2008;322:249–89. PubMed Central PMCID: PMCPMC2864707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pratt LA, Kolter R. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol Microbiol. 1998;30(2):285–93. [DOI] [PubMed] [Google Scholar]
- 21.Newell PD, Yoshioka S, Hvorecny KL, Monds RD, O'Toole GA. Systematic analysis of diguanylate cyclases that promote biofilm formation by Pseudomonas fluorescens Pf0-1. J Bacteriol. 2011;193(18):4685–98. PubMed Central PMCID: PMCPMC3165641. 10.1128/JB.05483-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:2006 0008. PubMed Central PMCID: PMCPMC1681482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97(12):6640–5. PubMed Central PMCID: PMCPMC18686. 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palanisamy A, Friese MB, Cotran E, Moller L, Boyd JD, Crosby G, et al. Prolonged Treatment with Propofol Transiently Impairs Proliferation but Not Survival of Rat Neural Progenitor Cells In Vitro. PLoS One. 2016;11(7):e0158058 PubMed Central PMCID: PMCPMC4933334. 10.1371/journal.pone.0158058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patteson AE, Gopinath A, Goulian M, Arratia PE. Running and tumbling with E. coli in polymeric solutions. Sci Rep. 2015;5:15761 PubMed Central PMCID: PMCPMC4938119. 10.1038/srep15761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tart AH, Wolfgang MC, Wozniak DJ. The alternative sigma factor AlgT represses Pseudomonas aeruginosa flagellum biosynthesis by inhibiting expression of fleQ. J Bacteriol. 2005;187(23):7955–62. PubMed Central PMCID: PMCPMC1291279. 10.1128/JB.187.23.7955-7962.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coffey BM, Anderson GG. Biofilm formation in the 96-well microtiter plate. Methods Mol Biol. 2014;1149:631–41. 10.1007/978-1-4939-0473-0_48 [DOI] [PubMed] [Google Scholar]
- 28.O'Toole GA. Microtiter dish biofilm formation assay. J Vis Exp. 2011;(47). PubMed Central PMCID: PMCPMC3182663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keseler IM, Mackie A, Peralta-Gil M, Santos-Zavaleta A, Gama-Castro S, Bonavides-Martinez C, et al. EcoCyc: fusing model organism databases with systems biology. Nucleic Acids Res. 2013;41(Database issue):D605–12. PubMed Central PMCID: PMCPMC3531154. 10.1093/nar/gks1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gadsby DC. Ion channels versus ion pumps: the principal difference, in principle. Nat Rev Mol Cell Biol. 2009;10(5):344–52. PubMed Central PMCID: PMCPMC2742554. 10.1038/nrm2668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blattner FR, Plunkett G 3rd, Bloch CA, Perna NT, Burland V, Riley M, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277(5331):1453–62. [DOI] [PubMed] [Google Scholar]
- 32.Yuki K, Eckenhoff RG. Mechanisms of the Immunological Effects of Volatile Anesthetics: A Review. Anesth Analg. 2016;123(2):326–35. 10.1213/ANE.0000000000001403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hunte C, Screpanti E, Venturi M, Rimon A, Padan E, Michel H. Structure of a Na+/H+ antiporter and insights into mechanism of action and regulation by pH. Nature. 2005;435(7046):1197–202. 10.1038/nature03692 [DOI] [PubMed] [Google Scholar]
- 34.Appel M, Hizlan D, Vinothkumar KR, Ziegler C, Kuhlbrandt W. Conformations of NhaA, the Na/H exchanger from Escherichia coli, in the pH-activated and ion-translocating states. J Mol Biol. 2009;386(2):351–65. 10.1016/j.jmb.2008.12.042 [DOI] [PubMed] [Google Scholar]
- 35.Lee C, Yashiro S, Dotson DL, Uzdavinys P, Iwata S, Sansom MS, et al. Crystal structure of the sodium-proton antiporter NhaA dimer and new mechanistic insights. J Gen Physiol. 2014;144(6):529–44. PubMed Central PMCID: PMCPMC4242812. 10.1085/jgp.201411219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Padan E, Kozachkov L, Herz K, Rimon A. NhaA crystal structure: functional-structural insights. J Exp Biol. 2009;212(Pt 11):1593–603. 10.1242/jeb.026708 [DOI] [PubMed] [Google Scholar]
- 37.Armitage JP. Bacterial motility and chemotaxis. Sci Prog. 1992;76(301–302 Pt 3–4):451–77. [PubMed] [Google Scholar]
- 38.Luzar MA, Thomassen MJ, Montie TC. Flagella and motility alterations in Pseudomonas aeruginosa strains from patients with cystic fibrosis: relationship to patient clinical condition. Infect Immun. 1985;50(2):577–82. PubMed Central PMCID: PMCPMC261995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mahenthiralingam E, Campbell ME, Speert DP. Nonmotility and phagocytic resistance of Pseudomonas aeruginosa isolates from chronically colonized patients with cystic fibrosis. Infect Immun. 1994;62(2):596–605. PubMed Central PMCID: PMCPMC186146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Amiel E, Lovewell RR, O'Toole GA, Hogan DA, Berwin B. Pseudomonas aeruginosa evasion of phagocytosis is mediated by loss of swimming motility and is independent of flagellum expression. Infect Immun. 2010;78(7):2937–45. PubMed Central PMCID: PMCPMC2897393. 10.1128/IAI.00144-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lovewell RR, Hayes SM, O'Toole GA, Berwin B. Pseudomonas aeruginosa flagellar motility activates the phagocyte PI3K/Akt pathway to induce phagocytic engulfment. Am J Physiol Lung Cell Mol Physiol. 2014;306(7):L698–707. PubMed Central PMCID: PMCPMC3962627. 10.1152/ajplung.00319.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Davies D. Understanding biofilm resistance to antibacterial agents. Nat Rev Drug Discov. 2003;2(2):114–22. 10.1038/nrd1008 [DOI] [PubMed] [Google Scholar]
- 43.Lister JL, Horswill AR. Staphylococcus aureus biofilms: recent developments in biofilm dispersal. Front Cell Infect Microbiol. 2014;4:178 PubMed Central PMCID: PMCPMC4275032. 10.3389/fcimb.2014.00178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prindle A, Liu J, Asally M, Ly S, Garcia-Ojalvo J, Suel GM. Ion channels enable electrical communication in bacterial communities. Nature. 2015;527(7576):59–63. 10.1038/nature15709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Von Dossow V, Baur S, Sander M, Tonnesen H, Marks C, Paschen C, et al. Propofol increased the interleukin-6 to interleukin-10 ratio more than isoflurane after surgery in long-term alcoholic patients. J Int Med Res. 2007;35(3):395–405. [DOI] [PubMed] [Google Scholar]
- 46.Wu Y, Wu M, He G, Zhang X, Li W, Gao Y, et al. Glyceraldehyde-3-phosphate dehydrogenase: a universal internal control for Western blots in prokaryotic and eukaryotic cells. Anal Biochem. 2012;423(1):15–22. 10.1016/j.ab.2012.01.012 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Swimming ring pictures for E. coli and P. aeruginosa were shown. Radius of each plate = 4.25 cm.
(TIFF)
Expression of flagellin in ion transporter deletion mutants was probed using anti-flagellin antibody. fliC deletion mutant was used as a negative control. GAPDH was used as a loading control.
(TIFF)
The percentage of swimming reduction in E. coli ion transporter deletion mutants by isoflurane exposure was calculated. Statistical analysis was performed using one-way analysis of variance with Bonferroni post hoc analysis. * denotes p < 0.05 versus parent strain.
(TIFF)
(A) The area of swimming holos from E. coli K12 parent strain, the double and triple ion transporter deletion mutants. Data are shown as mean +/- S.D. of 4–8 replicates. Statistical analysis was performed using one-way analysis of variance with Bonferroni post hoc analysis. ** denotes p < 0.01 versus parent strain. (B) The percentage reduction in E. coli triple deletion mutant by 3% isoflurane was shown. Data are shown as mean +/- S.D. of 8 replicates. Statistical analysis was performed using unpaired student t test. ** denotes p < 0.01 versus parent strain.
(TIFF)
The growth of the E. coli K12 parent strain and its ion transporter deletion mutants is shown. Data are shown as mean +/- S.D. of 4 replicates. Statistical analysis was performed using one-way analysis of variance with Bonferroni post hoc analysis. No statistical significance was observed. n.s. = not significant.
(TIFF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.