Abstract
Germline mutations in the DNA mismatch repair (MMR) genes MSH2 and MLH1 are responsible for the majority of hereditary non-polyposis colorectal cancer (HNPCC), an autosomal-dominant early-onset cancer syndrome. Genetic testing of both MSH2 and MLH1 from individuals suspected of HNPCC has revealed a considerable number of missense codons, which are difficult to classify as either pathogenic mutations or silent polymorphisms. To identify novel MLH1 missense codons that impair MMR activity, a prospective genetic screen in the yeast Saccharomyces cerevisiae was developed. The screen utilized hybrid human-yeast MLH1 genes that encode proteins having regions of the yeast ATPase domain replaced by homologous regions from the human protein. These hybrid MLH1 proteins are functional in MMR in vivo in yeast. Mutagenized MLH1 fragments of the human coding region were synthesized by error-prone PCR and cloned directly in yeast by in vivo gap repair. The resulting yeast colonies, which constitute a library of hybrid MLH1 gene variants, were initially screened by semi-quantitative in vivo MMR assays. The hybrid MLH1 genes were recovered from yeast clones that exhibited a MMR defect and sequenced to identify alterations in the mutagenized region. This investigation identified 117 missense codons that conferred a 2-fold or greater decreased efficiency of MMR in subsequent quantitative MMR assays. Notably, 10 of the identified missense codons were equivalent to codon changes previously observed in the human population and implicated in HNPCC. To investigate the effect of all possible codon alterations at single residues, a comprehensive mutational analysis of human MLH1 codons 43 (lysine-43) and 44 (serine-44) was performed. Several amino acid replacements at each residue were silent, but the majority of substitutions at lysine-43 (14/19) and serine-44 (18/19) reduced the efficiency of MMR. The assembled data identifies amino acid substitutions that disrupt MLH1 structure and/or function, and should assist the interpretation of MLH1 genetic tests.
INTRODUCTION
DNA mismatch repair (MMR) is a multi-protein intracellular process for recognizing and repairing non-native DNA structures. These incorrect structures arise primarily following DNA replication and during recombination [reviewed in (1,2)]. In the absence of normal MMR, the cellular mutation rate increases and this deficiency is evidenced by high levels of microsatellite instability (MSI), i.e. insertions and deletions in repetitive DNA sequences, and an accumulation of single nucleotide alterations. In agreement with the ‘mutator hypothesis’ (3), which predicts an association between the increased mutation rates and tumorigenesis, mice that carry MMR gene deficiencies display elevated levels of MSI and have an increased susceptibility to cancer (4–7). In humans, heterozygosity for mutations in MMR genes results in predisposition to hereditary non-polyposis colorectal cancer (HNPCC), an autosomal-dominant syndrome characterized by early-onset colorectal cancer and other cancers (8–10). Depending on the population and clinical criteria used, recent studies suggest that HNPCC may comprise 2–8% of all colorectal cancers worldwide (11,12).
The basic mechanisms and protein components of MMR are conserved in a broad range of species, including Escherichia coli, the yeast Saccharomyces cerevisiae and humans. In E.coli, a protein complex composed of two MutS proteins recognizes and binds to single nucleotide mispairs and insertion–deletion loops in post-replicative DNA. Homodimers composed of two MutL proteins subsequently interact with the MutS:DNA complex and coordinate subsequent repair events involving the endonuclease MutH and several other proteins that excise the non-native DNA strand and synthesize a corrected version (13). In yeast and human cells, there exist multiple homologs of the E.coli MutS and MutL proteins and these interact in several combinations to effect MMR (1,2). The MutS homologs (MSH2, MSH3 and MSH6) form the heterodimers MSH2–MSH6 (MutSα) and MSH2–MSH3 (MutSβ), which bind to mispaired DNA in an ‘error-specific’ manner. Heterodimers composed of the MutL homologs [MLH1, MLH3 and PMS1 (PMS2 in humans)] then interact with the MSH2-containing complexes and coordinate downstream repair events. The heterodimer MutLα, composed of MLH1 and PMS1 (PMS2 in humans), interacts with both MutSα and MutSβ, while the heterodimer MutLβ, composed of MLH1 and MLH3, appears restricted to interactions with only MutSβ (14,15). The MLH1-containing heterodimers are essential for signaling downstream MMR events, which appear to involve the exonuclease EXO1 (16,17), DNA replication protein A (18,19) and proliferating cell nuclear antigen/POL30, a component of DNA polymerase δ (20–22). In addition, certain MMR proteins have been implicated in cellular functions apart from MMR. These include mitotic recombination (23,24), meiotic crossing over (25,26), immunoglobulin class-switch recombination (27,28) and the induction of apoptosis in response to DNA damage (29–33).
The MLH1 protein (MLH1p) is essential for MMR in both yeast and human cells (34–36). Structure-function studies of MLH1p have revealed the importance of an ATPase domain in the N-terminal region (37–40). Crystal structures of the conserved N-terminal portion of E.coli MutL has provided insights into the workings of the ATPase domain, which is highly conserved in the GHL (gyrase b/hsp90/mutL) family of ATPases (41,42). As with the E.coli MutL protein, binding and hydrolysis of ATP triggers a conformational change in MLH1p which appears to mediate downstream MMR events (38–40). These events appear to be dependent on binding of yMLH1 to yPMS1 (PMS2 in humans) through a domain in the C-terminal end of MLH1p (43,44). Interestingly, recent studies have shown the importance of additional amino acid residues in the N-terminal end for proper yMLH1–yPMS1 interaction and hMutLα–hMutSα complex formation (37,44–46). Taken together, it appears that the N-terminal end of MLH1p may have multiple functions in the process of MMR.
Germline genetic testing of individuals suspected of HNPCC has revealed over 300 different alterations in MMR genes with the vast majority (∼90%) of these being in the MLH1 and MSH2 genes (8,10). Approximately one-third of the MLH1 and MSH2 alterations are missense codons whose functional consequence is not immediately obvious. These ‘variants of uncertain significance’ require further investigation to determine whether each may be a pathogenic mutation or a silent polymorphism. In some cases, the consequence of the human alteration was tested yeast using a dominant mutator effect of the human MLH1 gene (47) or the loss of binding to other MMR proteins (43–45). However, these investigations do not assay the biological activity of the protein. The use of in vivo MMR assays using yeast genes with an alteration identical to the human alteration has allowed a functional analysis of many MLH1 and MSH2 missense codons (43,48–50). In a recent investigation of 18 previously observed human alterations, it was shown that missense codons may lead to expression of a MMR protein with no reduction in the MMR efficiency (‘silent polymorphisms’), complete elimination of MMR function (‘inactivating mutations’) or a protein that functions in MMR with a reduced efficiency (‘efficiency polymorphisms’) (51). The development of hybrid human-yeast MLH1 genes, which encode proteins that are functional in MMR in yeast cells, has made possible an analysis of human missense codons in the context of the native human MLH1 gene sequence (51).
The aim of the current investigation is to identify novel MLH1 missense codons that impair MMR function. To obtain functional information relevant to the native human MLH1 gene, a prospective genetic screen using hybrid human-yeast genes was developed. The human coding sequence of hybrid MLH1 genes was mutagenized by error-prone PCR, and these fragments were cloned by in vivo gap repair transformation in yeast to generate a library of clones containing a high frequency of mutations in the human coding sequence. The clones were subjected to in vivo MMR assays to identify transformants with a MMR deficiency. Sequence analysis of these mutant MLH1 genes led to the identification of 117 unique single missense codons that eliminated or reduced the efficiency of MMR compared to the parental hybrid molecule. In addition, all 19 possible amino acid substitutions at human MLH1p residues 43 (lysine-43) and 44 (serine-44) were tested for function in vivo. The majority of substitutions at lysine-43 (14/19) and serine-44 (18/19) impaired MMR function. Collectively, the results provide information that may assist in the interpretation of HNPCC genetic tests when ‘variants of uncertain significance’ are observed.
MATERIALS AND METHODS
Yeast expression vectors
Plasmid pMETc [p413MET25, (52)] contains a HIS3 selectable marker, a centromere sequence (CEN6) for mitotic stability, an ARS4 origin of DNA replication, the ampicillin-resistance gene for positive selection in E.coli and a multicloning site between the MET25 promoter and CYC1 terminator. Plasmid pMLH1, a derivative of pMETc lacking the MET25 promoter, contains a 3.8 kb genomic DNA fragment from S.cerevisiae strain S288C including the MLH1 gene coding sequence and 1.5 kb of 5′ flanking sequence (51). Plasmids pMLH1_h(41-86) and pMLH1_h(77-134) are identical to pMLH1 but contain codons encoding human MLH1p amino acid residues 41–86 and 77–134, respectively, in place of the homologous codons of yeast MLH1 (51). Plasmid pSH91 contains a TRP1 selectable marker, a centromere sequence (CEN11), an ARS1 origin of replication, the ampicillin-resistance gene and the URA3 coding sequence preceded by an in-frame (GT)16G tract (53).
Yeast strains, growth conditions and transformations
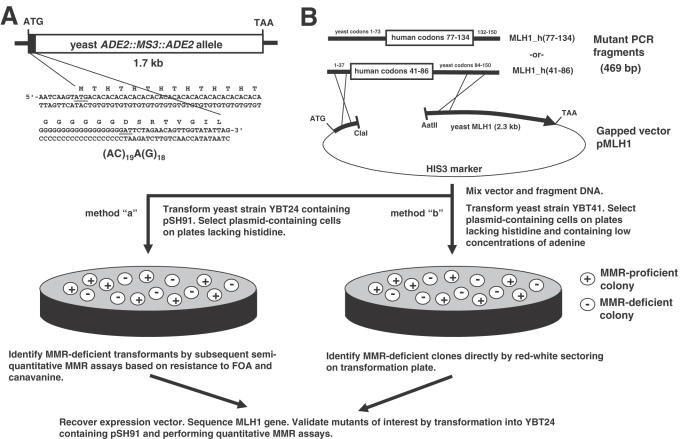
The strains used in this study were derived from S.cerevisiae YPH500 (MATα ade2-101 his3-Δ200 leu2-Δ1 lys2-801 trp1-Δ63 ura3-52) (54). The strain YBT24 contains a deletion of the entire MLH1 coding sequence and has the genotype MATα ade2-101 his3-Δ200 leu2-Δ1 lys2-801 trp1-Δ63 ura3-52 mlh1Δ::LEU2 (51). The strain YBT41 was derived from YBT24 and has the genotype MATα ADE2::MS3::ADE2 his3-Δ200 leu2-Δ1 lys2-801 trp1-Δ63 ura3-52 mlh1Δ::LEU2 where MS3 refers to a cloned synthetic oligonucleotide introducing an in-frame (AC)19A(G)18 microsatellite between the first and second codons of the ADE2 coding sequence (Figure 1A). The strain was constructed as follows: the yeast ADE2 translation initiation codon and 644 bp 5′ flanking sequence was PCR amplified from S.cerevisiae S288C DNA using the primers d(CGC GAT CGA TCA TGC TTA TGG GTT AGC) (H374-4) and d(GTG TGT GTG TGT GTG TGT GTG TGT GTC ATA CTT GAT TGT TTT GTC CG) [1-51-10]. The ADE2 coding sequence from codon 2 to 36 bp 3′ to the termination codon was PCR amplified from S.cerevisiae S288C DNA using the primers d(ACA CAC ACA CAC ACA CAC ACA CAC ACA CAC ACA CAC ACA GGG GGG GGG GGG GGG GGG GAT TCT AGA ACA GTT GGT ATA TTA G) [1-51-9] and d(TTA TTT GCT GTA CAA GTA TAT CAA TAA AC) [1-51-13]. The 673 and 1808 bp DNA fragments were mixed in approximately equimolar amounts and subjected to overlap extension PCR amplification (55) using primers H374-4 and 1-51-13. The predominant amplification product was the 2452 bp overlap extension PCR product. The DNA was purified (Wizard™ PCR Preps DNA Purification kit, Promega) and transformed into YBT24 containing pSH91 selecting for adenine prototrophs. The yeast strain YBT41 was shown to have the native ADE2 chromosomal gene replaced by the ADE2::MS3::ADE2 allele by PCR amplification of chromosomal DNA using ADE2-specific and microsatellite-specific primers (data not shown). The strain YBT24 containing pSH91 was grown in synthetic dextrose (SD) medium supplemented with adenine, histidine and lysine. The strain YBT41 containing pSH91 was grown in SD medium supplemented with histidine and lysine. The strains YBT24 and YBT41 containing pSH91 were transformed with pMLH1-derived expression vectors or the parental vector pMETc and histidine prototrophs were selected. All transformations were performed using the polyethylene glycol–lithium acetate method (56).
Figure 1.
(A) Sequence of the 5′ end of the ADE2::MS3::ADE2 reporter allele. The microsatellite (AC)19A(G)18 was inserted between the ATG initiator codon and the second codon (GAT) of the native ADE2 gene. The ADE2::MS3::ADE2 allele was introduced into haploid yeast strain YBT24 (mlh1Δ), replacing the native ADE2 allele to generate strain YBT41. (B) Schematic representation of the screen for inactivating mutations in MLH1. Fragments of human-yeast hybrid genes pMLH1_h(41-86) and pMLH1_h(77-134) were generated by error-prone PCR, mixed with a ClaI–AatII-digested pMLH1 expression vector and transformed into strains YBT24 (Method ‘a’) or YBT41 (Method ‘b’). Circularized plasmids were formed in vivo by homologous recombination between the PCR product and gapped vector. Yeast transformants with a MMR-deficient phenotype (i.e. containing a mutant mlh1 gene) are identified phenotypically as described in the figure. Plasmids containing the mutant mlh1 gene are recovered by shuttling into E.coli and sequenced to determine the nucleotide alteration(s) present in the mutagenized gene.
Error-prone PCR and in vivo gap repair cloning
Pools of mutant MLH1 gene fragments were generated by error-prone PCR using Mutazyme™ (a component of the GeneMorph PCR mutagenesis kit, Stratagene) or Taq (Promega) DNA polymerases, which have different misincorporation biases (57). The use of both enzymes should ensure that pools of mutagenized DNA are representative of all possible base substitutions. The XhoI-linearized plasmids pMLH1_h (41-86) and pMLH1_h(77-134) were used as templates in PCR mixes containing the buffers, nucleotides and enzyme concentrations recommended by the manufacturer of each DNA polymerase. The upstream and downstream primers were d(GCT GCA GGT GAG ATC ATA ATA TCC) (E124-1) and d(TCA ACT AGG ATC GTG GTA C) (D545-9), respectively, which amplify a 401 bp fragment spanning the human portion of each hybrid MLH1 gene. In preliminary experiments, the upstream primer d(GCG CGG ATC CAT AGA CCT ATC AAT AAG C) (R472-1) was used to generate a fragment of 475 bp. The protocol for temperature cycling was: 94°C for 2 min, 33 cycles of 94°C for 36 s, 55°C for 1 min, 72°C for 2 min and 72°C for 10 min. Conditions of high and low fidelity were manipulated by varying the amount of template DNA (3–74 ng) in reactions containing Mutazyme and the MgCl2 concentration (1.5–2.5 mM) in reactions containing Taq DNA polymerase. The PCR fragments were purified with Wizard™ PCR Preps (Promega) and used for in vivo gap repair cloning in yeast (58–60). Briefly, 0.5 μg purified PCR product was combined with 0.4 μg ClaI–AatII digested pMLH1 vector and the DNA mixture was co-transformed into YBT24 or YBT41 containing pSH91. Yeast cells in which fragment and vector recombine were stable transformants converted to histidine prototrophs due to the presence of the HIS3 marker gene on the pMLH1 expression vector. This process typically yielded ∼500 transformants (i.e. colonies) per plate; while equivalent transformations performed with restricted vector alone exhibited very few (<5) colonies per plate.
Identification of MMR-deficient transformants
Method ‘a’:
When gap repair cloning was carried out in YBT24 containing pSH91, transformants were assayed sequentially using a spot test for 5-fluoroorotic acid (FOA) resistance and a patch test for canavanine resistance. Briefly, individual clones from the transformation were grown in 3 ml SD (0.67% yeast nitrogen base without amino acids, 2% dextrose) medium containing adenine and lysine (day 1 culture), and the next day 120 μl of the saturated culture was subinoculated into 3 ml fresh SD medium containing adenine, lysine and uracil. The addition of uracil in the medium allows growth of cells containing a ura3 mutation arising from a frameshift in the (GT)16G-tract of pSH91. These ura3 mutants exhibit a FOA-resistant phenotype (48,53). Following 24 h growth, 4 μl of the culture was spotted in duplicate on SD plates containing adenine, lysine, uracil and 1 mg/ml FOA (Toronto Research Chemicals Inc., ON, Canada). The plates were incubated at 30°C for 48 h and then scored by counting the number of FOA-resistant colonies on each spot. Transformants that exhibited few colonies (<15, typically 0–5) per spot were scored as having low levels of MSI (i.e. MMR proficient) and were not analyzed further. Transformants that exhibited many colonies (≥15, typically 20–50) per spot were scored as having high levels of MSI (i.e. MMR deficient) and were arrayed on a master plate by applying 25 μl the day 1 cultures to SD plates containing adenine and lysine. These clones were subjected to a secondary assay based on spontaneous forward mutations in the arginine permease gene (CAN1), which cause resistance to canavanine. A 1 μl loop of cells from the arrayed transformants were patched out on SD plates containing adenine, lysine and 60 μg/ml canavanine. Plates were incubated 3 days at 30°C and scored by counting the number of canavanine-resistant colonies. Yeast clones that exhibited few colonies (<15) were scored as having low levels of genetic instability (i.e. normal in MMR) and were not analyzed further. Clones that exhibited many colonies (≥15, typically 30–100) were selected for further analysis.
Method ‘b’:
When in vivo gap repair was carried out in YBT41, the transformed cells were plated directly on SD plates containing low concentrations (4 μg/ml) of adenine and incubated for 4–5 days. As described previously (60), the cells that do not express the ADE2 gene form red colonies due to the accumulation of an intermediate in adenine biosynthesis, while cells expressing a wild-type ADE2 gene form white colonies. When the ADE2::MS3::ADE2 allele is unstable (i.e. mutates to ade2- at a high frequency due to instability of the MS3 microsatellite), the strain forms a white colony with red sectors on plates containing low adenine (see Results). In Method ‘b’, after gap repair transformation and growth on plates containing 4 μg/ml adenine, colonies that exhibit abundant red–white sectoring were selected for further analysis.
Preparation of yeast DNA and isolation of mutant MLH1 expression vectors
Total yeast DNA was prepared from 15 ml liquid cultures using the glass-bead method (61) and resuspended in 50 μl H2O. To recover mutant plasmids from the yeast strain, a 15 μl aliquot of each DNA sample was digested with BamHI, which restricts the pSH91 expression vector but not the MLH1 expression vector, and shuttled into E.coli strain DH5α by electroporation using a BTX ECM399 system (Genetronics, Inc.). Bacterial colonies were selected by growth on Luria–Bertani plates containing 50 μg/ml ampicillin and plasmid DNA was purified using the Wizard Plus SV Minipreps kit (Promega).
Mutagenesis of human MLH1 codons 43 and 44
For mutagenesis of human codon 43 (lysine-43), a degenerate primer with the sequence d(CTG TAT CGA TGC ANN NTC CAC AAG TAT TCA AGT G) (E124-1.7), where ‘N’ represents any of the four nucleotides A, C, G or T, was used in combination with primer d(CAG TTT AGA CGT CGT GAA CCT TTC ACA TAC) (E466-1) to PCR amplify an ∼140 bp fragment from human MLH1 cDNA clone (accession no. 217884) (American Type Culture Collection). Amplification of the fragment utilized Pfu DNA polymerase (Stratagene) and the recommended buffers and nucleotide concentrations. Cycling conditions were as follows: 95°C for 2 min; 33 cycles of 95°C for 36 s, 55°C for 1 min, 72°C for 2 min, and 72°C for 10 min. The resulting fragment was digested with ClaI and AatII and ligated into ClaI–AatII digested pMLH1 vector replacing the corresponding portion of the native yeast MLH1 gene. Transformation into E.coli strain DH5α yielded ampicillin-resistant colonies that contained plasmids identical to pMLH1_h(41-86) except for the mutant codon at position 43. Plasmid DNA was purified from individual colonies and prepared for DNA sequencing to determine the identity of codon 43 and also verify the sequence of the cloned PCR fragment. Plasmids containing codons for 13 of the 20 possible amino acid substitutions were identified in this way. Plasmids containing codons for the seven remaining amino acid substitutions were generated by direct cloning of PCR products. These variants were generated using primers identical to E124-1.7 except for the following triplets at codon 43: ‘TGT’ (K43C), ‘GAA’ (K43E), ‘CAT’ (K43H), ‘AAG’ (K43K), ‘CCT’ (K43P), ‘CAA’ (K43Q) and ‘TGG’ (K43W). For mutagenesis of human codon 44 (serine-44), a degenerate primer with the sequence d(CTG TAT CGA TGC AAA ANN NAC AAG TAT TCA AGT G) (E124-8) was used in combination with primer E466-1 to amplify a 122 bp portion of the hMLH1 gene. Cloning into ClaI–AatII digested pMLH1 vector was the same as described above for mutagenesis of codon 43. Plasmids identical to pMLH1_h(41-86), but containing codons for 14 of the 20 possible amino acid substitutions at codon 44, were identified in this way. In addition, a hybrid gene containing the stop codon ‘TGA’ at codon 44 was identified. Plasmids containing codons for the remaining six amino acid substitutions were generated by direct cloning using primers identical to E124-8 except for the following triplet at codon 43: ‘GAC’ (S44D), ‘GAG’ (S44E), ‘GGC’ (S44G), ‘AAG’ (S44K), ‘ATG’ (S44M) and ‘AAC’ (S44N).
DNA sequencing
DNA sequencing was performed at commercial facilities using dye-terminator chemistry and automated sequencers (ABI models 377 and 3700, Applied Biosystems). Chromatogram and text files were analyzed with Chromas (version 1.45, http://technelysium.com.au/chromas.html) and GeneRunner (version 3.04, Hastings Software Inc.) software, respectively. Sequencing was carried out in both the forward and reverse directions using primers d(GGA GTT CTC GAA GAC GAG) (D806-1), D545-9 and/or d(CGG GTG AAC GTT AAC ATC) (D545-10).
Quantitative in vivo MMR assays
Standardized MMR assays based on mutation to ura3 FOAr were performed as described previously (48,51). Briefly, YBT24 transformants containing an MLH1 expression vector and pSH91 were cultured overnight in medium lacking uracil and subcultured in liquid media containing adenine, lysine and uracil, which allows growth of ura3 FOAr mutants [which arise as a result of slippage in the (GT)16G-tract]. After 24 h in culture, optical density (OD595) measurements were taken and an aliquot was plated on SD plates containing adenine, lysine, uracil and 1 mg/ml FOA. Mutation frequencies were calculated as described previously (51), except that the concentration (CFU/ml) of total cells was determined from OD595 readings using the determined value 1 OD595 = 1.1 × 107 CFU/ml. The MMR defect is defined as the ratio of the mutation frequency in the test strain divided by that observed in the appropriate MMR-proficient control strain. Forward mutation rates to canavanine resistance were determined by fluctuation analysis using the method of the median (62). Individual colonies (YBT24 transformed with the MLH1 expression vector of interest) were expanded in liquid SD media containing the appropriate supplements. After 24 h in culture, OD595 measurements were taken and an aliquot was plated on SD plates containing the appropriate supplements and 60 μg/ml canavanine. Canavanine-resistant colonies were counted after 2–3 days growth at 30°C. Mutation frequencies were determined by dividing the concentration (CFU/ml) of canavanine-resistant colonies by the concentration (CFU/ml) of total cells. The MMR defect is defined as the ratio of the mutation frequency in the test strain divided by that observed in the same strain complemented by the appropriate control gene.
MLH1p accession numbers, alignment and mutation databases
Homo sapiens, NP_000240; Mus musculus, Q9JK91; Rattus norvegicus, NP_112315; Drosophila melanogaster, NP_477022; Saccharomyces cerevisiae, NP_013890; Schizosaccharomyces pombe, NP_596199; Arabidopsis thaliana, NP_567345; Caenorhabditis elegans, NP_499796; Escherichia coli, NP_418591 and Staphylococcus aureus, Q93T05. Sequences were retrieved from the Protein Database of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov) and aligned using ClustalW (http://www.ebi.ac.uk/clustalw/). The human MLH1 alterations were reported in the literature (63–66) or the following public mutation databases: International Collaborative Group on HNPCC (http://www.nfdht.nl), Human Gene Mutation Database (http://www.uwcm.ac.uk) and Swiss-protein (http://www.expasy.ch). The databases were last examined in May 2004.
RESULTS
Functional analysis of hybrid human-yeast MLH1 genes
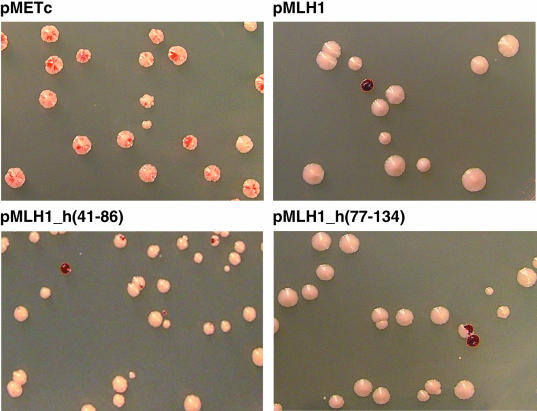
The human-yeast hybrid genes MLH1_h(41-86) and MLH1_h(77-134) encode chimeric MLH1 proteins that contain 46 and 58 amino acid regions, respectively, of human MLH1p replacing the homologous region of yeast MLH1p. When expressed in haploid yeast cells containing a deletion of the chromosomal MLH1 gene, these hybrids were active in MMR in a standardized in vivo assay that measures the frequency of frameshift mutations in an in-frame (GT)16G microsatellite preceding the URA3 gene (48,51,53). In the present study, the function of MLH1_h(41-86) and MLH1_h(77-134) was confirmed and extended using in vivo MMR assays that employ two other reporter genes. The first assay involved transformation of haploid yeast strain YBT41, which contains a chromosomal mlh1-null deletion and the ADE2::MS3:: ADE2 allele (Figure 1A) in place of the native ADE2 allele on chromosome XV. The strain YBT41 was transformed with pMLH1, pMLH1_h(41-86), pMLH1_h(77-134) or pMETc (the expression vector lacking an MLH1 gene), and histidine prototrophs were selected on plates containing low concentrations (4 μg/ml) of adenine (Figure 2 and Table 1). The ADE2::MS3::ADE2 allele allowed a determination of MMR proficiency based on the presence or absence of red–white sectoring in individual colonies. When strain YBT41 was transformed with pMETc, and thus did not express MLH1p, >95% of the colonies were red–white sectored (Table 1, ‘None’). This sectoring is likely due to the extreme instability of the in-frame (AC)19A(G)18 microsatellite resulting in frameshift mutations in the ADE2 gene. In contrast, when strain YBT41 was transformed with pMLH1, <2% of the colonies were sectored (Table 1). The integrity of the ADE2::MS3::ADE2 allele is maintained by the MMR process such that >98% of the colonies appeared white. When strain YBT41 was transformed with hybrids MLH1_h(41-86) and MLH1_h(77-134), the percentage of colonies exhibiting a red–white sectored appearance was ∼14% and <2%, respectively (Table 1). These results indicate that MLH1_h(41-86) and pMLH1_h(77-134) encode MLH1 proteins that efficiently maintain the stability of the ADE2::MS3::ADE2 allele. The second MMR assay was based on forward mutation to canavanine resistance, which detects mainly base substitutions and frameshift mutations in mononucleotide tracts in the arginine permease (CAN1) gene (67). The MLH1-deletion strain YBT24 was transformed with the MLH1 expression vectors, nine individual transformants were grown overnight in liquid culture and an aliquot was plated on solid growth media containing canavanine. The strain YBT24 containing the empty expression vector pMETc exhibited a CAN1 mutation frequency of 3.1 × 10−4, while the strain expressing the native yeast MLH1 gene (pMLH1) exhibited a mutation frequency of 7.1 × 10−6 (Table 1). This represents a mutation defect of 44 for yeast cells lacking MLH1p. The mutation frequencies of yeast cells expressing pMLH1_h(41-86) and pMLH1_h(77-134) were 2.8 × 10−5 and 1.7 × 10−5, respectively, which corresponded to mutation defects of 4.0 and 2.4. These results demonstrate that MLH1 proteins encoded by MLH1_h(41-86) and MLH1_h(77-134) are functional in the repair of a variety of DNA mismatch structures. Although the mutation frequencies exhibited by cells expressing the human-yeast hybrid genes are slightly elevated compared to those levels exhibited by cells expressing the native yeast MLH1 gene, the mutation frequencies conferred by the hybrids are at least 10-fold lower than those levels exhibited by yeast cells lacking any functional MLH1p. The complementation efficiencies for MLH1_h(41-86) and MLH1_h(77-134) are consistent with previous studies [(51); Table 1], and show that MLH1_h(77-134) is more proficient than MLH1_h(41-86) in MMR.
Figure 2.
Appearance of YBT41 colonies following transformation with different MLH1 expression vectors. YBT41 colonies were grown on plates containing low adenine (4 μg/ml) after the introduction of plasmids pMETc, the parental expression vector without an MLH1 gene, pMLH1, pMLH1_h(41-86) and pMLH1_h(77-134) (as indicated). Colonies with red–white sectoring indicate a high level of instability in the ADE2::MS3::ADE2 allele, i.e. mutation to ade2 (mutant cells appear red due to the accumulation of an intermediate in adenine biosynthesis). In all transformations, a background of 10% red colonies was consistently observed and may be a result of mutation in the ADE2::MS3::ADE2 allele prior to, or shortly after, transformation.
Table 1. Functional analysis of human-yeast hybrid MLH1 genes.
| MLH1 gene | ADE2::MS3::ADE2a | CAN1b | (GT)16G::URA3c | |
|---|---|---|---|---|
| Sectored CFU (% of total) | White CFU (% of total) | Mutation frequency (mutation defect) | Mutation frequency (mutation defect) | |
| None | >95 | <5 | 3.1 × 10−5 (44) | 1.9 × 10−3 (75) |
| MLH1 | <2 | >98 | 7.1 × 10−7 (1) | 2.5 × 10−5 (1) |
| MLH1_h(41-86) | 14 | 86 | 2.8 × 10−6 (4.0) | 1.2 × 10−4 (4.8) |
| MLH1_h(77-134) | <2 | >98 | 1.7 × 10−6 (2.4) | 5.4 × 10−5 (2.1) |
aYeast strain YBT41, which contains an MLH1 deletion and the ADE2::MS3::ADE2 allele, was transformed with expression vectors carrying the indicated MLH1 gene or the parental expression vector pMETc lacking an MLH1 gene (‘None’) and cells were plated on SD medium lacking histidine and containing 4 μg/ml adenine. Colonies (CFU), which are transformants since they grow without added histidine, were counted and visually inspected for red–white sectoring. In all transformations a background of ∼10% red colonies was consistently observed (see Figure 2) and these colonies were excluded from our analysis. The origin of these colonies are presumably host cells in which the ADE2 gene had mutated prior to, or shortly after, the transformation. Percentages were based on counts of 200–1200 colonies.
bMutation frequencies were based on forward mutation to canavanine resistance and were determined for the MLH1-deletion strain YBT24 harboring the indicated MLH1 gene or the parental expression vector pMETc (‘None’). The median value of nine independent cultures is shown. Mutation defects were calculated with respect to the mutation frequency conferred by the wild-type MLH1 gene.
cMutation frequencies were determined using a URA3 reporter gene preceded by an in-frame (GT)16G microsatellite. Values are from (51).
Prospective screen for mutations in the MLH1 ATPase domain
To generate random mutations in MLH1, 5′ end fragments of the MLH1_h(41-86) and MLH1_h(77-134) genes were synthesized by error-prone PCR and cloned directly in yeast by in vivo gap repair (Figure 1B). About 300–1000 yeast colonies, each representing a cloned PCR fragment, were obtained under the conditions utilized. As shown in Figure 1B, two methods were employed to identify colonies having a deficiency in MMR. In initial experiments, strain YBT24 containing pSH91 was used for in vivo gap repair (see Materials and Methods, Method ‘a’), and transformants were screened for resistance to FOA (ura3) by a semi-quantitative spot test. Those that exhibited a high number of FOA-resistant colonies compared to transformants containing unmutagenized hybrid plasmid were then subjected to canavanine patch analysis, and transformants scored as mutants in both screens subjected to further analysis. The second screening method utilized strain YBT41, which contains the ADE2::MS::ADE2 allele, for in vivo gap repair (see Materials and Methods, Method ‘b’). This method allowed single-step cloning and identification of MMR-deficient transformants since MMR-deficient cells exhibit red–white sectoring directly on transformation plates.
Mutations identified
Hybrid human-yeast MLH1 expression plasmids were isolated from 387 transformants that exhibited a deficiency in MMR. DNA sequencing revealed that 60 of the transformants harbored hybrid MLH1 genes that were identical to the unmutagenized parental gene. In preliminary experiments, several of these molecules were tested functionally by re-introduction into yeast strain YBT24. The results of these quantitative MMR assays demonstrate that the encoded proteins complemented the repair defect as well as the original parental hybrid (data not shown). These findings indicate that the clones without an alteration in the mutagenized region were indeed false positives arising from the selection strategy and do not represent molecules with spontaneous mutations in other regions of the DNA. This number of false positives was not surprising considering the observation that yeast expressing the parental hybrid MLH1 genes exhibits a mutator phenotype at a low frequency (Table 1 and Figure 2). The remaining 327 sequenced genes exhibited at least one alteration in the mutagenized region. There were 24 (7.3%) hybrid MLH1 genes that contained a frameshift mutation and 16 (4.9%) that contained a termination codon (Table 2). The identification of these types of mutations validated the screening strategy because they would be expected to encode truncated MLH1 proteins that lack MMR function. There were 129 (39%) plasmids that contained multiple (2 or more) alterations in the hybrid MLH1 genes and these were not analyzed further. Finally, there were 158 (48%) hybrid MLH1 genes that contained a single missense codon; these represented the most abundant type of alteration found in the screen.
Table 2. Termination codons identified in hybrid human-yeast MLH1 genes.
| Hybrid gene | MLH1 codon (species/region of hybrid)a | Screening methodb | Codon alteration | Consequence | Number of times isolated |
|---|---|---|---|---|---|
| MLH1_h (41-86) | 34 (Yeast) | a | GAG→TAG | E34-Term | 1 |
| 52 (Human) | a,b,b | AAA→TAA | K52-Term | 3 | |
| 53 (Human) | b,b | GAG→TAG | E53-Term | 2 | |
| 57 (Human) | a | AAG→TAG | K57-Term | 1 | |
| 71 (Human) | a | GAA→TAA | E71-Term | 1 | |
| 77 (Human) | b | TGT→TGA | C77-Term | 1 | |
| 120 (Yeast) | a | AGA→TGA | R120-Term | 1 | |
| 142 (Yeast) | a | AAA→TAA | K142-Term | 1 | |
| 77 (Human) | a | CGA→TGA | C77-Term | 1 | |
| MLH1_h (77-134) | 91 (Human) | a | TTA→TAA | L91-Term | 1 |
| 97 (Human) | a | TAT→TAA | Y97-Term | 1 | |
| 100 (Human) | a | CGA→TGA | R100-Term | 1 | |
| 104 (Human) | a | TTG→TAG | L104-Term | 1 |
aCodon numbering is relative to the yeast or human portion of the hybrid MLH1 proteins (see Figure 1B).
bProspective screening methods utilized yeast strain YBT24 for semi-quantitative patch assays (Method ‘a’) or YBT41 for the colorimetric assay (Method ‘b’) as described in the Materials and Methods.
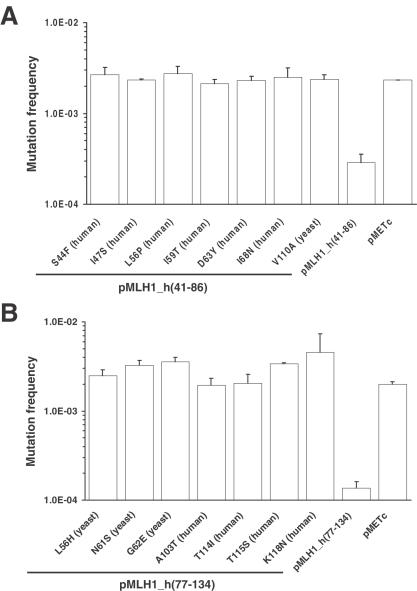
To verify the missense codons as mutations causing loss-of-MMR function, plasmids containing each variant gene were re-introduced into the parental strain (YBT24) for quantitative MMR assays based on stability of the (GT)16G microsatellite in pSH91. The mutation frequency of YBT24; pSH91 containing each cloned MLH1 variant was determined and compared to those levels exhibited by YBT24; pSH91 containing the appropriate parental hybrid gene or the empty expression vector pMETc. Importantly, we compared the mutation frequency of each variant to the mutation frequency of the corresponding parental hybrid to validate a loss-of-function because the two parental hybrids gave slightly different baseline values. Therefore, any effect on function is due to the amino acid substitution in the context of the appropriate hybrid human-yeast molecule. The results of two representative MMR assays, which tested missense variants in both pMLH1_h(41-86) and pMLH1_h(77-134), are shown in Figure 3. Mutation frequencies of 2.1–2.7 × 10−3 were exhibited by yeast cells carrying MLH1_h(41-86) genes encoding S44F, I47S, L56P, I59T, D63Y and I68N substitutions in the human and a V110A substitution in the yeast portion of the hybrid (Figure 3A). These mutation frequencies were similar to the mutation frequency of 2.3 × 10−3 conferred by the pMETc expression vector (lacking an MLH1 gene) and represent mutation defects of 7.2 or more compared to the parental hybrid MLH1_h(41-86). Mutation frequencies of 2.0–4.6 × 10−3 were exhibited by yeast cells carrying MLH1_h(77-134) genes encoding A103T, T114I, T115S and K118N substitutions in the human and L56H, N61S and G62E substitutions in the yeast portion of the hybrid (Figure 3B). These mutation frequencies were similar to the mutation frequency of 2.0 × 10−3 conferred by pMETc and represent mutation defects of 20 or more compared to the parental hybrid MLH1_h(77-134). The results demonstrate a substantial loss-of-MMR function for the MLH1 variants and further validate the screening procedure.
Figure 3.
Yeast strain YBT24 containing pSH91 was transformed with pMLH1_h(41-86), pMLH1_h(77-134), variants of these plasmids isolated in the prospective screen and the expression vector pMETc lacking an MLH1 gene. Mutation frequencies were determined using the standardized quantitative MMR assay as described in Materials and Methods. The mean mutation frequency ± SD of two independent cultures is shown. The species (noted in parenthesis) indicate the portion of the hybrid protein containing the amino acid alteration. (A) Mutation frequencies of pMLH_h(41-86) variants and controls: pMLH1_h(41-86) S44F (human), 2.7 × 10−3; I47S (human), 2.3 × 10−3; L56P (human), 2.7 × 10−3; I59T (human), 2.1 × 10−3; D63Y (human), 2.3 × 10−3; I68N (human), 2.5 × 10−3; V110A (yeast), 2.4 × 10−3; pMLH1_h(41-86), 2.9 × 10−4; pMETc, 2.3 × 10−3. (B) Mutation frequencies of pMLH_h(77-134) variants and controls: pMLH1_h(77-134) L56H (yeast), 2.5 × 10−3; N61S (yeast), 3.2 × 10−3; G62E (yeast), 3.6 × 10−3; A103T (human), 2.0 × 10−3; T114I (human), 2.0 × 10−3; T115S (human), 3.4 × 10−3; K118N (human), 4.6 × 10−3; pMLH1_h(77-134), 1.4 × 10−4; pMETc, 2.0 × 10−3.
Each of the 158 MLH1 variants containing a missense codon was tested in quantitative MMR assays as described above. To confirm the loss-of-MMR function, we assigned a level of 2 or greater for the mutation defect. This level represents a mutation frequency twice as high as the parental MLH1_h(41-86) and MLH1_h(77-134) hybrids and exceeds the maximal levels typically observed for these hybrids (Tables 3 and 4, footnotes). The results of the quantitative MMR assays demonstrated that 151 of the isolated variants (representing 124 non-redundant alterations) exhibited a mutation defect of 2.1 or more. As listed in Tables 3 and 4 [for variants of MLH1_h(41-86) and MLH1_h(77-134), respectively], a range of MMR defects was apparent and the vast majority of missense codons conferred a substantial loss-of-MMR function (Tables 3 and 4; ++ and +++). In addition to amino acid substitutions that impaired MMR activity, seven amino acid substitutions conferred little-to-no loss-of-MMR function in quantitative assays and were classified as functionally inert alterations (Table 5). Variants containing these alterations probably arose as false positives in the prospective screen. A comparison of the amino acid substitutions which had a deleterious effect on MMR activity revealed four alterations [D41G (human)/D38G (yeast), T45I (human)/T42I (yeast), E53V (human)/E50V (yeast) and I68N (human)/I65N (yeast)] that predict identical amino acid substitutions in the equivalent human and yeast residues. Additionally, three alterations [I37T (yeast), F80L (human) and G144S (yeast)] were isolated in the same codon using different hybrids. Identification of equivalent mutations in different hybrids further supports the notion that these substitutions confer detrimental effects on MMR function. In total, 117 unique amino acid substitutions in the N-terminal end of MLH1p were shown to cause a loss-of-MMR function in this study. As compared to an alignment of MLH1p orthologs, the majority of these substitutions occur at highly conserved amino acid residues (Figure 4). Interestingly, 10 substitutions (corresponding to human MLH1p I19F, N38D, N38S, S44F, V49E, N64S, G67E, I68N, C77R and K84E) have been previously reported as possible pathogenic mutations in the human population.
Table 3. MLH1 missense mutations identified in human-yeast hybrid MLH1_h(41-86).
| MLH1 gene or variant codon # | Screening methoda | Missense mutation | Consequence | Corresponding human residue | Mutation defectb | Number of times isolated |
|---|---|---|---|---|---|---|
| Yeast codon | ||||||
| 8 | a | CTT→CAC | L8H | L11 | ++ | 1 |
| 16 | a | ATT→TTT | I16F | I19 | ++ | 1 |
| 26 | a | GTA→ATA | V26I | A29 | ++ | 1 |
| 35 | a | AAT→GAT | N35D | N38 | +++ | 1 |
| a | AAT→ACT | N35T | N38 | ++ | 1 | |
| 37 | a,a | ATC→ACC | I37T | L40 | +++ | 2 |
| b | ATC→AAC | I37N | L40 | +++ | 1 | |
| Human codon | ||||||
| 41 | a | GAT→GGT | D41G | — | ++ | 1 |
| 42 | a | GCA→ACA | A42T | — | +++ | 1 |
| b | GCA→GAA | A42E | — | +++ | 1 | |
| b | GCA→GTA | A42Vc | — | +++ | 1 | |
| 44 | a | TCC→TTC | S44F | — | +++ | 1 |
| 45 | a,b | ACA→ATA | T45I | — | +++ | 2 |
| 46 | a | AGT→ACT | S46T | — | ++ | 1 |
| 47 | b | ATT→ACT | I47T | — | ++ | 1 |
| a | ATT→AGT | I47S | — | +++ | 1 | |
| 48 | a | CAA→TAT | Q48Y | — | +++ | 1 |
| 49 | b | GTG→GAG | V49E | — | ++ | 1 |
| a | GTG→ATG | V49M | — | ++ | 1 | |
| a | GTG→GCG | V49A | — | +++ | 1 | |
| 51 | a,b | GTT→GAT | V51D | — | ++ | 2 |
| a | GTT→GCT | V51A | — | ++ | 1 | |
| 52 | a,a,b | AAA→ATA | K52I | — | + | 3 |
| 53 | a,a,b | GAG→GTG | E53V | — | ++ | 3 |
| 54 | a | GGA→AGA | G54R | — | + | 1 |
| 55 | a,a,b | GGC→GAC | G55D | — | + | 3 |
| a | GGC→AGC | G55S | — | ++ | 1 | |
| 56 | a | CTG→ATG | L56M | — | + | 1 |
| a | CTG→CCG | L56P | — | +++ | 1 | |
| 57 | a | AAG→GAG | K57Ec | — | + | 1 |
| b | AAG→AAC | K57N | — | +++ | 1 | |
| 59 | b | ATT→AAT | I59N | — | +++ | 1 |
| a,a | ATT→TTT | I59F | — | +++ | 2 | |
| a | ATT→ACT | I59T | — | +++ | 1 | |
| 60 | a | CAG→CCG | Q60P | — | ++ | 1 |
| 61 | a | ATC→AAC | I61N | — | ++ | 1 |
| 63 | a | GAC→TAC | D63Y | — | +++ | 1 |
| 64 | b | AAT→ATT | N64I | — | ++ | 1 |
| 65 | b | GGC→GTC | G65V | — | +++ | 1 |
| a | GGC→GCC | G65A | — | +++ | 1 | |
| a | GGC→GAC | G65D | — | ++ | 1 | |
| a | GGC→AGC | G65S | — | ++ | 1 | |
| 67 | a,a,b | GGG→GAG | G67E | — | ++ | 3 |
| a | GGG→GTG | G67V | — | +++ | 1 | |
| 68 | a | ATC→AAC | I68N | — | +++ | 1 |
| a,b | ATC→TTC | I68F | — | ++ | 2 | |
| b | ATC→AGC | I68Sc | — | ++ | 1 | |
| 70 | a | AAA→AAT | K70N | — | +++ | 1 |
| a | AAA→ATA | K70I | — | +++ | 1 | |
| 72 | a,b | GAT→GGT | D72G | — | ++ | 2 |
| a,b | GAT→GTT | D72V | — | + | 2 | |
| 73 | b | CTG→ATG | L73M | — | ++ | 1 |
| a | CTG→CCG | L73P | — | ++ | 1 | |
| a | CTG→CAG | L73Q | — | ++ | 1 | |
| 76 | b | GTA→GAA | V76E | — | +++ | 1 |
| 77 | a | TGT→GGT | C77G | — | ++ | 1 |
| b | TGT→TCT | C77S | — | ++ | 1 | |
| 79 | b | AGG→TGG | R79Wc | — | ++ | 1 |
| 80 | a | TTC→TCC | F80Sc | — | ++ | 1 |
| b | TTC→CTC | F80L | — | +++ | 1 | |
| b | TTC→ATC | F80I | — | +++ | 1 | |
| 81 | a | ACG→ATG | T81M | — | + | 1 |
| 82 | a,a | ACG→TCG | T82S | — | + | 2 |
| a,b | ACG→AAG | T82K | — | +++ | 2 | |
| a | ACG→ATG | T82M | — | +++ | 1 | |
| 83 | a,b | TCC→CCC | S83P | — | ++ | 2 |
| a | TCC→TTC | S83F | — | +++ | 1 | |
| 84 | a | AAA→GAA | K84R | — | ++ | 1 |
| a | AAA→AGA | K84E | — | ++ | 1 | |
| 85 | a | TTA→TCA | L85S | — | ++ | 1 |
| Yeast codon | ||||||
| 86 | a | GAA→GGA | E86Gc | E89 | ++ | 1 |
| 88 | a | TTG→GTG | L88V | L91 | ++ | 1 |
| 99 | b | GAA→GGA | E99G | E102 | ++ | 1 |
| 108 | a | GCA→CCA | A108P | A111 | +++ | 1 |
| 110 | a | GTC→GCC | V110Ac | V113 | +++ | 1 |
| 112 | b | GTA→GAA | V112E | I115 | ++ | 1 |
| 113 | b | ACG→GCG | T113A | T116 | ++ | 1 |
| 144 | a | GGT→AGT | G144S | G147 | +++ | 1 |
aMMR-deficient transformants were identified by (‘a’) qualitative patch assays using YBT24 or (‘b’) colorimetric assay using YBT41 as described in the Materials and Methods section.
bYeast strain YBT24 containing pSH91 was transformed with pMLH1_h(41-86) containing the indicated missense mutations. Mutation frequencies were determined using a standardized MMR assay based on instability of the GT-tract in pSH91 [Ellison et al., (51)]. To calculate the mutation defect, the mean mutation frequency conferred by each variant was divided by the mutation frequency conferred by the parental MLH1_h(41-86) gene. +, Mutation defect of 2.1 to 3.9 (18–33% loss-of-MMR function relative to the mutation frequency of the MLH1-null strain YBT24); ++, Mutation defect of 4.0–7.6 (34–66% loss-of-MMR function); +++, Mutation defect of 7.8 or greater (>67% loss-of-MMR function). The mean mutation frequency conferred by pMLH1_h(41-86) was 2.7 × 10−4 (range: 1.1–4.4 × 10−4) The mean mutation frequency conferred by the empty expression vector pMETc was 3.2 × 10−3 (range: 1.9–7.0 × 10−3) (mutation defect = 11.7).
cIn addition to the indicated missense mutation, the following silent alterations were observed (mutation/silent alteration): A42V/F85F; K57E/T45T; I68S/I47I and I75I; R79W/D143D; F80S/L73L; E86G/T82T and K142K; V110A/T66T.
Table 4. MLH1 missense mutations identified in human-yeast hybrid MLH1_h(77-134).
| MLH1 gene or variant codon # | Screening methoda | Missense mutation | Consequence | Corresponding human residue | Mutation defectb | Number of times isolated |
|---|---|---|---|---|---|---|
| Yeast codon | ||||||
| 30 | a | AAA→AAT | K30N | K33 | +++ | 1 |
| 35 | a | AAT→AGT | N35S | N38 | +++ | 1 |
| 37 | a | ATC→TTC | I37F | L40 | ++ | 1 |
| a | ATC→ACC | I37T | L40 | +++ | 1 | |
| 38 | a,a,b | GAT→GGT | D38G | D41 | +++ | 3 |
| b | GAT→GAA | D38E | D41 | +++ | 1 | |
| b | GAT→ATT | D38N | D41 | +++ | 1 | |
| 40 | b | AAT→ATT | N40Ic | K43 | +++ | 1 |
| 41 | a,a | GCT→GTT | A41V | S44 | ++ | 2 |
| 42 | a | ACA→ATA | T42I | T45 | +++ | 1 |
| 45 | b | GAT→GGT | D45G | Q48 | + | 1 |
| 46 | b | ATT→AAT | I46N | V49 | +++ | 1 |
| 49 | a | AAG→GAG | K49E | K52 | ++ | 1 |
| 50 | a | GAA→GTA | E50V | E53 | + | 1 |
| 52 | a,a | GGA→AGA | G52R | G55 | + | 2 |
| 56 | b | CTT→CAT | L56H | I59 | +++ | 1 |
| 58 | a,b | ATA→AAA | I58K | I61 | +++ | 2 |
| 60 | a | GAT→GGT | D60G | D63 | ++ | 1 |
| 61 | b | AAC→AGC | N61S | N64 | +++ | 1 |
| 62 | a,b | GGA→GAA | G62E | G65 | +++ | 2 |
| a,a | GGA→AGA | G62R | G65 | ++ | 2 | |
| 65 | a | ATT→AAT | I65N | I68 | +++ | 1 |
| 71 | a | CCA→CTA | P71L | D74 | ++ | 1 |
| Human codon | ||||||
| 77 | a | TGT→CGT | C77R | — | ++ | 1 |
| 78 | a | GAG→GTG | E78V | — | ++ | 1 |
| 80 | a,a | TTC→CTCd | F80Lc | — | +++ | 2 |
| 89 | a | GAG→GTG | E89V | — | + | 1 |
| 99 | a | TTT→ATT | F99I | — | +++ | 1 |
| 99 | a | TTT→CTT | F99L | — | ++ | 1 |
| 100 | b | CGA→CAA | R100Q | — | ++ | 1 |
| 101 | a | GGT→GAT | G101Dc | — | +++ | 1 |
| 103 | a | GCT→GTT | A103V | — | ++ | 1 |
| a,b | GCT→ACT | A103T | — | ++ | 2 | |
| a | GCT→CCT | A103P | — | ++ | 1 | |
| 111 | a | GCT→ACT | A111T | — | +++ | 1 |
| 114 | a | ACT→ATT | T114I | — | ++ | 1 |
| 115 | b | ATT→AGT | I115Sc | — | +++ | 1 |
| b | ATT→AAT | I115N | — | +++ | 1 | |
| b | ATT→TTT | I115F | — | ++ | 1 | |
| 116 | a | ACA→TCA | T116S | — | + | 1 |
| 118 | b | AAA→AAT | K118N | — | +++ | 1 |
| a | AAA→ATA | K118I | — | + | 1 | |
| 133 | a | GGA→GAA | G133E | — | ++ | 1 |
| Yeast codon | ||||||
| 136 | a | CCC→CAC | P136H | P139 | + | 1 |
| 140 | a | GCT→GTT | A140V | A143 | ++ | 1 |
| 144 | a | GGT→AGT | G144S | G147 | ++ | 1 |
aMMR-deficient transformants were identified by (Method ‘a’) qualitative patch assays using YBT24 or (Method ‘b’) colorimetric assay using YBT41 as described in the Materials and Methods.
bYeast strain YBT24 containing pSH91 was transformed with pMLH1_h(77-134) containing the indicated missense mutations. Mutation frequencies were determined using a standardized MMR assay based on instability of the GT-tract in pSH91 [Ellison et al., (51)]. To calculate the mutation defect, the mean mutation frequency conferred by each variant was divided by the mutation frequency conferred by the parental MLH1_h(77-134) gene. +, Mutation defect of 2.5–9.0 (9–33% loss-of-MMR function relative to the mutation frequency of the MLH1-null strain YBT24); ++, Mutation defect of 9.1–17.9 (34–66% loss-of-MMR function); +++, Mutation defect of 18.0 or greater (>67% loss-of-MMR function). The mean mutation frequency conferred by pMLH1_h(77-134) was 1.2 × 10−4 (range: 0.6–2.4 × 10−4). The mean mutation frequency conferred by the empty expression vector pMETc was 3.3 × 10−3 (range: 1.8–7.0 × 10−3) (mutation defect = 27.5).
cIn addition to the indicated missense mutation the following silent alterations were observed (mutation/silent alteration): N40I/K134K; F80L/A92A; G101D/K54K; I115S/T116T.
dThe missense mutation TTC→TTA was also identified.
Table 5. MLH1p amino acid substitutions conferring minimal loss-of-MMR functiona.
| MLH1 gene and variant codon | Screening methodb | Missense mutation | Consequence | Corresponding human residue | Tolerated in other species?c |
|---|---|---|---|---|---|
| MLH1_h(41-86) | |||||
| 62 (Human) | b | CAA→CGA | Q62R | — | Yes |
| 64 (Human) | a | AAT→GAT | N64D | — | Yes |
| 71 (Human) | a | GAA→GAT | E71D | — | Yes |
| MLH1_h(77-134) | |||||
| 33 (Yeast) | a | ATG→TTG | M33L | I36 | Yes |
| 72 (Yeast) | a | ATC→ACC | I72T | I75 | No |
| 95 (Human) | a | TCT→ACT | S95T | — | No |
| 133 (Yeast) | a | TTG→TCG | L133S | K136 | Yes |
aMutation frequencies were measured using the standardized GT-tract instability assay as described in Materials and Methods. Mutation frequencies were MLH1_h(41-86) Q62R, 3.6 × 10−4; MLH1_h(41-86) N64D, 2.1 × 10−4; MLH1_h(41-86) E71D, 3.2 × 10−4; MLH1_h(77-134) M33L, 4.0 × 10−5; MLH1_h(77-134) I72T, 1.0 × 10−4; MLH1_h(77-134) S95T, 4.6 × 10−5 and MLH1_h(77-134) L133S, 1.5 × 10−4. These values represent mutation defects of 1.4, 0.8, 1.2, 0.3, 0.9, 0.4 and 1.3, respectively, compared to the appropriate parental hybrid gene.
bMMR-deficient yeast colonies were identified by qualitative patch assays using YBT24 (Method ‘a’) or colorimetric assay using YBT41 (Method ‘b’) as described in Materials and Methods.
cIndicates whether the variant (new) residue is present at the corresponding position in other species (Figure 4).
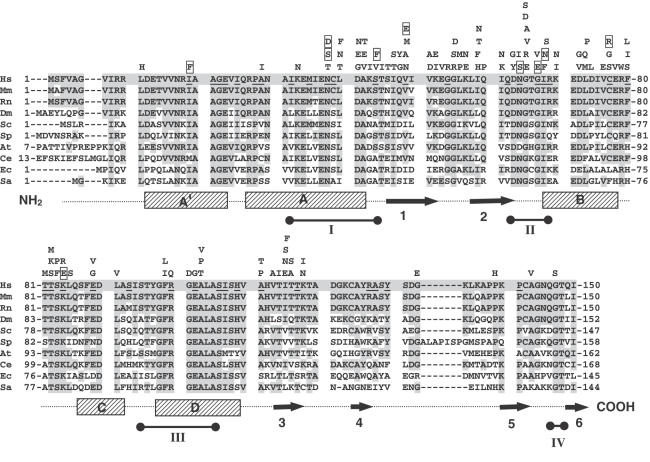
Figure 4.
Amino acid alignment of MLH1p orthologs. The 117 amino acid substitutions causing loss-of-MMR function are noted above the appropriate residue in the human MLH1p sequence. MLH1p sequences from human (Hs, H.sapiens), mouse (Mm, M.musculus), rat (Rn, R.norvegicus), fruit fly (Dm, D.melanogaster), yeast (Sc, S.cerevisiae and Sp, S.pombe), plant (At, A.thaliana), flatworm (Ce, C.elegans) and bacteria (Sa, S.aureus and Ec, E.coli) were aligned using ClustalW (http://www.ebi.ac.uk/clustalw/). Conserved residues are highlighted. Structural features, including α-helices (stippled boxes) and β-strands (arrows), in the E.coli MutL polypeptide (41) are indicated below the alignment. Barbells represent the location of the ATP binding motifs (I–IV), which are conserved in GHL ATPases (37,41). Underlined residues have been previously reported, as a result of HNPCC genetic testing, to exhibit missense alterations. Boxed residues (substitutions) were isolated in this study and are equivalent to substitutions found in the human population and associated with HNPCC (see Materials and Methods).
Saturation mutagenesis at human MLH1 codons 43 and 44
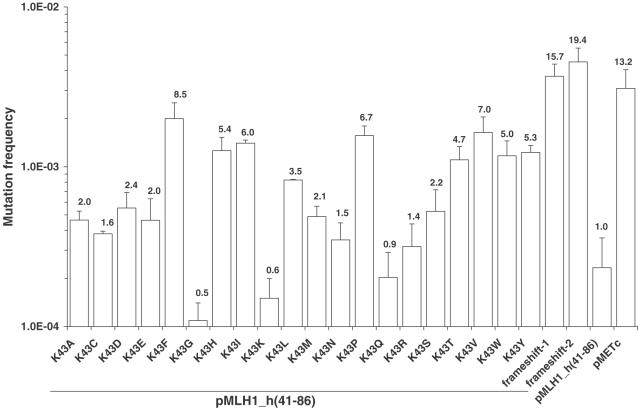
To investigate the functional consequence of all possible amino acid substitutions at a specific residue, comprehensive mutagenesis was carried out on human MLH1 codons 43 and 44 in hybrid MLH1_h(41-86). The codons 43 and 44 were chosen because they lie in a region known to be functionally important (34,41,51,43), but variable amongst species (Figure 4). We also focused on these adjacent residues because S44F was one of the first human alterations identified as being strongly associated with the development of HNPCC (34). Moreover, the functional and biochemical effects of a comparable change in yeast (corresponding to A41S and A41F) has been widely studied (37,68,69) and these results could be used as a basis for comparison. In native human MLH1p, residue 43 is lysine (K43), a positively-charged hydrophilic residue, and residue 44 is serine (S44), an uncharged small polar residue. Mutagenized genes were introduced into YBT24 containing pSH91 and tested in the standardized quantitative MMR assay. Mutation frequencies were compared to those conferred by the parental hybrid MLH1_h(41-86) and the same strain carrying the empty expression vector (pMETc). As expected, a silent K43K (AAA→AAG) alteration in MLH1_h(41-86) did not disrupt the MMR function compared to the parental hybrid MLH1_h(41-86), while spontaneous deletions in codons 43 (‘frameshift-1’) and 45 (‘frameshift-2’) exhibited mutation frequencies that were similar to that conferred by the empty expression vector pMETc (Figure 5). Of the 19 possible substitutions at codon 43, 14 (K43A, K43D, K43E, K43F, K43H, K43I, K43L, K43M, K43P, K43S, K43T, K43V, K43W and K43Y) conferred mutation frequencies in the range of 4.6 × 10−4–2.0 × 10−3. These values represent mutation defects of 2.0–8.5 compared to the parental hybrid (Figure 5). The remaining five alterations (K43C, K43G, K43N, K43Q and K43R) conferred mutation frequencies of 1.1 × 10−4–3.8 × 10−4, and values that represent a mutation defect of 1.6 or less thus have little-to-no effect on protein function.
Figure 5.
Mutation frequencies conferred by missense substitutions at human MLH1 codon 43 (K43). Yeast strain YBT24 containing pSH91 was transformed with pMLH1_h(41-86), variants of this plasmid containing the indicated amino acid substitution, and the expression vector pMETc lacking an MLH1 gene. Mutation frequencies were determined using the standardized quantitative MMR assay as described in Materials and Methods. The mean mutation frequency ± SD of two to nine independent cultures is shown. The cells containing the parental hybrid pMLH1_h(41-86) exhibited a mutation frequency of 2.3 × 10−4. The mutation defect (shown above each bar) for each variant and control was calculated by dividing the mutation frequency of cells expressing the variant by the mutation frequency of cells expressing parental hybrid pMLH1_h(41-86). MLH1_h(41-86) genes containing spontaneous nucleotide deletions in codon 43 (an A-deletion) and 45 (a CA-deletion) are referred to as ‘frameshift-1’ and ‘-2’, respectively.
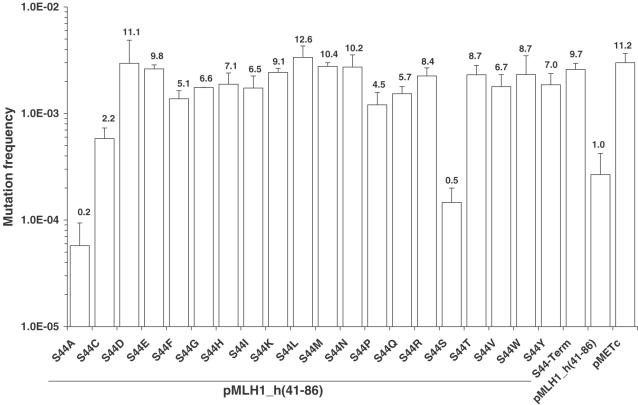
Amino acid alterations at residue 44 showed that 18 of the 19 possible substitutions gave a pronounced loss-of-MMR function compared to the parental hybrid MLH1_h(41-86) (Figure 6). As expected, a silent S44S (TCC→TCA) alteration in MLH1_h(41-86) did not disrupt the MMR function compared to the parental hybrid MLH1_h(41-86), while a hybrid containing a termination codon at position 44 (‘S44-Term’) exhibited a mutation frequency that was similar to that conferred by the empty expression vector pMETc (Figure 6). Eighteen (S44C, S44D, S44E, S44F, S44G, S44H, S44I, S44K, S44L, S44M, S44N, S44P, S44Q, S44R, S44T, S44V, S44W and S44Y) of the nineteen possible substitutions at codon 44 conferred a pronounced loss-of-MMR function with mutation frequencies in the range 5.8 × 10−4–3.4 × 10−3 (Figure 6), representing mutation defects of 2.2–12.6. The remaining alteration (S44A) conferred a mutation frequency of 5.8 × 10−5, which corresponds to a mutation defect of 0.2. While substitutions at residue S44 tended to be more severe (conferring a mutation defect of 5.0 or greater) than those at residue K43, the majority of substitutions at both codons impaired MMR activity to some degree. Interestingly, for both residues K43 and S44, the range of substitutions that resulted in little-to-no loss-of-MMR function closely mirrored the variability observed in nature (Figure 4).
Figure 6.
Mutation frequencies conferred by missense substitutions at human MLH1 codon 44 (S44). Yeast strain YBT24 containing pSH91 was transformed with pMLH1_h(41-86), variants of this plasmid containing the indicated amino acid substitution, and the expression vector pMETc lacking an MLH1 gene. Mutation frequencies were determined using the standardized quantitative MMR assay as described in Materials and Methods. The mean mutation frequency ± SD of two to twelve independent cultures is shown. The cells containing the parental hybrid pMLH1_h(41-86) exhibited a mutation frequency of 2.7 × 10−4. The mutation defect (shown above each bar) for each variant and control was calculated by dividing the mutation frequency of cells expressing the variant by the mutation frequency of cells expressing parental hybrid pMLH1_h(41-86). A MLH1_h(41-86) gene containing a termination codon at position 44 is referred to as ‘S44-Term’.
DISCUSSION
In this study, a prospective genetic screen in yeast was developed to identify codon changes in MLH1 causing loss-of-MMR function. A key feature of this screen was the use of functional human-yeast hybrid genes (51), which allowed isolation of mutations in the ATPase domain of the human MLH1p. In earlier studies, prospective genetic screens have been performed in E.coli (70) and yeast (71–74) in order to investigate the structure and function of the native MMR proteins in those organisms. However, a prospective genetic screen for mutations specific to human MLH1 coding sequences has not been previously reported. In total, we have assembled functional information on 162 amino acid substitutions in the ATPase domain of MLH1p. The data consists of 117 replacements that impair MMR function, seven functionally silent alterations and the effects of all amino acid substitutions at residues K43 and S44 on protein function. Importantly, previously reported HNPCC-associated alterations [see MLH1 mutational databases and other reports (64,65,75,76)] exhibited considerable overlap with the loss-of-MMR function mutations reported here. Of 33 distinct HNPCC-associated substitutions which have been reported in the N-terminal end of human MLH1p, 10 identical alterations were isolated in our prospective screen (see Figure 4). The novel mutations identified in this study may aid in the interpretation of HNPCC genetic tests if the same variation is observed clinically.
This study describes a novel method for the identification of human MMR gene sequence alterations that impair function of the encoded protein in MMR. If such variants occur in humans, individuals harboring such alleles may have an elevated mutation rate and a predisposition to develop cancer. The method employs the yeast S.cerevisiae, which has been used previously for the identification of mutant MMR genes. For example, Jeyaprakash et al. (77) used genetic complementation experiments and then direct cloning and DNA sequencing to ascertain the identity of the mutant gene in yeast strains with preexisting defects in microsatellite stability. More recent reports describe global mutagenesis of yeast, selection of yeast strains for those having alterations in MMR gene activity followed by cloning and DNA sequencing (71–74). It should be noted that these studies were focused on finding variants of the native yeast proteins. Indeed, if reported at all, expression of the human MMR proteins in yeast has either no detectable biological activity (MSH2, MSH3 and MSH6) or induces a dominant negative mutator phenotype (MLH1 and the MSH2–MSH6 heterodimers) (47,78). Previous studies have attempted to bypass these impediments by using, e.g. an hMSH2–ADE2 fusion gene to screen for stop codons in the hMSH2 coding sequence or assays based on gain or loss of the dominant mutator phenotype (47,78,79). However, these assays do not measure the native biological activity of the protein. The approach described in this study, employing hybrid human-yeast MMR proteins that are functional in MMR (51), has allowed functional analyses of substitutions in the native human MMR gene sequence in yeast.
In this investigation, 27 codon alterations were isolated on two or more occasions (see Tables 3 and 4). Interestingly, only 4 of these 27 duplicates could have been due to cloning of the same amplified mutant from a single PCR fragment pool (data not shown). Instead, the majority of duplications were isolated in different gap repair screens using different PCR fragment pools and therefore represent independent generation of the same mutation. Thus, an important source of duplicity must be that (i) certain nucleotides are more prone to mutagenesis, i.e. ‘mutational hotspots’ (although two different polymerases and PCR amplification conditions were employed; see Materials and Methods) or (ii) the screen is reaching its limits in terms of the number of loss-of-function mutations possible. These possibilities are not mutually exclusive and the duplications may, in fact, result from a combination of these factors.
The results of this investigation raise an important question. How many loss-of-MMR function mutations might be expected in the human MLH1 region that was mutagenized? To estimate this number, we compared the experimental results to all possible single nucleotide missense codons at human codons 41–86 and 77–134. This analysis was restricted to the human portion of each hybrid since the crossover site sites between yeast sequences in the vector and mutagenized fragment during in vivo gap repair will vary slightly between clones. The first estimate was based on the number of possible termination codons actually isolated in the prospective screen (Table 2). For hybrids MLH1_h(41-86) and MLH1-h(77-134), 5 of 16 (31%) and 5 of 21 (23%) of possible stop codons, respectively, were isolated. A second estimate is based on the results obtained by making all possible amino acid substitutions at human residues K43 and S44. At position 43, six different single base substitution codon changes (K43E, K43I, K43N, K43Q, K43R and K43T) are possible. Three of these exhibit 2-fold or greater loss-of-MMR function in vivo (Figure 5; K43E, K43I and K43T). Only a K43I codon was isolated in the prospective screen. At position 44, six different single base substitution codon changes (S44A, S44C, S44F, S44P, S44T and S44Y) are possible. Five of these confer loss-of-MMR function (Figure 6; S44C, S44F, S44P, S44T and S44Y), and one (S44F) was isolated in the prospective screen (Table 3). Thus, at human codons K43 and S44, 33% (1 of 3) and 20% (1 of 5), respectively, of all single base substitution mutations were isolated. Cumulatively, this analysis suggests that we have identified ∼25% of all single base substitution missense mutations in this region, implying that the total number of MLH1-inactivating amino acid substitutions in the region of MLH1 subjected to mutagenesis is ∼460.
The majority of amino acid substitutions that conferred a loss-of-MMR function were isolated at highly-conserved residues (Figure 4). Only 3 of the 117 substitutions that conferred loss-of-MMR function (hMLH1 K52E, K118I and P139H) occurred as the native amino acid at the equivalent position in any MLH1p from nine other species. In contrast, five of seven substitutions that exhibited little-to-no loss-of-MMR function (Table 5) occurred naturally at the equivalent position in other species. There were several amino acid substitutions whose functional consequence may not have been predicted based on either the variation in nature or the biochemical properties of the alternate residues. For example, we found the biochemically conservative alterations hMLH1p D38E (both acidic), S46T (both uncharged polar) and N64S (both uncharged polar) resulted in loss-of-MMR function. Some alterations that may have been expected to confer detrimental effects on MMR function gave little-to-no loss of function. Thus, the substitutions I72T, K43C and K43G, which considerably alter the charge, polarity and size of the residue, did not substantially impair MMR function. Clearly, the functional consequence of any substitution requires direct experimental evaluation. This point is particularly relevant for the interpretation of HNPCC genetic tests, which often reveal novel missense codons in MLH1 and other MMR genes.
As summarized in Figure 4, amino acid substitutions causing loss-of-MMR function were identified at 61 residues throughout the N-terminal ATPase domain of MLH1p. Previous structural models of E.coli MutL (41,42) and biochemical studies using yeast and human MLH1p (37–40) provide a foundation for understanding as to why substitutions at certain residues may impair MMR function. Three substitutions causing loss-of-MMR function were identified at the residue corresponding to hMLH1p N38, which helps to position a centrally located Mg2+ ion presumed to be important for ATP binding (38,39,42). The amino acid substitutions identified (corresponding to hMLH1p N38D, N38S and N38T) may either abolish Mg2+ interactions or alter the placement of this critical ion. Substitutions were also identified at residues corresponding to hMLH1p D63, T82, S83, F99 and R100, which appear to have a direct role in ATP binding (38,39,42). These substitutions are likely to perturb ATP binding and/or hydrolysis and, thus, prevent the conformational changes in MLH1p which are predicted to signal downstream MMR events. Most of the substitutions causing loss-of-MMR function were localized in the region corresponding to hMLH1p 41 through 86, which make up a large portion of the ATP-binding pocket and ‘ATP lid’ (41,42). Although this region is certainly important for DNA MMR, we suspect that many mutations were localized here because, as depicted in Figure 1B, this portion of the mutant MLH1 gene originates solely from recombination with a mutagenized PCR fragment. A mutational analysis of other regions of MLH1 will require different gapped vector DNAs and the use of additional human-yeast hybrid MLH1 genes.
Based on the crystal structure of E.coli MutL, human MLH1p residues K43 and S44 are expected to lie adjacent to helix αA within a conserved ATP-binding motif (motif I)(41,42). It has been predicted that mutations in and around helix Aα dislocate a conserved glutamic acid residue, corresponding to hMLH1p E34, which is important for ATP binding and hydrolysis (37,38,42). Specific biochemical functions have not been assigned to K43 or S44, but as shown in this investigation alterations in the size, charge and polarity of these residues result in distinct functional consequences for MLH1p. Previous investigations using the native yeast MLH1p have shown that substitutions mimicking a human HNPCC alteration (human MLH1p S44F) (34,68,69) result in a loss-of-MMR function in vivo (51,43). We confirmed this finding using the human-yeast hybrid gene MLH1_h(41-86) and showed that all amino acid substitutions except S44A impaired MMR function. At position 44, it appears that the small, uncharged side chains on serine and alanine are critical for proper MLH1p structure and/or function. At position K43, the systematic substitution of all possible amino acids resulted in a variety of functional consequences. In general, the introduction of amino acids with bulky and/or hydrophobic side chains tended to result in loss-of-MMR activity, while the introduction of amino acids with hydrophilic side chains were functionally tolerated. In addition, the results of this systematic analysis of all possible amino acid substitutions is consistent with our previous investigations, in which it was shown that substitutions in the human population can be either silent polymorphisms, inactivating mutations, or give rise to proteins that reduce the efficiency of MMR relative to the parental molecule (51).
The goal of this investigation is to identify MLH1p amino acid substitutions which impair DNA MMR in vivo. As noted, both human-yeast hybrid MLH1 proteins are functional in MMR but at a reduced efficiency compared to the native yeast protein. This is presumably due to the replacement of the yeast coding sequence with the conserved, but not identical, homologous human coding sequence. There is a formal possibility, therefore, that some of the missense codons we identified are specific to the hybrid protein and might not be observed when introduced to a native protein. Extrapolation of the results of this study to native proteins should consider the caveat that a subset of the variants may be specific to the hybrid molecule. At this time, it is not possible to conclude whether the missense mutations identified perturb other functions of MLH1p, such as its role in meiotic crossing-over (25,26), and signaling of apoptosis in response to DNA damage (31,32). However, based on the results of a recent investigation (80), it appears that the majority of the mutations the ATPase domain that inactivate MMR also inactivate meiotic crossing-over. Further studies will be required to determine whether the ATPase activity of MLH1p is critical in other functions of the protein. Further studies will also be required to determine whether the substitutions have detrimental effects on the stability and/or solubility of MLH1p, protein–protein interactions and/or specific steps in ATP binding or hydrolysis (17,37,41,44).
Understanding the functional consequence of missense codons in MLH1 is critical for increasing the utility of genetic tests for HNPCC. As described in this investigation, a rapid and biologically relevant strategy may be to accumulate functional information concerning putative disease-causing mutations in a prospective manner. Extending the prospective screen through additional portions of human MLH1 and other MMR genes could result in a comprehensive catalog of missense mutations that cause loss-of-MMR function.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Samantha Bohanon for technical assistance in the early stages of this project. This work was supported by a grant from the National Institutes of Health to G.A.B. (R44 CA081965).
REFERENCES
- 1.Kolodner R.D. and Marsischky,G.T. (1999) Eukaryotic DNA mismatch repair. Curr. Opin. Genet. Dev., 9, 89–96. [DOI] [PubMed] [Google Scholar]
- 2.Harfe B.D. and Jinks-Robertson,S. (2000) DNA mismatch repair and genetic instability. Annu. Rev. Genet., 34, 359–399. [DOI] [PubMed] [Google Scholar]
- 3.Loeb L.A. (1991) Mutator phenotype may be required for multistage carcinogenesis. Cancer Res., 51, 3075–3079. [PubMed] [Google Scholar]
- 4.de Wind N., Dekker,M., Berns,A., Radman,M. and te Riele,H. (1995) Inactivation of the mouse Msh2 gene results in mismatch repair deficiency, methylation tolerance, hyperrecombination, and predisposition to cancer. Cell, 82, 321–330. [DOI] [PubMed] [Google Scholar]
- 5.Reitmair A.H., Schmits,S., Ewel,A., Bapat,B., Redson,M., Mitri,A., Waterhouse,P., Mittrucker,H.-W., Wakeham,A., Liu,B., Thomason,A., Griesser,H., Gallinger,S., Ballhausen,W.G., Fishel,R. and Mak,T.W. (1995) MSH2 deficient mice are viable and susceptible to lymphoid tumors. Nature Genet., 11, 64–70. [DOI] [PubMed] [Google Scholar]
- 6.Prolla T.A., Baker,S.M., Harris,A.C., Tsao,J.-L., Yao,X., Bronner,C.E., Zheng,B., Gordon,M., Reneker,J., Arnheim,N., Shibata,D., Bradley,A. and Liskay,R.M. (1998) Tumor susceptibility and spontaneous mutation in mice deficient in Mlh1, Pms1 and Pms2 DNA mismatch repair. Nature Genet., 18, 276–279. [DOI] [PubMed] [Google Scholar]
- 7.Edelmann W., Yang,K., Umar,A., Heyer,J., Lau,K., Fan,K., Liedtke,W., Cohen,P.E., Kane,M.F., Lipford,J.R., Yu,N., Crouse,G.F., Pollard,J.W., Kunkel,T., Lipkin,M., Kolodner,R. and Kucherlapati,R. (1997) Mutation in the mismatch repair gene Msh6 causes cancer susceptibility. Cell, 91, 467–477. [DOI] [PubMed] [Google Scholar]
- 8.Peltomaki P. and de la Chapelle,A. (1997) Mutations predisposing to hereditary nonpolyposis colorectal cancer. Adv. Cancer Res., 71, 93–119. [DOI] [PubMed] [Google Scholar]
- 9.Jiricny J. and Nystrom-Lahti,M. (2000) Mismatch repair defects in cancer. Curr. Opin. Genet. Dev., 10, 157–161. [DOI] [PubMed] [Google Scholar]
- 10.Peltomaki P. (2001) Deficient DNA mismatch repair: a common etiologic factor for colon cancer. Hum. Mol. Genet., 10, 735–740. [DOI] [PubMed] [Google Scholar]
- 11.Lynch H.T. and de la Chappelle,A. (1999) Genetic susceptibility to non-polyposis colorectal cancer. J. Med. Genet., 36, 801–818. [PMC free article] [PubMed] [Google Scholar]
- 12.Mitchell R.J., Farrington,S.M., Dunlop,M.G. and Campbell,H. (2002) Mismatch repair genes hMLH1 and hMSH2 and colorectal cancer: a HuGE review. Am. J. Epidemiol., 156, 885–902. [DOI] [PubMed] [Google Scholar]
- 13.Modrich P. (1994) Mismatch repair, genetic stability, and cancer. Science, 266, 1959–1960. [DOI] [PubMed] [Google Scholar]
- 14.Lipkin S.M., Wang,V., Jacoby,R., Banerjee-Basu,S., Baxevanis,A.D., Lynch,H.T., Elliott,R.M. and Collins,F.S. (2000) MLH3: a DNA mismatch repair gene associated with mammalian microsatellite instability. Nature Genet., 24, 27–35. [DOI] [PubMed] [Google Scholar]
- 15.Flores-Rozas H. and Kolodner,R.D. (1998) The Saccharomyces cerevisiae MLH3 gene functions in MSH3-dependent suppression of frameshift mutations. Proc. Natl Acad. Sci. USA, 95, 12404–12409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tishkoff D.X., Boerger,A.L., Bertrand,P., Filosi,N., Gaida,G.M., Kane,M.F. and Kolodner,R.D. (1997) Identification and characterization of Saccharomyces cerevisiae EXO1, a gene encoding an exonuclease that interacts with MSH2. Proc. Natl Acad. Sci. USA, 94, 7487–7492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tran P.T., Simon,J.A. and Liskey,R.M. (2001) Interactions of Exo1p with components of MutLα in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 98, 9760–9765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin Y.L., Shivji,M.K., Chen,C., Kolodner,R., Wood,R.D. and Dutta,A. (1998) The evolutionarily conserved zinc finger motif in the largest subunit of human replication protein A is required for DNA replication and mismatch repair but not for nucleotide excision repair. J. Biol. Chem., 273, 1453–1461. [DOI] [PubMed] [Google Scholar]
- 19.Ramilo C., Gu,L., Guo,S., Zhang,X., Patrick,S.M., Turchi,J.J. and Li,G.-M. (2002) Partial reconstitution of human DNA mismatch repair in vitro: characterization of the role of human replication protein A. Mol. Cell. Biol., 22, 2037–2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson R.E., Kovvali,G.K., Guzder,S.N., Amin,N.S., Holm,C., Habraken,Y., Sung,P., Prakash,L. and Prakash,S. (1996) Evidence for involvement of yeast proliferating cell nuclear antigen in DNA mismatch repair. J. Biol. Chem., 271, 27987–27990. [DOI] [PubMed] [Google Scholar]
- 21.Umar A., Buermeyer,A.B., Simon,J.A., Thomas,D.C., Clark,A.B., Liskay,R.M. and Kunkel,T.A. (1996) Requirement for PCNA in DNA mismatch repair at a step preceding DNA resynthesis. Cell, 87, 65–73. [DOI] [PubMed] [Google Scholar]
- 22.Flores-Rozas H., Clark,D. and Kolodner,R.D. (2000) Proliferating cell nuclear antigen and Msh2p-Msh6p interact to form an active mispair recognition complex. Nature Genet., 26, 375–378. [DOI] [PubMed] [Google Scholar]
- 23.Datta A., Adjiri,A., New,L., Crouse,G.F. and Jinks-Robertson,S. (1996) Mitotic crossovers between diverged sequences are regulated by mismatch repair proteins in Saccharomyces cerevisiae. Mol. Cell. Biol., 16, 1085–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rizki A. and Lundblad,V. (2001) Defects in mismatch repair promote telomerase-independent proliferation. Nature, 411, 713–716. [DOI] [PubMed] [Google Scholar]
- 25.Hunter N. and Borts,R.H. (1997) Mlh1 is unique among mismatch repair proteins in its ability to promote crossing-over during meiosis. Genes Dev., 11, 1573–1582. [DOI] [PubMed] [Google Scholar]
- 26.Wang T.-F., Kleckner,N. and Hunter,N. (1999) Functional specificity of MutL homologs in yeast: evidence for three Mlh1-based heterocomplexes with distinct roles during meiosis in recombination and mismatch correction. Proc. Natl Acad. Sci. USA, 96, 13914–13919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ehrenstein M.R. and Neuberger,M.S. (1999) Deficiency in Msh2 affects the efficiency and local sequence specificity of immunoglobulin class-switch recombination: parallels with somatic hypermutation. EMBO J., 18, 3484–3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schrader C.E., Edelmann,W., Kuperlapati,R. and Stavnezer,J. (1999) Reduced isotype switching in splenic B cells from mice deficient in mismatch repair enzymes. J. Exp. Med., 190, 307–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gradia S., Acharya,S. and Fishel,R. (1997) The human mismatch recognition complex hMSH2-hMSH6 functions as novel molecular switch. Cell, 91, 995–1005. [DOI] [PubMed] [Google Scholar]
- 30.Hickman M.J. and Samson,L.D. (1999) Role of DNA mismatch repair and p53 signaling in induction of apoptosis by alkylating agents. Proc. Natl Acad. Sci. USA, 96, 10764–10769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gong J.G., Costanzo,A., Yang,H.-Q., Melino,G., Kaelin,W.G., Levrero,M. and Wang,J.Y.J. (1999) The tyrosine kinase c-Abl regulates p73 in apoptotic response to cisplatin-induced DNA damage. Nature, 399, 806–809. [DOI] [PubMed] [Google Scholar]
- 32.Wu J., Gu,L., Wang,H., Geacintov,N.E. and Li,G.-M. (1999) Mismatch repair processing of carcinogen-DNA adducts triggers apoptosis. Mol. Cell. Biol., 19, 8292–8301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gradia S., Acharya,S. and Fishel,R. (2000) The role of mismatched nucleotides in activating the hMSH2–hMSH6 molecular switch. J. Biol. Chem., 275, 3922–3930. [DOI] [PubMed] [Google Scholar]
- 34.Bronner C.E., Baker,S.M., Morrison,P.T., Warren,G., Smith,L.G., Lescoe,M.K., Kane,M., Earabino,C., Lipford,J., Lindblom,A., Tannergard,P., Bollag,R.J., Godwin,A.R., Ward,D.C., Nordenskjold,M., Fishel,R., Kolodner,R. and Liskay,R.M. (1994) Mutation in the DNA mismatch repair gene homologue hMLH1 is associated with hereditary non-polyposis colon cancer. Nature, 368, 258–261. [DOI] [PubMed] [Google Scholar]
- 35.Papadopoulos N., Nicoolaides,N.C., Wei,Y.-F., Ruben,S.M., Carter,K.C., Rosen,C.A., Hazeltine,H.A., Fleischman,R.D., Fraser,C.M., Adams,M.A., Venter,J.C., Hamilton,S.R., Petersen,G.M., Watson,P., Lynch,H.T., Peltomaki,P., Mecklin,J.-P., de la Chapelle,A., Kinzler,K.W. and Vogelstein,B. (1994) Mutation of a mutL homolog in hereditary colon cancer. Science, 263, 1625–1629. [DOI] [PubMed] [Google Scholar]
- 36.Prolla T.A., Christie,D.-M. and Liskay,R.M. (1994) Dual requirement in yeast DNA mismatch repair for MLH1 and PMS1, two homologs of the bacterial mutL gene. Mol. Cell. Biol., 14, 407–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tran P.T. and Liskay,M.R. (2000) Functional studies on the candidate ATPase domains of Saccharomyces cerevisiae MutL. Mol. Cell. Biol., 20, 6390–6398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hall M.C., Shcherbakova,P.V. and Kunkel,T.A. (2002) Differential ATP binding and intrinsic ATP hydrolysis by amino-terminal domains of the yeast Mlh1 and Pms1 proteins. J. Biol. Chem., 277, 3673–3679. [DOI] [PubMed] [Google Scholar]
- 39.Raschle M., Dufner,P., Marra,G. and Jiricny,J. (2002) Mutations within the hMLH1 and hPMS2 subunits of the human MutLα mismatch repair factor affect its ATPase activity, but not its ability to interact with hMutSα. J. Biol. Chem., 277, 21810–21820. [DOI] [PubMed] [Google Scholar]
- 40.Tomer G., Buermeyer,A.B., Nguyen,M.M. and Liskay,R.M. (2002) Contribution of human Mlh1 and Pms2 ATPase activities to DNA mismatch repair. J. Biol. Chem., 277, 21801–21809. [DOI] [PubMed] [Google Scholar]
- 41.Ban C. and Yang,W. (1998) Crystal structure and ATPase activity of MutL: implications for DNA repair and mutagenesis. Cell, 95, 541–552. [DOI] [PubMed] [Google Scholar]
- 42.Ban C., Junlop,M. and Yang,W. (1999) Transformation of MutL by ATP binding and hydrolysis: a switch in DNA mismatch repair. Cell, 97, 85–97. [DOI] [PubMed] [Google Scholar]
- 43.Pang Q., Prolla,T.A. and Liskay,R.M. (1997) Functional domains of the Saccharomyces cerevisiae Mlh1p and Pms1p DNA mismatch repair proteins and their relevance to human hereditary nonpolyposis colorectal cancer-associated mutations. Mol. Cell. Biol., 17, 4465–4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guerette S., Acharya,S. and Fishel,R. (1999) The interaction of the human MutL homologues in hereditary nonpolyposis colon cancer. J. Biol. Chem., 274, 6336–6341. [DOI] [PubMed] [Google Scholar]
- 45.Kondo E., Suzuki,H., Horii,A. and Fukushige,S. (2003) A yeast two-hybrid assay provides a simple way to evaluate the vast majority of hMLH1 germ-line mutations. Cancer Res., 63, 3302–3308. [PubMed] [Google Scholar]
- 46.Plotz G., Raedle,J., Brieger,A., Trojan,J. and Zeuzem,S. (2003) N-terminus of hMLH1 confers interaction of hMutLα and hMutLβ with hMutSα. Nucleic Acids Res., 31, 3217–3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shimodaira H., Filosi,N., Shibata,H., Suzuki,T., Radice,P., Kanamaru,R., Friend,S.H., Kolodner,R.D. and Ishioka,C. (1998) Functional analysis of human MLH1 mutations in Saccharomyces cerevisiae. Nature Genet., 19, 384–389. [DOI] [PubMed] [Google Scholar]
- 48.Polaczek P., Putzke,A.P., Leong,K. and Bitter,G.A. (1998) Functional genetic tests of DNA mismatch repair protein activity in Saccharomyces cerevisiae. Gene, 213, 159–167. [DOI] [PubMed] [Google Scholar]
- 49.Drotschmann K., Clark,A.B. and Kunkel,T.A. (1999) Mutator phenotypes of common polymorphisms and missense mutations in MSH2. Curr. Biol., 9, 907–910. [DOI] [PubMed] [Google Scholar]
- 50.Shcherbakova P.V. and Kunkel,T. (1999) Mutator phenotypes conferred by MLH1 overexpression and by heterozygosity for mlh1 mutations. Mol. Cell. Biol., 19, 3177–3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ellison A.R., Lofing,J. and Bitter,G.A. (2001) Functional analysis of human MLH1 and MSH2 missense variants and hybrid human-yeast MLH1 proteins in Saccharomyces cerevisiae. Hum. Mol. Genet., 10, 1889–1900. [DOI] [PubMed] [Google Scholar]
- 52.Mumberg D., Muller,R. and Funk,M. (1994) Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Mol. Gen. Genet., 22, 5767–5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Strand M., Prolla,T.A., Liskay,R.M. and Petes,T.D. (1993) Destabilization of tracts of simple repetitive DNA in yeast by mutations affecting DNA mismatch repair. Nature, 365, 274–276. [DOI] [PubMed] [Google Scholar]
- 54.Sikorski R.S. and Hieter,P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bitter G.A. (1998) Function of hybrid human-yeast cyclin-dependent kinases in Saccharomyces cerevisiae. Mol. Gen. Genet., 260, 120–130. [DOI] [PubMed] [Google Scholar]
- 56.Ito H., Fukuda,Y., Murata,K. and Kimura,A. (1983) Transformation of intact yeast cells treated with alkali cations. J. Bacteriol., 153, 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cline J. and Hogrefe,H. (2000) GeneMorph™ PCR mutagenesis kit produces a unique mutational spectrum. Stratagies Newsletter, 13, 157–161. [Google Scholar]
- 58.Scharer E. and Iggo,R. (1992) Mammalian p53 can function as a transcription factor in yeast. Nucleic Acids Res., 20, 1539–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ishioka C., Frebourg,T., Yan,Y.X., Vidal,M., Friend,S.H., Schmidt,S. and Iggo,R. (1993) Screening patients for heterozygous p53 mutations using a functional assay in yeast. Nature Genet., 5, 124–129. [DOI] [PubMed] [Google Scholar]
- 60.Bitter G.A., Schaeffer,T.N. and Ellison,A.E. (2002) Reporter gene regulation in Saccharomyces cerevisiae by the human p53 tumor suppressor protein. J. Mol. Microbiol. Biotechnol., 4, 539–550. [PubMed] [Google Scholar]
- 61.Hoffman C.S. and Winston,F. (1987) A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene, 57, 267–272. [DOI] [PubMed] [Google Scholar]
- 62.Lea D.E. and Coulson,C.A. (1949) The distribution of the numbers of mutants in bacterial populations. J. Genet., 49, 264–285. [DOI] [PubMed] [Google Scholar]
- 63.Scott R.J., McPhillips,M., Meldrum,C.J., Fitzgerald,P.E., Adams,K., Spigelman,A.D., du Sart,D., Tucker,K., Kirk,J. and Service,H.F.C. (2001) Hereditary nonpolyposis colorectal cancer in 95 families: differences and similarities between mutation-positive and mutation-negative kindreds. Am. J. Hum. Genet., 68, 118–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Terdiman J.P., Gum,J.R.,Jr, Conrad,P.G., Miller,G.A., Weinberg,V., Crawley,S.C., Levin,T.R., Reeves,C., Schmitt,A., Hepburn,M., Sleisenger,M.H. and Kim,Y.S. (2001) Efficient detection of hereditary nonpolyposis colorectal cancer gene carriers by screening for tumor microsatellite instability before germline testing. Gastroenterology, 120, 21–30. [DOI] [PubMed] [Google Scholar]
- 65.Kurzawski G., Suchy,J., Kladny,J., Safranow,K., Jakubowska,A., Elsakov,P., Kucinskas,V., Gardovski,J., Irmejs,A., Sibul,H., Huzarski,T., Byrski,T., Debniak,T., Cybul,C., Gronwald,J., Oszurek,O., Clark,J., Gozdz,S., Niepsuj,S., Slomski,R., Plawski,A., Lacka-Wojciechowska,A., Rozmiarek,A., Fiszer-Maliszewska,L., Bebenek,M., Sorokin,D., Stawicka,M., Godlewski,D., Richter,P., Brozek,I., Wysocka,B., Jawien,A., Banaszkiewicz,Z., Kowalczyk,J., Czudowska,D., Goretzki,P.E., Moeslein,G. and Lubinski,J. (2002) Germline MSH2 and MLH1 mutational spectrum in HNPCC families from Poland and the Baltic States. J. Med. Genet., 39, e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Woods M.O., Green,J.S., Robb,D., Pollett,A., Younghusband,B., Gallinger,S., Parfrey,P.S., McLaughlin,J.R. and Bapat,B. (2003) American Association for Cancer Research, 94th Annual Meeting. Cadmus Professional Communications, Toronto, Ontario, Canada, Vol. 44, pp. 1367.
- 67.Marsischky G.T., Filosi,N., Kane,M.F. and Kolodner,R. (1996) Redundancy of Saccharomyces cerevisiae MSH3 and MSH6 in MSH2-dependent mismatch repair. Genes Dev., 10, 407–420. [DOI] [PubMed] [Google Scholar]
- 68.Tannergard P., Lipford,J.R., Kolodner,R., Frodin,J.E., Nordenskjold,M. and Lindblom,A. (1995) Missense mutations in the MLH1gene in Swedish HNPCC families. Cancer Res., 55, 6092–6096. [PubMed] [Google Scholar]
- 69.Hackman P., Tannergard,P., Osei-Mensa,S., Chen,J., Kane,M.F., Kolodner,R., Lambert,B., Hellgren,D. and Lindblom,A. (1997) A human compound heterozygote for two MLH1 missense mutations. Nature Genet., 17, 135–136. [DOI] [PubMed] [Google Scholar]
- 70.Aronshtam A. and Marinus,M.G. (1996) Dominant negative mutator mutations in the mutL gene of Escherichia coli. Nucleic Acids Res., 24, 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Studamire B., Price,G., Sugawara,N., Haber,J. and Alani,E. (1999) Separation-of-function mutations in Saccharomyces cerevisiae MSH2 that confer mismatch repair defects but do not affect nonhomologous-tail removal during recombination. Mol. Cell. Biol., 19, 7558–7567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Amin N.S., Nguyen,M.-N., Oh,S. and Kolodner,R.D. (2001) Exo-1 dependent mutator mutations: model systems for studying functional interactions in mismatch repair. Mol. Cell. Biol., 21, 5142–5155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sia E.A., Dominska,M., Stefanovic,L. and Petes,T.D. (2001) Isolation and characterization of point mutations in mismatch repair genes that destabilize microsatellites in yeast. Mol. Cell. Biol., 21, 8157–8167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Argueso J.L., Smith,D., Yi,J., Waase,M., Sarin,S. and Alani,E. (2002) Analysis of conditional mutations in the Saccharomyces cerevisiae MLH1 gene in mismatch repair and in meiotic crossing over. Genetics, 160, 909–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nakahara M., Yokozaki,H., Yasui,W., Dohi,K. and Tahara,E. (1997) Identification of concurrent germ-line mutations in hMSH2 and/or hMLH1 in Japanese hereditary nonpolyposis colorectal cancer kindreds. Cancer Epidemiol. Biomarkers Prev., 6, 1057–1064. [PubMed] [Google Scholar]
- 76.Syngal S., Fox,E.A., Li,C., Dovidio,M., Eng,C., Kolodner,R.D. and Garber,J.E. (1999) Interpretation of genetic test results for hereditary nonpolyposis colorectal cancer: implications for clinical predisposition testing. JAMA, 282, 247–253. [DOI] [PubMed] [Google Scholar]
- 77.Jeyaprakash A., Gupta,R.D. and Kolodner,R. (1996) Saccharomyces cerevisiae pms2 mutations are alleles of MLH1, and pms2-2 corresponds to a hereditary nonpolyposis colorectal carcinoma-causing missense mutation. Molecular and Cellular Biology, 16, 3008–3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Clark A.B., Cook,M.E., Tran,H.T., Gordenin,D.A., Resnick,M. and Kunkel,T.A. (1999) Functional analysis of human MutSα and MutSβ complexes in yeast. Nucleic Acids Res., 27, 736–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Andreutti-Zaugg C., Scott,R.J. and Iggo,R. (1997) Inhibition of nonsense-mediated messenger RNA decay in clinical samples facilitates detection of human MSH2 mutations with an in vivo fusion protein assay and conventional techniques. Cancer Res., 57, 3288–3293. [PubMed] [Google Scholar]
- 80.Argueso J.L., Kijas,W., Sarin,S., Heck,J., Waase,M. and Alani,E. (2003) Systematic mutagenesis of the Saccharomyces cerevisiae MLH1 gene reveals distinct roles for Mlh1p in meiotic crossing over and in vegetative and meiotic mismatch repair. Mol. Cell. Biol., 23, 873–886. [DOI] [PMC free article] [PubMed] [Google Scholar]