Abstract
A C→U mutation (rdn5) in the conserved sarcin/ricin domain of yeast 25S rRNA has been shown to cause translational suppression and paromomycin resistance. It also separates the killing from the misreading effect of this antibiotic. We confirm these findings and provide in vitro evidence that rdn5 causes a 3-fold increase in translational errors and resistance to paromomycin. The role of this 25S rRNA domain in ribosome's decoding function was further demonstrated when 60S subunits from rdn5 cells were combined with 40S subunits from cells carrying an error-prone mutation in the eukaryotic accuracy center ribosomal protein S23, an homologue of Escherichia coli S12. These hybrids exhibited an error frequency similar to that of rdn5 alone, despite the error-prone mutation in S23. This was accompanied by extreme resistance to paromomycin, unlike the effects of the individual mutations. Furthermore, rdn5 lowers peptidyltransferase activity measured as a second-order rate constant (kcat/Ks) corresponding to the rate of peptide bond formation. This mutation was also found to affect translocation. Elongation factor 2 (EF2)-dependent translocation of Ac-Phe-tRNA from the A- to P-site was achieved at an EF2 concentration 3.5 times lower than in wild type. In conclusion, the sarcin/ricin domain of 25S rRNA influences decoding, peptide bond formation and translocation.
INTRODUCTION
Accurate and efficient translation of mRNA is a critical step in the process of protein synthesis and is essential for cell viability. Although the decoding center is basically, like the peptidyltransferase center, an RNA machine (1,2), ribosomal proteins have been shown to play an important role too (3,4).
Moreover, we and other workers have provided biochemical evidence that the two ribosomal subunits interact with each other in order to optimize translational fidelity. Changes in the one subunit affect the activity of the other. Thus, large ribosomal subunit protein L39 affected, among others, the fidelity of translation (5).
Another region of the large ribosomal subunit involved in an activity of the small ribosomal subunit is the sarcin/ricin domain of 25S yeast rRNA. The sarcin/ricin domain constitutes an important part of the binding site for the elongation factors of protein synthesis (6–8), and it is the site of action of the ribotoxins sarcin, the A chain of ricin and its analogue pokeweed antiviral protein (PAP) [for a review, see (9)]. These toxins confer their toxicity by disrupting binding of EF-G and of the EF-Tu ternary complex to the ribosome (6). The sarcin/ricin domain has a GAGA tetraloop that is closed by an invariant Watson–Crick C2658–G2663 pair (9). Liu and Liebman (10) provided genetic evidence that mutation C2658U (rdn5) of this region caused translational suppression and antibiotic resistance in eukaryotic cells. The location of the rdn5 mutation is analogous to Escherichia coli 2658 in 23S rRNA, and is in the universally conserved sarcin/ricin domain of 25S rRNA. The sarcin/ricin domain has been shown to interact with the decoding center in prokaryotes. Specifically, site-directed mutations G2661C, T in the GAGA tetraloop of E.coli 23S rRNA decreased nonsense suppression and +1 frameshifting, and G2661C was a synthetic lethal in combination with an antisuppressor mutation in E.coli ribosomal protein S12 (11,12). Alterations at specific residues of this E.coli protein have been found to cause hyperaccurate translation (13). Recently, it was shown that domain III of eukaryotic elongation factor 2 (EF2) interacts with both the sarcin/ricin loop of 25S rRNA and with ribosomal protein S23, the yeast homologue of E.coli S12 (14). Also, mutations in S23, have been found to affect fidelity in opposite directions (4,15).
Synthetic oligonucleotides with the sarcin/ricin loop (SRL) sequence mimic the form and function of the SRL in the ribosome. SRL mimics have served as a minimal substrate for EF-G binding, for sarcin and ricin activity and for other structural studies. The SRL RNA folds into two motifs: a GAGA tetraloop and a bulged-G motif, separated by an invariant Watson–Crick C2658–G2663 pair. Biochemical and structural studies suggest that the elongation factor binding site includes the major groove face of both motifs and may therefore include the intervening Watson–Crick C2658–G2663 pair. Juxtaposition of these motifs separated by one invariant Watson–Crick base pair, presents a unique site on the ribosome surface that is recognized by IF2, EFs and toxins alike (16).
In the present work, we provide biochemical evidence that rdn5 mutation in the SRL increases general misreading, not just translational suppression. In addition, we show that rdn5 decreases peptidyltransferase activity and makes the translocation process of protein synthesis more efficient.
In an effort to further elucidate the role of the sarcin/ricin domain in the decoding process, we combine mutation rdn5 either with an error-prone mutation in Lys-62 of ribosomal protein S23, or with an error-restrictive mutation in the same site and demonstrate that elements of both ribosomal subunits work in concert to control decoding and resistance to aminoglycoside antibiotic paromomycin.
MATERIALS AND METHODS
Plasmids, preparation of mutants and yeast transformations
The previously constructed strains rdnwt (L1494) and rdn5 (L1548) (10,17) were used in the present study (Table 1). Both strains are isogenic to wt strain (L1489), except that they contain a complete deletion of the chromosomal 9 kb rDNA units (RDN) including regulatory sequences, rRNA genes and a new multicopy plasmid pRDN carrying in tandem all the genes for 18S, 25S, 5.8S and 5S rRNAs as well as regulatory sequences. Strain L1548 carries in addition mutation C2658U (rdn5) of the sarcin/ricin domain introduced to plasmid pRDN (1,10). This strain, L1548, was studied in conjunction with strain L1494 not carrying the mutation. Ribosomal protein S23 mutants were prepared either carrying the chromosomal genes or by deletion of chromosomal RPS23A, one of the two genes encoding ribosomal protein S23, followed by introduction of a centromere-based plasmid pRS313 carrying either the wild-type allele of RPS23A gene (pS23A-wt) or mutant alleles of RPS23A (pS23A-R, pS23A-N). These plasmids were identical to pS23A-wt except for replacement of Lys-62 with Arg or Asn, respectively (4,18). The strains carrying chromosomal RPS23 genes together with one of these plasmids are designated as C/pS23A-wt, C/pS23A-R and C/pS23A-N (Table 1). The strains carrying a deletion of chromosomal RPS23A gene together with one of these plasmids are designated as E/pS23A-wt (data not shown), E/pS23A-R (Table 3) and E/pS23A-N (data not shown).
Table 1. Genotypes and growth rates of yeast strains.
| Strain | Genotype | Mutations | Growth rates (min) |
|---|---|---|---|
| wt (L1489) | MATa ade1–14(UGA) his 7-1(UAA) leu2-3, 112 lys2-L864 (UAG) trp1-Δ1 ura3-52 | None | 96 |
| rdnwt (L1494) | MATa ade 1-14(UGA)his7-1(UAA) lys2-L864(UAG)leu2-3, 112 trp1Δ1ura3-52RDNΔ [pRDN-wt-TL] | None | 102 |
| rdn5 (L1548) | MATa ade 1-14(UGA)his7-1(UAA) lys2-L864(UAG)leu2-3, 112 trp1Δ1ura3-52RDNΔ [pRDN-rdn5-TL] | C2658U in 25S rRNA | 140 |
| rdn5/pS23A-R | same as rdn5 plus pS23A-R | C2658U in 25S rRNA and Lys-62→Arg on plasmid rpS23 | 162 |
| rdn5/pS23A-N | same as rdn5 plus pS23A-N | C2658U in 25S rRNA and Lys-62→Asn on plasmid rpS23 | 155 |
| C/pS23A-wt | MATa met8-1 leu2-1 trp1-a lys2-BBδ his3-Δ1 ura3-52 [pS23A-wt] | None | 98 |
| C/pS23A-R | MATa met8-1 leu2-1 trp1-a lys2-BBδ his3-Δ1 ura3-52 [pS23A-R] | Lys-62→Arg on plasmid rpS23 | 120 |
| C/pS23A-N | MATa met8-1 leu2-1 trp1-a lys2-BBδ his3-Δ1 ura3-52 [pS23A-N] | Lys-62→Asn on plasmid rpS23 | 115 |
Table 3. Translational accuracy of hybrid ribosomes in the absence or presence of 50 μM paromomycin (PM).
| 60S origin | Genotype | 40S origin | Genotype | PM (μM) | Error frequency ± SE |
|---|---|---|---|---|---|
| E/pS23A-R | ΔrpS23A::TRP1/RPS23B/ pS23A-Ra | rdn5 | pRDN-rdn5b | — | 0.0040 ± 0.0010 |
| 50 | 0.0132 ± 0.0002 | ||||
| E/pS23A-R | ΔrpS23A::TRP1/RPS23B/ pS23A-Ra | E/pS23A-R | ΔrpS23A::TRP1/RPS23B/ pS23A-Ra | — | 0.0105 ± 0.0007c |
| 50 | 0.0222 ± 0.0034d | ||||
| rdn5 | pRDN-rdn5b | rdn5 | pRDN-rdn5b | — | 0.0133 ± 0.0008c |
| 50 | 0.0167 ± 0.0009d | ||||
| rdn5 | pRDN-rdn5b | E/pS23A-R | ΔrpS23A::TRP1/RPS23B/ pS23A-Ra | — | 0.0130 ± 0.0017c |
| 50 | 0.0207 ± 0.0027d |
aThe full genotype is: MATa met8-1 leu2-1 trp1-a lys2-BBδ his3-Δ1 ura3-52 ΔrpS23A::TRP1/RPS23B [pS23A-R].
bThe full genotype is: MATa ade 1-14(UGA)his7-1(UAA) lys2-L864(UAG)leu2-3, 112 trp1Δ1ura3-52RDNΔ [pRDN-rdn5-TL].
cStatistically different (one-way analysis of variance, F-Scheffè test, p < 0.1) from the wild-type hybrids in the absence of PM (line 1).
dStatistically different (one-way analysis of variance, F-Scheffè test, p < 0.1) from the wild-type hybrids in the presence of 50 μM PM (line 2).
Double mutants were constructed when rdn5 cells were transformed as described by Itoh et al. (19) with pS23A-wt, pS23A-R or pS23A-N (Table 1).
Antibiotic activity disc assay
For the antibiotic activity assay, approximately 107 cells were spread evenly on YPD (1% yeast extract, 2% peptone, 2% d-glucose, 2% agar) plates with the aid of glass beads. Paromomycin or cycloheximide at the appropriate concentrations were pipetted onto 5 mm Whatmann (3MM) paper discs placed evenly on the surface of the YPD medium. The plates were incubated at 30°C for 3–6 days.
Biochemical procedures
Experiments to measure the sensitivity of cells to paromomycin, the preparation of a yeast cell-free system for translation in vitro, the translation of poly(U) templates in vitro, the P- and A-site binding studies as well as the time course measurements of polyphenylalanine synthesis, and the sucrose gradient analysis were all performed as described recently (5).
Preparation of ribosomal subunits
To prepare ribosomal subunits for hybridization experiments, the S30 was centrifuged for 3 h at 125 000 g. The 125 000 g pellet was resuspended in a high-salt buffer containing 0.9 M KCl and 12 mM MgCl2, and recentrifuged through a 15–40% linear sucrose gradient at 125 000 g for 6 h. The fractions containing each of the ribosomal subunits were pooled and subjected to a final centrifugation at 125 000 g for 16 h, and the two pellets were resuspended in low-salt (100 mM KCl) buffer prior to use.
Formation, isolation and extraction of complex C
Unwashed or, alternatively, high-salt-washed ribosomes and soluble protein factors from wild-type or mutant yeast cells as well as Ac-[3H]Phe-tRNA were prepared according to recently described methodology (20). The donor Ac-[3H]Phe-tRNA was 245 A260/ml and contained 14 pmol of [3H]Phe charged/1 A260 tRNA and 180 000 c.p.m. incorporated/1 A260 tRNA. Complex C, i.e. the Ac-[3H]Phe-tRNA•poly(U)•80S ribosome complex, was formed from unwashed or high-salt-washed ribosomes in a binding mixture (0.2 ml) containing the following: 80 mM Tris–HCl pH 7.4, 160 mM ammonium chloride, 11 mM magnesium acetate, 2 mM spermidine, 6 mM β-mercaptoethanol (binding buffer), as well as 0.4 mM GTP, 30 A260/ml of 80S ribosomes, 0.4 mg/ml poly(U) and 16 A260/ml Ac-[3H]Phe-tRNA from yeast. Following incubation for 16 min at 30°C, the reaction was stopped by placing the binding mixture in ice. The solution was immediately filtered through a cellulose nitrate filter disc under vacuum with three 4 ml portions of binding buffer. The filter discs with the adsorbed complex C were gently shaken for 30 min at 5°C in binding buffer containing 0.05% Zwittergent 3–12 (ZW) (1.8 ml/whole disc). The extraction was stopped by removing the filters, and the complex C-containing ZW extract was used in the puromycin reaction.
Puromycin reaction
The reaction between the Ac-[3H]Phe-tRNA of complex C in the ZW extract, and puromycin (puromycin reaction) was carried out as described recently (20). ZW extract (0.9 ml) was preincubated for 5 min at 30°C. Then, puromycin (0.1 ml) at the desired concentrations was added, the puromycin reaction proceeded for the indicated time intervals and stopped with 1.0 ml of 1.0 N NaOH. If N0 represents the total radioactivity of bound donor (e.g. Ac-[3H]Phe-tRNA), the percentage (x) of the bound donor that was converted to product P (Ac-[3H]Phe-puromycin) was calculated by dividing P by N0 and multiplying by 100. Each percentage (x) was corrected, first, by dividing its value with factor A (A = C/C0, where C and C0 are the amounts of surviving complex C in binding buffer at the end of each incubation period and at zero time, respectively), and, second, with the extent factor α. The extent factor is determined when complex C is allowed to react completely, at any concentration of puromycin. By the first correction (x/A), the parallel inactivation of complex C during the puromycin reaction is subtracted, while by the second correction, the intervention of any species other than complex C is erased as if 100% of the bound Ac-[3H]Phe-tRNA were reactive toward puromycin. Thus, the experimental values of x were corrected by factor Aα, i.e. x′ = x/Aα.
Translocation
The reaction mixture for translocation (0.1 ml) was composed from the same binding buffer as that used for the formation of complex C, as well as 0.4 mM GTP, 30 A260/ml of ribosomes, 0.4 mg/ml of poly(U), and tRNAPhe at a ratio of 4:1 to ribosomes. At this ratio, all P-sites were occupied by tRNA after 30 min at 30°C. Then, 13 A260/ml of yeast Ac-[3H]Phe-tRNA were added and magnesium acetate was raised to 15 mM. Reincubation followed for 10 min at 30°C to form a poly(U)-programmed ribosomal complex in which P-sites were filled with tRNA and A-sites with Ac-[3H]Phe-tRNA.
The reaction was stopped by placing the binding mixture in ice. Filtering, washing and extraction of the ribosomal complex into binding buffer containing 0.05% ZW at 5°C were carried out as described previously (20). ZW extract was preincubated for 2 min at 30°C. Also, eukaryotic EF2 prepared from rabbit reticulocytes (21) or soluble protein factors (SPF) prepared from yeast (20) were preincubated with 1 mM GTP, 40 mM phosphocreatine and 40 μg/ml creatine phosphokinase for 15 min at 37°C. The enzymatic translocation reaction was started when GTP-activated EF2 at 0.02 μM or SPF at 0.05 mg/ml were added to the pretranslocation ZW complex and allowed to react for the indicated time intervals. Single-turnover translocation (EF2:active ribosomes = 9:1) was monitored by reaction with 2 mM puromycin (3×Ks) for 4 min (7 half-lives) at 30°C. The puromycin solution contained cycloheximide at a final concentration of 8 μM to ensure that no translocation takes place during the puromycin reaction. Nonenzymatic translocation was measured in the absence of elongation factors.
For concentration dependence experiments, increasing concentrations of EF2 or SPF were added to the pretranslocation ZW complex and reacted for 1 min after which translocation was again monitored by reaction with puromycin.
Statistics
The data variability was estimated using one-way analysis of variance (ANOVA) procedure. We used the F-Scheffè test to determine which means were significantly different from each other. All statistical tests were performed using an SPSS program for MS Windows, Release 6.0.
RESULTS
Growth rate of rdn5 mutants
In order to investigate further the properties of the sarcin/ricin domain of 25S rRNA of the yeast Saccharomyces cerevisiae, two previously constructed strains were used. The first, rdnwt (L1494), contains plasmid pRDN with only wild-type rRNA whereas the second, rdn5 (L1548), contains plasmid pRDN with essentially rRNA carrying only the C2658U mutation (Table 1). These strains are isogenic to wt strain (L1489), which carries the chromosomal RDN genes (10). No significant difference in the growth rate was observed between L1489 and L1494 showing that the deletion of the chromosomal RDN locus, and its replacement by plasmid pRDN does not affect cell growth (Table 1). However, the rdn5 mutant strain was characterized by an almost 40% reduction of cell growth: the doubling time of rdn5 was increased to 140 min compared to 102 min for rdnwt in complete (YPD) medium.
Suppression and drug resistance associated with rdn5
Paromomycin is an aminoglycoside antibiotic known to bind directly to the A-site of E.coli 16S rRNA, and to interfere with translational decoding, including misreading (22). This drug also causes translational misreading in yeast (1,23). The loss of translational accuracy has been linked to hypersensitivity of the mutant strains to antibiotics such as paromomycin.
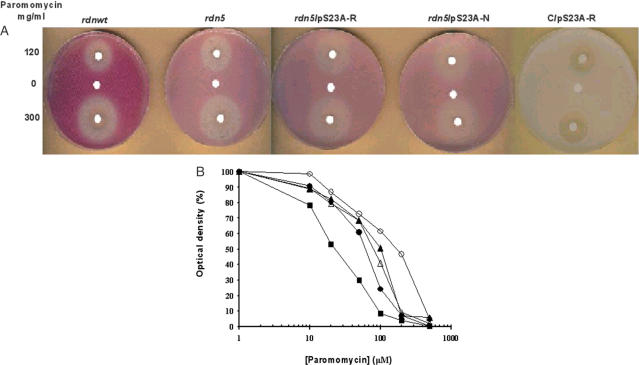
Figure 1A shows levels of suppression and paromomycin resistance. The red color of cells on YPD shows lack of suppression of the ade 1-14 (UGA) marker while white indicates strong suppression. Thus, rdn5 (lighter red color) caused intermediate suppression.
Figure 1.
Inhibition of growth in the presence of paromomycin. (A) Antibiotic disc assay. Cells (∼107) from rdnwt or rdn5 strains were spread on YPD plates and grown at 30°C. Paromomycin at the indicated concentrations or water as control (5 μl) were pipetted onto filter paper discs. Lighter background color shows suppression by rdn5. Clear and white circles around the discs indicate killing and phenotypic suppression, respectively, caused by the antibiotic paromomycin. The C/pS23A-R plate is used as control and it does not contain the ade 1-14 (UGA) marker, therefore it does not indicate suppression; the large clear circles are a measure of sensitivity to paromomycin. (B) Cells were grown in YPD at 30°C to an A660 of 0.9, which was taken as 100%. Concentrations of paromomycin are as indicated. Solid circles, rdnwt strain; solid triangles, rdn5 strain. The other strains are indicated as follows: open circles, rdn5/pS23A-R; open triangles, rdn5/pS23A-N; and solid squares, C/pS23A-R.
Next, we looked at paromomycin resistance associated with rdn5. By comparing antibiotic-induced zones of growth inhibition, Liu and Liebman (10) showed that L1548 cells containing essentially all rdn5 rRNA are more resistant to paromomycin than L1494 cells containing only wild-type rRNA. Our results confirmed this finding (Figure 1A); in a 9 cm diameter YPD plate, the rdn5 cells (L1548) had an area of growth inhibition (clear circles), in which the distance of the cells from the disc soaked with 5 μl of 300 mg/ml paromomycin was 5 mm compared with 7.5 mm for isogenic wild-type cells (L1494). When the disc was soaked with 5 μl of 120 mg/ml paromomycin, the distance of the cells from the disc was 3 mm compared with 5 mm, respectively.
Although the rdn5 mutation caused resistance to paromomycin, an antibiotic that induces misreading, it did not eliminate antibiotic-induced (phenotypic) suppression of the red color associated with the ade1-14 (UGA) nonsense mutation in L1548, in accordance with earlier findings (10). In fact, the white circle around the disc is larger in the plate containing rdn5 cells compared with wild-type cells (Figure 1A).
The higher resistance of rdn5 mutants towards paromomycin was confirmed in another experiment, in which cells were grown in YPD at 30°C to an A660 of 0.9 that was taken as 100%. The growth of the wild-type strain L1494 was inhibited by 50% at 66 μM paromomycin, while that of the rdn5 strain L1548 at 108 μM paromomycin (Figure 1B).
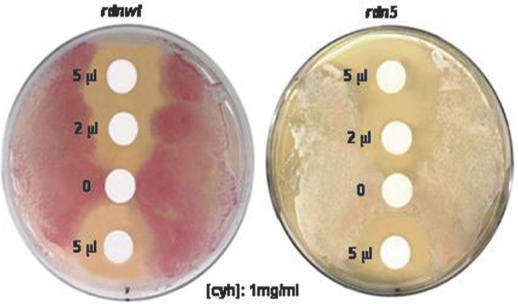
The sarcin/ricin domain is involved in the binding of the elongation factors, and it has been suggested that a conformational change in the structure of the domain might drive translocation (24). We therefore tested the effect of rdn5 mutation on the sensitivity towards cycloheximide, an antibiotic that acts on large ribosomal subunits of eukaryotic cells and inhibits the translocation step of translation (25). In contrast to the resistance towards paromomycin described above, rdn5 cells are more sensitive to cycloheximide. The rdn5 cells (L1548) had a zone of growth inhibition somewhat bigger than isogenic wild-type cells (L1494) around a disc with either 2 or 5 μl of 1 mg/ml cycloheximide (Figure 2).
Figure 2.
Inhibition of growth in the presence of cycloheximide with the antibiotic disc assay. Cells (∼107) from rdnwt or rdn5 strains were spread on YPD plates and grown at 30°C. Two or five microliters of 1 mg/ml cycloheximide were pipetted onto filter paper discs. Again, lighter background color shows suppression by rdn5. Larger clear circles around the rdn5 discs indicate sensitivity towards the antibiotic cycloheximide.
Polyphenylalanine synthesis activity of wild-type and mutant ribosomes
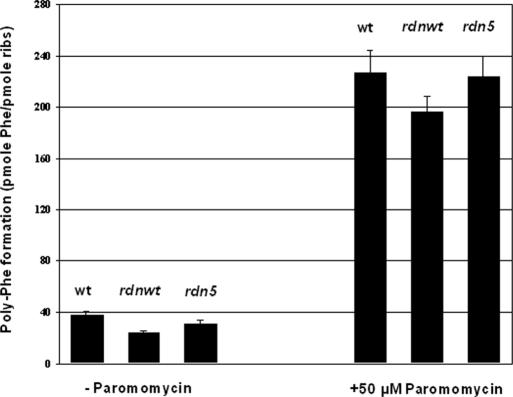
Using in vitro poly(U)-dependent polyphenylalanine synthesis, we determined the activity of wild-type and mutant ribosomes. All ribosomes were dependent on the presence of soluble protein factors. In their absence, the ribosomes did not polymerize phenylalanine. We found that the complete deletion of RDN and the introduction of plasmid pRDN is not entirely innocuous for the cell; it reduced protein synthesis activity from 38 pmol phenylalanine incorporated per pmole of ribosomes for the strain L1489 (wt), carrying the chromosomal RDN locus, to 24 pmol for its isogenic L1494 (rdnwt) containing the RDN deletion and pRDN-wt (Figure 3). However, the activity of rdn5 mutants (L1548) was slightly increased compared with that of L1494, from 24 to 31 pmol phenylalanine incorporated per pmole of ribosomes.
Figure 3.
Polyphenylalanine formation of wt, rdnwt and rdn5 ribosomes in the absence and in the presence of 50 μM paromomycin. Polyphenylalanine formation in the absence of the soluble protein factors was ∼2 and 9 pmol phenylalanine/pmol ribosomes, respectively.
When in vitro poly(U)-dependent polyphenylalanine synthesis was conducted in the presence of the aminoglycoside antibiotic paromomycin, a 7- to 8-fold increase in polyphenylalanine formation was observed in all three strains, with the rdn5 strain maintaining a somewhat higher activity than rdnwt (Figure 3). Thus, the polyphenylalanine synthesis was increased to 226, 196 and 223 pmol phenylalanine incorporated per pmole of ribosomes for the wt, rdnwt and rdn5 strains, respectively.
Effect of the sarcin/ricin domain on translational accuracy
The cell-free system also allowed measurement of the misincorporation of the near cognate amino acid leucine with poly(U) as template. The accuracy of translation was determined by the error frequency. The error frequency is defined as the ratio of the incorporation of [3H]leucine to the combined incorporation of [3H]leucine plus [3H]phenylalanine. It represents the number of errors per translated codon. As shown in Table 2, the accuracy of translation in rdn5 mutants was impaired; there was a 3-fold increase in the misincorporation of leucine from 0.0026 to 0.0087, i.e. from 26 to 87 errors every 104 codons. This is in vitro evidence for the participation of the sarcin/ricin loop in maintaining translational accuracy and agrees with previous genetic evidence (10) confirmed here (Figure 1A) that rdn5 causes nonsense suppression. It should be noted that the error rate of the wild-type strain is slightly higher than that reported for a wild-type strain with a different genotype (4), but it is still within the range reported in the literature (26).
Table 2. Translational error frequencies in vitro of wild-type and mutant ribosomes in the absence or presence of 50 μM paromomycin (PM).
| Strain | PM (μM) | No. of experiments | Error frequency ± SE |
|---|---|---|---|
| rdnwt | — | 5 | 0.0026 ± 0.0007 |
| 50 | 5 | 0.0079 ± 0.0012 | |
| rdn5 | — | 5 | 0.0087 ± 0.0005a |
| 50 | 5 | 0.0115 ± 0.0004b | |
| rdn5/pS23A-R | — | 5 | 0.0095 ± 0.0005a |
| 50 | 5 | 0.0121 ± 0.0003b | |
| rdn5/pS23A-N | — | 4 | 0.0028 ± 0.0004 |
| 50 | 4 | 0.0044 ± 0.0007 | |
| C/pS23A-wt | — | 4 | 0.0013 ± 0.0005 |
| 50 | 4 | 0.0057 ± 0.0011 | |
| C/pS23A-R | — | 4 | 0.0071 ± 0.0003c |
| 50 | 4 | 0.0170 ± 0.0005d | |
| C/pS23A-N | — | 4 | 0.0010 ± 0.0002 |
| 50 | — | n.d. |
aStatistically different (one-way analysis of variance, F-Scheffè test, p < 0.1) from the rdnwt or rdn5/pS23A-R strains in the absence of PM.
bStatistically different (one-way analysis of variance, F-Scheffè test, p < 0.1) from the rdnwt or rdn5/pS23A-R strains in the presence of 50 μM PM.
cStatistically different (one-way analysis of variance, F-Scheffè test, p < 0.1) from the C/pS23A-wt or C/pS23A-N strains in the absence of PM.
dStatistically different (one-way analysis of variance, F-Scheffè test, p < 0.1) from the C/pS23A-wt strain in the presence of 50 μM PM.
The aforementioned experiments were repeated under the closer to in vivo conditions of 2.5 mM Mg+2 and 3 mM spermidine, instead of 11 mM Mg+2. Under these conditions, polyphenylalanine synthesis dropped by as much as three times whereas the error frequencies remained remarkably similar for both wild-type and rdn5 strains (data not shown).
Effect of paromomycin on translational accuracy in vitro in rdn5 mutants
Paromomycin, an error-inducing antibiotic, increased the error frequencies of both rdnwt (wild-type) and rdn5 mutant ribosomes (Table 2). The results of Figure 1 and Table 2 show that rdn5 is a mutation which, in the presence of paromomycin, shows increased levels of suppression and mistranslation, while it confers resistance to paromomycin. Thus, it clearly separates the dual effects of paromomycin on suppression/mistranslation and killing of the cells. It should be noted that the increases in error frequencies in the presence of paromomycin, depicted in Table 2, have by definition taken into account the increases in polyphenylalanine synthesis activities shown in Figure 3.
Yeast ribosome function in the simultaneous presence of two mutations, rdn5 in 25S rRNA and another in ribosomal protein S23
In E.coli, there is a link between the two ribosomal subunits involving the sarcin/ricin domain on the 50S and ribosomal protein S12 on the 30S subunit in order to define the proper binding site for the EF-Tu•tRNA•GTP ternary complex (11). In eukaryotes, it was shown recently that domain III of EF2 interacts with the decoding center in 40S and with ribosomal protein S23, the equivalent to E.coli S12, which is adjacent to the decoding center (14). We and others have shown previously that ribosomal protein S23 affects translational accuracy (4,18). Site-directed mutations in Lys-62 of ribosomal protein S23 either decrease (Lys-62→Arg) or increase (Lys-62→Asn) translational accuracy. Since domain III of EF2 interacts with the sarcin/ricin loop also (14), this contact could be part of an information relay system between the decoding center and the sarcin/ricin loop. Consequently, we examined the possibility of an interplay in translational accuracy control between the sarcin/ricin domain of 25S rRNA and ribosomal protein S23 mutants, i.e. between the 60S and 40S ribosomal subunits.
Cells from the rdn5 strain carrying the two chromosomal genes for rpS23 were transformed with plasmids pS23A-wt, pS23A-R carrying mutation Lys-62→Arg or pS23A-N carrying mutation Lys-62→Asn. We have shown previously that the disruption of RPS23B, which is one of the two genes encoding the essential rpS23 has a slight or no effect on translational accuracy, and on the resistance towards paromomycin (4). The rdn5/pS23A-R or N double mutant cells were viable with only a relatively small increase in doubling time in each case. The rdn5/pS23A-R double mutant had a doubling time of 162 min and the rdn5/pS23A-N 155 min compared to 140 min for rdn5 (Table 1).
The levels of suppression by paromomycin and resistance to paromomycin of the rdn5/pS23A-R and rdn5/pS23A-N double mutants were tested with the antibiotic activity disc assay. Each double mutant showed slightly increased suppression by paromomycin compared with wild type, indicated by the slightly larger white circle around each disc (Figure 1A). Each suppression is similar to that shown in rdn5, despite the fact that Lys-62→Arg in C/pS23A-R strain is a suppressor mutation and Lys-62→Asn in C/pS23A-N strain is an antisuppressor mutation.
Subsequently, we examined the sensitivity of the double mutants towards paromomycin. Individually, rdn5 is resistant towards paromomycin (Figure 1), while the C/pS23A-R mutant is sensitive to paromomycin both in vivo and in vitro and the C/pS23A-N mutant is resistant to paromomycin (4). As expected, the rdn5/pS23A-N double mutant was slightly more resistant to paromomycin than rdn5 alone. Unexpectedly, however, the rdn5/pS23A-R double mutant also proved to be more resistant to paromomycin in vivo than even rdn5 alone, as indicated by the smaller area of clear circles around the discs (Figure 1A), despite the presence of the paromomycin-sensitive Lys-62→Arg mutation.
In another experiment, in which paromomycin was added to liquid YPD cultures, the rdn5/pS23A-R double mutant displayed again a significantly higher degree of resistance to paromomycin than rdn5 alone and in complete reversal to the sensitivity of the C/pS23A-R mutant towards paromomycin; 50% inhibition of growth was observed at 176 μM paromomycin for the rdn5/pS23A-R double mutant compared to 108 μM for the rdn5 and to only 22 μM for the paromomycin-sensitive C/pS23A-R mutant, and 66 μM for the rdnwt (Figure 1B). The other double mutant, rdn5/pS23A-N, showed 50% inhibition of growth at 90 μM paromomycin (Figure 1B), i.e. it was resistant to paromomycin in accordance with the character of each mutation alone.
The rdn5/pS23A-R double mutant exhibited an error frequency that is similar to the individual error frequencies of rdn5 and Lys-62→Arg mutations; thus, the error frequencies of the individual mutations were not additive, indicating some kind of an interaction between them (Table 2). If there was no interaction, the expected error frequency of the double mutant should equal the combined error frequencies of the individual mutants, i.e. 0.0087 + (0.0071−0.0013) = 0.0145, instead of the actual 0.0095. These results indicate that the effect of one mutation is modulated in the presence of the other. The other double mutant rdn5/pS23A-N exhibited a level of translational accuracy similar to that of the wild type, in agreement with the opposing characters of the two mutations, one of which, rdn5, is error-prone while the other, Lys-62→Asn, is error-restrictive (Table 2). Nevertheless, the size of the overall effect was not representative of the size of the effect of each mutation.
In addition, the error frequency of the rdn5/pS23A-R double mutant in the presence of 50 μM paromomycin was 0.0121, very similar to 0.0115 for rdn5, at the same time that the Lys-62→Arg mutant alone shows the much higher error frequency of 0.0170 over the wild type (Table 2). The rdn5/pS23A-N double mutant inhibited misreading by paromomycin in vitro, in line with the character of each mutation.
Properties of hybrid ribosomes
The aforementioned results were obtained in the presence of the two chromosomal genes encoding S23, an essential ribosomal protein. In view of the interesting character of the rdn5/pS23A-R double mutant and in order to eliminate the possibility of an artifact caused by the presence of the chromosomal genes, we repeated the in vitro accuracy experiments using 60S ribosomal subunits from rdn5 cells and 40S ribosomal subunits from cells carrying a deletion of gene RPS23A, i.e. of one of the two genes encoding S23, followed by introduction of plasmid pS23A-R. The experiments were conducted both in the absence and presence of paromomycin and the results are shown in Table 3. Hybrid ribosomes composed of Lys-62→Arg 40S subunits and wild-type 60S subunits showed an error frequency of 0.0105, up from 0.0040 for the wild-type hybrid ribosomes, while hybrid ribosomes composed of rdn5 60S subunits and wild-type 40S subunits showed an error frequency of 0.0133. Hybrid ribosomes composed of rdn5 60S and Lys-62→Arg 40S showed an error frequency of 0.0130, similar to those shown by each mutation alone.
In the presence of paromomycin, the rdn5/pS23A-R double mutant showed an error frequency of 0.0207, i.e. only slightly higher than 0.0167 for rdn5 (Table 3). This shows again mild infidelity but lower than the sum of the individual increases, which would give an error frequency of 0.0257 [0.0167 + (0.0222 − 0.0132)]. Finally, the increases in the error frequencies of hybrid ribosomes carrying rdn5 mutation, were less pronounced than the increases in the error frequencies of the wild-type or the Lys-62→Arg ribosomes, a fact that is a further indication of rdn5 resistance to paromomycin.
Thus, hybrid ribosomes carrying mutation rdn5 in 25S rRNA of 60S, and mutation Lys-62→Arg in protein S23 in the absence of chromosomal RPS23A still possess the properties described earlier i.e. decreased suppression compared to that shown by Lys-62→Arg mutation alone and resistance to paromomycin.
Effect of rdn5 mutation on the binding of tRNA to the P- and A-sites of the ribosome
The interesting phenotype exhibited by rdn5, led us to explore further the properties of rdn5 in ribosomal functions other than translational accuracy and resistance to antibiotics. In the first of these experiments, ternary complex C, i.e. the Ac-[3H]Phe-tRNA•poly(U)•80S ribosome complex, was prepared as described in detail previously (5), immobilized on cellulose nitrate filter discs and its amount determined by the radioactively labeled Ac-[3H]Phe-tRNA in a liquid scintillation spectrometer. Under these conditions, Ac-Phe-tRNA was preferentially bound to the P-site as confirmed by the finding (5) that the ribosome-bound Ac-[3H]Phe-tRNA was fully reactive towards puromycin (1 mM, 16 min). Our results showed a 36% increase in the P-site binding of Ac-Phe-tRNA for rdn5 mutants compared to wild type (Table 4).
Table 4. Binding capacities of P- and A-sites and peptidyltransferase activity of rdnwt and rdn5 mutant ribosomes.
| Strain | P-site binding (% of control) | A-site binding (% of control) | kcat (min−1) | Ks (mM) | kcat/Ks (min−1 mM−1) |
|---|---|---|---|---|---|
| rdnwt | 100a | 100b | 1.64 ± 0.11 | 0.53 | 3.1 |
| rdn5 | 136 ± 5c | 130 ± 3c | 0.92 ± 0.07c | 0.55 | 1.7 |
a100% represents 0.180 molecules of Ac-[3H]Phe-tRNA bound to P-site per molecule of ribosome.
b100% represents 0.126 molecules of [3H]Phe-tRNA bound to A-site per molecule of ribosome.
cStatistically different (one-way analysis of variance, F-Scheffè test, p < 0.1) from the respective rdnwt.
A-site binding was determined as follows: first, all P-sites were filled with deacylated tRNA, under conditions described elsewhere (5), and in which the molar ratio of tRNAphe:ribosomes is 4:1. Under these conditions, the P-sites are completely filled with tRNAphe (27). We determined the A-site binding by adding Phe-tRNA and measuring the radioactivity of cellulose filter discs. Table 4 shows the A-site binding capacity of the rdn5 mutant strain expressed as percent of the radioactivity contained in the Phe-tRNA•80S•poly(U) complex of the wild-type strain L1494. Strain L1548, i.e. the rdn5 strain, showed an A-site binding 30% higher than the wild-type level. This is in agreement with the finding that rdn5 is also prone to translational errors.
Effect of rdn5 mutation on the catalytic activity of the ribosome
Complex C, i.e. the Ac-[3H]Phe-tRNA•poly(U)•80S ribosome complex, contained ∼43% of the input Ac-Phe-tRNA (3.2 A260 units or 44.8 pmol of Ac-Phe-tRNA/0.2 ml of binding mixture), since only 43% of the input of radioactivity was adsorbed on the cellulose nitrate filter disc. Thus, complex C contained 19.3 pmol Ac-Phe-tRNA. Assuming a 1:1 combination, this complex C also engages 18% of the ribosomes added (6.0 A260 units or 107.3 pmol). Hence, 0.180 molecules of Ac-Phe-tRNA are bound per molecule of poly(U)-programmed ribosomes.
The catalytic activity of eukaryotic peptidyltransferase was determined with the puromycin reaction:
![]()
An advantage of this reaction is that it is carried out in two steps. In this way, all reactions involved in the binding of the donor Ac-Phe-tRNA to the ribosomes, and the formation of ternary complex C (step 1) have been separated from peptide bond formation (step 2). The reaction between the zwittergent-extracted complex C (C) and excess puromycin (S) proceeded as a pseudo-first-order reaction in which the product (P) is Ac-Phe-puromycin and C is converted to C′, which cannot re-form complex C (Equation 1). Thus, each catalytic center of peptidyltransferase works only once. The product was substantially inhibited by sparsomycin (1 × 10−7 M), an inhibitor of peptide bond formation, showing that what is measured is the formation of peptide bonds (data not shown). The rate constant (k3) gives the reactivity of the P-site-bound donor Ac-Phe-tRNA, and it is equal to the catalytic rate constant (kcat) of peptidyltransferase. Complex C from wild-type or mutant strains was remarkably stable since less than 5% of the bound Ac-Phe-tRNA was dissociated after 16 min. Nevertheless, the amount of donor that was converted to product was corrected by dividing its value with dissociation factor A as well as with extent factor α (see Materials and Methods).
Typical first-order time plots for Ac-[3H]Phe-puromycin formation were linear up to more than 90% depletion over a wide range of puromycin concentrations. The slopes of these straight lines give the kobs values at each puromycin concentration (data not shown).
As previously described (28), the apparent rate constant kobs is a function of [S] and it is given by Equation 2:
![]()
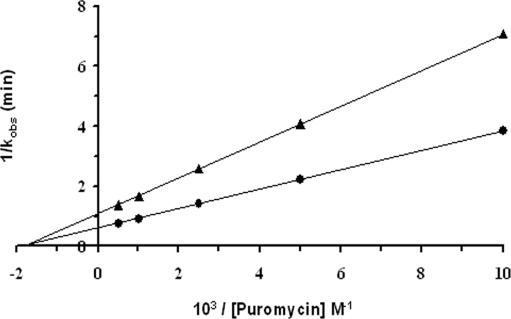
Equation 2 predicts that a plot of the reciprocal of the experimental kobs versus the reciprocal of the puromycin concentration (double reciprocal plot) should be linear. The values of kobs obtained from the wild-type strain were fitted in Equation 2 from which the double reciprocal plot gave kcat = 1.64 min −1 and Ks = 0.53 mM (Table 4 and Figure 4).
Figure 4.
Peptidyltransferase activity of rdnwt and rdn5 mutant ribosomes. Double reciprocal plots (1/kobs versus 1/[Puromycin]) for Ac-[3H]Phe-puromycin formation, using complex C prepared with ribosomes from the rdnwt strain (circles) and rdn5 strain (triangles). From these plots, the Ks and kcat values for each strain are determined.
The reaction of complex C from rdn5 mutant strain with puromycin also followed pseudo-first-order kinetics and the logarithmic time plots were linear (data not shown). The double reciprocal plot was also linear (Figure 4). From this plot, it was calculated that the first-order catalytic rate constants (kcat) for the rdn5 mutant is 0.92 min−1 (Table 4). The equilibrium constant Ks remained essentially the same. Ratio kcat/Ks is a second-order rate constant, which is an accurate measure of the ribosomal peptidyltransferase activity of each strain. Thus, the rdn5 mutant formed peptide bonds at about half the rate of the wild-type strain (kcat/Ks = 1.7 versus 3.1 min−1 mM−1). Thus, mutation C2658U lowers the peptidyltransferase activity of the ribosome. Both kcat and kcat/Ks are independent of the puromycin concentration and the percentage of active ribosomes, as explained previously (20).
Translocation studies
It is well documented that when the P-site is occupied with deacylated tRNA, the Ac-[3H]Phe-tRNA is exclusively present at the A-site and can participate in an EF-G-dependent translocation reaction at 15 mM Mg+2 (27). Translocation of yeast Ac-Phe-tRNA from the A- to P-site was monitored by reaction with 2 mM puromycin for 4 min at 30°C. Eukaryotic elongation factor-independent (spontaneous) translocation was slow and proceeded in a linear fashion at least up to 8 min reaching 20% in the rdn5 mutant compared to 16% in the wild type, if 100% indicates the extent of translocation in the presence of elongation factors.
In the presence of elongation factors and under conditions of single-round translocation, either elongation factor EF2 (0.1 μM) or the fraction containing the SPFs promoted rapid translocation that was complete within 20 s in both strains as measured by the puromycin reaction (data not shown). The puromycin reaction can detect changes in a reaction, which takes place in a time period of ≥5 s. No significant differences could be detected between the two strains either in the rate or the extent of enzymatic translocation.
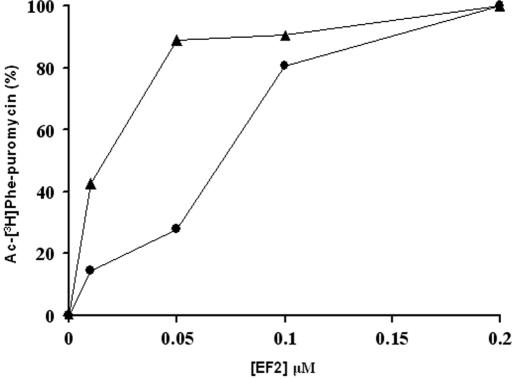
The efficiency of translocation in the wild type and the rdn5 mutant was tested further by examining the dependence of each strain for translocation on the concentrations of EF2 (Figure 5). To a fixed amount of pretranslocation complex, increasing concentrations of EF2 were added, and translocation was allowed to proceed for 1 min at 30°C. Then puromycin was added and the reaction was terminated after 4 min. As shown in Figure 5, the extent of translocation increased with EF2 concentration and reached a plateau as soon as peptidyl-tRNA is translocated on all active ribosomes in the reaction mixture. The concentration of EF2 at which 50% translocation was achieved was 3-fold lower for rdn5 than wild type. These results show that rdn5 ribosomes are more efficient in enzymatic translocation than wild-type ribosomes.
Figure 5.
Dependence of translocation on elongation factor concentration. Pretranslocation Zwittergent 3–12 complexes were incubated with different amounts of EF2 for 1 min at 30°C and the extent of translocation was determined by reaction with 2 mM puromycin for 4 min at 30°C. The concentrations of EF2 at which 50% translocation was achieved were 0.070 μM for the rdnwt strain (circles) and 0.025 μM for the rdn5 strain (triangles), respectively.
Effect of rdn5 on polysome profiles
The effect of rdn5 in the assembly of the ribosome was examined through sucrose gradient analysis. The ratio of 60S to 40S ribosomal subunits remained stable, indicating that the sarcin/ricin domain does not participate in the maturation of the 60S subunit. In contrast, the polysome profile of rdn5 indicated a significant decrease in the amount of 80S ribosomes. These results imply that the sarcin/ricin domain of 25S rRNA is involved in ribosomal subunit association. They are in accordance to earlier studies on C2663–C2658 that closes the GAGA tetraloop of the sarcin/ricin domain in 23S rRNA. The G2663C mutation, which disrupts the G2663–C2658 base pair does not affect the assembly of mutant 23S rRNA into 50S subunits, however the assembled particles are unable to form 70S ribosomes upon association with 30S subunits (29).
DISCUSSION
In the present study, we carried out a detailed biochemical analysis of the effect of mutation C2658U (rdn5) in the sarcin/ricin domain of yeast 25S rRNA on several parameters of protein synthesis. Mutation rdn5 has been previously found to affect translational fidelity, while causing increased resistance to paromomycin. We confirm these results and determine the in vitro error rate of rdn5 mutants as 3-fold higher than that of the wild type (Table 2). This was accompanied by a substantial increase of the A-site binding of Phe-tRNA, in agreement with an error-prone mutation, which allows easier access to the A-site. The increased error rate of rdn5, determined for the first time, is similar to the increased error rate observed in another element of the large ribosomal subunit, ribosomal protein L39 (5). Each of these two effects is in fact similar in size to the effect on translational accuracy of mutation Lys-62→Arg of ribosomal protein S23, which is located adjacent to the decoding center in the small ribosomal subunit. This is further proof of the interconnection between the two ribosomal subunits, which should be viewed as part of the whole and in continuous intercommunication.
Polyphenylalanine synthesis was increased by as much as 30% in rdn5 mutants compared with wild type (Figure 3). Since A-site binding is the rate-limiting step of the elongation cycle of protein synthesis, its similar increase in rdn5 mutants may explain the increase in polyphenylalanine synthesis.
Omnipotent suppressor mutations in yeast have generally been associated with hypersensitivity to misreading-inducing antibiotics, and antibiotic sensitivity has been suggested to result from an excess of errors during translation (10,30). However, a few omnipotent suppressors that are resistant to misreading-inducing antibiotics have been reported (31). Paromomycin induces misreading and also inhibits cell growth (32). Liu and Liebman (10) found that in the rdn5 omnipotent suppressor mutant, paromomycin caused phenotypic suppression even though the rdn5 mutation caused resistance to paromomycin. We confirm the separation of these two effects (Figure 1A) and provide additional evidence whereby 50% inhibition of growth of rdn5 mutant cells is observed at higher concentrations of paromomycin compared with wild type (Figure 1B). These results may be explained by the notion that the decoding function of the rdn5 ribosome remains accessible to paromomycin, allowing it to continue to cause phenotypic suppression. However, the rdn5 mutation may alter the normal interaction between the sarcin/ricin domain and the elongation factor EF-2, thereby antagonizing the killing effect of the antibiotic during translocation (10).
Paromomycin is an error-inducing antibiotic, which was also found to stimulate phenylalanine incorporation in wild-type and mutant ribosomes (4). In the experiments described here, paromomycin stimulated polyphenylalanine synthesis in all three strains almost by an order of magnitude (Figure 3). Significantly however, the increase in leucine misincorporation in the presence of paromomycin was even higher for each strain than the respective increase in polyphenylalanine formation (Table 2).
A possible mode of action of paromomycin is that it fixes the flexible nucleotides A1492 and A1493 (33) in a conformation similar to that induced by cognate tRNA-anticodon stem–loop (ASL), decreasing their entropy and slightly reducing the distance between helix 44 and the 30S shoulder (protein S12). This eliminates part of the cost of tRNA-dependent domain closure. With cognate tRNA-ASL, as in the case of Phe-tRNA and poly(U), this cost reduction results in increased activity of tRNA-ASL binding, whereas with near-cognate tRNA-ASL, such as Leu-tRNA and poly(U), it makes closure energetically favorable. Paromomycin affects the rates of almost every step during ribosomal aminoacyl-tRNA selection and, in particular, accelerates both GTP hydrolysis and accommodation. This ‘cost reduction’ caused by paromomycin, results in increased affinity of Phe-tRNA to the ribosome and, thus, in the increase of polyphenylalanine formation.
Recently, increased polyphenylalanine synthesis was also observed in E.coli ribosomes in the presence of the antibiotics pactamycin and streptomycin (34). Of the two antibiotics, streptomycin also increased the error frequency, while pactamycin decreased it. A third antibiotic, edeine, significantly enhanced misincorporation, but it reduced polyphenylalanine. To date, misreading antibiotics have been discovered that bind either to the decoding center of the A-site, like aminoglycosides such as paromomycin, or within the vicinity of helix 27 such as streptomycin, but not in proximity to the P-site, such as edeine. This finding is of particular interest, since it supports the idea that molecules acting on the P-site may affect A-site fidelity (34).
The changes observed in the rdn5/pS23A-R double mutant may arise from changes in the behavior of either the rdn5 mutation or the Lys-62→Arg mutation. This double mutant displays a highly increased resistance compared with the single mutants toward paromomycin in vivo (Figure 1A and B). From Figure 1B, we can determine that 50% inhibition of growth was achieved at 176 μM paromomycin for the double mutant compared with 108 μM for the rdn5 alone, and 22 μM paromomycin for the C/pS23A-R mutant. These results indicate a change in the function of Lys-62→Arg. This possibility is likely, since this site behaves as an antisuppressor and drug-resistant mutant when its equivalent E.coli Lys-42 is substituted by Arg, Asn, Thr or Gln as well as when Lys-62 of yeast is substituted by Asn, Thr or Gln. Only when Lys-62 of yeast is substituted by Arg, a polymorphism appears, which is characterized by suppression and drug sensitivity. These are partially reversed in the concomitant presence of rdn5. Thus, the importance of the Lys-62 site in the control of translational accuracy is again confirmed. The phenotypic ambiguity displayed by Lys-62 of S23 suggests that the accuracy of translation is highly dependent on this site. Anthony and Liebman (15) suggested that Lys-62 essentially divides S23 into two segments: all of their mutations that were N-terminal to Lys-62 in a region of S23 that is diverged from the prokaryotic homologues increased suppression and antibiotic sensitivity, while all those that were C-terminal to Lys-62 in a region well conserved with the prokaryotes, caused antisuppression and drug resistance. We suggest that even minor perturbations induced by the introduction of mutation rdn5 of the sarcin/ricin domain of 25S rRNA cause the Lys-62→Arg mutation to show decreased suppression compared to that shown by the Lys-62→Arg mutation alone, accompanied by drug resistance.
Mutation rdn5 reduced peptidyltransferase activity to 54% of the wild type, in agreement with the notion that the ribosome is a ribozyme. Nevertheless, this decrease was not as marked as in the cases of non-essential ribosomal proteins L24 and L41, deletions of which caused net 3-fold decreases in the peptidyltransferase activity of the ribosome (20,35). Thus, the difference in size of the effects on peptidyltransferase activity may arise, among others, from the different positions of the sarcin/ricin loop and proteins L24 and L41, relative to the peptidyltransferase center.
The fact that peptidyltransferase activity was reduced, while polyphenylalanine synthesis was increased may be explained by the hypothesis that the two processes have different rate-limiting steps. The rate-limiting step of the elongation cycle is the occupation of the A-site and this is much slower than peptidyl transfer (36,37). For the puromycin reaction, however, the rate-limiting step is peptide bond formation and not the binding of puromycin to the A-site (38). As a result, a significant reduction in peptidyltransferase activity would strongly decrease the rate of puromycin reaction, while the rate of polyphenylalanine synthesis could be unaffected or even increased. Such differences between these two processes have been observed previously with a series of 23S rRNA mutations (39), with ribosomal protein L2 (38) or with yeast ribosomal protein L41 (35).
Our results showed that in the presence of rdn5 mutation, both P- and A-site binding capacities were increased (Table 4). Thus, the sarcin/ricin domain, which is a site for binding of elongation factors EF1 and EF2, interacts with the tRNA binding sites on the ribosome. The increase in P-site binding may be explained by the hypothesis that complex C is stabilized by mutation rdn5 and is subsequently less reactive with puromycin to form peptide bonds. Finally, it appears that increased rates of P- and A-site binding are not necessarily followed by optimal orientation of these substrates in order to form peptide bonds.
This mutation of 25S rRNA also affected another ribosomal activity, the translocation step of protein synthesis. This is catalyzed by elongation factor EF-G in prokaryotes and EF2 in eukaryotes. Mutation rdn5 of sarcin/ricin loop increased the efficiency of EF2-dependent translocation as shown by the lower amounts of EF2 needed to achieve 50% translocation (Figure 5). The molecular mechanism of translocation and its catalysis by EF-G/EF2 is largely unknown. Direct evidence that the sarcin/ricin domain is in close proximity to the EF-G catalytic center was provided recently (40); nucleotide G2655 of this domain was shown to be critical for the formation of a covalent bond, by ultraviolet irradiation, between EF-G and a sarcin/ricin domain oligoribonucleotide containing 5-iodouridine. The proximity suggests that the sarcin/ricin domain RNA has a role in the activation of GTP hydrolysis that leads to a transition in the conformation of the factor and to its release from the ribosome (40). The contribution of the invariant C2658–G2663 base pair to the binding of elongation factors and the function of the ribosome has been evaluated by constructing mutations in the nucleotides and determining their phenotype (29). It has been shown that ribosomes carrying either the single transversion mutation G2663C that disrupts the Watson–Crick pair that closes the sarcin/ricin tetraloop or the double transversion mutations C2658G–G2663C and C2658A–G2663U that reverse the polarity of the pyrimidine and the purine but restore the potential to form a canonical pair, exhibit a severe effect in the binding of EF-G·GTP binary complex, inhibit the activity of ribosomes, and cause lethality. In contrast, a double transition mutation, C2658U–G2663A, which maintains nucleotide polarity but changes their identity, has no effect on the binding of the factor to the ribosome, on ribosome function or on cell growth (29). Comparison of the atomic structures of wild type, C2658U–G2663A and C2658G–G2663C 27 nt mimics of the sarcin/ricin loop from E.coli 23S rRNA suggested that differences between accessible functional groups of the lethal mutation and those of the viable mutation and wild-type sarcin/ricin loop may account for the impaired elongation factor binding to ribosomes with the C2658G–G2663C mutation, and may underlie the lethal phenotype (16). These functional groups are two Hoogsteen edge groups, C5 of C2658 and N7 of G2663, which are identical in the wild-type and the viable mutation but are different in the lethal mutation. They are part of the putative EF binding surface and binding studies indicate that the C2658G–G2663C mutation disrupts binding of EF-G to the SRL RNA (16). Based on the above, there are two possible explanations for the effects of rdn5, a C2658U mutation that is viable; either the Hoogsteen edge group of C2658U forms energetically significant contacts to EF-2 or complex formation with EF-2 induces the SRL structure to adopt a different conformation where differences in stacking interactions in which C2658U participates may play a role. Based on the above variety and unpredictability of the effects of mutations in the closing pair of the sarcin/ricin domain on ribosome function, it becomes clear that a better understanding of the effect of rdn5 mutation on EF-2 catalyzed translocation should await the crystal structure of eukaryotic ribosomes carrying this mutation.
Acknowledgments
ACKNOWLEDGEMENTS
We are indebted to Professor Susan Liebman for making yeast strains L1494 and L1548 available to us. We thank Dr Joo Hong for helping us with the cycloheximide resistance experiment. We also thank Dr Dimitrios Kalpaxis for critical reading of the manuscript. J.D. was a recipient of a 3-year scholarship by the Greek State Foundation of Scholarships (I.K.Y.). This work was supported in part by Grant 2471 from the Karatheodoris 2000 Research Program of the University of Patras.
REFERENCES
- 1.Velichutina I.V., Dresios,J., Hong,J.Y., Li,C., Mankin,A., Synetos,D. and Liebman,S.W. (2000) Mutations in helix 27 of the yeast Saccharomyces cerevisiae 18S rRNA affect the function of the decoding center of the ribosome. RNA, 6, 1174–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moore P.B. and Steitz,T.A. (2002) The involvement of RNA in ribosome function. Nature, 418, 229–235. [DOI] [PubMed] [Google Scholar]
- 3.Liebman S.W., Chernoff,Y.O. and Liu,R. (1995) The accuracy center of a eukaryotic ribosome. Biochem. Cell Biol., 73, 1141–1149. [DOI] [PubMed] [Google Scholar]
- 4.Synetos D., Frantziou,C.P. and Alksne,L.E. (1996) Mutations in yeast ribosomal proteins S28 and S4 affect the accuracy of translation and alter the sensitivity of the ribosomes to paromomycin. Biochim. Biophys. Acta, 1309, 156–166. [DOI] [PubMed] [Google Scholar]
- 5.Dresios J., Derkatch,I.L., Liebman,S.W. and Synetos,D. (2000) Yeast ribosomal protein L24 affects the kinetics of protein synthesis and ribosomal protein L39 improves translational accuracy, while mutants lacking both remain viable. Biochemistry, 39, 7236–7244. [DOI] [PubMed] [Google Scholar]
- 6.Hausner T.P., Atmadja,J. and Nierhaus,K.H. (1987) Evidence that the G2661 region of 23S rRNA is located at the ribosomal binding sites of both elongation factors. Biochimie, 69, 911–923. [DOI] [PubMed] [Google Scholar]
- 7.Moazed D. and Noller,H.F. (1987) Interaction of antibiotics with functional sites in 16S ribosomal RNA. Nature, 327, 389–394. [DOI] [PubMed] [Google Scholar]
- 8.Marchant A. and Hartley,M.R. (1994) Mutational studies on the alpha-sarcin loop of Escherichia coli 23S ribosomal RNA. Eur. J. Biochem., 226, 141–147. [DOI] [PubMed] [Google Scholar]
- 9.Wool I.G., Correll,C.C. and Chan,Y.-L (2000) Structure and function of the sarcin/ricin domain. In Garrett,R.A., Douthwaite,S.R., Liljas,A., Matheson,A.T., Moore,P.B. and Noller,H.F. (eds), The Ribosome: Structure, Function, Antibiotics, and Cellular Interactions. American Society for Microbiology Press, Washington, DC, pp. 461–473. [Google Scholar]
- 10.Liu R. and Liebman,S.W. (1996) A translational fidelity mutation in the universally conserved sarcin/ricin domain of 25S yeast ribosomal RNA. RNA, 2, 254–263. [PMC free article] [PubMed] [Google Scholar]
- 11.Tapprich W.E. and Dahlberg,A.E. (1990) A single base mutation at position 2661 in E. coli 23S ribosomal RNA affects the binding of ternary complex to the ribosome. EMBO J., 9, 2649–2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Melancon P., Tapprich,W.E. and Brakier-Gingras,L. (1992) Single-base mutations at position 2661 of Escherichia coli 23S rRNA increase efficiency of translational proofreading. J. Bacteriol., 174, 7896–7901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gorini L. (1970) Informational suppression. Annu Rev Genet., 4, 107–134. [DOI] [PubMed] [Google Scholar]
- 14.Spahn C.M., Gomez-Lorenzo,M.G., Grassucci,R.A., Jorgensen,R., Andersen,G.R., Beckmann,R., Penczek,P.A., Ballesta,J.P. and Frank,J. (2004) Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation. EMBO J., 23, 1008–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anthony R.A. and Liebman,S.W. (1995) Alterations in ribosomal protein RPS28 can diversely affect translational accuracy in Saccharomyces cerevisiae. Genetics, 140, 1247–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Correll C.C., Beneken,J., Plantinga,M.J., Lubbers,M. and Chan,Y.L. (2003) The common and the distinctive features of the bulged-G motif based on a 1.04 A resolution RNA structure. Nucleic Acids Res., 31, 6806–6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chernoff Y.O., Vincent,A. and Liebman,S.W. (1994) Mutations in eukaryotic 18S ribosomal RNA affect translational fidelity and resistance to aminoglycoside antibiotics. EMBO J., 13, 906–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alksne L.E., Anthony,R.A., Liebman,S.W. and Warner,J.R. (1993) An accuracy center in the ribosome conserved over 2 billion years. Proc. Natl Acad. Sci. USA, 90, 9538–9541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Itoh H., Fukada,Y., Murata,K. and Kimura,A. (1983) Transformation of intact yeast cells treated with alkali cations. J. Bacteriol., 153, 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dresios J., Panopoulos,P., Frantziou,C.P. and Synetos,D. (2001) Yeast ribosomal protein deletion mutants possess altered peptidyltransferase activity and different sensitivity to cycloheximide. Biochemistry, 40, 8101–8108. [DOI] [PubMed] [Google Scholar]
- 21.Merrick W.C., Kemper,W.M., Kantor,J.A. and Anderson,W.F. (1975) Purification and properties of rabbit reticulocyte protein synthesis elongation factor 2. J. Biol. Chem., 250, 2620–2625. [PubMed] [Google Scholar]
- 22.Puglisi J.D., Blanchard,S.C. and Green,R. (2000) Approaching translation at atomic resolution. Nature Struct. Biol., 10, 855–861. [DOI] [PubMed] [Google Scholar]
- 23.Palmer E., Wilhelm,J.M. and Sherman,F. (1979) Variation of phenotypic suppression due to the psi+ and psi− extrachromosomal determinants in yeast. J. Mol. Biol., 128, 107–110. [DOI] [PubMed] [Google Scholar]
- 24.Wool I.G., Gluck,A. and Endo,Y. (1992) Ribotoxin recognition of ribosomal RNA and a proposal for the mechanism of translocation. Trends Biochem. Sci., 17, 266–269. [DOI] [PubMed] [Google Scholar]
- 25.Gale E.F., Cundliffe,E., Reynolds,P.E. and Richmond,M.H. (1981) The Molecular Basis of Antibiotic Action, 2nd edn. John Wiley and Sons Ltd, Bristol, UK. [Google Scholar]
- 26.Kurland C.G., Jörgensen,F., Richter,A., Ehrenberg,M., Bilgin,N. and Rojas,A.M. (1990) Through the accuracy window. In Hill,W.E., Moore,P.B., Dahlberg,A., Schlessinger,D., Garrett,R.A. and Warner,J.R. (eds), The Ribosome: Structure, Function and Evolution. American Society for Microbiology, Washington, DC, pp. 513–526. [Google Scholar]
- 27.Rheinberger H.J. and Nierhaus,K.H. (1983) Testing an alternative model for the ribosomal peptide elongation cycle. Proc. Natl Acad. Sci. USA, 80, 4213–4217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Synetos D. and Coutsogeorgopoulos,C. (1987) Studies on the catalytic rate constant of ribosomal peptidyltransferase. Biochim. Biophys. Acta, 923, 275–285. [DOI] [PubMed] [Google Scholar]
- 29.Chan Y.L., Sitikov,A.S. and Wool,I.G. (2000) The phenotype of mutations of the base-pair C2658.G2663 that closes the tetraloop in the sarcin/ricin domain of Escherichia coli 23S ribosomal RNA. J. Mol. Biol., 298, 795–805. [DOI] [PubMed] [Google Scholar]
- 30.Surguchov A.P., Smirnov,V.N., Ter-Avanesyan,M.D., Inge-Vechtomov,S.G. (1984) Ribosomal suppression in eukaryotes. Phys. Chem. Biol., 4, 147–206. [Google Scholar]
- 31.Wakem L.P. and Sherman,F. (1990) Isolation and characterization of omnipotent suppressors in the yeast Saccharomyces cerevisiae. Genetics, 124, 512–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moazed D. and Noller,H.F. (1987) Interaction of antibiotics with functional sites in 16S ribosomal RNA. Nature, 327, 389–394. [DOI] [PubMed] [Google Scholar]
- 33.Ogle J.M., Carter,A.P. and Ramakrishnan,V. (2003) Insights into the decoding mechanism from recent ribosome structures. Trends Biochem. Sci., 28, 259–266. [DOI] [PubMed] [Google Scholar]
- 34.Dinos G., Wilson,D.N., Teraoka,Y., Szaflarski,W., Fucini,P., Kalpaxis,D. and Nierhaus,K.H. (2004) Dissecting the ribosomal inhibition mechanisms of edeine and pactamycin: the universally conserved residues G693 and C795 regulate P-site RNA binding. Mol. Cell, 13, 113–124. [DOI] [PubMed] [Google Scholar]
- 35.Dresios J., Panopoulos,P., Suzuki,K. and Synetos,D. (2003) A dispensable yeast ribosomal protein optimizes peptidyltransferase activity and affects translocation. J. Biol. Chem., 278, 3314–3322. [DOI] [PubMed] [Google Scholar]
- 36.Bilgin N., Ehrenberg,M. and Kurland,C. (1988) Is translation inhibited by noncognate ternary complexes? FEBS Lett., 233, 95–99. [DOI] [PubMed] [Google Scholar]
- 37.Schilling-Bartetzko S., Bartetzko,A. and Nierhaus,K.H. (1992) Kinetic and thermodynamic parameters for tRNA binding to the ribosome and for the translocation reaction. J. Biol. Chem., 267, 4703–4712. [PubMed] [Google Scholar]
- 38.Diedrich G., Spahn,C.M., Stelzl,U., Schafer,M.A., Wooten,T., Bochkariov,D.E., Cooperman,B.S., Traut,R.R. and Nierhaus,K.H. (2000) Ribosomal protein L2 is involved in the association of the ribosomal subunits, tRNA binding to A and P sites and peptidyl transfer. EMBO J., 19, 5241–5250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spahn C.M., Schäfer,M.A., Krayevsky,A.A. and Nierhaus,K.H. (1996) Conserved nucleotides of 23S rRNA located at the ribosomal peptidyltransferase center. J. Biol. Chem., 271, 32857–32862. [DOI] [PubMed] [Google Scholar]
- 40.Chan Y.L., Correll,C.C. and Wool,I.G. (2004) The location and the significance of a cross-link between the sarcin/ricin domain of ribosomal RNA and the elongation factor-G. J. Mol. Biol., 337, 263–272. [DOI] [PubMed] [Google Scholar]