Abstract
Apurinic/apyrimidinic (AP) sites are alkali labile lesions that, when encountered during DNA replication, can block polymerases or potentially result in mutagenic events. Owing to the instability of 2-deoxyribose lesions (AP), a chemically stable tetrahydrofuran analog (F) is often used as a model of abasic sites. A comparison of the two lesions in Saccharomyces cerevisiae revealed that the model lesion and 2-deoxyribose have distinct in vivo effects. Comprehensive comparative analyses of F and AP have not been carried out in Escherichia coli. We conducted a side-by-side investigation of F and AP in E.coli to compare their biological effects and interactions with SOS polymerases. Both lesions were examined in SOS-induced and uninduced cells. Our studies reveal that in uninduced E.coli the effects of individual polymerases in the replication of plasmids containing F or AP are distinct. However, when cells are SOS-induced, the biological effects of F and AP are similar.
INTRODUCTION
In the cell, spontaneous depurination and enzymatic processing of damaged nucleotides produce thousands of apurinic/apyrimidinic (AP) sites each day, making them one of the most frequently generated DNA lesions (1,2). Although base excision repair enzymes remove a majority of 2-deoxyribose sites (AP), those that are not repaired are poorly bypassed by DNA polymerases (3–5). When successful translesional synthesis (TLS) occurs, it is often mutagenic due to the non-coding nature of AP lesions. The ‘natural’ 2-deoxyribose site is heat and alkali labile (Figure 1) (6). Consequently, in vitro and in vivo studies to characterize abasic sites have frequently been carried out using a structurally similar but chemically stable tetrahydrofuran lesion (F). A comparison of F and AP conducted in Saccharomyces cerevisiae showed that nucleotide incorporation opposite each lesion is distinct, and prompted us to compare the biological effects of F and AP in Escherichia coli (7). We have examined the lethality and mutagenicity of F and AP in E.coli and investigated the role of SOS polymerases in the bypass of each abasic site. By examining both sites concurrently and in the same sequence contexts, we have determined how well F models the cellular effects of a 2-deoxyribose lesion (AP).
Figure 1.
Structure of abasic lesions 2-deoxyribose (AP) and tetrahydrofuran (F).
Inefficient bypass of F and AP makes these lesions highly lethal in E.coli and S.cerevisiae (7–11). Independent studies of the lesions in different sequence contexts suggest that AP and its model are bypassed with comparable efficiency. When F is introduced into E.coli on a single-stranded plasmid, TLS across the lesion occurs 0.8% to ∼6% of the time (10,11). The SOS-induction of E.coli elevates expression of specialized lesion-bypass polymerases (pol II, pol IV and pol V), resulting in a significant increase in TLS. Bypass of an AP lesion was found to occur 0.1–0.7% of the time in uninduced E.coli, but increased to 5–7% with SOS-induction (8).
There have been no comparative studies examining the effects of individual SOS-induced polymerases on F and AP in E.coli. However, analysis of N-2-acetylaminofluorene, a bulky DNA adduct, revealed that all three SOS polymerases are employed for bypass (12). In vitro, pol II, pol IV and pol V bypass F. Nucleotide incorporation by pol V follows the A-rule, but pol IV bypass results almost exclusively in −2 deletions (9,13,14). Although in vitro analyses of nucleotide incorporation opposite abasic sites have not been reported for pol II, studies in E.coli revealed that pol II and pol V preferentially incorporate dA opposite AP and F, respectively (9,15). In contrast, E.coli studies of pol IV lesion bypass show that deletions are the major product. However, when substitution products are formed, there is a strong preference for dC opposite F (9).
Owing to the effects of sequence context on nucleotide incorporation frequencies and deletion formation, results from published E.coli studies with F and AP cannot be directly compared. However, side-by-side examination of F and AP in S.cerevisiae revealed that nucleotide incorporation frequencies opposite the model site and 2-deoxyribose differ (7). Rev-dependent preferential incorporation of dC opposite AP is observed, whereas dA is most frequently incorporated opposite F. Nucleotide incorporation opposite abasic sites is variable in E.coli (8,9,11,16). Often there is preferred insertion of dA opposite F and AP (8,9,16). However, nearly equal incorporation of T and dA opposite F has been reported previously (11). Furthermore, depending upon sequence context and the polymerases available, TLS results in a significant amount of dC opposite AP (16). Replication of plasmids containing F and AP in E.coli, S.cerevisiae and mammalian cells also produces single-nucleotide deletions (8,16–18). In addition, a small amount of 2 nt deletion product results from pol IV TLS across F in E.coli. However, nearly all bypass by pol IV results in 2 nt deletions in vitro (9).
The variety of polymerases present and nucleic acid sequences in which the abasic lesions can reside create a complex matrix. The data referred to above illustrate how these factors make it difficult to compare the replication of AP and F using purified E.coli polymerases, as well as in the organism itself. For these reasons, we sought to carry out a comprehensive comparison of the replication of the abasic sites in E.coli in order to determine how appropriate a model F is of AP.
MATERIALS AND METHODS
General methods
Oligonucleotides were synthesized on an Applied Biosystems Incorporated 394 DNA synthesizer using standard protocols. DNA synthesis reagents were purchased from Glen Research. Oligonucleotides containing AP sites were generated by uracil DNA glycosylase (UDG) treatment of 2′-deoxyuridine, and the tetrahydrofuran analog (F) of an AP site was incorporated into oligonucleotides using commercial reagents (19). DNA was purified by 20% polyacrylamide denaturing gel electrophoresis [5% crosslink, 45% urea (by weight)]. DNA manipulation, including enzymatic labeling, was carried out using standard procedures (20). T4 polynucleotide kinase, BbsI, HaeIII and UDG were obtained from New England Biolabs. The T4 DNA polymerase was obtained from USB. [γ-32P]ATP was purchased from Amersham Pharmacia Biotech. The E.coli cells deficient in individual SOS polymerases were prepared as described previously (21). Quantification of radiolabeled oligonucleotides was carried out using a Molecular Dynamics Storm 840 PhosphorImager equipped with ImageQuant version 5.1 software.
Construction of M13 genomes
The M13 genomes were generated in triplicate for each 16mer insert as described previously (16,22). Briefly, lesion-containing oligonucleotide 16mers were 5′-phosphorylated and ligated into M13mp7(L2) plasmid using two scaffolds d(GGT CTT CCA CTG AAT CAT GGT CAT AGC) and d(AAA ACG ACG GCC AGT GAA TTG GAC GC). After ligation, the scaffolds were digested by reacting each ligation mixture with T4 DNA polymerase. The polymerase was removed by phenol extraction, and the DNA was purified by Centricon 100 filtration. The ligation efficiency was determined by using 1% TBE agarose gel analysis.
Preparation of E.coli for electroporation
Wild-type (K16), polymerase II [STL1336 (SpcR)], polymerase IV [Xs-1 (KanR)], polymerase V [SR1157U (CamR)] and SOS-polymerase triple knockout cells [SF2108 (SpcR, KanR and CamR)] were grown overnight from a genetic stock in 10 ml Luria–Bertani (LB) at 37°C, with orbital shaking (270 r.p.m.). The SOS-induced and uninduced cells were prepared as described previously (23). The cells to be SOS-induced were grown in LB to an OD600 of 0.3, while cells to be electroporated uninduced were grown to an OD600 of 0.5. To induce the cells, the E.coli were pelleted, resuspended in 0.1 M MgSO4 (50 ml) and then irradiated at 45 J/m2 with 254 nm light. The SOS-induced cells were then added to 50 ml of 2 × YT and grown for 40 min at 37°C with orbital shaking (270 r.p.m.). Both SOS-induced and uninduced cells were then pelleted, resuspended in ice-cold H2O, pelleted again and then resuspended in ice-cold 10% glycerol (2 ml). The prepared cells (100 μl) were mixed with 1 pmol of ligated M13 plasmid genome on ice, electroporated (∼2.5 kV, 4.74 ms) and then plated with X-Gal and isopropyl-β-d-thiogalactopyranoside. All experiments involving specific plasmids and cell types were carried out in triplicate.
Determining the percent bypass and mutagenicity F and AP in E.coli
Percent bypass was determined by comparing the number of plaques formed from plasmid with an insert containing F or AP to plasmid with an insert containing T instead of a lesion (16). The number of plaques observed from replication of plasmids containing F or AP was adjusted for false positives, as described previously (16). Typical plaque counts for thymidine containing controls (1c, 2c) were between 1200 and 1400. The numbers of plaques observed upon plating cells electroporated with plasmids containing F or AP ranged from 64 to 161 (before correction as noted above) under uninduced conditions and increased to between 302 and 395 under SOS-induced conditions. Analysis of mutations and deletions was carried out using the restriction endonuclease and postlabeling (REAP) assay (16,22). The REAP method has the advantage of providing statistically meaningful data rapidly, as well as allowing for the identification and analysis of insertion, substitution and deletion products in the same experiment. Briefly, a QIAprep Spin M13 Kit was used to isolate replicated plasmids. The DNA was then amplified by PCR using forward and reverse amino modified primers d(TTT CAC ACA GGA AAC AGC TAT GAC CAT G) and d(CAG GGT TTT CCC AGT CAC GAC GTT GTA A), respectively. The PCR products were purified by phenol extraction, and then treated with BbsI and shrimp alkaline phosphatase. The digested DNA was radiolabeled, digested with HaeIII, and the fragments were separated on a 20% denaturing gel. The appropriate product band(s) was excised from the gel, eluted and desalted on Sephadex G25 columns. The DNA was digested with P1 nuclease, and then 0.5 μl of each sample was spotted onto a cellulose TLC plate that was eluted for 10 h in 198 ml of buffer [180 sat (NH4)2HPO4 and 18 ml H3PO4]. The dried plates were exposed to a phosphorimaging screen for ∼24 h, and the relative amounts of each nucleotide were determined using ImageQuant 5.1.
RESULTS AND DISCUSSION
Plasmid construction and lesion bypass
A single F or AP lesion was site-specifically incorporated into single-stranded M13mp7(L1) plasmid and introduced into E.coli by electroporation as described previously (16,22). Briefly, 5′-phosphorylated oligonucleotides containing either F or AP were ligated into linearized plasmid using appropriate scaffolds to direct orientation. Chemically synthesized oligonucleotides containing F were characterized by electrospray ionization mass spectrometry (ESI-MS) (see Supporting information). Oligonucleotides containing AP sites were produced from chemically synthesized biopolymers in which 2′-deoxyuridine was incorporated during the solid phase synthesis. The AP sites were introduced immediately prior to plasmid preparation using UDG. The extent of the deglycosylation reaction was analyzed by 5′-32P-labeling an aliquot of the product (and precursor) and subjecting the oligonucleotide to mild alkaline hydrolysis (0.1 M NaOH at 55°C for 20 min) (see Supporting information). Both lesions were analyzed in two sequence contexts (Table 1). F and AP were studied in wild-type and SOS polymerase single knockout cells to ascertain the effects of specific polymerases. Since earlier studies of abasic sites have been conducted in E.coli both with and without induction, one sequence (5′-dC) from each lesion was analyzed in both SOS-induced and uninduced cells.
Table 1. Oligonucleotide plasmid inserts.
| d(GAA GAC CCN GGC GTC C) |
|---|
| 1a–c |
| a N = F |
| b N = AP |
| c N = T |
| d(GAA GAC CTN GGC GTC C) |
| 2a–c |
| a N = F |
| b N = AP |
| c N = T |
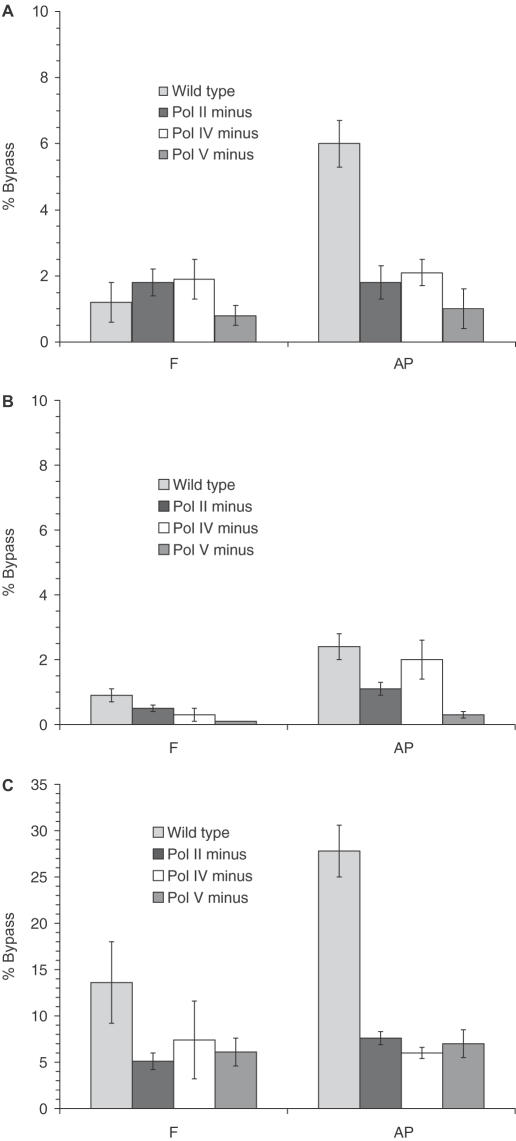
Bypass of F in uninduced cells ranged from 0.1 to 1.9% depending on the cell line, and increased to 5.1–13.6% (3- to 11-fold increase) with SOS-induction (Figure 2A–C). Plasmids containing AP show a similar increase in bypass upon SOS-induction. However, when wild-type cells are SOS-induced, TLS across AP is twice as efficient as bypass of F (Figure 2C). This is analogous to TLS in S.cerevisiae, where AP is bypassed more efficiently than F (7). Essentially, no bypass of F or AP occurred when cells were lacking all three SOS polymerases (data not shown). This illustrates the necessity of SOS polymerases even at low constitutively expressed levels for abasic site TLS.
Figure 2.
Percent bypass of F (1a, 2a) and AP (1b, 2b). (A) Uninduced (1a, b), (B) Uninduced (2a, b) and (C) SOS-induced (1a, b).
In uninduced cells, removal of pol V results in the most significant average decrease in TLS. This is consistent with earlier work that attributes most abasic site bypass to pol V (9,24). However, SOS-induced cells show approximately the same decrease in bypass with the removal of any of the three SOS polymerases (Figure 2C). This indicates that when sufficient quantities of pol II and/or pol IV are present, they can compensate for the elimination of pol V expression.
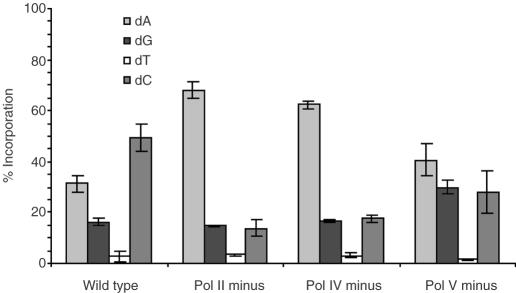
Mutations resulting from TLS
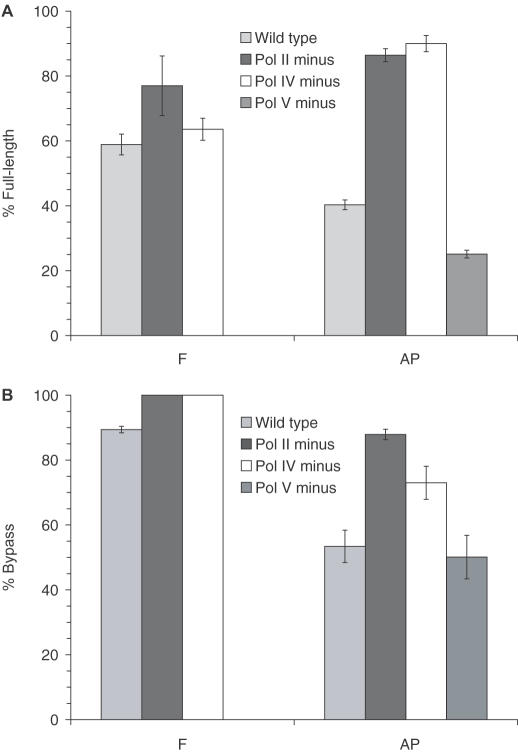
With and without SOS-induction, single-nucleotide deletions frequently result from bypass of F and AP (8,9,11). In uninduced cells, we found that the ratio of substitution to deletion products produced from bypass of each lesion is affected differently by SOS polymerases (Figure 3A and B). In the absence of pol V, bypass of F results in all single-nucleotide deletion, whereas TLS across AP produces 25.1–50.1% full-length product (Figure 3A and B). Bypass of F produces more deletion product when dC, rather than T, is 5′ to the lesion in all cell lines. TLS across AP shows this same sequence effect only in the absence of pol V. F and AP also respond differently to the removal of pol II or pol IV. TLS across AP using E.coli deficient in either of these polymerases results in much more full-length product than is produced in wild-type E.coli (Figure 3A and B). In contrast, TLS across F produces a comparable amount of full-length product in wild-type, pol II deficient and pol IV deficient cells. Therefore, in uninduced wild-type cells, pol II and pol IV are used much more often to bypass AP than F, and these enzymes are responsible for single-nucleotide deletions.
Figure 3.
Percent full-length bypass product in uninduced cells. (A) F (1a) and AP (1b). (B) F (2a) and AP (2b).
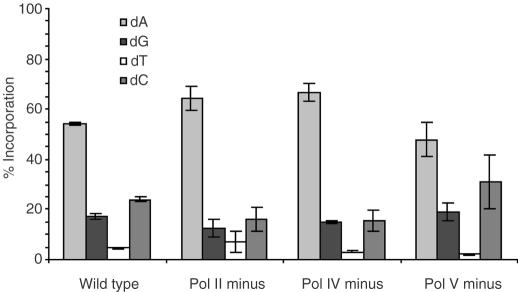
Bypass of F and AP in SOS-induced cells results in more deletion product than is produced from TLS in uninduced E.coli (Figures 3A, B and 4). However, in contrast to bypass in uninduced cells, the induced conditions show approximately the same quantity of deletions and comparable polymerase effects upon deletion formation for F and AP (Figure 4). There is an equivalent decrease in full-length product observed in pol V deficient cells for each lesion. Furthermore, the removal of either pol II or pol IV results in a substantial increase in full-length product compared with wild-type cells for both F and AP. This suggests that pol II and pol IV work in concert with one another to perform lesion bypass. Earlier studies in S.cerevisiae revealed that pol δ and pol ζ work cooperatively to efficiently bypass an abasic lesion (25). Polymerase δ inserts dA opposite the AP site and pol ζ extends from the inserted nucleotide (25). If polymerases II and IV act in a fashion similar to pol δ and pol ζ, then the removal of either polymerase would result in an increase in pol V bypass of the lesions and, consequently, the increase in full-length product observed during TLS in pol II or pol IV deficient cells.
Figure 4.
Percent full-length bypass product of F (1a) and AP (1b) in SOS-induced cells.
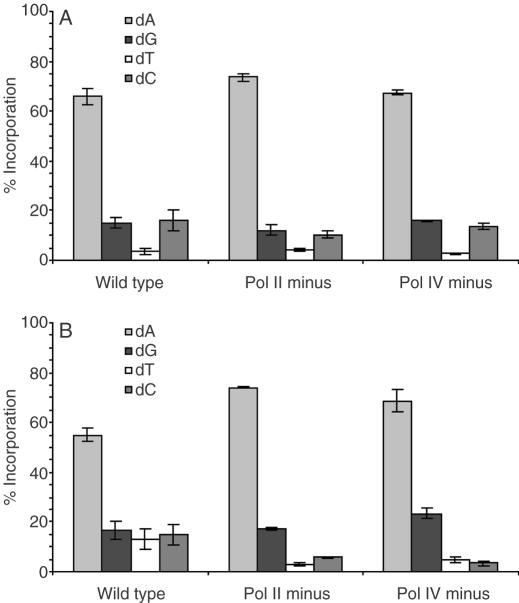
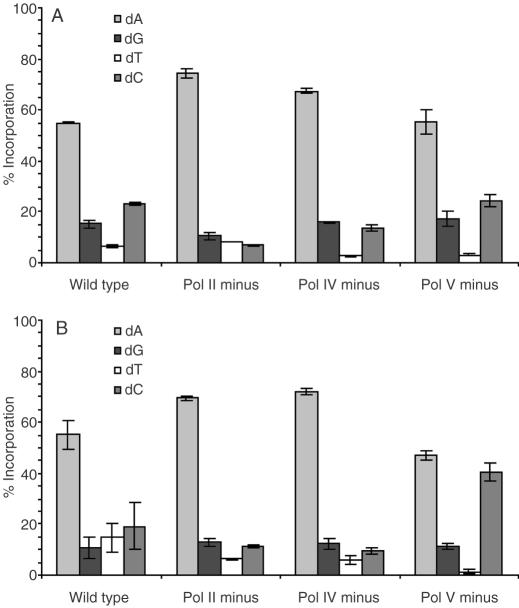
Nucleotide incorporation opposite F and AP
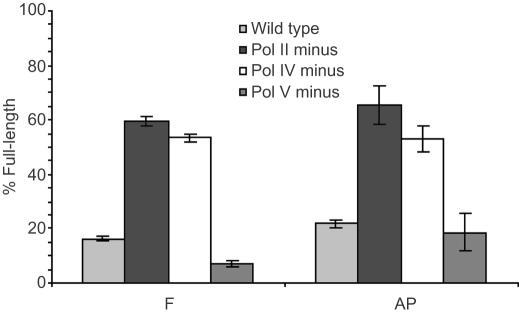
The nucleotide most frequently inserted opposite F and AP in all uninduced cell lines is dA (Figures 5A, B and 6A, B). However, pol V deficient cells show a large increase in dC incorporation opposite AP. As was seen in uninduced cells, bypass of both F and AP in pol II, pol IV and pol V deficient E.coli results in preferential incorporation of dA following SOS induction (Figures 7 and 8). The frequency of dC incorporation opposite both molecules increases in the absence of pol V under SOS conditions. One clear difference between F and AP following SOS-induction occurs in wild-type cells, where dC is preferentially incorporated opposite F, while AP still follows the A-rule (Figures 7 and 8). This is the only significant distinction between F and AP observed in SOS-induced cells. The increased dC incorporation opposite F and AP in pol V deficient cells suggests that pol II and/or pol IV are responsible for the increased dC incorporation opposite F in wild-type induced cells. This is consistent with earlier published data that show pol IV preferentially incorporates dC opposite F in SOS-induced E.coli (9). In addition, crystallographic studies of a pol IV homolog (Dpo4) from Sulfolobus solfataricus reveal that dC insertion allows for favorable hydrogen bonding with a 3′-adjacent template dG (26). The fact that dA is still preferred opposite AP suggests that pol V bypasses 2-deoxyribose more often than F in wild-type SOS-induced cells.
Figure 5.
Nucleotide incorporation opposite F in uninduced cells. (A) 1a, (B) 2a.
Figure 6.
Nucleotide incorporation opposite AP in uninduced cells. (A) 1b, (B) 2b.
Figure 7.
Nucleotide incorporation opposite F (1a) in SOS-induced cells.
Figure 8.
Nucleotide incorporation opposite AP (1b) in SOS-induced cells.
CONCLUSIONS
A comprehensive comparison of F and AP with respect to lethality, mutagenicity and SOS-polymerase interactions revealed that F is a good model for AP in SOS-induced cells, but the lesions are biologically distinct in uninduced E.coli. When cells are not induced, F, but not AP, requires pol V for full-length bypass product. In addition, in uninduced wild-type cells pol II and/or pol IV bypass AP more often than F. Therefore, replication of plasmids containing AP results in more deletion product than bypass of F. In SOS-induced cells, F accurately models AP with the exception of nucleotide selectivity opposite the molecules in wild-type cells. Pol V appears to play a smaller role in bypassing F than AP, resulting in preferential dC incorporation opposite F, while dA is still most often inserted opposite AP. Both with and without induction, pol II and pol IV appear to act cooperatively to bypass F and AP. Our studies conclude that if F is to be used as a model for 2-deoxyribose in E.coli, then the cells should be SOS-induced to ensure the model is accurately reflecting the effects of AP.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online. ESI-MS of 1a and 2a. Phosporimage demonstrating reactivity of 1b and 2b with 0.1 M NaOH.
Acknowledgments
ACKNOWLEDGEMENT
We are grateful for support of this research from the National Institute of General Medical Sciences (GM-063028) to M.M.G. and ES-012259 to M.F.G.
REFERENCES
- 1.Loeb L.A., Preston,B.D., Snow,E.T. and Schaaper,R.M. (1986) Apurinic sites as common intermediates in mutagenesis. Basic Life Sci., 38, 341–347. [DOI] [PubMed] [Google Scholar]
- 2.Lindahl T. (1982) DNA repair enzymes. Ann. Rev. Biochem., 51, 61–87. [DOI] [PubMed] [Google Scholar]
- 3.Kunkel T.A., Shearman,C.W. and Loeb,L.A. (1981) Mutagenesis in vitro by depurination of PhiX174 DNA. Nature, 291, 349–351. [DOI] [PubMed] [Google Scholar]
- 4.Sagher D. and Strauss,B. (1983) Insertion of nucleotides opposite apurinic/apyrimidinic sites in deoxyribonucleic acid during in vitro synthesis: uniqueness of adenine nucleotides. Biochemistry, 22, 4518–4526. [DOI] [PubMed] [Google Scholar]
- 5.Hevroni D. and Livneh,Z. (1988) Bypass and termination at apurinic sites during replication of single-stranded DNA in vitro: a model for apurinic site mutagenesis. Proc. Natl Acad. Sci. USA, 85, 5046–5050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takeshita M., Chang,C.-N., Johnson,F., Will,S. and Grollman,A.P. (1987) Oligodeoxynucleotides containing synthetic abasic sites. Model substrates for DNA polymerases and apurinic/apyrimidinic endonucleases. J. Biol. Chem., 262, 10171–10179. [PubMed] [Google Scholar]
- 7.Otsuka C., Sanadai,S., Hata,Y., Okuto,H., Noskov,V.N., Loakes,D. and Negishi,K. (2002) Difference between deoxyribose- and tetrahydrofuran-type abasic sites in the in vivo mutagenic responses in yeast. Nucleic Acids Res., 30, 5129–5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lawrence C.W., Borden,A., Banerjee,S.K. and LeClerc,J.E. (1990) Mutation frequency and spectrum resulting from a single abasic site in a single-stranded vector. Nucleic Acids Res., 18, 2153–2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maor-Shoshani A., Hayashi,K., Ohmori,H. and Livneh,Z. (2003) Analysis of translesion replication across an abasic site by DNA polymerase IV of Escherichia coli. DNA Repair, 2, 1227–1238. [DOI] [PubMed] [Google Scholar]
- 10.Henderson P.T., Delaney,J.C., Gu,F., Tannenbaum,S.R. and Essigmann,J.M. (2002) Oxidation of 7,8-dihydro-8-oxoguanine affords lesions that are potent sources of replication errors in vivo. Biochemistry, 41, 914–921. [DOI] [PubMed] [Google Scholar]
- 11.Shimizu H., Yagi,R., Kimura,Y., Makino,K., Terato,H., Ohyama,Y. and Ide,H. (1997) Replication bypass and mutagenic effect of α-deoxyadenosine site-specifically incorporated into single-stranded vectors. Nucleic Acids Res., 25, 597–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Napolitano R., Janel-Bintz,R., Wagner,J. and Fuchs,R.P.P. (2000) All three SOS-inducible DNA polymerases (Pol II, Pol IV, and Pol V) are involved in induced mutagenesis. EMBO J., 19, 6259–6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang M., Pham,P., Shen,X., Taylor,J.-S., O'Donnell,M., Woodgate,R. and Goodman,M.F. (2000) Roles of E.coli polymerases IV and V in lesion-targeted and untargeted SOS mutagenesis. Nature, 404, 1014–1018. [DOI] [PubMed] [Google Scholar]
- 14.Paz-Elizur T., Takeshita,M., Goodman,M.F., O'Donnell,M. and Livneh,Z. (1996) Mechanism of translesion DNA synthesis by DNA polymerase II. J. Biol. Chem., 271, 24662–24669. [DOI] [PubMed] [Google Scholar]
- 15.Tessman I. and Kennedy,M.A. (1994) DNA polymerase II of Escherichia coli in the bypass of abasic sites in vivo. Genetics, 136, 439–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kroeger K.M., Jiang,Y.L., Kow,Y.-W., Goodman,M.F. and Greenberg,M.M. (2004) Mutagenic effects of 2-deoxyribonolactone in Escherichia coli. An abasic lesion that disobeys the A-rule. Biochemistry, 43, 6723–6733. [DOI] [PubMed] [Google Scholar]
- 17.Gibbs P.E.M. and Lawrence,C.W. (1995) Novel mutagenic properties of abasic sites in Saccharomyces cerevisiae. J. Mol. Biol., 251, 229–236. [DOI] [PubMed] [Google Scholar]
- 18.Avkin S., Adar,S., Blander,G. and Livneh,Z. (2002) Quantitative measurement of translesion replication in human cells: evidence for bypass of abasic sites by a replicative DNA polymerase. Proc. Natl Acad. Sci. USA, 99, 3764–3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shibutani S., Takeshita,M. and Grollman,A.P. (1997) Translesional synthesis on DNA templates containing a single abasic site. J. Biol. Chem., 272, 13916–13922. [DOI] [PubMed] [Google Scholar]
- 20.Maniatis T., Fritsch,E.F. and Sambrook,J. (1982) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 21.Yeiser B., Pepper,E.D., Goodman,M.F. and Finkel,S.E. (2002) SOS-induced DNA polymerases enhance long-term survival and evolutionary fitness. Proc. Natl Acad. Sci. USA, 99, 8737–8741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Delaney J.C. and Essigmann,J.M. (1999) Context-dependent mutagenesis by DNA lesions. Chem. Biol., 6, 743–753. [DOI] [PubMed] [Google Scholar]
- 23.Delaney J.C., Henderson,P.T., Helquist,S.A., Morales,J.C., Essigmann,J.M. and Kool,E.T. (2003) High-fidelity in vivo replication of DNA base shape mimics without Watson–Crick hydrogen bonds. Proc. Natl Acad. Sci. USA, 100, 4469–4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goodman M.F. (2000) Coping with replication ‘train wrecks’ in Escherichia coli using Pol V, Pol II and Rec A proteins. Trends Biochem. Sci., 25, 189–195. [DOI] [PubMed] [Google Scholar]
- 25.Haracska L., Unk,I., Johnson,R.E., Johansson,E., Burgers,P.M.J., Prakash,S. and Prakash,L. (2001) Roles of yeast DNA polymerses δ and ζ and of Rev1 in the bypass of abasic sites. Genes Dev., 15, 945–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ling H., Boudsocq,F., Woodgate,R. and Yang,W. (2004) Snapshots of replication through an abasic lesion structural basis for base substitutions and frameshifts. Mol. Cell, 13, 751–762. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.