Abstract
Mediator is a large, modular protein complex remotely conserved from yeast to man that conveys regulatory signals from DNA-binding transcription factors to RNA polymerase II. In Saccharomyces cerevisiae, Mediator is thought to be composed of 24 subunits organized in four sub-complexes, termed the head, middle, tail and Cdk8 (Srb8-11) modules. In this work, we have used screening and pair-wise two-hybrid approaches to investigate protein–protein contacts between budding yeast Mediator subunits. The derived interaction map includes the delineation of numerous interaction domains between Mediator subunits, frequently corresponding to segments that have been conserved in evolution, as well as novel connections between the Cdk8 (Srb8-11) and head modules, the head and middle modules, and the middle and tail modules. The two-hybrid analysis, together with co-immunoprecipitation studies and gel filtration experiments revealed that Med31 (Soh1) is associated with the yeast Mediator that therefore comprises 25 subunits. Finally, analysis of the protein interaction network within the Drosophila Mediator middle module indicated that the structural organization of the Mediator complex is conserved from yeast to metazoans. The resulting interaction map provides a framework for delineating Mediator structure–function and investigating how Mediator function is regulated.
INTRODUCTION
In eukaryotes, transcription of protein-coding genes requires, in addition to the RNA polymerase II (Pol II), the general transcription factors (GTFs) TFIIA, TFIIB, TFIID, TFIIE, TFIIF and TFIIH (1) for the proper assembly of the pre-initiation complex through the recognition of class-II gene promoters. Altogether the Pol II initiation machinery is constituted of some 45 different polypeptides with a combined mass of more than 2.2 MDa. However, this huge ensemble is unable to support activated transcription by DNA-binding transcriptional activators in vitro. This observation has led to the suggestion that co-activators are required to transmit the regulatory signals from the transcriptional activators to the Pol II initiation apparatus. In the yeast Saccharomyces cerevisiae, Kim et al. (2) identified and purified a 20 subunit complex, termed Mediator, that is required for Pol II response to gene-specific activators. Independently, Thompson et al. (3) identified several of these subunits plus four additional subunits (Srb8-11) in a genetic screen aimed at finding suppressors of a CTD-truncation mutant of the largest subunit of Pol II, Rpb1. Together, these 24 subunits form the Mediator complex. Some subunits of yeast Mediator (mainly the ones encoded by non-essential genes, such as Srb8-11) seem to have a more gene-specific function. However, a conditional mutation of the main component of the head-domain, Med17 (Srb4), has the same effect as the rpb1-1 allele that abolishes all Pol II transcription at 37°C (4), demonstrating that Mediator is required for transcription of virtually all protein-coding genes. Biochemical experiments with mammalian transcription systems led to the isolation of several Mediator-like complexes which were thought initially to have disparate subunit compositions (5–8). More thorough studies indicated that all subunits are present in at least one form of the Mediator in mammals (9). Recently, a systematic study looking both at primary sequences and secondary structure predictions of bona fide components of the purified Mediator-like complexes suggested that most S.cerevisiae Mediator subunits have been in fact conserved in other organisms making it a hallmark of the transcription machinery in the eukaryotic kingdom [(10); Table 1]. Still, the various forms of Mediator in multicellular organisms contain additional subunits compared to the yeast complex. Curiously one of these additional subunits, Med31 (Soh1), which is very well conserved from metazoans to yeast, has up to now never been detected in purified S.cerevisiae Mediator while it is present in the Drosophila and human complexes (11,12).
Table 1. Conservation of the Mediator subunits.
| Unified nomenclaturea | S.cerevisiaeb,c | D.melanogasterd | Homo sapiense |
|---|---|---|---|
| Cdk8 | Srb10/Ume5 | Cdk8 | CDK8 |
| CycC | Srb11/Ume3 | CycC | CycC |
| Med1 | Med1 | Trap220 | Med220 |
| Med2 | Med2 | ||
| Med3 | Med3/Pgd1/Hrs1 | ||
| Med4 | Med4 | Trap36 | Med36 |
| Med5 | Nut1 | ||
| Med6 | Med6 | Med6 | Med33 |
| Med7 | Med7 | Med7 | Med34 |
| Med8 | Med8 | Arc32 | Arc32 |
| Med9 | Med9/Cse2 | CG5134 | Med25 |
| Med10 | Med10/Nut2 | Nut2 | Med10 |
| Med11 | Med11 | Med21 | HSPC296 |
| Med12 | Srb8 | Kto | Med230 |
| Med13 | Srb9/Ssn2 | Skd | Med240 |
| Med14 | Rgr1 | Trap170 | Med150 |
| Med15 | Gal11 | Arc105 | Arc105 |
| Med16 | Sin4 | Trap95 | Med95 |
| Med17 | Srb4 | Trap80 | Med78 |
| Med18 | Srb5 | p28/CG14802 | p28b |
| Med19 | Rox3 | CG5546 | LCMR1 |
| Med20 | Srb2 | Trfp | hTRF |
| Med21 | Srb7 | Trap19 | Med17 |
| Med22 | Srb6 | Med24 | Surf5 |
aTaken from Bourbon et al. (50)
bComparisons were taken from Boube et al. (10) with the exceptions of Med9/Cse2 and its relatives which were taken from Tomori-sato et al. (49), and Med15 (Gal11) and its relatives which were taken from (51).
cFrom SGD.
dFrom Flybase.
eHuman Med acronyms are based on a unified nomenclature proposed by Rachez and Freedman (52).
Budding yeast Mediator is thought to be organized in four different sub-complexes. The Cdk8 (Srb8-11) module is composed of Cdk8 (Srb10), CycC (Srb11), Med12 (Srb8) and Med13 (Srb9). The Cdk8 module was found to have a protein kinase activity, to be relatively labile and to be absent from the Mediator complex when yeast cells enter diauxic shift (4,13,14). This module is mainly involved in transcription repression, notably through phosphorylation of the repeated C-terminal domain of Rpb1 Pol II subunit (4,13), but also through phosphorylation of Ste12, Gcn4 and Msn2 transcription activators (15,16). Conversely, phosphorylation of Gal4 by Cdk8 (Srb10) is required for proper transcription activation in response to galactose (17).
Electron microscopy and urea dissociation, as well as reconstitution experiments, identified three separate modules (constituting the head, middle and tail domains seen in electron microscopy analyses) in addition to the Cdk8 (Srb8-11) module (18–20). Various lines of evidence strongly suggest that the tail [or Med15 (Gal11)] module, constituted of Med2, Med3, Med14 (Rgr1), Med15 (Gal11) and Med16 (Sin4), is the main target for the transcriptional activators (21,22). According to structural studies using electron microscopy, the middle (or Med9–10) module, consisting of Med1, Med4, Med7, Med9, Med10, Med21 (Srb7), and possibly Med5 (Nut1), and the head module, consisting of Med6, Med8, Med11, Med17 (Srb4), Med18 (Srb5), Med19 (Rox3), Med20 (Srb2) and Med22 (Srb6), establish direct contacts with Pol II (23).
The protein–protein contacts within the head and middle modules have been explored using pull-down experiments from S.cerevisiae Mediator subunits expressed in baculovirus-infected insect cells and these approaches have led to a first protein interaction map (24–26). However, the structural organization of the tail and Cdk8 (Srb8-11) modules has not been investigated to date, neither have the interactions between subunits belonging to different modules.
In this work, we have obtained a detailed interaction map of yeast Mediator using two different two-hybrid approaches, and combined it with the one previously obtained by GST pull-down experiments (24–26) and proteome-wide two-hybrid screens (27,28). For the first time, we show connections between the different modules. In addition, the conserved Med31 (Soh1) protein was found to interact with middle module subunits and to be associated in vivo and in vitro with Mediator. Further, we have delineated several interaction domains between Mediator subunits. Significantly, these often correspond to segments that have been conserved during eukaryotic evolution. We have also found that some of the Drosophila melanogaster proteins, predicted to belong to an insect Mediator middle module equivalent, are engaged in interactions which are conserved. These data validate several interactions found in yeast Mediator and indicate that the metazoan subunits are structurally and functionally homologous to their yeast counterparts. Altogether, our results constitute a framework for future detailed Mediator structure–function analyses both in yeast and metazoan cells.
MATERIALS AND METHODS
D.melanogaster Mediator subunit cloning
The Drosophila Mediator subunit open-reading-frames (ORF) were isolated from cDNA clones made by the Berkeley Drosophila Genome Project (BDGP) and inserted into the Invitrogen Gateway entry vector pENTR1A or a home-made derivative, termed pGATEN. The latter was derived from pENTR1A by inserting a SalI–KpnI cloning adapter made of the two complementary oligonucleotides GATEN1 (TCGACTGGGCCTCCATGGCCCAATTGACTAGTAGCGGATCCGGAGGCCTCTACGTAGGTA) and GATEN2 (CTACGTAGAGGCCTCCGGATCCGCTACTAGTCAATTGGGCCATGGAGGCCCAG), between the unique SalI and KpnI sites. The entire Med1 (Trap220) ORF was inserted into the pGATEN vector as a 4.4 kb BamHI (partial)–BspHI (Klenow filled) fragment isolated from the SD26657 cDNA clone, between the BamHI and EcoRV sites. The entire Med4 (Trap36) ORF was inserted into the GATEN vector as a 884 bp BlpI (Mung bean nuclease treated)–XhoI fragment isolated from the LD46084 cDNA clone, between the StuI and XhoI sites. The entire Med7 ORF was inserted into the GATEN vector as a 899 bp SmaI–XhoI fragment generated from the bs44CO1 cDNA clone, between the StuI and XhoI sites. The entire Med21 (Trap19) ORF was inserted into the pGATEN derivative pGATENS7, as follows. First, a SfiI–MfeI cloning adapter, made of the two complementary oligonucleotides S7ADA1 (TGGCCATGGCGGATCGGCTTACAC) and S7ADA2 (AATTGTGTAAGCCGATCCGCCATGGCCATGG), was inserted into the pGATEN plasmid cut with SfII and MfeI. This resulted in the inclusion of the first six Med21 (Trap19) codons into the pGATENS7 vector. Second, the remaining Med21 (Trap19) ORF was inserted into the pGATENS7 vector as a 448 bp MfeI–DraI fragment generated from the GH07258 cDNA clone, between the MfeI and EcoRV sites. The entire Med10 (Nut2) ORF was inserted into the pGATEN vector as a 558 bp ApoI (partial)–XhoI fragment isolated from the SD24044 cDNA clone, between the MfeI and XhoI sites. Finally, the entire Med9 (CG5134) ORF was inserted into the pENTR1A vector as a 665 bp ApoI–HpaI fragment generated from the LD07740 cDNA clone, between the EcoRI and EcoRV sites. All constructs were sequence verified.
Plasmid constructions
pACT2 and pGBT9 (29) were modified to act as Invitrogen Gateway destination plasmids by introducing the RfB cassette at their unique SmaI site to obtain pACT2-Dest and pGBT9-Dest. The 24 yeast Mediator subunit genes were amplified using oligonucleotides matching the gene sequence just after the initiation codon for the 5′ forward primer and just before the stop codon for the 3′ reverse primer. The oligonucleotides were flanked with attB1 or attB2 sequences, respectively. The amplified sequence was cloned into pDONR201 (Invitrogen) using standard BP reaction. The recombinant plasmids were sequence verified. The 24 yeast and the 6 investigated Drosophila Mediator ORFs were then transferred into either pACT2-Dest or pGBT9-Dest by the LR reaction.
Protein expression
To verify the expression of the Gal4 DNA-binding domain (GBD) Mediator subunit fusion proteins, we constructed a pGBT9-Dest derivative that allowed the addition of the EGFP protein at the C-terminus of each GBD–Med fusion protein. The 24 Mediator genes were then transferred into pGBT9-Dest-EGFP by the LR reaction and transformed in yeast. The expression of all GBD–Med–EGFP proteins was confirmed by illuminating the yeast colonies grown on plates at 302 nm.
Yeast two-hybrid assays
The two-hybrid assays were performed in Y187 (MATα gal4 gal80 his3 trp1-901 ade2-101 ura3-52 leu2-3, 112 URA3::GAL1::lacZ LYS2::GAL4(UAS)::HIS3 cyhR), Y190 (MATa gal4 gal80 his3 trp1-901 ade2-101 ura3-52 leu2-3, 112 URA3::GAL1::lacZ LYS2::GAL4(UAS)::HIS3 cyhR) or their diploids (30).
The interaction of each pair of yeast Mediator subunits was tested by individually conjugating Y187 transformed with each of the pGBT9-Med plasmids with Y190 transformed with each of the pACT2-Med plasmids. Conjugations were performed at 30°C overnight in YPD in 96 well plates. Diploids were selected on SC–Leu–Trp plates. The β-galactosidase activity was revealed by an X-Gal overlay assay (31). The absence of growth of the haploids and the lack of wells cross-contamination were verified. The whole experiment was duplicated.
Each Mediator subunit was also screened against the FRYL2 library of yeast genomic DNA cloned in pACT2ΔΔ (32) as previously reported (33).
Immunoprecipitations
YPH500 strain derivatives were constructed such that Med17 (Srb4) was tagged with an epitope suitable for tandem affinity purification, TAP (34), or Med31 (Soh1) was tagged with a 3 HA epitope, or both proteins were tagged in the same strain. Tag coding sequences were introduced at the 3′ end of ORFs at the chromosome, using the standard one-step PCR protocol (35). The insertion of the tag was checked by PCR and western blotting with appropriate antibodies (12CA5 or PAP).
Immunopreciptations were performed as follows: 100 ml of cells growing exponentially in YPD medium were collected washed twice with water, then twice with extraction buffer [10 mM Tris–HCl pH 7.5, 150 mM NaCl, 20% glycerol, 0.1% Nonidet P-40, 1 mM DTT, protease inhibitor cocktail (Roche)] and resuspended in 1 ml of the same buffer. All subsequent steps were performed at 4°C. The cells were broken with glass beads, and the debris were eliminated by centrifugation. For Med17–TAP (Srb4–TAP) purification, IgG–Sepharose beads (Amersham Biosciences) were used, while Protein A Sepharose beads were used for Med31 (Soh1–HA) purification. An aliquot of 25 μl of Sepharose beads was washed with 1 ml of PBS 0.1% BSA. Protein A Sepharose beads were incubated 30 min at 30°C with 3 μg of 12CA5 antibodies in 100 μl of PBS 0.1% BSA and then washed with 1 ml PBS 0.1% BSA. An aliquot of 150 μl of protein extract was added on the beads and incubated under agitation for 2 h at 10°C. The beads were centrifuged, and the supernatant was collected. The beads were then washed with 40 volumes of extraction buffer and resuspended in 50 μl of the same buffer. Bound proteins were eluted from IgG–Sepharose by boiling. In experiments with Protein A Sepharose coupled to 12CA5, bound proteins were released by incubation in extraction buffer with the HA peptide (PB250; 0.5 mg per ml) for 30 min at 30°C under agitation.
Exclusion chromatography
The Med31–HA (Soh1–HA) Med17–TAP (Srb4–TAP) strain extract immunopurified as described above was run through a 2.3 ml Superose 6 column in the extraction buffer without glycerol, at a flow rate of 50 μl per min. Sixty microlitres of fractions were collected. Protein molecular weight standards were run on the same column (Catalase, Thyroglobulin and Albumin from Pharmacia). Fractions were analysed by western blotting with PAP and 12CA5 antibodies. The western blot was revealed using chemiluminescent ECL and signals were measured in Multimage light cabinet with fluorchem software (Alpha Innotech corporation).
RESULTS
Pair-wise two-hybrid analysis of Mediator subunits interactions
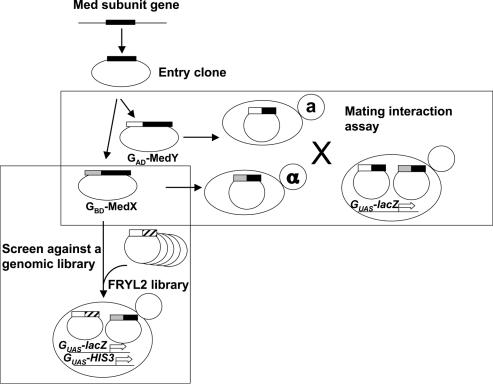
To test for interactions between the 24 yeast Mediator subunits, each subunit was cloned as a fusion protein to the Gal4 DNA-binding (GBD) or activation (GAD) domain under the control of a strong ADH1 promoter (see Figure 1 for the experimental scheme). The GBD or GAD fusion protein expressing vectors were transformed in the yeast Y187 or Y190 tester strains that bear the GAL1::lacZ and GALUAS::HIS3 reporter genes. The possible auto-activation of each Mediator protein fused to GBD was first tested. The Y187 strains expressing the GBD–Med2, –Med3, –Med4, –Med13 (Srb9) or –Med15 (Gal11) protein fusion showed strong expression of β-galactosidase and were thus eliminated from further analysis. The 19 remaining Y187 strains transformed with GBD fusion proteins were crossed individually to a Y190 strain containing each of the 24 GAD fusion proteins and the 456 resulting diploids were tested for β-galactosidase activity, revealing reporter gene activation. An example of the mating assay is shown in Figure 2. The interactions that were found using the conjugation procedure were confirmed by co-transformation of the two Mediator subunits expressing vectors in Y190 and testing for the expression of β-galactosidase. The 19 interactions that were confirmed were considered as positives (black and white circles in Figure 3 lower left part).
Figure 1.
Experimental scheme. Yeast Mediator subunit genes were individually cloned into a Gateway entry vector. Each Mediator ORF was then fused to the Gal4 DNA-binding (GBD–MedX) or activation domain (GAD–MedY). Each GBD–MedX fusion protein was tested for two-hybrid interactions in a mating assay against each GAD–MedY fusion protein. The GBD–MedX fusion proteins were also screened against the FRYL2 library of yeast genomic fragments fused to the GAD domain. GUAS-lacZ and GUAS-HIS3 represent the GAL1::lacZ and GAL4(UAS)::HIS3 reporter genes, respectively.
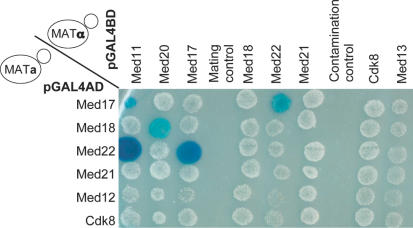
Figure 2.
Mating assay for Mediator subunit interactions. An example of the mating assay is shown where Y190 strains transformed by plasmids encoding fusions of the GBD with Med12 (Srb8), Med17 (Srb4), Med18 (Srb5), Med21 (Srb7), Med22 (Srb6) or Cdk8 (Srb10) [Med13 (Srb9) was omitted since it activated the transcription of the reporter gene] were crossed with Y187 derivatives transformed with plasmids encoding fusions of the GAD with Med11, Med12 (Srb8), Med13 (Srb9), Med17 (Srb4), Med18 (Srb5), Med20 (Srb2), Med21 (Srb7) or Med22 (Srb6). Patches of cells were overlaid with X-Gal agarose (31) to reveal β-galactosidase activity (blue colour). The interactions between Med11 and Med17 (Srb4), Med11 and Med22 (Srb6), Med18 (Srb5) and Med20 (Srb2), as well as between Med17 (Srb4) and Med22 (Srb6) can be observed. The mating control indicated that haploid parents did not grow. A contamination control was included to detect carry over from separate samples in the 96 well plates.
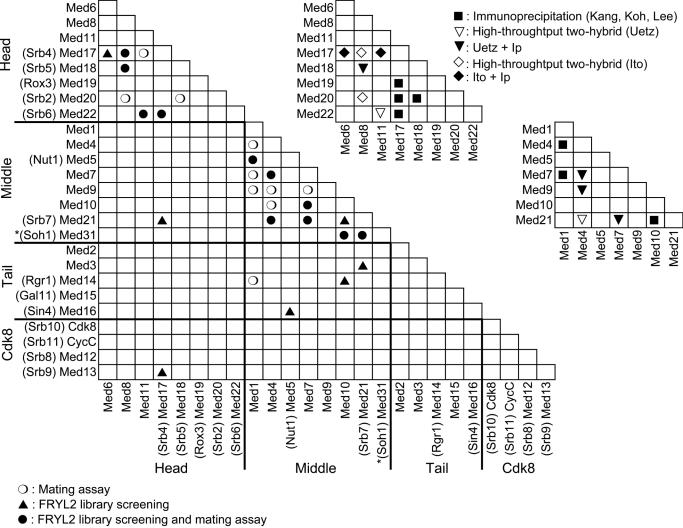
Figure 3.
Two-hybrid interactions between yeast Mediator subunits. The two-hybrid interactions between the yeast Mediator subunits are indicated on the left-hand side half-matrix. The interactions that were only found in two-hybrid screens with the FRYL2 library are indicated by an upward pointing closed triangle, those that were only observed in the mating assays are indicated by an open circle, and those that were found in both assays are indicated by a closed circle. The asterisk on Med31 (Soh1) indicates that it was only tested as a GBD fusion after it was found in the Med10 and Med21 (Srb7) screens, since Med31 (Soh1) was not taken into account initially because it was not previously considered as a yeast Mediator subunit. Results from previously published screens are indicated in the upper right half of the figure. Black squares represent GST pull-down results (24–26), downward pointing open triangles indicate two-hybrid interactions from Uetz et al. (27), open diamonds those from Ito et al. (28), black triangles interactions observed both by GST pull-down and Uetz et al. (27), and black diamonds interactions observed by GST pull-down and Ito et al. (28).
Most of the detectable interactions concerned subunits that belong to the same module. For instance, 11 interactions connected components of the middle module of Mediator and 7 contacts linked head module subunits. Surprisingly, no interactions were detected within the tail module. However, it is noteworthy that 3 out of the 5 auto-activating Mediator subunits belong to the tail module, which might explain at least in part the observed bias. Interestingly, Med1 was found to interact with Med14 (Rgr1) connecting the middle module to the Mediator tail.
Screening the Mediator subunits with a genomic library
As a first step toward structure–function analyses, we sought to further identify interaction domains between the yeast Mediator proteins. Moreover, our previous experience with subunits of RNA polymerases revealed that some interactions cannot be uncovered using complete proteins (33). To address these issues, we screened the Mediator subunits (except the auto-activating ones) as previously described (33) with a library (FRYL2) that fused random S.cerevisiae genomic DNA fragments to the GAD-encoding sequence [(32), Figure 1]. Supplementary Table S1 lists the fragments of the Mediator subunits selected in the library screens. Altogether, 17 interactions between the 24 known Mediator subunits were found in the screens with the FRYL2 library, 10 of which were previously found in the mating assays (black triangles and dots in Figure 3 lower left part). Thus, 7 additional protein–protein contacts were identified in the screens, 5 of which concerned interactions between modules. These latter interactions are represented by a contact between Med17 (Srb4) and Med21 (Srb7), connecting the head and middle modules, and interactions between Med5 (Nut1) and Med16 (Sin4), Med3 and Med21 (Srb7), and Med10 and Med14 (Rgr1), linking the middle and tail modules. In addition, we observed a protein–protein interaction between Med13 (Srb9) and Med17 (Srb4) connecting the CDK8 and head modules. Importantly, fragments of Med31 (Soh1) were selected in our Med10 and Med21 (Srb7) screens suggesting that Med31 (Soh1) belongs to yeast Mediator.
Med31 (Soh1) is associated with yeast Mediator in vivo and in vitro
Med31 (Soh1) was initially isolated in yeast as a multicopy suppressor of the hyper-recombination phenotype of hpr1 mutants which are also altered in transcription elongation (36,37). Med31 (Soh1) homologues have been consistently found in Mediator-like complexes purified from multicellular eukaryotes but never in yeast Mediator (11,12). We thus decided to investigate if Med31 (Soh1) was a novel S.cerevisiae Mediator subunit.
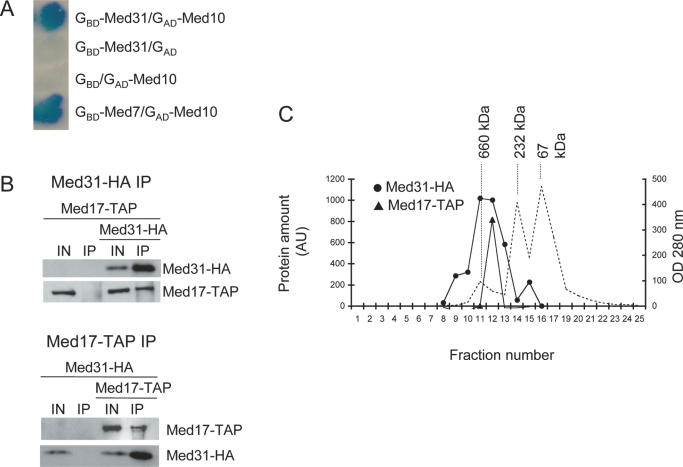
The complete Med31 (Soh1) subunit was tested as a fusion with the GBD against all 24 Mediator subunits and itself fused to the GAD. Only GAD–Med10 and GAD–Med21 (Srb7) interacted, suggesting that Med31 (Soh1) is a middle module subunit (Figures 3 and 4A). To confirm that Med31 (Soh1) was part of Mediator, we co-immunoprecipitated (CoIP) Med17 (Srb4), a well-established Mediator subunit, and Med31 (Soh1), since the two proteins do not seem to contact each other, and belong to the head and middle module, respectively. For that purpose, Med17 (Srb4) was tagged with TAP epitope and Med31 (Soh1) with HA. As can be seen in Figure 4B, the immunoprecipitation (IP) of Med31–HA (Soh1–HA) from yeast extracts resulted in the co-immunoprecipitation of Med17–TAP (Srb4–TAP). Conversely, the IP of Med17–TAP (Srb4–TAP) allowed the co-immunoprecipitation of Med31–HA suggesting that Med31 (Soh1) was part of the Mediator complex. To confirm that Med31 (Soh1) is indeed associated with Mediator, the material immunoprecipitated with anti-HA antibodies, as it was prepared in Figure 4B, top panel, was chromatographed on a Superose 6 sizing column (Figure 4C). Med17–TAP (Srb4–TAP) and Med31–HA (Soh1–HA) co-eluted in a single high molecular weight peak, indicating that the proteins belonged to a single stable complex in vitro. Altogether, our observations and the strong conservation of Med31 (Soh1) in multicellular eukaryotes (12) indicate that Med31 (Soh1) is a bona fide Mediator subunit interacting with Med10 and Med21 (Srb7).
Figure 4.
Med31 (Soh1) is associated with yeast Mediator. (A) Med31 (Soh1) two-hybrid interaction with Med10. Patches of cells were treated as in Figure 2. GAD and GBD indicate empty vectors used as negative controls. GBD–Med7 was used as a positive control for Med10 interaction. (B) Med31 (Soh1) and Med17 (Srb4) co-immunoprecipitate. After immunoprecipitation, the proteins were revealed by western blotting using 12CA5 or PAP antibodies, binding to the HA or TAP tag, respectively. Top panel: anti-HA IP; bottom panel: PAP IP. Negative controls were strains expressing Med17–TAP (Srb4–TAP) or Med31–HA (Soh1–HA) only. (C) Gel filtration analysis of extracts from strains expressing Med31 (Soh1) and Med17 (Srb4) after anti-HA immunoprecipitation. The immunoprecipitation extract was run on a Superose 6 gel filtration column calibrated with molecular weight markers. The presence of Med31–HA (Soh1–HA) or Med17–TAP (Srb4–TAP) was revealed by western blotting as in (B). The amount of protein was measured by densitometric analyses of films and is expressed in arbitrary units on the left scale. Measuring UV absorbance during chromatography (right scale) assessed the position of the markers. The dashed line indicates the UV trace. The molecular weight of the markers is indicated above their respective peak abundance.
Comparison with other data
Proteome-wide two-hybrid screens performed by Ito et al. (28) and Uetz et al. (27) identified 10 protein–protein contacts between the 24 previously known yeast Mediator subunits that are confined to the head or middle modules (Figure 3, upper part right). All these contacts were also detected in our screens. In addition, we found 16 new interactions.
Med10 and Med21 (Srb7) clones were selected repeatedly in the Med31 (Soh1) screen performed by Ito et al. (28). Nevertheless, it was difficult to assess whether or not these contacts were specific since in these screens Med31 (Soh1) also interacted with Med4, Med6, Med7, Med8, Med9 and Med17 (Srb4) and 61 other potential partners. Given the high likelihood of representing false-positives, these interactions are thus not represented in Figure 3.
Interactions between some of the yeast Mediator subunits have also been previously explored using co-expression of full-size proteins in a baculovirus system followed by a GST pull-down assay (24–26). These investigations have been limited to subunit–subunit contacts within the head or within the middle module (black or white triangles in the upper right part of Figure 3). Five out of seven contacts in the head module are identical to those defined here, since we have missed the interaction between Med17 (Srb4) and Med20 (Srb2) and that between Med17 (Srb4) and Med19 (Rox3). In the middle module, the six interactions found previously constitute a subset of the 12 contacts observed here.
Analysis of interaction domains
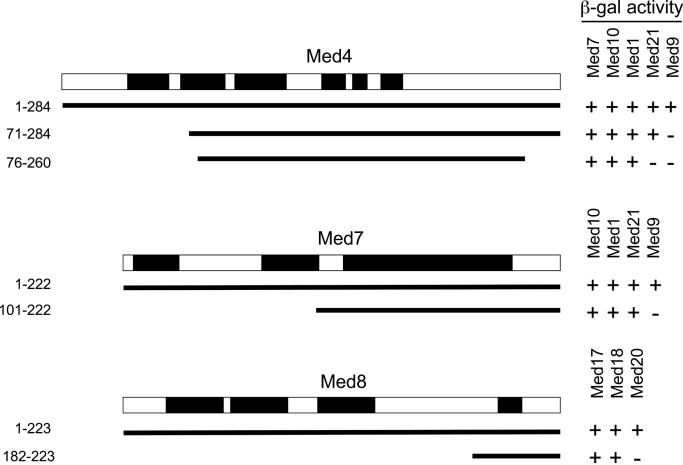
A usual concern with the use of the two-hybrid system for predicting interactions between yeast proteins is that a third endogenous partner might act as a bridge between the GAD and GBD fusion proteins and lead to the activation of the reporter gene. For example, Med4 interacted with Med1, Med7, Med9, Med10 and Med21 (Srb7), i.e. all the other middle module subunits except Med5 (Nut1). Moreover, several of the Med4 partners interacted with each other, raising the formal possibility that some of the two-hybrid interactions with Med4 might be due to a bridging partner. To address this issue, we have taken advantage of the selection of several prey fragments to devise a method providing strong evidence for direct interactions in the two-hybrid system. As indicated in Figure 5, an N-terminal truncation of Med4 removing the first 70 amino acids did not interact anymore with Med9 indicating that they were essential (but not necessarily sufficient) for the interaction between the two subunits. Since the interactions with Med1, Med7, Med10 and Med21 (Srb7) were still maintained, the interaction seen between Med4 and Med9 was not mediated by the other partners of Med4, thus is likely to be direct. Similarly, further truncation of six amino acids at the N-terminus of Med4 and of 16 amino acids at its C-terminus resulted in the loss of Med21 (Srb7) interaction but not of Med1, Med7 and Med10 interaction. This observation supports a direct interaction between Med4 and Med21 (Srb7). Using a similar reasoning, we could demonstrate that the Med7–Med9 and Med8–Med20 (Srb2) protein–protein contacts were direct.
Figure 5.
Analysis of direct interactions of yeast Med4, Med7 and Med8 with their partners. Med4 protein fragments used to analyse the interactions of Med4, Med7 and Med8 with their partners are represented. + indicates GAL1::lacZ activation; − indicates absence of activation. The Mediator subunit conserved domains defined according to Boube et al. (10) are indicated by black boxes. The conservation of the C-terminal domain of Med8 was not reported previously. An alignment is shown in supplementary Figure S1.
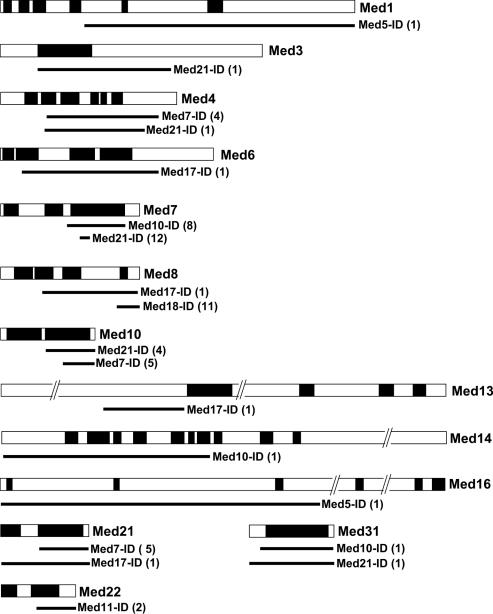
Since various fragments of the Mediator subunits were usually selected in the library screens (Table S1), we could delineate, sometimes quite precisely, the interaction domains (IDs; Figure 6). We looked at the position of the IDs relative to the conserved domains of Mediator subunits, as previously defined by Boube et al. (10). As shown in Figure 6, most of the IDs mapped to conserved domains suggesting that they are evolutionarily constrained due to their functional importance in maintaining the structural integrity of Mediator.
Figure 6.
Interaction domains map to conserved regions of the Mediator subunits. The conserved domains of the Mediator proteins for which interaction domains could be mapped are represented by black boxes. The solid lines indicate the extent of the interaction domains (10) (Figure S1). The number of independent clones selected in the screens is indicated between parenthesis.
Two-hybrid protein interaction map of the putative Drosophila Mediator middle module equivalent
A recent bioinformatics analysis of several eukaryotic genomes has suggested that nearly all the budding yeast Mediator subunits might be distantly conserved in metazoans and plants (10). Primary sequence conservation is often restricted to relatively short segments of 20–30 amino acids long. Moreover, even if several homology blocks can be defined, almost no amino acids are strictly conserved in the analysed eukaryotic sequences. If the function of the predicted homologous Mediator subunits has been conserved during evolution, then one would expect a selective pressure to maintain the structural determinants involved in direct contacts between subunits. In addition, a two-hybrid interaction found between metazoan Mediator subunit homologues is likely to be direct since the divergence of primary sequence should prevent yeast subunits to act as a bridge in vivo.
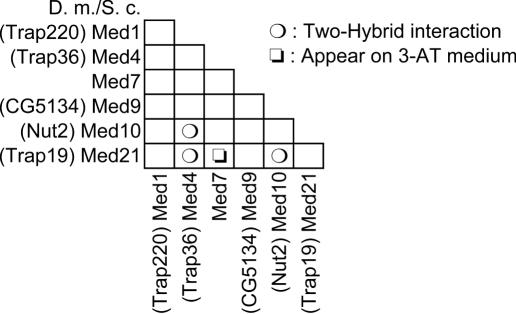
As our data provided us with a detailed protein–protein interaction map for the yeast Mediator middle module, we have next investigated the connections between the Drosophila Mediator subunit homologues predicted to belong to a metazoan middle module counterpart. For this purpose, we cloned in two-hybrid vectors the D.melanogaster ORFs coding for the predicted Med1, Med4, Med7, Med9, Med10 and Med21 (Srb7) fly homologues and tested for their interactions, as previously done for the yeast proteins. The Drosophila Med7 and Med21 (Srb7) counterparts, which are relatively well conserved during evolution, readily interacted as observed for the yeast proteins. Similarly, Drosophila Med4 and Med10, which are remotely related to their yeast cousins, also interacted with each other and with Med21 (Srb7) as the yeast Mediator subunits did (Figure 7). Concerning the Drosophila Med1 or Med9 homologues, no interaction could be detected with any of the tested proteins. Taken together, these observations strongly suggest that the structure of the middle module has been conserved during evolution and that the interactions previously seen between the yeast Med4, Med10 and Med21 (Srb7) proteins are direct.
Figure 7.
Interactions between the D.melanogaster Mediator middle module subunits The two-hybrid interactions between the predicted D.melanogaster Mediator middle module subunits are indicated in the half-matrix. CG5134 was predicted by Tomomori-Sato et al. (49) to be homologous to Med9. The other homology predictions are those of Boube et al. (10). The activation of the GAL1::lacZ reporter gene to reveal the interaction between Med7 and Med21 (Srb7) required cultivation of the transformed strain on 3-AT medium.
DISCUSSION
Using two alternative two-hybrid approaches, we have obtained a detailed protein–protein interaction network for 20 out of the 25 subunits of the yeast Mediator complex (Figure 8). This map encompasses and extends the one that has been established previously using a pull-down approach (24–26). We have found novel interactions that link the three modules of the core Mediator complex. In several cases, we have delineated interacting domains between subunits and provide evidence that the interactions are direct. In addition, two-hybrid interactions, co-immunoprecipitation and gel filtration experiments indicated that Med31 (Soh1) is associated with the Mediator complex in yeast. Investigating the putative Drosophila Mediator middle module counterpart, we showed here that several predicted metazoan homologues of yeast subunits are engaged in conserved interactions suggesting that, even though the primary sequences of Mediator subunits have extensively diverged, the overall structure of eukaryotic Mediator has been conserved during evolution.
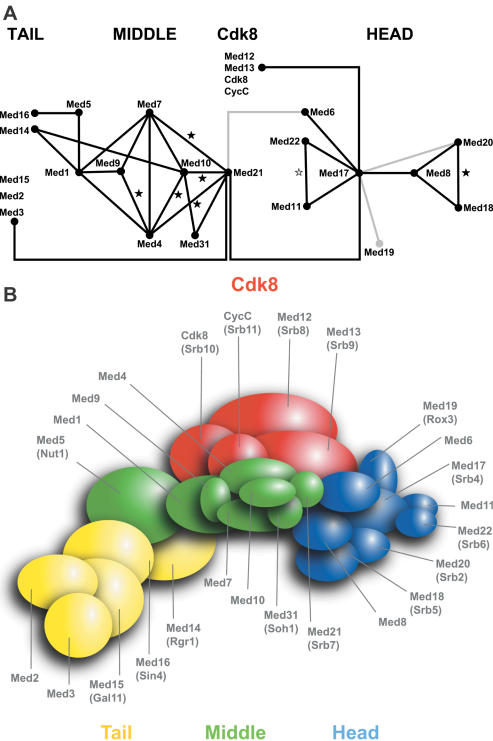
Figure 8.
Integrated interaction map of the yeast Mediator. (A) Connection map of Mediator subunits. The direct links between the different Mediator subunits found in this work, by Kang et al. (24–26), Ito et al. (28) and Uetz et al. (27) are combined to produce the integrated Mediator map. The interactions between Med17 (Srb4) and Med19 (Rox3) or Med20 (Srb2) (24) and between Med6 and Med21 (38) that were not found or confirmed in our screens or mating assays are indicated by grey lines. Black stars indicate the interactions that were found with the Mediator middle module homologous subunits from Drosophila. The grey star indicates a conserved contact between the worm Med22 (MDT-22/ZK970.3) and Med11 (MDT-11/R144.9) homologues (44). (B) Topological organization of yeast Mediator. This model was made taking into account all data mentioned in (A) and the relative size of the Mediator subunits.
In this work, we draw an interaction map of budding yeast Mediator, connecting most of its subunits (Figure 8). Our results are in agreement with a general topology of Mediator composed of a head, middle and tail module (18–20). New interactions were found to connect these head, middle and tail modules. Med14 (Rgr1) was previously shown to connect the tail and middle modules (19). In this study, we found that Med14 (Rgr1) interacts with Med1 and Med10, thereby identifying its middle module partners. Our mapping data also revealed that Med5 (Nut1) interacts with both Med16 (Sin4) and Med1, thereby determining the position of Med5 (Nut1) within the Mediator complex. Moreover, we showed an interaction between Med21 (Srb7) and Med3 connecting the middle and tail module. Similarly, we identified a link between the middle and head modules through the Med21 (Srb7)–Med17 (Srb4) interaction. In addition to the contacts identified between the Cdk8 and middle module (24), we found that the Cdk8 module is connected with the head module via an interaction between Med13 (Srb9) and Med17 (Srb4).
Since it has been reported that interactions with transcriptional regulators or the CTD of Rpb1 can change the conformation of the Mediator complex and alter the interaction between mediator subunits (38–40), it should be noted that the interactions reported in this study may not occur all at the same time, but may be dependent on the transcriptional state of the Mediator complex. Such conformational changes have been proposed to play a pivotal role in regulating Mediator function (39,40). In this light, it is interesting that most of the interactions we find between the individual submodules were only found by screening the genomic library, which was designed to contain fragments, rather than by directly testing the interactions between full-length proteins. Such submodule interactions are of interest for understanding the two different global conformations of Mediator and the observation that most of these are not picked up when examining full-length proteins indicates the possibility of a regulatory mechanism that involves masking certain domains. Whatever the mechanism, it is clear that the submodule interactions which are demonstrated here are of highest interest for further investigating how Mediator function is regulated through the conformational switch.
Med31 (Soh1) is a very well-conserved protein that co-purifies with metazoan Mediator (11,12). In yeast, Med31 (Soh1) was genetically associated with transcription but was not found in highly purified Mediator complex (41). Here, Med31 (Soh1) fragments were selected in two-hybrid screens with Med10 and Med21 (Srb7), in keeping with Ito et al. data (28) who used Med31 (Soh1) as bait and selected Med10 and Med21 as major interactants among others. To buttress the conclusion that Med31 (Soh1) is a bona fide yeast Mediator subunit, we showed that Med31 (Soh1) co-immunopurified with Med17 (Srb4) and vice versa. Additionally, Med31 (Soh1) and Med17 (Srb4) co-eluted in a single high molecular weight fraction on a gel filtration column. Several possibilities could explain why Med31 (Soh1) was not previously identified as a yeast Mediator subunit. Med31 (Soh1) might be a labile subunit that is lost during purification. Alternatively, Med31 (Soh1) may have escaped mass spectrometric identification since tryptic digestion of the protein is expected to produce peptides under 500 Da or over 2000 Da, except for one, outside the usual window of detection of the technique.
It was previously proposed that the majority of the yeast Mediator proteins is conserved in eukaryotic organisms (10). Significantly, some of the homologue assignations have been confirmed by the purification of Schizosaccharomyces pombe Mediator (42). Similarly, a large human Mediator-like complex has been reported to contain homologues of yeast Med11, Med18 (Srb5), Med19 (Rox3) and Med22 (Srb6) (43). Nevertheless, since the primary sequences of most Mediator subunits have widely diverged across the eukaryotic kingdom, their functional conservation could be questioned. To address these issues, we reasoned that if the yeast and Drosophila Mediator subunits interacted similarly in a two-hybrid assay, one could argue (i) that the homologue assignments were correctly predicted, (ii) that the interactions are likely direct since no bridging yeast proteins might interact in a stable manner with so-widely divergent proteins and (iii) that the structural organization of the subunits is conserved. The conservation of several interactions between the Drosophila and yeast middle module subunits thus strongly suggested that at least part of the Mediator middle module is structurally conserved across evolution. In addition, the detected interaction between the Med4 (TRAP36) and Med10 (Nut2) Drosophila proteins confirmed a direct contact between the yeast Med4 and Med10 proteins. Along the same line, in Caenorhabditis elegans, a proteome-wide two-hybrid analysis indicated a conserved contact between the worm Med22 (MDT-22/ZK970.3) and Med11 (MDT-11/R144.9) homologues (44).
We were unable to find interactions with Drosophila Med9 (CG5134), possibly due to the insertion of a hydrophobic linker between the Gal4 domains and the subunit. Nevertheless, a recent Drosophila proteome-wide screen detected a contact between the predicted fly Med9 and Med4 (45) strongly suggesting that their respective assignation was correct. Our inability to detect a protein–protein interaction of the fly Med1 homologue (i.e. Trap220) probably does not stem from a false assignation. Indeed, the five conserved segments found in Med1 (10) have been systematically found in the same order within putative ORFs from a larger spectrum of eukaryotic species (Henri-Marc Bourbon, unpublished observations). In addition, the formal possibility that the tested Drosophila Med1 is non-functional could be ruled out since the full-length cDNA used in the two-hybrid assays could fully rescue Trap220 loss-of-function mutants (Muriel Boube, unpublished data). At this stage, we rather favour the simple hypothesis that the Gal4–Med1 protein fusions were not appropriately folded, modified and/or expressed in yeast for interaction with its partners.
Only 9 out of 28 interactions were obtained both by the mating assay and by the library screening, indicating that each two-hybrid approach has its own bias. A number (10) of subunit interactions were found in the mating assays but not in the library screens. In some cases, the FRYL2 library may not have been exhaustively screened because of low transformation efficiencies due to the presence of a toxic bait or because the relevant interaction was weak in comparison with that of other preys. Another possible caveat is that clones encompassing the interaction domains may be absent from the tested library. Conversely, some of the interactions observed by screening were not found in the corresponding mating assay. It is possible that a Mediator subunit inhibitory domain, be it for protein folding, stability, or contact, has to be removed to observe the interaction since part of the protein is lacking in the selected fragments when a discrepancy between mating and screening occurs.
Five Mediator subunits out of 25 did not interact with any other partner. Two of these, subunits, Med2 and Med15 (Gal11), behaved as activators in the two-hybrid system and hence could only be tested as GAD fusions against the other subunits fused to the GBD and were not screened with the library, decreasing the probability that we could detect a protein contact. Interestingly, the three remaining subunits, Cdk8, CycC and Med12 (Srb8), all belong to the Cdk8 module which has an inhibitory role, raising the possibility that their binding to the reporter gene promoter through the GAD may have prevented its activation and thus the detection of an interaction. The lack of interaction of three of the four Cdk8 module subunits might also stem from poor representation of interacting fragments in the FRYL2 library, even though it has already been carefully prepared and successfully used in many other studies in addition to this one (32,33,46). We found previously that fusions of entire high molecular weight proteins, like the two large subunits of Pol I, II or III, may be unable to select interacting protein fragments in screens or to interact when tested directly with their partners (33) (Michel Werner, unpublished data). This observation might explain why we were unable to identify partners for Med15 (Gal11; 120 kDa) and Med12 (Srb8; 168 kDa). Nevertheless, we could find partners for 80% of Mediator subunits, a success rate that compares favourably with other approaches.
For the GST pull-down experiments, each possible pair within the head or middle module has been co-expressed in a baculovirus system (24–26). Checking the interactions that were found by these biochemical techniques against two-hybrid approaches could clearly (i) confirm interactions, (ii) reveal potential biases (iii) lead to the discovery of new interesting interactions, and (iv) delineate interacting domains. Strikingly, 11 out of the 20 two-hybrid interactions concerning intra-head or intra-middle module proteins were supported by co-immunoprecipitation data. Only the co-immunoprecipitations of Med17 (Srb4) with Med19 (Rox3) or Med20 (Srb2) were not confirmed by our two-hybrid assays.
When using the two-hybrid system to look for yeast protein interactions, there is the formal possibility that a third partner might bridge the two proteins under investigation leading to activation of the reporter gene. Thus, a two-hybrid interaction cannot be taken as an evidence of a direct contact. In our protein–protein interaction map, 11 of the 20 intra-module connections confirmed the Kang et al. (24) co-immunoprecipitation data and are thus direct. To investigate the nature of the contacts between the protein pairs that have not been found using the GST pull-down assay, we devised a method taking advantage of the availability of families of truncated fragments. These were tested against their Mediator subunit partners, based on the reasoning that if a single protein–protein contact was lost at a time, we could infer that it was direct. Using this test, we could show that the Med4–Med9, Med4–Med21 (Srb7), Med7–Med9 and Med8–Med20 (Srb2) contacts were direct. In retrospect, it is interesting to note that all the interactions between RNA polymerase subunits, predicted using the two-hybrid system (33), were indeed demonstrated to be direct in the crystallographic structure of Pol II (47,48). It is thus likely that most of the Mediator contacts reported here behave in the same way.
A two-hybrid screen with a library of genomic fragments has a number of advantages even though it is not exhaustive. This approach allows the identification of short interaction domains that are able to fold independently. It is notorious that the Mediator subunits are difficult to produce at high levels, hampering their crystallographic analyses. The definition of the interaction domains might help in that respect. In addition, coupled with phylogenetic analyses, these domains may also be used as targets for directed mutagenesis experiments to decipher the functional significance of intra- or extra-module contacts, both in yeast and metazoans.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank M. Fromont-Racine for providing us with the FRYL2 library, B. Termenon and D. Tourbiez for technical help, A. Sentenac and P. Thuriaux for careful reading of the manuscript. H.-M.B acknowledges David Cribbs for his constant support. B.G. was supported by grants from the Ministère de la Recherche et de la Technologie and from the Association pour la Recherche sur le Cancer (ARC). This work was supported from a grant (to H.-M.B.) from the ARC. N.B., F.H. and T.B. are supported by grants from the Netherlands Organization for Scientific Research (NWO).
NOTE ADDED IN PROOF
T. Linder and C. M. Gustafsson arrived independently at the conclusion that Med31 (Son1) is a subunit of S.cerevisiae and Schizosaccharomyces pombe Mediator (Linder,T. and Gustafsson,C.M. The Soh1/MED31 protein is an ancient component of Schizosaccharomyces pombe and Saccharomyces cerevisiae mediator. J. Biol. Chem., in press).
REFERENCES
- 1.Hampsey M. (1998) Molecular genetics of the RNA polymerase II general transcriptional machinery. Microbiol. Mol. Biol. Rev., 62, 465–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim Y.-J., Björklund,S., Li,Y., Sayre,M.H. and Kornberg,R.D. (1994) A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell, 77, 599–608. [DOI] [PubMed] [Google Scholar]
- 3.Thompson C.M., Koleske,A.J., Chao,D.M. and Young,R.A. (1993) A multisubunit complex associated with the RNA polymerase II CTD and TATA-binding protein in yeast. Cell, 73, 1361–1375. [DOI] [PubMed] [Google Scholar]
- 4.Holstege F.C.P., Jennings,E.G., Wyrick,J.J., Lee,T.I., Hengartner,C.J., Green,M.R., Golub,T.R., Lander,E.S. and Young,R.A. (1998) Dissecting the regulatory circuitry of a eukaryotic genome. Cell, 95, 717–728. [DOI] [PubMed] [Google Scholar]
- 5.Ito M., Yuan,C.X., Malik,S., Gu,W., Fondell,J.D., Yamamura,S., Fu,Z.Y., Zhang,X., Qin,J. and Roeder,R.G. (1999) Identity between TRAP and SMCC complexes indicates novel pathways for the function of nuclear receptors and diverse mammalian activators. Mol. Cell, 3, 361–370. [DOI] [PubMed] [Google Scholar]
- 6.Näär A.M., Beaurang,P.A., Zhou,S., Abraham,S., Solomon,W. and Tjian,R. (1999) Composite co-activator ARC mediates chromatin-directed transcriptional activation. Nature, 398, 828–832. [DOI] [PubMed] [Google Scholar]
- 7.Ryu S., Zhou,S., Ladurner,A.G. and Tjian,R. (1999) The transcriptional cofactor complex CRSP is required for activity of the enhancer-binding protein Sp1. Nature, 397, 446–450. [DOI] [PubMed] [Google Scholar]
- 8.Rachez C., Lemon,B.D., Suldan,Z., Bromleigh,V., Gamble,M., Näär,A.M., Erdjument-Bromage,H., Tempst,P. and Freedman,L.P. (1999) Ligand-dependent transcription activation by nuclear receptors requires the DRIP complex. Nature, 398, 824–828. [DOI] [PubMed] [Google Scholar]
- 9.Sato S., Tomomori-Sato,C., Parmely,T.J., Florens,L., Zybailov,B., Swanson,S.K., Banks,C.A., Jin,J., Cai,Y., Washburn,M.P. et al. (2004) A set of consensus Mammalian mediator subunits identified by multidimensional protein identification technology. Mol. Cell, 14, 685–691. [DOI] [PubMed] [Google Scholar]
- 10.Boube M., Joulia,L., Cribbs,D.L. and Bourbon,H.M. (2002) Evidence for a mediator of RNA polymerase II transcriptional regulation conserved from yeast to man. Cell, 110, 143–151. [DOI] [PubMed] [Google Scholar]
- 11.Park J.M., Gim,B.S., Kim,J.M., Yoon,J.H., Kim,H.S., Kang,J.G. and Kim,Y.J. (2001) Drosophila Mediator complex is broadly utilized by diverse gene-specific transcription factors at different types of core promoters. Mol. Cell. Biol., 21, 2312–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gu W., Malik,S., Ito,M., Yuan,C.X., Fondell,J.D., Zhang,X., Martinez,E., Qin,J. and Roeder,R.G. (1999) A novel human SRB/MED-containing cofactor complex, SMCC, involved in transcription regulation. Mol. Cell, 3, 97–108. [DOI] [PubMed] [Google Scholar]
- 13.Liao S.-M., Zhang,J., Jeffrey,D.A., Koleske,A.J., Thompson,C.M., Chao,D.M., Viljoen,M., van Vuuren,H.J.J. and Young,R.A. (1995) A kinase-cyclin pair in the RNA polymerase II holoenzyme. Nature, 374, 193–196. [DOI] [PubMed] [Google Scholar]
- 14.Hengartner C.J., Myer,V.E., Liao,S.-M., Wilson,C.J., Koh,S.S. and Young,R.A. (1998) Temporal regulation of RNA polymerase II by Srb10 and Kin28 cyclin-dependent kinases. Mol. Cell, 2, 43–53. [DOI] [PubMed] [Google Scholar]
- 15.Nelson C., Goto,S., Lund,K., Hung,W. and Sadowski,I. (2003) Srb10/Cdk8 regulates yeast filamentous growth by phosphorylating the transcription factor Ste12. Nature, 421, 187–190. [DOI] [PubMed] [Google Scholar]
- 16.Chi Y., Huddleston,M.J., Zhang,X., Young,R.A., Annan,R.S., Carr,S.A. and Deshaies,R.J. (2001) Negative regulation of Gcn4 and Msn2 transcription factors by Srb10 cyclin-dependent kinase. Genes Dev., 15, 1078–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirst M., Kobor,M.S., Kuriakose,N., Greenblatt,J. and Sadowski,I. (1999) GAL4 is regulated by the RNA polymerase II holoenzyme-associated cyclin-dependent protein kinase SRB10/CDK8. Mol. Cell, 3, 673–678. [DOI] [PubMed] [Google Scholar]
- 18.Dotson M.R., Yuan,C.X., Roeder,R.G., Myers,L.C., Gustafsson,C.M., Jiang,Y.W., Li,Y., Kornberg,R.D. and Asturias,F.J. (2000) Structural organization of yeast and mammalian mediator complexes. Proc. Natl Acad. Sci. USA, 97, 14307–14310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Y., Bjorklund,S., Jiang,Y.W., Kim,Y.-J., Lane,W.S., Stillman,D.J. and Kornberg,R.D. (1995) Yeast global transcriptional regulators Sin4 and Rgr1 are components of mediator complex/RNA polymerase II holoenzyme. Proc. Natl Acad. Sci. USA, 92, 10864–10686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee Y.C. and Kim,Y.J. (1998) Requirement for a functional interaction between mediator components Med6 and Srb4 in RNA polymerase II transcription. Mol. Cell. Biol., 18, 5364–5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han S.J., Lee,Y.C., Gim,B.S., Ryu,G.H., Park,S.J., Lane,W.S. and Kim,Y.J. (1999) Activator-specific requirement of yeast mediator proteins for RNA polymerase II transcriptional activation. Mol. Cell. Biol., 19, 979–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park J.M., Kim,H.S., Han,S.J., Hwang,M.S., Lee,Y.C. and Kim,Y.J. (2000) In vivo requirement of activator-specific binding targets of mediator. Mol. Cell. Biol., 20, 8709–8719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davis J., Takagi,Y., Kornberg,R. and Asturias,F. (2002) Structure of the yeast RNA polymerase II holoenzyme. Mediator conformation and polymerase interaction. Mol. Cell, 10, 409. [DOI] [PubMed] [Google Scholar]
- 24.Kang J.S., Kim,S.H., Hwang,M.S., Han,S.J., Lee,Y.C. and Kim,Y.J. (2001) The structural and functional organization of the yeast mediator complex. J. Biol. Chem., 276, 42003–42010. [DOI] [PubMed] [Google Scholar]
- 25.Koh S.S., Ansari,A.Z., Ptashne,M. and Young,R.A. (1998) An activator target in the RNA polymerase II holoenzyme. Mol. Cell, 1, 895–904. [DOI] [PubMed] [Google Scholar]
- 26.Lee T.I., Wyrick,J.J., Koh,S.S., Jennings,E.G., Gadbois,E.L. and Young,R.A. (1998) Interplay of positive and negative regulators in transcription initiation by RNA polymerase II holoenzyme. Mol. Cell. Biol., 18, 4455–4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uetz P., Giot,L., Cagney,G., Mansfield,T.A., Judson,R.S., Knight,J.R., Lockshon,D., Narayan,V., Srinivasan,M., Pochart,P. et al. (2000) A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature, 403, 623–627. [DOI] [PubMed] [Google Scholar]
- 28.Ito T., Chiba,T., Ozawa,R., Yoshida,M., Hattori,M. and Sakaki,Y. (2001) A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl Acad. Sci. USA, 98, 4569–4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bartel P.L., Chien,C.-T., Sternglanz,R. and Fields,S. (1993). Using the two hybrid system to detect protein–protein interactions. In Hartley,D.A. (ed.), Cellular Interactions in Development: A Practical Approach. Oxford University Press, Oxford, pp. 153–179. [Google Scholar]
- 30.Harper J.W., Adami,G.R., Wei,N., Keyomarsi,K. and Elledge,S.J. (1993) The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell, 75, 805–816. [DOI] [PubMed] [Google Scholar]
- 31.Werner M., Chaussivert,N., Willis,I.M. and Sentenac,A. (1993) Interaction between a complex of RNA polymerase III subunits and the 70 kDa component of TFIIIB. J. Biol. Chem., 268. [PubMed] [Google Scholar]
- 32.Fromont-Racine M., Rain,J.-C. and Legrain,P. (1997) Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nature Genet., 16, 277–282. [DOI] [PubMed] [Google Scholar]
- 33.Flores A., Briand,J.-F., Gadal,O., Andrau,J.-C., Rubbi,L., Van Mullem,V., Boschiero,C., Goussot,M., Marck,C., Carles,C. et al. (1999) A protein–protein interaction map of yeast RNA polymerase III. Proc. Natl Acad. Sci. USA, 96, 7815–7820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rigaut G., Shevchenko,A., Rutz,B., Wilm,M., Mann,M. and Seraphin,B. (1999) A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol., 17, 1030–1032. [DOI] [PubMed] [Google Scholar]
- 35.Longtine M.S., McKenzie,A.,III, Demarini,D.J., Shah,N.G., Wach,A., Brachat,A., Philipsen,P. and Pringle,J.R. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast, 14, 953–961. [DOI] [PubMed] [Google Scholar]
- 36.Fan H.-Y., Cheng,K.K. and Klein,H.L. (1996) Mutations in the RNA polymerase II transcription machinery suppress the hyperrecombination mutant hpr1Δ of Saccharomyces cerevisiae. Genetics, 142, 749–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chavez S., Beilharz,T., Rondon,A.G., Erdjument-Bromage,H., Tempst,P., Svejstrup,J.Q., Lithgow,T. and Aguilera,A. (2000) A protein complex containing Tho2, Hpr1, Mft1 and a novel protein, Thp2, connects transcription elongation with mitotic recombination in Saccharomyces cerevisiae. EMBO J., 19, 5824–5834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gromoller A. and Lehming,N. (2000) Srb7p is a physical and physiological target of Tup1p. EMBO J., 19, 6845–6852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Naar A.M., Taatjes,D.J., Zhai,W., Nogales,E. and Tjian,R. (2002) Human CRSP interacts with RNA polymerase II CTD and adopts a specific CTD-bound conformation. Genes Dev., 16, 1339–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taatjes D.J., Naar,A.M., Andel,I.F., Nogales,E. and Tjian,R. (2002) Structure, function, and activator-induced conformations of the CRSP coactivator. Science, 295, 1058–1062. [DOI] [PubMed] [Google Scholar]
- 41.Myers L.C. and Kornberg,R.D. (2000) Mediator of transcriptional regulation. Annu. Rev. Biochem., 69, 729–749. [DOI] [PubMed] [Google Scholar]
- 42.Samuelsen C.O., Baraznenok,V., Khorosjutina,O., Spahr,H., Kieselbach,T., Holmberg,S. and Gustafsson,C.M. (2003) TRAP230/ARC240 and TRAP240/ARC250 Mediator subunits are functionally conserved through evolution. Proc. Natl Acad. Sci. USA, 100, 6422–6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sato S., Tomomori-Sato,C., Banks,C.A., Sorokina,I., Parmely,T.J., Kong,S.E., Jin,J., Cai,Y., Lane,W.S., Brower,C.S. et al. (2003) Identification of mammalian Mediator subunits with similarities to yeast Mediator subunits Srb5, Srb6, Med11, and Rox3. J. Biol. Chem., 278, 15123–15127. [DOI] [PubMed] [Google Scholar]
- 44.Li S., Armstrong,C.M., Bertin,N., Ge,H., Milstein,S., Boxem,M., Vidalain,P.O., Han,J.D., Chesneau,A., Hao,T. et al. (2004) A map of the interactome network of the metazoan C.elegans. Science, 303, 540–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Giot L., Bader,J.S., Brouwer,C., Chaudhuri,A., Kuang,B., Li,Y., Hao,Y.L., Ooi,C.E., Godwin,B., Vitols,E. et al. (2003) A protein interaction map of Drosophila melanogaster. Science, 302, 1727–1736. [DOI] [PubMed] [Google Scholar]
- 46.Fromont-Racine M., Mayes,A.E., Brunet-Simon,A., Rain,J.C., Colley,A., Dix,I., Decourty,L., Joly,N., Ricard,F., Beggs,J.D. et al. (2000) Genome-wide protein interaction screens reveal functional networks involving Sm-like proteins. Yeast, 17, 95–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cramer P., Bushnell,D.A. and Kornberg,R.D. (2001) Structural basis of transcription: RNA polymerase II at 2.8 ångstrom resolution. Science, 292, 1863–1876. [DOI] [PubMed] [Google Scholar]
- 48.Cramer P., Bushnell,D.A., Fu,J., Gnatt,A.L., Maier-Davis,B., Thompson,N.E., Burgess,R.R., Edwards,A.M., David,P.R. and Kornberg,R.D. (2000) Architecture of RNA polymerase II and implications for the transcription mechanism. Science, 288, 640–649. [DOI] [PubMed] [Google Scholar]
- 49.Tomomori-Sato C., Sato,S., Parmely,T.J., Banks,C.A., Sorokina,I., Florens,L., Zybailov,B., Washburn,M.P., Brower,C.S., Conaway,R.C. et al. (2004) A mammalian Mediator subunit that shares properties with Saccharomyces cerevisiae Mediator subunit Cse2. J. Biol. Chem., 279, 5846–5851. [DOI] [PubMed] [Google Scholar]
- 50.Bourbon H.M., Aguilera,A., Ansari,A.Z., Asturias,F.J., Berk,A.J., Bjorklund,S., Blackwell,T.K., Borggrefe,T., Carey,M., Carlson,M. et al. (2004) A unified nomenclature for protein subunits of Mediator complexes linking transcriptional regulators to RNA polymerase II. Mol. Cell, 14, 553–557. [DOI] [PubMed] [Google Scholar]
- 51.Novatchkova M. and Eisenhaber,F. (2004) Linking transcriptional mediators via the GACKIX domain super family. Curr. Biol., 14, R54–R55. [DOI] [PubMed] [Google Scholar]
- 52.Rachez C. and Freedman,L.P. (2001) Mediator complexes and transcription. Curr. Opin. Cell Biol., 13, 274–280. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.