Abstract
Aryl hydrocarbon receptor nuclear translocator (ARNT) belongs to the basic helix–loop–helix Per-Arnt-Sim (bHLH PAS) protein which dimerizes with other PAS proteins. Although it has a transactivation domain (TAD), ARNT functions as an assistant partner of main factors, such as aryl hydrocarbon receptor and hypoxia-inducible factors, rather than acting as a straightforward transcription factor. However, ARNT may function as an active transcription factor using its TAD either in association with itself, single-minded protein 1, or trachealess protein. In the present study, we identified a novel ARNT partner, a HIF-1α variant, which is ubiquitously expressed in human tissues and cancer cell lines. The HIF-1α variant, designated HIF-1α417, bound to ARNT and, moreover, stimulated the transcription of the erythropoietin enhancer reporter gene. This stimulation was markedly augmented by ARNT but not by the ARNT603 mutant lacking the TAD. Thus, augmentation by ARNT suggests that ARNT determined the transcriptional activity. HIF-1α417 was found to be associated with ARNT and to bind to the hypoxia response element containing the E-box core. Moreover, HIF-1α417 promoted the nuclear translocation of ARNT, and conversely ARNT stabilized HIF-1α417. Taken together, our results suggest that HIF-1α417 is a novel partner that is required for transcription activity of ARNT.
INTRODUCTION
The basic helix–loop–helix Per-Arnt-Sim (bHLH PAS) proteins are critical transcription factors that control gene expression networks in many essential physiological processes, such as those involving xenobiotics, hypoxic responses, and neural development (1). These proteins have a conserved basic DNA-binding sequence adjacent to a helix–loop–helix domain, which allows homo- or heterodimerization between bHLH proteins. A compact four-helical bundle formed by two HLH domains facilitates the close interaction of the basic regions with DNA (2). They also have a conserved PAS region, which consists of two adjacent PAS domains, termed PAS A and PAS B (1). Although the roles of the PAS region are not understood fully, it appears to contribute to the specificity of DNA binding by controlling partner choice for dimerization (3).
The bHLH PAS superfamily is divided into two classes. The members of class I play dominant roles in gene transcription, and those of class II help in nuclear localization, protein stabilization, and the DNA binding of class I members. In general, to form a functional DNA-binding complex, one member in class I must dimerize with a member in class II (3). In particular, the aryl hydrocarbon receptor nuclear translocator (ARNT) belongs to class II. ARNT was first identified as a component of a heterodimeric DNA-binding protein that transactivates certain genes that encodes drug-metabolizing enzymes in response to environmental pollutants like dioxin (4). When such pollutants enter a cell and associate with aryl hydrocarbon receptor (AhR), AhR dissociates from HSP90 and translocates to the nucleus, and then dimerizes with ARNT. The AhR–ARNT heterodimer complex then binds to the xenobiotic responsive element (XRE) and activates the transcription of cytochrome P4501A1, quinine reductase, and glutathione S-transferase genes (5,6). In addition, ARNT participates in multiple partnerships with other bHLH PAS proteins, such as hypoxia-inducible factor 1 and 2 alpha (HIF-1α and HIF-2α) (7,8), single-minded protein 1 and 2 (SIM1 and SIM2) (9), and cardiovascular helix–loop–helix factor 1 (CHF1) (10). Of these, HIF-1α and HIF-2α have emerged as key transcription factors that function as master regulators of oxygen homeostasis. These alpha subunits in association with ARNT upregulate the expressions of proteins that increase oxygen delivery and allow cells to survive in oxygen-deficient conditions. To date, about 60 kinds of hypoxia-inducible genes have been found to be regulated by HIF at the gene level (11). Since ARNT was co-purified with HIF-1α, it has an alternative name, HIF-1β (12).
ARNT contains a conserved domain designated PAS that is shared by Drosophila PER (dPER) and Drosophila SIM (dSIM). It also contains a conserved bHLH domain, which is immediately N-terminal to the PAS domain. These domains are essential for DNA binding and dimerization with AhR or HIF-1α (13). In addition, ARNT contains a transactivation domain (TAD) at its C-terminus (14). Since AhR and HIF-1α also contain one or two TADs, AhR-ARNT and HIF-1α-ARNT dimers have multiple TADs. Many domain analyses have revealed that the TAD of ARNT is not essential for the transcriptional activities of these transcriptional complexes (15–17). Instead, AhR or HIF-1α is likely to work as a main transcription factor by utilizing their own TADs. Therefore, ARNT is considered to function as a general partner, which acts to aid the transcriptional function of main factors (3).
On the other hand, the TAD of ARNT may play a key role in the transcriptional activities of ARNT–ARNT homodimer (18–20) and ARNT–SIM1 heterodimer (21–23). Although no physiological role for ARNT homodimer has been defined yet, reporter gene assays have demonstrated that a putative ARNT homodimer can activate transcription by binding to the E-box element, as do other typical members of the bHLH PAS family. Moreover, dSIM is critical to the development of the midline cells of the central nervous system of Drosophila. SIM1 and SIM2 are mammalian homologs of dSIM, and both have repressive functions in gene expression (9,21). However, when SIM1 is overexpressed with ARNT, the SIM1–ARNT complex can enhance the expression of the CNS midline enhancer (CME)- or of erythropoietin enhancer (EpoE)-reporter genes, both of which contains the E-box core sequence, 5′-ACGTG-3′ (22,23). These results suggest that the SIM1–ARNT complex can function as a transactivator under certain situations. In this case, the transcriptional activity totally depends on the TAD of ARNT because SIM1 has no TAD. Taken together, ARNT functions as a general partner protein in association with transcription factors that have strong transcriptional activity, and as a main transcription factor in association with proteins without transcriptional activity.
In the present study, we found a novel partner whilst searching for novel HIF-1α variants. This protein was derived from the alternatively spliced mRNA of HIF-1α and its mRNA was found to be expressed ubiquitously in human tissues and cancer cell lines. We designated the HIF-1α variant, which is composed of 417 amino acids, as HIF-1α417. HIF-1α417 has the bHLH PAS domain that is essential for DNA- and ARNT-binding, but does not have TAD. Its expression was found to enhance the transcription of the EpoE-reporter gene, and this activity was markedly increased by ARNT co-transfection; moreover, this transcriptional activity was determined by the TAD of ARNT. It was also found that HIF-1α417 associates with ARNT and binds to the hypoxia response element. Moreover, HIF-1α417 helped ARNT enter into the nucleus, and ARNT stabilized HIF-1α417 by blocking its proteasomal degradation, thus demonstrating a functional partnership between these proteins. Taken together, we suggest that HIF-1α417 is an ARNT partner protein, which is required for the transcription activity of ARNT.
MATERIALS AND METHODS
Web-based search for putative splice variants of HIF-1α mRNA
To search for different cDNA variants expressed from the HIF1A gene, a nucleotide–nucleotide BLAST search was performed against the human dbEST (Expressed Sequence Tags) database at the GenBank Web server (www.ncbi.nlm.nih.gov/BLAST/). The HIF-1α cDNA sequence (GenBank # U22431) and the HIF1A gene sequence (GenBank # AH006957) were used to identify putative splice variants of HIF-1α mRNA.
Antibodies
Rabbit anti-hemagglutinin (HA) and goat anti-ARNT antibodies were purchased from BIODESIGN International (Saco, Maine) and Santa Cruz Biotechnology (Santa Cruz, CA), respectively. Anti-ODDD antibody was generated in rabbits against a bacterially expressed fragment that encompassed the ODDD of human HIF-1α (418–698), as described previously (24), and anti-HIF-1α417 antibody was generated in rats against a bacterially expressed His-tagged protein that encompassed the full length of the variant protein. The specificity of anti-HIF-1α417 antibody was examined by western blot using bacterial proteins expressed under uninduced or IPTG-induced conditions.
Cell culture
Most cancer cell lines were obtained from ATCC, and SNU601 cell line, a stomach cancer cell line, was obtained from the Korean Cell Line Bank (Seoul, Korea). Hep3B cells were cultured in α-modified Eagle's medium, and HEK293, HeLa, and SNU601 cells were cultured in Dulbecco's modified Eagle's medium. Both culture media were supplemented with 10% heat-inactivated fetal calf serum (FCS), 100 units/ml penicillin, and 100 μg/ml of streptomycin in a humidified atmosphere containing 5% CO2 at 37°C. Oxygen tensions in the incubator (Vision Sci Co., Korea) were either 140 mm Hg (20% O2, v/v, normoxia) or 7 mm Hg (1% O2, v/v, hypoxia). All culture media and sera were purchased from GIBCO/BRL (Grand Island, NY).
Identification and cloning of human HIF-1α cDNA variants
Reverse transcription–polymerase chain reactions (RT–PCRs) were performed to identify HIF-1α and its variant cDNAs. RNAs were isolated from cultured cells using TRIZOL (GIBCO/BRL), and were reverse-transcribed using the avian myeloblastosis virus reverse transcriptase system (Promega, Madison, WI). One microgram of total RNA was added to 50-μl of an RT–PCR reaction mixture that contained 5 μCi [α-32P]dCTP and 250 nM of each primer set. RT–PCR was performed using one cycle of reverse transcription at 44°C for 1 h and 20 cycles of: denaturation at 94°C for 30 s, annealing at 53°C for 30 s and elongation at 72°C for 30 s. The resulting PCR products (5 μl) of HIF-1α cDNA were electrophoresed in a 4% polyacrylamide gel at 120 V in a 0.3× TBE buffer at 4°C, and dried gels were autoradiographed. The PCR products were extracted from the gels and directly sequenced. The nucleotide sequence of the forward primer for exon 9 was 5′-CCTGATGCTTTAACTTTGCTG-3′, and the sequence of the reverse primer for exon 12 was 5′-TTCTAATGGTGACAACTGATC-3′.
To clone full length HIF-1α variants, the cDNAs were amplified by RT–PCR using two primers, the sequences of which were 5′-GTGAAGACATCGCGGGGAC-3′ for the forward primer for exon 1, and 5′- TAAGAAAAAGCTCAGTTAAC-3′ for a reverse primer for exon 15. The PCR products so obtained were cloned using a pCR2.1-TOPO cloning kit (Invitrogen, Carlsbad, CA). To select a colony of Escherichia coli transformed with the HIF-1α cDNA variant, colony PCRs using primers for exons 9 and 12 were performed. The resulting PCR products were electrophoresed in a 2% agarose gel and visualized with ethidium bromide. A plasmid containing the full length HIF-1α cDNA variant was amplified and purified, and the insert DNA sequence was analyzed.
Northern Blot and RT–PCR Assays
To investigate the tissue distribution of HIF-1α and its variant mRNA, northern blot analysis that used specific probes was performed using a human multiple tissue blot (FirstChoice™ Blot 4, Ambion Inc., Austin, TX). The RNA blot was prehybridized at 35°C for 4 h in the ULTRAhyb™ Oligo solution (Ambion Inc, Austin TX), and hybridized to a gene-specific oligonucleotide probe radiolabeled by using T4 polynucleotide kinase and [γ-32P]ATP (DNA 5′ end-labeling system, Promega), in the same solution at 35°C for 18 h. The blot was then washed twice at 35°C for 30 min in 2× SSC plus 0.05% SDS, and the membrane was autoradiographed using intensifying screens by exposure at −70°C. The probe sequence for the wild-type mRNA is 5′- CCTGAATCTGGGGCATGGTAAAAGAAAG-3′, which is specific for exon 10, and that of variant mRNA is 5′-CTGGGACTATTAGGCGTTGCTGCCAAAA-3′, which is specific for the junction of exons 9 and 11. To measure β-actin mRNA, the tissue blot was hybridized with the radiolabeled β-actin cDNA probe, which was generated using a set of PCR primers: 5′-CCAGATCATGTTTGAGACCT-3′ and 5′-TTGAAGGTAGTTTCGTGGAT-3′. mRNAs of erythropoietin and β-actin in Hep3B cells were quantified by semi-quantitative RT–PCR, as described in the previous section. The nucleotide sequences of the primer pairs were 5′-CTGGAGAGGTACCTCTTGGAGG-3′ and 5′-CCCCTGTGTACAGCTTCAGCTT-3′ for erythropoietin; 5′-CCAGATCATGTTGAGAC-CT-3′ and 5′-TTGAAGGTAGTTTCGTGGAT-3′ for β-actin.
Expression plasmids and transfection
HA-tagged HIF-1α and ARNT expression plasmids were generous gifts from Dr Eric Huang (NCI, Bethesda, MD). HA-tagged HIF-1α variant expression plasmid was made using a PCR-based mutagenesis kit (Stratagene, Cedar Creek, TX). Mutagenesis using specific oligonucleotides, 5′-CCTAATAGTCCCAGTGAATATTG-3′ and 5′-CGTTGCTGCCAAAATCTAAAGATATG-3′, was employed to delete nucleotides derived from exon 10. Untagged HIF-1α variant expression plasmid was constructed by directly ligating full-length PCR products into the pcDNA3 vector, and stable HIF-1α (HIF-1αS) expression plasmid was constructed by substitution of the nucleotides coding two proline residues (402 and 564) with the nucleotides coding alanine. The ARNT603 mutant plasmid was made using the mutagenesis kit. Mutagenesis with specific oligonucleotides, 5′-TTCCCTGATCTAACTATGTTTCCC-3′ and 5′-ATTCCTGAAATTCTCTGCCGG-3′, was employed to delete the transactivation domain at the C-terminus of ARNT. All constructs were verified by DNA sequencing.
For transient expression, about 40% of the confluent HEK293 cells in 60-mm cell culture dishes were transfected with 0.5–8 μg of DNAs using the calcium phosphate method. Transfected cells were incubated for 48 h in Dulbecco's modified Eagle's medium supplemented with 10% FCS and then used for experiments.
Reporter assays
Luciferase reporter genes, which contain the EPO enhancer or the HRE-mutated EPO enhancer, were generous gifts from Dr Eric Huang (NCI, Bethesda, MD). To assay reporter activity, HEK293 cells were co-transfected with 0.6 μg of luciferase reporter gene and 0.6 μg of plasmid cytomegalovirus-β-gal, and/or 0.6 μg of plasmid HIF-1α variant, and/or 0.6 μg of plasmid ARNT per dish, using the calcium phosphate method. pcDNA3 was added to ensure that the final DNA concentrations in both control and experimental dishes were at the same level. After stabilizing for 48 h, the cells were lysed and assayed for luciferase activity using a Lumat LB9507 luminometer (Berthold Technologies, Bad Wildbad, Germany). β-Gal assays were performed to normalize transfection efficiencies.
Extraction of nuclear proteins
Nuclear proteins were extracted as described previously (24). Scraped cells were centrifuged at 3000 r.p.m. for 5 min at 4°C and then washed twice with ice-cold phosphate-buffered saline. Cells were then resuspended in three packed cell volumes of a lysis buffer that consisted of 10 mM Tris, pH 7.8, 10 mM KCl, 0.1 mM EDTA, 1.5 mM MgCl2, and 0.2% NP-40, 0.5 mM DTT, 1 mM Na3VO4 and 0.4 mM phenylmethylsulfonyl fluoride (PMSF). Cells were then vortexed at medium speed for 10 s and incubated on ice for 5 min. Nuclei were then pelleted at 3000 r.p.m. for 5 min at 4°C. One packed volume of extract buffer, which consisted of 20 mM Tris, pH 7.8, 420 mM NaCl, 0.1 mM EDTA, 1.5 mM MgCl2, 20% glycerol, 0.5 mM dithiothreitol, 1 mM Na3VO4 and 0.4 mM PMSF, was added to the nuclei and vortexed at medium speed for 5 s per minute for 10 min. The nuclear extracts were then centrifuged at 20 000 g at 4°C for 5 min, aliquoted into chilled tubes, frozen quickly in liquid nitrogen, and stored at −70°C. Protein concentrations were measured using the bicinchoninic acid method (Bio-Rad). BSA was used as a standard.
Immunoblotting and immunoprecipitation
For immunoblotting, total proteins or nuclear proteins were separated on 6.5 or 10% SDS–polyacrylamide gel, and transferred to an Immobilon-P membrane (Millipore, Bedford, MA). Membranes were then blocked with 5% nonfat milk in Tris-buffered saline containing 0.1% Tween-20 (TTBS) at room temperature for 1 h and incubated overnight at 4°C with anti-HIF417, anti-HA, or anti-ARNT, diluted 1:5000 in 5% nonfat milk in TTBS. Horseradish peroxidase-conjugated anti-rabbit, anti-goat or anti-rat antiserum was used as a secondary antibody (1:5000 dilution in 5% nonfat milk in TTBS, 1 h incubation) and the antigen–antibody complexes were visualized using an Enhanced Chemiluminescence Plus kit (Amersham Biosciences Corp., Piscataway, NJ).
For immunoprecipitation, HEK293 cells were co-transfected with the pHA-tagged HIF-1α variant and pARNT. Forty-two hours after transfection, the cells were solubilized and the cell lysates were incubated with anti-HA antiserum or pre-immune serum, and then incubated with protein A–Sepharose beads (Amersham Pharmacia Biotech). After washing, the immunocomplexes were eluted in SDS sample buffer containing 10 mM DTT, and then subjected to SDS–PAGE and immunoblotting using anti-ARNT antibody.
Electrophoretic mobility gel-shift (EMSA) assay
The oligonucleotide probe used in the gel-shift assay had the sequence 5′-ACCGGCCCTACGTGCTGTCTCAC-3′. The 32P-labeled double-stranded probe was prepared and an EMSA assay was performed, as described previously (24). DNA–protein binding reactions were carried out for 20 min at 4°C and run on a 5% non-denaturing polyacrylamide gel. For supershift analysis, 1 μl of anti-HIF417, anti-ARNT, or anti-ODDD antiserum was added to the completed EMSA reaction mixture and incubated for 2 h at 4°C prior to loading.
RESULTS
Identification of a novel splice variant of HIF-1α
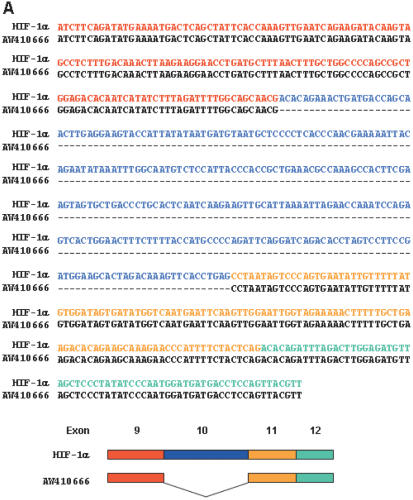
A nucleotide–nucleotide BLAST homology search for the HIF-1α cDNA against the human EST database was performed to screen for putative splice variants of HIF-1α. The resulting sequences were individually compared with the sequence of wild-type HIF-1α cDNA, and this showed that one EST (AW410666—derived from the human rhabdomyosarcoma NIH_MGC_17 cDNA library) contains the HIF-1α sequence lacking 287 nucleotides (Figure 1A). To determine whether this deletion represented a real variant, the EST sequence was compared to the structure of the HIF1A gene, which is ∼8 kb long and consists of 15 exons and 14 introns (25). The nucleotide segment missing in AW410666 EST matched exon 10 in the HIF1A gene, leaving exons 9 and 11 directly joined in the cDNA. To confirm that this variant mRNA is actually transcribed, a pair of PCR primers was designed to amplify the cDNA segment from exon 9 to exon 12. HIF-1α cDNAs in four human cell lines were reverse-transcribed and amplified by semi-quantitative RT–PCR using [α-32P]dCTP. Two distinct DNA bands of different sizes (537 and 250 bp) were reproducibly observed by gel electrophoresis (Figure 1B). DNA sequencing revealed that the lower band lacked only exon 10, and that the upper band consisted of all exons (9–12). These PCR fragments were quantified by excising the corresponding radioactive bands from dried gel and measuring the radioactivity of each band with a beta-counter. The relative amounts of HIF-1α and its variant fragment were then assessed from counted c.p.m. values. Figure 1C shows that the variant was present at about 10% of the wild-type level in Hep3B and HEK293 cells, and at a lower level in HeLa and SNU601 cells. To obtain a clone of the variant, we amplified full-length HIF-1α cDNA using the primers described in the Materials and Methods section, and inserted the cDNAs into pCR2.1 plasmid. The DNA sequencing of the plasmid containing the full-length cDNA of the variant revealed that there is no additional alternative site than exon 10.
Figure 1.
Identification of a novel spliced variant of HIF-1α cDNA. (A) A comparison of the nucleotide sequences of the human EST clone (AW410666) and human HIF-1α cDNA. Dashed lines represent nucleotides deleted in the EST clone. Exons containing the HIF-1α and the EST clone are illustrated schematically in the lower panel. (B) Total RNA was extracted from Hep3B, HEK293, HeLa and SNU601 cells. HIF-1α cDNAs were amplified and identified using a highly sensitive RT–PCR method. The illustration in the right panel shows the nucleotide sequences of the exon 9/10 and exon 9/11 junctions. (C) The relative amounts of the mRNAs were calculated using the equation (c.p.m. of 250 bp-fragment/c.p.m. of 537 bp fragment) × (222/99). The correction factor, 222/99, represents the ratio of G and C numbers in the 537 bp fragment versus that of the 250 bp fragment. The results are representative of three separate experiments. (D) Filter membranes that contained 2 μg of human poly(A) mRNAs in each lane were purchased from Ambion Inc. (Austin, TX). Northern blot analysis was performed using 32P-labeled specific probes for HIF-1α and its variant. β-actin mRNA was analyzed as a loading control.
We then performed northern blot to confirm whether the splice variant is expressed in normal human tissues. To generate specific probes for the wild-type or variant mRNA, we designed two oligo probes; the complimentary oligomer to bind exon 10 (for the wild type) and another to specifically bind to the junction of exon 9 and 11 (for the variant). The specificity of each oligo probe was confirmed using standard mRNA samples obtained from HEK293 cells transfected with plasmid HIF-1α/wild or plasmid HIF-1α/variant (data not shown). Wild-type HIF-1α mRNA of ∼4.0 kb was highly expressed in the small intestine, lung, kidney and prostate, and moderately expressed in all other tissues. The smaller variant mRNA of 3.5 kb was also highly expressed in the small intestine, and ubiquitously expressed in all human tissues examined (Figure 1D).
Structure of a novel splice variant, HIF-1α417
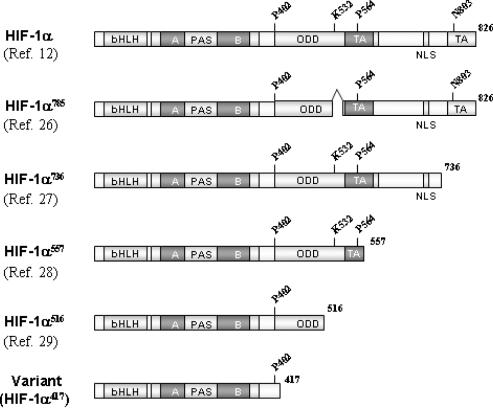
The full-length cDNA of the splice variant was cloned and sequenced as described in the Materials and Methods section, and its structure is summarized in Figure 2. In this mRNA, exons 9 and 11 were directly joined, thus altering the reading frame. This frame-shift may generate immediate termination of translation due to two sequential stop codons, as shown in Figure 1B. Consequently, this alternative splicing adds Ala417 following Asn416 at the C-terminus protein and produces a 417-amino acid polypeptide, designated here as HIF-1α417. Compared with HIF-1α, HIF-1α417 conserves only the bHLH and PAS domains, which are essential for binding between DNA and ARNT. However, it lacks all the other important domains of HIF-1α, such as, ODDD, TAD and NLS. Based on the structure of HIF-1α417, it is neither expected to be regulated by oxygen tension, nor to exhibit transcriptional activity. To date, four splice variants of human HIF-1α have been reported (26–29), since the structure of HIF-1α was identified (12). These variants commonly contain the NH2-side of HIF-1α, which contains bHLH and PAS domains, but have a variable COOH-side, as shown in Figure 2. In this new variant, the NH2-side is conserved but the COOH-side is deleted.
Figure 2.
Protein structures of the cloned HIF-1α variant and of other variants. The HIF-1α variant cloned in the present study is the shortest HIF-1α variant identified to date. This variant, composed of 417 amino acids, contains only the bHLH and PAS domains, which are essential for DNA and ARNT binding.
HIF-1α417 protein is constitutively expressed
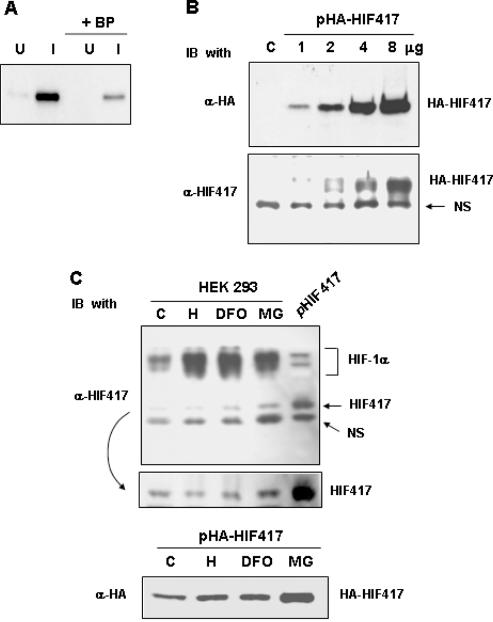
Anti-HIF-1α417 antibody was raised from rat and its specificity was confirmed using bacterial proteins expressed under uninduced or IPTG-induced conditions (Figure 3A). Moreover, the immune reaction was effectively blocked by an antigenic peptide. This antibody also detected HA-HIF-1α417 protein expressed in HEK293 (Figure 3B). HA-HIF-1α417 levels were double-checked using an anti-HA antibody in the same samples. To detect endogenous HIF-1α417 protein, western blotting using anti-HIF-1α417 antibody was performed in untransfected HEK293 cells. The lysates from HEK293 cells that expressed untagged HIF-1α417 protein was loaded on the last lane as a reference for HIF-1α417 identification. Three immunoreactive species were identified (Figure 3C, top). The upper one was wild-type HIF-1α, and the lower one was a protein non-specifically reacted with the antibody. The middle one was considered as endogenous HIF-1α417 because its intensity was specifically enhanced in cells overexpressing HIF-1α417. The HIF-1α417 protein was present even under normoxic conditions, but was not induced by hypoxia or desferrioxamine. However, its protein level slightly increased after the treatment with the proteasome inhibitor, MG132 (Figure 3C, middle). In contrast, the levels of wild-type HIF-1α were markedly enhanced by hypoxia, desferrioxamine, or MG132 treatment. The protein level of expressed HA-tagged HIF-1α417 was also increased by MG132 (Figure 3C, bottom), which suggests that HIF-1α417 is constitutively expressed regardless of oxygen or iron level, and that its stability is partly regulated by a proteasomal pathway.
Figure 3.
Generation of a specific antibody against HIF-1α417 and the identification of endogenous HIF-1α417 protein. (A) To identify the specificity of an antibody against HIF-1α417 protein, lysates (50 ng proteins) of IPTG-induced (I) or uninduced (U) bacteria expressing His-tagged HIF-1α417 protein were analyzed by western blotting using anti-HIF-1α417 antibody (α-HIF417). Blocking peptide (BP; 1 μg/ml), which was a bacterially expressed HIF-1α417 protein purified by nickel-affinity chromatography, was added to the primary antibody solution to confirm the antibody specificity. (B) HEK293 cells with various doses of pcDNA–HA-tagged HIF-1α417 (HA-HIF417) using the calcium phosphate method. Total cell lysates (50 μg protein) were analyzed by western blotting using an anti-HA antibody (α-HA) and α-HIF417. NS indicates a non-specific protein cross-reacted with α-HIF417 antiserum. (C) To identify the presence of endogenous HIF-1α417 protein (HIF417), the lysates of untransfected HEK293 cells were analyzed by western blotting using α-HIF417 (upper panel). HEK293 cells were cultured under normoxic (C) or hypoxic (H) conditions, or in the presence of 130 μM desferrioxamine (DFO) or 20 μM MG132 (MG) for 8 h. As a reference for the HIF417 protein, untagged HIF417 expressed in HEK293 cells was also blotted onto the same membrane (pHIF417). The ECL film was developed in a darker shade to compare the HIF417 levels (middle panel). HEK293 cells transfected with pHA-HIF417 were cultured under normoxic (C) or hypoxic (H) conditions, or in the presence of 130 μM desferrioxamine (DFO) or 20 μM MG132 (MG) for 8 h. The cell lysates were analyzed by western blotting using α-HA (lower panel).
The expression of HIF-1α417 strongly stimulates ARNT-dependent transcription
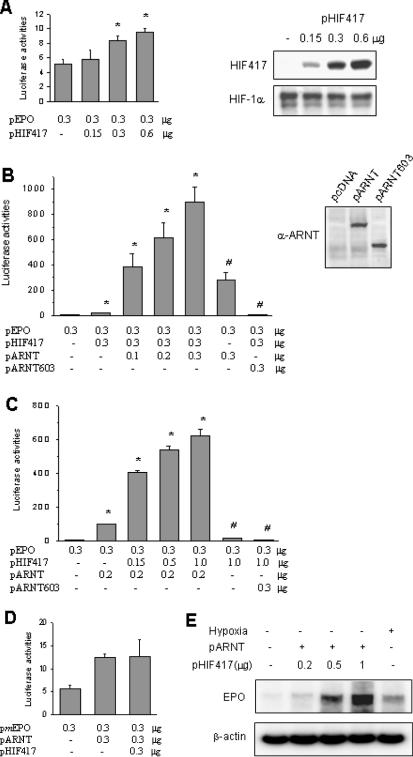
Since HIF-1α417 lacks both transactivation domains of HIF-1α, it might be expected that it would not function as a transcription factor. Unexpectedly, however, expressed HIF-1α417 stimulated EPO enhancer activity under normoxic conditions (Figure 4A). When we co-expressed ARNT with HIF-1α417, reporter activities were enhanced up to a 100-fold versus the untransfected control value (Figure 4B), and these reporter activities were increased by either ARNT or HIF-1α417 in a gene-dose dependent manner (Figure 4B and C). When TAD lacking ARNT603, instead of wild ARNT, was expressed, reporter activity was completely diminished even in the presence of HIF-1α417. In the case of the Epo-reporter vector containing a mutated HRE site, no significant enhancement of reporter activity was observed in HIF-1α417-ARNT-co-transfected cells (Figure 4D). To examine whether HIF-1α417 and ARNT stimulate the transcription of the EPO gene, EPO-producing Hep3B cells were transfected with HIF-1α417 and ARNT plasmids. HIF-1α417 in concert with ARNT enhanced the EPO mRNA expression under normoxic conditions in a gene-dose dependent manner (Figure 4E). These results suggest that HIF-1α417 is required for transcriptional activation by ARNT.
Figure 4.
Cooperation of HIF-1α417 and ARNT in HRE-reporter gene transcription. (A) A luciferase reporter plasmid containing the Epo enhancer was co-transfected into HEK293 cells with various doses of pHIF417. The reporter activity was measured in lysates from cells cultured under normoxic conditions. Western blotting in the right panel demonstrated that HIF417 was expressed in a gene dose-dependent manner and endogenous HIF-1α levels were not affected by HIF417 expression. (B) Various doses of pARNT (or pARNT603) and pHIF417 were co-transfected into cells using the EPO reporter. Western blotting in the right panel demonstrated the expression of ARNT and ARNT603 proteins. (C) Various doses of pHIF417 and pARNT (or pARNT603) were co-transfected into cells using the EPO reporter. (D) The mutated Epo reporter lacking the HRE core sequence was co-transfected into cells with pARNT and pHIF417. Bars represent the mean and SD of six or more experiments. Statistical differences were compared using the unpaired two-tailed Student's t-test. *: P < 0.05 versus the control group transfected with only the reporter plasmid. #: P < 0.05 versus the group co-transfected with both pHIF417 and pARNT. (E) Total RNAs were isolated from untransfected or transfected Hep3B cells subjected to normoxia (lanes 1–4) or 16 h hypoxia (lane 5), and erythropoietin (EPO) mRNA was analyzed by semi-quantitative RT–PCR. Cells were transfected with 0.5 μg of pARNT and various doses of pHIF417. β-actin mRNA was analyzed as a loading control.
The HIF-1α417/ARNT complex binds to HRE
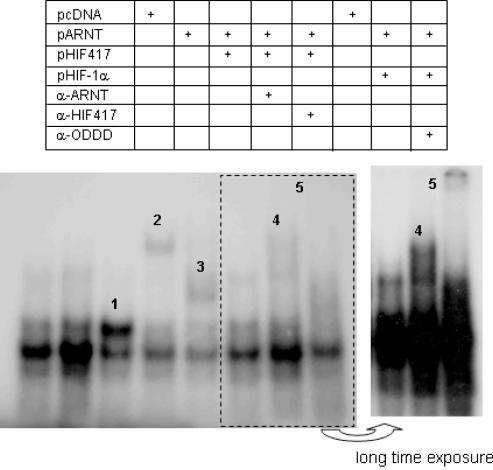
The stimulation of HRE reporter activities by HIF-1α417 suggests that HIF-1α417 dimerizes with ARNT and in turn binds to HRE, as it does to HIF-1α. To confirm this possibility, we performed an EMSA assay using an oligo probe containing the HIF-1-binding sequence of the EPO gene. The nuclear extract from normoxic HEK293 cells showed three constitutive DNA-binding species, the middle one of which was the densest (refer to the first lane of Figure 5). This is similar to the EMSA result reported previously by Maxwell et al. (30). When HIF-1α417 and ARNT were co-expressed, the intensity of the upper band became markedly stronger than that in the control cells (the third lane). Moreover, the mobility of this DNA-binding species was retarded by either ARNT antibody or HIF-1α417 antibody (the fourth and fifth lanes). In contrast, the co-expression of HIF-1α and ARNT produced a different DNA-binding species with lower mobility, which was super-shifted by HIF-1α/ODDD antibody (the seventh and eighth lanes). This result suggests that the HIF-1α417/ARNT complex binds to HRE and forms one constitutive DNA-binding species in EMSA gel.
Figure 5.
EMSA assay for the HRE binding activity of HIF-1α417 and ARNT. Binding to a radiolabeled oligonucleotide containing the HRE core sequence was observed in nuclear extracts prepared from HEK293 cells transfected with pARNT plus pHIF417 or pHIF-1α. For supershift analysis, 1 μl of ARNT, HIF-1α417 or HIF-1α-ODDD anti-serum was added to the completed EMSA reaction mixture. 1, DNA binding by HIF-1α417–ARNT co-expression; 2, supershift of ‘band 1’ by anti-ARNT antibody; 3, supershift of ‘band 1’ by anti-HIF-1α417 antibody; 4, DNA binding by HIF-1α–ARNT co-expression; 5, supershift of ‘band 4’ by anti-ODDD antibody. The EMSA film was exposed for a longer time to make denser 4 and 5 bands (the right panel).
HIF-1α417 associates with ARNT and enhances the nuclear translocation of ARNT
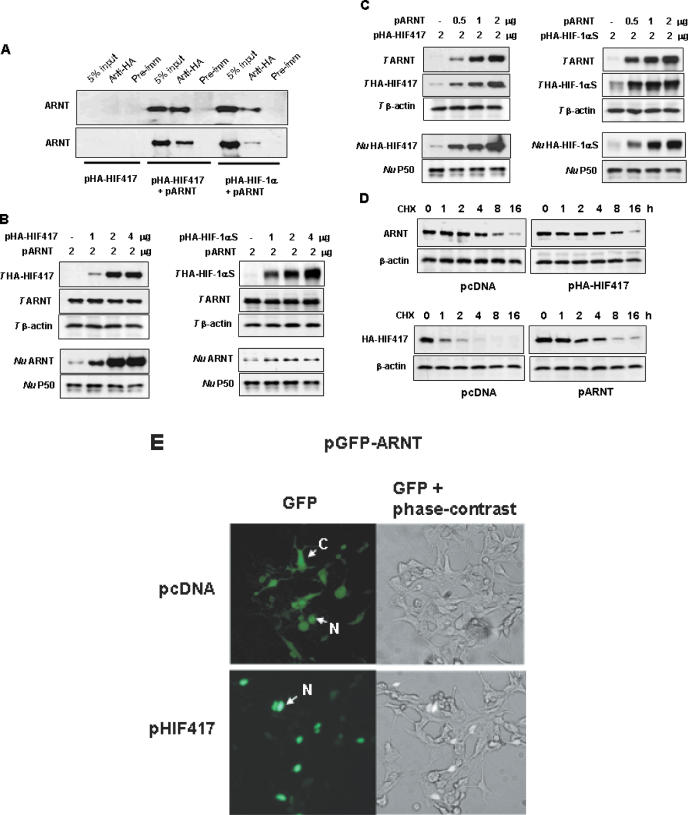
To confirm that HIF-1α417 forms a complex with ARNT, we observed the co-precipitation of HIF-1α417 and ARNT. As expected, ARNT co-precipitated with both HIF-1α417 and HIF-1α (Figure 6A). Moreover, HIF-1α417 increased the nuclear levels of ARNT without altering the total amount of ARNT (Figure 6B, left panel). ARNT is prone to leak from the nuclei during isolation of the nuclei in the absence of its partner proteins including HIF-1α, and its partner protein can prevent the loss of ARNT from the nucleus by forming a bigger, dimerized protein (31). To exclude the possibility that the nuclear accumulation of ARNT was shown as an artifact, we co-expressed ARNT and full-length HIF-1αS, which was stably expressed due to the mutation on Pro402 and Pro564 residues and bound with ARNT. However, the full-length HIF-1α did not increase the levels of nuclear ARNT as much when compared with HIF-1α417 (Figure 6B, right panel). This suggests that HIF-1α417 specifically enhances the translocation of ARNT to the nucleus.
Figure 6.
In vivo interaction between HIF-1α417 and ARNT. (A) Association of HIF-1α417 with ARNT. HEK293 cells were co-transfected with pARNT and either pHA-HIF417 or pHA-HIF-1α. Lysates were prepared and immunoprecipitations were performed with α-HA. The co-immunoprecipitation of ARNT with HIF-1α variants was identified by western blotting using α-ARNT. (B) Nuclear translocation of ARNT by HIF-1α417 or full-length HIF-1α. HEK293 cells were co-transfected with pARNT and various doses of pHA-HIF417 (left panel) or pHA-HIF-1αS (right panel). (C) ARNT-dependent expression of HIF-1α417 or full-length HIF-1α. HEK293 cells were co-transfected with various doses of pARNT and 2 μg of pHA-HIF417 (left panel) or pHA-HIF-1αS (right panel). Proteins in total cell lysates (T HIF417, T HIF-1αS, T ARNT, T β-actin) or in nuclear fractions (Nu HIF417, Nu HIF-1αS, Nu ARNT, Nu P50) were analyzed by western blotting using anti-HA and specific antibodies. β-Actin and NF-κB P50 proteins were analyzed as loading controls for total lysates and nuclear extracts, respectively. (D) Stabilization of HIF-1α417 protein by ARNT. HEK293 cells were transfected with pARNT and/or pHA-HIF417, and then treated with 60 μg/ml of cycloheximide (CHX). At the indicated time-points after CHX treatment, the cell lysates were analyzed by western blotting using α-ARNT or α-HA. (E) Nuclear translocation of ARNT by HIF-1α417. The GFP–ARNT expressing plasmid (pGFP–ARNT) was co-transfected into HEK293 cells with pcDNA or pHIF417, and the expression of GFP–ARNT protein was examined by fluorescence microscopy 48 h after transfection. The images in the left panel were captured under a green fluorescence filter, and those in the right panel were overlaid with green fluorescence and phase-contrast images. C, expression in the cytoplasm; N, expression in the nucleus.
Inversely, ARNT enhanced the amount of HIF-1α417 protein in total and nuclear extracts (Figure 6C, left panel). In addition, ARNT also enhanced the amount of the full-length HIF-1α in both extracts (Figure 6C, right panel), which was comparable to the result demonstrated by Forsythe et al. (32). Thus HIF-1α417 seems to be stabilized by associating with ARNT, as HIF-1α is. To examine protein stability, we monitored the protein level after blocking de novo protein synthesis with cycloheximide. HIF-1α417 was significantly stabilized by the co-expression of ARNT, whereas the degradation rate of ARNT was not altered by the co-expression of HIF-1α417 (Figure 6D). Next, to confirm that HIF-1α417 enhances the nuclear translocation of ARNT, we co-transfected a GFP–ARNT expression vector with pcDNA–HIF-1α417 into HEK293 cells. The expressed GFP–ARNT fusion protein was found to be distributed either to the nucleus or to the cytoplasm. However, GFP–ARNT protein co-expressed with HIF-1α417 was localized to the nucleus. Since GFP–ARNT protein was concentrated in the nucleus, the nuclei of the co-expressing cells appeared brighter than those of cells expressing GFP–ARNT only (Figure 6E).
DISCUSSION
In this paper, we identified and cloned a novel partner of ARNT, which is a novel splice variant of HIF-1α. As demonstrated by RT–PCR and northern blotting, its mRNA was ubiquitously expressed in human tissues and in cancer cell lines. The difference between this variant and HIF-1α is that it lacks the 10th exon, which introduces a frame-shift resulting in a 417-amino acid polypeptide, designated as HIF-1α417. We also confirmed that HIF-1α417 protein is translated from the variant mRNA by western blotting. Based on its structure, the variant could be expected to bind with either DNA or ARNT via the bHLH PAS domain, but not to function as a primary transcription factor, because it lacks TAD. Moreover, expressed HIF-1α417 enhanced the transcription of the Epo reporter gene in HEK293 cells, and this activity was markedly augmented by ARNT, but not by ARNT603 lacking TAD, which suggests that transcriptional activity is determined by the TAD of ARNT. EMSA assays revealed that the HIF-1α417–ARNT complex binds to HRE and forms a dense band at the same position as constitutive DNA binding. Immunoprecipitation also revealed a physical association between HIF-1α417 and ARNT. Moreover, HIF-1α417 promoted the nuclear translocation of ARNT, and conversely ARNT stabilized HIF-1α417. These results suggested that HIF-1α417 functions as a partner of ARNT and that it is required for transcription activity of ARNT.
The bHLH PAS superfamily contains the HIF family, which includes three members (HIF-1α, HIF-2α, and HIF-3α) that are derived from different gene loci, each of which includes several isoforms generated by alternative mRNA splicing (ref. 33 for review). To date, four isoforms of HIF-1α and five isoforms of HIF-3α have been reported. In particular, the structures and functions of HIF-1α isoforms have been extensively studied because HIF-1α functions as a master protein in adaptation to hypoxia and in tumor promotion. The first HIF-1α isoform loses exon 14 and translates into a 736-amino acid polypeptide (HIF-1α736), which is regulated by oxygen tension and transactivates VEGF promoter (27). The second isoform loses exon 12 and produces a short form of HIF-1α557, which was induced specifically by the zinc ion (28). The third loses both exon 11 and 12 and produces a shorter form of HIF-1α516 (29). Both HIF-1α557 and HIF-1α516 isoforms function as dominant–negative isoforms to block the dimerization of HIF-1α and ARNT by sequestering ARNT to the cytoplasm. The fourth isoform loses only exon 11 and produces a shorter form of HIF-1α785 (26). This isoform is markedly induced by PMA and reactive oxygen species, which are well-known tumor promoters, and enhanced HIF-1 activity in cancer cells. In addition, HIF-1α785 overexpression strikingly enhanced tumor growth in vivo. Thus, HIF-1α785 is considered to play an important role in tumor promotion (26). In the present study, we found a novel isoform of HIF-1α, the HIF-1α417, which is the shortest of the HIF-1α isoforms. This novel isoform is likely to play a role in the constitutive activation of ARNT. The proteins structures of HIF-1α isoforms are illustrated in Figure 2.
HIF-3α, like HIF-1α has a truncated isoform containing only the bHLH PAS domain, which was designated as Inhibitory PAS domain (IPAS) protein in the mouse, in which it is translated by an alternatively spliced variant of mouse HIF-1α mRNA (34). HIF-3α has no transactivation function due to its lack of a transactivation domain, but it does dominantly and negatively regulate HIF-mediated gene expression by sequestering HIF-1α. Moreover, IPAS expression in hepatoma cells selectively impairs the induction of hypoxia-inducible genes regulated by HIF-1, and results in retarded tumor growth and tumor vascular density in vivo (35). In mice, IPAS was selectively expressed in Purkinje cells of the cerebellum and in the corneal epithelium of the eye, and the expression of IPAS in the cornea correlates with low VEGF gene expression under hypoxic conditions (35). In terms of protein structure, HIF-1α417 and IPAS protein are similar. Both proteins have only the bHLH PAS domain and can dimerize with other members of the bHLH PAS superfamily. However, their functions are quite different because HIF-1α417 is a positive regulator of transcription and IPAS protein is a negative regulator. Moreover, their tissue distributions are different; HIF-1α417 is ubiquitously expressed in normal tissues and cancer cells, but IPAS protein is expressed exclusively in the cerebellum and the eye.
HIF-1α protein is tightly regulated by oxygen tension. In aerobic conditions, HIF-1-prolyl hydroxylases modify the two proline residues located at either end of ODDD (ref. 36 for review). Von Hippel–Lindau protein (pVHL), a part of the E3 ubiquitin ligase protein complex, then binds to the modified HIF-1α, which results in its ubiquitination and proteasomal degradation (30). Since the enzymatic reaction of prolyl hydroxylation requires oxygen as a substrate, hypoxia limits this hydroxylation, thereby precluding the binding of pVHL and leading to the stabilization of HIF-1α (36). Since HIF-1α in an ARNT-deficient Hepa1C4 cell line was also induced by hypoxia, ARNT is thought to be unnecessary for the hypoxic stabilization of HIF-1α protein (31). Under aerobic conditions, however, ARNT seems to participate in the stabilization of HIF-1α. Recently, Isaacs et al. (37) demonstrated that ARNT or its bHLH PAS domain peptide, binds with the bHLH PAS domain of HIF-1α, and that this results in the stabilization of HIF-1α under aerobic conditions. This strongly supports our finding that ARNT stabilizes HIF-1α417 protein. Although the stability of HIF-1α417 is not controlled by oxygen due to its lack of ODDD, HIF-1α417 nevertheless appears to be degraded by a proteasomal system regardless of oxygen tension because its level was enhanced after treatment with the proteasome inhibitor MG132 (Figure 3C). Moreover, in the presence of ARNT, the HIF-1α417 level was markedly enhanced (Figure 6C) and its half-life prolonged (Figure 6D). Therefore, there may be a novel degradation mechanism in targeting the bHLH PAS domain, whereby ARNT prevents HIF-1α417 from degradation by masking the domain. In addition, the counter effect of HIF-1α417 on ARNT was also evaluated by western blotting (Figure 6B) and fluorescence microscopy (Figure 6E). Both methods clearly revealed that HIF-1α417 enhanced the nuclear translocation of ARNT. Therefore, the reciprocal interaction between ARNT and HIF-1α417 appears to accelerate the heterodimerization of these proteins in the nucleus.
Heterodimerization with ARNT is obligatory for DNA binding by HIF-1/2alpha, SIM1/2, Trachealess (TRH, a Drosophila bHLH PAS protein), and AhR. Interestingly, the bHLH PAS proteins contain the conserved structure, KEKS(RD/KN)AAR(S/T)RR, in their basic regions, which binds DNA. Not surprisingly then, bHLH PAS proteins bind to similar DNA motifs, which contain 5′-RCGTG-3′ core sequence. Therefore, a close relationship between the DNA binding regions of the bHLH PAS proteins could constitute an overlapping of transcriptional functions. Indeed, the SIM1–ARNT complex stimulated the activity of the Epo enhancer, which is a target of HIF-1α–ARNT (23). As another example, TRH–ARNT stimulated the activity of the CNS midline enhancer, a target of SIM1–ARNT (38). In Drosophila, ectopic HIF-1α expression induced the expression of the BREATHLESS gene, a target of TRH–ARNT (39). This supports the possibility that the HIF-1α–ARNT dimer binds to the TRH–ARNT binding site of the BREATHLESS gene. Therefore, we cannot rule out the possibility that the HIF-1α417–ARNT dimer, as HIF-1 does, could cross-target genes that are regulated by other bHLH PAS protein–ARNT dimers. In the present study, however, little can be said of the exact physiological role of HIF-1α417.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr Eric Huang (NCI, USA) for generously donating essential plasmids and for his kind discussion. This work was supported by a grant (R04-2002-000-20006-0) from the basic research program of the Korea Science and Engineering Foundation and by the Seoul National University College of Medicine Research Fund 2003.
REFERENCES
- 1.Gu Y.Z., Hogenesch,J.B. and Bradfield,C.A. (2000) The PAS superfamily: sensors of environmental and developmental signals. Annu. Rev. Pharmacol. Toxicol., 40, 519–561. [DOI] [PubMed] [Google Scholar]
- 2.Ellenberger T., Fass,D., Arnaud,M. and Harrison,S.C. (1994) Crystal structure of transcription factor E47: E-box recognition by a basic region helix-loop-helix dimer. Genes Dev., 8, 970–980. [DOI] [PubMed] [Google Scholar]
- 3.Kewley R.J., Whitelaw,M.L. and Chapman-Smith,A. (2004) The mammalian basic helix-loop-helix/PAS family of transcriptional regulators. Int. J. Biochem. Cell Biol., 36, 189–204. [DOI] [PubMed] [Google Scholar]
- 4.Reyes H., Reisz-Porszasz,S. and Hankinson,O. (1992) Identification of the Ah receptor nuclear translocator protein (Arnt) as a component of the DNA binding form of the Ah receptor. Science, 256, 1193–1195. [DOI] [PubMed] [Google Scholar]
- 5.Matsushita N., Sogawa,K., Ema,M., Yoshida,A. and Fujii-Kuriyama,Y. (1993) A factor binding to the xenobiotic responsive element (XRE) of P-4501A1 gene consists of at least two helix-loop-helix proteins, Ah receptor and Arnt. J. Biol. Chem., 268, 21002–21006. [PubMed] [Google Scholar]
- 6.Mimura J. and Fujii-Kuriyama,Y. (2003) Functional role of AhR in the expression of toxic effects by TCDD. Biochim. Biophys. Acta., 1619, 263–268. [DOI] [PubMed] [Google Scholar]
- 7.Jiang B.H., Rue,E., Wang,G.L., Roe,R. and Semenza,G.L. (1996) Dimerization, DNA binding, and transactivation properties of hypoxia-inducible factor 1. J. Biol. Chem., 271, 17771–17778. [DOI] [PubMed] [Google Scholar]
- 8.Ema M., Taya,S., Yokotani,N., Sogawa,K., Matsuda,Y. and Fujii-Kuriyama,Y. (1997) A novel bHLH-PAS factor with close sequence similarity to hypoxia-inducible factor 1alpha regulates the VEGF expression and is potentially involved in lung and vascular development. Proc. Natl Acad. Sci. USA, 94, 4273–4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Probst M.R., Fan,C.M., Tessier-Lavigne,M. and Hankinson,O. (1997) Two murine homologs of the Drosophila single-minded protein that interact with the mouse aryl hydrocarbon receptor nuclear translocator protein. J. Biol. Chem., 272, 4451–4457. [DOI] [PubMed] [Google Scholar]
- 10.Chin M.T., Maemura,K., Fukumoto,S., Jain,M.K., Layne,M.D., Watanabe,M., Hsieh,C.M. and Lee,M.E. (2000) Cardiovascular basic helix loop helix factor 1, a novel transcriptional repressor expressed preferentially in the developing and adult cardiovascular system. J. Biol. Chem., 275, 6381–6387. [DOI] [PubMed] [Google Scholar]
- 11.Semenza G.L. (2001) HIF-1 and mechanisms of hypoxia sensing. Curr. Opin. Cell Biol., 13, 167–171. [DOI] [PubMed] [Google Scholar]
- 12.Wang G.L. and Semenza,G.L. (1995) Purification and characterization of hypoxia-inducible factor-1. J. Biol. Chem., 270, 1230–1237. [DOI] [PubMed] [Google Scholar]
- 13.Reisz-Porszasz S., Probst,M.R., Fukunaga,B.N. and Hankinson,O. (1994) Identification of functional domains of the aryl hydrocarbon receptor nuclear translocator protein (ARNT). Mol. Cell Biol., 14, 6075–6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jain S., Dolwick,K.M., Schmidt,J.V. and Bradfield,C.A. (1994) Potent transactivation domains of the Ah receptor and the Ah receptor nuclear translocator map to their carboxyl termini. J. Biol. Chem., 269, 31518–31524. [PubMed] [Google Scholar]
- 15.Fukunaga B.N., Probst,M.R., Reisz-Porszasz,S. and Hankinson,O. (1995) Identification of functional domains of the aryl hydrocarbon receptor. J. Biol. Chem., 270, 29270–29278. [DOI] [PubMed] [Google Scholar]
- 16.Ko H.P., Okino,S.T., Ma,Q. and Whitlock,J.P.,Jr (1996) Dioxin-induced CYP1A1 transcription in vivo: the aromatic hydrocarbon receptor mediates transactivation, enhancer-promoter communication, and changes in chromatin structure. Mol. Cell Biol., 16, 430–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H., Ko,H.P. and Whitlock,J.P. (1996) Induction of phosphoglycerate kinase 1 gene expression by hypoxia. Roles of Arnt and HIF1-alpha. J. Biol. Chem., 271, 21262–21267. [DOI] [PubMed] [Google Scholar]
- 18.Sogawa K., Nakano,R., Kobayashi,A., Kikuchi,Y., Ohe,N., Matsushita,N. and Fujii-Kuriyama,Y. (1995) Possible function of Ah receptor nuclear translocator (Arnt) homodimer in transcriptional regulation. Proc. Natl Acad. Sci. USA, 92, 1936–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swanson H.I. and Yang,J.H. (1999) Specificity of DNA binding of the c-Myc–Max and ARNT/ARNT dimers at the CACGTG recognition site. Nucleic Acids Res., 27, 3205–3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huffman J.L., Mokashi,A., Bachinger,H.P. and Brennan,R.G. (2001) The basic helix-loop-helix domain of the aryl hydrocarbon receptor nuclear transporter (ARNT) can oligomerize and bind E-box DNA specifically. J. Biol. Chem., 276, 40537–40544. [DOI] [PubMed] [Google Scholar]
- 21.Ema M., Morita,M., Ikawa,S., Tanaka,M., Matsuda,Y., Gotoh,O., Saijoh,Y., Fujii,H., Hamada,H., Kikuchi,Y. and Fujii-Kuriyama,Y. (1996) Two new members of the murine Sim gene family are transcriptional repressors and show different expression patterns during mouse embryogenesis. Mol. Cell Biol., 16, 5865–5875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moffett P. and Pelletier,J. (2000) Different transcriptional properties of mSim-1 and mSim-2. FEBS Lett., 466, 80–86. [DOI] [PubMed] [Google Scholar]
- 23.Woods S.L. and Whitelaw,M.L. (2002) Differential activities of murine single minded 1 (SIM1) and SIM2 on a hypoxic response element. Cross-talk between basic helix-loop-helix/per-Arnt-Sim homology transcription factors. J. Biol. Chem., 277, 10236–10243. [DOI] [PubMed] [Google Scholar]
- 24.Chun Y.S., Choi,E., Kim,G.T., Lee,M.J., Lee,M.J., Lee,S.E., Kim,M.S. and Park,J.W. (2000) Zinc induces the accumulation of hypoxia-inducible factor HIF-1alpha, but inhibits the nuclear translocation of HIF-1beta, causing HIF-1 inactivation. Biochem. Biophys. Res. Commun., 268, 652–656. [DOI] [PubMed] [Google Scholar]
- 25.Iyer N.V., Leung,S.W. and Semenza,G.L. (1998) The human hypoxia-inducible factor-1alpha gene: HIF1A structure and evolutionary conservation. Genomics, 52, 159–165. [DOI] [PubMed] [Google Scholar]
- 26.Chun Y.S., Lee,K.H., Choi,E., Bae,S.Y., Yeo,E.J., Huang,L.E., Kim,M.S. and Park,J.W. (2003) Phorbol ester stimulates the nonhypoxic induction of a novel hypoxia-inducible factor-1alpha isoform: implications for tumor promotion. Cancer Res., 63, 8700–8707. [PubMed] [Google Scholar]
- 27.Gothie E., Richard,D.E., Berra,E., Pages,G. and Pouyssegur,J. (2000) Identification of alternative spliced variants of human hypoxia-inducible factor-1alpha. J. Biol. Chem., 275, 6922–6927. [DOI] [PubMed] [Google Scholar]
- 28.Chun Y.S., Choi,E., Yeo,E.J., Lee,J.H., Kim,M.S. and Park,J.W. (2001) A new HIF-1 alpha variant induced by zinc ion suppresses HIF-1-mediated hypoxic responses. J. Cell Sci., 114, 4051–4061. [DOI] [PubMed] [Google Scholar]
- 29.Chun Y.S., Choi,E., Kim,T.Y., Kim,M.S. and Park,J.W. (2002) A dominant-negative isoform lacking exons 11 and 12 of the human hypoxia-inducible factor-1alpha gene. Biochem. J., 362, 71–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maxwell P.H., Wiesener,M.S., Chang,G.W., Clifford,S.C., Vaux,E.C., Cockman,M.E., Wykoff,C.C., Pugh,C.W., Maher,E.R. and Ratcliffe,P.J. (1999) The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature, 399, 271–275. [DOI] [PubMed] [Google Scholar]
- 31.Chilov D., Camenisch,G., Kvietikova,I., Ziegler,U., Gassmann,M. and Wenger,R.H. (1999) Induction and nuclear translocation of hypoxia-inducible factor-1 (HIF-1): heterodimerization with ARNT is not necessary for nuclear accumulation of HIF-1alpha. J. Cell Sci., 112, 1203–1212. [DOI] [PubMed] [Google Scholar]
- 32.Forsythe J.A., Jiang,B.H., Iyer,N.V., Agani,F., Leung,S.W., Koos,R.D. and Semenza,G.L. (1996) Activation of vascular endothelial growth factor gene transcription by hypoxia-inducible factor-1. Mol. Cell Biol., 16, 4604–4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee J.W., Bae,S.H., Jeong,J.W., Kim,S.H. and Kim,K.W. (2004) Hypoxia-inducible factor (HIF-1) alpha: its protein stability and biological functions. Exp. Mol. Med., 36, 1–12. [DOI] [PubMed] [Google Scholar]
- 34.Makino Y., Kanopka,A., Wilson,W.J., Tanaka,H. and Poellinger,L. (2002) Inhibitory PAS domain protein (IPAS) is a hypoxia-inducible splicing variant of the hypoxia-inducible factor-3alpha locus. J. Biol. Chem., 277, 32405–32408. [DOI] [PubMed] [Google Scholar]
- 35.Makino Y., Cao,R., Svensson,K., Bertilsson,G., Asman,M., Tanaka,H., Cao,Y., Berkenstam,A. and Poellinger,L. (2001) Inhibitory PAS domain protein is a negative regulator of hypoxia-inducible gene expression. Nature, 414, 550–554. [DOI] [PubMed] [Google Scholar]
- 36.Masson N. and Ratcliffe,P.J. (2003) HIF prolyl and asparaginyl hydroxylases in the biological response to intracellular O(2) levels. J. Cell Sci., 116, 3041–3049. [DOI] [PubMed] [Google Scholar]
- 37.Isaacs J.S. and Jung,Y.J. (2004) Aryl hydrocarbon nuclear translocator (ARNT) promotes oxygen-independent stabilization of hypoxia-inducible factor-1alpha by modulating an Hsp90-dependent regulatory pathway. J. Biol Chem., 279, 16128–16135. [DOI] [PubMed] [Google Scholar]
- 38.Ohshiro T. and Saigo,K. (1997) Transcriptional regulation of breathless FGF receptor gene by binding of TRACHEALESS/dARNT heterodimers to three central midline elements in Drosophila developing trachea. Development, 124, 3975–3986. [DOI] [PubMed] [Google Scholar]
- 39.Zelzer E., Wappner,P. and Shilo,B.Z. (1997) The PAS domain confers target gene specificity of Drosophila bHLH/PAS proteins. Genes Dev., 11, 2079–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]