Abstract
Multi-conformation continuum electrostatics (MCCE) was used to analyze various structures of the NS3 RNA helicase from the hepatitis C virus in order to determine the ionization state of amino acid side chains and their pKas. In MCCE analyses of HCV helicase structures that lacked ligands, several active site residues were identified to have perturbed pKas in both the nucleic acid binding site and in the distant ATP-binding site, which regulates helicase movement. In all HCV helicase structures, Glu493 was unusually basic and His369 was abnormally acidic. Both these residues are part of the HCV helicase nucleic acid binding site, and their roles were analyzed by examining the pH profiles of site-directed mutants. Data support the accuracy of MCCE predicted pKa values, and reveal that Glu493 is critical for low pH enzyme activation. Several key residues, which were previously shown to be involved in helicase-catalyzed ATP hydrolysis, were also identified to have perturbed pKas including Lys210 in the Walker-A motif and the DExD/H-box motif residues Asp290 and His293. When DNA was present in the structure, the calculated pKas shifted for both Lys210 and Asp290, demonstrating how DNA binding might lead to electrostatic changes that stimulate ATP hydrolysis.
INTRODUCTION
Helicases are molecular motors that move along nucleic acid chains to separate complementary strands, eliminate secondary structures, or displace nucleic acid binding proteins. The energy for helicase movement arises from the hydrolysis of ATP to ADP and inorganic phosphate. Helicases are classified into four large superfamilies based on genetic similarities (1), and many of them function as ring-like oligomers that encircle DNA (2), whereas others do not form rings. There is controversy over whether the non-ring helicases function as monomers or dimers. Generally, the non-ring helicases are members of genetic superfamilies I (SFI) and II (SFII), whereas the ring helicases are members of superfamilies III (SFIII) and IV (SFIV). Both ring and non-ring helicases share ATP-binding domains that resemble one, first seen in the RecA protein (3). In ring helicases, such as the T7 gene 4 protein (4–6), each subunit possesses a single RecA-like domain, whereas in non-ring helicases, such as Bacillus stearothermophilus PcrA (7–9) and the helicase encoded by the hepatitis C virus (HCV) (10–13), two RecA-like domains are present in each protein monomer.
Most models that explain how the chemical energy of ATP hydrolysis is converted to physical helicase movement involve conformational changes in the RecA-like domains that in turn produce a mechanical force leading to protein translocation (14). However, in a recent study on the pH dependence of the HCV helicase, Lam et al. (15) noted that the affinity of an enzyme–ATP complex for RNA varied over 100-fold in the physiological pH range, which suggests that electrostatic forces might also play pivotal roles in regulating helicase movement. Lam et al. proposed that the rotation of a negatively charged region towards RNA leads to the enzyme propelling itself along the nucleic acid backbone (15). Such an electromechanical model for helicase action, which is analogous to one recently proposed to explain the properties of myosin (16), is presently difficult to test because little is known about the electrostatic properties of helicases or the ionization state of their amino acid side chains.
In this study, we have performed electrostatic potential calculations using four previously published HCV helicase structures. The goal was to identify key amino acids that are critical for helicase–substrate interactions by using the software package Multi-conformation continuum electrostatics (MCCE) (17,18), which calculates the ionization states of all protein side chains in all possible conformations at various pH and uses this information to determine a theoretical pKa for each residue. By examining this data we sought to identify residues whose pKas were shifted relative to those of similar amino acids in solution. Residues with perturbed pKas frequently participate in enzymatic reactions because their protons are more available for acid–base catalysis or hydrogen bonding at physiological pH. This procedure is similar to the one recently described by Ondrechen et al. (19) called theoretical microscopic titration curves (THEMATICS), which uses a different software to generate titration curves. The MCCE method was chosen because it was found to accurately predict perturbed pKas that are often underestimated by other computational techniques. For example MCCE predicts a pKa for the active site glutamate in hen-egg-white lysozyme that is closer to the experimentally determined pKa than the pKas predicted by other programs (18).
While performing these calculations, we noticed that a large number of amino acid side chains in each structure had notably perturbed pKas, but only about one third of these perturbations would change the apparent ionization state of a residue in the biologically relevant pH range (pH 6–8). Upon DNA binding, electrostatic changes in perturbed residues were found not only in DNA binding sites but also in allosteric sites, which suggests that the ionization state of the protein side chains could be as important as major conformational changes in facilitating motor action.
To test the accuracy of the MCCE-predicted pKas, two ionizable amino acids in the RNA binding cleft of HCV helicase were changed by using site-directed mutagenesis, and the pH profiles of the altered proteins were examined. As mentioned above, the binding of RNA to an HCV helicase–ATP complex is highly pH dependent, and as a result HCV helicase rapidly unwinds RNA (or DNA) best at an acidic pH. Thus, an ionizable side chain with a pKa near 7 is probably critical for protein–nucleic acid interaction. Two obvious candidate residues are His369, which is about 3 Å from the phosphate backbone of DNA, and Glu493, which is about 6 Å from the DNA bases (11). Given this information alone, one would logically predict that the removal of the positive charge from His369 (with an expected pKa of 6) would lead to weaker binding to the negatively charged nucleic acid backbone. However, MCCE predicts a pKa of 4–4.6 (2.3 with DNA-bound) for His369, which suggests that it is always deprotonated between pH 6 and 8. On the other hand, MCCE predicts that Glu493 has an unusually high pKa at ∼7. A protonated Glu493 would lead to tighter binding at low pH by masking a negative charge that would repel RNA. Analysis of the mutant HCV helicases is consistent with the MCCE predictions and demonstrates that protonation of Glu493 below pH 7 leads to tighter nucleic acid binding and low-pH activation of HCV helicase.
MATERIALS AND METHODS
Reagents
The RNase-free reagents were purchased from Ambion (Austin, TX), and nucleotides and nucleic acids were treated with the RNAsecure reagent (Ambion). Poly(U) RNA with an average length of 2500 nt was purchased from Sigma (St. Louis, MO). DNA oligonucleotides were purchased from Integrated DNA Technologies (Coralville, IA), and their concentrations were determined from provided extinction coefficients.
Helicase structure selection, modification, and pKa calculations
There are presently four coordinate files deposited in the Protein Data Bank (PDB) that contain crystal structures of the HCV helicase. They have accession codes 1HEI (10), 1A1V (11), 8OHM (12) and 1CU1 (13). All these structures lack ligands except 1A1V, which contains a short oligonucleotide bound to the enzyme. Two of these studies, 1HEI and 1A1V, utilized virtually the same protein obtained from HCV genotype 1a. The other two structures utilized genotype 1b proteins. The coordinate files in 1HEI and 1CU1 each contain two molecules in the crystallographic asymmetric unit, whereas 1A1V and 8OHM contain only one each. Calculations were performed on each subunit in isolation because there is as yet no direct evidence that the interfaces seen in the files containing two helicase molecules are biologically relevant.
In cases where PDB files had missing side chain atoms, side chain reconstruction was performed by using Swiss PdbViewer, version 3.7 (20). Since the 1A1V PDB file contains coordinates for HCV helicase bound to a non-standard nucleotide sequence composed of deoxyuracil, the deoxyuracil was changed to deoxycytosine in order to allow MCCE to recognize the nucleotide sequence. The pKa calculations were then performed using the MCCE method described by Alexov and Gunner (17,21). The method involves the initial protonation of the protein structure followed by the generation of a library of protein conformers that contain amino acid residues of differing side chain positions. The program Delphi (22) is then used to calculate the electrostatics of each library conformer by using the Poisson–Boltzmann finite difference (PBFD) method. The PBFD method as implemented in Delphi, calculates the electrostatic potential of arbitrary shaped proteins given the coordinates of the protein's atoms. Pairwise Lennard–Jones interactions are then calculated between all conformers and added to the electrostatic interaction terms, determined in the Delphi runs. Monte Carlo sampling is then used to generate random protein microstates, and the free energy of each microstate was determined. The ionic strength that was used in the calculations was 0.15 M. The resultant free energy information is utilized in the determination of pKa values. MCCE and Delphi were obtained from Dr Barry Honig's laboratory and are available online at http://trantor.bioc.columbia.edu/. Calculated pKas were compared with the typical pKas of Asp = 3.65, Glu = 4.25, His = 6.0, Tyr = 10.06, Lys = 10.52, and Arg = 12.48 [data from The Merck Index, 13th edn (23)].
Mutagenesis
The quick-change site-directed mutagenesis kit (Stratagene, La Jolla, CA) was used to construct the altered helicase enzymes from the plasmid p33Helstop (24), which contains the helicase region of NS3 isolated from an infectious clone of HCV genotype 1a H77c (25) along with an N-terminal His-tag. The following oligonucleotides were utilized: H369K(+) 5′-GAC ATC TCA TCT TCT GCA AGT CAA AGA AGA AGT GCG-3′, H369K(−) 5′-CGC ACT TCT TCT TTG ACT TGC AGA AGA TGA GAT GTC-3′, H369A(+) 5′-GAC ATC TCA TCT TCT GCG CCT CAA AGA AGA AGT GCG, H369A(−) 5′-CGC ACT TCT TCT TTG AGG CGC AGA AGA TGA GAT GTC, E493K(+), 5′-CGT CCG TCC TCT GTA AGT GCT ATG ACG CGG GC-3′, E493K(−) 5′-GCC CGC GTC ATA GCA CTT ACA GAG GAC GGA CG-3′, E493Q(+) 5′-GAC TCG TCC GTC CTC TGT CAA TGC TAT GAC GCG GGC TGT G-3′, E493Q(−) 5′-CAC AGC CCG CGT CAT AGC ATT GAC AGA GGA CGG ACG AGT C-3′. The appropriate mutations were confirmed by sequencing the plasmid DNA. Recombinant proteins His–Hel, H396A, H369K, E493K, and E493Q were expressed and purified to apparent homogeneity as described previously (24).
Unwinding assay
To generate substrates for helicase assays, two synthetic oligonucleotides were annealed by heating them to 95°C and allowing them to cool slowly to room temperature. Before annealing, the shorter strand was [32P]-labeled by using polynucleotide kinase. The DNA substrate consisted of a shorter DNA oligonucleotide 5′-[32P]-GCC TCG CTG CCG TCG CCA-3′ annealed to a longer DNA oligonucleotide 5′-TGG CGA CGG CAG CGA GGC TTT TTT TTT TTT TTT TTT TT-3′. The DNA substrate (1 nM) and 100 nM HCV helicase were incubated in reaction buffer (25 mM MOPS, 3 mM MgCl2, and 0.1% Tween-20) for 30 min. Reactions were initiated by the addition of 5 mM ATP, terminated with EDTA after 10 min, and analyzed by non-denaturing PAGE as described previously (24,26,27).
ATPase assay
Reactions were performed at 37°C in 25 mM MOPS buffer at various pH, 3 mM MgCl2, 0.1% Tween-20, 5 mM ATP, and helicase. To determine the constant KRNA, defined as the concentration of nucleic acids that supports a half maximum rate of catalysis, reactions were performed in the presence of 4 mM ATP with various concentrations of poly(U) RNA (average length 2500 nt). Data were fit into Equation 1,
![]()
where vo is the initial rate of ATP hydrolysis, [NA] is the nucleic acid concentration (expressed as total nucleotide concentration), and [E]T is the total molar concentration of enzyme monomers.
RESULTS
This study was initiated to examine the electrostatic properties of HCV helicase in order to examine the ionic interactions between the enzyme, ATP, and its nucleic acid substrates. The goal was to answer three questions. First, can electrostatic analysis identify active site residues in helicase structures that lack any ligands? Second, does nucleic acid binding influence the proton affinity of side chains in the ATP binding site? Third, are absolute values for pKas calculated in silico accurate? The HCV helicase studied here was selected because numerous coordinate data files for it are available, both in the presence and absence of DNA, and because HCV helicase has been recently shown to unwind duplex nucleic acids in a pH dependent manner (15). This pH effect allowed us to test the accuracy of the predictions using site-directed mutagenesis.
MCCE structure analysis
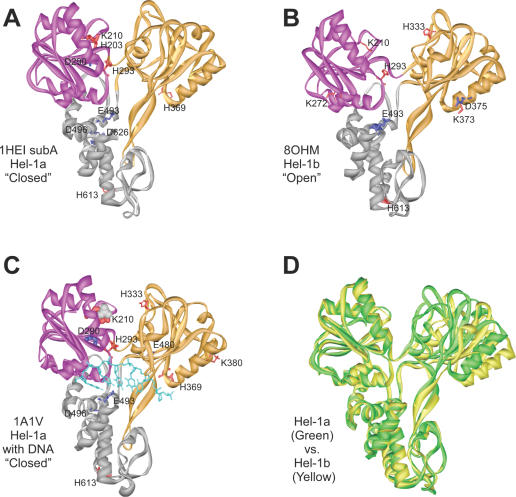
Only PDB file 1A1V contains coordinates for HCV helicase that is bound to an oligonucleotide. Five structures of the HCV helicase in the absence of ligands are present in the Protein Data Bank. Two of the structures are found in the coordinate file 1HEI, which contains two crystallographically independent molecules (subunits A and B). 1CU1 also contains two molecules in the unit cell, and the fifth structure is in the file 8OHM. The five structures differ primarily with regard to the rotation of domain 2 of the helicase, which can pivot relative to domains 1 and 3 (10). The protein can be described to be in an ‘open’ conformation when the cleft separating domains 1 and 2 is the widest and in a ‘closed’ conformation when the cleft is narrow (Figure 1D). By this definition, subunit A in file 1HEI, subunits A and B in 1CU1, and the protein in 1A1V are in a closed conformation, while subunit B in 1HEI and the protein in 8OHM are both in an open conformation.
Figure 1.
HCV helicase amino acids with perturbed pKas. The amino acids listed in Table 1 with pKas perturbed by over 2 pH units are shown as sticks on the corresponding helicase structure. Residues with higher than expected pKas are colored blue and residues with pKas more acidic than expected are colored red. The stick width corresponds to the magnitude of the perturbation undergone by the residue. Structurally similar RecA-like domains 1 and 2 are colored purple and orange respectively. (A) HCV helicase from genotype 1a (Hel-1a) without DNA in the closed conformation (PDB file 1HEI subunit A). (B) HCV helicase from genotype 1B (Hel-1b) in the open conformation (PDB file 8OHM) (C) Hel-1a with DNA (PDB file 1A1V). DNA ligands are colored light blue, and in this structure, a sulfate ion (gray, space-fill) occupies the ATP binding site. (D) Comparison of the closed and open HCV helicase conformations. The structures of Hel-1a (green) and Hel-1b (yellow) are aligned along domains 1 and 3 to show the relative rotation of domain 2.
When the individual coordinate files for HCV helicase were analyzed using MCCE, significantly perturbed pKas were found for a surprisingly high number of residues. A significantly perturbed pKa is defined here as one that is altered more than 2 pH units from its typical value (28). About 20% of all ionizable HCV helicase residues had pKas that were perturbed by over 2 pH units. Furthermore, for ∼10–20% of the ionizable residues in each coordinate file, MCCE did not report a pKa because all conformations of the residues were either ionized or not ionized at all. Identical pKas were reported by MCCE when datasets were repeatedly analyzed.
The majority of the perturbed residues were acids that were more acidic or bases that were more basic and would therefore not change the ionization state of the residues at neutral pH. Therefore, we devised a simple procedure to screen out significantly perturbed pKas that would not influence a residue's net charge under biological conditions. In the screening procedure, Asp and Glu residues whose pKas increased by 2 pH units, Tyr, Arg or Lys residues whose pKa were 2 pH units lower than expected, and all His residues that had pKas perturbed by more than 2 pH units (in either direction), were selected. After screening, ∼10% of the ionizable amino acid side chains in each structure were found to have ‘physiologically’ perturbed pKas.
The residues identified in 1HEI (subunit A), 8OHM, and 1A1V are listed in Table 1, and their locations are noted in Figure 1. These three structures represent the protein without DNA in a closed conformation (1HEI subunit A), in an open conformation (8OHM), and in a closed conformation with DNA bound (1A1V). Also noted in Table 1 are comments regarding the location of the perturbed residue, whether it is part of a known helicase conserved sequence signature motif (29), and references to studies verifying its role in helicase action using site-directed mutagenesis.
Table 1. HCV helicase residues with unusual ionization states at physiological pH.
| 1HEI subA Hel-1a ‘closed’ pKa | 8OHM Hel-1b ‘open’ pKa | 1A1V Hel-1a:DNA ‘closed’ pKa | Comments | |
|---|---|---|---|---|
| His203 | 0.8 | 5.3 | 5.5 | SFII helicase motif I:HAPT |
| Lys210 | 4.9 | 8.1 | 1.7 | SFII helicase motif I:GSGK |
| K210A (37–39,50) | ||||
| K210Q (37) | ||||
| K210N (36) | ||||
| K210E (40,41) | ||||
| Lys272 | 8.6 | 8.4 | 10.3 | TxGx motif (33) |
| Asp290 | 7.7 | 3.4 | 10.3 | SFII helicase motif II: DECH |
| D290A (38,39) | ||||
| D290N (36) | ||||
| His293 | 2.2 | 3.7 | 2.5 | SFII helicase motif II:DECH |
| H293A (36,37) | ||||
| H293K (36) | ||||
| H293Q (36) | ||||
| His333 | 4.8 | 3.6 | 3.7 | Conserved in all HCV isolates (27) |
| His369 | 4.0 | 4.6 | 2.3 | SFII helicase motif IV: HSKKKCD |
| H369A, H369K (this study) | ||||
| Lys373 | 9.9 | 8.0 | 8.8 | SFII helicase motif IV: HSKKKCD |
| Conserved in all HCV isolates (27) | ||||
| Asp375 | 5.0 | 6.4 | 4.2 | SFII helicase motif IV: HSKKKCD |
| Lys380 | 10.7 | 10.1 | 8.4 | Conserved in all HCV isolates (27) |
| Glu480 | 4.0 | 3.9 | 8.0 | Conserved in all HCV isolates (27) |
| Glu493 | 6.8 | 7.1 | 7.6 | pH dependent- DNA binding |
| E493Q, E493K (this study) | ||||
| Asp496 | 5.7 | 4.4 | 5.9 | Conserved in all HCV isolates (27) |
| His613 | 3.5 | 3.5 | 3.4 | |
| Asp626 | 10.3 | n.d. | n.d. |
The pKas for the enzyme without ligands in the closed conformation were calculated by using the coordinates for subunit A in PDB file 1HEI. The values for HCV helicase in the open conformation were obtained by analyzing file 8OHM, and values for the DNA bound enzyme were obtained using coordinates in PDB file 1A1V. Known roles and/or site-directed mutants of each residue are noted in the last column. Major changes are underlined. The MCCE program did not determine a pKa for residues labeled n.d.
In all HCV helicase structures that lacked nucleic acid, MCCE analysis identified several amino acid side chains with perturbed pKas that are near DNA in 1A1V. Notably, in all datasets, residue Glu493 had an unusually high pKa, and residue His369 had an unusually low pKa. As seen in the structure of HCV helicase co-crystallized with ssDNA, His369 is most intimately associated with DNA and lies about 3 Å from the nucleic acid backbone (Figure 2A). Glu493 and Asp496 are part of a helix that links domain 2 with a critical hydrophobic residue (Trp501), which stacks between the nucleic acid bases and is thought to act like a bookend, and prevents the helicase from sliding in the absence of ATP (30–32). When the protein assumes a more open conformation, a few more residues are perturbed in the DNA binding cleft (Figure 1B). These residues include Lys272, Lys373 and Asp375. Lys373 and Asp375 are near His369 and part of the helicase motif IV. Lys272 is the fourth residue in a conserved TxGx motif whose conserved Thr contacts the DNA backbone (33).
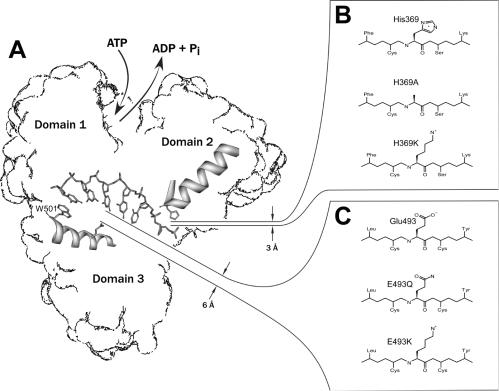
Figure 2.
Site-directed mutagenesis of HCV helicase. (A) Diagram showing the locations of Glu493 and His369 in the DNA binding cleft. (B) Substitutions made for His369. (C) Substitutions made for Glu493.
In each dataset, several amino acids in conserved helicase motifs, which cluster around the putative ATP-binding cleft between domains 1 and 2, were also found to have perturbed pKas (Table 1, Figure 1). In the closed conformation that lack ligands (1HEI subunit A), three key residues in the putative ATP binding site were identified by MCCE analysis: Lys210, Asp290, and His293. Although a structure of HCV helicase with an ATP analogue has not yet been reported, alignments with other helicases co-crystallized with nucleotides [such as PcrA (8,9)] indicate that these residues interact intimately with ATP. Lys210 is a key residue in the A motif of the classical nucleotide binding site that was first described by John Walker (34). Asp290 is part of the Walker B motif (34) and is the first residue of the common DExD/H-box sequence (35). His293 is the last residue in the DExD/H-box.
Although the pKa of His293 was similar in all HCV helicase structures that were analyzed, pKas for both Lys210 and Asp290 were not. When DNA was bound in structure 1A1V, Lys210 was even more acidic than it was in 1HEI subunit A, and Asp290 was more basic (Table 1). On the other hand, when the protein assumed an open conformation, both Lys210 and Asp290 had pKas that were much closer to ‘normal’. In the open structures such as that seen in 8OHM, Lys210 was appropriately basic and Asp290 was more acidic than it was in any closed structure (Table 1).
The results for subunit B of 1HEI and 1CU1 are not reported here in detail. Both datasets revealed perturbed pKas similar to those in Table 1 for both His369 and Glu493. Most pKas that were predicted by MCCE for 1HEI subunit B, which is in the open conformation, resembled those predicted for 8OHM. For example, in MCCE results for subunit B of 1HEI, Lys210 had a pKa of 9.8, and Asp290 had a pKa of 5.0. However, 1HEI lacks amino acids 233–261 in domain 1, and this difference could affect the reliability of the MCCE results. Similarly, the MCCE results for 1CU1 could be skewed by the fact that the protein in 1CU1 differs significantly from the others. Instead of consisting of only the helicase portion of the NS3, the protein in 1CU1 consists of the entire NS3 protein with its serine protease covalently tethered to part of the NS4A protease cofactor. Since the protease domain affects rates of helicase action and ATP hydrolysis (24), comparison of the results obtained from 1CU1 with the others in Table 1 might not be reasonable. It should be noted, however, that some additional perturbed amino acids were found in the protease domain (in 1CU1) including His57 (pKa = 1.3), which is part of the catalytic triad of the NS3 serine protease active site.
Site-directed mutagenesis to test pKa prediction accuracy
We attempted to test the accuracy of some of the pKas predicted by MCCE by examining the pH dependence of helicase activity. Previously, we showed that when pH is lowered from 7.5, the amount of DNA (or RNA) that is needed to stimulate the ATPase activity of HCV helicase to its maximum decreased by over 200-fold (15). This effect correlates with a similar decrease in the enzyme's affinity for ssDNA in the presence of the non-hydrolysable ATP analog ADP(BeF3), which suggests that the residues involved in low-pH activation probably interact with ssDNA. The diagram in Figure 2 shows two ionizable residues in the ssDNA-binding cleft that could be involved in activation. One is in domain 2, His369, and the other is in domain 3, Glu493. Whereas His369 is positioned so that it can form a hydrogen bond with the phosphate backbone, Glu493 is more distant, but could directly interact more with DNA if the protein changes conformation upon ATP binding (Figure 2). Of the two residues, His369 seems more likely to be critical for activation because His369 is closer to DNA than Glu493 and because histidines are normally less acidic than glutamates. However, MCCE predicts pKas of 4 (1HEIsubA) and 2.3 (1A1V) for His369 and pKas of 6.8 (1HEIsubA) and 7.6 (1A1V) for Glu493, which suggests that Glu493 would be the activating residue.
If His369 is the activating residue, then its protonation would permit tighter DNA binding at a low pH. Tighter binding would result from either formation of additional hydrogen bonds or ionic attraction of the positively charged histidine to the negatively charged DNA backbone. To test this hypothesis, His369 was changed to either an uncharged alanine (H369A) or a lysine (H369K), which should retain a positive charge at all pHs below 10. If His369 were critical for low-pH activation, then H369A should be inactive from pH 6.5 to 7.5, while H369K should be active throughout the entire pH range.
Alternatively, if Glu493 is responsible for the low-pH activation of HCV helicase, then its protonation would help attract DNA. In other words, a negatively charged form would repel DNA whereas its neutral form would allow for the tighter binding that is necessary for processive DNA unwinding. To test this idea, Glu493 was changed to either a glutamine, E493Q, or a lysine, E493K. If titration of 493 is the cause of activation, then E493Q and E493K should retain activity at an elevated pH.
The above mutations were made in a recombinant HCV helicase that was isolated from the HCV genotype 1a infectious clone H77C (25). In this helicase construct, the protease domain is replaced with an N-terminal poly-histidine tag (24). After the four mutant proteins were purified, their ability to unwind DNA was compared with that of the parent protein, designated here as His–Hel. In general, mutation of Glu493 led to a helicase that unwound duplex nucleic acids faster than the wild type even at a high pH, where the wild type was not active. Conversely, His369 mutants were pH sensitive like the wild type.
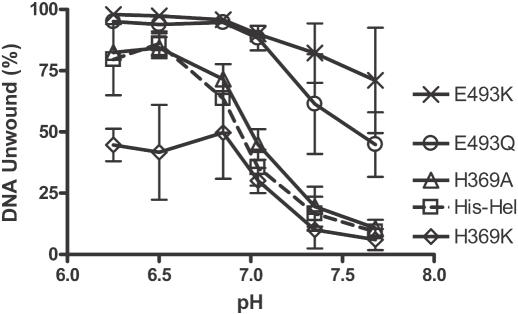
In the typical unwinding experiment shown in Figure 3, changing His369 to alanine (H369A) seemed to have no apparent effect at any pH. However, changing His369 to a positively charged lysine caused the enzyme to unwind more slowly at a low pH. At higher pH, like the wild type and H369A, H369K lost most of its ability to unwind DNA. In contrast, when a possible negative charge is eliminated from position 493, the DNA unwinding activity is clearly less pH-dependent than the wild type. Unlike the wild-type protein (His–Hel), both E493Q and E493K retain most of their ability to unwind DNA above pH 7. Similar results were obtained when these experiments were repeated, either with different length duplex substrates or in the presence of an enzyme trap (i.e. single turnover conditions).
Figure 3.
Activity of the HCV helicase and site-directed mutants at various pH. DNA unwinding assays were performed in MOPS buffers containing either His–Hel (squares), H369A (triangles), H369K (diamonds), E493Q (circles), or E493K (×). All reactions contained 1 nM of duplex [32P]DNA substrate, 100 nM protein, were initiated with ATP, incubated at 23°C for 10 min, terminated, and analyzed by using native polyacrylamide gels. The average rates obtained from three separate reactions are shown with the standard deviations as error bars.
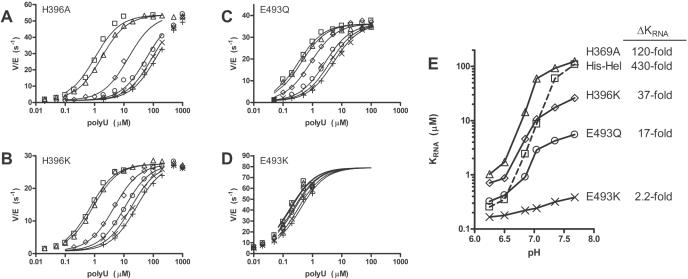
It is difficult to pinpoint exactly which step is pH dependent because the unwinding reaction is very complex. Nevertheless, based on previous work (15), we suspect that at a low pH, the helicase-ATP complex binds nucleic acids more tightly than it does at a high pH. To determine the pH dependence of the affinity of a helicase–ATP complex for RNA, the initial rates of ATP hydrolysis were measured at different concentrations of RNA and at different pH (Figure 4). The concentration of RNA leading to 50% ATPase stimulation, defined as KRNA, was then determined from each titration and plotted versus pH for His–Hel and the four mutants (Figure 4E). Previously, we have shown that KRNA determined in this manner, correlates with an equilibrium binding constant of a helicase–ATP complex for RNA, as determined in direct binding experiments with a non-hydrolyzable ATP analog (15).
Figure 4.
Effect of RNA and DNA on HCV helicase catalyzed ATP hydrolysis at different pH. (A–D) Initial rates of ATP hydrolysis at various concentrations of poly(U) RNA (average length 2500 nt). Reactions were performed in MOPS buffers at pH 6.25 (squares), pH 6.5 (triangles), pH 6.85 (diamonds), pH 7.04 (circles), pH 7.35 (×), and pH 7.68 (+). The enzyme used was either H369A (A), H369K (B), E493Q (C), or E493K (D). The data were fit into Equation 1 to determine the concentration necessary to support a half maximum rate of ATP hydrolysis (KRNA). (E) Each titration was repeated two to five times, and the average KRNAs determined at each pH for either His–Hel (squares), H369A (triangles), H369K (diamonds), E493Q (circles), or E493K (×) are plotted. In repeat titrations, KRNAs varied by no more than 40%. Data for His–Hel were published previously (15).
The results show that changing His369 to Ala has the least effect on the pH dependence of KRNA. H369K has a lower KRNA than H369A, as would be expected from charge attraction to RNA, but the KRNA for H369K is still highly pH dependent, albeit 12-fold less dependent than wild type. Interestingly, at a low pH, H369K binds RNA 3-fold less tightly than wild type. Thus, it is unlikely that a positive charge at NS3 position 369 is needed to tightly tether the protein to nucleic acid or that His369 adopts a proton in this pH range. A pKa of 4 or below for His369, as was determined by MCCE, therefore seems quite reasonable.
In stark contrast, when a potential negative charge is removed from NS3 position 493 (E493Q) the KRNA is 25-fold less dependent on pH. At a low pH, His–Hel and E493Q bind RNA with similar affinities, which suggests that at pH 6.5, Glu493 is normally protonated and thus similar to a Gln. The remaining pH dependence of E493Q's KRNA can be explained by the fact that there are other acidic residues with unusually high pKas, in the vicinity, such as Asp496 (Table 1, Figure 1). When Glu493 is changed to a positively charged residue (E493K), KRNA is lower than that seen for the wild type (even at a low pH), and almost all pH dependence is eliminated. In this case, the positive charge at position 493 could help neutralize other negatively charged residues in the region. The data support an elevated pKa for Glu493 as was calculated by MCCE and identify Glu493 as a residue critical for the low-pH activation of the HCV helicase.
DISCUSSION
When the perturbed residues are mapped on the various helicase structures (Figure 1), two key points are immediately apparent. First, the MCCE program successfully predicted the DNA binding sites from protein structures that lacked DNA. We tested whether two of these perturbed amino acids were involved in nucleic acid binding by using site-directed mutagenesis. Our results confirmed the importance of His369 and Glu493 and support the accuracy of the MCCE predicted pKas. Second, electrostatic changes occur at distant sites upon ligand binding. For example, the pKas of residues in the ATP binding site change upon DNA binding. These allosteric effects might explain some aspects of the helicase mechanism, such as the stimulation of ATP hydrolysis when ssDNA binds the enzyme.
The identification of the importance of Glu493 for regulating low pH activation explains why this residue is conserved in all HCV genotypes that are sequenced to date [see figure 1 in ref. (27)]. Only one isolate has been reported to lack Glu at NS3 position 493. Interestingly, that isolate (Genbank accession no. AF207759), which contains an Asp at position 493, was obtained from a patient with remarkably less chronic liver damage than typical HCV patients. Although the data presented here supports the elevated pKa predicted by MCCE, it should be noted that the experiments were performed at an ionic strength considerably lower than that used in the calculations. This was necessary because the NS3 fragment that was utilized here lacks the protease domain and unwinds DNA poorly under normal saline conditions.
Although His369 was shown here not to be responsible for the low pH activation of HCV helicase, the data here confirm its intimate association with DNA. The data in Figure 4 show that in the presence of ATP, H369A binds RNA about four times weaker than the wild-type protein at optimal pH. H369K binds RNA 3-fold weaker at low pH, but ∼5-fold tighter than wild type at high pH. A similarly increased affinity of H369K for DNA was also noted in electrophoretic mobility shift assays (gel-shifts) that were conducted in the absence of ATP (A. Lam, unpublished results). We speculated whether the increased affinity of H369K for DNA under certain conditions could impede its translocation along nucleic acids. Alternatively, a Lys at position 369 could interact with a nearby charged residue (such as Lys371 or Lys372) that might be critical for helicase action, thereby causing H369K to unwind DNA more slowly than the wild type under optimal conditions.
In addition to identifying the nucleic acid binding site, MCCE analysis also successfully identified three residues that are known to play a key role in ATP binding and hydrolysis: Lys210, Asp290, and His293. The importance of all three has been extensively confirmed by using site-directed mutagenesis. In 1HEI subunit A, both Lys210 and His293 are abnormally acidic, and Asp290 is very basic, which suggests that they all are uncharged at neutral pH. However, there is considerable experimental evidence that ionizable residues are needed in these positions. Substitution of Lys210 with a non-ionizable residue results in a helicase that is unable to unwind DNA or hydrolyze ATP (36–39). On the other hand, substitution of Lys210 with an acidic side chain (K210E) yields a protein that, although lacks helicase activity, retains RNA-stimulated ATPase activity (40,41). Similarly, substitution of Asp290 with a non-ionizable residue results in an inactive helicase with no ability to hydrolyze ATP (36,38,39), and a virus with this substitution is no longer viable (42). Non-ionizable amino acids are better tolerated at position 293. H293A and H293Q mutants retain some ability to hydrolyze ATP (36,40), although with a Lys in this position (H293K) the enzyme hydrolyzes ATP almost half as well as the wild type.
Interestingly, DNA binding brings about changes in the ATP binding cleft. These allosteric effects can be seen by comparing the MCCE result subunit A of 1HEI with those obtained by using 1A1V (Table 1). Notably, these perturbations occurred in the Walker A motif residue Lys210, and the Walker-B motif (and DExD/H box) residue Asp290. The significance of this lies in the fact that DNA binding has been demonstrated to stimulate the ATPase activity of the helicase (43). It is not unprecedented that ligand binding leads to electrostatic changes at distant sites. For example, recent studies with aspartic proteases have revealed that proton uptake and release occurs at various distant sites upon inhibitor binding (44,45).
One simple explanation for the allosteric effect seen at the helicase ATP binding site is that DNA binding to the protein leads to a subtle rotation of domain 2 of the enzyme such that the electrostatic environment around the ATP binding cleft is altered. In support of this idea is the fact that when domain 2 rotates toward domain 1 (the closed conformation), the pKa of both Lys210 and Asp290 becomes perturbed (compare 1HEI subunit A and 8OHM, Table 1). Lys210 is significantly more acidic than normal both in the closed structure and when DNA is bound. Asp290 is more basic in the closed structure and when DNA is bound. The possibility that pKa differences between 1HEI subunit A and 8OHM could be due to the fact that the two proteins (Hel-1a and Hel-1b) were derived from different viral genotypes is unlikely, because pKas similar to those obtained with 8OHM were seen in subunit B of 1HEI, which represents Hel-1a in the open conformation (data not shown). We speculate that DNA stabilizes the closure of the cleft between domains 1 and 2, forcing Lys210 to become more acidic and Asp290 to become more basic. Thus, upon DNA binding (or cleft closure) Lys210 could lose a proton whereas Asp290 could gain a proton. Since Asp290 and Lys290 are only about 3–4 Å apart, proton transfer could occur even between the residues. The change in the ionization state of these two side chains could then accelerate the rate of ATP hydrolysis. For example, charge removal could reposition the catalytically essential Mg2+ that is needed to catalyze ATP hydrolysis. Alternatively, Asp290 could act as a catalytic base to extract a proton from the water molecule that subsequently engages in a nucleophilic attack of ATP.
We have tried to test this hypothesis by measuring the rates of HCV helicase-catalyzed ATP hydrolysis both in the presence and absence of DNA and RNA at various pH [see figure 4 in ref. (15)], but since rates are basically constant from pH 6 to 10, it is difficult to determine whether differences outside this range are due to ionization or protein stability. To examine this in more detail we planned to use site-directed mutagenesis to alter the electrostatic environment around Lys210 (and/or Asp290) so that its theoretical pKa (as determined by MCCE) is between pH 6 and 8. If such a protein then displays pH dependent ATP-hydrolysis, it could be very valuable for elucidating the precise mechanism of ATP hydrolysis.
To see if the MCCE method could also identify active site residues on other helicases, we have also analyzed structures of both the PcrA helicase and the T7 helicase. Although these results will be difficult to verify because there is as yet no pH dependent effects reported for the PcrA and T7 helicases, the results support the general conclusions made by using the HCV structures. MCCE successfully identified several key DNA binding residues in the ligand-free PcrA structure (PDB file 1PJR). These include His565 and His587, both of which lie within 5 Å of DNA in PcrA structures that contain bound DNA (9). To examine a ring helicase, T7 gene 4 helicase structures were analyzed with MCCE. As with the HCV helicase, MCCE, located residues in both the ATP and DNA binding sites in a structure of T7 that lacked ligands [PDB file 1E0K (5)]. The most critical residue identified was the Walker A motif residue Lys318, which is analogous to Lys210 in HCV helicase. Again this residue was highly acidic and its importance has been extensively confirmed by using site-directed mutagenesis (46,47). Most of the other perturbations that were noted in the T7 structure were located either in regions that are thought to interact with DNA [such as His475 (48)] or regions that link the subunits of the ring together.
The advent of structural genomics has brought about a need for the development of techniques that are able to determine functional characteristics of proteins from structural information alone. As has been pointed out by others, one way to locate enzyme active sites is to search protein structures for residues that have an unusual ionization state under physiological conditions (49). This study has illustrated that the MCCE method (17,18) identifies critical residues in helicases and accurately predicts pKas. Even though many of the residues identified were shown in this and other studies, to be required for proper helicase function, the precise roles of these residues in helicase action and their electrostatic interrelationships observed here warrant further investigation.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by a Liver Scholar Award from the American Liver Foundation and National Institutes of Health grant AI052395.
REFERENCES
- 1.Gorbalenya A.E. and Koonin,E.V. (1993) Helicases: amino acid sequence comparisons and structure-function relationships. Curr. Opin. Struct. Biol., 3, 419–429. [Google Scholar]
- 2.Patel S.S. and Picha,K.M. (2000) Structure and function of hexameric helicases. Annu. Rev. Biochem., 69, 651–697. [DOI] [PubMed] [Google Scholar]
- 3.Story R.M. and Steitz,T.A. (1992) Structure of the recA protein–ADP complex. Nature, 355, 374–376. [DOI] [PubMed] [Google Scholar]
- 4.Sawaya M.R., Guo,S., Tabor,S., Richardson,C.C. and Ellenberger,T. (1999) Crystal structure of the helicase domain from the replicative helicase- primase of bacteriophage T7. Cell, 99, 167–177. [DOI] [PubMed] [Google Scholar]
- 5.Singleton M.R., Sawaya,M.R., Ellenberger,T. and Wigley,D.B. (2000) Crystal structure of T7 gene 4 ring helicase indicates a mechanism for sequential hydrolysis of nucleotides. Cell, 101, 589–600. [DOI] [PubMed] [Google Scholar]
- 6.Toth E.A., Li,Y., Sawaya,M.R., Cheng,Y. and Ellenberger,T. (2003) The crystal structure of the bifunctional primase-helicase of bacteriophage T7. Mol. Cell, 12, 1113–1123. [DOI] [PubMed] [Google Scholar]
- 7.Subramanya H.S., Bird,L.E., Brannigan,J.A. and Wigley,D.B. (1996) Crystal structure of a DExx box DNA helicase. Nature, 384, 379–383. [DOI] [PubMed] [Google Scholar]
- 8.Soultanas P., Dillingham,M.S., Velankar,S.S. and Wigley,D.B. (1999) DNA binding mediates conformational changes and metal ion coordination in the active site of PcrA helicase. J. Mol. Biol., 290, 137–148. [DOI] [PubMed] [Google Scholar]
- 9.Velankar S.S., Soultanas,P., Dillingham,M.S., Subramanya,H.S. and Wigley,D.B. (1999) Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell, 97, 75–84. [DOI] [PubMed] [Google Scholar]
- 10.Yao N., Hesson,T., Cable,M., Hong,Z., Kwong,A.D., Le,H.V. and Weber,P.C. (1997) Structure of the hepatitis C virus RNA helicase domain. Nature Struct. Biol., 4, 463–467. [DOI] [PubMed] [Google Scholar]
- 11.Kim J.L., Morgenstern,K.A., Griffith,J.P., Dwyer,M.D., Thomson,J.A., Murcko,M.A., Lin,C. and Caron,P.R. (1998) Hepatitis C virus NS3 RNA helicase domain with a bound oligonucleotide: the crystal structure provides insights into the mode of unwinding. Structure, 6, 89–100. [DOI] [PubMed] [Google Scholar]
- 12.Cho H.S., Ha,N.C., Kang,L.W., Chung,K.M., Back,S.H., Jang,S.K. and Oh,B.H. (1998) Crystal structure of RNA helicase from genotype 1b hepatitis C virus. A feasible mechanism of unwinding duplex RNA. J. Biol. Chem., 273, 15045–15052. [DOI] [PubMed] [Google Scholar]
- 13.Yao N., Reichert,P., Taremi,S.S., Prosise,W.W. and Weber,P.C. (1999) Molecular views of viral polyprotein processing revealed by the crystal structure of the hepatitis C virus bifunctional protease-helicase. Structure Fold. Des., 7, 1353–1363. [DOI] [PubMed] [Google Scholar]
- 14.Delagoutte E. and von Hippel,P.H. (2002) Helicase mechanisms and the coupling of helicases within macromolecular machines. Part I: Structures and properties of isolated helicases. Q. Rev. Biophys., 35, 431–478. [DOI] [PubMed] [Google Scholar]
- 15.Lam A.M.I., Rypma,R.S. and Frick,D.N. (2004) Enhanced nucleic acid binding to ATP-bound hepatitis C virus NS3 helicase at low pH activates RNA unwinding. Nucleic Acids. Res., 32, 4060–4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Masuda T. (2003) An electromechanical model of myosin molecular motors. J. Theor. Biol., 225, 507–515. [DOI] [PubMed] [Google Scholar]
- 17.Alexov E.G. and Gunner,M.R. (1997) Incorporating protein conformational flexibility into the calculation of pH-dependent protein properties. Biophys. J., 72, 2075–2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Georgescu R.E., Alexov,E.G. and Gunner,M.R. (2002) Combining conformational flexibility and continuum electrostatics for calculating pK(a)s in proteins. Biophys. J, 83, 1731–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ondrechen M.J., Clifton,J.G. and Ringe,D. (2001) THEMATICS: a simple computational predictor of enzyme function from structure. Proc. Natl Acad. Sci. USA, 98, 12473–12478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guex N. and Peitsch,M.C. (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis, 18, 2714–2723. [DOI] [PubMed] [Google Scholar]
- 21.Alexov E.G. and Gunner,M.R. (1999) Calculated protein and proton motions coupled to electron transfer: electron transfer from QA- to QB in bacterial photosynthetic reaction centers. Biochemistry, 38, 8253–8270. [DOI] [PubMed] [Google Scholar]
- 22.Rocchia W., Sridharan,S., Nicholls,A., Alexov,E., Chiabrera,A. and Honig,B. (2002) Rapid grid-based construction of the molecular surface and the use of induced surface charge to calculate reaction field energies: applications to the molecular systems and geometric objects. J. Comput. Chem., 23, 128–137. [DOI] [PubMed] [Google Scholar]
- 23.O'Neil M.J. (2001) The Merck Index, 13th edn. Merck Research Laboratories, Whitehouse Station, NJ. [Google Scholar]
- 24.Frick D.N., Rypma,R.S., Lam,A.M. and Gu,B. (2004) The nonstructural protein 3 protease/helicase requires an intact protease domain to efficiently unwind duplex RNA. J. Biol. Chem., 279, 1269–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yanagi M., Purcell,R.H., Emerson,S.U. and Bukh,J. (1997) Transcripts from a single full-length cDNA clone of hepatitis C virus are infectious when directly transfected into the liver of a chimpanzee. Proc. Natl Acad. Sci. USA, 94, 8738–8743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lam A.M.I., Keeney,D., Eckert,P.Q. and Frick,D.N. (2003) Hepatitis C virus NS3 ATPases/helicases from different genotypes exhibit variations in enzymatic properties. J. Virol., 77, 3950–3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lam A.M., Keeney,D. and Frick,D.N. (2003) Two novel conserved motifs in the hepatitis C virus NS3 protein critical for helicase action. J. Biol. Chem., 278, 44514–44524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harris T.K. and Turner,G.J. (2002) Structural basis of perturbed pKa values of catalytic groups in enzyme active sites. IUBMB Life, 53, 85–98. [DOI] [PubMed] [Google Scholar]
- 29.Hall M.C. and Matson,S.W. (1999) Helicase motifs: the engine that powers DNA unwinding. Mol. Microbiol., 34, 867–877. [DOI] [PubMed] [Google Scholar]
- 30.Kim J.W., Seo,M.Y., Shelat,A., Kim,C.S., Kwon,T.W., Lu,H.H., Moustakas,D.T., Sun,J. and Han,J.H. (2003) Structurally conserved amino acid W501 is required for RNA helicase activity but is not essential for DNA helicase activity of hepatitis C virus NS3 protein. J. Virol., 77, 571–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Preugschat F., Danger,D.P., Carter,L.H., 3rd, Davis,R.G. and Porter,D.J. (2000) Kinetic analysis of the effects of mutagenesis of W501 and V432 of the hepatitis C virus NS3 helicase domain on ATPase and strand-separating activity. Biochemistry, 39, 5174–5183. [DOI] [PubMed] [Google Scholar]
- 32.Paolini C., Lahm,A., De Francesco,R. and Gallinari,P. (2000) Mutational analysis of hepatitis C virus NS3-associated helicase. J. Gen. Virol., 81, 1649–1658. [DOI] [PubMed] [Google Scholar]
- 33.Lin C. and Kim,J.L. (1999) Structure-based mutagenesis study of hepatitis C virus NS3 helicase. J. Virol., 73, 8798–8807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walker J.E., Saraste,M., Runswick,M.J. and Gay,N.J. (1982) Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J, 1, 945–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tanner N.K. and Linder,P. (2001) DExD/H box RNA helicases: from generic motors to specific dissociation functions. Mol. Cell., 8, 251–262. [DOI] [PubMed] [Google Scholar]
- 36.Tai C.L., Pan,W.C., Liaw,S.H., Yang,U.C., Hwang,L.H. and Chen,D.S. (2001) Structure-based mutational analysis of the hepatitis c virus ns3 helicase. J. Virol., 75, 8289–8297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heilek G.M. and Peterson,M.G. (1997) A point mutation abolishes the helicase but not the nucleoside triphosphatase activity of hepatitis C virus NS3 protein. J. Virol., 71, 6264–6266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Levin M.K. and Patel,S.S. (1999) The helicase from hepatitis C virus is active as an oligomer. J. Biol. Chem., 274, 31839–31846. [DOI] [PubMed] [Google Scholar]
- 39.Min K.H., Sung,Y.C., Choi,S.Y. and Ahn,B.Y. (1999) Functional interactions between conserved motifs of the hepatitis C virus RNA helicase protein NS3. Virus Genes, 19, 33–43. [DOI] [PubMed] [Google Scholar]
- 40.Kim D.W., Kim,J., Gwack,Y., Han,J.H. and Choe,J. (1997) Mutational analysis of the hepatitis C virus RNA helicase. J. Virol., 71, 9400–9409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chang S.C., Cheng,J.C., Kou,Y.H., Kao,C.H., Chiu,C.H., Wu,H.Y. and Chang,M.F. (2000) Roles of the AX(4)GKS and arginine-rich motifs of hepatitis C virus RNA helicase in ATP- and viral RNA-binding activity. J. Virol., 74, 9732–9737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kolykhalov A.A., Mihalik,K., Feinstone,S.M. and Rice,C.M. (2000) Hepatitis C virus-encoded enzymatic activities and conserved RNA elements in the 3′ nontranslated region are essential for virus replication in vivo. J. Virol., 74, 2046–2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suzich J.A., Tamura,J.K., Palmer-Hill,F., Warrener,P., Grakoui,A., Rice,C.M., Feinstone,S.M. and Collett,M.S. (1993) Hepatitis C virus NS3 protein polynucleotide-stimulated nucleoside triphosphatase and comparison with the related pestivirus and flavivirus enzymes. J. Virol., 67, 6152–6158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alexov E. (2004) Calculating proton uptake/release and binding free energy taking into account ionization and conformation changes induced by protein-inhibitor association: application to plasmepsin, cathepsin D and endothiapepsin–pepstatin complexes. Proteins, 56, 572–584. [DOI] [PubMed] [Google Scholar]
- 45.Lee A.Y., Gulnik,S.V. and Erickson,J.W. (1998) Conformational switching in an aspartic proteinase. Nature Struct. Biol., 5, 866–871. [DOI] [PubMed] [Google Scholar]
- 46.Notarnicola S.M. and Richardson,C.C. (1993) The nucleotide binding site of the helicase/primase of bacteriophage T7. Interaction of mutant and wild-type proteins. J. Biol. Chem., 268, 27198–27207. [PubMed] [Google Scholar]
- 47.Patel S.S., Hingorani,M.M. and Ng,W.M. (1994) The K318A mutant of bacteriophage T7 DNA primase-helicase protein is deficient in helicase but not primase activity and inhibits primase-helicase protein wild-type activities by heterooligomer formation. Biochemistry, 33, 7857–7868. [DOI] [PubMed] [Google Scholar]
- 48.Notarnicola S.M., Park,K., Griffith,J.D. and Richardson,C.C. (1995) A domain of the gene 4 helicase/primase of bacteriophage T7 required for the formation of an active hexamer. J. Biol. Chem., 270, 20215–20224. [DOI] [PubMed] [Google Scholar]
- 49.Ringe D., Wei,Y., Boino,K.R. and Ondrechen,M.J. (2004) Protein structure to function: insights from computation. Cell Mol. Life Sci., 61, 387–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wardell A.D., Errington,W., Ciaramella,G., Merson,J. and McGarvey,M.J. (1999) Characterization and mutational analysis of the helicase and NTPase activities of hepatitis C virus full-length NS3 protein. J. Gen. Virol., 80, 701–709. [DOI] [PubMed] [Google Scholar]