Abstract
To study the molecular basis for predator-prey coevolution, we investigated how Caenorhabditis elegans responds to the predatory fungus Arthrobotrys oligospora. C. elegans and other nematodes were attracted to volatile compounds produced by A. oligospora. Gas-chromatographic mass-spectral analyses of A. oligospora-derived volatile metabolites identified several odors mimicking food cues attractive to nematodes. One compound, methyl 3-methyl-2-butenoate (MMB) additionally triggered strong sex- and stage-specific attraction in several Caenorhabditis species. Furthermore, when MMB is present, it interferes with nematode mating, suggesting that MMB might mimic sex pheromone in Caenorhabditis species. Forward genetic screening suggests that multiple receptors are involved in sensing MMB. Response to fungal odors involves the olfactory neuron AWCs. Single-cell RNA-seq revealed the GPCRs expressed in AWC. We propose that A. oligospora likely evolved the means to use olfactory mimicry to attract its nematode prey through the olfactory neurons in C. elegans and related species.
DOI: http://dx.doi.org/10.7554/eLife.20023.001
Research Organism: C. elegans, Other
Introduction
Predation, in which individuals of one species (predators) kill and consume the biomass of individuals of another species (prey) (Abrams, 2000), imposes a strong selective pressure on both predators and prey. To minimize the risk of being eaten, prey have often evolved specific behaviors and strategies such as camouflage, avoidance, mimicry, and tonic immobility to increase their chance of survival. In turn, predators also evolved enhanced predatory strategies to secure food sufficient to survive and reproduce, giving rise to an evolutionary arms race between predator and prey (Dawkins and Krebs, 1979).
While most carnivores are animals, examples of predacious plants and fungi exist. Venus flytraps, sundews and pitcher plants acquire some or most of their nutrients from trapping and consuming insects, a feature that evolved in response to nitrogen limitation (Darwin, 1875). Many fungi that grow in nitrogen-poor environments have likewise evolved carnivorism and nematodes, being the most numerically abundant animals on earth, conveniently became the prey (Barron, 1977; Nordbring-Hertz, 1988). This predatory lifestyle has independently evolved multiple times among different fungal lineages including Zygomycetes, Ascomycetes and Basidiomycetes (Barron, 1977; Liou and Tzean, 1997; Yang et al., 2007). Nematode-trapping fungi depend on their elaborate traps to prey on nematodes. However, the majority of the nematode-trapping fungi do not constitutively generate these trapping structures; trap-morphogenesis is only triggered by the presence of nematodes (Pramer and Stoll, 1959). This suggests that trap-formation might be a highly energy-consuming process and that, to conserve energy, these fungi have evolved to sense signals from nematodes, which indicate the presence of prey. One such signal is the group of nematode pheromones, ascarosides. These pheromones regulate various aspects of behavior and development in C. elegans and are sufficient to induce trap-morphogenesis in several species of nematode-trapping fungi (Hsueh et al., 2013). Since the nematode-trapping fungi have clearly evolved the ability to eavesdrop on nematode communication, we wondered whether the nematodes also sense and respond to their fungal predators.
C. elegans is known to respond to pathogenic bacteria. For example, Pseudomonas aeruginosa triggers an aversive learning behavior, while another pathogen Bacillus nematocida attracts C. elegans (Niu et al., 2010; Zhang et al., 2005). Bacterial secondary metabolites are able to modulate the signaling and the protective lawn-avoidance behavior in C. elegans, demonstrating that the sensory neurons of C. elegans are critical for its defensive responses against pathogens (Meisel et al., 2014; Pradel et al., 2007; Zhang et al., 2005). By contrast, little is known about how C. elegans responds to its natural predators such as the nematode-trapping fungi, with only an early study showing that the nematode Panagrellus redivivus was attracted to a species of nematode-trapping fungi (Balan and Gerber, 1972). Many prey are known to begin fleeing when they sense a possible predator, while many predators are also known to attract their prey (Haynes et al., 2002; Rosier, 2011). Therefore, we investigated the behavioral and molecular basis for how C. elegans responds to Arthrobotrys oligospora, one of the most common nematode-trapping fungal species and one that we had previously found to eavesdrop on ascarosides produced by C. elegans (Hsueh et al., 2013).
Here, we show that C. elegans and other nematodes are attracted to A. oligospora and the attraction observed for C. elegans is mediated by the two AWC olfactory neurons. We identified several odorants produced by A. oligospora and found that many of the odorants attracted C. elegans, and appear to represent olfactory mimics of the food and sex cues. Through genetic screens and single-cell transcriptome analyses of the AWC neuron, we identified the potential chemosensory receptors that were expressed in this neuron, which is involved in sensing some of these fungal odors. Our study shows that in order to catch its nematode prey, A. oligospora has evolved to lure the nematodes by producing olfactory mimicry of the food and sex cues that are attractive to Caenorhabditis nematodes.
Results
Nematode-trapping fungus A. oligospora attracts C. elegans and other nematodes
To study how C. elegans responds to one of its natural predators, the nematode-trapping fungus A. oligospora, we tested the chemotaxis behavior of C. elegans in the presence of the fungal culture. We designed a four-point chemotaxis assay. In this assay, A. oligospora cultures were grown in two quadrants of a plate, following which synchronized adult C. elegans were placed in the middle of the plates and allowed to crawl freely until they reached a source of sodium azide in each quadrant that paralyzed the worms (Figure 1A). The number of immobilized worms in each of the quadrants was counted and their chemotaxis index (Bargmann et al., 1993) was determined as described in Figure 1A. We found that the adult C. elegans were consistently attracted to A. oligospora, suggesting that the fungus might lure the worms by producing compounds that are attractive to them (Figure 1B). This strong attraction was only observed in the adult animals; dauers and the L1 larvae exhibit much weaker attraction to A. oligospora (Figure 1B). C. elegans moves at a similar speed on A. oligospora culture; thus we think the attraction is unlikely due to a sedative secreted by the fungus or a mechanical cue that retained the worms in the fungal quadrants (Figure 1C).
Figure 1. A. oligospora attracts many nematode species, including C. elegans.
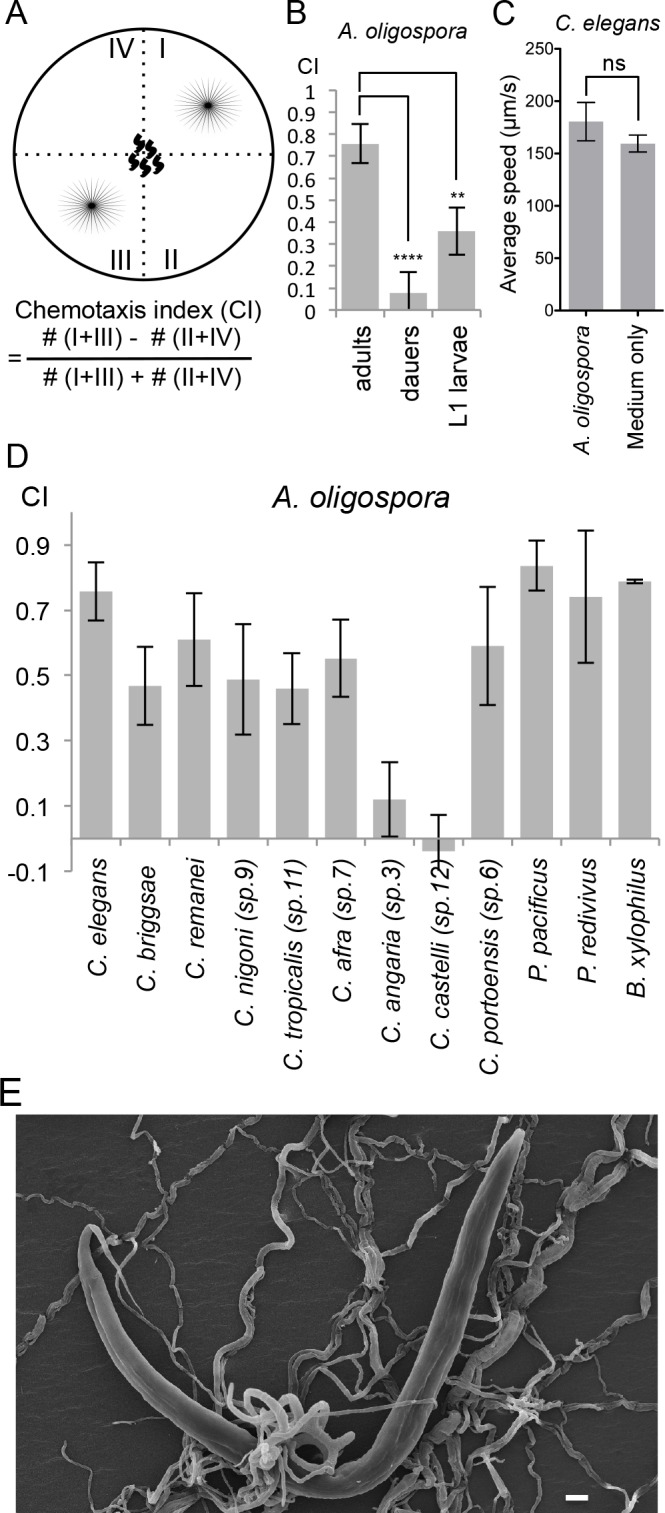
(A) Schematic representation of the 4-point chemotaxis assays and the formulation used to calculate the chemotaxis index (CI). (B) C. elegans adults are attracted to A. oligospora, but the dauers and the L1 larvae have much weaker attraction (Mean ± SD, n = 4–12 trials). (C) Speed of wild-type C. elegans on A. oligospora culture or medium only control. Locomotion of the animals was tracked by the commercial WormLab system (MBF bioscience). (D) Many Caenorhabditis nematodes and the more distant Panagrellus redivivus, Pristionchus pacificus, and Bursaphelenchus xylophilus were attracted to A. oligospora (Mean ± SD, n = 4–12 trials). (E) SEM image of C. elegans trapped in A. oligospora. Scale bar = 10 μm.
To investigate whether the attraction towards A. oligospora is a conserved behavior observed across different nematode species or if it is specific to C. elegans, we tested the chemotaxis behavior of eight additional Caenorhabditis species (Félix et al., 2014). We observed significant attraction toward A. oligospora in six other Caenorhabditis species, though not in C. angaria and C. castelli (Figure 1D). We thus further tested three additional nematode species that are more phylogenetically distant from C. elegans: Pristionchus pacificus, Panagrellus redivivus, and Bursaphelenchus xylophilus. Like C. elegans, P. pacificus and P. redivivus are both free-living bacterivorous nematodes, whereas B. xylophilus is a fungal feeder and a parasite of pine trees (Futai, 2013; Sommer, 2006; Srinivasan et al., 2013). We found that all three of these additional species were also attracted to A. oligospora, indicating that A. oligospora produces diffusible compounds that are attractive to a variety of nematodes to help lure the prey and induce trap-morphogenesis (Figure 1E).
A. oligospora produces volatile compounds to attract C. elegans
To determine whether these diffusible nematode-attracting compounds were soluble or volatile, we designed another version of the chemotaxis assay using three-division Petri dishes. In this assay, A. oligospora was grown in one of the three sectors and C. elegans was placed in a separate sector, where the worms could only sense the volatile compounds, but not the soluble ones produced by A. oligospora (Figure 2A). We let the nematodes move freely for 2 hr, after which their chemotaxis index was determined by counting the number of worms that had migrated to the sectors with or without the fungal culture (Figure 2A). We still observed significant attraction of C. elegans toward A. oligospora (Figure 2B), suggesting that A. oligospora likely produces volatile compound(s) that are attractive to C. elegans.
Figure 2. C. elegans are attracted to volatile compounds produced by A. oligospora.

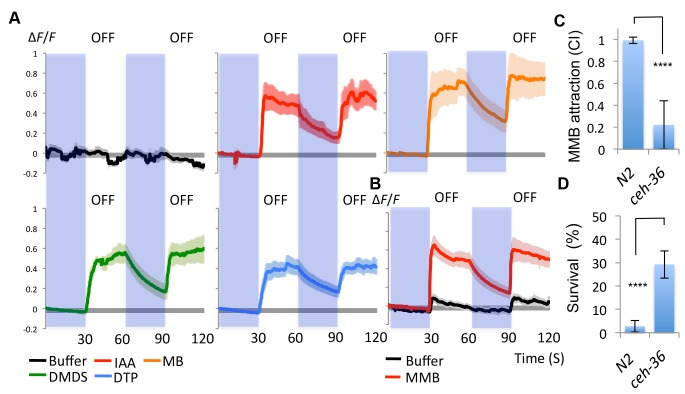
(A) Schematic representation of the volatile chemotaxis assays in which A. oligospora were grown in compartmentalized Petri dishes that contained three sections. The CI was calculated based on the formula presented. (B) C. elegans adults preferentially migrate to the section that was inoculated with A. oligospora (Mean ± SD, n = 6–7 trials). (C) A. oligospora attraction is reduced in several mutants defective in AWC neuron function (Mean ± SD, n = 3–4 trials). (D) Laser ablation of the AWC neurons reduced attraction to A. oligospora.
The olfactory AWCs neurons mediate A. oligospora-attraction in C. elegans
We next sought to identify the C. elegans sensory neurons responsible for sensing odors emanating from A. oligospora and for mediating the attraction. There are 32 sensory neurons present in the amphid, phasmid and inner labial organs of C. elegans. For some of these neurons, their functions have been studied in detail (Bargmann, 2006). For example, the amphid ASE neurons are required for chemotaxis to water-soluble attractants, whereas AWA and AWC neurons mediate chemotaxis to volatile attractants (Bargmann et al., 1993; Bargmann and Horvitz, 1991).
To determine whether genes known to play a role in chemotaxis are required for sensing A. oligospora, we first tested several known mutants for A. oligospora attraction. We found that che-1, a gene required for ASE neuron identity and function, was dispensable for A. oligospora attraction but that genes known to play a role in AWC function, such as odr-1, odr-3, tax-2 and tax-4, were required (Figure 2C). We thus laser-ablated both AWC neurons in C. elegans and analyzed these animals individually for chemotaxis to A. oligospora. Approximately 70% of the trials of mock-ablated control animals were attracted to A. oligospora, whereas only 20% of the trials of the AWC-ablated animals were attracted, indicating that AWCs play a major role in A. oligospora attraction (Figure 2D).
Identification of A. oligospora-derived odorants that attract C. elegans
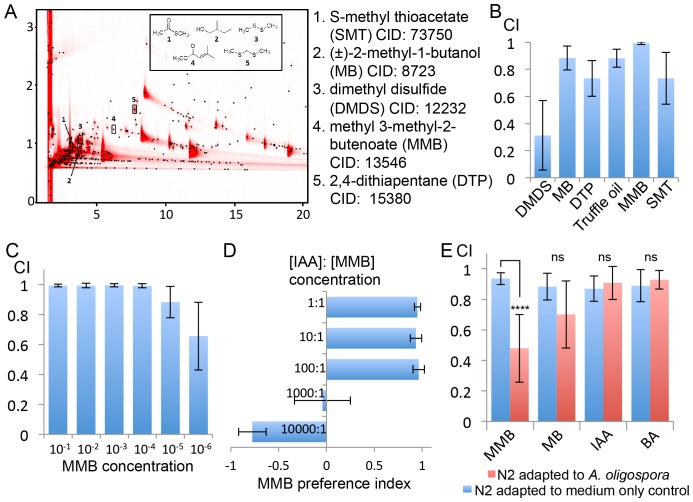
To understand how A. oligospora attracts nematodes, we decided to identify A. oligospora-derived odorants. Solid-phase microextraction (SPME) was used to sample headspace volatiles from vials containing medium inoculated with A. oligospora and from control vials containing only medium. The samples then were subjected to comprehensive two-dimensional gas chromatographic mass spectral analyses (GC x GC-TOFMS). Five compounds present only in samples containing A. oligospora culture were identified: dimethyl disulfide (DMDS), (±)−2-methyl-1-butanol (MB), 2,4-dithiapentane (DTP), methyl 3-methyl-2-butenoate (MMB), and S-methyl thioacetate (SMT; Figure 3A and Supplementary file 1). To examine whether these A. oligospora-derived odorants attracted C. elegans, we tested the individual compounds in chemotaxis assays. We found that DMDS was only moderately attractive to C. elegans, but that all the other compounds, especially MMB, were highly attractive to C. elegans (Figure 3B). DMDS has been identified from many bacteria (Tomita et al., 1987) and we have also detected DMDS in another two ascomycete fungi, Aspergillus terreus and A. niger (Supplementary file 1). These observations suggest that DMDS is a relatively common odor, and it may be sensed as a food cue by the bacterivorous C. elegans. DMDS, MB, and DTP have been previously identified as odors that contribute to truffle aroma (Díaz et al., 2002; Pacioni et al., 2014; Pennazza et al., 2013); we therefore tested C. elegans chemotaxis in response to commercial truffle oil. We found that truffle oil was very attractive to C. elegans (Figure 3B). These results indicate that several A. oligospora-derived odorants might mimic food signals to attract nematodes.
Figure 3. GC x GC-TOFMS analyses identified A. oligospora-derived odorants that are attractive to C. elegans.
(A) Representative two-dimensional gas chromatogram obtained from SPME sampling of headspace above A. oligospora cultures. The total detector response is indicated by the intensity of red in the chromatogram. Black dots represent discrete elution peaks identified during data processing (shown for a signal-to-noise threshold of 100). The elution peaks labeled 1–5 represent compounds that were detected only in A. oligospora cultures, and not in the medium-only controls. (B) Chemotaxis assays of A. oligospora-derived odorants. Chemicals were tested at a 10−2 dilution and truffle oil was tested undiluted (Mean ± SD, n = 6 trials). (C) N2 adults were strongly attracted to MMB across a wide-range of concentrations (Mean ± SD, n = 6 trials). (D) C. elegans prefers MMB over isoamyl alcohol (IAA; Mean ± SD, n = 3–7 trials). The MMB preference index was calculated according to the formula in Figure 1. (E) Chemotaxis plot showing C. elegans chemotactic response to different odorants after 2 hr adaptation to A. oligospora culture (red) or the medium-only control (blue; Mean ± SD, n = 4–10 trials).
Attraction to A. oligospora-derived chemicals diminished upon adaptation to the fungal culture
One of the identified odorants, MMB, was highly attractive to C. elegans. When varying doses were tested for elicited responses, MMB attracted C. elegans across a 106 fold concentration range (Figure 3C). Moreover, when C. elegans were given a choice between isoamyl alcohol (IAA, a previously described attractant) and MMB, the worms exhibited a strong preference for MMB (Figure 3D). To examine whether MMB is indeed a genuine molecule naturally produced by A. oligospora, and not an artifact of GC-MS analysis, we exposed C. elegans to A. oligospora cultures for two hours, and measured the chemotaxis behavior of these A. oligospora-adapted animals. If the A. oligospora cultures were emitting MMB, we expected to subsequently observe significantly decreased attraction toward MMB as a result of olfactory adaptation, a behavior that has been well-characterized in C. elegans (Colbert and Bargmann, 1995). Indeed, we found that when C. elegans were exposed to A. oligospora volatiles for two hours, their chemotactic response to MMB was strongly reduced (Figure 3E). This reduction in chemotaxis was odorant-specific, as opposed to a global reduction in chemotaxis, as A. oligospora-adapted animals were still attracted to IAA and benzealdehyde (BA), two odorants that were not detected in the A. oligospora volatiles and are known to attract C. elegans (Bargmann et al., 1993) (Figure 3E).
A. oligospora-derived odorants activate the AWCon neuron
Having demonstrated that the AWC neurons are required for attraction towards A. oligospora, we examined whether they could be directly activated by A. oligospora-derived odorants. The gene str-2 encodes a predicted G-protein coupled receptor (GPCR) that is randomly and asymmetrically expressed in one of two bilateral AWC chemosensory neurons, called AWCon (Troemel et al., 1999). We thus transgenically expressed the calcium indicator GCaMP6 in AWC by driving it with the str-2 promoter. We then measured the AWCon activity in response to different odorants via a microfluidic device (the olfactory chip) (Chronis et al., 2007). The AWCon neurons are known to respond to odor removal (Chalasani et al., 2007), so we tested whether the AWCon neurons responded to the removal of the A. oligospora-derived odorants. We observed a strong activity in the AWCon neuron triggered by several A. oligospora-derived odorants (Figure 4A). To test if the activation of the AWCon neuron was a primary response, we also imaged the AWCon activity in an unc-13 background. Mutations of unc-13 impair the priming of presynaptic vesicles for release, either impairing or completely blocking synaptic transmission (Richmond et al., 1999). AWCon neurons were still strongly activated by MMB removal in an unc-13 mutant background, suggesting that this activation is likely to be a direct chemosensory response to MMB (Figure 4B). Compared to wild-type animals, the AWC- mutant (ceh-36) had a strong defect in MMB attraction (Figure 4C).
Figure 4. AWCon neurons respond to A. oligospora-derived odorants.
(A) GCaMP6 measurements of the AWCon neurons in response to A. oligospora-derived odorants (10−3) and buffer control (n = 4–7 trials for each odorant tested). (B) GCaMP6 measurements of the AWCon neurons in response to MMB in the unc-13 mutant background (n = 11 and 14 for MMB and buffer control respectively). (C) The AWC- mutant is defective in MMB attraction. Chemotaxis response to 10−4 MMB was measured for the wild-type (N2) or AWC- mutant (ceh-36). (D) The AWC- mutant is more likely to survive predation by A. oligospora. Y axis indicates % of nematodes that were still alive after 16 hr incubation with A. oligospora culture (Mean ± SD, n = 10–12 trials).
AWC-defective mutants exhibit lower predation when encountering A. oligospora
Our data suggest that the AWC neurons sense A. oligospora-derived odorants and mediate attraction. We thus hypothesized that mutants defective in AWC neuron functioning would exhibit lower predation by A. oligospora. To address this hypothesis, A. oligospora was inoculated at one side of a 9 cm Petri dish, and unc-119 nematodes, which are strongly defective in locomotion, were added onto the A. oligospora culture to induce trap formation. After traps had emerged from A. oligospora hyphae, AWC+ (wild-type) or AWC- (ceh-36) animals were then placed at the opposite side of the 9 cm Petri dish and allowed to freely explore. Survival rates of C. elegans were determined after an overnight incubation. We found that the majority of the wild-type animals were trapped by A. oligospora (with less than 3% of these animals surviving), but ~30% of the AWC- nematodes survived (Figure 4D). These results indicate that C. elegans with decreased attraction to A. oligospora have a better chance to survive under these conditions.
MMB triggers sex- and developmental stage-specific attraction and interferes with mating in Caenorhabditis species
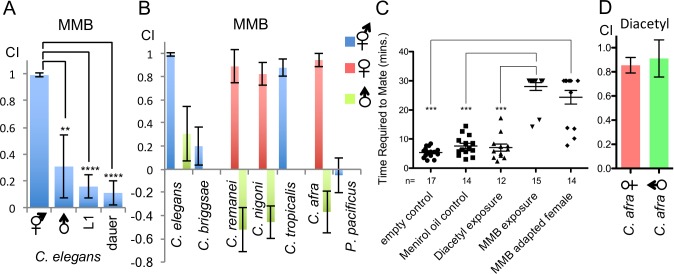
MMB potently attracted C. elegans adult hermaphrodites. To investigate why adult hermaphrodites were so attracted to this odor, we further examined the chemotaxis behavior in different developmental stages of C. elegans. We found that attraction to MMB was both sex- and developmental stage-dependent, as L1 larvae, dauers, and the male animals did not exhibit strong attraction to this odor (Figure 5A). Furthermore, when we tested MMB attraction in other Caenorhabditis species, we found that MMB was highly attractive to adult females but repulsive to males in the gonochoristic species such as C. remanei, C. nigoni (sp. 9), and C. afra (sp.7; Figure 5B). These observations prompted us to hypothesize that MMB-triggered attraction might be related to sex pheromone-triggered attraction. Therefore, we monitored the mating behavior of C. afra in the presence or absence of MMB to examine whether MMB might interfere with mate seeking. We chose this species as we found that it has a highly robust and reproducible mating behavior (Figure 5C). When one female and two males were placed on a 3 cm NG plate without food, the nematodes mated within average of ~5 min. However, if 1 μl MMB was placed on the lid of the Petri dishes, in a majority of the trials, the animals failed to mate within the 30 min tracking time, suggesting that MMB interferes with mating (Figure 5C). Furthermore, if the female animals were pre-exposed to MMB odor for an hour before testing, mating efficiency also dramatically decreased (Figure 5C). In contrast, if mineral oil (solvent control) or diacetyl (another food odor attractive to C. elegans and both sexes of C. afra; Figure 5D) was present, mating was not affected. These results suggested that MMB interfered with mating in the gonochoristic species C. afra and are consistent with the hypothesis that MMB might mimic a sex pheromone that specifically and strongly attracted hermaphroditic and female nematodes.
Figure 5. MMB triggers a sex- and lifestage-specific attraction in Caenorhabditis species and interferes with mating in C. afra.
(A) Responses of C. elegans hermaphrodites, males, L1 larvae and dauers to a 10−4 dilution of MMB (Mean ± SD, n = 4–10 trials). (B) Responses of Caenorhabditis sp. and P. pacificus to a 10−4 dilution of MMB (Mean ± SD, n = 4–10 trials). (C) Mating behavior of C. afra was recorded. Y axis indicates the time required for the nematodes to mate; if no mating was observed within the 30 min tracking period, data points were represented as 30 min. (D) Chemotaxis of C. afra male and female nematodes in response to 10−2 diacetyl.
Genetic screening suggests redundancy for the MMB receptor
We conducted a genetic screen to identify genes that encode a receptor for MMB. The C. elegans genome contains ~1300 genes encoding putative chemoreceptors, but only a few of their chemical ligands have been identified (Thomas and Robertson, 2008). The diacetyl receptor, odr-10, was the first C. elegans chemoreceptor identified through a genetic screen and more recently, three pairs of GPCRs (srbc-64/srbc-66, srg-36/srg-37, and daf-37/daf-38) have been shown to encode receptors that sense ascarosides (Kim et al., 2009; Park et al., 2012; Sengupta et al., 1996). The design of our genetic screen followed an approach similar to that used in the screen that identified the diacetyl receptor, odr-10 (Sengupta et al., 1996). In brief, mutagenized animals were given a choice between MMB and IAA (another odor sensed by the AWC neurons). Under this condition, more than 95% of the wild-type N2 animals preferred MMB. We expected that mutants with specific defects in sensing MMB, but with otherwise intact AWC function, would exhibit an abnormal preference for IAA in our screen. After two rounds of behavioral screen enrichment, mutant lines were isolated for further analyses. In total, we isolated thirteen mutants with varying degrees of chemotactic deficiency to MMB. Two mutants (sy837 and sy838) showed dramatic decreases in MMB attraction, whereas eleven other mutant lines only exhibited minor decreases. Diminished MMB attraction seemed to be specific, as attraction to other odorants such as IAA and BA remained at wild-type levels (Figure 6A). SNP mapping with the Hawaiian strain revealed the mutations mapped to chromosome X for both strongly defective mutants (Figure 6B). Complementation assays revealed that the two mutants failed to complement each other, suggesting that the phenotype was likely caused by a mutation in the same gene (Figure 6C). By whole-genome sequencing of the two mutant lines (PS7117 and PS7118), we found that both lines had mutations in the odr-7 gene, causing premature stop codons at codon 83 and codon 188 respectively (Figure 6—figure supplement 1). A genomic rescue line generated with the fosmid WRM0639bF05 that contains the odr-7 gene restored the chemotaxis defects (Figure 6—figure supplement 2). ODR-7 is a nuclear receptor that modulates gene expression in the AWA and AWC neurons, including that of two GPCR genes, odr-10 and str-2 (Colosimo et al., 2003; Sengupta et al., 1994). We identified two independent odr-7 mutants in our genetic screen but not any other mutants that exhibited a strong reduction in MMB chemotaxis, as one might expect if MMB attraction was mediated via a single chemosensory receptor. Thus, the results from our genetic screen suggest that MMB is more likely to be sensed by multiple redundant receptors in C. elegans.
Figure 6. Forward genetic screens isolated two mutants with strong defects in MMB chemotaxis.
(A) Response of the two mutant lines to MMB, IAA, and Benzaldehyde (BA; n = 4–8 trials for each odorant tested). The mutants showed strong defects in MMB chemotaxis but remained attracted to two other odorants, IAA and BA. (B) SNP mapping with the Hawaiian polymorphic strain CB4856 revealed that the mutations in both mutants were located on chromosome X. Mutants #7–22 and #18–10 were crossed to CB4856 and three F2 progeny from each mutant line that had MMB chemotaxis defects were analyzed for eight SNP markers on each chromosome. (C) Mutants #7–22 and #18–10 failed to complement. Responses of the F1 progeny from a cross between the two mutants to 10−4 dilution of MMB. (Mean ± SD, n = 5–8 trials).
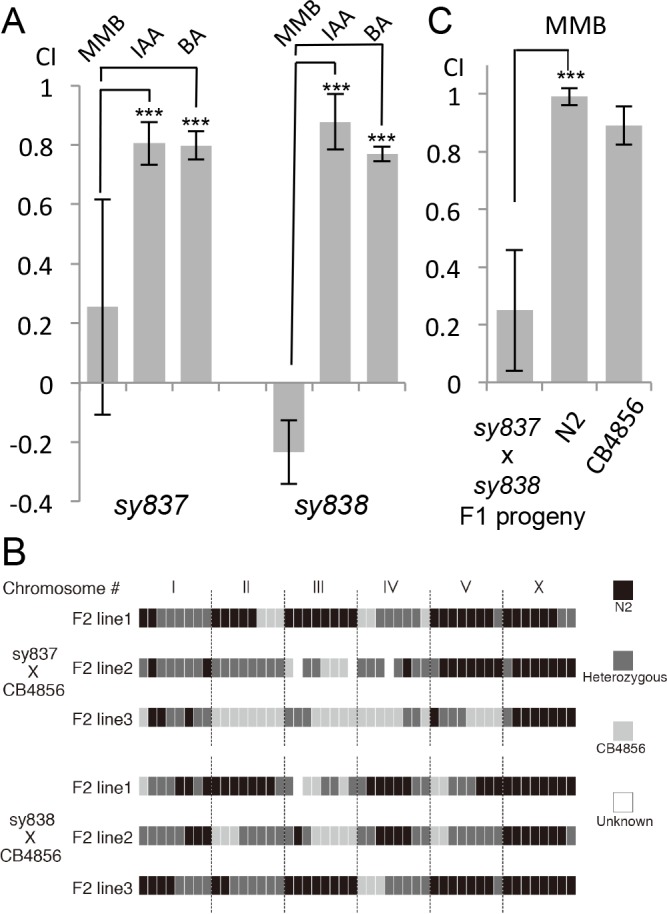
Figure 6—figure supplement 1. Gene structure of odr-7.
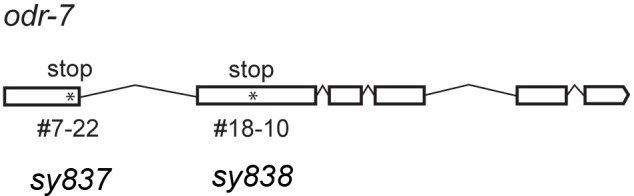
Figure 6—figure supplement 2. Fosmid clone WRM0639bF05 which contains odr-7 rescues the MMB attraction defect of odr-7 (sy837).
Single-cell RNA-seq identifies AWC-expressed GPCR candidates that might sense the A. oligospora-derived odors
To identify candidate GPCRs expressed in the AWC neurons, we microdissected GFP-labeled AWCon neurons (str-2::GFP) and performed single-cell RNA-seq (Schwarz et al., 2012). Nine individual AWC neurons were dissected, separately amplified for their transcripts with RT-PCR, and equal aliquots from the nine separate purified products were sequenced as a single AWC RT-PCR pool. In order to obtain independent biological replicates for statistical analysis, we then also sequenced five aliquots of five individual RT-PCR products from five individually dissected AWC neurons.
We detected expression of 5894 protein-coding genes and 43 non-protein-coding genes that had a minimum estimated expression level of at least 0.1 transcripts per million (TPM) with a credibility interval of 99% (minTPM). The overall pattern of gene activity in AWC neurons is shown in Figure 7; details of observed gene activity, with summaries of gene function are given in Figure 7—source data 1. This AWC single-cell RNA-seq data set is not perfect; for example, some of the known AWCon markers such as str-2 were not detected in our analyses while other genes were not known to express in the AWCon were found in our RNA-seq data set, which is likely due to that the RNAseq analysis having a higher sensitivity than the GFP reporter assays.
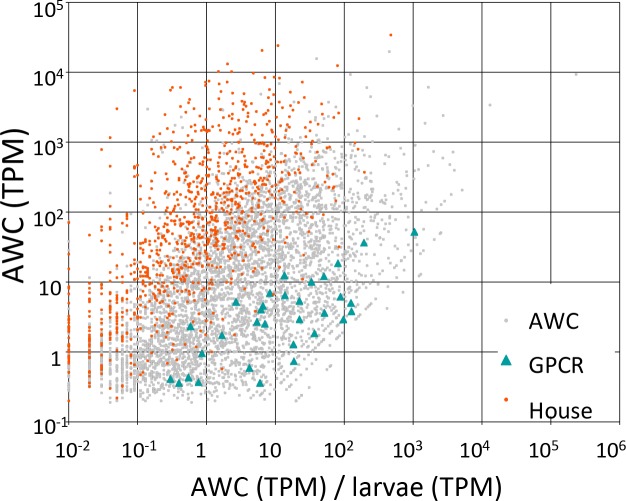
Figure 7. Candidate GPCRs expressed in AWC neurons.
Gene expression in AWC neurons. Expression data are shown for 5326 C. elegans genes (5294 protein-coding and 32 non-coding RNAs) that exhibited above-background expression in our pooled AWC RNA-seq (i.e., that had a minimum expression level, in a 99% credibility interval, of ≥0.1 TPM). The x-axis shows AWC-specificity, computed as the ratio of AWC gene expression (in TPM) to larval gene expression (in TPM) for each gene. The y-axis shows the absolute magnitude of AWC gene activity in TPM. Among most genes (labeled ‘AWC’ and shown as gray dots) are highlighted 32 genes encoding G-protein coupled receptors shown as green triangles and 984 genes encoding housekeeping functions shown as red dots. The housekeeping genes were identified in a previous single-cell RNA-seq analysis (Schwarz et al., 2012). Full identities, annotations, and expression data for all C. elegans genes are provided in Figure 7—source data 1.
DOI: http://dx.doi.org/10.7554/eLife.20023.010
Gene Ontology analysis showed that genes specifically upregulated in AWC neurons preferentially encoded a number of functions, including G-protein-coupled receptor activity [GO:0004930] and olfactory learning [GO:0008355] (Figure 7—source data 3). Among the 5894 genes, we identified 48 GPCRs, of which 34 encoded one of the putative chemosensory receptor PFAM domains (Figure 7—source data 4). In an earlier analysis of a subset of our RNA-seq data by older computational methods, we identified 46 GPCRs, of which 35 encoded a chemosensory PFAM domain (Figure 7—source data 4 and Figure 7—source data 2). We speculated that some of these receptors might function as the chemoreceptors for the odors produced by A. oligospora that evoke a strong response in the AWC neurons.
Discussion
For a non-motile predator like A. oligospora, catching fast-moving nematode prey is a major task. The fact that in A. oligospora trap morphogenesis is usually only triggered by the presence of nematodes makes it even more challenging. Since trap morphogenesis takes more than 12 hr, there is a long period during which nematodes could escape before the functional traps become fully developed. In this study, we have shown that A. oligospora may overcome this problem by luring nematodes to stay nearby, by producing several volatile compounds that mimic nematode food and sex cues.
In nature, Caenorhabditis nematodes are most commonly associated with rotten fruits and plants where they feast on the bacterial blooms in these decomposing materials (Frézal and Félix, 2015). Several of the odors we identified from A. oligospora, including the sulfurous compounds DMDS, DTP and SMT, have pungently overripe or rotten smells, suggesting that these odors might be sensed as food cues by nematodes and, as a result, are attractive to C. elegans. Such olfactory mimicry is also employed by various plants to attract pollinators. For example, the dead horse arum (Helicodiceros muscivorus) and the infamous corpse flower (Amorphophallus titanium) are known to produce rotten smells to attract insects; in both cases, one of the odors identified was DMDS (Lim, 2012; Shirasu et al., 2010; Stensmyr et al., 2002). Similarly, carnivorous plants such as the Venus flytraps, sundews and pitcher plants produce volatiles that mimic fruits and flowers to attract insect prey (Di Giusto et al., 2010; Jurgens et al., 2009). These examples demonstrate that olfactory mimicry of food might be a common strategy used by carnivorous plants and fungal species to lure their prey.
MMB, the most attractive compound we identified from A. oligospora volatiles, elicited a strong sex- and developmental stage-specific attraction for many Caenorhabditis nematodes. In the gonochoristic Caenorhabditis species (C. remanei, C. nigoni and C. afra), MMB was highly attractive to the females but repulsive to the males. In the hermaphroditic Caenorhabditis species including C. elegans and C. tropicalis, MMB was highly attractive to the adult hermaphrodites, but not to the males, dauers, or larvae. The presence of MMB strongly interfered with mating in C. afra, which is in contrast to the presence of diacetyl, another attracting chemical that is likely to be a food cue. If the female nematodes were adapted to MMB, mating was also strongly affected. Therefore, we hypothesize that MMB might be an olfactory mimic of a sex pheromone produced by male nematodes.
Multiple species have been reported to produce pheromone mimics to attract prey or pollinators. For example, the American bolas spider Mastophora hutchinsoni produces odorants that mimic female moth sex pheromones to attract different moth species (Haynes et al., 2002). Orchids such as Ophrys sphegodes, O. exaltata and Mormolyca ringens produce volatile compounds that greatly resemble the chemical composition of female bee pheromones in order to attract the male bees as pollinators (Flach et al., 2006; Mant et al., 2005; Schiestl et al., 2000). Whether male Caenorhabditis nematodes produce any volatile sex pheromones is currently unknown, but a recent study has found that male pinewood nematodes Bursaphelenchus xylophilus produce an as yet unidentified volatile pheromone that attracts the females (Shinya et al., 2015). Our study suggests that MMB produced by A. oligospora might function as an olfactory mimic of a sex pheromone produced by male nematodes to attract hermaphrodites or females. Therefore, it would be of interest to screen male Caenorhabditis nematodes for the production of any volatile compounds that could be functionally or structurally related to MMB. Why A. oligospora produces an odor that only attracts the females and the hermaphrodites is open to speculation. We suspect that in the niches inhabited by A. oligospora, there is a larger population of female and hermaphroditic nematodes than males. Furthermore, males are potently attracted to females and hermaphrodites so they might be also lured to the trapped female and hermaphroditic nematodes. C. elegans dauers are not attracted to MMB or A. oligospora, which might enable them to better survive A. oligospora predation. MMB attraction only seems to be conserved in Caenorhabditis species; P. pacificus was attracted to A. oligospora, but not to MMB, suggesting that A. oligospora attraction is likely mediated via other odorants in P. pacificus.
A. oligospora has evolved a strategy of olfactory mimicry to efficiently attract prey, but is this strategy common among other species of predacious fungi? Most known nematode-trapping fungi, like A. oligospora, only generate traps upon sensing the presence of nematodes, so they may benefit by attracting their prey. We believe that the olfactory mimicry strategy used by A. oligospora likely represents a conserved mechanism that is employed by many other predacious fungi. In fact, an earlier study reporting that the nematode Panagrellus redivivus was attracted to another Arthrobotrys species supports this hypothesis (Balan and Gerber, 1972).
In C. elegans, the olfactory neuron AWCs are known to mediate chemoattraction to certain volatile compounds (Bargmann et al., 1993; Bargmann and Horvitz, 1991). AWC neurons have also recently been shown to function in thermosensation and to respond to several other types of sensory stimuli, indicating that this neuron plays multiple roles in sensing environments and modulating C. elegans behavior (Kuhara et al., 2008; Zaslaver et al., 2015). We believe that our single-cell RNA-seq analysis of AWC neurons provides a useful transcriptomic dataset with which the C. elegans community can study diverse functions of AWC. Our study has demonstrated that AWC neurons can sense different odors produced by A. oligospora (Figure 4A), and the response is likely to be primary, as strong activation of the AWCon neuron was still observed in the unc-13 mutant (Figure 4B). We have identified at least 34 chemosensory GPCRs expressed in the AWC neurons, many of which might be candidates for receptors of the A. oligospora odors. A. oligospora attracts a broad range of nematode species, and we suspect that this is due to the production of a multitude of attractive odorants that are attractive to different nematode species and are sensed by multiple chemoreceptors in the nematodes. As a consequence, attraction to A. oligospora has been maintained in nematodes throughout evolution despite the strong selective pressure against it.
In conclusion, our data has shown that A. oligospora has evolved to produce olfactory mimics of food and sex cues to lure its nematode prey, revealing a novel molecular mechanism for microscopic predator-prey interactions (Figure 8). We expect that this example of coevolution will provide a foundation for future work that will further elucidate how different nematodes interact with different predatory fungi, while having broader implications for understanding other types of predator-prey interactions and their coevolution.
Figure 8. A. oligospora produce volatiles that mimic food and sex cues to attract C. elegans via its olfactory neuron AWCs.
Materials and methods
Strains
All nematodes used in this study were maintained under standard conditions at 20°C. C. elegans strains used in this study included N2, CB4856 (Hawaiian), CX2065 odr-1(n1936), CX2205 odr-3(n2150), CB3329 che-10(e1809), PR678 tax-4(p678), CX3085 tax-2(ks31); tax-4(p678), CX4 odr-7(ky4), PR813 osm-5(p813), NL333 gpa-1(pk15), PS6663 syEx1324[Pstr-2::GCaMP6s::unc-54 3'UTR; Pofm-1::RFP], PS6712 syEx1336[Pstr-2::GCaMP6s::unc-54 3'UTR; Pofm-1::RFP] unc-13(e51); CX3695 kyIs140[str-2::GFP + lin-15(+)], PS7117 odr-7(sy837), PS7117 odr-7(sy837), and TWN1 odr-7(sy837); yphEx1[WRM0639bF05+ ofm-1::RFP]. Different nematode species used in the chemotaxis assays were: PS1010 C. angaria (sp. 3), EG4788 C. portoensis (sp. 6), JU1199 C. afra (sp. 7), JU1325 C. nigoni (sp. 9), JU1373 C. tropicalis (sp. 11), JU1426 C. castelli (sp. 12), AF16 C. briggsae, PB4641 C. remanei, PS312 Pristionchus pacificus, and MT8872 Panagrellus redivivus. The fungal strains used in this study were Arthrobotrys oligospora (CBS115.81), Aspergillus terreus (NIH2624) and Aspergillus niger (ATCC11414).
Chemotaxis assays
Unless noted otherwise, chemotaxis assays were population assays performed with synchronized adult animals in the presence of sodium azide as described previously (Bargmann et al., 1993). The responses of different nematode species to A. oligospora were evaluated with a 4-point assay as described in Figure 1A. Chemotactic responses to different chemicals were tested using the standard 2-point assays (Bargmann et al., 1993). Chemicals were diluted 10−2 for chemotaxis assays unless specified otherwise. The results were subjected to standard t-tests for statistical significance.
Headspace sampling
Headspace volatiles were sampled using solid-phase microextraction (SPME). Prior to collection, each SPME fiber (divinylbenzene/carboxen/polydimethylsiloxane, 50/30 mm; Supelco, Bellefonte, PA) was pre-conditioned for 10 min at 275°C in the GC injector. The SPME fibers were inserted through the vial septa and exposed for a time period of 12–48 hr at room temperature, and then analyzed immediately.
GC X GC TOF MS analyses
Comprehensive two-dimensional gas chromatographic mass spectral analyses (GC x GC-TOFMS) were carried out using a Pegasus 4D (Leco Corp., ST. Joseph, MI) equipped with an Rxi-5ms primary column (5% diphenyl/95% dimethyl polysiloxane; 30m length x 250 mm i.d. x 0.25 mm film thickness; Restek, Bellefonte, PA) and an Rxi-17 secondary column (50% diphenyl/50% dimethyl polysiloxane; 0.8 m length x 180 mm i.d. x 0.18 mm film thickness; Restek, Bellefonte, PA). The primary column oven temperature was held at 40°C for 1 min, then increased at a rate of 3 °C/min to a temperature of 150°C, after which it was increased at a rate of 12 °C/min to a final temperature of 275°C. The secondary column oven followed a temperature program that maintained a + 15°C offset from the primary column oven. A 3.5 s modulation period (0.80 hot pulse; 0.95 cold pulse) was used with a modulator temperature program which initiated at 105°C for 1 min then increased at 5 °C/min to a final temperature of 275°C. Splitless injection was used, with the split valve opening at 20s. The carrier gas flow rate was 1.3 mL/min.
Calcium imaging
Calcium imaging was performed in a microfluidic olfactory chip designed by the Bargmann lab (Chronis et al., 2007). Adult animals expressing GCaMP6 under the str-2 promoter in the wild-type (N2) or the unc-13(e51) background were used for imaging. Plasmid pSM was used as the backbone plasmid for cloning Pstr-2 ::GCaMP6 (Mccarroll, 2005). Animals were presented with alternating streams of S-basal buffer and different fungal odorants diluted in S-complete buffer. Image analysis was done using a custom script written in Python that calculated fluorescence intensity from an individual region of interest (ROI) (Cho and Sternberg, 2014).
Mating behavior of C. afra
C. afra was maintained on OP50 and NG medium as for C. elegans. To track the mating behavior of the nematodes, L4 male and female animals were picked onto separate plates 24 hr before tracking. One female and two male adults were then placed on the opposite sides of a 3 cm chemotaxis plate and recorded immediately until mating occurred or 30 min if no mating behavior was observed.
Genetic screen, SNP mapping and whole-genome sequencing
N2 animals were treated with ethyl methanesulfonate (EMS) following a standard protocol (Kutscher and Shaham, 2014). Semi-synchronized F2 populations were subjected to the chemotaxis screens. Mutants that chemotacted to IAA, instead of MMB, were collected and subjected to another round of assays. Approximately 40,000 genomes were screened, and about a dozen mutants were isolated. Two mutants that exhibited a strong phenotype were subjected to further analyses. SNP mapping was done using methods developed by the Jorgensen lab (Davis et al., 2005). Whole-genome sequencing data were analyzed by the CloudMap pipeline developed by the Hobert lab (Minevich et al., 2012).
Single cell transcriptomic analyses of the AWC neuron
GFP-labeled AWCon cells from the C. elegans strain CX3695 (provided by the Bargmann lab) were micro-dissected and were frozen immediately in liquid nitrogen. Cells were lysed gently and subjected to 3’-tailed RT-PCR based on a method developed to amplify cDNA from a single cell (Schwarz et al., 2012). We dissected nine individual AWC neurons, separately amplified their transcripts with RT-PCR, and purified and quantitated the RT-PCR products before sequencing them. We first removed equal aliquots from the nine separate purified products into a single AWC RT-PCR mixture, and sequenced the mixture as a single AWC RT-PCR pool. In order to obtain independent biological replicates for statistical analysis, we then also sequenced five aliquots of five individual RT-PCR products from five individually dissected AWC neurons.
As a control for detecting the activities of genes with housekeeping and other functions not specific to AWC, we reused two published RNA-seq read sets, generated by RT-PCR of two separate dilute aliquots from a single preparation of C. elegans whole animal RNA (Schwarz et al., 2012). The preparation was obtained from starved mixed-stage hermaphrodites, mostly L3 and L4 stage larvae, with some young adults and L2-stage larvae, and a few L1-stage larvae and eggs; as before, we briefly describe it as ‘larval RNA’. This larval RNA had previously been quantitated and tested for quality with a Bioanalyzer, and successfully used for standard full-length RNA-seq (Mortazavi et al., 2010). The two RNA-seq read sets counted as technical rather than biological replicates, because their starting RNA aliquots derived from a single whole larval RNA stock. However, single-cell RT-PCR amplification is an inherently noisy procedure for both technical and biological reasons (Davidson et al., 2014), and the different expression values for genes in the two larval RNA-seq read sets therefore provided important information about the noisiness of comparisons between expression levels of genes in AWC neurons versus whole animals. We thus treated these two larval read sets as independent replicates in our statistical analysis (below), while pooling them into a single data set for estimates of gene activity in whole larvae.
We did two rounds of RNA-seq analysis. Our initial round of RNA-seq analysis was done by methods essentially identical to those in Schwarz et al. (2012). In particular, we mapped raw RNA-seq reads to C. elegans protein-coding genes from the WS190 release of WormBase with bowtie (Langmead et al., 2009), and used ERANGE 3.1 (Mortazavi et al., 2008) to quantitate their mapped read counts per gene and their gene expression levels in Reads per Kilobase of exon model per Million mapped reads (RPKM). We carried out this initial analysis using only the RNA-seq data from our pool of nine AWC cells, along with our previously published whole-larval RNA-seq data as a control for housekeeping and non-specific genes. RPKM values from this analysis (mapped to current gene names in WormBase), along with current gene annotations, are given in Figure 7—source data 2.
Subsequently, as we added more data, both our sequencing technologies for the AWC RT-PCRs and our RNA-seq analysis methods became more advanced than they had been in Schwarz et al. (2012). For RNA-seq, although the first round of AWC RNA-seq had raw 38-nt sequencing reads as before, the second round had raw 50-nt reads instead, and their FastQ quality coding scale (Cock et al., 2010) changed between the first and second AWC sequencing runs. To allow coherent use of all AWC reads as a single pooled data set for quantitating AWC gene expression, we recoded the quality scale of the older AWC read set (fastq-illumina, phred score with offset of 64) to that of the newer (fastq-sanger, phred score with offset of 33) using seqret from EMBOSS 6.5.7 (Rice et al., 2000) before further analysis.
After RNA-seq of the AWC cells, we likewise used more advanced methods to analyze the RNA-seq data. All reads were quality-filtered as follows: AWC reads that had failed Chastity filtering were discarded (Chastity filtering had not been available for the larval reads); raw 38-nt reads were trimmed 1 nt to 37 nt; all reads were trimmed to remove any indeterminate (‘N’) residues or residues with a quality score of less than 3; and raw 38-nt reads that had been trimmed below 37 nt were deleted, as were raw 50-nt AWC reads that had been trimmed below 50 nt. This left us with a total of 131,614,334 filtered AWC reads for analysis, of which 96,433,929 reads could be partitioned into biologically independent replicates (Supplementary file 2A).
Proceeding to RNA-seq analysis proper, we used RSEM (version 1.2.17; Li and Dewey, 2011, PMID 21816040) with bowtie2 (Langmead and Salzberg, 2012) and SAMTools (Li et al., 2009) to map filtered reads to a C. elegans gene index and generate readcounts and gene expression levels in transcripts per million (TPM). To create the C. elegans gene index, we ran RSEM's rsem-prepare-reference with the arguments ‘–bowtie2 –transcript-to-gene-map’ upon a collection of coding DNA sequences (CDSes) from both protein-coding and non-protein-coding C. elegans genes in WormBase release WS245 (Harris et al., 2014). The CDS sequences were obtained from ftp://ftp.sanger.ac.uk/pub2/wormbase/releases/WS245/species/c_elegans/PRJNA13758/c_elegans.PRJNA13758.WS245.mRNA_transcripts.fa.gz and ftp://ftp.sanger.ac.uk/pub2/wormbase/releases/WS245/species/c_elegans/PRJNA13758/c_elegans.PRJNA13758.WS245.ncRNA_transcripts.fa.gz.
To analyse overall gene expression levels in AWC neurons, we pooled all 132 million RNA-seq reads from both the nine-cell pool and the five individual cells into a single AWC read set before mapping them and computing their genewise TPMs. Similarly, to analyse overall gene expression levels in whole larvae for comparison to AWC neurons, we pooled all 23.4 million RNA-seq reads from both larval data sets into a single larval data set before mapping it and computing its TPMs.
Conversely, to analyze the statistical significance for differences of gene expression between AWC neurons and whole larvae, we used only the read sets from the individual AWC neurons. Our past experience with single-cell RNA-seq data sets (Schwarz et al., 2012) led us to expect that gene expression data from single AWCs would be highly noisy, but that this noise would be canceled out by pooling non-overlapping subsets of the individual AWCs and analyzing them as independent biological replicates. We thus mapped and quantitated the single-cell AWC read sets as two pools (cells 1, 2, and 4 for pool 1; cells 3 and 5 for pool 2). Cells were chosen for the two pools so that the total number of reads in the two pools would be as close to equal as possible (45.7 million and 50.7 million reads for pools 1 and 2; Supplementary file 2A).
For each RNA-seq data set of interest, we computed mapped reads and expression levels per gene by running RSEM's rsem-calculate-expression with the arguments "--bowtie2 -p 8 --no-bam-output --calc-pme --calc-ci --ci-credibility-level 0.99 --fragment-length-mean 200 ‑‑fragment-length-sd 20 --estimate-rspd --ci-memory 30000". These arguments, in particular '--estimate-rspd', were aimed at dealing with single-end data from 3'-biased RT-PCR reactions; the arguments '--phred33-quals' and '--phred64-quals' were also used for the AWC and larval reads, respectively. We computed posterior mean estimates both for read counts and for gene expression levels, and rounded PMEs of read counts down to the nearest lesser integer. We also computed 99% credibility intervals (CIs) for expression data, so that we could use the minimum value in the 99% CI for TPM as a reliable minimum estimate of a gene's expression (minTPM). We defined non-zero, above-background expression for a gene in a given RNA-seq data set by that gene having an minimum estimated expression level of at least 0.1 transcripts per million (TPM) in a credibility interval of 99% (minTPM). The most relevant results from RSEM analysis are given in Figure 7—source data 1. The numbers of genes being scored as expressed in AWC above background levels, for various data sets, are given in Supplementary file 2B.
For individual genes, we computed the significance for differences of gene activity between AWC and whole larvae with DESeq2 (Love et al., 2014). DESeq2 was run in R 3.2.3, using default parameters to compute p-values that had been adjusted for multiple testing (Noble, 2009) by the collective false discovery rate (FDR) formula (Benjamini and Hochberg, 1995). We computed these FDR-adjusted p-values (‘padj’) by testing two pools of individual AWC read sets against two larval sets, at an α threshold of 0.01; genes for which this gave significant changes are listed in Figure 7—source data 1. We defined genes expressed significantly more strongly in AWC than in whole larvae as those genes with padj ≤ 0.01 and AWC/larvae expression ratios of >1.
Data availability
RNA-seq reads for the wild-type AWC neurons are available in the NCBI Sequence Read Archive (SRA), under accession number SRP074082 (http://trace.ncbi.nlm.nih.gov/Traces/sra/?study=SRP074082). RNA-seq reads for the two pools of whole C. elegans mixed-stage wild-type N2 larvae were previously published by Schwarz et al. (2012), and are available in the NCBI SRA under accession number SRP015688 (http://trace.ncbi.nlm.nih.gov/Traces/sra/?study=SRP015688).
Acknowledgements
We thank Marie-Anne Felix (IBENS), Cori Bargmann (The Rockefeller University), Ryoji Shinya (Sternberg lab), Vivian Chiu (then in Sternberg lab) and the Caenorhabditis Genetics Center (University of Minnesota) for providing the nematode strains used in this study. We also thank Valerie Knowlton (NCSU) for acquiring the SEM image and Clay Wang (USC) for providing the Aspergillus strains. Michael Milligan (SUNY Fredonia) provided invaluable assistance with GC x GC-TOFMS analyses and Chris Cronin and Sreekanth Chalasani (Salk Institute) provided advices for the calcium imaging set-up. We thank Gladys Medina, Sarah Kim, Chiang Yu Shi and Ka-Naam Heung for providing technical assistance and members of the Sternberg laboratory for helpful discussion. This work was supported by HHMI, with which PWS is an investigator, by the NIH K99 award 1 K99GM108867-1 to YPH, by NIH GM084389 to PWS, and startup funds from Academia Sinica to YPH. RN was supported by T32-GM007616.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Funding Information
This paper was supported by the following grants:
National Institute of General Medical Sciences 1K99GM108867-1 to Yen-Ping Hsueh.
Academia Sinica to Yen-Ping Hsueh.
National Institutes of Health T32-GM007616 to Paul W Sternberg.
Howard Hughes Medical Institute 047-101 to Paul W Sternberg.
National Institutes of Health GM084389 to Paul W Sternberg.
Additional information
Competing interests
The authors declare that no competing interests exist.
Author contributions
Y-PH, Conceptualization, Data curation, Formal analysis, Supervision, Funding acquisition, Validation, Investigation, Writing—original draft, Writing—review and editing.
MRG, Data curation, Formal analysis, Writing—original draft.
EMS, Data curation, Formal analysis, Methodology, Writing—original draft, Writing—review and editing.
RDN, Data curation, Formal analysis.
C-HL, Data curation, Formal analysis.
SG, Data curation, Validation.
FCS, Formal analysis, Supervision, Writing—review and editing.
PWS, Conceptualization, Resources, Supervision, Funding acquisition, Writing—review and editing.
Additional files
Major datasets
The following dataset was generated:
Hsueh YP,Gronquist MR,Schwarz EM,Nath RD,Lee CH,Gharib S,Schroeder FC,Sternberg PW,2016,Single-cell RNA-seq of AWC[on] chemosensory neurons of Caenorhabditis elegans,https://trace.ncbi.nlm.nih.gov/Traces/sra/?study=SRP074082,Publicly available at the NCBI Sequence Read Archive (accession no. SRP074082)
The following previously published datasets were used:
Schwarz EM,Kato M,Sternberg PW,2012,C. elegans linker cell transcriptome,https://trace.ncbi.nlm.nih.gov/Traces/sra/?study=SRP015688,Publicly available at NCBI Sequence Read Archive (accession no. SRP015688)
References
- Abrams PA. The evolution of predator-prey interactions: theory and evidence. Annual Review of Ecology and Systematics. 2000;31:79–105. doi: 10.1146/annurev.ecolsys.31.1.79. [DOI] [Google Scholar]
- Balan J, Gerber NN. Attraction and killing of nematode Panagrellus redivivus by predaceous fungus Arthrobotrys dactyloides. Nematologica. 1972;18:163–173. doi: 10.1163/187529272X00403. [DOI] [Google Scholar]
- Bargmann CI, Hartwieg E, Horvitz HR. Odorant-selective genes and neurons mediate olfaction in C. elegans. Cell. 1993;74:515–527. doi: 10.1016/0092-8674(93)80053-H. [DOI] [PubMed] [Google Scholar]
- Bargmann CI, Horvitz HR. Chemosensory neurons with overlapping functions direct chemotaxis to multiple chemicals in C. elegans. Neuron. 1991;7:729–742. doi: 10.1016/0896-6273(91)90276-6. [DOI] [PubMed] [Google Scholar]
- Bargmann CI. Chemosensation in C. elegans. WormBook. 2006:1–29. doi: 10.1895/wormbook.1.123.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barron GL. Topics in Mycobiology No. 1. Canada: Canadian Biological Publications Ltd; 1977. The nematode-destroying fungi. [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate - a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B-Methodological. 1995;57:289–300. doi: 10.2307/2346101. [DOI] [Google Scholar]
- Chalasani SH, Chronis N, Tsunozaki M, Gray JM, Ramot D, Goodman MB, Bargmann CI. Dissecting a circuit for olfactory behaviour in Caenorhabditis elegans. Nature. 2007;450:63–70. doi: 10.1038/nature06292. [DOI] [PubMed] [Google Scholar]
- Cho JY, Sternberg PW. Multilevel modulation of a sensory motor circuit during C. elegans sleep and arousal. Cell. 2014;156:249–260. doi: 10.1016/j.cell.2013.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chronis N, Zimmer M, Bargmann CI. Microfluidics for in vivo imaging of neuronal and behavioral activity in Caenorhabditis elegans. Nature Methods. 2007;4:727–731. doi: 10.1038/nmeth1075. [DOI] [PubMed] [Google Scholar]
- Cock PJ, Fields CJ, Goto N, Heuer ML, Rice PM. The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Research. 2010;38:1767–1771. doi: 10.1093/nar/gkp1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colbert HA, Bargmann CI. Odorant-specific adaptation pathways generate olfactory plasticity in C. elegans. Neuron. 1995;14:803–812. doi: 10.1016/0896-6273(95)90224-4. [DOI] [PubMed] [Google Scholar]
- Colosimo ME, Tran S, Sengupta P. The divergent orphan nuclear receptor ODR-7 regulates olfactory neuron gene expression via multiple mechanisms in Caenorhabditis elegans. Genetics. 2003;165:1779–1791. doi: 10.1093/genetics/165.4.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin C. Insectivorous Plants. D. Appleton and Company; 1875. [Google Scholar]
- Davidson B, Abeler VM, Førsund M, Holth A, Yang Y, Kobayashi Y, Chen L, Kristensen GB, Shih I, Wang TL. Gene expression signatures of primary and metastatic uterine leiomyosarcoma. Human Pathology. 2014;45:691–700. doi: 10.1016/j.humpath.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis MW, Hammarlund M, Harrach T, Hullett P, Olsen S, Jorgensen EM. Rapid single nucleotide polymorphism mapping in C. elegans. BMC Genomics. 2005;6:118. doi: 10.1186/1471-2164-6-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawkins R, Krebs JR. Arms races between and within species. Proceedings of the Royal Society B: Biological Sciences. 1979;205:489–511. doi: 10.1098/rspb.1979.0081. [DOI] [PubMed] [Google Scholar]
- Di Giusto B, Bessière JM, Guéroult M, Lim LBL, Marshall DJ, Hossaert-McKey M, Gaume L. Flower-scent mimicry masks a deadly trap in the carnivorous plant Nepenthes rafflesiana. Journal of Ecology. 2010;98:845–856. doi: 10.1111/j.1365-2745.2010.01665.x. [DOI] [Google Scholar]
- Díaz P, Señoráns FJ, Reglero G, Ibañez E. Truffle aroma analysis by headspace solid phase microextraction. Journal of Agricultural and Food Chemistry. 2002;50:6468–6472. doi: 10.1021/jf025609t. [DOI] [PubMed] [Google Scholar]
- Félix MA, Braendle C, Cutter AD. A streamlined system for species diagnosis in Caenorhabditis (Nematoda: Rhabditidae) with name designations for 15 distinct biological species. PLoS One. 2014;9:e94723. doi: 10.1371/journal.pone.0094723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer EL, Tate J, Punta M. Pfam: the protein families database. Nucleic Acids Research. 2014;42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flach A, Marsaioli AJ, Singer RB, Amaral MC, Menezes C, Kerr WE, Batista-Pereira LG, Corrêa AG. Pollination by sexual mimicry in Mormolyca ringens: a floral chemistry that remarkably matches the pheromones of virgin queens of Scaptotrigona sp. Journal of Chemical Ecology. 2006;32:59–70. doi: 10.1007/s10886-006-9351-1. [DOI] [PubMed] [Google Scholar]
- Frézal L, Félix M-A. C. elegans outside the Petri dish. eLife. 2015;4:e05849. doi: 10.7554/eLife.05849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futai K. Pine wood nematode, Bursaphelenchus xylophilus. Annual Review of Phytopathology. 2013;51:61–83. doi: 10.1146/annurev-phyto-081211-172910. [DOI] [PubMed] [Google Scholar]
- Harris TW, Baran J, Bieri T, Cabunoc A, Chan J, Chen WJ, Davis P, Done J, Grove C, Howe K, Kishore R, Lee R, Li Y, Muller HM, Nakamura C, Ozersky P, Paulini M, Raciti D, Schindelman G, Tuli MA, Van Auken K, Wang D, Wang X, Williams G, Wong JD, Yook K, Schedl T, Hodgkin J, Berriman M, Kersey P, Spieth J, Stein L, Sternberg PW. WormBase 2014: new views of curated biology. Nucleic Acids Research. 2014;42:D789–D793. doi: 10.1093/nar/gkt1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes KF, Gemeno C, Yeargan KV, Millar JG, Johnson KM. Aggressive chemical mimicry of moth pheromones by a bolas spider: how does this specialist predator attract more than one species of prey? Chemoecology. 2002;12:99–105. doi: 10.1007/s00049-002-8332-2. [DOI] [Google Scholar]
- Hobert O. The neuronal genome of Caenorhabditis elegans. WormBook. 2013:1–106. doi: 10.1895/wormbook.1.161.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsueh YP, Mahanti P, Schroeder FC, Sternberg PW. Nematode-trapping fungi eavesdrop on nematode pheromones. Current Biology. 2013;23:83–86. doi: 10.1016/j.cub.2012.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jürgens A, El-Sayed AM, Suckling DM. Do carnivorous plants use volatiles for attracting prey insects? Functional Ecology. 2009;23:875–887. doi: 10.1111/j.1365-2435.2009.01626.x. [DOI] [Google Scholar]
- Kim K, Sato K, Shibuya M, Zeiger DM, Butcher RA, Ragains JR, Clardy J, Touhara K, Sengupta P. Two chemoreceptors mediate developmental effects of dauer pheromone in C. elegans. Science. 2009;326:994–998. doi: 10.1126/science.1176331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhara A, Okumura M, Kimata T, Tanizawa Y, Takano R, Kimura KD, Inada H, Matsumoto K, Mori I. Temperature sensing by an olfactory neuron in a circuit controlling behavior of C. elegans. Science. 2008;320:803–807. doi: 10.1126/science.1148922. [DOI] [PubMed] [Google Scholar]
- Kutscher LM, Shaham S. Forward and reverse mutagenesis in C. elegans. WormBook. 2014:1–26. doi: 10.1895/wormbook.1.167.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Käll L, Krogh A, Sonnhammer EL. A combined transmembrane topology and signal peptide prediction method. Journal of Molecular Biology. 2004;338:1027–1036. doi: 10.1016/j.jmb.2004.03.016. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nature Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biology. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup The sequence alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim G. What made ‘Wee Stinky’ stink – and when. The Spadix Speaks: Cornell’s Titan Arum Blog. 2012 http://titanarum.cals.cornell.edu/2012/03/25/what-made-wee-stinky-stink-and-when/
- Liou GY, Tzean SS. Phylogeny of the genus Arthrobotrys and allied nematode-trapping fungi based on rDNA sequences. Mycologia. 1997;89:876–884. doi: 10.2307/3761108. [DOI] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A. Prediction and analysis of coiled-coil structures. Methods in Enzymology. 1996;266:513–525. doi: 10.1016/S0076-6879(96)66032-7. [DOI] [PubMed] [Google Scholar]
- Mant J, Brändli C, Vereecken NJ, Schulz CM, Francke W, Schiestl FP. Cuticular hydrocarbons as sex pheromone of the bee Colletes cunicularius and the key to its mimicry by the sexually deceptive orchid, Ophrys exaltata. Journal of Chemical Ecology. 2005;31:1765–1787. doi: 10.1007/s10886-005-5926-5. [DOI] [PubMed] [Google Scholar]
- Mccarroll S. The Statistical and Molecular Logic of Gene Expression Patterns in Caenorhabditis Elegans. San Francisco: University of California; 2005. [Google Scholar]
- Meisel JD, Panda O, Mahanti P, Schroeder FC, Kim DH. Chemosensation of bacterial secondary metabolites modulates neuroendocrine signaling and behavior of C. elegans. Cell. 2014;159:267–280. doi: 10.1016/j.cell.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minevich G, Park DS, Blankenberg D, Poole RJ, Hobert O. CloudMap: a cloud-based pipeline for analysis of mutant genome sequences. Genetics. 2012;192:1249–1269. doi: 10.1534/genetics.112.144204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Schwarz EM, Williams B, Schaeffer L, Antoshechkin I, Wold BJ, Sternberg PW. Scaffolding a Caenorhabditis nematode genome with RNA-seq. Genome Research. 2010;20:1740–1747. doi: 10.1101/gr.111021.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Niu Q, Huang X, Zhang L, Xu J, Yang D, Wei K, Niu X, An Z, Bennett JW, Zou C, Yang J, Zhang KQ. A trojan horse mechanism of bacterial pathogenesis against nematodes. PNAS. 2010;107:16631–16636. doi: 10.1073/pnas.1007276107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble WS. How does multiple testing correction work? Nature Biotechnology. 2009;27:1135–1137. doi: 10.1038/nbt1209-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordbring-Hertz B. Nematophagous fungi: strategies for nematode exploitation and for survival. Microbiological Sciences. 1988;5:108–116. [PubMed] [Google Scholar]
- Pacioni G, Cerretani L, Procida G, Cichelli A. Composition of commercial truffle flavored oils with GC-MS analysis and discrimination with an electronic nose. Food Chemistry. 2014;146:30–35. doi: 10.1016/j.foodchem.2013.09.016. [DOI] [PubMed] [Google Scholar]
- Park D, O'Doherty I, Somvanshi RK, Bethke A, Schroeder FC, Kumar U, Riddle DL. Interaction of structure-specific and promiscuous G-protein-coupled receptors mediates small-molecule signaling in Caenorhabditis elegans. PNAS. 2012;109:9917–9922. doi: 10.1073/pnas.1202216109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennazza G, Fanali C, Santonico M, Dugo L, Cucchiarini L, Dachà M, D'Amico A, Costa R, Dugo P, Mondello L. Electronic nose and GC-MS analysis of volatile compounds in tuber magnatum Pico: evaluation of different storage conditions. Food Chemistry. 2013;136:668–674. doi: 10.1016/j.foodchem.2012.08.086. [DOI] [PubMed] [Google Scholar]
- Powell S, Szklarczyk D, Trachana K, Roth A, Kuhn M, Muller J, Arnold R, Rattei T, Letunic I, Doerks T, Jensen LJ, von Mering C, Bork P. eggNOG v3.0: orthologous groups covering 1133 organisms at 41 different taxonomic ranges. Nucleic Acids Research. 2012;40:D284–D289. doi: 10.1093/nar/gkr1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradel E, Zhang Y, Pujol N, Matsuyama T, Bargmann CI, Ewbank JJ. Detection and avoidance of a natural product from the pathogenic bacterium Serratia marcescens by Caenorhabditis elegans. PNAS. 2007;104:2295–2300. doi: 10.1073/pnas.0610281104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pramer D, Stoll NR. Nemin: a morphogenic substance causing trap formation by predaceous fungi. Science. 1959;129:966–967. doi: 10.1126/science.129.3354.966. [DOI] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. EMBOSS: the european molecular biology open software suite. Trends in Genetics. 2000;16:276–277. doi: 10.1016/S0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Richmond JE, Davis WS, Jorgensen EM. UNC-13 is required for synaptic vesicle fusion in C. elegans. Nature Neuroscience. 1999;2:959–964. doi: 10.1038/14755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosier RL, Langkilde T. Behavior under risk: how animals avoid becoming dinner. Nature Education Knowledge. 2011;2:8 [Google Scholar]
- Schiestl FP, Ayasse M, Paulus HF, Löfstedt C, Hansson BS, Ibarra F, Francke W. Sex pheromone mimicry in the early spider orchid (ophrys sphegodes): patterns of hydrocarbons as the key mechanism for pollination by sexual deception. Journal of Comparative Physiology A: Sensory, Neural, and Behavioral Physiology. 2000;186:567–574. doi: 10.1007/s003590000112. [DOI] [PubMed] [Google Scholar]
- Schwarz EM, Kato M, Sternberg PW. Functional transcriptomics of a migrating cell in Caenorhabditis elegans. PNAS. 2012;109:16246–16251. doi: 10.1073/pnas.1203045109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta P, Chou JH, Bargmann CI. odr-10 encodes a seven transmembrane domain olfactory receptor required for responses to the odorant diacetyl. Cell. 1996;84:899–909. doi: 10.1016/S0092-8674(00)81068-5. [DOI] [PubMed] [Google Scholar]
- Sengupta P, Colbert HA, Bargmann CI. The C. elegans gene odr-7 encodes an olfactory-specific member of the nuclear receptor superfamily. Cell. 1994;79:971–980. doi: 10.1016/0092-8674(94)90028-0. [DOI] [PubMed] [Google Scholar]
- Shinya R, Chen A, Sternberg PW. Sex attraction and mating in bursaphelenchus okinawaensis and B. xylophilus. Journal of Nematology. 2015;47:176–183. [PMC free article] [PubMed] [Google Scholar]
- Shirasu M, Fujioka K, Kakishima S, Nagai S, Tomizawa Y, Tsukaya H, Murata J, Manome Y, Touhara K. Chemical identity of a rotting animal-like odor emitted from the inflorescence of the titan arum (Amorphophallus titanum) Bioscience, Biotechnology, and Biochemistry. 2010;74:2550–2554. doi: 10.1271/bbb.100692. [DOI] [PubMed] [Google Scholar]
- Sommer RJ. Pristionchus pacificus. WormBook. 2006:1–8. doi: 10.1895/wormbook.1.102.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan J, Dillman AR, Macchietto MG, Heikkinen L, Lakso M, Fracchia KM, Antoshechkin I, Mortazavi A, Wong G, Sternberg PW. The draft genome and transcriptome of Panagrellus redivivus are shaped by the harsh demands of a free-living lifestyle. Genetics. 2013;193:1279–1295. doi: 10.1534/genetics.112.148809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stensmyr MC, Urru I, Collu I, Celander M, Hansson BS, Angioy AM. Pollination: Rotting smell of dead-horse arum florets. Nature. 2002;420:625–626. doi: 10.1038/420625a. [DOI] [PubMed] [Google Scholar]
- Thomas JH, Robertson HM. The Caenorhabditis chemoreceptor gene families. BMC Biology. 2008;6:42. doi: 10.1186/1741-7007-6-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita B, Inoue H, Chaya K, Nakamura A, Hamamura N, Ueno K, Watanabe K, Ose Y. Identification of dimethyl disulfide-forming bacteria isolated from activated sludge. Applied and Environmental Microbiology. 1987;53:1541–1547. doi: 10.1128/aem.53.7.1541-1547.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troemel ER, Sagasti A, Bargmann CI. Lateral signaling mediated by axon contact and calcium entry regulates asymmetric odorant receptor expression in C. elegans. Cell. 1999;99:387–398. doi: 10.1016/S0092-8674(00)81525-1. [DOI] [PubMed] [Google Scholar]
- Wootton JC. Non-globular domains in protein sequences: automated segmentation using complexity measures. Computers & Chemistry. 1994;18:269–285. doi: 10.1016/0097-8485(94)85023-2. [DOI] [PubMed] [Google Scholar]
- Yang Y, Yang E, An Z, Liu X. Evolution of nematode-trapping cells of predatory fungi of the Orbiliaceae based on evidence from rRNA-encoding DNA and multiprotein sequences. PNAS. 2007;104:8379–8384. doi: 10.1073/pnas.0702770104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaslaver A, Liani I, Shtangel O, Ginzburg S, Yee L, Sternberg PW. Hierarchical sparse coding in the sensory system of Caenorhabditis elegans. PNAS. 2015;112:1185–1189. doi: 10.1073/pnas.1423656112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Lu H, Bargmann CI. Pathogenic bacteria induce aversive olfactory learning in Caenorhabditis elegans. Nature. 2005;438:179–184. doi: 10.1038/nature04216. [DOI] [PubMed] [Google Scholar]