Abstract
Genomic regions containing trinucleotide repeats (TNRs) are highly unstable, as the repeated sequences exhibit a high rate of mutational change, in which they undergo either a contraction or an expansion of repeat numbers. Although expansion of TNRs is associated with several human genetic diseases, the expansion mechanism is poorly understood. Extensive studies in model organisms have indicated that instability of TNRs occurs by several mechanisms, including replication slippage, DNA repair and recombination. In all models, the formation of secondary structures by disease-associated TNRs is a critical step in the mutation process. In this report, we demonstrate that TNRs and inverted repeats (IRs) both of which have the potential to form secondary structures in vivo, increase spontaneous unequal sister-chromatid exchange (SCE) in vegetatively growing yeast cells. Our results also show that TNR-mediated SCE events are independent of RAD50, MRE11 and RAD51, whereas IR-stimulated SCEs are dependent on the RAD52 epistasis-group genes. We propose that many TNR expansion mutations occur by SCE.
INTRODUCTION
Most eukaryotic genomes contain microsatellite DNA, in which a number of bases are repeated several times. Such genomic regions are highly unstable, because the repeated sequences undergo frequent changes in their tract length: either a contraction or an expansion of the number of repeat units. The instability of microsatellites results in polymorphic alleles, allowing them to be extremely useful as markers for mapping studies in humans and other eukaryotes. In addition, changes in the repeat number can alter the function or the expression pattern of a gene, depending on the position of the repeat tract.
Several studies have indicated that alteration of the dinucleotide repeat-tract length is characteristic of certain types of human cancer that are associated with mismatch-repair defects (1). Similarly, expansion of trinucleotide repeats (TNRs) is known to cause at least 14 hereditary diseases in humans, including Huntington's disease, fragile X syndrome, myotonic dystrophy and Friedreich's ataxia (2,3). Among all of the possible triplet repeats, only three (CAG.CTG, CGG.GGC and GAA.TTC; hereafter, CAG, CGG and GAA, respectively) are known to be associated with repeat disorders and fragile sites. TNR expansion mutations are dynamic, in that the mutant allele can undergo further expansion in subsequent generations or during the lifespan of an individual. Recent studies have indicated that germ-line instability occurs throughout development, during both meiosis and mitosis (4–7). The mechanisms of these expansion mutations are poorly understood.
Extensive studies in model organisms, including bacteria, yeast and mice, have revealed that multiple DNA transaction pathways, including errors during DNA replication, DNA damage repair and meiotic recombination, can lead to repeat-tract length expansions (6–24). Although mechanistic details vary among the models, it is generally believed that secondary-structure formation by disease-associated TNRs is a critical step in the expansion process. It has been shown in vitro that single-stranded DNA containing TNRs can form secondary structures [reviewed in (25)]. For example, CAG and CGG repeats can form hairpin structures (25,26). GAA repeats have been shown to form triplex structures, although one report suggested that GAA and TTC repeats can form hairpin structures during in vitro DNA synthesis (27,28). Studies with diploid yeast strains containing heterozygous TNR-insertion mutations have indicated that TNRs in single-stranded DNA are likely to form hairpin structures in vivo (29). Hairpin structures are also the substrates for several nucleases in vivo. An endonucleolytic cleavage of such a secondary structure would produce a double-stranded break (DSB) in the DNA. Accordingly, long CAG tracts have been shown to induce DSBs within the repeated sequences during meiosis in yeast, and they have also been shown to act as fragile sites in mitotic yeast cells (22,30).
Formation of secondary structures is also known to compromise DNA replication in vitro (31,32), and in vivo studies have shown that disease-associated TNRs attenuate replication-fork progression (33,34). Replication blockage is believed to cause transient dissociation and reassociation of the replication fork, facilitating occasional misalignment between the template and the newly synthesized DNA strand. During reassociation, the nascent strand, instead of pairing with the original template, can switch the template and pair with the sister chromatid or the homolog, resulting in a homologous recombination event. Therefore, one would expect that the presence of TNRs will increase the frequency of sister-chromatid exchange (SCE). To explore this possibility, we monitored CAG-repeat-stimulated SCE in haploid yeast cells. Our results show that both CAG repeats and inverted repeats (IRs) stimulate spontaneous unequal SCE during vegetative growth in the yeast Saccharomyces cerevisiae. The results also indicate that the occurrence of IR-stimulated SCEs is dependent on RAD52 group genes, whereas TNR-stimulated SCEs are RAD51 independent.
MATERIALS AND METHODS
Yeast strains and media
The genotypes of all yeast strains used in this study are shown in Table 1. Yeast strains used in this study were derived from AS13 (35). All genetic manipulations were carried out following standard procedures, and media used in this study are described in (36). Three different sister-chromatid substrates were used in this study. The control sister-chromatid substrate (his3-SCScontrol) had a 120-bp Ho-cut site inserted into the BglII site within the his3-Δ3′ construct (37). The other two substrates had either a 140-bp IR (his3-SCSpal140), or 71 copies of the CAG repeat tract (his3-SCSCAG71) inserted within the his3-Δ3′ construct. The his3 sister-chromatid substrates were introduced at the ARG4 locus by a two-step transplacement procedure, using derivatives of the plasmid pDN121. The rad50- and mre11-mutant alleles were introduced by one-step transplacement procedure using EcoRI-digested pNKY83 and BamHI-digested pKJ112-5 plasmids, respectively. The rad52- and rad51- mutant alleles were introduced using BamHI-digested plasmids pSM22 (38) and pΔRAD51 (39), respectively.
Table 1. Yeast strains used in this study.
| Strain | Genotype | Source or reference |
|---|---|---|
| AS13 | MATa lue2-Bst ura3 ade6 | (35) |
| DNY378 | AS13 lys2 arg4 his3Δ arg4::his3-SCSCAG71 | This study |
| DNY380 | AS13 lys2 arg4 his3Δ arg4::his3-SCScontrol | This study |
| DNY381 | DNY380 rad50 | This study |
| DNY382 | DNY380 mre11 | This study |
| DNY384 | DNY378 rad50 | This study |
| DNY385 | DNY378 mre11 | This study |
| DNY393 | AS13 lys2 arg4 his3Δ arg4::his3-SCSpal140 | This study |
| DNY397 | DNY380 rad51 | This study |
| DNY398 | DNY380 rad52 | This study |
| DNY399 | DNY378 rad52 | This study |
| DNY400 | DNY378 rad51 | This study |
| DNY401 | DNY393 rad51 | This study |
| DNY402 | DNY393 rad52 | This study |
| DNY405 | DNY393 rad50 | This study |
| DNY406 | DNY393 mre11 | This study |
Plasmids
Standard molecular biology procedures were used for all plasmid constructions. The his3 sister-chromatid substrates were constructed as follows. A 1.8-kb BamHI fragment containing the entire HIS3 gene was cloned into the BamHI site of pUC19. The resulting plasmid, pDN116, was digested with BclI, present within the HIS3 coding sequence, and ligated with a 32-bp palindromic oligonucleotide containing a BamHI linker at the end, to yield pDN118. A BamHI fragment containing a 114-bp IR (40) was then inserted into the BamHI site present within the IR in pDN118 to generate pDN120. The plasmid pDN120 contains a 140-bp IR at the BclI site within the HIS3 coding sequence.
The his3-Δ3′ construct-containing plasmid, pDN127, was generated by insertion of a KpnI fragment from pDN120 into the KpnI site of pUC19; the orientation is such that the 3′end of the truncated his3 is near the plasmid KpnI site. A filled-in BamHI fragment containing the his3-Δ5′ construct, obtained from pNN287 (41), was then inserted into the filled-in XbaI site of pDN127, to generate pDN133, containing the complete his3-SCSpal140. The his3-Δ5′ construct in pDN133 has about 508 bp of sequence homology with the his3-Δ3′ construct.
An EcoRI–SalI fragment containing his3-SCSpal140 from pDN133 was then inserted into the blunt-ended BclI–BglII digested pDN121, to create pDN135. The plasmid pDN121 is pMLC28ARG4Pst6 (42), containing the URA3 gene at the BamHI site. A PstI fragment containing the complete ARG4 gene is present in pDN121. The his3 sister-chromatid substrate in pDN135 is oriented such that the his3-Δ3′ is near the ARG4 promoter.
The chromosomal HIS3 gene was deleted in a two-step transplacement procedure using BglII-linearized pDN126, which was constructed as described below. The plasmid pDN116 was digested with BstBI and XhoI, blunt-ended with the DNA polymerase Klenow fragment, and self-ligated to generate pDN123, in which the entire HIS3 coding region is deleted. An Asp-718–XbaI fragment from pDN123 containing the his3-deletion fragment was ligated with XbaI- and Asp-718-digested pRS306 to create pDN126.
The sister-chromatid substrate with the Ho-cut site (his3-SCScontrol) was generated using plasmid pMF102 (37), a gift from Michael Fasullo, Ordway Research Institute, Albany, NY. An EcoRI fragment containing the control his3 sister-chromatid substrate was blunt-ended with the Klenow fragment of Escherichia coli DNA polymerase I and was then ligated with the filled-in BglII–BclI-digested pDN121, to generate pDN134. The Ho-cut site is present at the BglII site of the his3-Δ3′ construct.
The his3 sister-chromatid substrate containing CAG repeats (his3-SCSCAG71) was constructed as follows. A KpnI fragment containing his3-Δ3′ was inserted into the KpnI site of pUC19. The resulting plasmid, pDN136, was digested with BglII and ligated with a DNA fragment having BamHI ends and containing 71 copies of CAG repeats to generate pMS1. The triplet repeat-containing fragment was generated by PCR using primers TRP3 (5′-CGCGGATCCATGAAGGCCTTCGAGTCCCTCAAGTCCTTC-3′) and TRP4 (5′-CGCGGATCCGGCGGCTGAGGAAGCTGAGGA-3′), and CJY39 yeast cells, which contain 79 copies of CAG repeats at the SalI site within the HIS4 coding region (22).
Determination of rates of spontaneous sister-chromatid recombination
The rates (events per cell per cell division) of spontaneous SCE were determined by the method of median. A single colony was inoculated into 3 ml of YPD broth. After overnight growth at 30°C, the culture was diluted and added to 13 tubes, each containing 3 ml of YPD broth. Each tube received about 10–20 cells. After 2 days (3 days in the case of repair mutants) of growth at 30°C, the cells were centrifuged, suspended in water, briefly sonicated and then they were plated at suitable dilutions on complete synthetic medium (CSM) to measure the number of viable cells, and onto complete synthetic medium lacking histidine (CSM-His) plates to measure the number of His+ recombinants. Colonies were counted after 4 days of growth at 30°C for wild-type strains, and after 7 days for repair-deficient mutants. At least three independent rate calculations were done for each strain, and the significance was measured by Student's t-test.
RESULTS
Experimental system to monitor sister-chromatid recombination
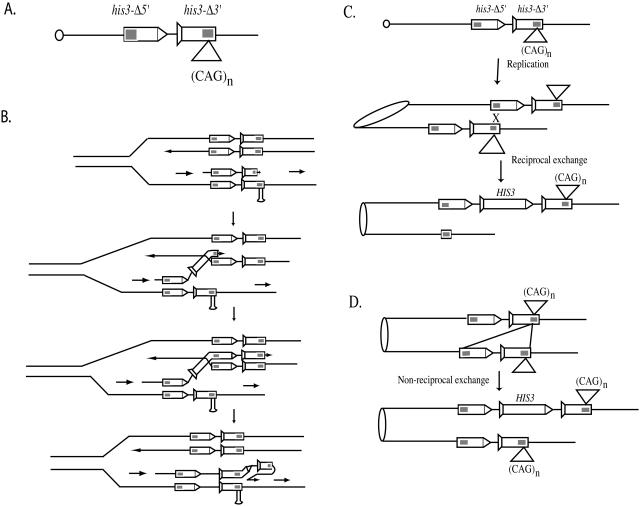
We are interested in knowing whether TNR instability occurs by SCE. Given that disease-associated TNRs attenuate replication-fork progression (31–34), TNRs should increase the frequency of SCE events, if replication pausing stimulates SCE by template switching. Since equal SCE is difficult to follow because of the identical nature of the two sister chromatids, we chose to monitor unequal SCE. As an alternative to the template-switching pathway, the secondary structures that may form due to the presence of the TNRs can generate DSBs; these DSBs can then lead to unequal exchange or non-reciprocal gap repair, resulting in an increased frequency of SCE events (Figure 1).
Figure 1.
Unequal sister-chromatid recombination assay used in this study. (A) The his3 unequal sister-chromatid recombination substrate. The 3′-deletion construct is marked with a tail, and the 5′-deletion substrate is marked with an arrowhead. The shaded region indicates the region that is homologous between the 3′- and 5′-deletion constructs. (B–D) Generation of a wild-type HIS3 gene by sister-chromatid exchange. (B) A wild-type gene can be formed by template switching due to attenuation of the replication-fork progression. The large loop must either be preserved from mismatch repair until the next round of replication, or be repaired in favor of loop retention. A wild-type gene can also be formed by unequal exchange (C) or by a non-reciprocal gene conversion event (D).
To determine whether TNRs increase SCE, we constructed a his3 sister-chromatid recombination substrate (SCS) (Figure 1), which is similar to the design of Fasullo and Davis (41). The his3-SCS consists of a tandem pair of the truncated his3 gene with 508 bp of overlapping homology (Figure 1). We introduced the his3-SCS into our laboratory strain AS13 (35) at the ARG4 locus to generate DNY378. In vegetative cells, sister chromatids are preferred over homologs for recombinational repair (43). The his3-Δ3′ construct also contains 71 copies of CAG repeats within the region of homology (see Materials and Methods). As a control, we constructed a similar substrate containing a 120-bp non-repeated DNA fragment (Ho-cut site) at the same site within the his3-Δ3′ construct to generate DNY380.
CAG repeats and inverted DNA repeats stimulate spontaneous unequal SCE
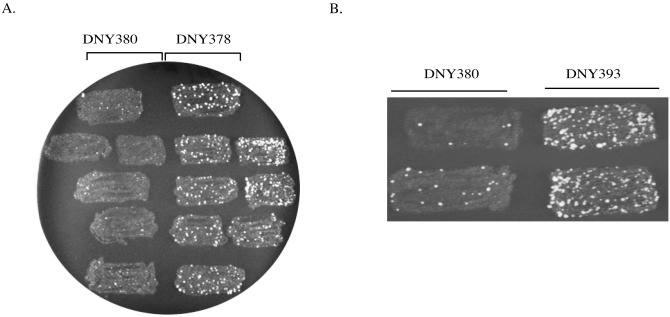
To determine whether the CAG repeats modulate unequal SCE, we patched the haploid colonies onto rich YPD medium. After 2 days at 30°C, the patches were replica-plated onto minimal plates lacking histidine, and incubated for 3 days at 30°C. The results are shown in Figure 2A. There were more His+ papillae on DNY378 patches than on DNY380 patches, suggesting that CAG repeats stimulate unequal SCE.
Figure 2.
Formation of His+ recombinants by spontaneous unequal SCE. (A) DNY380 has an insertion of a non-repeated sequence within the his3-Δ3′ construct, and DNY378 has 71 copies of CAG repeats. (B) DNY393 has an insertion of a 140-bp palindromic sequence.
As noted earlier, CAG repeats have the potential to form hairpin structures in vivo. Thus, if the increase in SCE is related to the formation of secondary structures, then one would expect that inverted DNA repeats, which also have the potential to form secondary structures in vivo, will increase SCE. To test this possibility, we constructed a his3-SCS containing a 140-bp palindromic sequence within the his3-Δ3′ construct. The his3-SCS containing the palindromic sequence (his3-SCSpal140) was inserted at the ARG4 locus into our strain background to generate DNY393. The qualitative patch assay was then performed to determine whether IRs stimulate SCE. Our results showed that both CAG repeats and IRs increase spontaneous unequal SCE during vegetative growth (Figure 2).
We also compared the rates of His+ recombinant formation in strains containing his3-SCS with non-repeated sequences, IRs or CAG repeats, by plating cells on synthetic medium lacking histidine. The rate of spontaneous unequal recombination in DNY380 was 7.6 × 10−7, and the rates of SCE in DNY378 and DNY393 were nearly 2- (P < 0.001) and 12-fold (P < 0.001) higher, respectively, than the rate in DNY380 (Table 2). This again indicates that CAG repeats and IRs both increase spontaneous unequal SCE.
Table 2. Rates of spontaneous unequal SCE in wild type and in mutants defective in RAD52 epistasis-group genes.
| his3 substrate | Genotype | Rate of recombination (× 106) | Fold difference in rate, relative to wild type |
|---|---|---|---|
| his3-SCScontrol | Wild type | 0.76 ± 0.19 | 1 |
| his3-SCScontrol | rad50 | 2.63 ± 0.44 | 3.4 ↑ |
| his3-SCScontrol | mre11 | 3.40 ± 0.20 | 4.4 ↑ |
| his3-SCScontrol | rad51 | 1.36 ± 0.38 | 1.8 ↑ |
| his3-SCScontrol | rad52 | 0.16 ± 0.24 | 0.21 ↓ |
| his3-SCSCAG71 | Wild type | 1.37 ± 0.15 | 1 |
| his3-SCSCAG71 | rad50 | 3.20 ± 0.24 | 2.3 ↑ |
| his3-SCSCAG71 | mre11 | 4.12 ± 0.56 | 3.0 ↑ |
| his3-SCSCAG71 | rad51 | 1.73 ± 0.31 | 1.3 ↑ |
| his3-SCSCAG71 | rad52 | 0.09 ± 0.56 | 0.06 ↓ |
| his3-SCSpal140 | Wild type | 9.25 ± 1.67 | 1 |
| his3-SCSpal140 | rad50 | 4.67 ± 0.74 | 0.5 ↓ |
| his3-SCSpal140 | mre11 | 2.83 ± 0.47 | 0.3 ↓ |
| his3-SCSpal140 | rad51 | 2.70 ± 0.20 | 0.29 ↓ |
| his3-SCSpal140 | rad52 | 0.58 ± 0.28 | 0.06 ↓ |
It is likely that the increase in SCE by IRs and by CAG repeats is due to hairpin formation by the repeated sequences. During DNA replication, when a secondary structure is formed ahead of the replication fork, the hairpin structures can be cleaved by structure-specific nucleases, generating DSBs. The DSB could be repaired, using the sister chromatid as a template; the replication fork would thereby be reconstituted. Alternatively, the stalled nascent 3′ end can separate from the template; the resulting single-stranded DNA with a free 3′ end can invade already replicated homologous regions on the sister chromatid and continue DNA synthesis, followed by denaturation and reannealing with the original template; such continuation of DNA synthesis will result in the formation of a complete HIS3 gene (Figure 1B).
RAD50 and MRE11 influence IR-stimulated SCE, but show no apparent control over CAG- and non-repeat-mediated SCE
IR-mediated genetic instability has been studied extensively in bacteria, yeast and mice (35,40,42,44–48). In all organisms studied, long IRs have been shown to stimulate genome rearrangements. The secondary-structure formation in single-stranded DNAs, most likely during DNA replication, is believed to be the mechanism of IR-mediated genome rearrangements, such as deletions and inter-chromosomal recombination between heteroalleles. In E.coli, although long IRs are highly unstable, they are relatively more stable in sbcC and sbcD mutants. The SbcCD complex of E.coli has both endonuclease and exonuclease activities, and it can cleave stem–loop structures in vitro (49).
One way of elucidating the mechanism of TNR- and IR-stimulated SCEs in yeast is by determining the genetic requirements for IR- and TNR-mediated SCE events. For example, if the wild-type HIS3 is generated by DSB repair, then the increase in SCE should depend on genes required for DSB repair. In yeast, RAD52 epistasis-group genes are required for DSB repair [reviewed in (50)]. RAD52 group genes are broadly divided into two subgroups: the RAD50, MRE11 and XRS2 subgroup, and the RAD51, RAD52, RAD54, RAD55, RAD57, RAD59 and RDH54/TID1 subgroup.
Rad50 and Mre11 of eukaryotes exhibit significant homology to the SbcC and SbcD proteins, respectively (51). Mre11, Rad50 and Xrs2/Nbs1 form a tight complex (MRX) that has been shown to play multiple roles in maintaining genome integrity [reviewed in (50)]. Because of the homology between Mre11 and Rad50, and SbcC and SbcD, it is likely that the MRX complex is involved in the processing of hairpin structures in yeast and humans. In vitro, Mre11 has been shown to possess exonuclease and endonuclease activities; in addition, the yeast Mre11/Rad50 complex can cleave and process secondary structures (52).
In yeast, the IR-mediated inter-chromosomal recombination is highly dependent on MRX and the Sae2 protein (53); IR-mediated recombination is reduced by 99% in mutants devoid of these proteins. Like IRs, CAG-stimulated intrachromosomal recombination is also RAD50 dependent (30). To determine whether the MRX complex is required for CAG- and IR-stimulated SCE, we measured His+ recombination rates in rad50 and mre11 mutants, and the results are shown in Table 2. The rate of unequal SCE for the control substrate, containing a non-repeated DNA sequence, was increased from 7.6 × 10−7 to 2.63 × 10−6 (P < 0.001) in the rad50 mutant, and to 3.4 × 10−6 (P < 0.001) in the mre11 mutant. Similarly, the rate of spontaneous SCE with the CAG-containing substrate was increased from 1.37 × 10−6 to 3.2 × 10−6 in the rad50 background, and to 4.12 × 10−6 in the mre11 background (P < 0.001). Although the difference in SCE rates between his3-SCScontrol and his3-SCSCAG71 in the rad50 background appears to be significant (P = 0.034), the rates in the mre11 background are similar (P = 0.05). These results suggest that spontaneous SCE is independent of the MRX-complex activity, and that deficiency in MRX activity has no effect on CAG-mediated SCEs.
However, IR-stimulated SCE was nearly 2- to 3-fold reduced in mutants that lack MRX activity. The rate of spontaneous SCE decreased from 9.25 × 10−6 to 4.67 × 10−6 (P < 0.001) in rad50 cells, and to 2.83 × 10−6 (P < 0.001) in mre11 cells, suggesting that (as observed with inter-chromosomal heteroallelic recombination) spontaneous IR-stimulated SCEs require the MRX complex. These results also suggest that CAG-stimulated and IR-mediated SCE events involve separate mechanistic pathways.
IR-stimulated spontaneous SCE events, but not CAG-mediated SCEs, are RAD51 dependent
The RAD51-group genes are generally required for conservative DSB repair, resulting in gene conversion events (50). RAD52 is required for nearly all homology-dependent recombination, including TNR-mediated intrachromosomal recombination events. An understanding of the relationship between the TNR- and IR-stimulated SCEs and the RAD51-group genes may shed some light on the mechanism of repeat sequence-stimulated SCE events.
Previous studies indicated that spontaneous SCE requires some, but not all, RAD51-group genes; spontaneous SCE is reduced 10- to 20-fold in the rad52 background, whereas the rad51 mutation has no effect on spontaneous SCEs (43,54). However, the ionizing radiation-associated SCEs are eliminated in rad51 and rad52 mutants (43,54). These results suggest that spontaneous and damage-associated SCEs involve different mechanistic pathways, and that spontaneous SCE probably does not result from DSB repair. To determine whether TNR- and IR-stimulated SCEs are due to DSB repair, we introduced rad51 and rad52 mutations into haploid strains bearing the sister-chromatid substrates, and we then measured the rate of His+ recombinant formation.
The rate of His+ prototroph formation by his3-SCScontrol was reduced by ∼5-fold in the rad52 background (Table 2). As previously observed, spontaneous SCE remained unaffected by the rad51 mutation. The his3-SCSCAG71 behaved similarly to the sister-chromatid substrate with a non-repeated DNA sequence: the rate of recombination was ∼15-fold reduced in the rad52 background, but remained unaffected by the rad51 mutation. However, the rate of SCE in strains containing the his3-SCSpal140 substrate was reduced 3-fold (P < 0.001) in the rad51 background, and 15-fold in the rad52 mutant (Table 2), suggesting that IR-stimulated SCE occurs by a pathway that is different from that of CAG-mediated SCEs. In addition, SCEs due to IRs are likely to occur by a DSB repair mechanism since both RAD51 and RAD52 exert strong influence on the rate of spontaneous unequal SCE.
DISCUSSION
Genomic regions containing IRs and TNRs are highly unstable since the presence of these sequence arrangements causes frequent alterations of the DNA in and surrounding the repeated sequences. In this report, we showed that TNRs and IRs, both of which have the potential to form secondary structures in vivo, increase spontaneous unequal SCE. Spontaneous SCE is believed to occur by a variety of mechanisms, including template switching, DSB repair and replication-associated repair (the latter is also known as break-induced replication). The results presented above suggest that normal spontaneous SCE does not involve DSB repair events. IRs increase SCE by a mechanism that is different from that for TNRs and for non-repeated DNA sequences, as IR-stimulated SCEs were highly dependent on both RAD51 and RAD52, whereas TNR- and non-repeat-mediated SCEs were independent of RAD51.
The rate of SCE by his3-SCSCAG71 was nearly 2-fold higher than the rate in his3-SCScontrol, but it was 7-fold lower than the rate in the IR-containing substrate (his3-SCSpal140). While SCEs caused by his3-SCSCAG71 and his3-SCScontrol showed significant increases in rad50 and mre11 cells, IR-mediated SCE was reduced to 30–50% in mutant cells (Table 2). In the rad51 mutant, his3-SCSpal140 showed a nearly 3-fold reduction in the rate of SCE, but the same rad51 mutation had little or no effect on the rate of SCE by his3-SCSCAG71 (P = 0.03) and his3-SCScontrol (P = 0.005) substrates (Table 2). These results suggest that TNR- and IR-stimulated SCEs occur by two different pathways. It is possible that IR- and CAG-stimulated SCEs occur by the same mechanism; however, the effect of mutations in RAD52-epistasis group genes on the CAG-mediated SCEs could not be detected due to the small difference in SCE rates between the control and CAG-containing substrates.
One explanation for the small increase in SCEs by the CAG-containing substrate over the non-repeat containing substrate is that CAG-stimulated SCEs depend on the length of the repeat tract, because the instability of TNRs is related to the repeat-tract length: the longer the repeat tract, the greater the possibility for expansion (2,26). The his3-SCSCAG71 had 71 CAGs; a tract length of >71 CAGs may generate a higher level of SCEs than that observed with his3-SCSCAG71. Another explanation of the low level of SCE by CAGs is that the cloning of CAG repeats within the his3-Δ3′ construct also included non-repeated DNA sequences (which were used as a primer to amplify the CAG repeats by PCR; see Materials and Methods) flanking the CAG repeats; these sequences are non-homologous to HIS3 sequences. Such non-homologous sequences are not present in the IR construct. The presence of the non-homologous sequences could reduce the frequency of homologous recombination leading to SCE, since these non-homologous sequences must be removed from the free 3′ end before successful pairing with the new template can occur. However, the latter possibility seems unlikely, because an oligonucleotide containing 30 CAG repeats but lacking the flanking non-homologous sequences exhibited, when inserted at the same site, an SCE rate (1.40 ± 0.17 × 10−6) similar to that of his3-SCSCAG71.
Previous studies showed that spontaneous SCE is independent of RAD50 and MRE11: there is almost no difference in SCE rates between the wild-type and rad50- and mre11-mutant strains (54,55). The mre11-mutant strains fail to increase SCEs after exposure to ionizing radiation, whereas spontaneous SCEs remain unaffected in both wild-type and mutant cells (55), suggesting that spontaneous SCE and ionizing radiation-induced DNA damage-associated SCE occur by separate mechanisms. Mutations in MRX genes give rise to an enhanced level of spontaneous mitotic heteroallelic recombination (50). Since DNA lesions in diploids at the G2 stage are preferentially repaired using the sister chromatid, instead of the homolog as a template (43), it has been suggested that Rad50, Mre11 and Xrs2 are generally involved in sister-chromatid recombination, and that the increase in heteroallelic recombination in the rad50, mre11 and xrs2 mutants is due to a shifting of the repair pathway, from involvement of sister chromatids to interactions between the homologs (50,55).
In our strain background, the rates of unequal SCE in strains containing his3-SCScontrol or his3-SCSCAG71, as in heteroallelic recombination, were several-fold higher in rad50 or mre11 mutants than in the wild-type strains (Table 2), suggesting that MRX activity is not required for the spontaneous SCE. In addition, these results suggest that MRX activity does not influence CAG-stimulated SCEs. However, as mentioned before, it is possible that CAG-stimulated SCEs do depend on MRX activity, but that such an effect could not be detected in our assay system due to the fact that the rate of TNR-induced SCE is only 2-fold higher than the rate for the control strain (DNY380).
IR-stimulated SCEs were affected by mre11 and rad50 mutations. Unlike other types of mitotic recombination, IR-induced mitotic heteroallelic recombination is highly dependent on MRX activity (53). Mitotic heteroallelic recombination by IRs appears to be DSB mediated, as it has been demonstrated that long IRs generate a low level of DSBs during vegetative growth in yeast (53). Physical analysis indicated that these DSBs are likely to be generated at secondary structures. DSBs were observed in rad50, rad50S, mre11 and sae2 mutants, suggesting that it is the processing of DSBs—rather than their formation—that is dependent on the MRX activity. These studies also indicate that the Mre11 hairpin cleavage activity is not required for IR-mediated DSB formation. These conclusions are consistent with our results for sister-chromatid recombination, indicating that IR-stimulated SCEs are dependent on RAD50, MRE11, RAD51 and RAD52, and that most IR-stimulated SCEs are due to DSB repair.
It is possible that stalling of DNA replication forks, at the secondary structures generated by IRs, leads to DSB formation due to cleavage of hairpin structures by structure-specific nucleases. The DSB is then repaired by SCE, and thereby reestablishes the replication fork. Dependence of IR-stimulated SCE on RAD51 is also in agreement with the previous observation that most replication-associated DSB repair is RAD51 dependent (56). Both equal and unequal SCE can result from DSB repair events (57). The assay system described here can recover only a portion of the DSB repair events.
The above results, taken with the results of previous studies on TNR- and IR-mediated genetic instability, suggest that two options are open to a replication fork that is stalled at the secondary structure. In one pathway, the replication fork may denature and then reanneal with homologous sequences present on the same DNA, on the sister chromatid or on the homolog, resulting in replication slippage or homologous recombination events. Alternatively, cleavage of secondary structures results in DSBs, which are then repaired by homologous recombination, with the sister or the homolog used as a template. In the case of IRs, replication slippage on the same DNA would generate deletion events, whereas for TNRs, either contractions or expansions would ensue.
If both IRs and TNRs can generate secondary structures, then why do CAG-stimulated SCEs not involve DSBs? It is possible that CAG repeats, like IRs, generate DSBs at the stalled replication forks. However, most of these DSBs are repaired by intramolecular single-strand annealing events, using the complementary sequences on the newly synthesized strand and on the broken template strand, after exonucleolytic processing of the broken template strand in the 5′–3′ direction. A minor proportion of DSBs are repaired using the sister chromatid as a template. The above model also requires that secondary structures be formed not involving the entire repeat tract. The intramolecular repair process would result in a contraction event, consistent with the observations that most tract-length alterations in yeast are contractions. We strongly believe that both IRs and TNRs generate DSBs at the stalled replication fork, based on the following observations. First, as mentioned above, a CAG repeat tract, containing 30 repeats without the flanking non-homologous sequences, exhibited an SCE rate that is similar to that of his3-SCSCAG71. Second, long CAG repeat tracts also act as fragile sites in mitotic yeast cells (30,58). Finally, in yeast, contractions are more frequent than expansions. DSB repair involving sister chromatids can lead to both equal and unequal SCE events. It is possible that repeat-tract expansions occur during equal SCE that cannot be detected by our assay system. A similar intermolecular DSB-repair mechanism was also proposed for Spo11-mediated meiotic expansion mutations observed in yeast (22).
In summary, both TNRs and IRs act as major sources of genetic instability. Several DNA transaction pathways can lead to IR- and TNR-stimulated genome rearrangements. Our results suggest that errors during replication of IR- and TNR-containing sequences are likely to lead to different outcomes, depending on how the stalled replication fork is processed at these sequences. It will be interesting to determine whether defects in the replication apparatus can result in an increased rate of SCEs, and whether other disease-associated TNRs also stimulate SCE.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Mike Fasullo for providing reagents and for stimulating discussions throughout the course of this work. We also thank Manuela Sironi for constructing the plasmid containing 30 copies of CAG repeats. We thank the Wadsworth Center Molecular Genetics Core facility for synthesizing oligonucleotides and for DNA sequencing services.
REFERENCES
- 1.Modrich P. and Lahue,R. (1996) Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu. Rev. Biochem., 65, 101–133. [DOI] [PubMed] [Google Scholar]
- 2.Wells R.D. and Warren,S.T. (1998) Genetic Instabilities and Hereditary Neurological Diseases. Academic Press, San Diego. [Google Scholar]
- 3.Cummings C.J. and Zoghbi,H.Y. (2000) Fourteen and counting: unraveling trinucleotide repeat diseases. Hum. Mol. Genet., 9, 909–916. [DOI] [PubMed] [Google Scholar]
- 4.Yoon S.-R., Dubeau,L., de Young,M., Wexler,N.S. and Arnheim,N. (2003) Huntington disease expansion mutations in humans can occur before meiosis is completed. Proc. Natl Acad. Sci. USA, 100, 8834–8838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Savouret C., Garcia-Cordier,C., Megret,J., te Riele,H., Junien,C. and Gourdon,G. (2004) MSH2-dependent germinal CTG repeat expansions are produced continuously in spermatogonia from DM1 transgenic mice. Mol. Cell. Biol., 24, 629–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kovtun I.V. and McMurray,C.T. (2001) Trinucleotide expansion in haploid germ cells by gap repair. Nature Genet., 27, 407–411. [DOI] [PubMed] [Google Scholar]
- 7.Nag D.K. (2003) Trinucleotide repeat expansions: timing is everything. Trends Mol. Med., 9, 455–457. [DOI] [PubMed] [Google Scholar]
- 8.Sia E.A., Jinks-Robertson,S. and Petes,T.D. (1997) Genetic control of microsatellite stability. Mutat. Res., 383, 61–70. [DOI] [PubMed] [Google Scholar]
- 9.Freudenreich C.H., Stavenhagen,J.B. and Zakian,V.A. (1997) Stability of CTG/CAG trinucleotide repeat in yeast is dependent on its orientation in the genome. Mol. Cell. Biol., 17, 2090–2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maurer D.J., O'Callaghan,B.L. and Livingston,D.M. (1996) Orientation dependence of trinucleotide CAG repeat instability in yeast. Mol. Cell. Biol., 16, 6617–6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miret J.J., Pessoa-Brandao,L. and Lahue,R.S. (1998) Orientation-dependent and sequence-specific expansions of CTG/CAG trinucleotide repeats in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 95, 12438–12443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kang S., Jaworski,A., Ohshima,K. and Wells,R.D. (1995) Expansion and deletion of CTG repeats from human disease genes are determined by the direction of replication in E. coli. Nature Genet., 10, 213–218. [DOI] [PubMed] [Google Scholar]
- 13.Cleary J.D., Nichol,K., Wang,Y.H. and Pearson,C.E. (2002) Evidence of cis-acting factors in replication-mediated trinucleotide instability in primate cells. Nature Genet., 31, 37–46. [DOI] [PubMed] [Google Scholar]
- 14.Fortune M.T., Vassilopoulos,C., Coolbaugh,M.I., Siciliano,M.J. and Monckton,D.G. (2000) Dramatic, expansion-biased, age-dependent, tissue-specific somatic mosaicism in a transgenic mouse model of triplet repeat instability. Hum. Mol. Genet., 9, 439–445. [DOI] [PubMed] [Google Scholar]
- 15.Kennedy L. and Shelbourne,P. (2000) Dramatic mutation ability in HD mouse stratium: does polyglutamine load contribute to cell specific vulnerability in Huntington's disease? Hum. Mol. Genet., 9, 2539–2544. [DOI] [PubMed] [Google Scholar]
- 16.Lia A.-S., Seznec,H., Hofmann-Radvanyi,H., Radvanyi,F., Duros,C., Saquet,C., Blanche,M., Junien,C. and Gourdon,G. (1998) Somatic instability of the CTG repeat in mice transgenic for the myotonic dystrophy region is age dependent but not correlated to the relative intertissue transcription levels and proliferative capacities. Hum. Mol. Genet., 8, 1285–1291. [DOI] [PubMed] [Google Scholar]
- 17.Manley K., Shirley,T.L., Flaherty,L. and Messer,A. (1999) Msh2 deficiency prevents in vivo somatic instability of the CAG repeat in Huntington disease transgenic mice. Nature Genet., 23, 471–473. [DOI] [PubMed] [Google Scholar]
- 18.van den Broek W.J.A.A., Nelen,M.R., Wansink,D.G., Coerwinkel,M.M., te Riele,H., Groenen,P.J.T.A. and Wieringa,B. (2002) Somatic expansion behavior of the (CTG)n repeat in myotonic dystrophy knock-in mice is differentially affected by Msh3 and Msh6 mismatch-repair proteins. Hum. Mol. Genet., 2, 191–198. [DOI] [PubMed] [Google Scholar]
- 19.Jakupciak J.P. and Wells,R.D. (1999) Genetic instabilities in (CTG.CAG) repeats occur by recombination. J. Biol. Chem., 274, 23468–23479. [DOI] [PubMed] [Google Scholar]
- 20.Sarkar P.S., Chang,H.C., Boudi,F.B. and Reddy,S. (1998) CTG repeats show bimodal amplification in E. coli. Cell, 95, 531–540. [DOI] [PubMed] [Google Scholar]
- 21.Richard G.-F., Goellner,G.M., McMurray,C.T. and Haber,J.E. (2000) Recombination-induced CAG trinucleotide repeat expansions in yeast involve the MRE11-RAD50-XRS2 complex. EMBO J., 19, 2381–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jankowski C., Nasar,F. and Nag,D.K. (2000) Meiotic instability of CAG repeat tracts occurs by double-strand break repair in yeast. Proc. Natl Acad. Sci. USA, 97, 2134–2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cohen H., Sears,D.D., Zenvirth,D., Heiter,P. and Simchen,G. (1999) Increased instability of human CTG repeat tracts on yeast artificial chromosomes during gametogenesis. Mol. Cell. Biol., 19, 4153–4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schweitzer J.K., Reinke,S. and Livingston,D.M. (2001) Meiotic alterations in CAG repeat tracts. Genetics, 159, 1861–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sinden R.R., Potaman,V.D., Oussatcheva,E.A., Pearson,C.E., Lyubchenko,Y.L. and Shlyakhtenko,L.S. (2002) Triplet repeat DNA structures and human genetic disease: dynamic mutations from dynamic DNA. J. Biosci., 27, 53–65. [DOI] [PubMed] [Google Scholar]
- 26.McMurray C.T. (1995) Mechanisms of DNA expansion. Chromosoma, 104, 2–13. [DOI] [PubMed] [Google Scholar]
- 27.Sakamoto N., Chastain P.D., Parniewski,P., Ohshima,K., Pandolfo,M., Griffith,J.D. and Wells,R.D. (1999) Sticky DNA: self-association properties of long GAA.TTC repeats in R.R.Y triplex structures from Friedreich's ataxia. Mol. Cell, 3, 465–475. [DOI] [PubMed] [Google Scholar]
- 28.Heidenfelder B.L., Makhov,A.M. and Topal,M.D. (2003) Hairpin formation in Friedreich's ataxia triplet repeat expansion. J. Biol. Chem., 278, 2425–2431. [DOI] [PubMed] [Google Scholar]
- 29.Moore H., Greenwell,P.W., Liu,C.P., Arnheim,N. and Petes,T.D. (1999) Triplet repeats form secondary structures that escape DNA repair in yeast. Proc. Natl Acad. Sci. USA, 96, 1504–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Freudenreich C.H., Kantrow,S.M. and Zakian,V.A. (1998) Expansion and length-dependent fragility of CTG repeats in yeast. Science, 279, 853–856. [DOI] [PubMed] [Google Scholar]
- 31.Usdin K. and Woodford,K.J. (1995) CGG repeats associated with DNA instability and chromosome fragility from structures that block DNA synthesis in vitro. Nucleic Acids Res., 23, 4202–4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kang S., Ohshima,K., Shimizu,M., Amirhaeri,S. and Wells,R.D. (1995) Pausing of DNA synthesis in vitro at specific loci in CTG and CGG triplet repeats from human hereditary disease genes. J. Biol. Chem., 270, 27014–27021. [DOI] [PubMed] [Google Scholar]
- 33.Pelletier R., Krasilnikovam M.M., Samadashwily,G.M., Lahue,R. and Mirkin,S.M. (2003) Replication and expansion of trinucleotide repeats in yeast. Mol. Cell. Biol., 23, 1349–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krasilnikova M.M. and Mirkin,S.M. (2004) Replication stalling at Friedreich's ataxia (GAA)n repeats in vivo. Mol. Cell. Biol., 24, 2286–2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nag D.K., White,M.A. and Petes,T.D. (1989) Palindromic sequences in heteroduplex DNA inhibits mismatch repair in yeast. Nature, 340, 318–320. [DOI] [PubMed] [Google Scholar]
- 36.Rose M.D., Winston,F. and Heiter,P. (1990) Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 37.Fasullo M., Bennet,T. and Koudelik,J. (1998) The Saccharomyces cerevisiae RAD9 checkpoint reduces the DNA-damage-associated stimulation of directed translocations. Mol. Cell. Biol., 18, 1190–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nag D.K. and Petes,T.D. (1993) Physical detection of heteroduplexes during meiotic recombination in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol., 13, 2324–2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shinohara A., Ogawa,H. and Ogawa,T. (1992) Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell, 69, 457–470. [DOI] [PubMed] [Google Scholar]
- 40.Nag D.K. and Kurst,A. (1997) A 140-base-pair long palindromic sequence induces double-strand breaks during meiosis in the yeast Saccharomyces cerevisiae. Genetics, 146, 835–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fasullo M.T. and Davis,R.W. (1987) Recombination substrates designed to study recombination between unique and repetitive sequences in vivo. Proc. Natl Acad. Sci. USA, 84, 6215–6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nasar F., Jankowski,C. and Nag,D.K. (2000) Long palindromic sequences induce double-strand breaks during meiosis in yeast. Mol. Cell. Biol., 20, 3449–3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kadyk L.C. and Hartwell,L.H. (1992) Sister chromatids are preferred over homologs as substrates for recombinational repair in Saccharomyces cerevisiae. Genetics, 132, 387–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leach D.R.F. (1994) Long DNA palindromes, cruciform structures, genetic instability and secondary structure repair. Bioassays, 16, 893–900. [DOI] [PubMed] [Google Scholar]
- 45.Gordenin D.A., Lobachev,K.S., Degtyareva,N.P., Malkova,A.L., Perkins,E. and Resnick,M.A. (1993) Inverted DNA repeats: a source of eukaryotic genomic instability. Mol. Cell. Biol., 13, 5315–5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Farah J.A., Hartsuiker,E., Mizuno,K., Ohta,K. and Smith,G.R., (2002) A 160 bp palindrome is Rad50.Rad32-dependent mitotic recombination hotspot in Schizosaccharomyces pombe. Genetics, 161, 461–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rattray A.J., McGill,C.B., Shafer,B.K. and Strathern,J.N. (2001) Fidelity of mitotic double-strand-break repair in Saccharomyces cerevisiae: a role for SAE2/COM1. Genetics, 158, 109–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Akgun E., Zahn,J., Baumes,S., Brown,G., Liang,F., Romanienko,P.J., Lewis,S. and Jasin,M. (1997) Palindrome resolution and recombination in the mammalian germ line. Mol. Cell. Biol., 17, 5559–5570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Connelly J.C., Kirkham,L.A. and Leach,D.R.F. (1998) The SbcCD nuclease of Escherichia coli is a structural maintenance of chromosomes (SMC) family protein that cleaves hairpin DNA. Proc. Natl Acad. Sci. USA, 95, 7969–7974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Symington L.S. (2002) Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev., 66, 630–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sharples G.J. and Leach,D.R.F. (1995) Structural and functional similarities between the SbcCD proteins of Escherichia coli and the Rad50 and Mre11 (Rad32) recombination and repair proteins of yeast. Mol. Microbiol., 17, 1215–1220. [DOI] [PubMed] [Google Scholar]
- 52.Trujillo K.M. and Sung,P. (2001) DNA structure-specific nuclease activities in the Saccharomyces cerevisiae Rad50*Mre11 complex. J. Biol. Chem., 276, 35458–35464. [DOI] [PubMed] [Google Scholar]
- 53.Lobachev K.S., Gordenin,D.A. and Resnick,M.A. (2002) The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell, 108, 183–193. [DOI] [PubMed] [Google Scholar]
- 54.Dong Z. and Fasullo,M. (2003) Multiple recombination pathways for sister chromatid exchange in Saccharomyces cerevisiae: role of RAD1 and the RAD52 epistasis group genes. Nucleic Acids Res., 31, 2576–2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bressan D.A., Baxter,B.K. and Petrini,J.H. (1999) The Mre11-Rad50-Xrs2 protein complex facilitates homologous recombination-based double-strand break repair in Saccharomyces cerevisiae. Mol. Cell. Biol., 19, 7681–7687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Davis A.P. and Symington,L.S. (2004) RAD51-dependent break-induced replication in yeast. Mol. Cell. Biol., 24, 2344–2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gonzalez-Barrera S., Cortes-Ledesma,F., Wellinger,R.E. and Aguilera,A. (2003) Equal sister chromatid exchange is a major mechanism of double-strand break repair in yeast. Mol. Cell, 11, 1661–1671. [DOI] [PubMed] [Google Scholar]
- 58.Callahan J.L., Andrews,K.J., Zakian,V.A. and Freudenreich,C.H. (2003) Mutations in yeast replication proteins that increase CAG/CTG expansions also increase repeat fragility. Mol. Cell. Biol., 21, 7849–7860. [DOI] [PMC free article] [PubMed] [Google Scholar]