Abstract
KpnBI is a restriction-modification (R-M) system recognized in the GM236 strain of Klebsiella pneumoniae. Here, the KpnBI modification genes were cloned into a plasmid using a modification expression screening method. The modification genes that consist of both hsdM (2631 bp) and hsdS (1344 bp) genes were identified on an 8.2 kb EcoRI chromosomal fragment. These two genes overlap by one base and share the same promoter located upstream of the hsdM gene. Using recently developed plasmid R-M tests and a computer program RM Search, the DNA recognition sequence for the KpnBI enzymes was identified as a new 8 nt sequence containing one degenerate base with a 6 nt spacer, CAAANNNNNNRTCA. From Dam methylation and HindIII sensitivity tests, the methylation loci were predicted to be the italicized third adenine in the 5′ specific region and the adenine opposite the italicized thymine in the 3′ specific region. Combined with previous sequence data for hsdR, we concluded that the KpnBI system is a typical type I R-M system. The deduced amino acid sequences of the three subunits of the KpnBI system show only limited homologies (25 to 33% identity) at best, to the four previously categorized type I families (IA, IB, IC, and ID). Furthermore, their identity scores to other uncharacterized putative genome type I sequences were 53% at maximum. Therefore, we propose that KpnBI is the prototype of a new ‘type IE’ family.
INTRODUCTION
Type I restriction endonucleases and the corresponding methylases were originally found in enteric bacteria such as Escherichia coli and Salmonella typhimurium (1,2) and later, identified in Citrobacter freundii (1) and Bacillus subtilis (3). Many homologous sequences that correspond to each subunit of type I enzymes have also been found from various bacterial genome projects including Archaebacteria (4). Similarly, type III enzymes were originally found in E.coli bacteriophage P1 and P15 (5,6) but more recently identified in several other bacteria (4,7). Both type I and type III restriction enzymes possess distinct gene structures, but share a few common characteristics, which include the presence of all subunits for restriction activity.
Two R-M systems were recognized in Klebsiella species and designated as KpnAI and KpnBI in our laboratory. The entire KpnAI system was cloned and characterized as the first type I R-M system in Klebsiella (8). However, only the hsdR subunit of KpnBI had been cloned and characterized (9). The genetic evidence from mutation studies suggests that KpnBI is either a type I or type III R-M system (9). To further characterize this system, we have cloned and sequenced the remaining modification genes in this paper.
Typical type I recognition sequences consist of a 5′ specific region of 3–4 bp, a non-specific spacer region of 6–8 bp, and a 3′ specific region of 4–5 bp (10). To facilitate the discovery of new restriction enzymes and their recognition sequences, we developed a simple plasmid R-M test (11). This test is based on the observation that, upon transformation, plasmids with unmodified recognition sites (positive plasmids) are restricted, whereas plasmids without a recognition site (negative plasmids) are not. A computer program (RM search) was developed to identify a DNA recognition sequence that was common to all the positive plasmids (12). This method was first used to identify several new restriction enzymes in clinical E.coli strains (11). Subsequently, this method was used to identify the recognition sequence of KpnAI and three previously reported Salmonella R-M systems (SEA, SEN and SG) (13,14). Here, we applied this method to determine the DNA recognition sequence for the KpnBI enzymes.
MATERIALS AND METHODS
Bacterial strains, bacteriophages and plasmids
Table 1 shows details of the strains and plasmids that were used. The KpnBI modification genes were isolated from wild-type GM236 and cloned in E.coli DH5α. The expression of the KpnBI R-M activities was measured by newly developed plasmid R-M tests (11) as well as by the traditional lambda phage assay (2). Plasmid pVC1 is the hsdM-hsdSKpnBI clone obtained in this study. To overcome the incompatibility problem in the complementation experiment, plasmid pJR61 was constructed by subcloning the original 3.9 kb EcoRI fragment (hsdRKpnBI gene) from pNLB1 (9) into pACYC184. The bacteria were grown in L-broth at 37°C with vigorous aeration. Plasmids were isolated using Rapid RPM (Qbiogen, Carlsbad, CA).
Table 1. Bacteria, bacteriophages and plasmids.
| Strains or plasmids | Relevant genotype, phenotype or description | Source or reference |
|---|---|---|
| Bacteria | ||
| K.pneumoniae | ||
| GM236 | R+KpnBI M+KpnBI | (9,32) |
| GM236R | R−KpnBI M+KpnBI | (9) |
| GM238 | R−KpnBI M−KpnBI | (9,32) |
| E.coli | ||
| DH5α | R−KI M+KI, Dam+ | Lab Stock |
| Bacteriophage | ||
| λ vir | cIts857S7 | (33) |
| Plasmids | ||
| pL17 | lambda 2.3 kb HindIII clone in pMECA | (11) |
| pL17a | 1.4 kb EcoRI subclone of pL17 | This study |
| pJR61 | hsdR+KpnBI, 3.9 kb EcoRI subclone of pNLB1 in pACYC184 | (9), This study |
| pVC1 | hsdM+KpnBI hsdS+KpnBI. 8.2 kb EcoRI fragment in pL17a | This study |
| pEVC1 | hsdM+KpnBI hsdS+KpnBI. 8.2 kb EcoRI fragment in pMECA | This study |
| pSVC1 | 5 kb SphI subclone of pVC1 | This study |
| pSVC2 | 3.2 kb SphI subclone of pEVC1 | This study |
| pSVC3 | 0.6 kb HincII subclone of pVC1 | This study |
| pSVC6 | 0.8 kb HincII subclone of pSVC1 in pMECA | This study |
| pSVC7 | 0.5 kb HincII subclone of pSVC1 in pMECA | This study |
| pSVC8 | 1.7 kb HincII subclone of pVC1 | This study |
| pSVC9 | 7.4 kb HindIII subclone of pVC1 | This study |
| pKpnBIA | KpnBI oligonucleotide in pMECA | This study |
| pKpnBIG | KpnBI oligonucleotide in pMECA | This study |
| pKpnBIH1A | KpnBI oligonucleotide in pMECA | This study |
| pKpnBIH1G | KpnBI oligonucleotide in pMECA | This study |
| pKpnBIH2A | KpnBI oligonucleotide in pMECA | This study |
| pKpnBIH2G | KpnBI oligonucleotide in pMECA | This study |
| pKpnBIDam | KpnBI oligonucleotide in pMECA | This study |
| pKpnBIC | KpnBI oligonucleotide in pMECA | This study |
Cloning strategy and vector search
KpnBI modification genes were cloned from GM236 chromosomal DNA using the ‘modification expression screening’ strategy (15). This method is based on the observation that plasmids with methylated recognition sequences are not subject to restriction, whereas plasmids with unmethylated recognition sequences are good substrates for restriction.
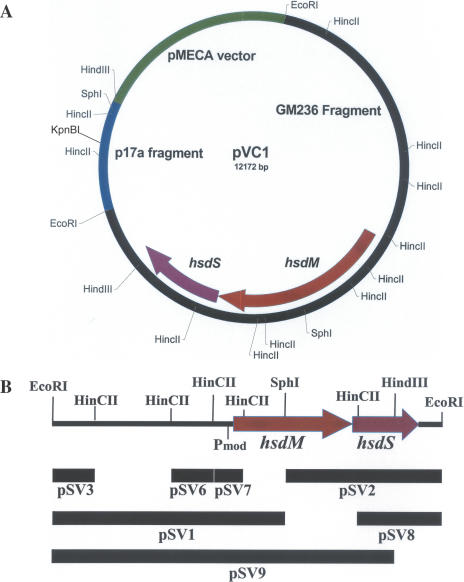
Our result shows that there is no KpnBI recognition site in pMECA, since pMECA was transformed into GM236R (R−) and GM236 (R+) at a similar frequency. We then searched for a plasmid candidate that has a KpnBI recognition site in the phage lambda subclones (14) and identified a plasmid (pL17), which showed low transformation frequency (EOT = 10−2). This plasmid contains a 2.3 kb HindIII fragment in plasmid pMECA. An EcoRI subclone that contains only a 1.4 kb portion of the above 2.3 kb fragment was derived from pL17 and designated as pL17a. This plasmid was used here as a cloning vector for the KpnBI modification genes, and it was later found that this 1.4 kb insert contains one KpnBI site (Figure 1A). The details of the cloning methods were described previously (11).
Figure 1.
(A) Restriction map of pVC1 that contains modification genes (hsdM and hsdS) of KpnBI. The vector contains one KpnBI site. (B) The 8.2 kb EcoRI fragment and the subclones used for DNA sequencing. Pmod indicates the position of the promoter.
Oligonucleotide synthesis and DNA sequencing
Oligonucleotide synthesis and DNA sequencing were performed in our core facility at the Center for Molecular Biology and Gene Therapy (Loma Linda, CA). Initially, an 8.2 kb EcoRI fragment that showed KpnBI modification activity was obtained (Figure 1A) and subsequently, a series of subclones (Figure 1B) were derived. The primer walking method was also used for DNA sequencing. A total of 37 sequencing reactions were performed to cover this 8.2 kb chromosome fragment in both directions. The DNA sequence data (Accession no. AY279080) were assembled using the Sequencher™ program (Genecode, Ann Arbor, MI).
KpnBI recognition sequence study
For the plasmid restriction tests, pL and pE series plasmids (11,14) were used. The computer program RM search (12) was used to find sequences that were common to all positive plasmids and sequences that were absent in negative plasmids. The candidate sequences were synthesized, cloned in pMECA and were subjected to plasmid restriction and modification tests (11).
KpnBI methylation site
HindIII and Dam methylase were used for the methylation sensitivity method described previously (14). HindIII does not cut its methylated recognition sequence m6AAGCTT but cuts Am6AGCTT (16). The Dam methylase methylates adenine and yields Gm6ATC (17).
RESULTS
Cloning the KpnBI modification genes
The GM236 chromosomal DNA was digested completely by EcoRI and ligated to the single EcoRI site in pL17a that contains a KpnBI site. The plasmid mixture was transformed into E.coli strain DH5α and a chromosomal EcoRI library that contains ∼5000 EcoRI fragments was constructed. When this plasmid library (10 ng each) was transferred into K.pneumoniae GM236 (R+KpnBI) and GM236R (R−KpnBI), only 25 AmpR transformants survived in GM236, whereas about 3000 AmpR transformants were obtained from GM236R. Thus, a ∼100-fold enrichment was accomplished. One of the first five AmpR clones tested from the 25 AmpR transformants listed above, contained an ∼8 kb chromosomal fragment, which expressed KpnBI modification activity and was named pVC1. The restriction map of this plasmid is shown in Figure 1A.
To confirm the modification status of the plasmid pVC1, the plasmid was recovered from DH5α and transferred to GM236 and GM236R. A similar number of AmpR transformants were obtained from both strains (EOT = 0.6). These results indicate that the original KpnBI site in the 1.4 kb lambda fragment is modified in DH5α and confirms the enrichment strategy described above.
Complementation tests
Both type I and type III systems contain hsdR subunits that express restriction activity only when they are combined with the corresponding modification subunit(s) (18). Thus, the expression of restriction activity was confirmed by using a complementation test with newly cloned pVC1 (M+KpnBI) and pJR61 (R+KpnBI). The plasmid pJR61 was transferred to a DH5α strain that already contains pVC1. Phage lambda was used to test complementation and the parental DH5α strain was used as a control. When lambda was challenged with DH5α containing both pVC1 and pJR61, the phage was severely restricted (EOP = 10−6), whereas modified phage was not restricted (EOP = 1.0). These data indicate that the R-M subunits were combined to show typical R-M activity. Similar results were obtained when this complementation test was performed in K.pneumoniae strain GM238, an R−M− derivative of GM236.
DNA sequencing and analysis of pVC1
When two SphI–EcoRI fragments of pVC1 (pSV1 and pSV2 in Figure 1B.) were subcloned in pMECA, neither subclone showed any modification activity. Subsequently, a series of HinCII and HindIII subclones were obtained for DNA sequencing purposes (Figure 1B, Table 1). Analysis of these DNA sequence data (Accession no. AY279080) shows only two large open reading frames (ORFs) of 2.6 kb and 1.3 kb. The first 2.6 kb ORF was translated and subjected to a BLAST search, which revealed homologies to many other type I hsdM genes. Similarly, the second 1.4 kb ORF revealed homologies to type I hsdS genes. We concluded that these two ORFs code hsdM and hsdS genes that are necessary components for the KpnBI methyltransferase.
Further analysis showed that the putative hsdM and hsdS genes overlap by 1 bp, possibly allowing translational coupling as described in EcoKI (19). A putative promoter (TTGATT-N17-TATCTG, Pmod) and the Shine–Dalgarno sequence (AGAATG) were found upstream of the hsdM gene but no candidate promoters were found for hsdS. This suggests that both hsdM and hsdS genes are, like other type I modification genes, transcribed from the same Pmod promoter.
The putative KpnBI HsdM subunit contains 877 amino acids, which has a molecular weight of 97 550 D. The composition of this protein is similar to other type I HsdM proteins and is negatively charged (133 negative and 100 positive amino acids). The length of the KpnBI HsdM subunit is, however, much longer than typical HsdM subunits (∼500 amino acids). The KpnBI HsdM protein contains two conserved motifs, ‘XFXGXG’ and ‘AVANPPF’, that are involved in S-adenosylmethionine binding and catalysis, respectively (10,20,21).
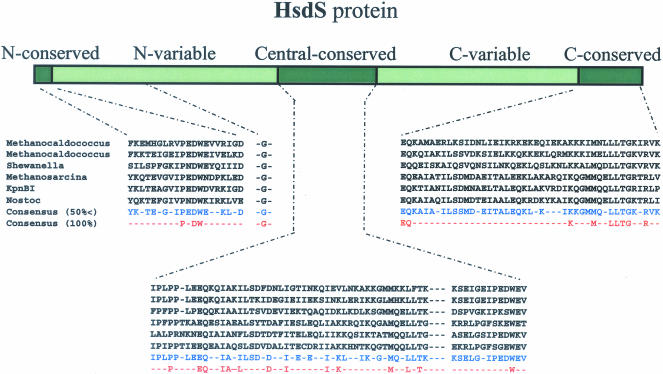
The putative KpnBI HsdS subunit contains 448 amino acids, (mw. 49 666 D) (Figure 2). This is about the average size of type I HsdS proteins, which range from 400 to 600 amino acids. All the HsdS subunits in each type I family contain a characteristic conserved region involved in subunit interaction as well as variable regions involved in DNA recognition (22). Determination of the conserved region and the variable region of the KpnBI HsdS protein was not possible due to the lack of homology with any preexisting Type I family members. However, we have identified possible conserved regions from homology to putative R-M systems (see discussion). Interestingly, this HsdSKpnBI protein contains four different amino acid sequence repeats, each appearing twice within the sequence (Figure 2). The repeats could be remnants of past recombination events. The longest repeat (MQQLLTGRTRLP) contains one mismatch (T/I) between the two repeats. A portion of this long repeat sequence (LLTGRT) was aligned with various kinases, but since no other hsdS subunits contain this sequence, the biological significance of this repeat is not clear.
Figure 2.
KpnBI HsdS protein sequence deduced from the nucleotide sequence. Several repeats are shown in bold and underlined.
Determination of the KpnBI recognition site
To determine the KpnBI recognition sequence, a total of 42 plasmids with various sizes were used for plasmid R-M tests (11) using wild-type strain GM236 (R+) and GM236R (R−). EOT values <10−1 were used again as the criteria for positive plasmids (11). This test shows whether the plasmid contains the recognition site (positive plasmid) or not (negative plasmid) (Table 2). All positive plasmids were subjected to modification tests to confirm that the modified plasmids are no longer restricted by GM236 (EOT = 0.8 ∼ 1.2).
Table 2. Plasmid R-M tests for KpnBI.
| Positive plasmidsa | pL2, pL3, pL4, pL6, pL9, pL10, pL16, pL17, pL26, pL37, pE18, pE19, pE26, pE29, pE32, pE33 |
| Negative plasmidsa | pL1, pL8, pL12, pL13, pL19, pL24, pL28 pE1, pE3, pE4, pE9, pE10, pE11, pE12, pE14, pE15, pE16, pE17, pE24, pE25, pE27, pE28, pE38, pE41, pE44, pMECA |
aPositive plasmids contain KpnBI site(s) (EOT < 10−1), whereas negative plasmids do not (EOT = ∼1.0).
When these data were put into the RM search program (12), only one possible candidate sequence with a degenerate base, CAAA (6N) RTCA was retrieved. To confirm this recognition sequence, two candidate oligonucleotides (with the R position as either G or A) were synthesized and cloned into pMECA (Figure 3A). Both clones were severely restricted (EOT = 2 × 10−2). As expected, the modified plasmids obtained from the surviving colonies were no longer subjected to restriction (EOT = 1.0). A more severe restriction (EOT = 1 × 10−3) was observed with a clone containing three tandem inserts of the recognition sequence. The DNA sequences surrounding these recognition sites (−20 to +20 bases as well as the center six random nucleotide region) were completely random. Therefore, we concluded that those two sequences are the only KpnBI recognition sequences.
Figure 3.
(A) Oligonucleotides used for the confirmation of the KpnBI site and (B) determination of the methylated adenines. KpnBI recognition sequences are shown in bold. All the oligonucleotides contain half of the EcoRV sites (ATCGAT) at each end for blunt-end cloning. Other recognition sites are underlined. Note that the B5 and B6 pair contains G/C differences in the sequence, whereas the rest of the oligonucleotide pairs (A1–A2, B1–B2 and B3–B4) contain A/G differences. Plasmids containing these oligonucleotide sequences are shown in parenthesis.
Methylation locus of KpnBI recognition site
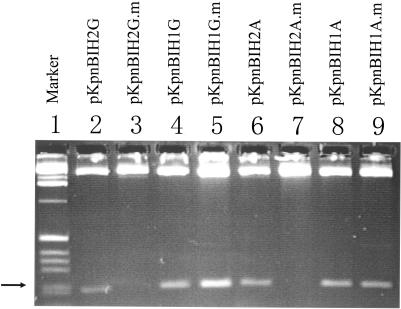
Type I methylase modifies only an adenine in each strand of the recognition sequences (18). The predicted KpnBI recognition sequence (5′CAAANNNNNNRTCA3′) contains three adenines in the 5′ specific region and only one adenine (complementary to thymine) in the 3′ specific region. To determine the specific adenine that was methylated in the 5′ region, a total of four different oligonucleotides were designed (Figure 3B, 1–4) and cloned into pMECA. These oligonucleotides contain HindIII sites that overlaps with the KpnBI recognition sequences. To modify (methylate) these sequences, the pMECA clones were transformed into the wild-type strain GM236 and were subsequently recovered. The modified and unmodified plasmids were then digested with HindIII in vitro (Figure 4). The digestion patterns show that both modified pKpnBIH1G and modified pKpnBIH1A plasmids produced 209 bp bands (Figure 4, lanes 5 and 9), whereas neither modified pKpnBIH2G nor modified pKpnBIH2A plasmids produced this 209 bp band (Figure 4, lanes 3 and 7). When unmodified, all these four plasmids produced 209 bp bands (Figure 4, lanes 2, 4, 6 and 8). These digestion experiments indicate that the plasmid DNA are protected only when the HindIII sequences in the B3 and B4 oligonucleotides (Figure 3) are modified. Since HindIII does not cut the DNA when the first adenine in the HindIII recognition sequence is methylated, whereas it cuts the DNA when the second adenine is methylated (16), we concluded that one of the target adenines is the third adenine in the KpnBI recognition sequence italicized in the 5′ component region (CAAA).
Figure 4.
HindIII digestion of KpnBI modified and unmodified plasmids. Modified plasmids contain a symbol (.m). The various oligonucleotides carried by each plasmid are shown in Figure 3. The arrow points to the 209 bp bands.
There is only one candidate adenine in the 3′ component region complementary to thymine and the methylation of this adenine was confirmed using Dam methylase which methylates adenine in the sequence GATC. Two additional oligonucleotides that contained the KpnBI recognition sequence were designed, one with a Dam site and another without, as a control (Figure 3B-5 and B-6). These plasmids were first modified in the Dam+ strain (DH5α). The Dam− strain (GM272) was used as a control. The modified plasmids were then subjected to the plasmid restriction test (11). The results clearly show that the plasmid containing the B-5 sequence (Figure 3) was resistant to restriction (EOT = 1.0), whereas the plasmid containing the B-6 sequence was restricted (EOT = 5 × 10−2). These results indicate that the single adenine in the 3′ component region is the target adenine for KpnBI methylation.
DISCUSSION
The grouping of type I enzymes into families is important in considering the origin and subsequent evolutionary changes of type I enzymes. Historically, type I enzymes were found in several enteric bacteria including E.coli, Salmonella, Citrobacter and Klebsiella species (1,8,23). Complementation tests, DNA–DNA hybridization, protein homology and antibody cross-reactivity have been used as criteria for determining family members, (24–26) and four different families (IA to ID) have so far been recognized. Recent genome projects have revealed that there are many DNA sequences which show amino acid homology to preexisting type I enzymes (4). These putative enzymes remain to be categorized into logical groups.
We have examined protein homology scores among type I family members. Both HsdR and HsdM subunits usually share >90% identity with the same family members and always <30% identity with members of other families. The only exceptions are the HsdR subunit identity between EcoAI and EcoEI (77%, type IB) and HsdM subunit identity between EcoAI and StySKI (88%, type IB). Similarly, amino acid sequences of the central conserved region of the HsdS subunits share >80% identity within the same family. The interpretation of homology among the HsdS proteins is more complicated because of the two target recognition domains (TRD), which contain variable amino acid sequences. The N terminal, central and C-terminal HsdS regions, however, are usually well-conserved within each family (22,27).
Our present study shows that the KpnBI R-M system is a typical type I system with a new specificity. We have compared the predicted amino acid sequences of HsdMKpnBI and HsdSKpnBI subunits as well as the previously reported HsdRKpnBI sequence (9) with all other type I enzymes. Homology scores with prototype members of each family are shown in Table 3A. The identity scores are only within the 20 to 30% range for each subunit. Based on the comparison of family members within a single family for type IA, IB, IC and ID, the criteria of having at least 70% identity scores to members of the same family, seems to be considered reasonable at this time for both HsdR and HsdM subunits, and 80% local homology, at various conserved regions for HsdS proteins. Using those criteria, the KpnBI system clearly does not share enough homology with any preexisting type I family members (Table 3A). We have, therefore, concluded that KpnBI represents the prototype of a new type I family, designated type IE.
Table 3. KpnBI amino acid sequence homology.
| R-M system | Amino acid length | Identity (%) | Positive (%) | |
|---|---|---|---|---|
| Total | Aligned | |||
| A. Established type I families | ||||
| HsdR (KpnBI) | (1013) | |||
| Type IA EcoKI | 1188 | 641 | 19 | 35 |
| Type IB EcoAI | 813 | NS | — | — |
| Type IC Eco124I | 1033 | 632 | 24 | 42 |
| Type ID StySBLI | 1088 | 790 | 25 | 42 |
| HsdM (KpnBI) | (877) | |||
| Type IA EcoKI | 529 | 390 | 27 | 44 |
| Type IB EcoAI | 489 | 247 | 26 | 40 |
| Type IC Eco124I | 520 | 499 | 31 | 50 |
| Type ID StySBLI | 539 | 493 | 26 | 46 |
| HsdS (KpnBI) | (448) | |||
| Type IA EcoKI | 464 | 444 | 20 | 40 |
| Type IB EcoAI | 589 | 168 | 33 | 56 |
| Type IC Eco124I | 409 | NS | — | — |
| Type ID StySBLI | 401 | 228 | 21 | 42 |
| B. BLAST search to NCBI protein database | ||||
| HsdR (KpnBI) | (1013) | |||
| 1. MmaGORF2294P | 1042 | 1028 | 48 | 65 |
| 2. Hpy99ORF1423P | 991 | 1004 | 47 | 66 |
| 3. HpyAORF1403P | 993 | 1005 | 47 | 65 |
| 4. Tde ATCC35405 | 1039 | 1039 | 46 | 64 |
| 5. XcaCORF2902P | 1096 | 1096 | 28 | 44 |
| HsdM (KpnBI) | (877) | |||
| 1. M.MmaGORF2294P | 808 | 838 | 54 | 68 |
| 2. [Nostoc sp. PCC 7120] | 657 | 684 | 53 | 69 |
| 3. M.HpyAORF1403P | 817 | 835 | 51 | 67 |
| 4. M.Hpy99ORF1423P | 815 | 833 | 51 | 67 |
| 5. M.TdeORFC815P | 871 | 896 | 46 | 61 |
| HsdS (KpnBI) | (448) | |||
| 1. S.SonORF383P | 439 | 201 | 40 | 63 |
| 2. S.MmaGORF2294P | 406 | 404 | 30 | 47 |
| 3. [Nostoc sp. PCC 7120] | 427 | 415 | 29 | 43 |
| 4. S.Mja GI:15669403 | 425 | 417 | 26 | 44 |
| 5. S.Mja GI:2129238 | 425 | 417 | 25 | 44 |
NS, no significant similarity was found.
Are there any other type IE family members in the natural environment? The BLAST search results (28) for each KpnBI subunit are shown in Table 3B. Among the five best candidates, all of the KpnBI subunits share high identity scores (40 to 53%) with the putative sequences from Archaeon Methanosarcina mazei Go1 (29). Homologies are also shared with other methano-bacteria as well as Helicobacter pylori strains. It is noteworthy that all these candidates are putative proteins obtained from various genome projects and their enzyme activities are yet to be demonstrated.
Because the HsdMKpnBI protein (877 amino acids) is much larger than average modification subunits (∼500 amino acids), the BLAST search indicated that the HsdMKpnBI protein shares marked homologies with other proteins mainly in the first half of the amino acid sequence. Therefore, when the amino acid sequence of only the first 50–520 amino acids of the HsdMKpnBI protein was used for BLAST search, the identity scores of the best five candidates in Table 3B increased by ∼10%, resulting in values between 61 and 69%.
During the BLAST search for HsdRKpnBI, higher identity scores such as 67–75% (positive scores, 80–85%) were obtained from comparisons of environmentally obtained DNA fragment samples from the Sargasso Sea (30). These sequence data were derived from the shotgun sequence strategy and comprised many short DNA sequences. Therefore, the amino acid homologies in these cases are localized to various locations of HsdRKpnBI and the lengths vary from 249 to 519 amino acids and never cover the entire KpnBI hsdR protein (1013 amino acids). Similar high identity scores such as 59–77% (positive scores, 73–86%) were also obtained for the HsdMKpnBI subunit at several localized regions. No comparable high scores were obtained for the HsdSKpnBI subunit sequence.
However, when the HsdS sequences obtained from the closest five candidates (Table 3B) were aligned, possible conserved regions unique to HsdS proteins became apparent (Figure 5). These sequences consist of a short weakly conserved N-constant region and two variable regions of ∼150 amino acids as well as two (central and c-terminal) constant regions of ∼50–60 amino acids. A single glycine (G) was found to be conserved in the N-variable region but its function, if any, remains to be determined.
Figure 5.
Estimation of the conserved and variable regions of the HsdSKpnBI sequence and its homologs. The five putative HsdS homolog sequences (Table 3B) were aligned using the ClustalW program. Amino acid sequences with more than 50% homology as well as 100% homology are also shown. Amino acid sequences used are from top to bottom: M.jannaschii DSM2661 [originally called Methanococcus jannaschii which contains two HsaS proteins (34)], Shewanella oneidensis MR-1 (35), Methanosarcina mazei Go1 (29), KpnBI (this study) and Cyanobacteria Nostoc sp. PCC7120 (36).
Another aspect of the KpnBI R-M system that must be clarified is the relative position of hsdR and hsdM-hsdS genes. Two different gene orders have been reported for type I systems: one, hsdR-hsdM-hsdS (IA and IB), and the other, hsdM-hsdS-hsdR (IC and ID) (31). The order reflects the order of transcription as well. The distance between two transcriptional units is quite small. The furthest distance reported is in EcoKI, which has 0.5 kb between hsdR and hsdM genes (19). We first assumed that the newly cloned hsdM and hsdS genes of KpnBI are located next to the previously cloned hsdR (9). However, we found no sequence overlap between the clone, pVC1(hsdM-hsdS), obtained in this study, and the previously cloned pKPB1 (hsdR)(9). This implies that these two transcriptional units exist at least 2 kb apart and the correct distance between them as well as their relative positions still remains to be elucidated.
Acknowledgments
ACKNOWLEDGEMENTS
We thank J. Kasarjian for useful discussion throughout this study and also for reviewing the manuscript. This work was supported by grant DAMD17-97-2-7016 from the Department of the Army. The content of the information does not necessarily reflect the position or the policy of the federal government or of the National Medical Technology Testbed, Inc.
DDBJ/EMBL/GenBank accession no. AY279080
REFERENCES
- 1.Daniel A.S., Fuller-Pace,F.V., Legge,D.M. and Murray,N.E. (1988) Distribution and diversity of hsd genes in Escherichia coli and other enteric bacteria. J. Bacteriol., 170, 1775–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bullas L.R., Colson,C. and Neufeld,B. (1980) Deoxyribonucleic acid restriction and modification systems in Salmonella: chromosomally located systems of different serotypes. J. Bacteriol., 141, 275–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu G., Willert,J., Kapfer,W. and Trautner,T.A. (1995) BsuCI, a type-I restriction-modification system in Bacillus subtilis. Gene, 157, 59. [DOI] [PubMed] [Google Scholar]
- 4.Roberts R.J., Vincze,T., Posfai,J. and Macelis,D. (2003) REBASE: restriction enzymes and methyltransferases. Nucleic Acids Res., 31, 418–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hadi S.M., Bachi,B., Shepherd,J.C., Yuan,R., Ineichen,K. and Bickle,T.A. (1979) DNA recognition and cleavage by the EcoP15 restriction endonuclease. J. Mol. Biol., 134, 655–666. [DOI] [PubMed] [Google Scholar]
- 6.Hadi S.M., Bachi,B., Iida,S. and Bickle,T.A. (1983) DNA restriction–modification enzymes of phage P1 and plasmid p15B. Subunit functions and structural homologies. J. Mol. Biol., 165, 19–34. [DOI] [PubMed] [Google Scholar]
- 7.De Backer O. and Colson,C. (1991) Identification of the recognition sequence for the M.StyLTI methyltransferase of Salmonella typhimurium LT7: an asymmetric site typical of type-III enzymes. Gene, 97, 103–107. [DOI] [PubMed] [Google Scholar]
- 8.Lee N.S., Rutebuka,O., Arakawa,T., Bickle,T.A. and Ryu,J. (1997) KpnAI, a new type I restriction-modification system in Klebsiella pneumoniae. J. Mol. Biol., 271, 342–348. [DOI] [PubMed] [Google Scholar]
- 9.Valinluck B., Lee,N.S. and Ryu,J. (1995) A new restriction-modification system, KpnBI, recognized in Klebsiella pneumoniae. Gene, 167, 59–62. [DOI] [PubMed] [Google Scholar]
- 10.Murray N.E. (2000) Type I restriction systems: sophisticated molecular machines (a legacy of Bertani and Weigle). Microbiol. Mol. Biol. Rev., 64, 412–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kasarjian J.K.A., Iida,M. and Ryu,J. (2003) New restriction enzymes discovered from Escherichia coli clinical strains using a plasmid transformation method. Nucleic Acids Res., 31, e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ellrott K.P., Kasarjian J.K.A., Jiang,T. and Ryu,J. (2002) Restriction enzyme recognition sequence search program. Biotechniques, 33, 1322–1326. [DOI] [PubMed] [Google Scholar]
- 13.Bullas L.R. and Colson,C. (1975) DNA restriction and modification systems in Salmonella. III. SP, a Salmonella potsdam system allelic to the SB system in Salmonella typhimurium. Mol. Gen. Genet., 139, 177–188. [PubMed] [Google Scholar]
- 14.Kasarjian J.K.A., Masumi Hidaka, Takashi Horiuchi, Masatake Iida and Junichi Ryu (2004) The recognition and modification sites for the bacterial type I restriction systems KpnAI, StySEAI, StySENI and StySGI. Nucleic Acids Res., 32, e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hammond A.W., Gerard,G.F. and Chatterjee,D.K. (1991) Cloning the KpnI Restriction-Modification System in Escherichia coli. Gene, 97, 97–102. [DOI] [PubMed] [Google Scholar]
- 16.McClelland M., Nelson,M. and Raschke,E. (1994) Effect of site-specific modification on restriction endonucleases and DNA modification methyltransferases. Nucleic Acids Res., 22, 3640–3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palmer B.R. and Marinus,M.G. (1994) The dam and dcm strains of Escherichia coli—a review. Gene, 143, 1–12. [DOI] [PubMed] [Google Scholar]
- 18.Bickle T.A. and Kruger,D.H. (1993) Biology of DNA restriction. Microbiol. Rev., 57, 434–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loenen W.A., Daniel,A.S., Braymer,H.D. and Murray,N.E. (1987) Organization and sequence of the hsd genes of Escherichia coli K-12. J. Mol. Biol., 198, 159–170. [DOI] [PubMed] [Google Scholar]
- 20.Dryden D.T., Willcock,D.F. and Murray,N.E. (1995) Mutational analysis of conserved amino-acid motifs in EcoKI adenine methyltransferase. Gene, 157, 123–124. [DOI] [PubMed] [Google Scholar]
- 21.Willcock D.F., Dryden,D.T. and Murray,N.E. (1994) A mutational analysis of the two motifs common to adenine methyltransferases. EMBO J., 13, 3902–3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cowan G.M., Daniel,A.S., Gann,A.A., Kelleher,J.E. and Murray,N.E. (1988) Defining domains in type-I restriction and modification enzymes. Gene, 74, 239–241. [DOI] [PubMed] [Google Scholar]
- 23.Fuller-Pace F.V., Cowan,G.M. and Murray,N.E. (1985) EcoA and EcoE: alternatives to the EcoK family of type I restriction and modification systems of Escherichia coli. J. Mol. Biol., 186, 65–75. [DOI] [PubMed] [Google Scholar]
- 24.Murray N.E., Gough,J.A., Suri,B. and Bickle,T.A. (1982) Structural homologies among type I restriction-modification systems. EMBO J., 1, 535–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Titheradge A.J., King,J., Ryu,J. and Murray,N.E. (2001) Families of restriction enzymes: an analysis prompted by molecular and genetic data for type ID restriction and modification systems. Nucleic Acids Res., 29, 4195–4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ryu J., Rajadas,P.T. and Bullas,L.R. (1988) Complementation and hybridization evidence for additional families of type I DNA restriction and modification genes in Salmonella serotypes. J. Bacteriol., 170, 5785–5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cowan G.M., Gann,A.A. and Murray,N.E. (1989) Conservation of complex DNA recognition domains between families of restriction enzymes. Cell, 56, 103–109. [DOI] [PubMed] [Google Scholar]
- 28.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J.H., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deppenmeier U., Johann,A., Hartsch,T., Merkl,R., Schmitz,R.A., Martinez-Arias,R., Henne,A., Wiezer,A., Baumer,S., Jacobi,C. et al. (2002) The genome of Methanosarcina mazei: Evidence for lateral gene transfer between bacteria and archaea. J. Mol. Microbiol. Biotechnol., 4, 453–461. [PubMed] [Google Scholar]
- 30.Venter J.C., Remington,K., Heidelberg,J.F., Halpern,A.L., Rusch,D., Eisen,J.A., Wu,D.Y., Paulsen,I., Nelson,K.E., Nelson,W. et al. (2004) Environmental genome shotgun sequencing of the Sargasso Sea. Science, 304, 66–74. [DOI] [PubMed] [Google Scholar]
- 31.Wilson G.G. (1991) Organization of restriction-modification systems. Nucleic Acids Res., 19, 2539–2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Satta G., Debbia,E., Pruzzo,C. and Calegari,L. (1978) The peculiar behaviour of coliphage P1vir mutants on restricting hosts. Microbios, 22, 93–102. [PubMed] [Google Scholar]
- 33.Sanger F., Coulson,A.R., Hong,G.F., Hill,D.F. and Petersen,G.B. (1982) Nucleotide sequence of bacteriophage lambda DNA. J. Mol. Biol., 162, 729–773. [DOI] [PubMed] [Google Scholar]
- 34.Bult C.J., White,O., Olsen,G.J., Zhou,L., Fleischmann,R.D., Sutton,G.G., Blake,J.A., FitzGerald,L.M., Clayton,R.A., Gocayne,J.D. et al. (1996) Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science, 273, 1058–1073. [DOI] [PubMed] [Google Scholar]
- 35.Heidelberg J.F., Paulsen,I.T., Nelson,K.E., Gaidos,E.J., Nelson,W.C., Read,T.D., Eisen,J.A., Seshadri,R., Ward,N., Methe,B. et al. (2002) Genome sequence of the dissimilatory metal ion-reducing bacterium Shewanella oneidensis. Nat. Biotechnol., 20, 1118–1123. [DOI] [PubMed] [Google Scholar]
- 36.Kaneko T., Nakamura,Y., Wolk,C.P., Kuritz,T., Sasamoto,S., Watanabe,A., Iriguchi,M., Ishikawa,A., Kawashima,K., Kimura,T. et al. (2001) Complete genomic sequence of the filamentous nitrogen-fixing Cyanobacterium anabaena sp strain PCC 7120. DNA Res., 8, 205–213. [DOI] [PubMed] [Google Scholar]