Abstract
Screening for large gene rearrangements is established as an important part of molecular medicine but is also challenging. A variety of robust methods can detect whole-gene deletions, but will fail to detect more subtle rearrangements that may involve a single exon. In this paper, we describe a new, versatile and robust method to assess exon copy number, called multiplex PCR/liquid chromatography assay (MP/LC). Multiple exons are amplified using unlabeled primers, then separated by ion-pair reversed-phase high-performance liquid chromatography (IP-RP-HPLC), and quantitated by fluorescent detection using a post-column intercalation dye. The relative peak intensities for each target directly reflect exon copy number. This novel technique was used to screen a panel of 121 unrelated retinoblastoma patients who were tested previously using a reference strategy. MP/LC correctly scored all deletions and demonstrated a previously undetected RB1 duplication, the first to be described. MP/LC appears to be an easy, versatile, and cost-effective method, which is particularly relevant to denaturing HPLC (DHPLC) users since it broadens the spectrum of available applications on a DHPLC system.
INTRODUCTION
Rearrangement-based disorders, also termed genomic disorders, are caused by an alteration of the genome, which can lead to the loss or gain of a dosage-sensitive gene or to disruption of a gene (1). Such alterations range from extra copy of a complete chromosome(s) to deletion or duplication of a single exon. Duchenne/Becker muscular dystrophy is a striking example, as ∼70% of cases are due to large deletions or duplications (2,3). Since the first comprehensive review on this topic (4), the number of characterized genomic disorders has continued to expand (5), which is why screening for gross rearrangements has been established as an important part of molecular diagnosis, although it requires dedicated tools, as conventional PCR-based methods are mainly designed for point mutation detection. As a result, such rearrangements go undetected because they are hidden by the normal allele or the other altered allele in dominant or recessive diseases, respectively.
Chromosomal duplications or deletions can be detected using standard and molecular cytogenetic approaches such as high resolution karyotyping, fluorescent in situ hybridization (FISH), or color bar coding on combed DNA(6). Southern-blot and pulse-field gel electrophoresis (PFGE) are also commonly used to identify gene rearrangements. These techniques are being challenged by array-CGH that uses PAC and BAC clones, which has a higher throughput than FISH and PFGE and is particularly useful to detect rearrangements that are not visible by standard karyotypic analysis (7). However, these techniques are labor intensive and/or difficult to implement and fail to detect more discrete rearrangements that may involve a single exon. Hemizygosity studies based on segregation analysis of microsatellite markers require sampling of the parents and also depend on the informativeness of the markers. Transcript-based analysis may be an attractive alternative but is limited by RNA availability and could result in false-negatives due to nonsense mediated decay (8). Real-time PCR strategies on genomic DNA are methods of choice for DNA quantitation and therefore gene dosage. However, throughput remains limited by the small number of dyes currently available, making these strategies inappropriate for routine detection of exon deletion and duplication in large genes such as NF1, BRCA1 or RB1. An elegant multiplex ligation-dependent probe assay (MLPA) has been recently described, which is related to the previously described multiplex amplifiable probe hybridization (9). It consists of relative quantification of up to 40 different DNA sequences in a single tube. Following probe hybridization, ligation, and PCR, amplification products are detected and quantified by capillary electrophoresis (10). This technique is easy to use when available as a manufactured kit for the gene of interest but proves to be time-consuming and difficult to implement for other applications (11).
This problem was addressed by designing a `cloning-free' MLPA but this promising assay is currently described for cases in which the number of patients to be tested is limited (12).
Lastly, various semiquantitative multiplex fluorescent PCR protocols have been used to detect gross rearrangements (13), culminating in the so-called quantitative multiplex PCR of short fluorescent fragments (QMPSF) (14). QMPSF has been successfully adapted to different genes (15,16) but requires purified fluorescent labeled oligonucleotides, with the attendant high costs.
In this paper, we describe a new, versatile and robust method to assess gene copy number. This method, called multiplex PCR/liquid chromatography assay (MP/LC), is based on the well-known properties of ion-pair reversed-phase high performance liquid chromatography (IP-RP-HPLC) for separation and quantitation of nucleic acids (17). First, a multiplex PCR with unlabeled primers enables simultaneous amplification of multiple exons (up to 12 exons in this case) under semiquantitative conditions. Second, PCR products are separated by non-denaturing IP-RP-HPLC and, lastly, are quantitated by fluorescent detection using a post-column intercalation dye. The exceptional sensitivity of the dye allows the use of limiting PCR conditions to remain in the exponential phase of amplification. Consequently, relative peak intensities for each amplicon directly reflect exon copy number. Following an optimization step, this novel technique was used to screen a panel of 121 unrelated retinoblastoma patients who were tested previously using a reference strategy consisting of a combination of QMPSF and FISH analyses (18). Gross rearrangement screening is mandatory in RB1 analysis, as an average 13% of the mutational spectrum consists of large deletions (19). All rearrangements that were found using the reference strategy were correctly scored using MP/LC and, more interestingly, MP/LC also demonstrated an RB1 duplication that is not detected by the reference strategy. This is the first RB1 duplication to be described in the literature.
MATERIALS AND METHODS
Patients and reference strategy
We studied 121 consecutively diagnosed cases for constitutional RB1 rearrangement screening. The diagnosis of retinoblastoma was established on the basis of current ophthalmologic and—when tumor was available—histologic criteria. We examined 58 probands with bilateral and/or familial retinoblastoma and 63 probands with unilateral, sporadic retinoblastoma. Individual written consent for molecular analysis was obtained from all sampled individuals or their legal guardian(s). Forty-six deleterious point mutations [(18) and C. Houdayer, C. Dehainault, M. Gauthier-Villars and D. Stoppa-Lyonnet, unpublished data] and three rearrangements were identified in this sample, using a previously described strategy (18). Briefly, our reference strategy for rearrangement screening consists of a combination of QMPSF, FISH, long-range PCR and RT–PCR analyses. The rearrangements found consisted of two large deletions (encompassing promoter to exon 2 and exons 21–22, respectively), and one single-exon deletion (exon 18). For MP/LC optimization, we used a panel of seven DNAs carrying various RB1 deletions and five DNAs from normal controls. Mutated samples included a whole-gene deletion, five multi-exon deletions, i.e. promoter to exon 2 (two samples), exons 1–17, exons 3–17, exons 18–27, and one single-exon deletion (exon 3).
DNA extraction
DNA was extracted from 5 ml of whole blood samples collected on EDTA using a modified perchlorate/chloroform procedure, as described previously (20). DNA quantity for each sample was determined using a PicoGreen assay (Molecular Bioprobes) and a Genios microplate reader (Tecan) according to the manufacturer's recommendations. Multiplex PCR was performed with 50 ng/μl aliquots of DNA.
MP/LC
Multiplex PCR
All 27 exons and the promoter were amplified in five multiplex reactions with minor modifications of a previously described protocol (18). Primers were designed using Amplify 2.1 software (University of Wisconsin, WI) and positions that contained reported polymorphic sites were avoided to prevent false-positives resulting from amplification of a single allele. All primers were standard, i.e. desalted, unlabeled, without tail, and purchased from Proligo (Table 1). PCR was performed using 100 ng of genomic DNA with the GeneAmp 10× PCR Gold buffer, then run on a GeneAmp PCR system 9700 (Applied Biosystems). Multiplexes 1, 2 and 3 were performed in a 25 μl final volume using 0.4 mM of each dNTP, 4 mM MgCl2 and 1.5 U of AmpliTaq Gold DNA Polymerase (Applied Biosystems). Five percent and 7.5% dimethyl sulfoxide (DMSO) were added to multiplexes 3 and 1, respectively. Multiplexes 4 and 5 were performed in a 20 μl final volume using 0.4 mM of each dNTP, 6 mM MgCl2 and 1 unit AmpliTaq Gold DNA Polymerase (Applied Biosystems). Cycling conditions for multiplexes 1 and 3 consisted of a first denaturation step of 95°C for 5 min, followed by 25 cycles of denaturation at 95°C for 20 s, annealing at 60–44°C for 20 s (touchdown protocol, see Table 2) and extension at 72°C for 40 s, followed by a final extension at 72°C for 5 min. Cycling conditions for multiplexes 2, 4 and 5 consisted of a first denaturation step of 95°C for 5 min, followed by 23 cycles of denaturation at 95°C for 30 s, annealing at 55°C for 30 s and extension at 72°C for 45 s, followed by a final extension at 72°C for 5 min. Amplicons derived from the BRCA1 gene were included in each of the five multiplexes as internal controls of exon copy number. Samples that harbored complete RB1 exon deletions were routinely used as references of 50% copy number.
Table 1. Primer sets for MP/LC.
| Exon/Promoter | Primer sequence (5′ to 3′) | Concentrationa (μM) | Amplicon size (bp) | |
|---|---|---|---|---|
| Forward | Reverse | |||
| Multiplex 1 | ||||
| Exon 1 | AAGGCGTCATGCCGCCCAAAAC | AAGGCGTCATGCCGCCCAAAAC | 0.6 | 176 |
| BRCA1-exon 11 | TGTATTTTTTTAATGACAATTCAG | CTCCACATGCAAGTTTGAAAC | 1.0 | 225 |
| Promoter | CTGGACCCACGCCAGGTTTC | CCCGACTCCCGTTACAAAAAT | 0.6 | 259 |
| Multiplex 2 | ||||
| Exon 2 | CATTTGGTAGGCTTGAGTTTG | ATCCTTACCAATACTCCATCC | 0.8 | 165 |
| BRCA1-exon 11 | TGTATTTTTTTAATGACAATTCAG | CTCCACATGCAAGTTTGAAAC | 0.5 | 225 |
| Exon 10 | TCTTTAATGAAATCTGTGCC | GATATCTAAAGGTCACTAAG | 0.8 | 295 |
| Exon 13 | CTTATGTTCAGTAGTTGTGG | TATACGAACTGGAAAGATGC | 0.8 | 343 |
| Multiplex 3 | ||||
| Exon 18 | TAAACAATCAAAGGACCGAG | TTTGCTTACATATCTGCTGC | 0.06 | 125 |
| Exon 22 | TCCTCAGACATTCAAACGTG | AGGATACTTTTGACCTACCC | 0.06 | 159 |
| Exon 20 | CTTATTCCCACAGTGTATCG | TCTTACTTGGTCCAAATGCC | 0.34 | 185 |
| BRCA1-exon 11 | TGTATTTTTTTAATGACAATTCAG | CTCCACATGCAAGTTTGAAAC | 0.30 | 225 |
| Exon 19 | ATAATCTGTGATTCTTAGCC | AAGAAACATGATTTGAACCC | 0.10 | 273 |
| Multiplex 4 | ||||
| Exon 26 | AGTAAGTCATCGAAAGCATC | AACGAAAAGACTTCTTGCAG | 0.05 | 209 |
| Exon 9 | TGCATTGTTCAAGAGTCAAG | AGTTAGACAATTATCCTCCC | 0.12 | 223 |
| Exon 27 | CGCCATCAGTTTGACATGAG | CAGTCACATCTGTGAGAGAC | 0.08 | 238 |
| Exon 7 | CCTGCGATTTTCTCTCATAC | ATGTTTGGTACCCACTAGAC | 0.08 | 257 |
| Exon 3 | CAGTTTTAACATAGTATCCAG | AGCATTTCTCACTAATTCAC | 0.20 | 282 |
| Exon 12 | GGCAGTGTATTTGAAGATAC | AACTACATGTTAGATAGGAG | 0.13 | 311 |
| BRCA1-exon 18 | GGGAGGCTCTTTAGCTTCTTAGGACAGC | GAGACCCATTTTCCCAGCATCACCAGC | 0.07 | 358 |
| Exon 8 | AGTAGTAGAATGTTACCAAG | TACTGCAAAAGAGTTAGCAC | 0.13 | 382 |
| Multiplex 5 | ||||
| Exon 5 | GCATGAGAAAACTACTATGAC | CTAACCCTAACTATCAAGATG | 0.07 | 196 |
| Exon 24 | GTATTTATGCTCATCTCTGC | TGAGGTGTTTGAATAACTGC | 0.06 | 212 |
| Exon 6 | CACCCAAAAGATATATCTGG | ATTTAGTCCAAAGGAATGCC | 0.06 | 224 |
| Exon 11 | GAGACAACAGAAGCATTATAC | CGTGAACAAATCTGAAACAC | 0.09 | 247 |
| Exon 14 | AGCCTGGGCAAAACAGTGAG | AGCCTGGGCAAAACAGTGAG | 0.05 | 268 |
| Exon 23 | ATCTAATGTAATGGGTCCAC | CTTGGATCAAAATAATCCCC | 0.07 | 289 |
| Exon 25 | GGTTGCTAACTATGAAACAC | AGAAATTGGTATAAGCCAGG | 0.07 | 299 |
| Exon 4 | GTAGTGATTTGATGTAGAGC | CCCAGAATCTAATTGTGAAC | 0.12 | 307 |
| Exon 21 | AAACCTTTCTTTTTTGAGGC | TACATAATAAGGTCAGACAG | 0.09 | 330 |
| Exon 17 | AAAAATACCTAGCTCAAGGG | TGTTAAGAAACACCTCTCAC | 0.07 | 349 |
| BRCA1-exon 18 | GGGAGGCTCTTTAGCTTCTTAGGACAGC | GAGACCCATTTTCCCAGCATCACCAGC | 0.07 | 358 |
| Exons 15 and 16 | CAATGCTGACACAAATAAGG | AGCATTCCTTCTCCTTAACC | 0.14 | 368 |
aIn the final PCR volume; forward and reverse equimolar.
Table 2. Touchdown protocol for multiplexes 1 and 4.
| Multiplex 1 | ||||||||
| Denaturation (°C) | 95 | 95 | 95 | 95 | 95 | 95 | 95 | 95 |
| Annealing (°C) | 60 | 59 | 58 | 57 | 56 | 55 | 54 | 53 |
| Elongation (°C) | 72 | 72 | 72 | 72 | 72 | 72 | 72 | 72 |
| Number of cycles | 2 | 2 | 2 | 3 | 3 | 4 | 4 | 5 |
| Multiplex 4 | ||||||||
| Denaturation (°C) | 95 | 95 | 95 | 95 | 95 | 95 | 95 | 95 |
| Annealing (°C) | 51 | 50 | 49 | 48 | 47 | 46 | 45 | 44 |
| Elongation (°C) | 72 | 72 | 72 | 72 | 72 | 72 | 72 | 72 |
| Number of cycles | 2 | 2 | 2 | 3 | 3 | 4 | 4 | 5 |
Liquid chromatography
Aliquots of 5 μl of PCR products were injected for 0.1 min on a semi-automated 3500 Wave HS system (Transgenomic), incorporating a HSD (high sensitivity detector). HSD uses post-column intercalation chemistries, i.e. SYBR Green 1 (Molecular Bioprobes) and fluorescence detection to provide sensitivity enhancement for the detection of DNA (21). Separation was performed using a DNAsep cartridge (Transgenomic) and a constant oven temperature of 50°C to ensure non-denaturing conditions. The mobile phase was 0.1 M triethylammonium acetate (TEAA) solution in water, pH 7 (WAVE Optimized buffer A, Transgenomic) and PCR products were eluted at a flow rate of 0.9 ml/min with an acetonitrile gradient. The gradient was created by mixing buffer A and 0.1 M TEAA solution in water, 25% acetonitrile, pH 7 (WAVE Optimized buffer B, Transgenomic). Three different gradients were used depending on the multiplex to be analyzed (Table 3).
Table 3. Gradients used for RB1 analysis.
| Method name | Fragment sizes (bp) | Gradient | ||
|---|---|---|---|---|
| %B, initial | %B, final | Duration (min.) | ||
| Multiplex 1 | 176, 225, 259 | 54.3 | 59.2 | 7.2 |
| Multiplex 2 | 165, 225, 295, 343 | 47.3 | 61.8 | 6.3 |
| Multiplex 3 | 125, 159, 185, 225, 273 | 47.3 | 61.8 | 6.3 |
| Multiplex 4 | 209, 223, 238, 257, 282, 311, 358, 382 | 56.6 | 62.7 | 8.4 |
| Multiplex 5 | 196, 212, 224, 247, 268, 289, 299, 307, 330, 349, 358, 368 | 54.3 | 60.6 | 9.6 |
Following separation, eluted PCR products were mixed with the Wave HS Staining Solution I that contained SYBR Green 1 by using the HSX accessory, and were then detected with the FDX fluorescence detector (Transgenomic). Between runs, the column was flushed for 0.5 min with 75% acetonitrile in water and re-equilibrated for 0.7 min at the starting conditions of the gradient. Data were analyzed using Navigator 1.5.3 software (Transgenomic). Briefly, electrophoregrams from patients were first superposed to those of a normal control and a mutated control that harbored a whole-gene deletion. Fluorescence intensities were then normalized by adjusting the peaks that were obtained for the control amplicons to the same height. The yield of each amplicon in the various samples was evaluated and deletions or duplications of one or more amplicons were revealed by a 2-fold decrease or by a 1.5-fold increase of the corresponding peak(s), respectively. High-throughput data analysis was made possible by automated profile normalization using the Mutation Calling option of the software. An alternative solution to profile superposition was to export chromatographic data to Excel (Microsoft) for ratio calculation.
RESULTS
A HPLC-based technique was designed to identify RB1 rearrangements in 121 unrelated retinoblastoma patients. An optimization step that was performed on various mutated samples (see Materials and Methods section) was necessary before use in routine screening.
Optimization step
Multiplex PCR
RB1 was screened in five multiplex reactions. For all experiments, we used standardized primers (unpurified, unlabeled and without tail). Multiplex systems were designed by combining various sets of primers for which the reaction conditions were determined separately. Primer sets were added sequentially with respect to amplicon sizes and the regions to be amplified. If necessary, reaction components and cycling conditions were adjusted to yield fluorescent peaks within the same range. Hence the 5′ part of the gene was amplified in a separate mix due to its high GC content and a touchdown protocol had to be used to achieve homogeneous peak heights for multiplexes 1 and 4. DNA quality and quantity must be carefully checked to ensure successful analysis. Phenol-based extraction must be avoided, due to the possibility of phenol residues that could alter PCR kinetics. We found that a genomic DNA quantity ranging from 50 to 150 ng could be used for MP/LC analysis and this is why we decided to work in the medium range, i.e. 100 ng.
Chromatographic conditions and data collection
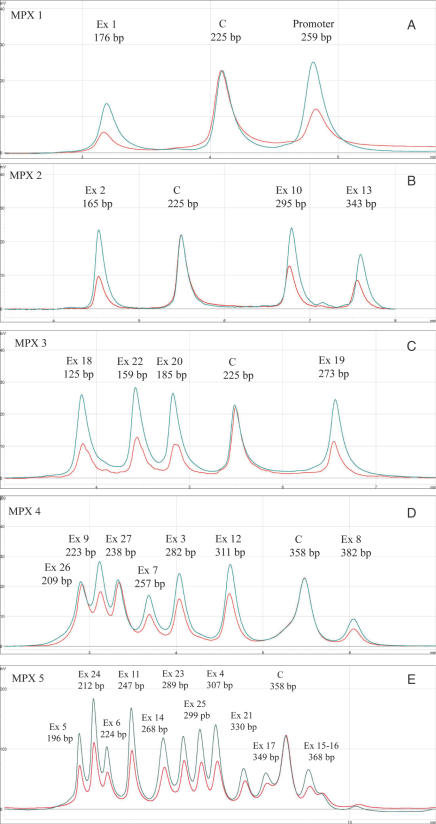
We used an empirical approach to find optimum fragment separation conditions. A successful gradient separation was developed by starting with standardized conditions as predicted by Navigator 1.5.3 software (Transgenomic) followed by fine-tuning to ensure best separation. Three variables were used to optimize gradient separation: the initial percentage of organic solvent (%B), the final percentage of organic solvent, and the gradient time or slope. In order to maintain optimum run times, we first defined the conditions that allowed adequate peak separation, although part of the gradient time was wasted. We then tried to reduce the gradient time while maintaining a constant gradient retention factor (i.e. selectivity was kept constant). Further adjustments were made to ensure a time window of 2.5 min between the injection peak and the first eluted peak to avoid variations in retention time for the first eluted peaks. An increase or decrease in the flow rate did not markedly improve separation and it was therefore kept at 0.9 ml/min. We also kept a constant temperature of 50°C that provides optimum resolution, as demonstrated previously (22). We therefore had to design five multiplexes to meet the requirements of successful multiplex PCR amplification and chromatographic separation (Figure 1). Relative gene dosage was obtained by comparing peak heights rather than peak areas, since the curves did not always reach the baseline between eluted peaks. This procedure provided us with sound results (see discussion).
Figure 1.
MP/LC chromatograms for the five different multiplexes used in RB1 analysis. x-axis: retention time in min; y-axis: fluorescence intensity. Exons under study are indicated with their size at the top of the corresponding peaks. The chromatograms in blue and red represent the normal and deleted controls, respectively. Profiles are superimposed and then normalized using the control amplicon C. A whole-gene deleted control is used except for MPX 4 for which a deletion from promoter to exon 17 was used. This figure shows the good resolution of the three first peaks that are situated close together.
Sample analysis
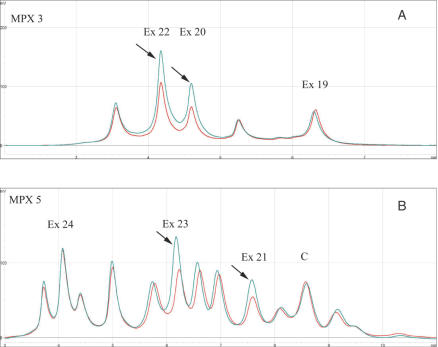
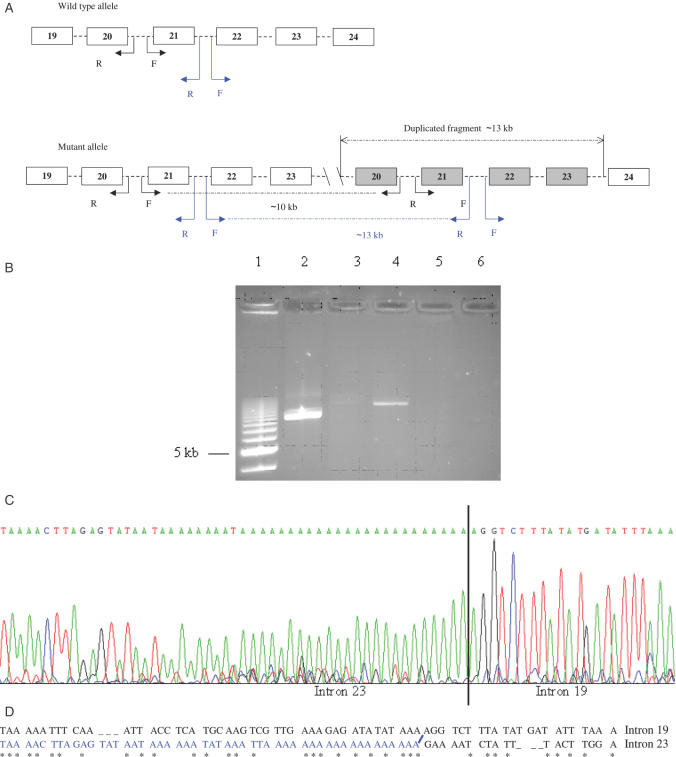
Following optimization, we blindly tested 121 consecutively diagnosed retinoblastoma patients who were tested previously by our reference strategy (see Materials and Methods section). Our novel MP/LC approach correctly identified the three deleted samples identified previously, i.e. two multi-exon deletions (encompassing promoter to exon 2 and exons 21–22, respectively), and one single-exon deletion (exon 18). All but one of the remaining samples were scored as normal. A duplication from exon 20 to 23 was also found in a bilaterally affected retinoblastoma proband (Figure 2A and B). This result was confirmed by long range PCR that used dedicated primers, allowing amplification from the sole duplicated allele (Figure 3A and B). These primers were chosen in opposite directions to avoid amplification of the wild-type allele but allowing the duplicated allele to generate an abnormal product that comprised the breakpoint. Further sequencing of this product allowed breakpoint identification (Figure 3C). Alignment of introns 19 and 23 using CLUSTAL W 1.8 software (European Bioinformatics Institute) showed blocks of similarity around the breakpoint that may explain the duplication by a recombination mechanism mediated by the poly(A) tracts (Figure 3D). It was noted that the breakpoint in intron 23 was located at the 3′end of a LINE-1 repeat element (23) that could play a role in this mechanism.
Figure 2.
MP/LC chromatograms for multiplexes 3 (A) and 5 (B). x-axis: retention time in minutes; y-axis: fluorescence intensity. The chromatograms in red and blue represent the normal and duplicated samples, respectively. Profiles are superimposed and then normalized using the control amplicon C. Duplicated exons are indicated by an arrow.
Figure 3.
Duplication of exons 20–23 of RB1. (A) PCR strategy for the amplification of the duplicated allele. RB1 exons 19–24 (not drawn to scale) are boxed and numbered, introns are indicated by dashed lines. F and R indicate forward and reverse primers, respectively. Two different pairs of primers were used (indicated in black and blue). These primers are in the opposite direction on the wild-type allele, but generate a 10 or a 13 kb fragment on the mutant duplicated allele. Exons in grey are duplicated. (B) Long range PCR using primers in (A): 10 and 13 kb PCR products (lanes 2 and 4, respectively), normal controls (lanes 3 and 5), blank (lane 6), 1 kb ladder (lane 1). (C) Breakpoint sequencing using the 10 kb product and primers located in introns 19 and 23. Sequence quality is poor due to the poly(A) stretch. (D) Alignment of breakpoint flanking sequences from introns 19 and 23, using CLUSTAL W 1.8 software. Sequence similarities are indicated with an asterisk. The rearranged sequence is in blue and results from a recombination between introns 19 and 23, possibly mediated by the poly(A) tracts.
DISCUSSION
In this paper, we describe a new technique based on semiquantitative MP/LC that is dedicated to high-throughput screening for gross rearrangements. We report its application to the analysis of the RB1 gene.
MP/LC design and performance
Ion-pair RP-HPLC on nonporous alkylated polystyrene-divinylbenzene particles, e.g. the DNAsep column (Transgenomic) that was used in this work, allowed the separation of the double-stranded DNA fragments based on size, irrespective of base composition. Retention is mainly determined by the charge of the polynucleotides, thus determining the number of ion pairs formed, so that smaller species elute prior to larger, more negatively charged molecules. This technique therefore overcomes the drawbacks of anion-exchange chromatography, for which retention is also sequence-dependent (22), and lends itself to PCR product separation and analysis (24). Moreover, IP-RP-HPLC has also been used for quantification of PCR products in a few gene expression studies (25,26) and in a so-called ‘walking quantitative DHPLC’ (16). Unfortunately, these attractive approaches remain limited by their lack of throughput and sensitivity, since a single amplicon is tested by ultraviolet (UV) analysis.
MP/LC makes use of the well-known robustness of IP-RP-HPLC for the separation and quantitation, combined with the high sensitivity provided by fluorescence detection of DNA. Following chromatographic separation, unlabeled eluted products are mixed with a staining solution that contains SYBR Green I. This fluorescent dye only fluoresces in a bound state in the presence of a nucleic acid complex, thereby allowing detection of eluted PCR products. SYBR Green I has been widely used for PCR product quantitation in multiple protocols that requires high sensitivity (27,28). Its exceptional sensitivity is particularly useful for applications in which the quantity of DNA is limited e.g. detection of low-yield DNA amplification products. We found that fluorescence detection increases the sensitivity to ∼100-fold compared with UV detection, while maintaining the same resolution. This allowed us to use limiting PCR conditions, i.e. a small number of cycles, to remain in the exponential phase of amplification. Signal strength is therefore directly dependent on the target copy number, so that relative gene dosage can be obtained by comparing peak height of the DNA test to those of normal and mutated controls. Repeatability and reproducibility are key issues in this assay, as reliable peak identification on the basis of retention time depends on the precision with which this position can be reproduced. Reproducibility is reported for the sophisticated 12-plex on 25 normal samples (Table 4). Since thresholds for deletions and duplications were set to the consensus values of 0.75 and 1.25, respectively, MPX5 was considered to be reliable. Quantitation was performed on the basis of peak height and peak area measurement and most often, similar results were obtained. However, in a few cases, peak height measurement gave better results (e.g. exons 5 and 15–16, Table 4). This is possibly due to baseline definition; therefore, at this point in time, we would recommend the use of peak height when the Navigator v 1.5.3 software (Transgenomic) is used for analysis. Due to the good reproducibility of the method, interpretation is straightforward with rapid analysis by groups of samples. Repeatability was also demonstrated by injection of 16 different PCR products that were derived from the same DNA sample, which gave identical results (data not shown). We also believe that the performance of MP/LC could be increased using a DNAsep HT cartridge (Transgenomic) that would give higher resolution.
Table 4. Statistical analyses of 25 normal samples for MPX5 (12-plex).
| Peak size (bp) | Peak height, range (SD) | Peak area, range (SD) | Retention time, range in min (SD) | Resolution,a range (SD) |
|---|---|---|---|---|
| Exon 5 (196) | 0.83–1.14 (0.10) | 0.79–1.2 (0.12) | 3.69–3.76 (0.02) | 0.45–0.69 (0.07) |
| Exon 24 (212) | 0.85–1 11 (0.08) | 0.84–1.10 (0.08) | 4.02–4.10 (0.02) | 0.78–0.82 (0.01) |
| Exon 6 (224) | 0.86–1.10 (0.08) | 0.90–1.10 (0.07) | 4.33–4.42 (0.02) | 1.04–1.06 (0.007) |
| Exon 11 (247) | 0.88–1.07 (0.05) | 0.88–1.09 (0.06) | 4.90–5.00 (0.02) | 1.20–1.24 (0.01) |
| Exon 14 (268) | 0.88–1.12 (0.06) | 0.89–1.11 (0.06) | 5.66–5.74 (0.02) | 0.89–0.93 (0.01) |
| Exon 23 (289) | 0.83–1.10 (0.09) | 0.81–1.12 (0.10) | 6.12–6.20 (0.02) | 0.95–0.99 (0.01) |
| Exon 25 (299) | 0.88–1.15 (0.08) | 0.88–1.12 (0.08) | 6.51–6.59 (0.02) | 0.79–0.81 (0.006) |
| Exon 4 (307) | 0.90–1.09 (0.05) | 0.90–1.07 (0.05) | 6.87–6.95 (0.03) | 1.17–1.20 (0.008) |
| Exon 21 (330) | 0.81–1.09 (0.09) | 0.81–1.10 (0.09) | 7.49–7.60 (0.03) | 1.04–1.10 (0.01) |
| Exon 17 (349) | 0.90–1.12 (0.06) | 0.90–1.11 (0.06) | 7.99–8.12 (0.03) | 0.90–0.93 (0.01)b |
| Exons 15–16 (368) | 0.92–1.09 (0.04) | 0.80–1.17 (0.12) | 8.98–9.10 (0.03) | 0.66–1.00 (0.15)b |
Peak height and area were normalized to 1.00, corresponding to a copy number of two.
aResolution between adjacent peaks is calculated as follows: 2 × (difference in retention time of the two peaks)/(width of peak 1 + width of peak 2).
bCalculated with the control peak.
However, as for all HPLC assays, these performances depend on careful maintenance and strict quality control of the system. The use of degraded columns could lead to unexpected results, such as a spurious increase or decrease of the peaks or variations in retention times. Therefore, before each run, the system integrity and performance must be checked by injecting increasing amounts of a sizing standard to ensure correct peak resolution and quantitation (Table 5). PCR using ultrapure water with high resistivity gave the best results for subsequent MP/LC analysis, probably because of the low ion content that decreases undesirable ionic interactions. Moreover, a total organic carbon (TOC) level <30 p.p.b. is recommended, as organic components are known to interact with the alkyl groups grafted on the column, thereby leading to loss of resolution (29). Lastly, a reliable multiplex PCR is obviously a key step for a successful MP/LC analysis. We experienced very few difficulties in multiplex design, as identification of the best multiplex was facilitated by testing various combinations of primers and, if necessary, by designing other primers in view of their negligible cost in this method.
Table 5. Performance of the system, based on serial dilutions (not diluted, 1/2, 1/4, 1/8, 1/16, 1/32) of a sizing standard (double-stranded fragments from a pUC18 HaeIII digest).
| Peak size (bp) | Based on peak height | Based on peak area | Analysis range (μg of PCR products) | Lower detection limit (μg of PCR products) | ||
|---|---|---|---|---|---|---|
| Regression equation | R2 | Regression equation | R2 | |||
| 80 | y = 124.52x + 0.2504 | 0.998 | y = 1 × 106x − 10 391 | 0.998 | 0.010–0.600 | 0.006 |
| 102 | y = 222.4x − 0.4394 | 0.999 | y = 3 × 106x − 12002 | 0.999 | 0.010–0.600 | 0.006 |
| 174 | y = 560.28x + 2.9036 | 0.999 | y = 7 × 106x + 68 670 | 0.997 | 0.010–0.600 | <0.001 |
| 257 | y = 924x + 17.953 | 0.996 | y = 1 × 107x + 169 236 | 0.996 | 0.050–0.600 | <0.001 |
| 267 | y = 933.03x + 19.397 | 0.996 | y = 1 × 107x + 176 685 | 0.997 | 0.050–0.600 | <0.001 |
| 298 | y = 949.3x + 22.866 | 0.994 | y = 1 × 107x + 187 837 | 0.996 | 0.050–0.600 | <0.001 |
| 434 | y = 1305x + 41.427 | 0.990 | y = 2 × 107x + 338 614 | 0.994 | 0.050–0.600 | <0.001 |
| 458 | y = 1269.6x + 42.151 | 0.990 | y = 2 × 107x + 293 285 | 0.997 | 0.050–0.600 | <0.001 |
| 587 | y = 1375.1x + 54.327 | 0.986 | y = 2 × 107x + 370 551 | 0.996 | 0.050–0.600 | <0.001 |
Regression equations, R2, analysis range and lower detection limit are indicated for each peak. The fluorescent signal increases in accordance with the sample load.
Following verification and optimization, MP/LC that constitutes a very powerful tool in our hands, allows a reliable and high-throughput screening of RB1 genomic rearrangements. MP/LC identified all deletions found with the reference method, but, more interestingly, also revealed an RB1 duplication. Surprisingly, this is the first duplication to be described, whereas an average of 13% of the RB1 mutational spectrum consists of large deletions. Duplications are generally not rare events since recent estimates suggest that as much as 5–10% of the human genome might be duplicated (1). As in the case of BRCA1 (30,31), a higher rate of duplications could have been expected for RB1, and the low rates observed could be due to the poor sensitivity of the methods used (19) and the absence of duplicated controls, since it is more difficult to detect a duplication than a deletion (1.5-fold increase versus a 2-fold decrease in signal strength, respectively), thereby demonstrating the sensitivity of MP/LC. We have subsequently optimized our QMPSF conditions to obtain more reproducible data and we are now able to detect this duplication using QMPSF (data not shown). The strength of MP/LC lies in the simplicity and robustness of the technologies that are used. PCR uses unlabeled primers thereby excluding possible effects of the 5′-fluorophore on primer hybridization, and since SYBR Green is added following chromatographic separation, it can neither modify PCR kinetics (32) nor alter resolution and retention time. Sensitivity is therefore enhanced while providing the same resolution obtained in dye-free experiments. Another interesting aspect is the very low cost of primers. MP/LC obviously requires a DHPLC system equipped with fluorescent detection because of the inadequate sensitivity of UV analysis. The staining solution (Transgenomic) costs entailed are estimated to be 0.35 euros per injection, but could be lowered by home-made dilutions of SYBR Green I (Molecular Probes). Moreover, multiple custom applications dealing with DNA copy number and requiring a variety of primer sets can be assayed by MP/LC because of its simplicity and cost-effectiveness. As an example, we used ‘walking MP/LC’ for breakpoint deletion mapping, e.g. we narrowed deletions in BRCA1 with markers spread along the suspected deleted region to finally obtain minimum fragment intervals amenable to sequencing (A. Laugé, C. Houdayer and D. Stoppa-Lyonnet, unpublished data). This application is particularly relevant in molecular diagnostic medicine since identification and analysis of the sequence context of rearrangement breakpoints may improve the design and efficacy of mutation search procedures in genomic disorders (33).
In summary, we describe a high-throughput MP/LC assay that can differentiate one, two or three copies of the target DNA. We believe that each gene involved in human diseases should be screened for gross rearrangements, as an increasing number of studies reveal the importance of genomic disorders in DNA pathology (1),(5). MP/LC is very easy to implement and versatile, and therefore allows multiple gene screening at a low cost. Finally, MP/LC is particularly relevant to DHPLC users, as it broadens the spectrum of available applications on a DHPLC system.
Acknowledgments
ACKNOWLEDGEMENTS
The authors would like to thank A. Idirian and B. Mehaye for helpful support during the course of this study. This work was supported by a grant from Retinostop.
REFERENCES
- 1.Stankiewicz P. and Lupski,J.R. (2002) Genome architecture, rearrangements and genomic disorders. Trends Genet., 18, 74–82. [DOI] [PubMed] [Google Scholar]
- 2.Koenig M., Hoffman,E.P., Bertelson,C.J., Monaco,A.P., Feener,C. and Kunkel,L.M. (1987) Complete cloning of the Duchenne muscular dystrophy (DMD) cDNA and preliminary genomic organization of the DMD gene in normal and affected individuals. Cell, 50, 509–517. [DOI] [PubMed] [Google Scholar]
- 3.Den Dunnen J.T., Grootscholten,P.M., Bakker,E., Blonden,L.A., Ginjaar,H.B., Wapenaar,M.C., van Paassen,H.M., van Broeckhoven,C., Pearson,P.L. and van Ommen,G.J. (1989) Topography of the Duchenne muscular dystrophy (DMD) gene: FIGE and cDNA analysis of 194 cases reveals 115 deletions and 13 duplications. Am. J. Hum. Genet., 45, 835–847. [PMC free article] [PubMed] [Google Scholar]
- 4.Lupski J.R. (1998) Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet., 14, 417–422. [DOI] [PubMed] [Google Scholar]
- 5.Shaw C.J. and Lupski,J.R. (2004) Implications of human genome architecture for rearrangement-based disorders: the genomic basis of disease. Hum. Mol. Genet., 13, R57–64. [DOI] [PubMed] [Google Scholar]
- 6.Gad S., Aurias,A., Puget,N., Mairal,A., Schurra,C., Montagna,M., Pages,S., Caux,V., Mazoyer,S., Bensimon,A. et al. (2001) Color bar coding the BRCA1 gene on combed DNA: a useful strategy for detecting large gene rearrangements. Genes Chromosomes Cancer, 31, 75–84. [DOI] [PubMed] [Google Scholar]
- 7.Vissers L.E., de Vries,B.B., Osoegawa,K., Janssen,I.M., Feuth,T., Choy,C.O., Straatman,H., van der Vliet,W., Huys,E.H., van Rijk,A. et al. (2003) Array-based comparative genomic hybridization for the genomewide detection of submicroscopic chromosomal abnormalities. Am. J. Hum. Genet., 73, 1261–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maquat L.E. (2004) Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat. Rev. Mol. Cell Biol., 5, 89–99. [DOI] [PubMed] [Google Scholar]
- 9.Armour J.A., Sismani,C., Patsalis,P.C. and Cross,G. (2000) Measurement of locus copy number by hybridisation with amplifiable probes. Nucleic Acids Res., 28, 605–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schouten J.P., McElgunn,C.J., Waaijer,R., Zwijnenburg,D., Diepvens,F. and Pals,G. (2002) Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res., 30, e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sellner L.N. and Taylor,G.R. (2004) MLPA and MAPH: new techniques for detection of gene deletions. Hum. Mutat., 23, 413–419. [DOI] [PubMed] [Google Scholar]
- 12.White S.J., Vink,G.R., Kriek,M., Wuyts,W., Schouten,J., Bakker,B., Breuning,M.H. and den Dunnen,J.T. (2004) Two-color multiplex ligation-dependent probe amplification: detecting genomic rearrangements in hereditary multiple exostoses. Hum. Mutat., 24, 86–92. [DOI] [PubMed] [Google Scholar]
- 13.Purroy J., Bisceglia,L., Jaeken,J., Gasparini,P., Palacin,M. and Nunes,V. (2000) Detection of two novel large deletions in SLC3A1 by semi-quantitative fluorescent multiplex PCR. Hum. Mutat., 15, 373–379. [DOI] [PubMed] [Google Scholar]
- 14.Casilli F., Di Rocco,Z.C., Gad,S., Tournier,I., Stoppa-Lyonnet,D., Frebourg,T. and Tosi,M. (2002) Rapid detection of novel BRCA1 rearrangements in high-risk breast-ovarian cancer families using multiplex PCR of short fluorescent fragments. Hum. Mutat., 20, 218–226. [DOI] [PubMed] [Google Scholar]
- 15.Le Meur N., Martin,C., Saugier-Veber,P., Joly,G., Lemoine,F., Moirot,H., Rossi,A., Bachy,B., Cabot,A., Joly,P. et al. (2004) Complete germline deletion of the STK11 gene in a family with Peutz-Jeghers syndrome. Eur. J. Hum. Genet., 12, 415–418. [DOI] [PubMed] [Google Scholar]
- 16.Audrezet M.P., Chen,J.M., Raguenes,O., Chuzhanova,N., Giteau,K., Le Marechal,C., Quere,I., Cooper,D.N. and Ferec,C. (2004) Genomic rearrangements in the CFTR gene: extensive allelic heterogeneity and diverse mutational mechanisms. Hum. Mutat., 23, 343–357. [DOI] [PubMed] [Google Scholar]
- 17.Xiao W. and Oefner,P.J. (2001) Denaturing high-performance liquid chromatography: a review. Hum. Mutat., 17, 439–474. [DOI] [PubMed] [Google Scholar]
- 18.Houdayer C., Gauthier-Villars,M., Lauge,A., Pages-Berhouet,S., Dehainault,C., Caux-Moncoutier,V., Karczynski,P., Tosi,M., Doz,F., Desjardins,L. et al. (2004) Comprehensive screening for constitutional RB1 mutations by DHPLC and QMPSF. Hum. Mutat., 23, 193–202. [DOI] [PubMed] [Google Scholar]
- 19.Richter S., Vandezande,K., Chen,N., Zhang,K., Sutherland,J., Anderson,J., Han,L., Panton,R., Branco,P. and Gallie,B. (2003) Sensitive and efficient detection of RB1 gene mutations enhances care for families with retinoblastoma. Am. J. Hum. Genet., 72, 253–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johns M.B. Jr and Paulus-Thomas,J.E. (1989) Purification of human genomic DNA from whole blood using sodium perchlorate in place of phenol. Anal Biochem., 180, 276–278. [DOI] [PubMed] [Google Scholar]
- 21.Bahrami A.R., Dickman,M.J., Matin,M.M., Ashby,J.R., Brown,P.E., Conroy,M.J., Fowler,G.J., Rose,J.P., Sheikh,Q.I., Yeung,A.T. et al. (2002) Use of fluorescent DNA-intercalating dyes in the analysis of DNA via ion-pair reversed-phase denaturing high-performance liquid chromatography. Anal Biochem., 309, 248–252. [DOI] [PubMed] [Google Scholar]
- 22.Huber C.G., Oefner,P.J., Preuss,E. and Bonn,G.K. (1993) High-resolution liquid chromatography of DNA fragments on non-porous poly(styrene-divinylbenzene) particles. Nucleic Acids Res., 21, 1061–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Toguchida J., McGee,T.L., Paterson,J.C., Eagle,J.R., Tucker,S., Yandell,D.W. and Dryja,T.P. (1993) Complete genomic sequence of the human retinoblastoma susceptibility gene. Genomics, 17, 535–543. [DOI] [PubMed] [Google Scholar]
- 24.Huber C.G., Oefner,P.J. and Bonn,G.K. (1995) Rapid and accurate sizing of DNA fragments by ion-pair chromatography on alkylated nonporous poly(styrene-divinylbenzene) particles. Anal Chem., 67, 578–585. [Google Scholar]
- 25.Hayward-Lester A., Oefner,P.J. and Doris,P.A. (1996) Rapid quantification of gene expression by competitive RT–PCR and ion-pair reversed-phase HPLC. Biotechniques, 20, 250–257. [DOI] [PubMed] [Google Scholar]
- 26.Robinson C.A., Hayward-Lester,A., Hewetson,A., Oefner,P.J., Doris,P.A. and Chilton,B.S. (1997) Quantification of alternatively spliced RUSH mRNA isoforms by QRT-PCR and IP-RP-HPLC analysis: a new approach to measuring regulated splicing efficiency. Gene, 198, 1–4. [DOI] [PubMed] [Google Scholar]
- 27.Morrison T.B., Weis,J.J. and Wittwer,C.T. (1998) Quantification of low-copy transcripts by continuous SYBR Green I monitoring during amplification. Biotechniques, 24, 954–958. [PubMed] [Google Scholar]
- 28.Zipper H., Brunner,H., Bernhagen,J. and Vitzthum,F. (2004) Investigations on DNA intercalation and surface binding by SYBR Green I, its structure determination and methodological implications. Nucleic Acids Res., 32, e103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mabic S. and Kano,I. (2003) Impact of purified water quality on molecular biology experiments. Clin. Chem. Lab. Med., 41, 486–491. [DOI] [PubMed] [Google Scholar]
- 30.Gad S., Caux-Moncoutier,V., Pages-Berhouet,S., Gauthier-Villars,M., Coupier,I., Pujol,P., Frenay,M., Gilbert,B., Maugard,C., Bignon,Y.J. et al. (2002) Significant contribution of large BRCA1 gene rearrangements in 120 French breast and ovarian cancer families. Oncogene, 21, 6841–6847. [DOI] [PubMed] [Google Scholar]
- 31.Barrois M., Bieche,I., Mazoyer,S., Champeme,M.H., Bressac-de Paillerets,B. and Lidereau,R. (2004) Real-time PCR-based gene dosage assay for detecting BRCA1 rearrangements in breast-ovarian cancer families. Clin. Genet., 65, 131–136. [DOI] [PubMed] [Google Scholar]
- 32.Nath K., Sarosy,J.W., Hahn,J. and Di Como,C.J. (2000) Effects of ethidium bromide and SYBR Green I on different polymerase chain reaction systems. J. Biochem. Biophys. Methods, 42, 15–29. [DOI] [PubMed] [Google Scholar]
- 33.Abeysinghe S.S., Stenson,P.D., Krawczak,M. and Cooper,D.N. (2004) Gross Rearrangement Breakpoint Database (GRaBD). Hum. Mutat., 23, 219–221. [DOI] [PubMed] [Google Scholar]