Abstract
Although the significance of human genetic polymorphisms in therapeutic outcomes is well established, the importance of our “second genome” (the microbiome) has been largely overlooked. In this Review, we highlight recent studies that shed light on the mechanisms linking the human gut microbiome to the efficacy and toxicity of xenobiotics, including drugs, dietary compounds and environmental toxins. Continued progress in this area could enable more precise tools for predicting patient responses and the development of a next generation of therapeutics based on or targeted at the gut microbiome. Indeed, the admirable goal of precision medicine may require us to first understand the microbial pharmacists within.
Recent advances in the culture-independent interrogation of microbial community structure1, 2 and function2–4, including advances in sequencing technologies and the development of bioinformatic tools, have spawned a veritable “microbiome renaissance”. Due to these advances, the microbiota is now often referred to as the “forgotten organ” due to our understanding and appreciation of its contributions to host physiology, metabolism, and disease5.
The gut microbiota is a diverse and dense microbial community, unparalleled when compared to other body habitats. It is estimated that the gut microbiota is composed of more than 100 trillion cells and 5 million unique genes, which outnumber our own host cells and genes by more than 3-fold and 100-fold, respectively6. Although the gut microbiota is predicted to be composed of thousands of species, the majority of them belong to six bacterial phyla: Firmicutes, Bacteroidetes, Actinobacteria, Proteobacteria, Fusobacteria, and Verrucomicrobia7. In addition to bacteria, the gut microbiota also includes fungi, Archaea, protozoa and viruses. The gut microbiota is highly dynamic and demonstrates significant inter- and intra-individual variation. The structure of this microbial community is tightly linked to environmental factors like diet and drug intake3, 8 (discussed below), but has also been associated with age9 and host genetics10.
The microbiota has key functions such as the breakdown of plant polysaccharides (i.e., fiber) that are indigestible to the host, the biosynthesis of essential vitamins and amino acids, the detoxification of xenobiotics, the resistance against pathogens, and immune system development. Indeed, the microbiome has now been linked to many areas previously considered unrelated to microorganisms, from the circadian rhythm11 to neuroscience12–14, cancer biology15, 16, forensics17, 18 and metabolic disease19, 20.
Despite this exponential rise in microbiome research, the links between the microbiota and pharmacology remain critically underexplored. The discovery that human gut microorganisms can metabolize drugs dates back nearly a century21. Experiments with the sulfonamide antibiotic prontosil, the first broad-spectrum and commercially available antibiotic, demonstrated a lack of antibacterial activity in vitro22. This is due to the fact that gut microorganisms activate prontosil by reducing its azo bond. This biotransformation impacts a wide range of compounds, from azo dyes23, which are commonly used as additives in foods, to sulfasalazine, which is used to treat ulcerative colitis and rheumatoid arthritis24, 25. Soon after these discoveries, it became clear that the biotransformation of drugs by the gut microbiota might be far more widespread than previously appreciated, but limited mechanistic insights were uncovered due in part to the difficulties in analyzing complex gut microbial communities using traditional culture-based techniques. To date, the fields of pharmacogenetics and pharmacogenomics still largely focus on variations in the human genome, rather than on the genes encoded by the microbiome26.
Multiple reviews have highlighted the role of the gut microbiome in pharmacology and precision medicine27–32. Here, we focus on recent studies that provide insight into the microbial and molecular mechanisms that are relevant to the prevention and treatment of human disease. Gut microorganisms can impact drug therapy through a variety of different mechanisms that can generally be grouped into direct or indirect effects (Fig. 1). Direct mechanisms include the biotransformation of drugs or their metabolites into products with altered bioactivities. Indirect mechanisms involve more complex host-microbial interactions that modulate host pathways for xenobiotic metabolism or transport. We also discuss other classes of xenobiotics, including dietary compounds, food additives, and environmental toxins. Finally, we briefly highlight the immediate translational implications of this research and discuss early progress towards microbiome-based diagnostics and co-therapies.
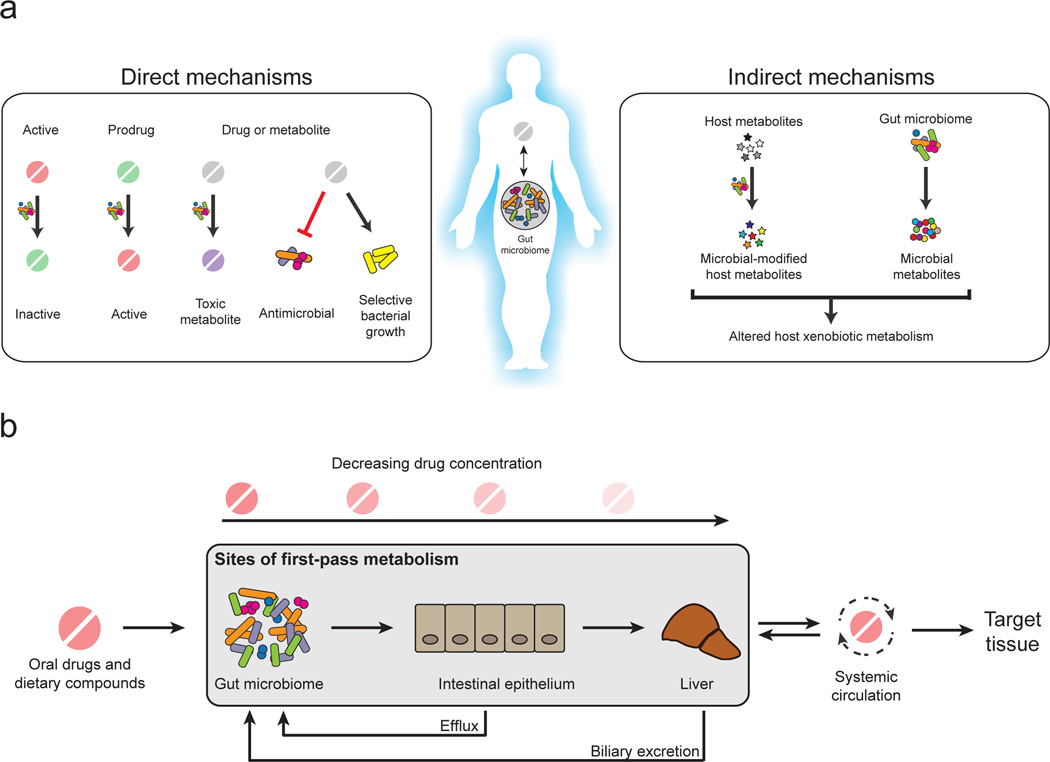
Figure 1. Mechanisms linking the gut microbiota and xenobiotic metabolism.
A. The gut microbiota can directly metabolize xenobiotics into active, inactive, or toxic metabolites. Xenobiotics may also shape the composition of the gut microbiota through antimicrobial activity or selective growth. The gut microbiota can indirectly influence xenobiotics through the modulation of host pathways for metabolism and transport. B. The gut microbiota can also influence xenobiotic metabolisms as a component of first-pass metabolism. Prior to entering systemic circulation and reaching the target tissue, orally ingested compounds are subject to metabolism in the intestine and liver, lowering the eventual systemic drug concentration. The gut microbiota may metabolize compounds prior to absorption, after efflux from the intestinal epithelium, or following biliary excretion from the liver.
The gut microbiota and pharmaceuticals
The gut microbiota can influence the metabolism of dozens of pharmaceuticals, in many cases changing their efficacy and/or side effect profiles. In this section, we highlight key examples of the direct and indirect mechanisms by which the gut microbiota influences drug therapy.
Microbial metabolism of drugs and their metabolites
The bioavailability of orally administered drugs depends on the extent of first-pass metabolism by intestinal and hepatic enzymes prior to reaching systemic circulation33. However, oral drugs may encounter the gut microbiota prior to reaching host tissues, representing another important site of first-pass metabolism (Fig. 1). In fact, there is already in vitro and/or in vivo evidence for the metabolism of 50 drugs by the gut microbiota28 (Supplementary information S1 (table)). This number is likely an underestimate given the lack of any systematic analyses of the gut microbial metabolism of drugs and the vast genetic diversity within the microbiome34. Furthermore, the rate of absorption likely has an important role in determining the extent of microbial metabolism due to the fact that the density of gut microorganisms increases dramatically in the distal small intestine (ileum) and colon. Drugs and their metabolites can also reencounter the gut microbiota via biliary excretion, at which point they can be further metabolized and reabsorbed via enterohepatic circulation.
Despite the diversity of chemical structures among the drugs that are known to be subject to gut microbial metabolism, two broad chemical transformation patterns have been repeatedly found: reduction and hydrolysis (Fig. 2; Supplementary information S1 (table)). These two reactions may reflect the energetic demands of the gut microbiota. The gut is largely anaerobic so gut microorganisms cannot rely on oxygen as a terminal electron acceptor for respiration35. Reductive xenobiotic metabolism may facilitate anaerobic respiration by expanding the range of alternative electron acceptors that are available for respiration. On the other hand, hydrolysis directly provides substrates for microbial growth. For example, many dietary components are glycosylated and their hydrolysis liberates sugars that could be shunted into glycolysis36.
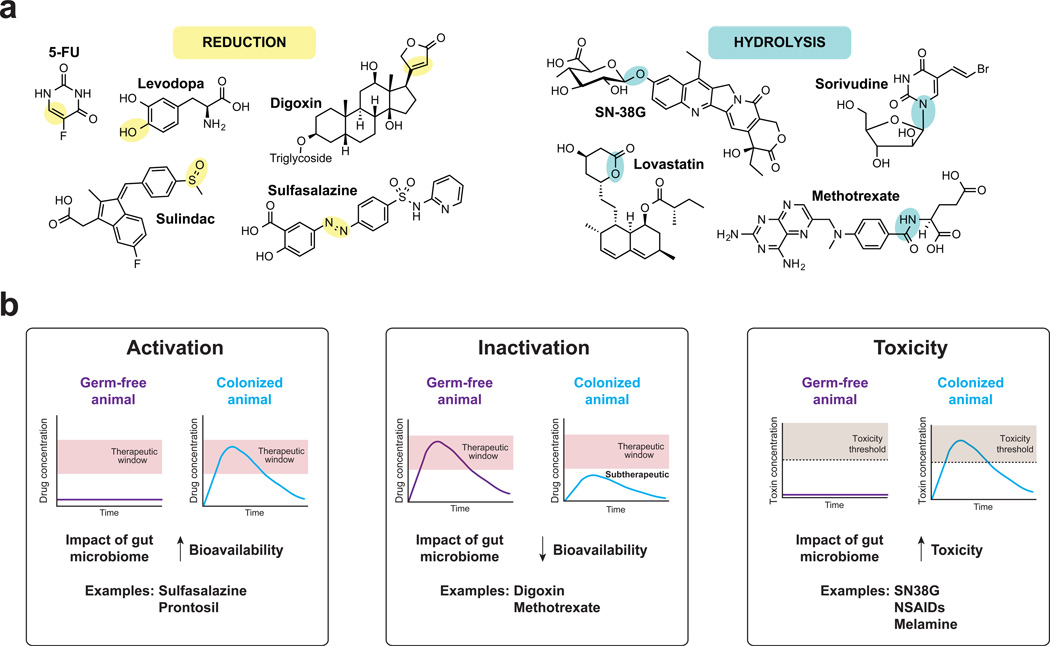
Figure 2. Major reaction types catalyzed by the gut microbiome and their pharmacological consequences.
A majority of known microbial biotransformations segregate into one of two reaction classes: reduction, in which compounds gain electrons from electron donors (part a), and hydrolysis, in which chemical bonds are cleaved through the addition of water (part b). The sites of modifications are highlight in yellow (reduction) and blue (hydrolysis). For a comprehensive list of drug biotransformations see Supplementary information S1 (Table). The microbial metabolism of pharmaceuticals can lead to their activation (part c), inactivation (part d) or result in the production of toxic compounds (part e); this is illustrated by the differential effects of the drugs in germ-free animals, compared to colonized animals. Activation refers to the conversion of a prodrug to its bioactive form, contributing to therapeutic concentrations. Examples include the prodrug sulfasalazine and prontosil. Inactivation refers to the conversion of an active metabolite to a downstream metabolite with reduced bioactivity. Examples include the cardiac drug digoxin and the anti-inflammatory drug methotrexate. Toxicity occurs due to the microbial production of metabolites that are toxic to the host. Examples include the hydrolysis of SN-38G to SN-38, the hydrolysis of glucuronidated NSAIDs to NSAIDs, and the metabolism of melamine to cyanuric acid.
The commonality of these two reaction types (reduction and hydrolysis) may also imply that there are core microbial species or gene families that impact a wide range of small molecules37. If so, the identification of the major players could serve as the basis for predicting the manner in which a novel drug will be modified by the gut microbiota. Such knowledge is likely to revolutionize drug development and precision medicine, similarly to the revolution that followed the discovery that cytochrome P450 enzymes (CYPs) are expressed in the intestine and liver, where they metabolize multiple xenobiotics38, 39. Chemical functional groups that are subject to microbial metabolism could be removed through rational design or exploited to control drug delivery.
The therapeutic effects of multiple prodrugs that contain azo bonds require bioactivation mediated by gut microorganisms. Following oral administration, the azo bond is reduced by microbial azoreductases, which liberate the biologically active compound. For instance, the above-mentioned antibacterial drug prontosil is cleaved by the microbiota, which results in the production of triaminobenzene and sulfanilamide21, a bacteriostatic antibiotic that inhibits folate metabolism. Based on these findings, azo bonds have been exploited in drug development. For example, sulfasalazine was strategically designed to treat rheumatoid arthritis, which at the time was believed to be the result of bacterial infections, by linking the sulfonamide sulfapyridine with the anti-inflammatory drug salicylic acid via an azo bond40, 41 (Fig. 2). Intact sulfasalazine can be recovered from the stool of antibiotic-treated or germ-free rats, but not in conventionally-raised animals42. Furthermore, a simplified gut microbiota composed of 4 bacterial strains (Bacteroides sp., Streptococcus faecalis, and 2 Lactobacillus sp.) is sufficient to restore sulfasalazine metabolism in gnotobiotic rats and the in vitro incubation of sulfasalazine with bacterial isolates from these animals results in drug cleavage42.
Azoreductases are widespread across multiple bacterial phyla found in the human gut28 and possess broad substrate compatibility43, 44; however, they metabolize azo compounds at different rates depending on the broader chemical structure of the molecule45. Gut microorganisms can also metabolize the downstream metabolites of azo reductions. For example, the bioactive component of sulfasalazine, 5-aminosalicylic acid, is inactivated by microbial arylamine N-acetyltransferases. The activity of these enzymes can vary up to 10-fold between individuals46, highlighting the considerable inter-individual differences in gut microbial metabolism that may contribute to variations in drug efficacy.
β-glucuronidases are another generalist enzyme family expressed by human-associated gut bacteria that influences the biological activity and toxicity of a wide range of drugs, dietary components, and endogenous metabolites47. Recent studies have uncovered the role of β-glucuronidase in the toxicity of drugs used to treat cancer and inflammation. In these examples, gut bacteria metabolize and interfere with drug metabolites generated by the host detoxification pathway of glucuronidation. UDP-glucuronosyltransferases expressed in the liver add glucuronic acid to multiple substrates, including drugs and endogenously produced compounds like hormones and bile acids48. This biotransformation typically interferes with the biological activity of the substrate and increases molecular weight and solubility, thereby enabling the elimination of these products in the urine (via renal excretion) or feces (via biliary excretion).
Biliary excretion provides another opportunity for drug metabolism by gut bacterial β-glucuronidases, which can re-activate the drug in the gut causing increased toxicity. An example of this phenomenon is irinotecan (CPT-11; Fig. 2), a widely used intravenous prodrug for the treatment of colorectal cancer (CRC). CPT-11 undergoes a complex metabolic transformation after administration: host carboxylesterases convert CPT-11 to the bioactive compound SN-3849; SN-38 is glucuronidated in the liver into the inactive metabolite SN-38G; SN-38G is then transported into the intestine via the biliary route; and microbial β-glucuronidases liberate the sugar moiety from SN-38G. The resulting SN-38 in the gut lumen exhibits toxicity towards intestinal epithelial cells and is thought to exacerbate the diarrhea found in up to 80% of CRC patients50, 51. These side effects can be ameliorated by reducing drug doses or halting drug administration, which in turn impedes effective treatment52. A similar mechanism contributes to the side effects of nonsteroidal anti-inflammatory drugs (NSAIDs), including diclofenac, indomethacin and ketoprofen53. Up to 70% of chronic NSAID users develop injury to the distal small intestine, indicated by damage to the mucosa, ulcerations and, in some cases, perforations. In the liver, NSAIDs are subject to host glucuronidation prior to biliary excretion into the gut lumen. Microbial β-glucuronidases can then liberate the glucuronide, allowing reabsorption of the aglycone into enterocytes. NSAIDs can be further metabolized into reactive metabolites by these gut epithelial cells, which causes mitochondrial and ER stress, compromising mucosal integrity and promoting inflammation51, 54.
These examples demonstrate how microbial metabolism can contribute to the side effects of treatment by interfering with host pathways for drug detoxification. β-glucuronidases are widely distributed across many gut bacterial species including members of the Proteobacteria, Firmicutes, and Actinobacteria phyla55–60. However, it remains unclear if these enzymes all exhibit a similarly broad substrate scope or if they are specialized for distinct niches (whether physical or occupational) within the gastrointestinal tract. More work is necessary to determine if the abundance and/or activity of these enzymes explain inter-individual variations in drug toxicity.
Microbial metabolism can also interfere with the bioavailability of drugs. A classic example of this phenomenon comes from the cardiac glycoside digoxin, used for cardiac arrhythmia (irregular heart beat) and heart failure26. Digoxin use is challenging due to its exceedingly narrow therapeutic range (0.5-2 ng/ml), making even minor changes to its concentration clinically relevant. In addition, approximately 10% of patients excrete high levels of an inactive metabolite of digoxin, dihydrodigoxin, which results from the bacterial reduction of the α,β-unsaturated lactone ring61, 62 (Fig. 2). In some cases more than 50% of the administered drug is inactivated63, substantially decreasing systemic drug concentrations. Seminal studies conducted in the 1980s suggested that only a single bacterial species, Eggerthella lenta, reduces digoxin64, but unfortunately neither the presence nor the abundance of this species predicts digoxin reductions64–66. This discrepancy appears to be driven by strain-level variations in the E. lenta population67. In E. lenta DSM2243, digoxin induces the expression of a 2-gene operon, referred to as the cardiac glycoside reductase (cgr) operon. The proteins encoded in this operon, Cgr1 and Cgr2, are homologous to enzymes involved in electron transport. Cgr1 shows similarity to cytochrome c reductases, which are membrane-bound proteins involved in shuttling electrons from quinones to an electron reductase partner. Cgr2, which shows similarity to FAD-binding fumarate reductases, is predicted to interact and accept electrons from Cgr1 and in turn reduce the lactone ring of digoxin37. The cgr operon is not found in the genomes of strains of E. lenta that lack the ability to reduce digoxin, suggesting that the cgr operon is necessary for the reduction of digoxin and providing an explanation for the difficulty in predicting digoxin reduction based only on the presence of E. lenta species. Furthermore, digoxin reduction is enhanced in the presence of a complex gut microbiome and suppressed by dietary protein67, suggesting that microbial drug metabolism is sensitive to both microbial and environmental interactions.
Microbial control of xenobiotic metabolism and absorption
Compounds that resist microbial metabolism can still be influenced by the gut microbiota through multiple mechanisms (Box 1). Comparisons of germ-free and colonized mice have revealed that the microbiota impacts the expression of multiple host genes involved in drug metabolism and transport68, 69. This influence on host gene expression by the gut microbiota can be local68, 70 (e.g., in the ileum) or distant, including impacting the most vital drug-metabolizing organ, the liver69, 71. In the liver, more than 100 genes are differentially expressed between germ-free and colonized mice69. One of the largest groups of differentially expressed genes is the CYPs39. An example that illustrates the importance of the microbiota on xenobiotic metabolism in the liver mediated by CYPs is the anesthetic pentobarbital. Pentobarbital is administered intravenously and is metabolized by CYPs in the liver. Germ-free animals, which show elevated expression of CYPs compared to colonized animals, are more efficient at metabolizing pentobarbital compared to colonized animals69.
Box 1. Microbial modulation of the immune system and drug therapy.
The microbiota has a critical role in the development and maintenance of the immune system150. However, it has only recently become clear that the microbiota helps to mediate the effects of drugs targeting the immune system, and that changes to the structure or function of the microbiota represent an unanticipated side effect of treatment.
Multiple studies have implicated the gut microbiome in the efficacy of drugs used for cancer. Treatment of mice with cyclophosphamide increased intestinal permeability, promoting the translocation of Gram-positive bacteria into secondary lymphoid organs151. This translocation is thought to contribute to the concomitant production of pathogenic T helper 17 (TH17) cells and memory TH1 immune responses required to limit tumor growth151, 152. Consistent with this model, the efficacy of cyclophosphamide was reduced in germ-free mice and in animals treated with broad-spectrum antibiotics, whereas the adoptive transfer of pathogenic TH17 cells restored cyclophosphamide efficacy. A similar dependence on the gut microbiota was found for CpG-oligonucleotide immunotherapy153. The response to CpG-oligonucleotides in germ-free mice and in animals treated with broad-spectrum antibiotics was poor, as evidenced by decreased cytokine production and tumor necrosis. Oxaliplatin, a platinum-based drug that induces apoptosis through the production of reactive oxygen species in the tumor154, was also dependent on the microbiota153. More recently, members of the Bifidobacterium genus were shown to enhance the immune response to tumors in a manner that increased the efficacy of α-PD-L1, an antibody that blocks immune inhibitory pathways155. Also, CTLA-4 blockade immunotherapy was shown to depend on particular Bacteroides species (B. thetaiotaomicon and B. fragilis)156. Together, these results indicate that the immune response to specific members of the gut microbiota may help set the stage for cancer treatment.
Conversely, recent studies have suggested that anti-inflammatory drugs used to treat inflammatory bowel disease may impact the gut microbiome. The ulcerative colitis-like phenotype of TRUC (T-bet−/− Rag2−/−) mice is dependent on the cytokine tumor necrosis factor (TNF)157. Treatment with blocking antibodies against TNF suppresses colitis in patients158. This therapy is also effective in the TRUC model and is accompanied by a significant increase in the abundance of Staphyloccocus159. However, more work is necessary to determine the functional consequences of the changes to the gut microbiota in response to anti-TNF and other related therapies.
A recent RNA-sequencing based study confirmed the differential expression of multiple genes involved in xenobiotic metabolism in the liver of germ-free and colonized mice72. Furthermore, this study reported a significant increase in the expression of the xenobiotic-sensing transcription factors aryl hydrocarbon receptor (AhR), constitutive androstane receptor (CAR), peroxisome proliferator-activated receptor α (PPARα), and nuclear factor erythroid 2-related factor (Nrf2) in germ-free mice. However, additional work is necessary to elucidate the mechanisms responsible for these differences in gene expression.
The microbiota also dramatically changes the serum metabolome. Comparisons of germ-free and colonized mice revealed that the gut microbiota not only alters the abundance of endogenous metabolites, with 10% of shared metabolites differing in abundance by at least 50%, but also contributes uniquely microbial compounds to systemic circulation73, 74. Some of these microbial metabolites are processed by the host in a manner analogous to xenobiotics (i.e., conjugation)74. This overlap between host response to drugs and microbial metabolites may have implications during drug therapy; for example, it may result in increased toxicity or half-life of xenobiotics due to competition between the drug and microbial metabolites for the same host enzymes that are involved in drug detoxification or elimination. An example of this type of interaction comes from acetaminophen (paracetamol), one of the most widely used drugs worldwide. Acetaminophen overdose can lead to severe and sometimes fatal hepatotoxicity75, and both drug metabolism and toxicity varies between individuals76, 77. Acetaminophen is metabolized in the liver and the predominant metabolites that result from its metabolism, acetaminophen-sulfate and acetaminophen-glucuronide, are inactive. However, another minor metabolite, N-acetyl-p-benzoquinone imine (NAPQI), causes toxicity in the liver. Based on these findings, a metabolomic study aimed to determine whether pre-dose urinary metabolite profiles could predict acetaminophen metabolism in humans78. Pre-dose levels of a microbe-related metabolite, p-cresol sulfate, were found to be inversely associated with the ratio of acetaminophen-sulfate to acetaminophen-glucuronide. The microbial metabolite p-cresol is an end-product of tyrosine and phenylalanine metabolism and has been demonstrated to be produced by a number of microorganisms, including those belonging to the Firmicutes (Clostridium difficile), Bacteroidetes, Actinobacteria and Fusobacterium phyla79, 80. Following absorption and circulation, p-cresol is metabolized in the liver to p-cresol sulfate. p-cresol and acetaminophen are both substrates of the human cytosolic sulfotransferase SULT1A181 (Fig. 3). This competition likely impedes the host’s ability to detoxify acetaminophen, increasing the likelihood of accumulating the toxic metabolite NAPQI.
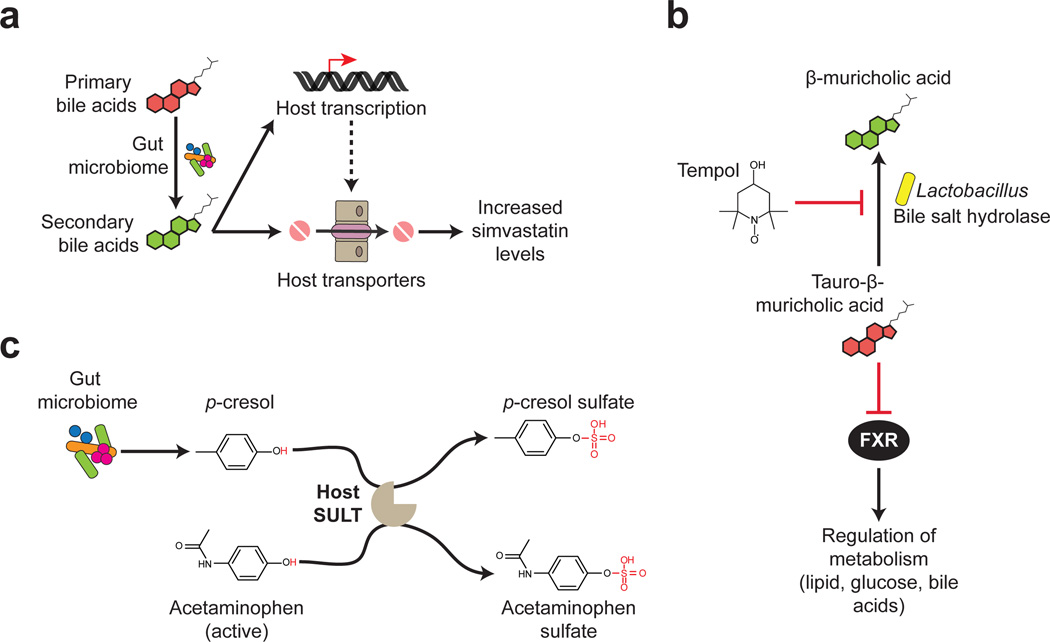
Figure 3. Host-microbiota interactions shape therapeutic outcomes.
A. Simvastatin drug levels in the host positively correlate with levels of secondary bile acids. The metabolism of bile acids by gut bacteria possibly contributes to the absorption of simvastatin through modulating the expression of host transporters or through directly competing with the transporter. B. The protective effects of tempol on diet-induced obesity are mediated through the gut microbiota. Tempol treatment reduces the abundance of Lactobacillus spp., which is involved in deconjugating taurine-conjugated bile acids into free bile acids via bile salt hydrolases (BSH). This results in elevated levels of taurine-conjugated bile acids, such as tauro-β-muricholic acid, a known antagonist of the metabolic regulator farnesoid X receptor (FXR). C. Microbial metabolites compete with drugs for host xenobiotic metabolism enzymes. The microbial product p-cresol, a product of tyrosine metabolism, and acetaminophen both serve as substrates for the same enzyme, the host sulfotransferase SULT. Therefore, elevated levels of p-cresol inhibit the conversion of acetaminophen-glucuronide (the active form) to acetaminophen-sulfate (the inactive form) by SULT.
Host-microbial interactions may also influence drug efficacy. Statins, which are cholesterol lowering drugs prescribed for coronary artery disease, display substantial inter-individual variations in efficacy, with up to 33% of patients failing to reach lipid-lowering targets82. In humans, the response to simvastatin treatment (indicated by LDL cholesterol levels) is positively associated with the pretreatment levels of three microbiota-produced secondary bile acids: lithocholic acid, taurolithocholic acid and glycolithocholic acid. Although the mechanism(s) responsible for this association remain unknown, one intriguing hypothesis is that primary bile acids may compete for the same intestinal transporters that enable the absorption of statins83 (Fig. 3). Thus, microbial bile acid metabolism may decrease this competition, priming the host for more effective statin therapy.
A similar type of interaction may influence the efficacy of tempol, an antioxidant that protects against diet-induced obesity in animal models84. Treatment of mice with tempol altered the relative abundance of the two dominant bacterial phyla in the distal gut, increasing the abundance of Bacteroidetes and decreasing the abundance of Firmicutes85. Within the Firmicutes phylum, the genus Lactobacillus was significantly reduced (Fig. 3). Multiple members of the Lactobacillus genus encode bile salt hydrolases, which produce free bile acids by deconjugating taurine-conjugated bile acids86. Consistent with these findings, tempol increases the intestinal concentration of multiple taurine-conjugated bile acids, including tauro-β-muricholic acid (T-β-MCA). T-β-MCA is an antagonist of the farnesoid X receptor (FXR), a master regulator of lipid, glucose, and bile acid metabolism85, 87, 88. Tempol does not further reduce adiposity in intestinal specific FXR-null mice85, consistent with the hypothesis that changes in microbial bile acid metabolism and subsequent signaling via the FXR pathway contribute to the mechanism of action of tempol. It is yet to be determined whether tempol has direct antimicrobial effects against members of the gut microbiota or whether the observed changes in microbial community structure are mediated through drug interactions with the host.
The gut microbiota and other xenobiotics
In addition to influencing drugs, the gut microbiota can metabolize numerous xenobiotic compounds found in our diet, including natural products and chemical additives. In some cases, these compounds have beneficial health effects that depend on microbial bioactivation. In other instances, the gut microbiota can produce toxic metabolites. In this section we highlight key examples of how gut microbial metabolism impacts the health effects of the foods that we consume.
Diet-derived bioactive compounds
Our diet is rich in small molecules that have important consequences for human health and disease. Many of these “diet-derived bioactive compounds” are metabolized by the gut microbiota (Fig. 4, Supplementary information S2 (table) and Supplementary information S3 (table)) and some are dependent on this transformation for activation and/or absorption47. Here, we highlight examples with considerable recent evidence elucidating their interaction with and dependence on the gut microbiota.
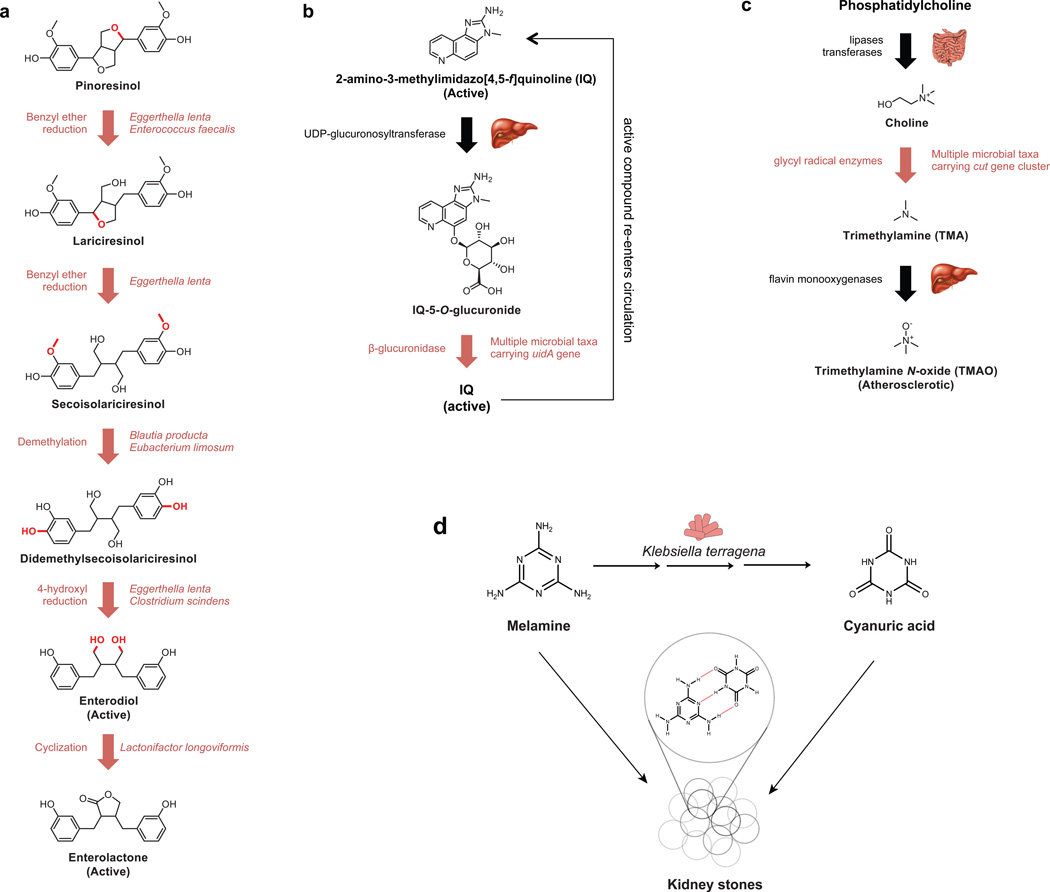
Figure 4. Microbial metabolism of dietary compounds.
A. The plant-derived dietary lignans pinoresinol and secoisolariciresinol are metabolized by several bacteria into the cancer-protective compounds enterodiol and enterolactone. B. The microbiota is responsible for the reactivation of the heterocyclic amine 2-amino-3-methylimidazo[4,5-f] quinoline after hepatic inactivation, which leads to delayed excretion of the carcinogenic compound. C. The microbial production of trimethylamine from choline-containing compounds represents a critical link between dietary phosphatidylcholine and the atherosclerotic metabolite trimethylamine N-oxide (TMAO). D. The metabolism of melamine by the gut microbiome leads to kidney stones. Klebsiella terragena converts melamine to cyanuric acid, which complexes with melamine into insoluble aggregates in the kidney.
Clinical and epidemiological studies suggest that dietary polyphenols such as anthocyanins (ACNs) and proanthocyanidins (PACs) protect against metabolic syndrome89, 90. Supplementation of high-fat diets with ACNs or PACs has been shown to suppress the expression of genes involved in fatty acid and triacylglycerol synthesis, the regulation of lipogenesis and cholesterol biosynthesis, and the assembly of very low density lipoproteins90–92. ACNs and PACs have also been argued to stem the development of insulin resistance by increasing insulin signaling, glycogen accumulation and adiponectin secretion in the presence of free fatty acids93. Intriguingly, rodent studies indicate that just 6–12% of radiolabeled polyphenols are metabolized and enter circulation during their passage through the gut94, 95, leaving open the question of how these compounds confer their protective effects. Recent studies of polyphenol extracts isolated from grapes20 and cranberries96 provide support for a mechanism that acts via the gut microbiota. Mice fed high-fat diets supplemented with polyphenols showed reduced diet-induced weight gain and adiposity, improved insulin sensitivity, and diminished markers of intestinal inflammation and oxidative stress compared to controls20, 96. These improvements were coupled to dramatic 7-to-10-fold blooms of Akkermansia muciniphila, a mucin-degrading bacterium argued to have an important role in the preservation of the integrity of the gut mucus layer, thus limiting the risk of systemic inflammation97. A. muciniphila abundance has been linked to reduced weight gain, adiposity, insulin resistance, and/or inflammatory markers in many contexts, including during pregnancy98, following gastric bypass surgery19, 99, in prebiotic or metformin treatment experiments100, 101, and in other polyphenol feeding experiments involving green or black tea102, 103 or a grape juice and red wine mixture103. Furthermore, the administration of live (but not heat-killed) A. muciniphila was sufficient to reduce host adiposity, inflammatory markers, and insulin resistance in diet-induced obese mice97, 101. Further work is needed to establish how polyphenols promote the expansion of A. muciniphila and whether this effect is direct or mediated through changes in host physiology. However, a recent in vitro study reported that exposure of a complex human fecal microbial community to black tea or grape-derived polyphenols can directly increase the abundance of A. muciniphila, suggesting a limited dependence on host factors in this process103.
Fruit-derived ellagitannins are believed to provide protective properties for the plant by preventing microbial decay104. Hydrolysis of ellagitannins releases ellagic acid, which can be metabolized by the gut microbiota into a number of structurally related urolithins that can reach high concentrations locally in the colon and systemically105. A number of in vitro studies have shown that urolithins have antioxidant, anticancer, anti-inflammatory and antimicrobial properties; however, there are currently a limited number of in vivo and mechanistic studies on urolithins. . A recent survey found that ellagic acid metabolism varied significantly among individuals but could be generally grouped into three categories depending on the metabolites generated, including a subset of individuals that did not produce any urolithins106, 107. This implies that the composition of an individual’s gut microbiota is a key determinant in whether beneficial products within a diet can be extracted or activated. Gut bacterial isolates capable of metabolizing ellagitannins have been identified, including members of the Gordonibacter genus (phylum: Actinobacteria)108, 109.
Two phytoestrogen classes, isoflavones and lignans, represent plant-derived chemicals that are metabolized by a diverse array of gut bacteria (such as Actinobacteria, Bacteroidetes and Firmicutes) to molecules that bind estrogen receptors and may evoke breast cancer-protective effects110–112. One such isoflavone, daidzin, is a glycosidic isoflavone found predominantly in soy products and is metabolized to equol by several species of gut-residing bacteria (e.g., Enterococcus faecium, Lactobacillus mucosae, Bifidobacterium sp., Coriobacteriaceae sp., Eggerthella sp.) via glycosidic cleavage and reduction of an α,β-unsaturated ketone113. Facile absorption introduces equol into systemic circulation where it demonstrates a high affinity for estrogen receptor β (ERβ). The biological effect of equol’s ERβ affinity may be particularly apparent in Asian populations, who traditionally consume diets rich in phytoestrogens, with consumption of approximately 10 mg of isoflavones per day being associated with lower incidence of breast cancer (risk reduced by 12%)114. This may be attributed to both a higher concentration of isoflavones (equol precursors) in the gut and the presence of gut bacteria that are able to generate equol115. By contrast, women in Western populations, who typically consume a much lower amount of isoflavones (approximately 0.3 mg per day), demonstrated no association between isoflavone consumption and breast cancer risk. While there are results that both support116 and discount117 that individuals producing equol may have a decreased breast cancer risk, further consideration and characterization of the equol-producing bacteria present in the microbiota will help to deconvolute these results.
The breast cancer-protective effects associated with consumption of plant lignans (found in flaxseed, sesame seeds, legumes, grains, berries, cruciferous vegetables and tea) are similarly dependent on metabolism by gut bacteria118, 119. In a multi-step pathway involving several gut bacteria (including Enterococcus faecalis, Eggerthella lenta, Blautia producta, Eubacterium limosum, Clostridium scindens, and Lactonifactor longoviformis), lignans such as pinoresinol and secoisolariciresinol are metabolized to the bioactive “mammalian lignans” enterodiol (END) and enterolactone (ENL)120 (Fig. 4). The protective effects of END and ENL in a chemically induced breast cancer model were assessed when germ-free rats or germ-free rats that had been colonized with a bacterial consortium that was demonstrated to convert secoisolariciresinol to END and ENL (composed of Clostridium saccharogumia, E. lenta, B. producta and L. longoviformis) were fed a flaxseed-rich diet121. For the colonized group, the number of breast tumors was 2.5 times lower and the tumor size and weight were ~2 times lower than was observed for the germ-free group. These findings highlight the importance of gut bacteria for actualizing the breast cancer-protective effects of lignans.
Conversely, microbial biotransformation may exacerbate the effect of harmful diet-derived compounds. For instance, microbial β-glucuronidase activity may contribute to associations between the risk of CRC and the intake of heterocyclic amines, compounds formed during the charring of meat. Multiple carcinogenic heterocyclic amines are detoxified through hepatic glucuronidation, including prevalent diet-derived compounds like 2-amino-3-methylimidazo[4,5-f]quinolone (IQ), 2-amino-1-methyl-6-phenylimidazo[4,5-b] pyridine (PhIP) and 2-amino-3,8-dimethylimidazo[4,5-f] quinoxaline (MeIQx)122 (Fig. 4). The glucuronidated compounds are excreted into the intestinal lumen via bile, at which point microbial glucuronidases could theoretically release the conjugate group, reactivating the toxic compound and thereby augmenting its genotoxicity. To this end, studies of 2-amino-3-methylimidazo[4,5-f]quinolone (IQ) have repeatedly observed more DNA damage and DNA adducts in conventional versus germ-free mice123, 124. Importantly, 2-amino-3-methylimidazo[4,5-f]quinolone (IQ) genotoxicity was assessed directly in gnotobiotic rats monoassociated with isogenic Escherichia coli strains either carrying or deficient in the gene uidA, which encodes β-glucuronidase125. Microbial β-glucuronidase increased the colonic genotoxicity of 2-amino-3-methylimidazo[4,5-f]quinolone (IQ) threefold and led to multiple peaks in urinary and fecal excretion of the compound, consistent with enterohepatic circulation.
In addition, analysis of a large clinical cohort recently found an association between risk of cardiovascular disease and microbial metabolites of choline-containing compounds, which are liberated in the intestine through the lipase-mediated breakdown of dietary phosphatidylcholine. In the colon, choline-containing compounds undergo metabolism by microbial glycyl radical enzymes126 to form the intermediate gas trimethylamine (TMA; Fig. 4). In turn, TMA is absorbed and oxidized by hepatic flavin monooxygenases, forming TMA N-oxide (TMAO), a metabolite linked to accumulation of cholesterol in macrophages and foam cell deposition127, and higher risk of a major adverse cardiac event128. A similar mechanism appears responsible for the link between atherosclerosis and dietary L-carnitine, a compound abundant in red meat129. The microbial choline utilization (cut) gene clusters responsible for the production of TMA have been detected in 20 members of the human gut microbiome, including representatives of the major Firmicutes, Proteobacteria, and Actinobacteria (but not Bacteroidetes) phyla126. Because choline utilization capability is unevenly distributed across the gut microbiome, inter-individual differences in gut microbial community composition could potentially serve as biomarkers for the strength of linkage between diet and cardiovascular outcomes.
Artificial sweeteners and emulsifiers
Many processed foods contain chemical additives that are meant to enhance flavor or maximize shelf life without any consequence for the consumer. However, a number of recent studies have begun to suggest that these dietary additives may have deleterious interactions with the gut microbiota.
Non-caloric artificial sweeteners (NAS) are widely used food additives designed to be resistant to host metabolism and provide a sweet flavor without caloric consequences. Yet, in some cases, the gut microbiota is still capable of modifying these compounds, converting them to bioactive metabolites. Cyclamate is a classic example. Many intestinal microorganisms, including those belonging to the genus Enterococcus, Clostridium, Corynebacterium, Campylobacter and Escherichia, have demonstrated the ability to convert cyclamate to cyclohexylamine130, which displays toxicity in animals. A fraction of animals dosed with high levels of cyclamate developed bladder tumors and, as a result, this compound has been banned from being included in any food and drugs in the US and UK since 1970131. Recent studies suggest that multiple other NAS alter the gut microbiota, including xylitol132 and saccharin133. Chronic NAS consumption in mice was shown to impact gut microbial community structure133, resulting in an increase in abundance of bacteria belonging to the Bacteroides genus and some members of the Clostridiales order. These differences appear to have a functional consequence, as germ-free mice colonized with gut microorganisms from NAS-treated mice or stool microorganisms exposed to NAS ex vivo develop glucose intolerance. Preliminary results in humans suggest that saccharin may only impact a subset of individuals133, potentially explaining why large-scale epidemiological analyses have failed to link NAS consumption to diabetes134. Further work is required to determine the mechanisms through which NAS shapes the structure and function of the gut microbiota, and whether these changes have implications for host glucose homeostasis.
The gut microbiota may also be affected by emulsifying agents. These additives are used in processed foods like ice cream to allow them to be stored for long periods of time without particles falling out of suspension. However, these compounds have detergent-like properties and may have an impact on the composition of the gut microbiota and on the integrity of host tissue. Controlled feeding of two emulsifying agents, carboxymethylcellulose and polysorbate-80 (Tween 80), to mice resulted in a reduction of the thickness of intestinal mucus and, as a result, microbial cells showed increased encroachment towards epithelial cells135. The composition of the gut microbiome was also impacted with decreased abundances of the Bacteroidales (phylum: Bacteroidetes) and increased levels of mucolytic bacteria, such as Ruminococcus gnavus135. Although the overall mucus layer is decreased in thickness in treated animals, the increase in abundance of mucolytic bacteria may reflect increased mucin accessibility by bacterial penetration. These shifts in microbial community structure were accompanied by low-grade inflammation, increased gut permeability, increased weight and adiposity, and development of metabolic syndrome. Interestingly, emulsifiers failed to have this effect on germ-free mice. However, transfer of the microbiota from emulsifier-treated animals to germ-free recipients was sufficient to induce the same symptoms (low-grade inflammation, increased gut permeability, increased weight and adiposity, and development of metabolic syndrome), even in the absence of further emulsifier feeding. This suggests that the composition of the gut microbiota is a key driver of metabolic syndrome and low-grade inflammation. Furthermore, this inflammation may be exacerbated among individuals that are predisposed to intestinal conditions, such as colitis, as emulsifier feeding of genetically sensitized animals that are prone to inflammation (Il10−/− and Tlr5−/− mice) promoted a colitis phenotype.
Toxicity caused by dietary contaminants
The industrial compound melamine and its microbial metabolite, cyanuric acid, form an insoluble complex that interferes with kidney function, leading to severe renal toxicity136. In 2008, melamine tainted milk in China caused over 50,000 infant hospitalizations and six deaths due to renal failure137, provoking scientific inquiry into the mechanisms responsible. Administration of melamine in combination with broad-spectrum antibiotics resulted in decreased kidney damage in animal models138, potentially due to the decreased microbial conversion of melamine into cyanuric acid139, 140. Colonization of melamine-fed animals with Klebsiella terrigena, which produces cyanuric acid, led to increased kidney damage138 (Fig. 4). Interestingly, K. terrigena is sparsely distributed in the gut of healthy individuals, being present in approximately 1% of the population141. Therefore, additional work is necessary to determine whether the gut microbiota is a major contributor to inter-individual differences in the toxicity of melamine and other dietary contaminants.
Moving towards microbiome-based medicine
The emerging appreciation that the gut microbiota influences pharmacology and nutrition has begun to reveal the immediate translational potential of this research28, 30, 142. Continued progress in this area could lead to approaches to improve drug outcomes by altering the gut microbiota and to predict drug outcomes by metabolite or genetic screening of the gut microbiota. In this section, we highlight recent studies that provide a proof-of-principle demonstration for each of these goals (Fig. 5). Furthermore, these studies may also provide additional information about the microbiome that can be used to harvest new drugs (Box 2).
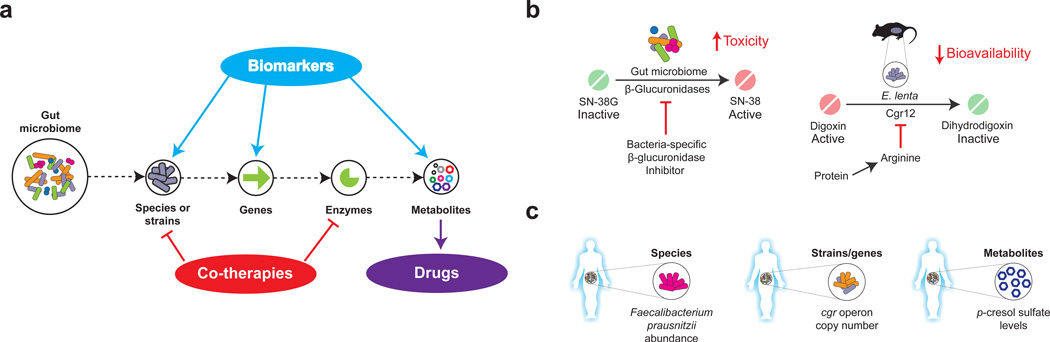
Figure 5. Translational implications of microbiome research in pharmacology.
A. Metagenomic and metabolomic approaches enable the dissection of microbial communities at multiple scales from complex communities to individual metabolites. This information can be used to find biomarkers, to develop co-therapies that target the microbiota or to identify novel drugs. B. Inhibiting microbial enzymes in the gut. Such examples include using small molecules to inhibit bacterial β-glucuronidase activity (left panel) and the dietary inhibition of cardiac drug inactivation by Eggerthella lenta (right panel). C. Microbiome-based diagnostics. Examples include measuring: the abundance of bacterial species that are associated with tacrolimus efficacy (left panel); the presence or absence of genes that are associated with the bioavailability of digoxin (middle panel); and the levels of the microbial metabolite p-cresol, which is associated with acetaminophen metabolism (right panel).
Box 2. Mining the microbiome for new drugs.
In addition to influencing drug outcomes, the gut microbiome may provide a rich source of novel therapeutics. A recent analysis of 2,430 reference genomes from human-associated microorganisms identified more than 14,000 biosynthetic gene clusters predicted to synthesize diverse small molecules from saccharides, nonribosomally-encoded peptides, polyketides, and ribosomally encoded and post-translationally modified peptides160. The gut and oral cavity represented the richest sources of gene clusters, with considerable variation in the number of gene clusters between individuals. Of note, gene clusters encoding antibacterial thiopeptides were found in every body site. A new class of thiopeptides, named lactocillin, was isolated from the vaginal isolate Lactobacillus gasseri JV-V03. Interestingly, lactocillin showed broad activity against Gram-positive pathogens, consistent with the activity observed for other thiopeptides, but lactocillin displayed no activity towards other vaginal Lactobacillus isolates160.
Bile acid metabolizing bacteria, or their metabolites, could represent another source of new drugs. Broad-spectrum antibiotics used in clinical practice can provide an opportunity for infection by enteric pathogens161. In patients undergoing bone marrow transplantation and in mice exposed to a panel of antibiotics, the abundance of Clostridium scindens was inversely associated with Clostridium difficile infection162. C. scindens was sufficient to protect mice from infection following antibiotic treatment due its unique ability to generate the secondary bile acids deoxycholate and lithocholate, which inhibit the growth of C. difficile162–164. Thus, bacteria that are able to metabolize bile acids, or the metabolites that result from these reactions, could represent a novel treatment regimen for C. difficile infection.
Targeting the microbiome for therapeutic benefit
Despite the numerous undesirable biotransformations catalyzed by the gut microbiota, our ability to manipulate gut microbial metabolism in a targeted fashion in order to prevent these biotransformations remains in its infancy. One approach would be to develop small molecule inhibitors that target the microbial enzymes responsible for undesirable xenobiotic transformations (Fig. 5). However, the complexity of the gut microbiota and its many redundant enzymes raises questions on whether these targets will truly be druggable. The answer appears to be yes for the bacterial β-glucuronidases, for which multiple inhibitors now exist that have minimal impact on the mammalian homolog143–146. This specificity towards the bacterial enzymes was revealed with the assistance of crystallography and bioinformatics, which demonstrated that these inhibitors interacted with a “bacterial loop” that is highly conserved and well distributed across the gut bacteria enzymes, but is absent from the mammalian enzyme146. However, not all bacterial glucuronidases contain this loop, including those from members of the Bacteroidetes phylum60.
Notably, β-glucuronidase inhibitors are capable of rescuing mice from drug toxicity. Mice receiving irinotecan along with a β-glucuronidase inhibitor showed significantly lower incidence of diarrhea and less damage to the gastrointestinal epithelium than mice receiving irinotecan alone146 (Fig. 5). Similarly, in animals exposed to the NSAIDs diclofenac, indomethacin, and ketoprofen, co-administration of the β-glucuronidase inhibitor reduced mucosal injury and enteropathy compared to control mice not receiving the inhibitor54, 147. These inhibitors may also be useful for minimizing the toxicity induced by other bacterial deglucuronidation events, such as the colonic reactivation of the heterocyclic amine 2-amino-3-methylimidazo[4,5-f] quinolone (IQ)125.
Dietary intervention represents an alternative strategy to control the microbial biotransformation of drugs (Fig. 5), as diet has been shown to rapidly and reproducibly alter the gut microbiota in humans and animal models8, 148. Research into the cardiac drug digoxin has provided an initial proof-of-principle for this approach. The amino acid arginine prevents digoxin inactivation by E. lenta in vitro, decreasing the expression and activity of the genes responsible (the cgr operon)67. In these experiments, germ-free mice were colonized with E. lenta and fed an identical diet only differing in the amount of total protein. Following the administration of digoxin, mice fed a high-protein diet showed significantly elevated serum and urinary digoxin levels compared to controls. Furthermore, dietary protein did not have an effect on mice colonized with a strain of E. lenta that lacks the cgr operon67. Therefore, these results highlight the potential to revise the nutritional guidelines for drugs based on their interaction with the gut microbiota.
Developing microbiome-based diagnostics
Another emerging area of interest is the development of diagnostic biomarkers that predict the optimal drug or dosage based on the gut microbiome (Fig. 5). While this could theoretically be used for any microbial metabolite, species, or gene family linked to the drug responsible (or even in an unbiased fashion), we highlight three examples of more targeted tests: the pain-reliever acetaminophen, the cardiac drug digoxin, and the immunosuppressant tacrolimus.
As discussed above, the pre-dose levels of p-cresol sulfate were found to be inversely associated with the ratio of acetaminophen sulfate to acetaminophen glucuronide. Therefore, it has been suggested that the concentration of p-cresol sulfate could serve as a predictive biomarker for drug detoxification, helping to minimize liver damage78 (Fig. 5).
Likewise, the variation in metabolic activity between distinct strains of E. lenta suggests a potential microbiome-based genetic test for drug bioavailability. Members of this species vary in whether or not they carry the genes responsible for digoxin reduction, the cgr operon67. Using quantitative PCR, human fecal samples were evaluated for their cgr ratio (the proportion of cgr abundance normalized to the abundance of the E. lenta species). Notably, the cgr ratio could be used to effectively discriminate microbial communities exhibiting low versus high digoxin reduction (Fig. 5). Such diagnostics might allow physicians to discriminate a priori which patients are likely to respond favorably to digoxin therapy.
The immunosuppressant drug tacrolimus has a very narrow therapeutic range and a fraction of patients receiving this therapy require an increase in dosing. A study examining kidney transplant patients found a positive correlation between patients that required an increase in tacrolimus dosing and the abundance of the gut bacterium Faecalibacterium prausnitzii149 (Fig. 5). Although the reason for the observed correlation between F. prausnitzii and tacrolimus dosing is unknown, the abundance of this bacterium may still serve as a useful biomarker for increased dosing requirements.
Finally, an alternative approach may be to identify surrogate biomarkers (e.g., proteins, metabolites, or nucleic acids) in the blood or urine that predict the abundance of clinically relevant microorganisms, which would enable the rapid and routine stratification of patients according to their predicted therapeutic outcomes.
Outlook
The studies discussed throughout this Review emphasize that the human gut microbiota has an important role in xenobiotic metabolism, influencing the efficacy and toxicity of drugs, dietary compounds, and environmental toxins. Gut microorganisms have evolved numerous enzymes that allow them to directly metabolize xenobiotics and their metabolites, as well as ill-defined mechanisms for controlling host xenobiotic metabolism and transport.
Given the recent resurgence in microbiome research, it is now an opportune time to consider a more comprehensive view of pharmacology that includes the membership, structure and function of our resident microbial communities and a deeper understanding of their interactions with each other, their host habitat and the nutritional milieu of the gastrointestinal tract. Continued progress will require concerted efforts to expand the scope of metagenomic and metabolomic surveys, while also developing complementary experimental and computational approaches to model gut microbial metabolism along the entire length of the gastrointestinal tract. This work will provide fundamental insights into poorly studied, yet clinically relevant, microbial taxa and enable the more complete annotation of the genetic “dark matter” of the microbiome. Studies of xenobiotic metabolism, and microbial metabolism in general, will be essential for the microbiome field to move beyond simply describing “who’s there” to interpreting “what they are doing”. The translational implications of this work are already becoming apparent, whether through the discovery of gut microbial signatures that predict drug outcomes, co-therapies that precisely target members of the gut microbiome, or new drugs harvested from the microbiome (Box 2). Together, these results emphasize that the microbiome will be a key component of a 21st century pharmacopoeia, as it provides a modifier, target and source for the drugs of the future.
Supplementary Material
Acknowledgments
The authors apologize to all of those colleagues whose work could not be included in this Review owing to space constraints. The authors also thank the reviewers for their comments and suggestions. This work was supported by the US National Institutes of Health (R01AT008618, R01HL122593 and F32DK101154), the Young Investigator Grant for Probiotics Research, the George Williams Hooper Research Foundation and the University of California San Francisco (UCSF) Department of Microbiology & Immunology. P.J.T. is a Nadia's Gift Foundation Innovator supported, in part, by the Damon Runyon Cancer Research Foundation (DRR-42-16).
Glossary
- Aglycone
The remaining compound following removal of a glycosyl moiety.
- Azo bond
A chemical bond composed of N=N.
- Bile Acids
Steroid acids produced in the liver that emulsifies fats during digestion.
- Biliary excretion
The transfer of xenobiotics and other compounds from the plasma to bile via hepatocytes, followed by release into the gut lumen.
- Bioavailability
The proportion of an administered compound that reaches systemic circulation and therefore has the potential to influence the intended target.
- Conjugation
The addition of a chemical unit (e.g., glucuronic acid, glutathione) to xenobiotics, increasing the solubility and molecular weight of the parent compound and facilitating elimination from the body.
- Cytochrome P450
A family of enzymes responsible for the oxidative biotransformation of xenobiotics and other compounds.
- Enterohepatic circulation
The circulation of xenobiotics and endogenous compounds that are absorbed from the intestines, transported to the liver, and then renter the intestine via the bile ducts, where they may be reabsorbed or metabolized by the gut microbiome.
- First-pass metabolism
The metabolism of orally ingested compounds prior to reaching general circulation.
- Folate
A B vitamin essential for DNA synthesis, DNA repair and other biological reactions.
- Germ-free
Animals devoid of microorganisms.
- Glucuronidation
The addition of a glucuronic acid to a substrate used as a mechanism of xenobiotic metabolism by the host.
- Gnotobiotics
The colonization of germ-free animals with individual microbes or defined microbial communities.
- Hydrolysis
A chemical reaction wherein a chemical bond is cleaved with a water molecule, which serves as the nucleophile.
- Metabolic syndrome
A collection of physiological and biochemical conditions resulting in impaired energy utilization and storage, defined as a combination of high blood pressure, elevated blood sugar levels, excess fat, and abnormal cholesterol levels. This syndrome increases the risk of heart disease, stroke, and diabetes.
- Metformin
An oral antidiabetic medication used to treat type 2 diabetes.
- Microbiota
The collection of all microorganisms (archaea, bacteria, microscopic fungi, parasites, and viruses) found in a given body habitat.
- Microbiome
The combined genetic material and metabolic activities of the microbiota.
- Pharmacogenetics
The study of how genetic factors influence therapeutic outcomes.
- Pharmacogenomics
The use of sequencing-based genomic methods to analyze the links between genetics and therapeutic outcomes.
- Pharmacopeia
A manual for the preparation and use of medicinal drugs. The name is derived from the Greek words pharmakon (drug) and –poios (making).
- Prodrug
A drug that is administered in an inactive form and becomes active when metabolized.
- Reduction
A chemical reaction wherein the oxidation state of a chemical bond is reduced. For example, a carbon-carbon bond modified to a carbon-hydrogen bond is a reductive transformation.
- Serum metabolome
The collection of all metabolites found in serum.
- Xenobiotics
Compounds foreign to a biological system. For humans, these include drugs, dietary bioactive compounds, food additives, and environmental toxins.
Footnotes
Competing interests statement
P.J.T. is on the Scientific Advisory Board for Seres Therapeutics and Whole Biome, has consulted for Pfizer in the past year and has current research support from MedImmune.
References
- 1.Faith JJ, et al. The long-term stability of the human gut microbiota. Science. 2013;341:1237439. doi: 10.1126/science.1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kuczynski J, et al. Experimental and analytical tools for studying the human microbiome. Nat. Rev. Genet. 2012;13:47–58. doi: 10.1038/nrg3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Maurice CF, Haiser HJ, Turnbaugh PJ. Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell. 2013;152:39–50. doi: 10.1016/j.cell.2012.10.052. This was the first study to develop methods to define the metabolically active set of gut bacteria and demonstrate that xenobiotics shape the structure and physiology of these bacteria.
- 4.Maurice CF, Turnbaugh PJ. Quantifying the metabolic activities of human-associated microbial communities across multiple ecological scales. FEMS Microbiol. Rev. 2013;37:830–848. doi: 10.1111/1574-6976.12022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O'Hara AM, Shanahan F. The gut flora as a forgotten organ. EMBO Rep. 2006;7:688–693. doi: 10.1038/sj.embor.7400731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qin J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eckburg PB, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.David LA, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yatsunenko T, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goodrich JK, et al. Human genetics shape the gut microbiome. Cell. 2014;159:789–799. doi: 10.1016/j.cell.2014.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thaiss CA, et al. Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell. 2014;159:514–529. doi: 10.1016/j.cell.2014.09.048. [DOI] [PubMed] [Google Scholar]
- 12.Diaz Heijtz R, et al. Normal gut microbiota modulates brain development and behavior. Proc. Natl. Acad. Sci. USA. 2011;108:3047–3052. doi: 10.1073/pnas.1010529108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foster JA, McVey Neufeld KA. Gut-brain axis: how the microbiome influences anxiety and depression. Trends Neurosci. 2013;36:305–312. doi: 10.1016/j.tins.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 14.Hsiao EY, et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell. 2013;155:1451–1463. doi: 10.1016/j.cell.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arthur JC, et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 2012;338:120–123. doi: 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kostic AD, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14:207–215. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fierer N, et al. Forensic identification using skin bacterial communities. Proc. Natl. Acad. Sci. USA. 2010;107:6477–6481. doi: 10.1073/pnas.1000162107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Franzosa EA, et al. Identifying personal microbiomes using metagenomic codes. Proc. Natl. Acad. Sci. USA. 2015 doi: 10.1073/pnas.1423854112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liou AP, et al. Conserved shifts in the gut microbiota due to gastric bypass reduce host weight and adiposity. Sci. Transl. Med. 2013;5:178ra141. doi: 10.1126/scitranslmed.3005687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Roopchand DE, et al. Dietary Polyphenols Promote Growth of the Gut Bacterium Akkermansia muciniphila and Attenuate High-Fat Diet-Induced Metabolic Syndrome. Diabetes. 2015;64:2847–2858. doi: 10.2337/db14-1916. This study suggests that the benefical effects of dietary polyphenols may be mediated by the gut microbiome.
- 21.Fuller AT. Is p-aminobenzenesulphonamide the active agent in protonsil therapy? The Lancet. 1937;229:194–198. [Google Scholar]
- 22.Colebrook L, Buttle GAH, O'Meara RAQ. The mode of action of p-aminobenzene sulphonamide and prontosil in hemolytic Streptococcal infections. The Lancet. 1936;228:1323–1326. [Google Scholar]
- 23.Radomski JL, Mellinger TJ. The absorption, fate and excretion in rats of the water-soluble azo dyes, FD&C Red No. 2, FD&C Red No. 4, and FD&C Yellow No. 6. J. Pharmacol. Exp. Ther. 1962;136:259–266. [PubMed] [Google Scholar]
- 24.Klotz U, Maier K, Fischer C, Heinkel K. Therapeutic efficacy of sulfasalazine and its metabolites in patients with ulcerative colitis and Crohn's disease. N. Engl. J. Med. 1980;303:1499–1502. doi: 10.1056/NEJM198012253032602. [DOI] [PubMed] [Google Scholar]
- 25.Plosker GL, Croom KF. Sulfasalazine: a review of its use in the management of rheumatoid arthritis. Drugs. 2005;65:1825–1849. doi: 10.2165/00003495-200565130-00008. [DOI] [PubMed] [Google Scholar]
- 26.Rocco TP, Fang JC. In: Goodman & Gilman's The Pharmacological Basis of Therapeutics. Brunton LL, Lazo JS, Parker KL, editors. New York: McGraw-Hill; 2011. [Google Scholar]
- 27.Grundmann O. The gut microbiome and pre-systemic metabolism: current stat and evolving research. J. Drug Metab. Toxicol. 2010;1:1–7. [Google Scholar]
- 28.Haiser HJ, Turnbaugh PJ. Developing a metagenomic view of xenobiotic metabolism. Pharmacol. Res. 2013;69:21–31. doi: 10.1016/j.phrs.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H, Jia W. Cometabolism of microbes and host: implications for drug metabolism and drug-induced toxicity. Clin. Pharmacol. Ther. 2013;94:574–581. doi: 10.1038/clpt.2013.157. [DOI] [PubMed] [Google Scholar]
- 30.Nicholson JK, Holmes E, Wilson ID. Gut microorganisms, mammalian metabolism and personalized health care. Nat. Rev. Microbiol. 2005;3:431–438. doi: 10.1038/nrmicro1152. [DOI] [PubMed] [Google Scholar]
- 31.Saad R, Rizkallah MR, Aziz RK. Gut Pharmacomicrobiomics: the tip of an iceberg of complex interactions between drugs and gut-associated microbes. Gut Pathog. 2012;4:16. doi: 10.1186/1757-4749-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tralau T, Sowada J, Luch A. Insights on the human microbiome and its xenobiotic metabolism: what is known about its effects on human physiology? Expert Opin. Drug Metab. Toxicol. 2015;11:411–425. doi: 10.1517/17425255.2015.990437. [DOI] [PubMed] [Google Scholar]
- 33.Pond SM, Tozer TN. First-pass elimination. Basic concepts and clinical consequences. Clin. Pharmacokinet. 1984;9:1–25. doi: 10.2165/00003088-198409010-00001. [DOI] [PubMed] [Google Scholar]
- 34.Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 35.Arkhipova OV, Akimenko VK. Unsaturated organic acids as terminal electron acceptors for reductase chains of anaerobic bacteria. Microbiology. 2005;76:725–737. [PubMed] [Google Scholar]
- 36.Novel G, Didier-Fichet ML, Stoeber F. Inducibility of beta-glucuronidase in wild-type and hexuronate-negative mutants of Escherichia coli K-12. J. Bacteriol. 1974;120:89–95. doi: 10.1128/jb.120.1.89-95.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haiser HJ, Seim KL, Balskus EP, Turnbaugh PJ. Mechanistic insight into digoxin inactivation by Eggerthella lenta augments our understanding of its pharmacokinetics. Gut Microbes. 2014;5:233–238. doi: 10.4161/gmic.27915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Groot MJ. Designing better drugs: predicting cytochrome P450 metabolism. Drug Discov. Today. 2006;11:601–606. doi: 10.1016/j.drudis.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 39.Zanger UM, Schwab M. Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol. Ther. 2013;138:103–141. doi: 10.1016/j.pharmthera.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 40.Bachrach WH. Sulfasalazine: I. An historical perspective. Am. J. Gastroenterol. 1988;83:487–496. [PubMed] [Google Scholar]
- 41.Svartz N. Sulfasalazine: II. Some notes on the discovery and development of salazopyrin. Am. J. Gastroenterol. 1988;83:497–503. [PubMed] [Google Scholar]
- 42.Peppercorn MA, Goldman P. The role of intestinal bacteria in the metabolism of salicylazosulfapyridine. J. Pharmacol. Exp. Ther. 1972;181:555–562. [PubMed] [Google Scholar]
- 43.Chen H, Wang RF, Cerniglia CE. Molecular cloning, overexpression, purification, and characterization of an aerobic FMN-dependent azoreductase from Enterococcus faecalis. Protein Expr. Purif. 2004;34:302–310. doi: 10.1016/j.pep.2003.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morrison JM, Wright CM, John GH. Identification, isolation and characterization of a novel azoreductase from Clostridium perfringens. Anaerobe. 2012;18:229–234. doi: 10.1016/j.anaerobe.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 45.Sousa T, et al. On the colonic bacterial metabolism of azo-bonded prodrugs of 5-aminosalicylic acid. J. Pharm. Sci. 2014;103:3171–3175. doi: 10.1002/jps.24103. [DOI] [PubMed] [Google Scholar]
- 46.Delomenie C, et al. Identification and functional characterization of arylamine N-acetyltransferases in eubacteria: evidence for highly selective acetylation of 5-aminosalicylic acid. J. Bacteriol. 2001;183:3417–3427. doi: 10.1128/JB.183.11.3417-3427.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carmody RN, Turnbaugh PJ. Host-microbial interactions in the metabolism of therapeutic and diet-derived xenobiotics. J. Clin. Invest. 2014;124:4173–4181. doi: 10.1172/JCI72335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wells PG, et al. Glucuronidation and the UDP-glucuronosyltransferases in health and disease. Drug Metab. Dispos. 2004;32:281–290. doi: 10.1124/dmd.32.3.281. [DOI] [PubMed] [Google Scholar]
- 49.Wiseman LR, Markham A. Irinotecan. A review of its pharmacological properties and clinical efficacy in the management of advanced colorectal cancer. Drugs. 1996;52:606–623. doi: 10.2165/00003495-199652040-00013. [DOI] [PubMed] [Google Scholar]
- 50.Stein A, Voigt W, Jordan K. Chemotherapy-induced diarrhea: pathophysiology, frequency and guideline-based management. Ther. Adv. Med. Oncol. 2010;2:51–63. doi: 10.1177/1758834009355164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mani S, Boelsterli UA, Redinbo MR. Understanding and modulating mammalian-microbial communication for improved human health. Annu. Rev. Pharmacol. Toxicol. 2014;54:559–580. doi: 10.1146/annurev-pharmtox-011613-140007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rothenberg ML, et al. Phase II trial of irinotecan in patients with progressive or rapidly recurrent colorectal cancer. J. Clin. Oncol. 1996;14:1128–1135. doi: 10.1200/JCO.1996.14.4.1128. [DOI] [PubMed] [Google Scholar]
- 53.Higuchi K, et al. Present status and strategy of NSAIDs-induced small bowel injury. J. Gastroenterol. 2009;44:879–888. doi: 10.1007/s00535-009-0102-2. [DOI] [PubMed] [Google Scholar]
- 54. Saitta KS, et al. Bacterial beta-glucuronidase inhibition protects mice against enteropathy induced by indomethacin, ketoprofen or diclofenac: mode of action and pharmacokinetics. Xenobiotica. 2014;44:28–35. doi: 10.3109/00498254.2013.811314. This study demonstrated that toxicity associated with NSAIDs can be alleviated by inhibiting bacterial enzyme activity with small molecule inhibitors.
- 55.Beaud D, Tailliez P, Anba-Mondoloni J. Genetic characterization of the beta-glucuronidase enzyme from a human intestinal bacterium, Ruminococcus gnavus. Microbiology. 2005;151:2323–2330. doi: 10.1099/mic.0.27712-0. [DOI] [PubMed] [Google Scholar]
- 56.Dabek M, McCrae SI, Stevens VJ, Duncan SH, Louis P. Distribution of beta-glucosidase and beta-glucuronidase activity and of beta-glucuronidase gene gus in human colonic bacteria. FEMS Microbiol. Ecol. 2008;66:487–495. doi: 10.1111/j.1574-6941.2008.00520.x. [DOI] [PubMed] [Google Scholar]
- 57.Flores R, et al. Association of fecal microbial diversity and taxonomy with selected enzymatic functions. PLOS One. 2012;7:e39745. doi: 10.1371/journal.pone.0039745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roy D, Ward P. Rapid detection of Bifidobacterium dentium by enzymatic hydrolysis of β-Glucuronide substrates. J. Food Protect. 1992;55:291–295. doi: 10.4315/0362-028X-55.4.291. [DOI] [PubMed] [Google Scholar]
- 59.Russell WM, Klaenhammer TR. Identification and cloning of gusA, encoding a new beta-glucuronidase from Lactobacillus gasseri ADH. Appl. Environ. Microbiol. 2001;67:1253–1261. doi: 10.1128/AEM.67.3.1253-1261.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wallace BD, et al. Structure and inhibition of microbiome beta-glucuronidases essential to the alleviation of cancer drug toxicity. Chem. Biol. 2015;22:1238–1249. doi: 10.1016/j.chembiol.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lindenbaum J, Rund DG, Butler VP, Jr, Tse-Eng D, Saha JR. Inactivation of digoxin by the gut flora: reversal by antibiotic therapy. N. Engl. J. Med. 1981;305:789–794. doi: 10.1056/NEJM198110013051403. [DOI] [PubMed] [Google Scholar]
- 62.Matzuk MM, Shlomchik M, Shaw LM. Making digoxin therapeutic drug monitoring more effective. Ther. Drug Monit. 1991;13:215–219. doi: 10.1097/00007691-199105000-00005. [DOI] [PubMed] [Google Scholar]
- 63.Peters U, Falk LC, Kalman SM. Digoxin metabolism in patients. Arch. Intern. Med. 1978;138:1074–1076. [PubMed] [Google Scholar]
- 64.Saha JR, Butler VP, Jr, Neu HC, Lindenbaum J. Digoxin-inactivating bacteria: identification in human gut flora. Science. 1983;220:325–327. doi: 10.1126/science.6836275. [DOI] [PubMed] [Google Scholar]
- 65.Mathan VI, Wiederman J, Dobkin JF, Lindenbaum J. Geographic differences in digoxin inactivation, a metabolic activity of the human anaerobic gut flora. Gut. 1989;30:971–977. doi: 10.1136/gut.30.7.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rowland IR. Factors affecting metabolic activity of the intestinal microflora. Drug Metab. Rev. 1988;19:243–261. doi: 10.3109/03602538808994135. [DOI] [PubMed] [Google Scholar]
- 67. Haiser HJ, et al. Predicting and manipulating cardiac drug inactivation by the human gut bacterium Eggerthella lenta. Science. 2013;341:295–298. doi: 10.1126/science.1235872. This was the first study to show that the bacterial inactivation drugs could be predicted with a genetic marker and prevented using dietary intervention.
- 68.Hooper LV, et al. Molecular analysis of commensal host-microbial relationships in the intestine. Science. 2001;291:881–884. doi: 10.1126/science.291.5505.881. [DOI] [PubMed] [Google Scholar]
- 69.Bjorkholm B, et al. Intestinal microbiota regulate xenobiotic metabolism in the liver. PLOS One. 2009;4:e6958. doi: 10.1371/journal.pone.0006958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lundin A, et al. Gut flora, Toll-like receptors and nuclear receptors: a tripartite communication that tunes innate immunity in large intestine. Cell. Microbiol. 2008;10:1093–1103. doi: 10.1111/j.1462-5822.2007.01108.x. [DOI] [PubMed] [Google Scholar]
- 71.Claus SP, et al. Colonization-induced host-gut microbial metabolic interaction. mBio. 2011;2:e00271-00210. doi: 10.1128/mBio.00271-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Selwyn FP, Cui JY, Klaassen CD. RNA-seq quantification of hepatic drug processing genes in germ-free mice. Drug Metab. Dispos. 2015;43:1572–1580. doi: 10.1124/dmd.115.063545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Claus SP, et al. Systemic multicompartmental effects of the gut microbiome on mouse metabolic phenotypes. Mol. Syst. Biol. 2008;4:219. doi: 10.1038/msb.2008.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wikoff WR, et al. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc. Natl. Acad. Sci. USA. 2009;106:3698–3703. doi: 10.1073/pnas.0812874106. This study demonstrated that the microbiome plays a significant role in the signature and abundance of circulating metabolites in mammalian blood.
- 75.Hodgman MJ, Garrard AR. A review of acetaminophen poisoning. Crit. Care Clin. 2012;28:499–516. doi: 10.1016/j.ccc.2012.07.006. [DOI] [PubMed] [Google Scholar]
- 76.Court MH, et al. Interindividual variability in acetaminophen glucuronidation by human liver microsomes: identification of relevant acetaminophen UDP-glucuronosyltransferase isoforms. J. Pharmacol. Exp. Ther. 2001;299:998–1006. [PubMed] [Google Scholar]
- 77.Harrill AH, et al. Mouse population-guided resequencing reveals that variants in CD44 contribute to acetaminophen-induced liver injury in humans. Genome Res. 2009;19:1507–1515. doi: 10.1101/gr.090241.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Clayton TA, Baker D, Lindon JC, Everett JR, Nicholson JK. Pharmacometabonomic identification of a significant host-microbiome metabolic interaction affecting human drug metabolism. Proc. Natl. Acad. Sci. USA. 2009;106:14728–14733. doi: 10.1073/pnas.0904489106. This was the first study to show the potential of using levels of microbial metabolites as predictive biomakers for drug metabolism.
- 79.Bone E, Tamm A, Hill M. The production of urinary phenols by gut bacteria and their possible role in the causation of large bowel cancer. Am. J. Clin. Nutr. 1976;29:1448–1454. doi: 10.1093/ajcn/29.12.1448. [DOI] [PubMed] [Google Scholar]
- 80.Selmer T, Andrei PI. p-Hydroxyphenylacetate decarboxylase from Clostridium difficile. A novel glycyl radical enzyme catalysing the formation of p-cresol. Eur. J. Biochem. 2001;268:1363–1372. doi: 10.1046/j.1432-1327.2001.02001.x. [DOI] [PubMed] [Google Scholar]
- 81.Gamage N, et al. Human sulfotransferases and their role in chemical metabolism. Toxicol. Sci. 2006;90:5–22. doi: 10.1093/toxsci/kfj061. [DOI] [PubMed] [Google Scholar]
- 82.Mangravite LM, Thorn CF, Krauss RM. Clinical implications of pharmacogenomics of statin treatment. Pharmacogenomics J. 2006;6:360–374. doi: 10.1038/sj.tpj.6500384. [DOI] [PubMed] [Google Scholar]
- 83.Kaddurah-Daouk R, et al. Enteric microbiome metabolites correlate with response to simvastatin treatment. PLOS One. 2011;6:e25482. doi: 10.1371/journal.pone.0025482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mitchell JB, et al. A low molecular weight antioxidant decreases weight and lowers tumor incidence. Free Radic. Biol. Med. 2003;34:93–102. doi: 10.1016/s0891-5849(02)01193-0. [DOI] [PubMed] [Google Scholar]
- 85.Li F, et al. Microbiome remodelling leads to inhibition of intestinal farnesoid X receptor signalling and decreased obesity. Nat. Commun. 2013;4:2384. doi: 10.1038/ncomms3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jiang J, et al. Diversity of bile salt hydrolase activities in different lactobacilli toward human bile salts. Ann. Microbiol. 2010;60:81–88. [Google Scholar]
- 87.de Wit NJ, et al. The role of the small intestine in the development of dietary fat-induced obesity and insulin resistance in C57BL/6J mice. BMC Med. Genomics. 2008;1:14. doi: 10.1186/1755-8794-1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sayin SI, et al. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 2013;17:225–235. doi: 10.1016/j.cmet.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 89.Tsuda T. Regulation of adipocyte function by anthocyanins; possibility of preventing the metabolic syndrome. J. Agric. Food Chem. 2008;56:642–646. doi: 10.1021/jf073113b. [DOI] [PubMed] [Google Scholar]
- 90.Quesada H, et al. Grape seed proanthocyanidins correct dyslipidemia associated with a high-fat diet in rats and repress genes controlling lipogenesis and VLDL assembling in liver. Int. J. Obes. (Lond) 2009;33:1007–1012. doi: 10.1038/ijo.2009.136. [DOI] [PubMed] [Google Scholar]
- 91.Tsuda T, Horio F, Uchida K, Aoki H, Osawa T. Dietary cyanidin 3-O-beta-D-glucoside-rich purple corn color prevents obesity and ameliorates hyperglycemia in mice. J. Nutr. 2003;133:2125–2130. doi: 10.1093/jn/133.7.2125. [DOI] [PubMed] [Google Scholar]
- 92.Baiges I, Palmfeldt J, Blade C, Gregersen N, Arola L. Lipogenesis is decreased by grape seed proanthocyanidins according to liver proteomics of rats fed a high fat diet. Mol. Cell. Proteomics. 2010;9:1499–1513. doi: 10.1074/mcp.M000055-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cefalu WT, et al. Botanicals and the metabolic syndrome. Am. J. Clin. Nutr. 2008;87:481S–487S. doi: 10.1093/ajcn/87.2.481S. [DOI] [PubMed] [Google Scholar]
- 94.Felgines C, et al. Radiolabelled cyanidin 3-O-glucoside is poorly absorbed in the mouse. Br. J. Nutr. 2010;103:1738–1745. doi: 10.1017/S0007114510000061. [DOI] [PubMed] [Google Scholar]
- 95.Abia R, Fry SC. Degradation and metabolism of 14C-labelled proanthocyanidins from carob (Ceratonia siliqua) pods in the gastrointestinal tract of the rat. J. Sci. Food Agr. 2001;81:1156–1165. [Google Scholar]
- 96.Anhe FF, et al. A polyphenol-rich cranberry extract protects from diet-induced obesity, insulin resistance and intestinal inflammation in association with increased Akkermansia spp. population in the gut microbiota of mice. Gut. 2015;64:872–883. doi: 10.1136/gutjnl-2014-307142. [DOI] [PubMed] [Google Scholar]
- 97.Everard A, et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. USA. 2013;110:9066–9071. doi: 10.1073/pnas.1219451110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Santacruz A, et al. Gut microbiota composition is associated with body weight, weight gain and biochemical parameters in pregnant women. Br. J. Nutr. 2010;104:83–92. doi: 10.1017/S0007114510000176. [DOI] [PubMed] [Google Scholar]
- 99.Zhang H, et al. Human gut microbiota in obesity and after gastric bypass. Proc. Natl. Acad. Sci. USA. 2009;106:2365–2370. doi: 10.1073/pnas.0812600106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Everard A, et al. Responses of gut microbiota and glucose and lipid metabolism to prebiotics in genetic obese and diet-induced leptin-resistant mice. Diabetes. 2011;60:2775–2786. doi: 10.2337/db11-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Shin NR, et al. An increase in the Akkermansia spp. population induced by metformin treatment improves glucose homeostasis in diet-induced obese mice. Gut. 2014;63:727–735. doi: 10.1136/gutjnl-2012-303839. This study suggests that the antidiabetic drug metformin may act in part by altering the gut microbiome.
- 102.Axling U, et al. Green tea powder and Lactobacillus plantarum affect gut microbiota, lipid metabolism and inflammation in high-fat fed C57BL/6J mice. Nutr. Metab. (Lond) 2012;9:105. doi: 10.1186/1743-7075-9-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kemperman RA, et al. Impact of polyphenols from black tea and red wine/grape juice on a gut model microbiome. Food Res. Int. 2013;53:659–669. [Google Scholar]
- 104.Haslam E. Natural polyphenols (vegetable tannins) as drugs: possible modes of action. J. Nat. Prod. 1996;59:205–215. doi: 10.1021/np960040+. [DOI] [PubMed] [Google Scholar]
- 105.Nunez-Sanchez MA, et al. Targeted metabolic profiling of pomegranate polyphenols and urolithins in plasma, urine and colon tissues from colorectal cancer patients. Mol. Nutr. Food. Res. 2014;58:1199–1211. doi: 10.1002/mnfr.201300931. [DOI] [PubMed] [Google Scholar]
- 106.Garcia-Munoz C, Vaillant F. Metabolic fate of ellagitannins: implications for health, and research perspectives for innovative functional foods. Crit Rev Food Sci Nutr. 2014;54:1584–1598. doi: 10.1080/10408398.2011.644643. [DOI] [PubMed] [Google Scholar]
- 107.Tomas-Barberan FA, Garcia-Villalba R, Gonzalez-Sarrias A, Selma MV, Espin JC. Ellagic acid metabolism by human gut microbiota: consistent observation of three urolithin phenotypes in intervention trials, independent of food source, age, and health status. J. Agric. Food Chem. 2014;62:6535–6538. doi: 10.1021/jf5024615. [DOI] [PubMed] [Google Scholar]
- 108.Garcia-Villalba R, Beltran D, Espin JC, Selma MV, Tomas-Barberan FA. Time course production of urolithins from ellagic acid by human gut microbiota. J. Agric. Food Chem. 2013;61:8797–8806. doi: 10.1021/jf402498b. [DOI] [PubMed] [Google Scholar]
- 109.Selma MV, Beltran D, Garcia-Villalba R, Espin JC, Tomas-Barberan FA. Description of urolithin production capacity from ellagic acid of two human intestinal Gordonibacter species. Food Funct. 2014;5:1779–1784. doi: 10.1039/c4fo00092g. [DOI] [PubMed] [Google Scholar]
- 110.Adlercreutz H. Lignans and human health. Crit. Rev. Clin. Lab Sci. 2007;44:483–525. doi: 10.1080/10408360701612942. [DOI] [PubMed] [Google Scholar]
- 111.Lampe JW. Is equol the key to the efficacy of soy foods? Am. J. Clin. Nutr. 2009;89:1664S–1667S. doi: 10.3945/ajcn.2009.26736T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Patisaul HB, Jefferson W. The pros and cons of phytoestrogens. Front. Neuroendocrinol. 2010;31:400–419. doi: 10.1016/j.yfrne.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Setchell KD, Clerici C. Equol: history, chemistry, and formation. J. Nutr. 2010;140:1355S–1362S. doi: 10.3945/jn.109.119776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wu AH, Yu MC, Tseng CC, Pike MC. Epidemiology of soy exposures and breast cancer risk. Br. J. Cancer. 2008;98:9–14. doi: 10.1038/sj.bjc.6604145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Setchell KD, Brown NM, Lydeking-Olsen E. The clinical importance of the metabolite equol-a clue to the effectiveness of soy and its isoflavones. J. Nutr. 2002;132:3577–3584. doi: 10.1093/jn/132.12.3577. [DOI] [PubMed] [Google Scholar]
- 116.Duncan AM, Merz-Demlow BE, Xu X, Phipps WR, Kurzer MS. Premenopausal equol excretors show plasma hormone profiles associated with lowered risk of breast cancer. Cancer Epidem. Biomar. 2000;9:581–586. [PubMed] [Google Scholar]
- 117.Virk-Baker MK, Barnes S, Krontiras H, Nagy TR. S-(-)equol producing status not associated with breast cancer risk among low isoflavone-consuming US postmenopausal women undergoing a physician-recommended breast biopsy. Nutr. Res. 2014;34:116–125. doi: 10.1016/j.nutres.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Mazur W, Adlercreutz H. Natural and anthropogenic environmental oestrogens: the scientific basis for risk assessment. Pure Appl. Chem. 1998;70:1759–1776. [Google Scholar]
- 119.Penalvo JL, Haajanen KM, Botting N, Adlercreutz H. Quantification of lignans in food using isotope dilution gas chromatography/mass spectrometry. J. Agric. Food Chem. 2005;53:9342–9347. doi: 10.1021/jf051488w. [DOI] [PubMed] [Google Scholar]
- 120.Clavel T, Borrmann D, Braune A, Dore J, Blaut M. Occurrence and activity of human intestinal bacteria involved in the conversion of dietary lignans. Anaerobe. 2006;12:140–147. doi: 10.1016/j.anaerobe.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 121. Mabrok HB, et al. Lignan transformation by gut bacteria lowers tumor burden in a gnotobiotic rat model of breast cancer. Carcinogenesis. 2012;33:203–208. doi: 10.1093/carcin/bgr256. This study determined that the microbial production of bioactive metabolites from dietary sources mediates anticancer effects for the host.
- 122.Kaderlik KR, et al. Glucuronidation of N-hydroxy heterocyclic amines by human and rat liver microsomes. Carcinogenesis. 1994;15:1695–1701. doi: 10.1093/carcin/15.8.1695. [DOI] [PubMed] [Google Scholar]
- 123.Hirayama K, et al. Effects of human intestinal flora on mutagenicity of and DNA adduct formation from food and environmental mutagens. Carcinogenesis. 2000;21:2105–2111. doi: 10.1093/carcin/21.11.2105. [DOI] [PubMed] [Google Scholar]
- 124.Kassie F, et al. Intestinal microflora plays a crucial role in the genotoxicity of the cooked food mutagen 2-amino-3-methylimidazo [4,5-f]quinoline. Carcinogenesis. 2001;22:1721–1725. doi: 10.1093/carcin/22.10.1721. [DOI] [PubMed] [Google Scholar]
- 125. Humblot C, et al. Beta-glucuronidase in human intestinal microbiota is necessary for the colonic genotoxicity of the food-borne carcinogen 2-amino-3-methylimidazo[4,5-f]quinoline in rats. Carcinogenesis. 2007;28:2419–2425. doi: 10.1093/carcin/bgm170. This study showed that microbial glucuronidation activity in the gut contributes to the carcinogenic effect of heterocyclic amines from charred meat.
- 126.Craciun S, Balskus EP. Microbial conversion of choline to trimethylamine requires a glycyl radical enzyme. Proc. Natl. Acad. Sci. USA. 2012;109:21307–21312. doi: 10.1073/pnas.1215689109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Wang Z, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. doi: 10.1038/nature09922. This study demonstrated the metabolism of dietary lipids by gut bacteria contributes to heart disease.
- 128.Tang WH, et al. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N. Engl. J. Med. 2013;368:1575–1584. doi: 10.1056/NEJMoa1109400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Koeth RA, et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 2013;19:576–585. doi: 10.1038/nm.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Drasar BS, Renwick AG, Williams RT. The role of the gut flora in the metabolism of cyclamate. Biochem. J. 1972;129:881–890. doi: 10.1042/bj1290881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Legator MS, Palmer KA, Green S, Petersen KW. Cytogenetic studies in rats of cyclohexylamine, a metabolite of cyclamate. Science. 1969;165:1139–1140. doi: 10.1126/science.165.3898.1139. [DOI] [PubMed] [Google Scholar]
- 132.Tamura M, Hoshi C, Hori S. Xylitol affects the intestinal microbiota and metabolism of daidzein in adult male mice. Int. J. Mol. Sci. 2013;14:23993–24007. doi: 10.3390/ijms141223993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Suez J, et al. Artificial sweeteners induce glucose intolerance by altering the gut microbiota. Nature. 2014;514:181–186. doi: 10.1038/nature13793. [DOI] [PubMed] [Google Scholar]
- 134.Brown RJ, de Banate MA, Rother KI. Artificial sweeteners: a systematic review of metabolic effects in youth. Int. J. Pediatr. Obes. 2010;5:305–312. doi: 10.3109/17477160903497027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Chassaing B, et al. Dietary emulsifiers impact the mouse gut microbiota promoting colitis and metabolic syndrome. Nature. 2015;519:92–96. doi: 10.1038/nature14232. This research showed that dietary supplements can disrupt mucus-bacterial interactions, promoting gut inflammation.
- 136.Hau AK, Kwan TH, Li PK. Melamine toxicity and the kidney. J. Am. Soc. Nephrol. 2009;20:245–250. doi: 10.1681/ASN.2008101065. [DOI] [PubMed] [Google Scholar]
- 137.Ingelfinger JR. Melamine and the global implications of food contamination. N. Engl. J. Med. 2008;359:2745–2748. doi: 10.1056/NEJMp0808410. [DOI] [PubMed] [Google Scholar]
- 138. Zheng X, et al. Melamine-induced renal toxicity is mediated by the gut microbiota. Sci. Transl. Med. 2013;5:172ra122. doi: 10.1126/scitranslmed.3005114. This study attributed the toxicity of dietary contaminants to gut microbial metabolism.
- 139.Shelton DR, Karns JS, McCarty GW, Durham DR. Metabolism of melamine by Klebsiella terragena. Appl. Environ. Microbiol. 1997;63:2832–2835. doi: 10.1128/aem.63.7.2832-2835.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Jutzi K, Cook AM, Hutter R. The degradative pathway of the s-triazine melamine. The steps to ring cleavage. Biochem. J. 1982;208:679–684. doi: 10.1042/bj2080679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Podschun R. Isolation of Klebsiella terrigena from human feces: biochemical reactions, capsule types, and antibiotic sensitivity. Zentralbl. Bakteriol. 1991;275:73–78. doi: 10.1016/s0934-8840(11)80769-3. [DOI] [PubMed] [Google Scholar]
- 142.Sonnenburg JL, Fischbach MA. Community health care: therapeutic opportunities in the human microbiome. Sci. Transl. Med. 2011;3:78ps12. doi: 10.1126/scitranslmed.3001626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ahmad S, et al. A high throughput assay for discovery of bacterial beta-glucuronidase inhibitors. Curr. Chem. Genomics. 2011;5:13–20. doi: 10.2174/1875397301105010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ahmad S, Hughes MA, Yeh LA, Scott JE. Potential repurposing of known drugs as potent bacterial beta-glucuronidase inhibitors. J. Biomol. Screen. 2012;17:957–965. doi: 10.1177/1087057112444927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Roberts AB, Wallace BD, Venkatesh MK, Mani S, Redinbo MR. Molecular insights into microbial beta-glucuronidase inhibition to abrogate CPT-11 toxicity. Mol. Pharmacol. 2013;84:208–217. doi: 10.1124/mol.113.085852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Wallace BD, et al. Alleviating cancer drug toxicity by inhibiting a bacterial enzyme. Science. 2010;330:831–835. doi: 10.1126/science.1191175. This was the first study to show that the toxicity associated with the microbial metabolism of cancer drugs in the gut can be mitigated using bacterial enzyme-specific small molecule inhibitors.
- 147.LoGuidice A, Wallace BD, Bendel L, Redinbo MR, Boelsterli UA. Pharmacologic targeting of bacterial beta-glucuronidase alleviates nonsteroidal anti-inflammatory drug-induced enteropathy in mice. J. Pharmacol. Exp. Ther. 2012;341:447–454. doi: 10.1124/jpet.111.191122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Carmody RN, et al. Diet dominates host genotype in shaping the murine gut microbiota. Cell Host Microbe. 2015;17:72–84. doi: 10.1016/j.chom.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Lee JR, et al. Gut microbiota and tacrolimus dosing in kidney transplantation. PLOS One. 2015;10:e0122399. doi: 10.1371/journal.pone.0122399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Viaud S, et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science. 2013;342:971–976. doi: 10.1126/science.1240537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Viaud S, et al. Cyclophosphamide induces differentiation of Th17 cells in cancer patients. Cancer Res. 2011;71:661–665. doi: 10.1158/0008-5472.CAN-10-1259. [DOI] [PubMed] [Google Scholar]
- 153.Iida N, et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science. 2013;342:967–970. doi: 10.1126/science.1240527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ozben T. Oxidative stress and apoptosis: impact on cancer therapy. J. Pharm. Sci. 2007;96:2181–2196. doi: 10.1002/jps.20874. [DOI] [PubMed] [Google Scholar]
- 155.Sivan A, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 2015;350:1084–1089. doi: 10.1126/science.aac4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Vetizou M, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 2015;350:1079–1084. doi: 10.1126/science.aad1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Garrett WS, et al. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 2007;131:33–45. doi: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Neurath MF. New targets for mucosal healing and therapy in inflammatory bowel diseases. Mucosal Immunol. 2014;7:6–19. doi: 10.1038/mi.2013.73. [DOI] [PubMed] [Google Scholar]
- 159.Rooks MG, et al. Gut microbiome composition and function in experimental colitis during active disease and treatment-induced remission. ISME J. 2014;8:1403–1417. doi: 10.1038/ismej.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Donia MS, et al. A systematic analysis of biosynthetic gene clusters in the human microbiome reveals a common family of antibiotics. Cell. 2014;158:1402–1414. doi: 10.1016/j.cell.2014.08.032. This study revealed the potential of the human microbiome as a rich source of bioactive natural products, including antibiotics.
- 161.Rupnik M, Wilcox MH, Gerding DN. Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat. Rev. Microbiol. 2009;7:526–536. doi: 10.1038/nrmicro2164. [DOI] [PubMed] [Google Scholar]
- 162.Buffie CG, et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature. 2015;517:205–208. doi: 10.1038/nature13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Sorg JA, Sonenshein AL. Inhibiting the initiation of Clostridium difficile spore germination using analogs of chenodeoxycholic acid, a bile acid. J. Bacteriol. 2010;192:4983–4990. doi: 10.1128/JB.00610-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Wilson KH. Efficiency of various bile salt preparations for stimulation of Clostridium difficile spore germination. J. Clin. Microbiol. 1983;18:1017–1019. doi: 10.1128/jcm.18.4.1017-1019.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.