Abstract
Extracellular vesicles released from cells are under intense investigation for their roles in cell-cell communication and cancer progression. However, individual vesicles have been difficult to probe as their small size renders them invisible by conventional light microscopy. However, as a consequence of their small size these vesicles possess highly curved lipid membranes that offer an unconventional target for curvature-sensing probes. In this article, we present a strategy for using peptide-based biosensors to detect highly curved membranes and the negatively charged membrane lipid phosphatidylserine, we delineate several assays used to validate curvature- and lipid-targeting mechanisms, and we explore potential applications in probing extracellular vesicles released from sources such as apoptotic cells, cancer cells, or activated platelets.
Keywords: membrane curvature, curvature sensing, lipid targeting, peptide probes, extracellular vesicles
Introduction
Membranes, membrane curvature, and phosphatidylserine
A close-knit relationship exists between regulated membrane morphology and many cellular processes. Intracellular vesicle trafficking requires production of highly curved membrane vesicles, such as transport vesicles that bud from the endoplasmic reticulum and golgi via coat proteins and fuse with target membranes by the activity of the universal membrane fusion machinery SM/SNARE proteins (Südhof and Rothman, 2009). The endoplasmic reticulum itself is highly curved and dynamic as well, with regulated morphology essential for communication between organelles by structured endoplasmic reticulum-organelle contacts (Westrate et al., 2015). One less understood aspect of membrane trafficking occurs through extracellular vesicles (EVs), a group divided by a blurry line into exosomes (~30–100 nm) and microvesicles (~100–1000 nm). Exosomes possess high membrane curvature as a function of their small radius. This curvature is produced by neutral sphingomyelinase-catalyzed ceramide synthesis (Trajkovic et al., 2008) or by ESCRT-mediated endosomal membrane deformation leading to inward budding of intraluminal vesicles in multivesicular endosomes (Colombo et al., 2014). When these endosomes fuse with the plasma membrane, exosomes are excreted into extracellular space. While the mechanism of microvesicle shedding is less understood, it appears to require phosphatidylserine (PS) externalization and ARF6 activity (Muralidharan-Chari et al., 2009).
PS is normally sequestered on the inner leaflet of the plasma membrane by ATP-dependent lipid translocases (also called flippases). The abundant negatively charged lipid has been found to ‘moonlight’, performing a number of functions including regulation of membrane charge and targeting positively-charged signaling proteins to the plasma membrane (and to a lesser extent to endomembranes) which redistribute during transient membrane depolarization caused by calcium signaling (Yeung et al., 2008). In its best-characterized function, PS externalization occurs when ATP-dependent lipid translocases are inactivated and energy-independent scramblases are activated, providing an ‘eat-me’ signal for clearance of aging red blood cells or apoptotic cells. PS externalization also occurs in activated platelets, providing a surface to catalyze the formation of blood clotting protein complexes to initiate the blood coagulation cascade. While this asymmetry disappears permanently in apoptosis, transient PS externalization can be caused by calcium signaling (Balasubramanian et al., 2007) or intracellular infection by viruses or bacteria (Zwaal et al., 2005). Platelet-derived EVs, also called platelet microparticles, have likewise been recognized for their role in cell-cell communication, coagulation, and cancer (Italiano et al., 2010) and are often identified by PS externalization after platelet activation (Morel et al., 2010). However, much less is known about PS localization on endomembranes. One tool used to study organelle localization of PS comes from supplementing cells with fluorophore-labeled PS (Devaux et al., 2002), which is complicated by potential functional artifacts caused by labeled lipids. As such, chemical probes provide another approach to visualizing PS.
Methods to visualize intracellular membrane curvature rely on electron microscopy for static images or fluorescence microscopy for dynamic images, but even with the advent of superresolution microscopy which allows detection as small as the 20–40 nm scale (Thompson et al., 2012), EV structure and regions of membrane curvature will fall below the limit of detection for fluorescence microscopy. A recent study discovering the ability of neural stem cell EVs to activate immune signaling by EV-associated interferon-γ/interferon-γ receptor complexes (Cossetti et al., 2014) also demonstrated that super-resolution microscopy can be used to visualize internalized EVs and determine their localization within subcellular compartments of target cells. Even using super-resolution microscopy, however, it is difficult to distinguish between individual EVs. Proteins use a number of membrane recognition domains to target specific membranes and membrane subdomains (Lemmon, 2008) that can be exploited to investigate membrane dynamics. For example, a genetically encoded PS sensing protein fused with a fluorescent protein has been used to investigate PS localization by fluorescence microscopy (Yeung et al., 2008).
Cells regulate membrane curvature using protein-lipid interactions (McMahon and Gallop, 2005); at the same time, membrane curvature allosterically modulates membrane protein function (Tonnesen et al., 2014). A common feature of highly curved membranes, lipid-packing defects, arise from a mismatch between individual lipid geometry and global membrane curvature (Antonny, 2011). Protein-lipid interactions regulate MP clustering (van den Bogaart et al., 2011; Aimon et al., 2014) while curvature-mediated attractions between proteins may lead to higher-order clustering (Reynwar et al., 2007). Detecting curved membranes has been difficult as protein-lipid interactions that allow peripheral membrane proteins to interact with membranes are not well understood, and consequent lipid-lipid interactions also affect membrane properties such as curvature, fluidity, and thickness.
Extracellular Vesicles
Cells from every domain of life (Brown et al., 2015) produce EVs that can transport bioactive molecules including RNA (Valadi et al., 2007), proteins (Shen et al., 2011), and lipids (Subra et al., 2007) (Figure 1A). EVs have received particular attention for their role in cancer, particularly in regards to what is known as the ‘seed and soil’ hypothesis (Hood et al., 2011) where cancer cell EVs prepare the metastatic niche, creating a microenvironment that facilitates cancer cell invasion and growth. Recently, intravital microscopy experiments demonstrated that cancer EV-mediated cell-cell communication occurs in vivo (Zomer et al., 2015) ‘seeding’ tumor metastasis, while mass spectrometry analyses identified specific EV proteins as potential cancer biomarkers (Melo et al., 2015). EV membranes, especially those of exosomes, are highly curved as a function of the small vesicle radius. As a result of their high global membrane curvature, the membranes of small EVs will contain lipid-packing defects (Vanni et al., 2014) that could be targeted by membrane curvature sensors (Figure 1A).
Figure 1. MARCKS-ED binds EVs by targeting lipid membranes.
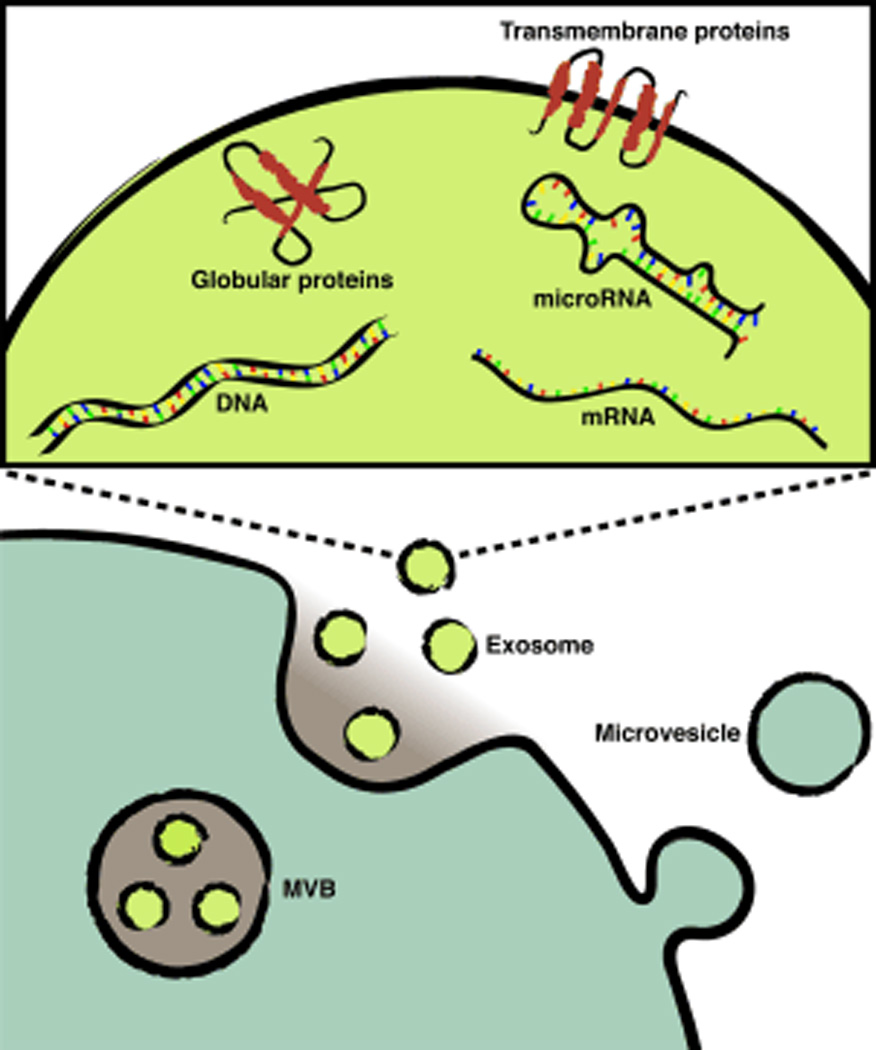
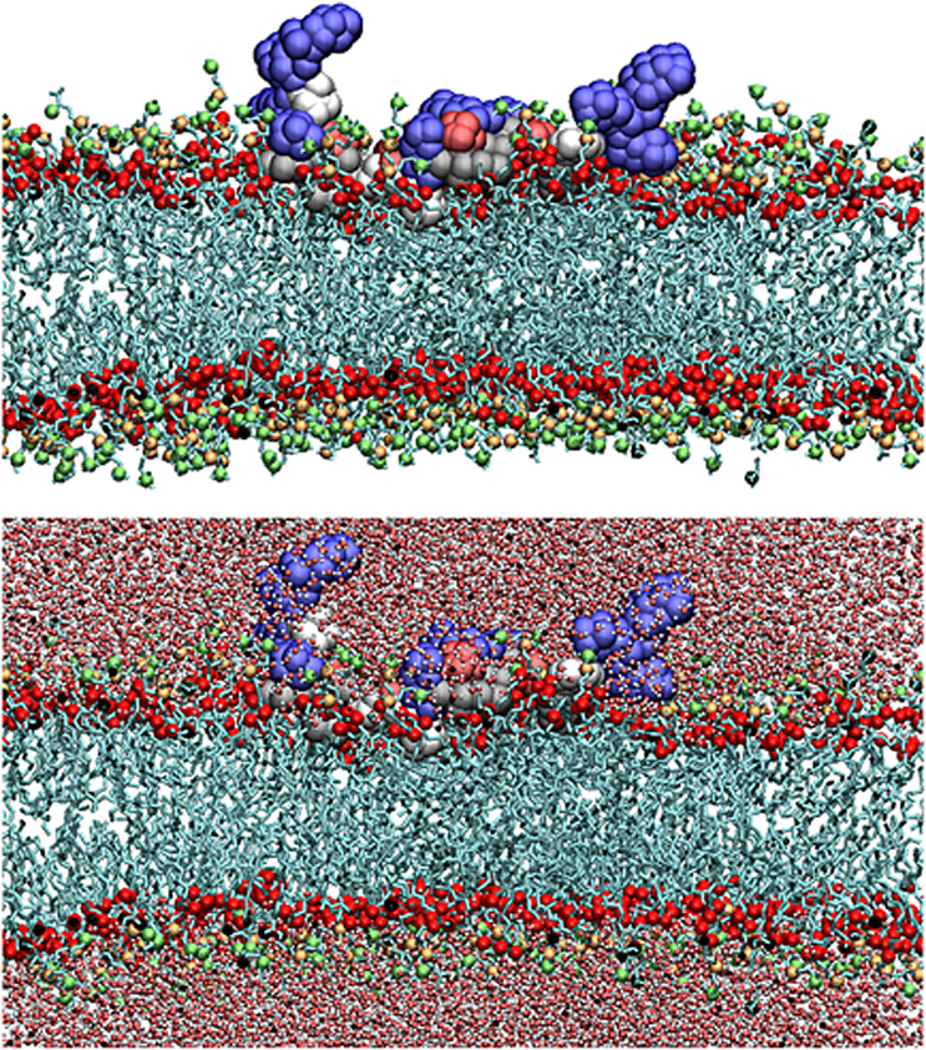
(a) Schematic illustrating cellular production of EVs including exosomes and microvesicles, adapted with permission from (Kastelowitz and Yin, 2014). (b) A representative snapshot of fully atomistic molecular dynamic simulations illustrating d-MARCKS-ED adopting a boat-like conformation during membrane binding, adapted with permission from (Yan et al., 2015).
Exogenous peptide probes
The most commonly used exogenous label for PS, annexin A5 (also known as annexin V), binds to PS-containing membranes in the presence of high calcium concentrations (Tait and Gibson, 1992) and then forms supramolecular clusters in a two-dimensional network over the membrane surface (Andree et al., 1992). This requires a relatively high amount of PS for binding. Clustered annexin A5 also induces negative membrane curvature, leading to internalization when bound to the plasma membrane (Kenis et al., 2004), which may complicate potential in vivo uses.
Proteins such as annexin A5 offer one route to membrane targeting, but recently peptide and peptidomimetic drugs have attracted considerable attention as potential novel therapeutics with the advantages of low manufacturing cost and ease of synthesis, including the ability to incorporate non-standard amino acids. Compared to protein therapeutics, peptides are also usually less immunogenic, often possess higher activity at similar masses, and can penetrate into tissues due to their smaller sizes (Vlieghe et al., 2010). Mirror image d-peptides and peptidomimetics have the additional advantage of protease resistance. Proteins and peptides which bind more weakly to curved membranes cannot induce membrane curvature, but act as sensors of membrane curvature (Antonny, 2011). However, there are still few lipid-targeting molecules available to investigate membrane biology (Gao and Zheng, 2013). There is a need for fluorescent small molecule and peptide probes in precision medicine, particularly in oncology (Garland et al., 2016), and membrane-targeting peptides represent a novel approach.
An emerging method to target lipid membranes is to design peptides based on known membrane-interacting proteins. Myristoylated alanine-riche C-kinase substrate (MARCKS) is a peripheral membrane protein that sequesters the second messenger lipid PI(4,5)P2. Upon signaling events leading to phosphorylation in the effector domain by protein kinase C or interaction with calcium-bound calmodulin, MARCKS dissociates from the membrane and PI(4,5)P2 is able to diffuse laterally (McLaughlin and Murray, 2005). The first investigations of the effector domain (ED; residues 151–175) of human MARCKS came from Cafiso and coworkers (Rauch et al., 2002). MARCKS-ED is a lysine-rich peptide; lysine-rich motifs are a common feature of proteins capable of sensing membrane PS. For example, the C. elegans PS receptor PSR-1 recognizes PS through a lysine-rich motif (Yang et al., 2015). These lysine-rich regions may form parts of basic-aromatic clusters, which are unstructured domains found in several lipid-interacting peripheral membrane proteins (Zhang et al., 2003). MARCKS-ED also possesses several serine and phenylalanine residues capable of participating in membrane interactions.
MARCKS-ED has been shown in vivo to bind to apoptotic cells in C. elegans (Morton et al., 2013). Compared to annexin A5, MARCKS-ED and other MARCKS-derived peptides would have the advantage of being positive curvature sensors rather than negative curvature inducers, potentially avoiding complications associated with induction of endocytosis. Biophysical validation of the peptide-lipid interactions utilized by MARCKS-ED was accomplished through a combination of atomistic molecular dynamic simulations, fluorescence anisotropy, fluorescence enhancement, and electron paramagnetic resonance studies (Morton et al., 2014). Using in vitro liposome models (Morton et al., 2012), these experiments showed MARCKS-ED binds to membranes through a composition of (a) electrostatic interactions between positively charged lysine residues and negatively charged PS headgroups and (b) hydrophobic insertion of bulky phenylalanine residues into lipid-packing defects inherent to the highly curved lipid membranes of small vesicles. MARCKS-ED also remains unstructured when membrane-bound, adopting a boat-like conformation where terminal lysines face the solvent (Figure 1B). The binding is stereo-independent, as both l- and d-MARCKS-ED bind in a similar mode (Morton et al., 2014; Yan et al., 2015).
Methods for detecting extracellular vesicles with peptide probes
Extracellular vesicle isolation
The most commonly used approaches to EV isolation rely on several ultracentrifugation steps (Théry et al., 2006). These methods require specialized equipment and may not give adequate yields, as many smaller vesicles will be lost in early steps. Another popular approach is ExoQuick™ (SBI, Mountain View, CA), a polymer-based reagent which precipitates EVs without ultracentrifugation and with smaller initial sample volumes. These can be characterized by nanoparticle tracking analysis or by transmission electron microscopy. Although less commonly used, cryoelectron microscopy allows researchers to characterize the contents of isolated EVs.
Fluorescently labeled peptides
Chemical synthesis of peptides can be accomplished by standard solid phase Fmoc chemistry (Merrifield, 1963), and N-terminal labeling with a fluorophore such as 4-chloro-7-nitrobenzo-2-oxa-1,3-diazole (NBD) can be achieved via a flexible linker such as ε-aminohexanoic acid using the same method (Morton et al., 2013).
Fluorometry
In order to monitor the binding of MARCKS-derived peptides to membranes, there are several possible methods. One approach for detecting membrane curvature and PS is fluorometry, or fluorescence spectroscopy. This is accomplished by conjugating a fluorescent small molecule with desired spectral properties to the N-terminal end of the peptide during synthesis.
Two methods encompassed by this are fluorescence anisotropy and fluorescence enhancement, which in the context of this discussion measure specific interactions between lipid-targeting peptides and curved vesicles using a spectrofluorometer. Fluorescence anisotropy binding assays rely on the polarized emission of fluorophore labels. The spectrofluorometer excites samples with polarized light and measures the intensity of emission in different polarization orientations. Unbound peptides will have depolarized emission due to tumbling. Upon binding of fluorophore-labeled peptide to EV, fluorophore tumbling and rotational diffusion decreases, increasing the polarization of emitted light. Titrating in varying concentration of labeled peptide can thus allow for determining the apparent affinity of the peptide for vesicles based on lipid concentration.
Another approach for measuring lipid membrane binding is through fluorescence enhancement assays. A solvatochromic fluorophore labels can report membrane binding by changes in fluorescence intensity. For example, NBD fluorescence is increased and blue-shifted in nonpolar environments, allowing for measurement of peptide-membrane interactions by fluorometry. Depending on the amino acid composition of the peptide, aromatic residues can occasionally undergo measurable changes in intrinsic fluorescence upon binding. This is most commonly observed with tryptophan, as tyrosine and phenylalanine are much more weakly fluorescent. For example, substituting tryptophan residues into MARCKS-related protein (Arbuzova et al., 2002) allowed for analysis of binding of the protein to membrane and to calmodulin.
Nanoparticle tracking analysis
Although standard fluorescence microscopy is challenging, one way to visualize EVs is to use nanoparticle tracking analysis (NTA). For example, the NanoSight LM10 (Malvern Instruments, Malvern, UK) is capable of characterizing EVs across the ~30–1000 nm range. Samples in liquid suspension are incubated in a sample chamber, and a laser beam is passed through the chamber. Laser light scattered from particles are recorded by a charge-coupled device video camera attached to an optical microscope. The NTA software creates particle tracks from recorded movies and uses the Stokes-Einstein equation to calculate the diameter of each particle based on the rate of Brownian motion (Kastelowitz and Yin, 2014). NTA can also be used for peptide colocalization with small vesicles. NanoSight instruments equipped with filters may also be used to detect fluorophore-labeled peptides that are bound to EVs, where a filter blocks light from the excitation laser and permits light emitted from fluorophore-labeled particles. By comparing analyzed tracks of EVs captured in scatter mode versus fluorescent mode, it is possible to measure the size distribution of total EVs versus peptide-bound EVs. Accurate quantification of EVs still remains challenging. Analysis of EVs by nanoparticle tracking analysis (Gardiner et al., 2013) and by other methods such as transmission electron microscopy, flow cytometry, and resistive pulse sensing (van der Pol et al., 2014) illustrate inter-platform variability in EV quantification.
Applications
Following biochemical and biophysical validation of the lipid-targeting properties of MARCKS-derived peptides, there are a number of potential applications relevant to basic biology. MARCKS-ED may potentially be used to detect more transient PS externalization. For example, calcium signaling can activate scramblases leading to PS externalization associated with apoptosis (Suzuki et al., 2013). However, PS externalization can occur transiently without cells undergoing apoptosis (Fadeel and Xue, 2009). Annexin A5 staining is frequently used as an assay to detect apoptotic cells, but perhaps a more sensitive PS probe would find that decreases in PS asymmetry of short durations occur far more often than previously appreciated. Studies of PS externalization under various physiological conditions would benefit from complementation with established metal ion sensors (Carter et al., 2014) to compare the dynamics of cell signaling (e.g. calcium second messenger signaling) with PS dynamics.
MARCKS-ED may find a use as a peptide probe for EVs (Kastelowitz and Yin, 2014). Distinguishing PS+ from PS- EVs is difficult by analytical methods such as lipid mass spectrometry, which will not distinguish internal from external PS. Annexin A5 is a popular protein probe for PS+ vesicles, but it requires high concentrations of external PS for detection. A sensitive probe for externalized PS on EVs may also distinguish subpopulations with different properties or functions, such as differing source or recipient cells, or differing rates of clearance. MARCKS-ED probes could be used to investigate whether PS+ EVs represent transient PS externalization from EV-producing cells or distinguish between PS+ and PS− EVs in order to characterize EV heterogeneity.
There are a number of potential biomedical applications for lipid-targeting peptides. Another potential use for MARCKS-ED relies on sensing surface PS on EVs to inhibit functions of externalized PS. Platelet-derived microparticles, long studied in their own right, are increasingly appreciated as a member of the broader EV category. Externalized PS on platelet-derived microparticles causes thrombosis, or coagulation occurring within a blood vessel. Cancer is associated with a substantially elevated risk of thrombosis (Noble and Pasi, 2010), and EVs released by cancer cells may promote hypercoagulability. MARCKS-ED may act as an anticoagulant by blocking this externalized PS.
It remains to be seen whether MARCKS-ED also blocks the ‘eat-me’ signal to reduce uptake of PS-exposing cells or EVs, as has been observed for annexin A5 (Mulcahy et al., 2014). Recombinant annexin A5 has been investigated for its ability to reduce inflammation (Ewing et al., 2011). MARCKS-derived peptides also bind to PS-exposing membranes, so it stands to reason that if annexin A5 is able to reduce inflammation, MARCKS-derived peptides may do so as well. Externalized PS also can activate the complement cascade. The role of EVs in activating complement cascade is unclear, as many EVs express complement inhibitors on their surfaces (Clayton et al., 2003), although recent research has found increased complement activation from metastatic cancer cell EVs (Whitehead et al., 2015). It would be informative to investigate whether MARCKS-derived peptides inhibit activation of the complement cascade in the context of tumor microenvironmental physiology.
Although mechanisms of EV cellular uptake are still an area of active investigation, it is possible that by binding to specific EVs, MARCKS-ED-conjugated drugs might be endocytosed, constituting a targeted delivery system. Exosome-sized vesicles traffic between malaria-infected red blood cells in order to increase parasite survival under stress (Regev-Rudzki et al., 2013). If MARCKS-ED binds to these EVs, drug-peptide conjugates could potentially be used to prevent parasite communication with the upside of delaying drug resistance. It is likely that the role of EVs in pathology is only beginning to be understood.
To discover the minimal residues required for curvature sensing by MARCKS-ED, systematic truncation must be performed in order to determine the minimum active sequence. Further residue analysis and chemical optimization for curvature-sensing properties could be accomplished through deletion, alanine-scanning, and cyclization in order to increase binding affinity or specificity of MARCKS-derived peptides for different EV types. It would be informative to test the membrane binding of peptide derivatives of the ED of MARCKS-related protein, a MARCKS homolog with a nearly identical ED, with the notable difference of a serine to proline substitution. Whether the conformational rigidity of this residue affects membrane curvature sensing remains unknown.
In one further extension of the lipid-targeting labeled peptide approach, it may be possible to probe membrane morphology and PS dynamics within live cells using a genetically encoded MARCKS-derived peptide. A previously reported genetically encoded biosensor for membrane PS utilized a known membrane-binding protein fused to a fluorescent protein (Yeung et al., 2008). Understanding the dynamics of subcellular localization of PS using a small, sensitive peptide probe could provide insights into membrane protein localization and regulation. Using bioorthogonal ‘click’ chemistry (Jewett and Bertozzi, 2010) to label curvature probes with small molecule fluorophores could also provide an advanced method to visualize membrane curvature and lipid composition without relying on large fluorescent proteins. According to the ‘electrostatic switch’ model (McLaughlin and Aderem, 1995), highly positive peripheral membrane proteins such as MARCKS and KRAS localize to the cytosol upon reduction of their positive charge. However, as suggested by results showing moderate levels of PS on endomembranes, moderately positive membrane proteins may localize to endomembranes (Yeung et al., 2008). It would be possible to create MARCKS-derived phosphomimics by replacing serine residues with negatively charged aspartate or glutamate residues and comparing the membrane localization. If cytosolic-facing PS is found on other membranes, perhaps regulated PS flipping plays an underappreciated functional role in these contexts as well.
Outlook
Lipid-targeting peptide probes such as MARCKS-ED have proven to offer a novel approach to characterizing EVs by binding highly curved lipid membranes and probing activated platelets by binding externalized PS. It remains to be seen whether MARCKS-derived peptide sensors will be capable of distinguishing transient PS externalization in live cells. If MARCKS-ED indeed has a higher sensitivity for low levels of externalized PS than does annexin A5, it may be possible to detect more transient PS exposure on cell membranes. MARCKS-ED thus could be employed as fluorescent biosensors for externalized PS associated with calcium signaling, apoptosis, or other cellular activation; or for membrane curvature, allowing for EV detection. On the intracellular side of the plasma membrane, genetically encoded MARCKS-derived peptides may also act as sensors for PS, charge, or membrane curvature. MARCKS-derived peptides offer a novel approach to probing the dynamics of PS and membrane curvature in cellular signaling. These lipid-targeting strategies offer the possibility of probing EVs ex vivo or in vivo.
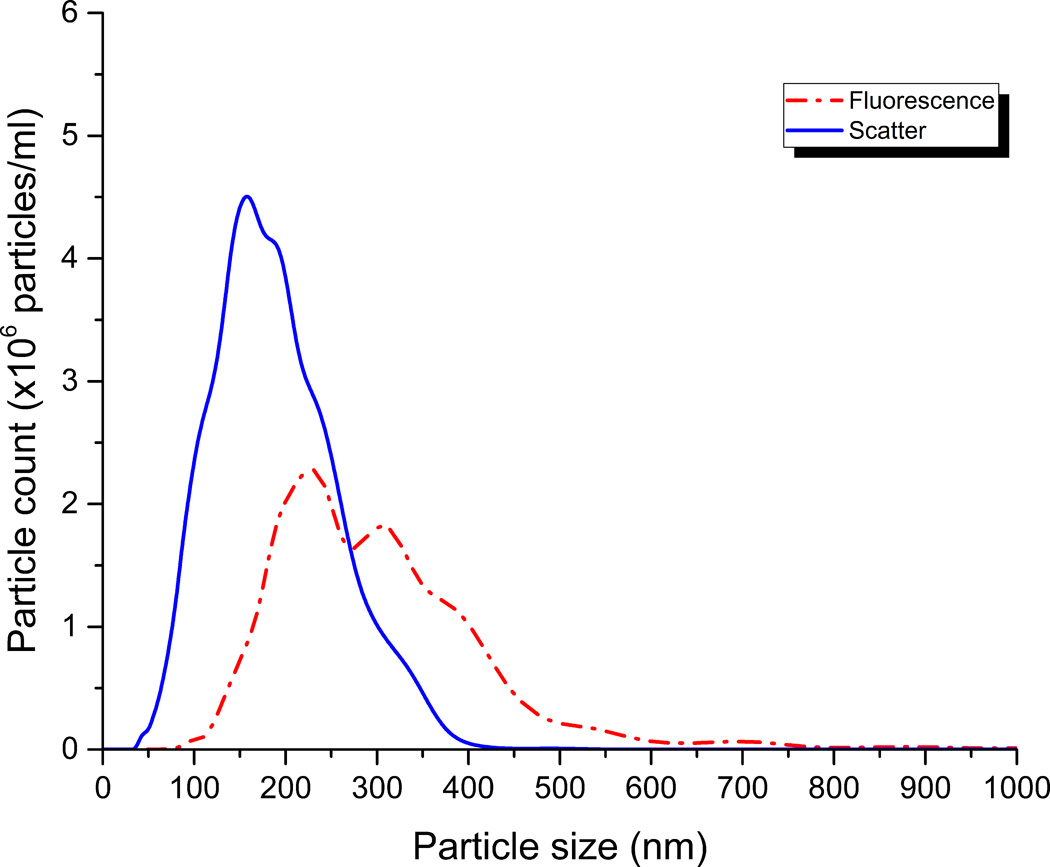
Figure 2. Representative size distributions of EVs and peptide-labeled EVs by nanoparticle tracking analysis.
Alexa 546-labeled MARCKS-ED peptide (110 nM) was incubated with pooled human plasma (1:2000 dilution in HEPES-buffered saline) and analyzed by nanoparticle tracking analysis (NanoSight). Size distributions from representative individual videos taken in scatter mode (blue) to report EV size distribution, and fluorescence mode (red) to report MARCKS-labeled EV size distribution. This illustrates the potential for using fluorescently labeled peptides to probe vesicles from complex ex vivo samples and suggests the presence of a subpopulation of PS+ vesicles.
Acknowledgments
We thank the National Institutes of Health (NIH R01GM103843) for financial support. A.F. is supported by the Signaling and Cellular Regulation Training Program (NIH T32GM08759). We thank Noah Kastelowitz for proofreading the manuscript.
Literature Cited
- Aimon S, Callan-Jones A, Berthaud A, Pinot M, Toombes GES, Bassereau P. Membrane shape modulates transmembrane protein distribution. Dev Cell. 2014;28:212–218. doi: 10.1016/j.devcel.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andree H, Stuart M, Hermens W, Reutelingsperger C, Hemker H, Frederik P, Willems G. Clustering of lipid-bound annexin V may explain its anticoagulant effect. J Biol Chem. 1992;267:17907–17912. [PubMed] [Google Scholar]
- Antonny B. Mechanisms of membrane curvature sensing. Annu Rev Biochem. 2011;80:101–123. doi: 10.1146/annurev-biochem-052809-155121. [DOI] [PubMed] [Google Scholar]
- Arbuzova A, Schmitz AAP, Vergères G. Cross-talk unfolded: MARCKS proteins. Biochem J. 2002;362:1–12. doi: 10.1042/0264-6021:3620001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian K, Mirnikjoo B, Schroit AJ. Regulated externalization of phosphatidylserine at the cell surface: implications for apoptosis. J Biol Chem. 2007;282:18357–18364. doi: 10.1074/jbc.M700202200. [DOI] [PubMed] [Google Scholar]
- Bogaart G van den, Meyenberg K, Risselada HJ, Amin H, Willig KI, Hubrich BE, Dier M, Hell SW, Grubmüller H, Diederichsen U, et al. Membrane protein sequestering by ionic protein-lipid interactions. Nature. 2011;479:552–555. doi: 10.1038/nature10545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown L, Wolf JM, Prados-Rosales R, Casadevall A. Through the wall: extracellular vesicles in Gram-positive bacteria, mycobacteria and fungi. Nat Rev Microbiol. 2015;13:620–630. doi: 10.1038/nrmicro3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter KP, Young AM, Palmer AE. Fluorescent sensors for measuring metal ions in living systems. Chem Rev. 2014;114:4564–4601. doi: 10.1021/cr400546e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton A, Harris CL, Court J, Mason MD, Morgan BP. Antigen-presenting cell exosomes are protected from complement-mediated lysis by expression of CD55 and CD59. Eur J Immunol. 2003;33:522–531. doi: 10.1002/immu.200310028. [DOI] [PubMed] [Google Scholar]
- Colombo M, Raposo G, Théry C. Biogenesis, Secretion, and Intercellular Interactions of Exosomes and Other Extracellular Vesicles. Annu Rev Cell Dev Biol. 2014;30:255–289. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- Cossetti C, Iraci N, Mercer TR, Leonardi T, Alpi E, Drago D, Alfaro-Cervello C, Saini HK, Davis MP, Schaeffer J, et al. Extracellular vesicles from neural stem cells transfer IFN-γ via Ifngr1 to activate Stat1 signaling in target cells. Mol Cell. 2014;56:193–204. doi: 10.1016/j.molcel.2014.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaux PF, Fellmann P, Hervé P. Investigation on lipid asymmetry using lipid probes. Chem Phys Lipids. 2002;116:115–134. doi: 10.1016/s0009-3084(02)00023-3. [DOI] [PubMed] [Google Scholar]
- Ewing MM, Vries MRde, Nordzell M, Pettersson K, Boer HC de, Zonneveld AJ van, Frostegård J, Jukema JW, Quax PHA. Annexin A5 therapy attenuates vascular inflammation and remodeling and improves endothelial function in mice. Arterioscler Thromb Vasc Biol. 2011;31:95–101. doi: 10.1161/ATVBAHA.110.216747. [DOI] [PubMed] [Google Scholar]
- Fadeel B, Xue D. The ins and outs of phospholipid asymmetry in the plasma membrane: roles in health and disease. Crit Rev Biochem Mol Biol. 2009 doi: 10.1080/10409230903193307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Zheng H. Illuminating the lipidome to advance biomedical research: peptide-based probes of membrane lipids. Future Med Chem. 2013;5:947–959. doi: 10.4155/fmc.13.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner C, Ferreira YJ, Dragovic RA, Redman CWG, Sargent IL. Extracellular vesicle sizing and enumeration by nanoparticle tracking analysis. J Extracell Vesicles. 2013;2 doi: 10.3402/jev.v2i0.19671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garland M, Yim JJ, Bogyo M. A Bright Future for Precision Medicine: Advances in Fluorescent Chemical Probe Design and Their Clinical Application. Cell Chem Biol. 2016;23:122–136. doi: 10.1016/j.chembiol.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood JL, San RS, Wickline SA. Exosomes Released by Melanoma Cells Prepare Sentinel Lymph Nodes for Tumor Metastasis. Cancer Res. 2011;71:3792–3801. doi: 10.1158/0008-5472.CAN-10-4455. [DOI] [PubMed] [Google Scholar]
- Italiano JE, Mairuhu ATA, Flaumenhaft R. Clinical relevance of microparticles from platelets and megakaryocytes. Curr Opin Hematol. 2010;17:578–584. doi: 10.1097/MOH.0b013e32833e77ee. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewett JC, Bertozzi CR. Cu-free click cycloaddition reactions in chemical biology. Chem Soc Rev. 2010;39:1272. doi: 10.1039/b901970g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastelowitz N, Yin H. Exosomes and microvesicles: identification and targeting by particle size and lipid chemical probes. Chembiochem. 2014;15:923–928. doi: 10.1002/cbic.201400043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenis H, Genderen H van, Bennaghmouch A, Rinia HA, Frederik P, Narula J, Hofstra L, Reutelingsperger CPM. Cell surface-expressed phosphatidylserine and annexin A5 open a novel portal of cell entry. J Biol Chem. 2004;279:52623–52629. doi: 10.1074/jbc.M409009200. [DOI] [PubMed] [Google Scholar]
- Lemmon MA. Membrane recognition by phospholipid-binding domains. Nat Rev Mol Cell Biol. 2008;9:99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- McLaughlin S, Aderem A. The myristoyl-electrostatic switch: a modulator of reversible protein-membrane interactions. Trends Biochem Sci. 1995;20:272–276. doi: 10.1016/s0968-0004(00)89042-8. [DOI] [PubMed] [Google Scholar]
- McLaughlin S, Murray D. Plasma membrane phosphoinositide organization by protein electrostatics. Nature. 2005;438:605–611. doi: 10.1038/nature04398. [DOI] [PubMed] [Google Scholar]
- McMahon HT, Gallop JL. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 2005;438:590–596. doi: 10.1038/nature04396. [DOI] [PubMed] [Google Scholar]
- Melo SA, Luecke LB, Kahlert C, Fernandez AF, Gammon ST, Kaye J, LeBleu VS, Mittendorf EA, Weitz J, Rahbari N, et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015;523:177–182. doi: 10.1038/nature14581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrifield RB. Solid Phase Peptide Synthesis. I. The Synthesis of a Tetrapeptide. J Am Chem Soc. 1963;85:2149–2154. [Google Scholar]
- Morel O, Jesel L, Freyssinet J-M, Toti F. Cellular Mechanisms Underlying the Formation of Circulating Microparticles. Arterioscler Thromb Vasc Biol. 2010;31:15–26. doi: 10.1161/ATVBAHA.109.200956. [DOI] [PubMed] [Google Scholar]
- Morton LA, Saludes JP, Yin H. Constant pressure-controlled extrusion method for the preparation of Nano-sized lipid vesicles. J Vis Exp. 2012:e4151. doi: 10.3791/4151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton LA, Tamura R, Jesus AJ de, Espinoza A, Yin H. Biophysical investigations with MARCKS-ED: dissecting the molecular mechanism of its curvature sensing behaviors. Biochim Biophys Acta. 2014;1838:3137–3144. doi: 10.1016/j.bbamem.2014.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton LA, Yang H, Saludes JP, Fiorini Z, Beninson L, Chapman ER, Fleshner M, Xue D, Yin H. MARCKS-ED peptide as a curvature and lipid sensor. ACS Chem Biol. 2013;8:218–225. doi: 10.1021/cb300429e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulcahy LA, Pink RC, Carter DRF. Routes and mechanisms of extracellular vesicle uptake. J Extracell Vesicles. 2014;3 doi: 10.3402/jev.v3.24641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralidharan-Chari V, Clancy J, Plou C, Romao M, Chavrier P, Raposo G, D’Souza-Schorey C. ARF6-regulated shedding of tumor cell-derived plasma membrane microvesicles. Curr Biol. 2009;19:1875–1885. doi: 10.1016/j.cub.2009.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble S, Pasi J. Epidemiology and pathophysiology of cancer-associated thrombosis. Br J Cancer. 2010;102(Suppl):S2–S9. doi: 10.1038/sj.bjc.6605599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pol E van der, Coumans FAW, Grootemaat AE, Gardiner C, Sargent IL, Harrison P, Sturk A, Leeuwen TG van, Nieuwland R. Particle size distribution of exosomes and microvesicles determined by transmission electron microscopy, flow cytometry, nanoparticle tracking analysis, and resistive pulse sensing. J Thromb Haemost. 2014;12:1182–1192. doi: 10.1111/jth.12602. [DOI] [PubMed] [Google Scholar]
- Rauch ME, Ferguson CG, Prestwich GD, Cafiso DS. Myristoylated alanine-rich C kinase substrate (MARCKS) sequesters spin-labeled phosphatidylinositol 4,5-bisphosphate in lipid bilayers. J Biol Chem. 2002;277:14068–14076. doi: 10.1074/jbc.M109572200. [DOI] [PubMed] [Google Scholar]
- Regev-Rudzki N, Wilson DW, Carvalho TG, Sisquella X, Coleman BM, Rug M, Bursac D, Angrisano F, Gee M, Hill AF, et al. Cell-cell communication between malaria-infected red blood cells via exosome-like vesicles. Cell. 2013;153:1120–1133. doi: 10.1016/j.cell.2013.04.029. [DOI] [PubMed] [Google Scholar]
- Reynwar BJ, Illya G, Harmandaris VA, Müller MM, Kremer K, Deserno M. Aggregation and vesiculation of membrane proteins by curvature-mediated interactions. Nature. 2007;447:461–464. doi: 10.1038/nature05840. [DOI] [PubMed] [Google Scholar]
- Shen B, Wu N, Yang J-M, Gould SJ. Protein targeting to exosomes/microvesicles by plasma membrane anchors. J Biol Chem. 2011;286:14383–14395. doi: 10.1074/jbc.M110.208660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subra C, Laulagnier K, Perret B, Record M. Exosome lipidomics unravels lipid sorting at the level of multivesicular bodies. Biochimie. 2007;89:205–212. doi: 10.1016/j.biochi.2006.10.014. [DOI] [PubMed] [Google Scholar]
- Südhof TC, Rothman JE. Membrane fusion: grappling with SNARE and SM proteins. Science. 2009;323:474–477. doi: 10.1126/science.1161748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki J, Fujii T, Imao T, Ishihara K, Kuba H, Nagata S. Calcium-dependent phospholipid scramblase activity of TMEM16 protein family members. J Biol Chem. 2013;288:13305–13316. doi: 10.1074/jbc.M113.457937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tait JF, Gibson D. Phospholipid binding of annexin V: effects of calcium and membrane phosphatidylserine content. Arch Biochem Biophys. 1992;298:187–191. doi: 10.1016/0003-9861(92)90111-9. [DOI] [PubMed] [Google Scholar]
- Théry C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006;Chapter 3(Unit 3):22. doi: 10.1002/0471143030.cb0322s30. [DOI] [PubMed] [Google Scholar]
- Thompson MA, Lew MD, Moerner WE. Extending microscopic resolution with single-molecule imaging and active control. Annu Rev Biophys. 2012;41:321–342. doi: 10.1146/annurev-biophys-050511-102250. [DOI] [PubMed] [Google Scholar]
- Tonnesen A, Christensen SM, Tkach V, Stamou D. Geometrical membrane curvature as an allosteric regulator of membrane protein structure and function. Biophys J. 2014;106:201–209. doi: 10.1016/j.bpj.2013.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, Wieland F, Schwille P, Brügger B, Simons M. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science. 2008;319:1244–1247. doi: 10.1126/science.1153124. [DOI] [PubMed] [Google Scholar]
- Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- Vanni S, Hirose H, Barelli H, Antonny B, Gautier R. A sub-nanometre view of how membrane curvature and composition modulate lipid packing and protein recruitment. Nat Commun. 2014;5:4916. doi: 10.1038/ncomms5916. [DOI] [PubMed] [Google Scholar]
- Vlieghe P, Lisowski V, Martinez J, Khrestchatisky M. Synthetic therapeutic peptides: science and market. Drug Discov Today. 2010;15:40–56. doi: 10.1016/j.drudis.2009.10.009. [DOI] [PubMed] [Google Scholar]
- Westrate LM, Lee JE, Prinz WA, Voeltz GK. Form Follows Function: The Importance of Endoplasmic Reticulum Shape. Annu Rev Biochem. 2015;84:150112144356008. doi: 10.1146/annurev-biochem-072711-163501. [DOI] [PubMed] [Google Scholar]
- Whitehead B, Wu L, Hvam ML, Aslan H, Dong M, Dyrskjøt L, Ostenfeld MS, Moghimi SM, Howard KA. Tumour exosomes display differential mechanical and complement activation properties dependent on malignant state: implications in endothelial leakiness. J Extracell Vesicles. 2015;4 doi: 10.3402/jev.v4.29685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Jesus AJ de, Tamura R, Li V, Cheng K, Yin H. Curvature sensing MARCKS-ED peptides bind to membranes in a stereo-independent manner. J Pept Sci. 2015 doi: 10.1002/psc.2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Chen Y-Z, Zhang Y, Wang X, Zhao X, Godfroy JI, Liang Q, Zhang M, Zhang T, Yuan Q, et al. A lysine-rich motif in the phosphatidylserine receptor PSR-1 mediates recognition and removal of apoptotic cells. Nat Commun. 2015;6:5717. doi: 10.1038/ncomms6717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung T, Gilbert GE, Shi J, Silvius J, Kapus A, Grinstein S. Membrane phosphatidylserine regulates surface charge and protein localization. Science. 2008;319:210–213. doi: 10.1126/science.1152066. [DOI] [PubMed] [Google Scholar]
- Zhang W, Crocker E, McLaughlin S, Smith SO. Binding of peptides with basic and aromatic residues to bilayer membranes: phenylalanine in the myristoylated alanine-rich C kinase substrate effector domain penetrates into the hydrophobic core of the bilayer. J Biol Chem. 2003;278:21459–21466. doi: 10.1074/jbc.M301652200. [DOI] [PubMed] [Google Scholar]
- Zomer A, Maynard C, Verweij FJ, Kamermans A, Schäfer R, Beerling E, Schiffelers RM, de Wit E, Berenguer J, Ellenbroek SIJ, et al. In Vivo Imaging Reveals Extracellular Vesicle-Mediated Phenocopying of Metastatic Behavior. Cell. 2015;161:1046–1057. doi: 10.1016/j.cell.2015.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwaal RFA, Comfurius P, Bevers EM. Surface exposure of phosphatidylserine in pathological cells. Cell Mol Life Sci. 2005;62:971–988. doi: 10.1007/s00018-005-4527-3. [DOI] [PubMed] [Google Scholar]