Abstract
Molecular recognition of carbohydrates plays vital roles in biology but has been difficult to achieve with synthetic receptors. Through covalent imprinting of carbohydrates in boroxole-functionalized cross-linked micelles, we prepared nanoparticle receptors for a wide variety of mono- and oligosaccharides. The boroxole functional monomer bound the sugar templates through cis-1,2-diol, cis-3,4-diol, and trans-4,6-diol. The protein-sized nanoparticles showed excellent selectivity for D-aldohexoses in water with submillimolar binding affinities and completely distinguished the three biologically important hexoses (glucose, mannose, and galactose). Glycosides with nonpolar aglycon showed stronger binding due to enhanced hydrophobic interactions. Oligosaccharides were distinguished based on their monosaccharide building blocks, glycosidic linkages, chain length, as well as additional functional groups that could interact with the nanoparticles.
Graphical abstract
INTRODUCTION
Carbohydrates occupy a unique place in biology. Unlike peptides and nucleic acids, they comprise entirely of hydrophilic building blocks and are thus solvated strongly by water. This feature implies that carbohydrates tend to cover the surface of a cell and represent the first line of interaction when other entities approach the cell. For this reason, it is not surprising that carbohydrates are involved in many important biological processes including fertilization, cell–cell interactions, immune response, and viral and bacterial infection.1–3 In addition, they are important sources of energy for most organisms and form parts of the backbone for DNAs and RNAs.
Lectins are protein receptors that perform molecular recognition of carbohydrates in nature. During the last several decades, chemists have devoted great efforts towards developing synthetic analogues of lectins that can bind sugars or their derivatives selectively.1–6 On the applied level, the research potentially can lead to tools useful in the study and intervention of carbohydrate-related biological processes. On the fundamental level, the research tackles one of the most difficult challenges in supramolecular chemistry.
Selective binding of carbohydrates in water is difficult for multiple reasons. Due to strong interactions between water and the hydroxyls of a carbohydrate, a supramolecular host in aqueous solution has to pay a tremendous amount of desolvation energy to bind its sugar guest. Unlike proteins and DNAs, carbohydrates do not adopt well-defined three-dimensional conformations, making the design of their complementary hosts difficult. Monosaccharides, the building blocks of more complex carbohydrates, differ minutely in structure, often by the stereochemistry of a single hydroxyl. Even with the same building block, slightly different connections between the monomers lead to oligo- and polysaccharides with completely different physical, chemical, and biological properties.
Molecular recognition of carbohydrates has progressed steadily in the last decades. Over the years, synthetic receptors moved from organic to aqueous solution; carbohydrate guests being studied transitioned from simple monosaccharides to functionalized oligosaccharides. Chemists nowadays are able to distinguish glucosides from their isomeric sugars by their all equatorial substitutions.7,8 Binding affinities for monosaccharides by synthetic receptors in water could approach those by natural lectins (binding constant Ka = 103–104 M−1).1,2 Despite these impressive accomplishments, however, a general method for molecular recognition of carbohydrates in water is still not available, due to the many challenges mentioned above.
Synthetic carbohydrate receptors can be classified in two groups, depending on whether noncovalent or covalent bonds are used for binding. The first group often utilizes strategically positioned hydrogen bonds in a relatively hydrophobic microenvironment to bind the guest.6–10 The second group largely relies on the fast and reversible boronate bonds formed between organic boronic acids and the diol functionalities on a sugar for the molecular recognition.5,11–15
We recently reported a method to construct molecularly imprinted nanoparticles (MINPs) with precisely positioned boronic acids to recognize monosaccharides in water.16 The MINP receptors could distinguish D-aldohexoses with remarkable selectivity. For example, MINP(glucose), i.e., MINP prepared with glucose as the template, bound glucose with Ka = 1.18 × 103 M−1. Any change in the C2, C4, or C6 hydroxyl essentially turned off the binding and inversion of the C3 hydroxyl weakened the binding by over two-fold.
Unfortunately, although the boronic acid-functionalized MINPs showed impressive binding for monosaccharides, the synthetic method could not be easily applied to oligosaccharides. Herein, we report that, by modifying the key ingredients in the MINP preparation (i.e., the cross-linkable surfactant, the cross-linker, and the sugar-binding functional monomer) and the imprinting procedure, we now can create nanoparticle receptors for oligosaccharides (and monosaccharides) directly in water. The generality and simplicity of the in situ imprinting are the highlights of this approach. The preparation and purification took about 2 days and required no special techniques, and thus could be potentially adopted by researchers without substantial training in chemistry. These receptors are soluble in water, resemble proteins in size, and displayed selectivity for monosaccharides and oligosaccharides that has not been achieved by previous synthetic materials.
RESULTS AND DISCUSSION
Design and Synthesis
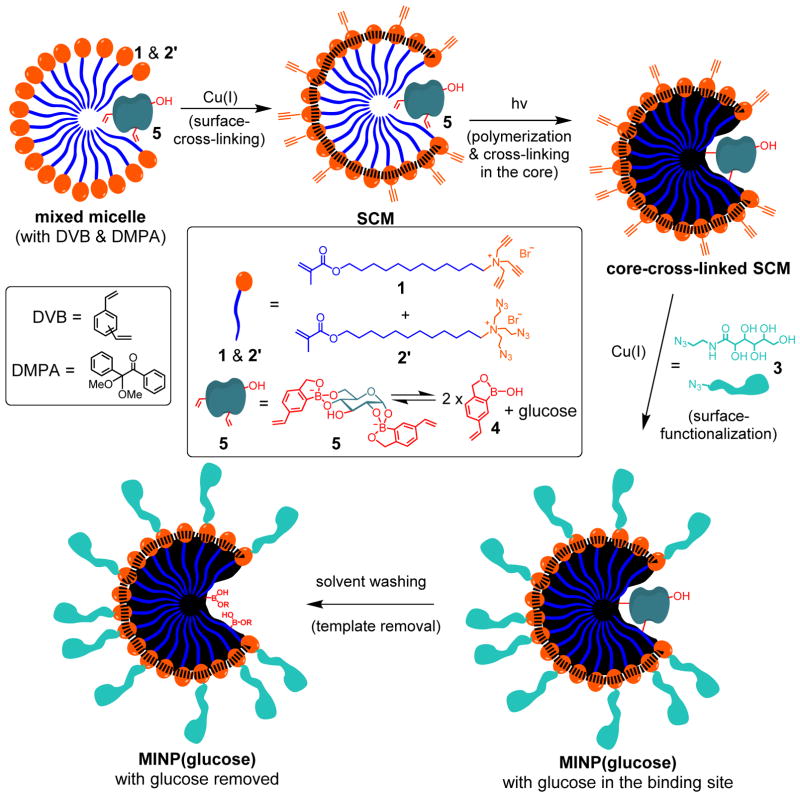
Molecular imprinting is a tremendously useful technique for creating guest-complementary binding sites in polymers or on surface.17–28 However, conventional imprinting often produces intractable highly cross-linked polymers, hindering their usage in biology. To make the imprinted materials soluble in water, we recently reported a process to imprint within cross-linked micelles. Because the polymerization and cross-linking took place within the micelle boundaries, the resulting nanoparticles become fully soluble in water due to their hydrophobic/hydrophilic core–shell structure.29,30
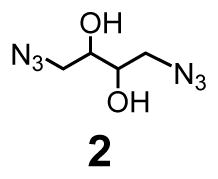
MINPs are generally prepared by first solubilizing a hydrophobic template molecule with the micelles of a cross-linkable surfactant such as 1 (see Scheme 1 for structure). The surfactant contains a propargylated headgroup and a methacrylate-containing hydrophobic tail that undergo orthogonal cross-linking chemistries. Cross-linking by a diazide cross-linker such as 2 yields alkyne-functionalized surface-cross-linked micelles (SCMs), which can be functionalized by another round of click reaction with an azide-containing ligand such as 3 (see Scheme 1 for structure). Afterwards, free radical core-cross-linking leads to the formation of a polymer matrix around the template within the SCM, and thus creates the binding site in the micellar core complementary to the template in size, shape, and binding functionality.
Scheme 1.
Preparation of boroxole-functionalized MINP(glucose).
The templates used in the D-aldohexose-binding MINPs were the boronate esters formed from the sugars and 4-vinylphenylboronic acid.16 They had to be synthesized in a separate step prior to the MINP preparation through azeotropic removal of water in dioxane at 88 °C.31 Because oligosaccharides generally have extremely low solubility in dioxane and many organic solvents, this method is not suitable for imprinting oligosaccharides. If we want to imprint more sensitive sugar derivatives such as glycoproteins in a longer term, organic solvents and high temperatures clearly have to be avoided.
In this work, we synthesized boroxole-containing functional monomer (FM) 432 and a new cross-linker 2′ to address the above challenges (Scheme 1). Benzoboroxole is known to bind 1,2- and 1,3-diols with higher affinities than phenylboronic acid33,34 and have been used to create sugar-binding polymers.35–43 We reasoned that the anionic boronate derivative formed (i.e., 5) might be especially stable in the cationic micelles of 1. (As will be shown later, the structure of 5 was inferred from our binding studies, as well as the binding property of boroxole.)33,34 If the complex can survive the surface- and core-cross-linking of the micelles, we would be able to imprint a sugar directly in the micellar solution. In situ imprinting is highly desirable because it eliminates the separate template preparation and may be more compatible with templates sensitive to organic solvents and/or high temperatures.
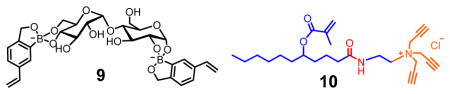
There are two considerations behind the design of cross-linker 2′. First, since a noncovalently formed FM•template complex (i.e., 5) is involved, we have to avoid other diol-containing molecules such as 2 in the MINP preparation, at least prior to the formation of the binding site. Second, 2′ is amphiphilic and expected to form mixed micelles with 1, enabling the alkyne and azide groups to be intimately mixed on the surface of the micelles and in close proximity to one another. As a result, the local concentrations of the reactive groups are exceedingly high on the micelle surface, making the surface cross-linking particularly facile.44,45
As usual, we solubilized DVB (a free radical cross-linker) and DMPA (a photoinitiator) in the (mixed) micelles prior to any cross-linking. The presence of DVB increases the cross-linking density of the core and was confirmed previously to be important to the molecular recognition of the final MINP.29 The 3:2 ratio of 1 and 2′ left the SCM with alkynyl groups on the surface.44–46
Normally, we perform surface-functionalization before core-cross-linking because it uses the same Cu(I) catalysts as the surface-cross-linking step and thus can be conveniently done right afterwards. However, because the surface ligand (3) contains many hydroxyls and is expected to compete with glucose for the boroxole binding group, we reversed the order and performed the core-cross-linking in the second step, via UV-initiated radical polymerization of 1, 2′, 5, and DVB.
At this point, the binding site was already formed inside the surface- and core- doubly-cross-linked micelles. Surface-functionalization with 4 using the click reaction afforded MINP(glucose) with the template still bound in the binding site. The sugar-derived ligand 4 was installed so that the final nanoparticles could be easily recovered by precipitation into acetone.29 The template molecules were removed by repeated washing using acetone/water, methanol/acetic acid, and acetone. The power obtained was completely soluble in water.
The reaction progress was generally monitored by 1H NMR spectroscopy.29,30 Dynamic light scattering (DLS) afforded the size and molecular weight of the MINP. The nanoparticles were typically 4–5 nm in diameter. In our experience, the DLS-determined size showed good agreement with the size obtained from transmission electron microscopy (TEM) for similarly cross-linked micelles.44
MINPs for Binding Monosaccharides
We examined the binding of the MINP by isothermal by isothermal titration calorimetry (ITC), a method of choice for studying intermolecular interactions.47 In addition to its accuracy, the method affords the number of binding sites per particle (N), as well as other thermodynamic binding parameters. We have demonstrated in several studies that (for fluorescently labeled guests) ITC gave very similar binding constants for MINPs as other spectroscopic methods.29,30,48
As shown in Table 1, MINP(glucose) prepared with template/FM = 1:2 bound glucose with Ka = 2.30 × 103 M−1 in 10 mM HEPES buffer at pH 7.4 (entry 1). Binding was somewhat weaker at pH 8.5 or 6.5 (entries 6 and 7). Reducing the template/FM ratio to 1:1 lowered the binding constant (entry 2). Having an excess of FM (thee equiv to the template) did not improve the binding (entry 3). Binding was negligible by the nonimprinted materials prepared without FM 3 and the glucose template (entry 4) or with FM 3 but without glucose (entry 5). These results demonstrated that molecular imprinting was clearly in operation and the optimal binding stoichiometry was 1:2 between the template and the boroxole.49
Table 1.
ITC binding data for monosaccharide guests.a
| Entry | Host | Guest | Ka (× 103 M−1) | −ΔG (kcal/mol) | N |
|---|---|---|---|---|---|
| 1 | MINP(glucose) | glucose | 2.30 ± 0.11 | 4.58 | 1.1 ± 0.1 |
| 2 | MINP(glucose)b | glucose | 0.95 ± 0.01 | 4.06 | 1.2 ± 0.1 |
| 3 | MINP(glucose)c | glucose | 2.33 ± 0.38 | 4.59 | 1.0 ± 0.1 |
| 4 | NINPd | glucose | <0.05e | - | - |
| 5 | NINPf | glucose | <0.05e | - | - |
| 6 | MINP(glucose) | glucoseg | 1.30 ± 0.16 | 4.24 | 1.0 ± 0.1 |
| 7 | MINP(glucose) | glucoseh | 0.52 ± 0.09 | 3.70 | 1.1 ± 0.1 |
| 8 | MINP(glucose) | allosei | 0.37 ± 0.09 | 3.51 | 0.8 ± 0.1 |
| 9 | MINP(mannose) | mannose | 1.90 ± 0.34 | 4.47 | 1.0 ± 0.3 |
| 10 | MINP(mannose) | altrosej | 0.50 ± 0.01 | 3.68 | 1.0 ± 0.1 |
| 11 | MINP(galactose) | galactosek | 3.37 ± 0.30 | 4.81 | 1.0 ± 0.1 |
| 12 | MINP(6) | 6 | 65.3 ± 8.8 | 6.56 | 1.1 ± 0.1 |
| 13 | MINP(6) | 7 | 11.0 ± 1.2 | 5.51 | 1.0 ± 0.1 |
| 14 | MINP(6) | 8 | 4.66 ± 0.39 | 5.00 | 1.1 ± 0.1 |
The FM/template ratio in the MINP synthesis was 1:2 unless otherwise indicated. The titrations were performed in 10 mM HEPES buffer at pH 7.4. The ITC titration curves are reported in the Supporting Information, including the binding enthalpy and entropy.
The template/FM ratio was 1:1.
The template/FM ratio was 1:3.
Prepared without FM 3 and the glucose template.
Binding was extremely weak. Because the binding constant was estimated from ITC, -ΔG and N are not listed in the table (Figure 62S in Supporting Information).
Prepared with FM 4 but without the glucose template.
The binding was in 10 mM HEPES buffer at pH 8.5.
The binding was in 10 mM HEPES buffer at pH 6.5.
The binding for other D-aldohexoses including mannose, galactose, altrose, gulose, talose, idose, and xylose was extremely weak, with estimated Ka <0.02 × 103 M−1 (Figure 66S and 67S).
The binding for other D-aldohexoses including glucose, allose, galactose, gulose, talose, and idose was extremely weak, with estimated Ka <0.02 × 103 M−1 (Figure S68).
The binding for other D-aldohexoses including glucose, mannose, allose, altrose, gulose, talose, and idose was extremely weak, with estimated Ka <0.05 × 103 M−1 (Figure 69S and 70S).
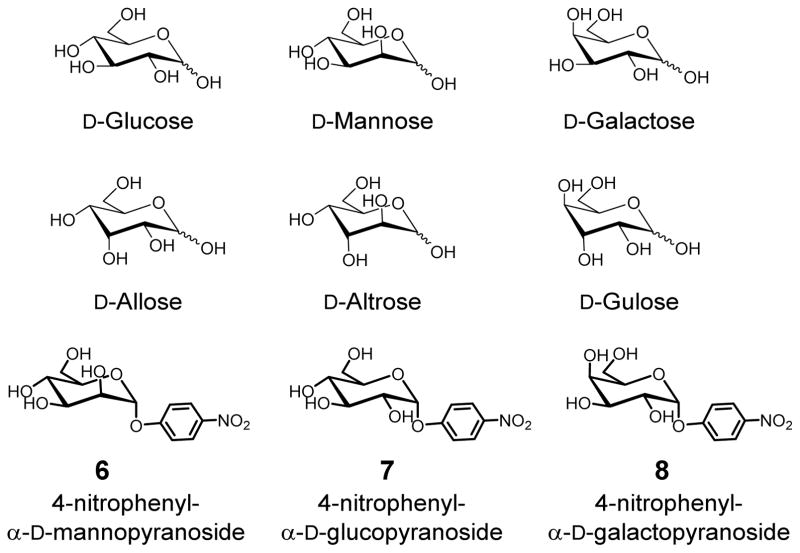
MINP(glucose) displayed excellent selectivity: among the seven isomeric sugars, only allose showed noticeable binding with Ka = 0.37 × 103 M−1, while the rest were not bound at all (Chart 1). Similar selectivity was found for MINP(mannose), which only bound altrose among the remaining seven D-aldohexoses.
Chart 1.
Structures of selected D-aldohexoses and glycosides.
The boroxole-functionalized MINP(glucose) and MINP(mannose) showed higher binding selectivity than the boronic acid-functionalized MINPs, but the trend remained the same.16 The selectivity suggests that the C2 and C4 hydroxyls were critical to the molecular recognition and any inversion at these positions shuts off the binding. The C6 hydroxyl was also essential, as xylose, lacking this hydroxyl, showed no binding. The C3 hydroxyl played a secondary role in the binding, with its inversion lowering Ka by 74–86% from the template sugar.
MINP(galactose), on the other hand, behaved distinctively differently. Among the eight D-aldohexoses, it bound only its template and achieved stronger binding (Ka = 3.37 × 103 M−1) than either MINP(glucose) or MINP(mannose) for its template (Table 1).
Hall and co-workers reported that benzoboroxole binds glucose in a 1:1 ratio in water, with Ka = 17 M−1.33,34 It is possible that the 2nd binding observed in our MINPs was weaker than the first one in bulk aqueous solution and simply not observed in Hall’s study. The hydrophobic and positive environment of the cationic micelle conceivably could stabilize the negatively charged boronate and enable the second, less stable adduct to form under our imprinting and binding conditions.
Benzoboroxole binds the methyl pyranosides of glucose, mannose, and galactose with Ka = 10–30 M−1,33,34 thus lacking intrinsic selectivity for these sugars. The much higher selectivity and binding affinity displayed by our MINPs must come from the microenvironment of the cross-linked micelle and the two-point binding as revealed in the binding studies. It is known that that benzoboroxole has a strong preference for trans-4,6-diol over trans-3,4-diol in glucosides, suggesting the C3 hydroxyl would not be involved in binding in glucose and mannose.50 Hall’s work also demonstrated that, for galatopyranosides, cis-3,4-diol is preferred by boroxole over cis-4,6-diol. This preference was also maintained by MINP(galactose), because gulose, which differs from galactose only by the C3 hydroxyl and contains the cis-4,6-diol, was not bound.51
For MINP(6) prepared with 4-nitrophenyl α-D-mannopyranoside 6 as the template, the aromatic aglycon was expected to create a complementary hydrophobic binding pocket in the MINP, as we have demonstrated in several recent studies.29,30,48 Indeed, a much stronger binding of Ka = 65.3 × 103 M−1 was obtained. Gratifyingly, excellent binding selectivity was maintained for this MINP. The Ka values for the corresponding glucoside 7 and galactoside 8 were ~1/6 and 1/14, respectively. Thus, inversion of one or two hydroxyl groups was easily distinguished in the glycosides as well.
By confining the polymerization/cross-linking largely within micelles, we not only made our materials water-soluble but also were able to control the number of binding sites on the nanosized MINP. This feature distinguishes our MINP from other molecularly imprinted nanoparticles in the literature.52–60 Our previous studies indicate that the SCM of 1 has roughly 50 cross-linked surfactants. With surfactant/template = 50/1 in the synthesis, the MINPs on average contained one binding site per nanoparticle (Table 1).61 As demonstrated recently, this number can be tuned easily through changing the surface/template ratio.29
MINPs for Binding Oligosaccharides
FM 4 not only afforded MINPs with higher binding affinity and selectivity than 4-vinylphenylboronuc acid but also enabled us to imprint oligosaccharides.
Maltose was the first oligosaccharide template used in our study and expected to form FM•template complex 9 based on the binding motifs identified in the monosaccharide-binding MINPs. Because numerous hydrogen-bonding groups exist in the complex, we hypothesized that the micelle/MINP should contain hydrogen-bonding groups that interact with 9 through hydrogen bonds, in addition to hydrophobic and electrostatic interactions present in the normal micelle/MINP. Amide-functionalized cross-linkable surfactant 10 was recently found to enhance the binding of guest through hydrogen bonds.62 To our delight, MINP(maltose) prepared with 10 as the cross-linkable surfactant bound maltose with Ka = 20.5 × 103 M−1, substantially higher than the value obtained (Ka = 3.50 × 103 M−1) for MINP prepared with surfactant 1 (Table 2, entries 1 and 2). When the template/FM ratio was varied (1:1, 1:2, and 1:3), 1:2 gave the highest Ka, supporting the 1:2 binding model shown in 9.
Table 2.
ITC binding data for oligosaccharide guests.a
| Entry | Host | Guest | Ka (103 M−1) | Krel | -ΔG (kcal/mol) | N |
|---|---|---|---|---|---|---|
| 1 | MINP(maltose) | maltose | 20.5 ± 3.2 | 1 | 5.88 | 1.0 ± 0.1 |
| 2 | MINP(maltose)b | maltose | 3.50 ± 0.23 | - | 4.83 | 1.2 ± 0.1 |
| 3 | MINP(maltose)c | maltose | 5.72 ± 0.61 | - | 5.12 | 1.2 ± 0.1 |
| 4 | MINP(maltose)d | maltose | 19.7 ± 2.5 | - | 5.85 | 1.0 ± 0.1 |
| 5 | MINP(maltose) | cellobiose | 7.99 ± 0.12 | 0.39 | 5.32 | 1.2 ± 0.1 |
| 6 | MINP(maltose) | gentiobiose | 4.37 ± 0.53 | 0.21 | 4.96 | 1.2 ± 0.1 |
| 7 | MINP(maltose) | maltulose | <0.05 | <0.002 | - | - |
| 8 | MINP(maltose) | lactose | 0.79 ± 0.16 | 0.04 | 3.95 | 0.8 ± 0.1 |
| 9 | MINP(maltose) | maltotriose | <0.05 | <0.002 | - | - |
| 10 | MINP(maltose) | glucose | 1.81 ± 0.22 | 0.09 | 4.44 | 0.9 ± 0.1 |
| 11 | MINP(maltose) | maltosee | 15.2 ± 2.0 | - | 5.70 | 0.8 ± 0.1 |
| 12 | MINP(maltose) | maltosef | 18.8 ± 2.7 | - | 5.83 | 0.9 ± 0.1 |
| 13 | MINP(cellobiose) | maltose | 9.45 ± 0.14 | 0.29 | 5.42 | 1.1 ± 0.1 |
| 14 | MINP(cellobiose) | cellobiose | 32.9 ± 5.9 | 1 | 6.16 | 1.1 ± 0.1 |
| 15 | MINP(cellobiose) | gentiobiose | 4.77 ± 0.67 | 0.14 | 5.01 | 1.1 ± 0.1 |
| 16 | MINP(cellobiose) | maltulose | <0.05 | <0.002 | - | - |
| 17 | MINP(cellobiose) | lactose | 1.29 ± 0.09 | 0.04 | 4.24 | 0.8 ± 0.1 |
| 18 | MINP(lactose) | maltose | 3.24 ± 0.42 | 0.06 | 4.79 | 1.0 ± 0.1 |
| 19 | MINP(lactose) | cellobiose | 6.83 ± 0.92 | 0.13 | 5.23 | 0.8 ± 0.1 |
| 20 | MINP(lactose) | gentiobiose | 11.6 ± 1.7 | 0.22 | 5.54 | 0.9 ± 0.1 |
| 21 | MINP(lactose) | maltulose | 0.50 ± 0.13 | 0.01 | 3.67 | 1.0 ± 0.1 |
| 22 | MINP(lactose) | lactose | 52.2± 9.5 | 1 | 6.43 | 1.3 ± 0.1 |
| 23 | MINP(maltotriose) | maltotriose | 52.8 ± 8.6 | 1 | 6.44 | 1.1 ± 0.1 |
| 24 | MINP(maltotriose) | maltose | 14.1 ± 2.0 | 0.27 | 5.66 | 1.0 ± 0.1 |
| 25 | MINP(maltotriose) | glucose | 0.56 ± 0.02 | 0.01 | 3.75 | 1.0 ± 0.1 |
The template/FM ratio in the MINP synthesis was 1:2 unless otherwise indicated. The cross-linkable surfactants were a 3:2 mixture of 10 and 2′ unless otherwise indicated. The titrations were performed in 10 mM HEPES buffer at pH 7.4. Krel is the binding constant of a guest relative to that of the template sugar for a particular MINP. The ITC titration curves are reported in the Supporting Information, including the binding enthalpy and entropy.
The cross-linkable surfactants were a 3:2 mixture of 1 and 2′.
The template/FM ratio was 1:1.
The template/FM ratio was 1:3.
The titration was performed in the presence of cellobiose in 10 mM HEPES buffer at pH 7.4. [MINP] = 15 μM. [cellobiose] = 75 μM.
The titration was performed in the presence of lactose in 10 mM HEPES buffer at pH 7.4. [MINP] = 15 μM. [lactose] = 75 μM.
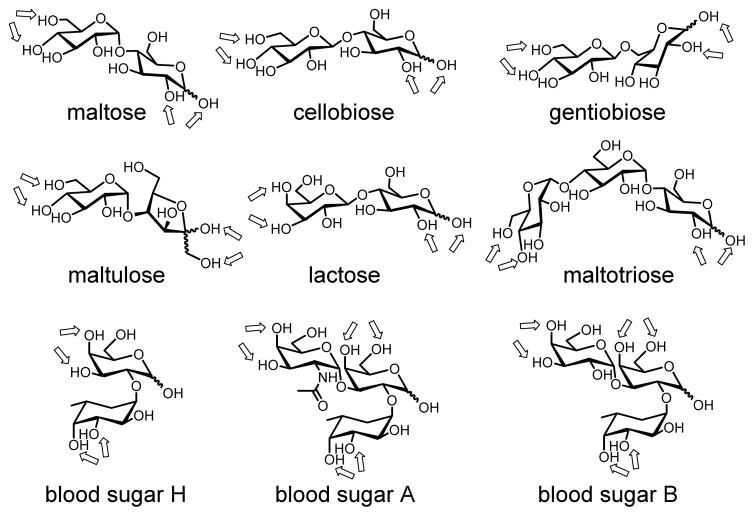
Binding of the oligosaccharides (Chart 2) worked fully as expected (Table 2). The selectivity of a particular MINP is indicated by Krel, which is the binding constant of a sugar guest relative to that of the template. Cellobiose and gentiobiose had a Krel value of 0.39 and 0.21 toward MINP(maltose), indicating that changing the α 1,4-glycosidic linkage to the β 1,4 or α 1,6 weakened the binding significantly. Replacing one of the two glucoses in maltose with fructose and galactose was even less tolerated, yielding Krel of <0.002 and 0.04 for maltulose and lactose, respectively. To probe the sensitivity, we also measured the binding of maltose by MINP(maltose) in the presence of 5 equiv of competing sugars (cellobiose and lactose). As shown by entries 11 and 12, the binding constant obtained was about 74% and 92%, respectively, of the original value (entry 1). These numbers were in line with the selectivity indicated by Krel.
Chart 2.
Structures of oligosaccharides used in this study. The arrows indicate the hydroxyls potentially involved in the boronate formation with FM 4.
Interestingly, shortening the chain length was better tolerated than lengthening the chain length, as glucose was bound with Krel = 0.09 but maltotriose with Krel <0.002. The result is reasonable because maltotriose should not fit into the binding pocket generated from the smaller maltose but glucose should be able to fit it, although only expected to bind one of the two boroxoles. Note that Ka (= 1.81 × 103 M−1) for glucose by MINP(maltose) was close to that (= 2.30 × 103 M−1) by MINP(glucose) in Table 1. It seems that the hydrogen-bonding interactions between the bound glucose and the amide-functionalized MINP nearly compensated for the loss of one boronate binding interaction.
We then created MINPs for all the other oligosaccharides and studied their binding. Good selectivity was generally obtained and each MINP always bound its own template sugar better than other sugars (Table 2 and Table 3S). As far as the absolute binding strength is concerned, gentiobiose, lactose, and maltotriose gave somewhat higher Ka values than the other sugars. The stronger binding for maltotriose could result from the additional hydroxyls on the template that interacted with the amide-functionalized MINP by hydrogen bonds. For MINP(maltotriose), as the guest became smaller (i.e., from maltotriose to maltose to glucose), binding expectedly weakened monotonously (Table 2, entries 21–23).
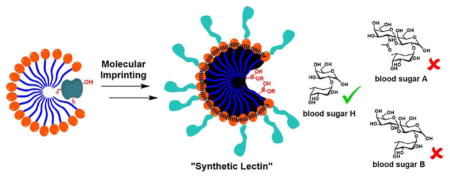
To test whether these boroxole-functionalized receptors could distinguish more challenging targets, we prepared MINPs for the three sugars that determine the human blood type: type O has sugar H on the surface of its blood cells, type A has A, type B has B, and type AB has both A and B.
As shown in Table 3, MINP(H), generated from sugar H, bound its template with Ka = 35.6 × 103 M−1 and showed no binding for the other two sugars. The difference between sugar A and B was extremely subtle: among the numerous functional groups, the only difference is a single acetoamido group in sugar A versus a hydroxyl in B (Chart 2). Impressively, MINP(A) was found to bind sugar A twice as strongly as sugar B, and MINP(B) displayed even higher selectivity. Meantime, sugar H showed weak binding to MINP(A) and MINP(B), with Krel = 0.13 in both cases.
Table 3.
ITC binding data for blood sugars.a
| Entry | Host | Guest | Ka (103 M−1) | Krel | -ΔG (kcal/mol) | N |
|---|---|---|---|---|---|---|
| 1 | MINP(H) | sugar H | 35.6 ± 5.2 | 1 | 6.2 | 1.0 ± 0.1 |
| 2 | MINP(H) | sugar A | <0.02 | <0.001 | - | - |
| 3 | MINP(H) | sugar B | <0.02 | <0.001 | - | - |
| 4 | MINP(A) | sugar H | 9.9 ± 0.1 | 0.13 | 5.45 | 1.0 ± 0.1 |
| 5 | MINP(A) | sugar A | 76.7 ± 1.2 | 1 | 6.66 | 1.0 ± 0.1 |
| 6 | MINP(A) | sugar B | 39.0 ± 4.8 | 0.51 | 6.26 | 1.1 ± 0.1 |
| 7 | MINP(B) | sugar H | 7.6 ± 0.8 | 0.13 | 5.29 | 1.1 ± 0.1 |
| 8 | MINP(B) | sugar A | 21.8 ± 4.3 | 0.38 | 5.91 | 1.0 ± 0.1 |
| 9 | MINP(B) | sugar B | 57.1 ± 7.5 | 1 | 6.48 | 1.1 ± 0.1 |
The template/FM ratio in the MINP synthesis was 1:2 for MINP(H) and 1:3 for MINP(A) and MINP(B). The cross-linkable surfactants were a 3:2 mixture of 10 and 2′. The titrations were performed in 10 mM HEPES buffer at pH 7.4. The ITC titration curves are reported in the Supporting Information, including the binding enthalpy and entropy.
CONCLUSIONS
In summary, we have reported a facile and general method to create protein-sized water-soluble nanoparticle receptors for a wide range of mono- and oligosaccharides. The in situ imprinting was enabled by the strong interactions between FM 4 and the appropriate diol functionalities on the sugar in the micellar environment. The number of binding sites on these “synthetic lectins” could be controlled easily. Importantly, the binding sites on the sugar can be identified prior to imprinting (namely, cis-1,2-diol, cis-3,4-diol, and trans-4,6-diol), making the molecular recognition highly predictable. Among the eight D-aldohexoses, glucose, mannose, and galactose are the most biologically relevant and can be distinguished completely. With the ability to differentiate oligosaccharides by their building blocks, chain length, and glycosidic linkages, we expect these “synthetic lectins” could become highly useful in biology and chemistry in the future.
Supplementary Material
Acknowledgments
We thank the National Institute of General Medical Sciences of the National Institutes of Health (R01GM113883) for financial support of the research.
Footnotes
The authors declare no competing financial interests.
Experimental details, ITC titration curves, and additional data. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Kamerling JP, Boons G-J. Comprehensive glycoscience: from chemistry to systems biology. 1. Elsevier; Amsterdam; Boston: 2007. [Google Scholar]
- 2.Wang B, Boons G-J. Carbohydrate recognition: biological problems, methods, and applications. Wiley; Hoboken, N.J: 2011. [Google Scholar]
- 3.Jin S, Cheng Y, Reid S, Li M, Wang B. Med Res Rev. 2010;30:171. doi: 10.1002/med.20155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davis AP, James TD. Carbohydrate Receptors. Wiley-VCH Verlag GmbH & Co; KGaA: 2005. [Google Scholar]
- 5.James TD, Phillips MD, Shinkai S. Boronic acids in saccharide recognition. RSC Publishing; Cambridge: 2006. [Google Scholar]
- 6.Ferrand Y, Crump MP, Davis AP. Science. 2007;318:619. doi: 10.1126/science.1148735. [DOI] [PubMed] [Google Scholar]
- 7.Barwell NP, Crump MP, Davis AP. Angew Chem Int Ed. 2009;48:7673. doi: 10.1002/anie.200903104. [DOI] [PubMed] [Google Scholar]
- 8.Ke C, Destecroix H, Crump MP, Davis AP. Nat Chem. 2012;4:718. doi: 10.1038/nchem.1409. [DOI] [PubMed] [Google Scholar]
- 9.Rauschenberg M, Bomke S, Karst U, Ravoo BJ. Angew Chem Int Ed. 2010;49:7340. doi: 10.1002/anie.201002847. [DOI] [PubMed] [Google Scholar]
- 10.Kobayashi K, Asakawa Y, Kato Y, Aoyama Y. J Am Chem Soc. 1992;114:10307. [Google Scholar]
- 11.Kim KT, Cornelissen JJLM, Nolte RJM, van Hest JCM. J Am Chem Soc. 2009;131:13908. doi: 10.1021/ja905652w. [DOI] [PubMed] [Google Scholar]
- 12.Pal A, Bérubé M, Hall DG. Angew Chem Int Ed. 2010;49:1492. doi: 10.1002/anie.200906620. [DOI] [PubMed] [Google Scholar]
- 13.Wu X, Li Z, Chen XX, Fossey JS, James TD, Jiang YB. Chem Soc Rev. 2013;42:8032. doi: 10.1039/c3cs60148j. [DOI] [PubMed] [Google Scholar]
- 14.Bull SD, Davidson MG, Van den Elsen JMH, Fossey JS, Jenkins ATA, Jiang YB, Kubo Y, Marken F, Sakurai K, Zhao JZ, James TD. Acc Chem Res. 2013;46:312. doi: 10.1021/ar300130w. [DOI] [PubMed] [Google Scholar]
- 15.Wulff G, Vesper W. J Chromatogr. 1978;167:171. [Google Scholar]
- 16.Awino JK, Gunasekara RW, Zhao Y. J Am Chem Soc. 2016;138:9759. doi: 10.1021/jacs.6b04613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wulff G. Angew Chem Int Ed Engl. 1995;34:1812. [Google Scholar]
- 18.Wulff G. Chem Rev. 2001;102:1. doi: 10.1021/cr980039a. [DOI] [PubMed] [Google Scholar]
- 19.Haupt K, Mosbach K. Chem Rev. 2000;100:2495. doi: 10.1021/cr990099w. [DOI] [PubMed] [Google Scholar]
- 20.Ye L, Mosbach K. Chem Mater. 2008;20:859. [Google Scholar]
- 21.Shea KJ. Trends Polym Sci. 1994;2:166. [Google Scholar]
- 22.Sellergren B. Molecularly imprinted polymers: man-made mimics of antibodies and their applications in analytical chemistry. Elsevier; Amsterdam: 2001. [Google Scholar]
- 23.Komiyama M. Molecular imprinting: from fundamentals to applications. Wiley-VCH; Weinheim: 2003. [Google Scholar]
- 24.Zimmerman SC, Lemcoff NG. Chem Commun. 2004:5. doi: 10.1039/b304720b. [DOI] [PubMed] [Google Scholar]
- 25.Yan M, Ramström O. Molecularly imprinted materials: science and technology. Marcel Dekker; New York: 2005. [Google Scholar]
- 26.Alexander C, Andersson HS, Andersson LI, Ansell RJ, Kirsch N, Nicholls IA, O’Mahony J, Whitcombe MJ. J Mol Recognit. 2006;19:106. doi: 10.1002/jmr.760. [DOI] [PubMed] [Google Scholar]
- 27.Sellergren B, Hall AJ. Supramol Chem. John Wiley & Sons, Ltd; 2012. [Google Scholar]
- 28.Haupt K, Ayela C. Molecular Imprinting. Springer; Heidelberg; New York: 2012. [Google Scholar]
- 29.Awino JK, Zhao Y. J Am Chem Soc. 2013;135:12552. doi: 10.1021/ja406089c. [DOI] [PubMed] [Google Scholar]
- 30.Awino JK, Zhao Y. Chem-Eur J. 2015;21:655. doi: 10.1002/chem.201404919. [DOI] [PubMed] [Google Scholar]
- 31.Wulff G, Schauhoff S. J Org Chem. 1991;56:395. [Google Scholar]
- 32.Kim H, Kang YJ, Kang S, Kim KT. J Am Chem Soc. 2012;134:4030. doi: 10.1021/ja211728x. [DOI] [PubMed] [Google Scholar]
- 33.Dowlut M, Hall DG. J Am Chem Soc. 2006;128:4226. doi: 10.1021/ja057798c. [DOI] [PubMed] [Google Scholar]
- 34.Bérubé M, Dowlut M, Hall DG. J Org Chem. 2008;73:6471. doi: 10.1021/jo800788s. [DOI] [PubMed] [Google Scholar]
- 35.Jay JI, Lai BE, Myszka DG, Mahalingam A, Langheinrich K, Katz DF, Kiser PF. Mol Pharmaceutics. 2010;7:116. doi: 10.1021/mp900159n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schumacher S, Katterle M, Hettrich C, Paulke BR, Hall DG, Scheller FW, Gajovic-Eichelmann N. J Mol Recognit. 2011;24:953. doi: 10.1002/jmr.1142. [DOI] [PubMed] [Google Scholar]
- 37.Mahalingam A, Geonnotti AR, Balzarini J, Kiser PF. Mol Pharmaceutics. 2011;8:2465. doi: 10.1021/mp2002957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schumacher S, Grüneberger F, Katterle M, Hettrich C, Hall DG, Scheller FW, Gajovic-Eichelmann N. Polymer. 2011;52:2485. doi: 10.1002/jmr.1142. [DOI] [PubMed] [Google Scholar]
- 39.Ellis GA, Palte MJ, Raines RT. J Am Chem Soc. 2012;134:3631. doi: 10.1021/ja210719s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim H, Kang YJ, Jeong ES, Kang S, Kim KT. ACS Macro Lett. 2012;1:1194. doi: 10.1021/mz3004192. [DOI] [PubMed] [Google Scholar]
- 41.Kotsuchibashi Y, Agustin RVC, Lu JY, Hall DG, Narain R. ACS Macro Lett. 2013;2:260. doi: 10.1021/mz400076p. [DOI] [PubMed] [Google Scholar]
- 42.Liu CT, Tomsho JW, Benkovic SJ. Bioorg Med Chem. 2014;22:4462. doi: 10.1016/j.bmc.2014.04.065. [DOI] [PubMed] [Google Scholar]
- 43.Kotsuchibashi Y, Ebara M, Sato T, Wang Y, Rajender R, Hall DG, Narain R, Aoyagi T. J Phys Chem B. 2015;119:2323. doi: 10.1021/jp506478p. [DOI] [PubMed] [Google Scholar]
- 44.Zhang S, Zhao Y. Macromolecules. 2010;43:4020. [Google Scholar]
- 45.Zhao Y. Langmuir. 2016;32:5703. doi: 10.1021/acs.langmuir.6b01162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang S, Zhao Y. J Am Chem Soc. 2010;132:10642. doi: 10.1021/ja103391k. [DOI] [PubMed] [Google Scholar]
- 47.Schmidtchen FP. Supramol Chem. John Wiley & Sons, Ltd; 2012. [Google Scholar]
- 48.Awino JK, Zhao Y. Chem Commun. 2014;50:5752. doi: 10.1039/c4cc01516a. [DOI] [PubMed] [Google Scholar]
- 49.Because boronate formation with FM 3 leads to a chiral center at the boron, multiple diastereomers could form in the template-functional monomer complex. Regardless of the number of diastereomers formed and their relative populations, the sugar can interact with the templated boroxole groups during rebinding through the same bonding motifs. The measured binding constant is thus a composite term of these binding processes. However, since all the bindings involve the same number of boronate esters and similar hydrogen bonds between the remaining hydroxyl groups and the MINP, they should have very similar binding affinities.
- 50.Since the binding selectivity for MINP(mannose) suggests that the cis-1,2-diol was invovled in the binding, the C3 hydroxyl could not have participated in in the binding with the (cis) C2 hydroxyl.
- 51.If the cis-4,6-diol had been involved in the binding, gulose should have displayed weaker but significant binding as allose and altrose did toward MINP(glucose) and MINP(mannose), respectively.
- 52.Li Z, Ding J, Day M, Tao Y. Macromolecules. 2006;39:2629. [Google Scholar]
- 53.Hoshino Y, Kodama T, Okahata Y, Shea KJ. J Am Chem Soc. 2008;130:15242. doi: 10.1021/ja8062875. [DOI] [PubMed] [Google Scholar]
- 54.Priego-Capote F, Ye L, Shakil S, Shamsi SA, Nilsson S. Anal Chem. 2008;80:2881. doi: 10.1021/ac070038v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cutivet A, Schembri C, Kovensky J, Haupt K. J Am Chem Soc. 2009;131:14699. doi: 10.1021/ja901600e. [DOI] [PubMed] [Google Scholar]
- 56.Yang KG, Berg MM, Zhao CS, Ye L. Macromolecules. 2009;42:8739. [Google Scholar]
- 57.Zeng ZY, Patel J, Lee SH, McCallum M, Tyagi A, Yan MD, Shea KJ. J Am Chem Soc. 2012;134:2681. doi: 10.1021/ja209959t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ma Y, Pan GQ, Zhang Y, Guo XZ, Zhang HQ. Angew Chem Int Ed. 2013;52:1511. doi: 10.1002/anie.201206514. [DOI] [PubMed] [Google Scholar]
- 59.Çakir P, Cutivet A, Resmini M, Bui BT, Haupt K. Adv Mater. 2013;25:1048. doi: 10.1002/adma.201203400. [DOI] [PubMed] [Google Scholar]
- 60.Zhang Y, Deng C, Liu S, Wu J, Chen Z, Li C, Lu W. Angew Chem Int Ed. 2015;54:5157. doi: 10.1002/anie.201412114. [DOI] [PubMed] [Google Scholar]
- 61.Single-sited receptors have been obtained by molecular imprinting within dendrimers, see: Zimmerman SC, Wendland MS, Rakow NA, Zharov I, Suslick KS. Nature. 2002;418:399. doi: 10.1038/nature00877.Zimmerman SC, Zharov I, Wendland MS, Rakow NA, Suslick KS. J Am Chem Soc. 2003;125:13504. doi: 10.1021/ja0357240.
- 62.Arifuzzaman MD, Zhao Y. J Org Chem. 2016;81:7518. doi: 10.1021/acs.joc.6b01191. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.