Abstract
About a third of all human cancers harbor mutations in one of the K-, N-, or HRAS genes that encode an abnormal RAS protein locked in a constitutively activated state to drive malignant transformation and tumor growth. Despite more than three decades of intensive research aimed at the discovery of RAS-directed therapeutics, there are no FDA approved drugs that are broadly effective against RAS-driven cancers. While RAS proteins are often said to be “undruggable”, there is mounting evidence suggesting it may be feasible to develop direct inhibitors of RAS proteins. Here we review this evidence with a focus on compounds capable of inhibiting the interaction of RAS proteins with their effectors that transduce the signals of RAS and which drive and sustain malignant transformation and tumor growth. These reports of direct-acting RAS inhibitors provide valuable insight for further discovery and development of clinical candidates for RAS-driven cancers involving mutations in RAS genes or otherwise activated RAS proteins.
Keywords: RAS, oncogene, inhibitors
Introduction
Cancer is a leading cause of death in the developed world with over one million people diagnosed and more than 500,000 deaths per year in the United States alone. It is estimated that at least one in three people will develop some form of cancer during their lifetime. Over 30% of all human tumors arise from mutations that encode a RAS protein essentially locked in a constitutively activated state to stimulate signaling cascades necessary for malignant transformation, including cellular proliferation, survival and invasiveness, tumor angiogenesis, and metastasis.
RAS is an abbreviation of “Rat sarcoma”, reflecting how the first members of the RAS gene family were discovered over 3 decades ago. The RAS family is composed of 36 human genes, but KRAS, NRAS and HRAS by far play the most prominent roles in human cancer (1). Hereafter, the term “RAS” will be used when two or more of the isoforms may be involved. RAS proteins are monomeric enzymes with modest GTPase activity, but which bind GTP and GDP with high affinity. The guanine nucleotide exchange factor SOS1 catalzyes the displacement of GDP, allowing RAS to bind the more abundant GTP, while p120GAP contributes an arginine residue to the catalytic site of RAS, leading to inactivation (2). The active, GTP bound form of RAS has been described as a “coiled spring” which in turn activates effector proteins such as RAF1 and BRAF or PI3K, activating the RAF/MEK/ERK or PI3K/AKT/MTOR cascades, respectively (Fig. 1A-D) (2). Thus, RAS proteins are important regulators of multiple aspects of normal cell growth and physiology, as well as malignant transformation (3).
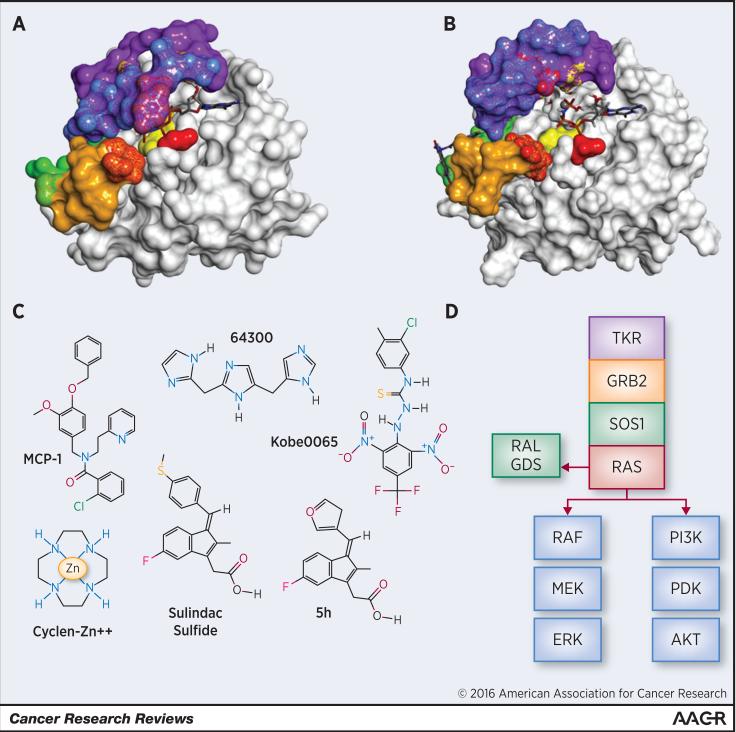
Fig. 1.
Surface model of HRAS-GTP interaction with ligands. The catalytic site of HRAS forms a shallow groove which contains a Mg+2 ion near the binding position of the terminal phosphate of GTP (A, B) and a hydrophobic slot at right accommodates the guanine moiety of GTP. The adjacent flexible loops switch I (blue) and switch II (mustard), make up a large part of the effector binding domain of HRAS. A and B show the markedly different topologies for the two states that GTP-bound HRAS adopts, state 1 and state 2, only the latter of which is inherently “active,” having dramatically higher affinity for complexing with its effector proteins (36). In state 2 of HRAS (A), both GTP and the Mg+2 ion bound to the catalytic site are obscured by the prominent Tyr32 in the flexible loop of switch I (blue; wild type HRAS, X-ray diffraction, PDB 5P21). In contrast, GTP and the Mg+2 ion are exposed in the more open state 1, where Tyr32 is retracted and RAS has higher affinity for the nucleotide exchange factor (Fig. 1B, T35S HRAS mutant, NMR, PDB 2LWI).
Binding surfaces for RAS–effector compounds are color coded: Green: Kobe 2601 binding, Yellow: sulindac/analog binding , Red: cyclen/metal binding, Purple: peptide binding, Blue: Switch I, Mustard: Switch II, (together, Switch I and II represent the binding site for intracellular antibody fragment. Mottling represents shared binding surfaces. A) Wild type HRAS (PDB 5P21) bound to GTP shows the state 2 closed configuration of the nucleotide binding pocket producing the effector binding form. B) GTP bound Mutant HRAS T35S (PDB 2LWI) with nucleotide binding site in the open state 1 nucleotide exchange factor binding configuration. Kobe2601 binds to a non-catalytic secondary site (green, left side) C) Structure of RAS- effector disrupting small molecules. D) Schematic of canonical signaling cascades associated with RAS isoforms, including upstream activation by tyrosine kinase receptor (TKR) and growth factor receptor bound protein 2 (GRB2) via guanine nucleotide exchange factor SOS1. RAS effector proteins RAF, PI3K, and RALGDS activate the MEK/ERK, PDK/AKT, and RALA/B pathways, respectively.
Activating mutations at codons 12, 13 or 61 of K-Ras occur de novo in approximately one third of all human cancers and are especially prevalent in pancreatic, colorectal, and lung tumors. These mutations affect the P-loop and switch-2 regions of the highly conserved N-terminal G-domain of RAS, decreasing p120GAP-mediated and intrinsic GTP hydrolysis rates. Functionally similar mutations in NRAS are more prevalent in hematologic cancers and metastatic melanoma, whereas HRAS mutations are less common, although with a few notable exceptions such as urothelial cell and thyroid carcinomas (4). RAS mutations also develop spontaneously in tumors that become resistant to radiation and/or chemotherapy, or targeted therapies, including receptor tyrosine kinase inhibitors that activate the RAS pathway (2). While RAS mutations are relatively infrequent in other tumor types, for example, breast cancer, RAS signaling can be pathologically activated by upstream growth factor receptors (e.g. ERBB2) that signal through RAS or in downstream pathway components (e.g. RAF1, BRAF; Fig. 1D) (5).
Despite numerous attempts over many years, there have been no drugs approved by the FDA that selectively inhibit the growth of RAS-driven tumors. Based on functional knockout studies, targeted therapies that inhibit RAS or RAS-mediated pathways would be expected to inhibit the proliferation, survival and spread of tumor cells with activated RAS. Multiple approaches have been undertaken to develop drugs to treat malignancies arising from RAS mutations including interfering with maturation, trafficking and localization to the plasma membrane, or inhibiting downstream signaling. While these approaches have been extensively reviewed by others (4), this review focuses on strategies to identify experimental compounds that directly bind RAS to disrupt signaling. This approach is considered to be the most challenging. In fact, RAS is often said to be “undruggable” due to the relative cellular abundance of its substrate, GTP, and the high affinity of RAS for binding GTP (6), as well as the apparent lack of suitable surfaces in critical regions necessary for small molecule binding.
Early approaches to inhibit RAS focused on disrupting post-translational lipid modifications of the C-terminal within the hypervariable region required for maturation and localization of RAS to the plasma membrane. Despite promising activity in preclinical tumor models involving mutant HRAS, farnesyl transferase (FT) inhibitors were ineffective in clinical trials, presumably because of alternative prenylation of the more commonly mutated KRAS isoform by geranylgeranyltransferases. This highlighted an important difference between RAS isoforms (6). Recent efforts have focused on the non-catalytic δ-subunit of the cyclic GMP phosphodiesterase (PDE) 6 isozyme, which functions as a chaperone binding the prenyl group and shuttling RAS to the plasma membrane. A small molecule inhibitor of PDEδ (deltarasin) inhibited KRAS signaling and pancreatic tumor cell growth in vitro and in vivo (7). However, its clinical utility has not yet been realized, possibly because of the potential for non-specific inhibition of other prenylated proteins, including other members of the Ras superfamily (1).
Targeting of downstream components of RAS signaling with inhibitors of RAF/MEK/ERK kinase or the PI3K/AKT pathways has been another strategy which may have unique effects in the context of different mutations of K-, N-, or HRAS isoforms, but is fraught with resistance mechanisms arising from complex feedback systems (6). However, there have been notable successes, such as combinations of MEK inhibitors with other targeted therapeutics in the case of malignant melanoma (4). The situation is made more complex with co-expression of mutations in receptor tyrosine kinases, BRAF, PTEN, and other signaling proteins as described in recent reviews (2, 4). Several other molecular targets have been identified by RNAi screening, which might provide new opportunities to inhibit the growth of RAS-driven tumors, including cyclin dependent kinase 4, cyclin D1, TIAM1, MYC, serine threonine kinase 33, and tank binding kinase 1, as well as several genes involved in mitosis (6). However, none have resulted in inhibitors that show sufficient selectivity for tumor cells with mutant RAS.
Other mutant RAS selective compounds have been identified targeting molecules outside the canonical RAS effector pathways. A high-throughput phenotypic screen of over 300,000 compounds identified a lead compound, ML210, that was synthetically lethal to cells expressing oncogenic HRAS. Though the specific molecular target of ML210 is unknown, it inhibited the growth of cells expressing mutant HRAS with an IC50 of 71 nM and was 4-fold selective versus cells lacking oncogenic HRAS (8). Another high-throughput screening strategy identified two compounds, RSL3 and RSL5 which induced non-apoptotic, MEK-dependent, oxidative cell death (9). RSL5, like a previously-identified Ras synthetic lethal compound, erastin, binds the voltage-dependent anion channel (VDAC3). Yet another small-molecule screen identified oncrasin, a compound selectively active against KRAS mutant cell lines. A highly potent analog, NSC-743380, showed anti-tumor activity in a preclinical model of KRAS driven renal cancer (10). A synthetic lethal screen using embryonic fibroblasts derived from mice expressing oncogenic KRAS (G12D) identified lanperisone which induced non-apoptotic cell death via oxidative stress (11). Collectively, the compounds identified by chemical synthetic lethal screens may open new avenues for the treatment of cancers with activated RAS, but further studies to define the underlying molecular target and chemical optimization to improve drug-like properties will be essential for their development as anticancer drugs. This review highlights approaches to inhibit direct interactions between the various isoforms of RAS and their immediate downstream effector proteins.
Disruption of RAS-effector interactions
Strong support for the concept of disrupting activated RAS-effector protein complexes as a therapeutic strategy came from studies using a membrane directed, single immunoglobulin antibody domain (iDab#6-memb) which can be expressed intracellularly by a retroviral vector. This antibody fragment was identified in a yeast two-hybrid screen of randomly encoded polypeptide sequences for binding to HRAS (G12V) bait protein. X-ray crystallography and biochemical studies demonstrated that this antibody fragment bound the switch I and II domains of GTP bound HRAS in a way predicted to be mutually exclusive of the amino terminal Ras binding domain of RAF1 (RAF-RBD), RAL guanine nucleotide dissociation stimulator (RALGDS), and PI3K. Phosphorylation of downstream signaling was robustly inhibited, which modestly suppressed the growth of colon tumor cells in vitro (12). In a transgenic mouse model of KRAS driven lung cancer, tumor initiation was dramatically reduced by expressing the antibody fragment. However, the tumor cells remained viable and resumed growth if antibody expression was halted. Whether this would hold true for disruption of RAS -effector interaction by small molecule inhibitors remains to be determined. The iDAb#6-memb selectively bound several RAS wild type and mutant isoforms including H, K, and NRAS in the GTP bound state, suggesting that a broad spectrum inhibitor of RAS-effector binding may be feasible for other therapeutic modalities. RAS-effector domain antibodies provide convincing proof of concept, but isolated antibody fragments are not currently a viable clinical approach for disrupting intracellular signaling.
Since the most critical residues involved in RAS-effector interaction have been identified, a therapeutically tractable peptide could be developed. A 7-amino acid peptide segment from the RAF-RBD potently blocked the association of RAS with either the isolated RAF-RBD or RALGDS (13). Similarly, Clark et al. identified regions of homology within the cysteine rich domain of RAF1 (RAF-CRD) and the GTPase activating domain of NF1 to design an 8 amino acid peptide, which inhibits the activation of ERK1/2 and competitively blocks interaction between RAS and the GTPase domain of NF1 (Fig.1A-B, purple region) (14). While unmodified peptides are generally not cell-permeant, recent advances in cell penetrating peptides along with novel approaches to enhance oral bioavailability of peptide therapeutics may enable RAS inhibitory peptides to become a more viable option for clinical development (15).
Small molecules
One of the earliest reports of a small molecule that selectively inhibited Ras-driven tumorigenesis, the nonsteroidal anti-inflammatory drug (NSAID), sulindac inhibited carcinomas harboring mutant HRAS in a rat model of chemically-induced breast tumorigenesis (16). Sulindac sulfide directly inhibits Ras activation of RAF1 and HRAS-induced focus formation (17). Further experiments showed that sulindac sulfide inhibits Ras-dependent activation of RAF1, but not RAF1 activity absent HRAS activation. RAS selectivity was independently confirmed in a report that sulindac sulfide inhibited focus formation by HRAS, but not by v-SRC or other oncogenic proteins, including those that signal through the cyclooxygenase (COX)/prostaglandin pathway (18). Biochemical support for these observations was described by other investigators who also showed the activity of other NSAIDs, including indomethacin, NS398, and aspirin, to disrupt RAS:RAF-RBD binding, albeit at appreciably higher concentrations (19). Although sulindac sulfide was reported to directly interact with RAS by equilibrium dialysis binding assays, other reports suggested different mechanisms involving the suppression of RAS-induced COX2 (17, 20). However, the non-COX inhibitory sulfone metabolite of sulindac also has selective effects on tumor cells with mutant RAS in vitro and in experimental models of tumorigenesis (16, 21).
To improve the RAS inhibitory and anticancer activity of sulindac and reduce the potential toxicities associated with COX inhibition, the sulindac scaffold was chemically modified. One derivative (7a) was found to have significantly reduced COX inhibitory activity, but improved potency to inhibit HRAS:RAF-RBD binding (22). Four derivatives more potently inhibited Ras-Raf binding than sulindac sulfide and selectively inhibited growth of HRAS-transformed Madin-Darby canine kidney cells. Another series of sulindac derivatives were reported to inhibit RAS-RAF binding more potently than sulindac sulfide and decrease ERK1/2 phosphorylation (23).
A recent report described an unexpected interaction between the polo-like kinase inhibitor, Rigosertib, with several RAS effector proteins including A-, B-, and c-RAF, as well as the RAS binding domains of RALGDS, PI3K-α, β, and γ. This enabled the compound to inhibit the interactions of both mutant and wild type isoforms of K- or N-RAS isoforms with critical downstream effectors (24). The compound inhibited RAS-mediated activation of ERK1/2 and PI3K signaling pathways via growth factor stimulation and activating mutation in NRAS. Multiple clinical trials of Rigosertib treatment for hematologic and solid malignancies are ongoing at this time.
Other approaches involving high throughput screening have been conducted to identify compounds that disrupt the HRAS-RAF1 protein-protein interactions. For example, a yeast two-hybrid screen identified a series of compounds exemplified by MCP1 which inhibits mutant or wild type HRAS-driven promoter/reporter constructs, HRAS-driven RAF1 activation, and mutant HRAS or NRAS-dependent phenotypes including anchorage independent growth and loss of cytoskeletal organization. Interestingly, the compounds did not interfere with RAS-induced activation of AKT (25). A derivative, MCP110, inhibited colon tumor cell growth and RAS-mediated signaling with moderate potency and inhibited colon tumor growth in mouse xenograft models alone and in combination with microtubule targeting agents (26). Studies of analogs (MCP-110, -116) confirmed inhibition of RAS-RAF-RBD binding , mutant HRAS-mediated recruitment of RAF1 to the plasma membrane, and downstream ERK phosphorylation (26). These compounds also disrupted RAS ortholog-driven (Let60) phenotypes in C. elegans. However, they also affected this phenotype when induced by constitutively active RAF ortholog, raising questions about whether the compounds act on the RAS or the RAF side of the interaction. With cross species activity, and implication that the compound may bind RAF, this family of compounds may act as a pan-RAS inhibitor. While MCP-110 strongly synergized with tubulin targeting agents in vitro, only modest synergy was observed with RAF or MEK inhibitors, which is inconsistent with a mechanism involving RAS inhibition. MCP-110 was modestly effective as a single agent in a human tumor xenograft mouse model (27). As predicted by in vitro studies, combination with paclitaxel enhanced MCP-110 antitumor activity. An in-depth evaluation of the SAR of this family of compounds raised new questions, but yielded only modest potency improvements in cell-based reporter, growth, and transformation assays (28). Despite encouraging results, there have been no new reports published on these compounds for over five years.
GTP-bound RAS adopts two forms, state 1 and state 2, only the latter of which is inherently “active,” having dramatically higher affinity for its effector proteins (29). Shima et al. took advantage of the preference of the P40D mutant form of the closely related GTPase, MRAS, to adopt the inactive state 1 conformation. They conducted an in silico screen of over 40,000 compounds to identify a compound (Kobe0065) which binds RAS in state 1 (29). Kobe0065, and a less potent analog (Kobe2602) identified by similarity searching, inhibited HRAS- and KRAS:RAF-RBD binding in vitro. The compounds also disrupted HRAS(G12V)-induced phosphorylation of MEK, ERK, AKT and formation of RALA-GTP with micromolar potency. NMR spectroscopy showed that the compounds bind near the effector domain of RAS, explaining the broad spectrum of activity, albeit with low potency. In vitro activity was more potent, inhibiting anchorage independent growth at low micromolar concentrations. However, efficacy as single agent in a KRAS-driven tumor xenograft model in nude mice was modest (29). The current state of development of this family of compounds is unknown.
Mutational studies of the p110α catalytic subunit of PI3K have demonstrated the importance of its RAS binding domain in the maintenance of K-RAS driven lung tumors, validating this protein-protein interaction as a therapeutic target (30). Few of the compounds which disrupt RAS-effector interactions have been reported to disrupt RAS-induced PI3K signaling. Kobe0065 and 2602 inhibit AKT phosphorylation, and modeling data predicts that they may affect RAS-PI3K interactions (29). Using a novel ensemble computational approach, compounds were identified by molecular dynamics simulation and virtual screening which inhibit signaling downstream of KRAS through a previously unappreciated allosteric mechanism. The goal was to identify allosteric inhibitory sites and compounds which in turn stabilize an inactive conformation of KRAS (31). Confirmed hit compounds, for example 64300, exhibit modest potency, inhibiting ERK phosphorylation in glioblastoma cells lacking RAS mutation at low micromolar concentrations. However, these compounds will require significant chemical optimization to improve drug-like properties.
A series of low molecular weight metal coordinating compounds have been described that bind and inhibit HRAS with low affinity. For example, cyclen, a polydentate compound coordinated with transition metals zinc or copper, preferentially bound to and stabilized HRAS in the non-productive state 1, lowering the affinity of HRAS for RAF1 (32). Interestingly, this compound placed the metal moiety close to the β and γ phosphates of GTP in the nucleotide pocket of HRAS, suggesting that this may be an approach to increase the intrinsic GTPase activity of mutant RAS isoforms by substituting for the arginine finger of p120RASGAP. Figs. 1A and 1B show the contact residues of Cyclen-Zn2+ (red) at the mouth of the nucleotide binding site, including Gly13 and residues of both switch I and II regions of the G-domain. A structurally distinct metal coordinating compound, Bis 2-picolylamine (BPA), similarly binds selectively to and stabilizes HRAS in the state 1 conformation(33). However, this compound class is unlikely to be further developed given its low affinity for binding HRAS.
In the unusual case of the G12C KRAS gene mutation, significant progress has been made to develop compounds that trap KRAS into an inactive, GDP bound, conformation. A fragment-based screen paired with crystallographic studies and molecular dynamics simulation identified compounds that irreversibly bind and inhibit KRAS in lung tumor cells harboring the G12C mutation (34). A separate group developed GDP analogs which also covalently bind the cysteine of KRAS(G12C) (35). Both classes should result in an inactive RAS-nucleotide complex, thus blocking downstream signal transduction. Covalent inhibitors can yield highly potent and selective compounds and are gaining favor as a drug discovery strategy. There are multiple examples of covalent inhibitors approved for various conditions which target cysteine proteases, hepatitis C virus (HCV) protease and kinases. However, the relatively low frequency of the G12C mutation may limit the utility of G12C selective compounds for cancers other than those of lung cancer, where this mutation is more prevalent than other KRAS mutations.
Conclusions
RAS proteins play an essential role in normal cell growth and malignant transformation. To date, no drugs have been approved by the FDA that directly binds Ras to selectively inhibit the oncogenic functions of RAS proteins, while sparing normal cell growth. A promising target represented by the interface between RAS proteins and downstream effectors has been validated by a synthetic antibody fragment. Few small molecules with the potential to disrupt RAS-effector interactions have been identified. Rigosertib and sulindac analogs, for example, have been reported to directly bind Ras and inhibit Ras-mediated signaling to inhibit transformation. Independent virtual screening campaigns have identified distinct compound classes which appear to act by disrupting RAS-effector interactions, albeit with modest potency and drug-like properties. Similar difficulties plague compounds identified via yeast two hybrid screening. While quite promising, covalent inhibitors targeting the reactive cysteine residue of KRAS G12C mutants likewise have hurdles of potency and drug-like properties which must be addressed before they are ready for clinical development.
Acknowledgements
We thank Drs. Andrzej Wierzbicki, Xi Chen, Bing Zhu (University of South Alabama) and Michael Boyd (ADT Pharmaceuticals) for insights and feedback in the conception of this review. Grant support: Research reported here was supported by the National Cancer Institute of the National Institutes of Health under Award Numbers: 1R01CA131378, 1R01CA148817, 1R01CA197147 and 1R01CA155638 to G.A. Piazza; and 1R21CA182941 to G.A. Piazza and B. Zhu. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Competing interests: The authors have declared no conflicts of interest.
References
- 1.Wennerberg K, Rossman KL, Der CJ. J Cell Sci. Vol. 118. England: 2005. The Ras superfamily at a glance. pp. 843–846. [DOI] [PubMed] [Google Scholar]
- 2.Gysin S, Salt M, Young A, McCormick F. Therapeutic strategies for targeting ras proteins. Genes Cancer. 2011;2:359–372. doi: 10.1177/1947601911412376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Downward J. Nat Rev Cancer. Vol. 3. England: 2003. Targeting RAS signalling pathways in cancer therapy. pp. 11–22. [DOI] [PubMed] [Google Scholar]
- 4.Cox AD, Fesik SW, Kimmelman AC, Luo J, Der CJ. Drugging the undruggable RAS: Mission possible? Nat Rev Drug Discov. 2014;13:828–851. doi: 10.1038/nrd4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eckert LB, Repasky GA, Ulku AS, McFall A, Zhou H, Sartor CI, Der CJ. Cancer Res. Vol. 64. United States: 2004. Involvement of Ras activation in human breast cancer cell signaling, invasion, and anoikis. pp. 4585–4592. [DOI] [PubMed] [Google Scholar]
- 6.Takashima A, Faller DV. Targeting the RAS oncogene. Expert Opin Ther Targets. 2013;17:507–531. doi: 10.1517/14728222.2013.764990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zimmermann G, Papke B, Ismail S, Vartak N, Chandra A, Hoffmann M, Hahn SA, Triola G, Wittinghofer A, Bastiaens PI, Waldmann H. Small molecule inhibition of the KRAS PDEdelta interaction impairs oncogenic KRAS signalling. Nature. 2013;497:638–642. doi: 10.1038/nature12205. [DOI] [PubMed] [Google Scholar]
- 8.Bittker JA, Weiwer M, Lewis T, Shimada K, Yang WS, MacPherson L, Dandapani S, Munoz B, Palmer M, Stockwell BR, Schreiber SL. Probe Reports from the NIH Molecular Libraries Program. National Center for Biotechnology Information; Bethesda MD: 2011. Screen for RAS-Selective Lethal Compounds and VDAC Ligands - Probe 2. [PubMed] [Google Scholar]
- 9.Yang WS, Stockwell BR. Chem Biol. Vol. 15. England: 2008. Synthetic lethal screening identifies compounds activating iron- dependent, nonapoptotic cell death in oncogenic-RAS-harboring cancer cells. pp. 234–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo W, Wu S, Wang L, Wei X, Liu X, Wang J, Lu Z, Hollingshead M, Fang B. PLoS One. Vol. 6. United States: 2011. Antitumor activity of a novel oncrasin analogue is mediated by JNK activation and STAT3 inhibition. p. e28487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shaw AT, Winslow MM, Magendantz M, Ouyang C, Dowdle J, Subramanian A, Lewis TA, Maglathin RL, Tolliday N, Jacks T. Selective killing of K-ras mutant cancer cells by small molecule inducers of oxidative stress. Proc Natl Acad Sci U S A. 2011;108:8773–8778. doi: 10.1073/pnas.1105941108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tanaka T, Rabbitts TH. Interfering with RAS-effector protein interactions prevent RAS- dependent tumour initiation and causes stop-start control of cancer growth. Oncogene. 2010;29:6064–6070. doi: 10.1038/onc.2010.346. [DOI] [PubMed] [Google Scholar]
- 13.Barnard D, Sun H, Baker L, Marshall MS. In vitro inhibition of Ras-Raf association by short peptides. Biochem Biophys Res Commun. 1998;247:176–180. doi: 10.1006/bbrc.1998.8746. [DOI] [PubMed] [Google Scholar]
- 14.Clark GJ, Drugan JK, Terrell RS, Bradham C, Der CJ, Bell RM, Campbell S. Peptides containing a consensus Ras binding sequence from Raf-1 and theGTPase activating protein NF1 inhibit Ras function. Proc Natl Acad Sci U S A. 1996;93:1577–1581. doi: 10.1073/pnas.93.4.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fosgerau K, Hoffmann T. Peptide therapeutics: current status and future directions. Drug Discov Today. 2015;20:122–128. doi: 10.1016/j.drudis.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 16.Thompson HJ, Jiang C, Lu J, Mehta RG, Piazza GA, Paranka NS, Pamukcu R, Ahnen DJ. Sulfone metabolite of sulindac inhibits mammary carcinogenesis. Cancer Res. 1997;57:267–271. [PubMed] [Google Scholar]
- 17.Herrmann C, Block C, Geisen C, Haas K, Weber C, Winde G, Moroy T, Muller O. Sulindac sulfide inhibits Ras signaling. Oncogene. 1998;17:1769–1776. doi: 10.1038/sj.onc.1202085. [DOI] [PubMed] [Google Scholar]
- 18.Gala M, Sun R, Yang VW. Cancer Lett. Vol. 175. Ireland: 2002. Inhibition of cell transformation by sulindac sulfide is confined to specific oncogenic pathways. pp. 89–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pan MR, Chang HC, Hung WC. Cell Signal. Vol. 20. England: 2008. Non-steroidal anti-inflammatory drugs suppress the ERK signaling pathway via block of Ras/c-Raf interaction and activation of MAP kinase phosphatases. pp. 1134–1141. [DOI] [PubMed] [Google Scholar]
- 20.Sheng GG, Shao J, Sheng H, Hooton EB, Isakson PC, Morrow JD, Coffey RJ, Jr., DuBois RN, Beauchamp RD. Gastroenterology. Vol. 113. United States: 1997. A selective cyclooxygenase 2 inhibitor suppresses the growth of H-ras-transformed rat intestinal epithelial cells. pp. 1883–1891. [DOI] [PubMed] [Google Scholar]
- 21.Lawson KR, Ignatenko NA, Piazza GA, Cui H, Gerner EW. Influence of K-ras activation on the survival responses of Caco-2 cells to the chemopreventive agents sulindac and difluoromethylornithine. Cancer Epidemiol Biomarkers Prev. 2000;9:1155–1162. [PubMed] [Google Scholar]
- 22.Karaguni IM, Glusenkamp KH, Langerak A, Geisen C, Ullrich V, Winde G, Moroy T, Muller O. New indene-derivatives with anti-proliferative properties. Bioorg Med Chem Lett. 2002;12:709–713. doi: 10.1016/s0960-894x(01)00839-3. [DOI] [PubMed] [Google Scholar]
- 23.Waldmann H, Karaguni IM, Carpintero M, Gourzoulidou E, Herrmann C, Brockmann C, Oschkinat H, Muller O. Sulindac-derived Ras pathway inhibitors target the Ras-Raf interaction and downstream effectors in the Ras pathway. Angew Chem Int Ed Engl. 2004;43:454–458. doi: 10.1002/anie.200353089. [DOI] [PubMed] [Google Scholar]
- 24.Athuluri-Divakar SK, Vasquez-Del Carpio R, Dutta K, Baker SJ, Cosenza SC, Basu I, Gupta YK, Reddy MV, Ueno L, Hart JR, Vogt PK, Mulholland D, Guha C, Aggarwal AK, Reddy EP. A Small Molecule RAS-Mimetic Disrupts RAS Association with Effector Proteins to Block Signaling. 2016;165:643–655. doi: 10.1016/j.cell.2016.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kato-Stankiewicz J, Hakimi I, Zhi G, Zhang J, Serebriiskii I, Guo L, Edamatsu H, Koide H, Menon S, Eckl R, Sakamuri S, Lu Y, Chen QZ, Agarwal S, Baumbach WR, Golemis EA, Tamanoi F, Khazak V. Proc Natl Acad Sci U S A. Vol. 99. United States: 2002. Inhibitors of Ras/Raf-1 interaction identified by two-hybrid screening revert Ras-dependent transformation phenotypes in human cancer cells. pp. 14398–14403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gonzalez-Perez V, Reiner DJ, Alan JK, Mitchell C, Edwards LJ, Khazak V, Der CJ, Cox AD. Genetic and functional characterization of putative Ras/Raf interaction inhibitors in C. elegans and mammalian cells. J Mol Signal. 2010;5:2. doi: 10.1186/1750-2187-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Skobeleva N, Menon S, Weber L, Golemis EA, Khazak V. In vitro and in vivo synergy of MCP compounds with mitogen-activated protein kinase pathway- and microtubule-targeting inhibitors. Mol Cancer Ther. 2007;6:898–906. doi: 10.1158/1535-7163.MCT-06-0602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu Y, Sakamuri S, Chen QZ, Keng YF, Khazak V, Illgen K, Schabbert S, Weber L, Menon SR. Solution phase parallel synthesis and evaluation of MAPK inhibitory activities of close structural analogues of a Ras pathway modulator. Bioorg Med Chem Lett. 2004;14:3957–3962. doi: 10.1016/j.bmcl.2004.05.048. [DOI] [PubMed] [Google Scholar]
- 29.Shima F, Yoshikawa Y, Ye M, Araki M, Matsumoto S, Liao J, Hu L, Sugimoto T, Ijiri Y, Takeda A, Nishiyama Y, Sato C, Muraoka S, Tamura A, Osoda T, Tsuda K, Miyakawa T, Fukunishi H, Shimada J, Kumasaka T, Yamamoto M, Kataoka T. In silico discovery of small-molecule Ras inhibitors that display antitumor activity by blocking the Ras-effector interaction. Proc Natl Acad Sci U S A. 2013;110:8182–8187. doi: 10.1073/pnas.1217730110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Castellano E, Sheridan C, Thin MZ, Nye E, Spencer-Dene B, Diefenbacher ME, Moore C, Kumar MS, Murillo MM, Gronroos E, Lassailly F, Stamp G, Downward J. Requirement for interaction of PI3-kinase p110alpha with RAS in lung tumor maintenance. Cancer Cell. 2013;24:617–630. doi: 10.1016/j.ccr.2013.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grant BJ, Lukman S, Hocker HJ, Sayyah J, Brown JH, McCammon JA, Gorfe AA. Novel allosteric sites on Ras for lead generation. PLoS One. 2011;6:e25711. doi: 10.1371/journal.pone.0025711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosnizeck IC, Graf T, Spoerner M, Trankle J, Filchtinski D, Herrmann C, Gremer L, Vetter IR, Wittinghofer A, Konig B, Kalbitzer HR. Stabilizing a weak binding state for effectors in the human ras protein by cyclen complexes. Angew Chem Int Ed Engl. 2010;49:3830–3833. doi: 10.1002/anie.200907002. [DOI] [PubMed] [Google Scholar]
- 33.Rosnizeck IC, Spoerner M, Harsch T, Kreitner S, Filchtinski D, Herrmann C, Engel D, Konig B, Kalbitzer HR. Metal-bis(2-picolyl)amine complexes as state 1(T) inhibitors of activated Ras protein. Angew Chem Int Ed Engl. 2012;51:10647–10651. doi: 10.1002/anie.201204148. [DOI] [PubMed] [Google Scholar]
- 34.Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature. 2013;503:548–551. doi: 10.1038/nature12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lim SM, Westover KD, Ficarro SB, Harrison RA, Choi HG, Pacold ME, Carrasco M, Hunter J, Kim ND, Xie T, Sim T, Janne PA, Meyerson M, Marto JA, Engen JR, Gray NS. Therapeutic targeting of oncogenic K-Ras by a covalent catalytic site inhibitor. Angew Chem Int Ed Engl. 2014;53:199–204. doi: 10.1002/anie.201307387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Geyer M, Schweins T, Herrmann C, Prisner T, Wittinghofer A, Kalbitzer HR. Conformational transitions in p21ras and in its complexes with the effector protein Raf-RBD and the GTPase activating protein GAP. Biochemistry. 1996;35:10308–10320. doi: 10.1021/bi952858k. [DOI] [PubMed] [Google Scholar]