Abstract
The evolutionary history and classification of epiphyllous cryptogams are still poorly known. Leptolejeunea is a largely epiphyllous pantropical liverwort genus with about 25 species characterized by deeply bilobed underleaves, elliptic to narrowly obovate leaf lobes, the presence of ocelli, and vegetative reproduction by cladia. Sequences of three chloroplast regions (rbcL, trnL‐F, psbA) and the nuclear ribosomal ITS region were obtained for 66 accessions of Leptolejeunea and six outgroup species to explore the phylogeny, divergence times, and ancestral areas of this genus. The phylogeny was estimated using maximum‐likelihood and Bayesian inference approaches, and divergence times were estimated with a Bayesian relaxed clock method. Leptolejeunea likely originated in Asia or the Neotropics within a time interval from the Early Eocene to the Late Cretaceous (67.9 Ma, 95% highest posterior density [HPD]: 47.9–93.7). Diversification of the crown group initiated in the Eocene or early Oligocene (38.4 Ma, 95% HPD: 27.2–52.6). Most species clades were established in the Miocene. Leptolejeunea epiphylla and L. schiffneri originated in Asia and colonized African islands during the Plio‐Pleistocene. Accessions of supposedly pantropical species are placed in different main clades. Several monophyletic morphospecies exhibit considerable sequence variation related to a geographical pattern. The clear geographic structure of the Leptolejeunea crown group points to evolutionary processes including rare long‐distance dispersal and subsequent speciation. Leptolejeunea may have benefitted from the large‐scale distribution of humid tropical angiosperm forests in the Eocene.
Keywords: ancestral area estimation, bryophyte, cryptic speciation, divergence time estimation, epiphyte, Leptolejeunea, phylogeny
1. Introduction
Range estimation is a challenging theme in morphologically little differentiated groups of organisms and suitable to improve understanding of species diversity and evolution. Many bryophyte genera belong to these critical groups and are in need of thorough reinvestigation including integrative molecular–morphological approaches; however, to date, only a limited number of studies is available (Dong et al., 2012; Forrest, Salazar‐Allen, Gudiño, Korpelainen, & Long, 2011; Hedenäs et al., 2014; Heinrichs et al., 2015; Renner et al., 2013; Vanderpoorten, Patiño, Dirkse, Blockeel, & Hedenäs, 2015; Vigalondo et al., 2016). These studies identified numerous morphologically not or weakly differentiated bryophyte species of which many have rather narrow ranges.
Prior to the advent of DNA‐based investigations, many bryophyte species were considered to have broad, often intercontinental ranges equivalent to the ranges of angiosperm genera (Shaw, 2001; Vanderpoorten, Gradstein, Carine, & Devos, 2010). The intercontinental distributions of these species were interpreted as a vicariant pattern within species of ancient origin (Schuster, 1983) some of which were thought to date back to the Jurassic (Stotler & Crandall‐Stotler, 1974). However, inferences of Mesozoic ages of bryophyte species have been contradicted by DNA‐based divergence time estimates that have identified crown‐group diversification events within the Cenozoic in many lineages (Cooper, Henwood, & Brown, 2012; Feldberg et al., 2014; Laenen et al., 2014; Wilson, Heinrichs, Hentschel, Gradstein, & Schneider, 2007). Divergence time estimates suggest long‐distance dispersal (LDD) is more likely than vicariance as the process resulting in extant intercontinental ranges (Devos & Vanderpoorten, 2009; Dong et al., 2012; Hartmann, Wilson, Gradstein, Schneider, & Heinrichs, 2006; Scheben, Bechteler, Lee, Pócs, Schäfer‐Verwimp, & Heinrichs, 2016; Sun, He, & Glenny, 2014). Divergence time estimates also suggested an important role of angiosperm‐dominated forests in shaping the diversity of epiphyllic cryptogams (Feldberg et al., 2014).
Leptolejeunea (Spruce) Steph. is a pantropical genus of nearly exclusively epiphyllous leafy liverworts that grow in lowland and lower montane rainforests, occasionally also in high montane rainforests up to ca. 3,000 m (Bischler, 1969). The genus includes both local endemics (Shu, Zhu, & Pócs, 2016) and intercontinentally distributed species such as L. elliptica, L. epiphylla, and L. maculata (Grolle, 1976; Pócs & Lye, 1999; Schuster, 1980; Zhu & So, 2001). Leptolejeunea is characterized by its minute size, deeply bilobed underleaves with two widely divergent and subulate lobes, elliptic to narrowly obovate leaf lobes often with dentate margins, the presence of one to several ocelli in leaf lobes, and vegetative reproduction by cladia (Figure 1). Several species show a tendency for dry leaves to become elevated and produce monoterpenes that emit a strong fragrance (Gradstein, Churchill, & Salazar‐Allen, 2001) meaning the genus can be readily identified even in the field; yet identification of species is notoriously difficult. Söderström et al. (2016) accepted 48 species but indicated knowledge problems or serious doubts about the taxonomic value of many. An earlier study estimated global diversity at 25 species (Gradstein et al., 2001). So far, only a few accessions have been included in molecular phylogenetic studies (Ahonen, Muonen, & Piippo, 2003; Heinrichs et al., 2014; Wilson, Gradstein, Schneider, & Heinrichs, 2007). Results from these studies rejected a previously hypothesized close relationship between Leptolejeunea and Drepanolejeunea based on shared underleaf shape and the presence of ocelli in leaves of both genera (Gradstein, 2013), and resolved Leptolejeunea in a relatively isolated position within Lejeuneaceae subf. Lejeuneoideae (Heinrichs et al., 2014). Lejeuneaceae subtribe Leptolejeuneinae was established as a result to accommodate Leptolejeunea (Heinrichs et al., 2014). However, molecular phylogenetic investigations conducted to date have not improved current morphology‐based species concepts nor resolved biogeographic patterns.
Figure 1.

Images of two species of Leptolejeunea. (a) Habitus of dried herbarium specimen of Leptolejeunea convexistipa showing epiphyllous growth on a fern leaf. (b) Leaf of Leptolejeunea epiphylla with four ocelli in a broken row indicated by red stars. (c) Part of shoot of Leptolejeunea convexistipa focusing on a leaf with one basal ocellus (red star). Note the characteristic underleaf of the genus Leptolejeunea at the bottom left corner (black arrowhead)
Currently, in contradiction to more traditional views of morphological species, widespread Leptolejeunea species are believed to be the result of recent LDD out of Asia, a hypothesis promoted by Schuster (1983: 618): “taxa such as Leptolejeunea elliptica… have shown dispersal, clearly in geologically recent times, well out from Asia into the Pacific, to South America, Central and southern North America.” Here, we extend the sampling of Heinrichs et al. (2014) and test previous hypotheses on origins and extant distribution of Leptolejeunea species. We provide evidence for a Cenozoic origin of the Leptolejeunea crown group and reject pantropical species ranges.
2. Materials and Methods
2.1. Taxon sampling, DNA extraction, PCR amplification, sequencing, and alignment
Tissue for DNA extraction was isolated from Leptolejeunea specimens from the herbaria EGR, GOET, SP, and Schäfer‐Verwimp (SV). Specimens were revised based on literature and considering results from phylogenetic analyses. Total genomic DNA was isolated using the Invisorb Spin Plant Mini Kit (Stratec Molecular GmbH, Berlin, Germany). Four markers were amplified: the nuclear ribosomal internal transcribed spacer region (ITS1‐5.8S‐ITS2), the chloroplast rbcL gene, the trnL‐trnF region, and the psbA gene together with the psbA‐trnH intergenic spacer. PCR amplification of the first three markers follows Bechteler, Lee, Schäfer‐Verwimp, Pócs, et al. (2016). The psbA/psbA‐trnH region was amplified using the PCR program and primers (trnK2F, 510F, 576R, trnHR) described in Forrest and Crandall‐Stotler (2004). This protocol was modified as follows: 0.4 μL of MyTaq Polymerase (Bioline Reagents Ltd., UK), 11 μL of reaction buffer, 1 μL of upstream primer, 1 μL of downstream primer, and 1 μL of template DNA. The mix was filled up with double‐distilled water to a total volume of 50 μL. Representatives of Pycnolejeunea and Xylolejeunea were chosen as outgroups following phylogenetic hypotheses of Wilson, Gradstein, et al. (2007), Bechteler, Lee, Schäfer‐Verwimp, Pócs, et al. (2016) and Bechteler, Lee, Schäfer‐Verwimp, Renner, et al. (2016). Corresponding sequences were downloaded from GenBank (http://www.ncbi.nlm.nih.gov/genbank/), in addition to published sequences of Leptolejeunea. The resulting dataset comprised 66 specimens of Leptolejeunea and three specimens each of Pycnolejeunea and Xylolejeunea (Table 1). All sequences were aligned manually with bioedit 7.1.3.0 (Hall, 1999), and ambiguous sites were excluded.
Table 1.
Taxa used in this study, including information about the geographical origin, voucher details, as well as GenBank accession numbers. Accession numbers in bold were obtained from GenBank
| Taxon | Origin | Collector, voucher number, and herbarium | GenBank accession numbers | |||
|---|---|---|---|---|---|---|
| rbcL | trnLF | psbA | nrITS | |||
| Leptolejeunea amphiophthalma Zwickel | Malaysia | Pócs et al. 13168/AA (EGR) | KX808754 | KX808806 | KY006551 | KX808704 |
| Leptolejeunea astroidea (Mitt.) Steph. | Príncipe Island | Shevock 40015A (EGR) | KX808792 | KX808851 | KY006539 | KX808742 |
| L. astroidea | Uganda | Pócs et al. 97108/O (EGR) | KX808791 | KX808850 | – | KX808741 |
| Leptolejeunea balansae Steph. | Malaysia | Pócs et al. 13184/F (EGR) | KX808777 | KX808832 | KY006538 | KX808725 |
| Leptolejeunea brasiliensis Bischl. | Brazil (I) | Peralta & Carmo 14222 (SP) | KX808758 | KX808810 | KY006502 | KX808708 |
| L. brasiliensis | Brazil (II) | Yano 28424 (SP) | KX808756 | KX808808 | KY006500 | KX808706 |
| L. brasiliensis | Brazil (III) | Peralta & Guiglota 13863 (SP) | KX808757 | KX808809 | KY006501 | KX808707 |
| Leptolejeunea convexistipa Bischl. | Dominican Republic | Schäfer‐Verwimp & Verwimp 27206/B (SV) | KX808800 | – | KY006540 | – |
| L. convexistipa | Ecuador (I) | Schäfer‐Verwimp 24419/C (SV) | KX808799 | – | – | KX808748 |
| L. convexistipa published as elliptica (Lehm. & Lindenb.) Schiffn. | Ecuador (II) | Wilson et al. 04‐18 (GOET) | DQ983698 | – | EF011862 | DQ987375 |
| L. convexistipa | Ecuador (III) | Schäfer‐Verwimp et al. 24407/E (SV) | KX808798 | KX808856 | KY006533 | KX808747 |
| L. convexistipa | Panama (I) | Schäfer‐Verwimp & Verwimp 30861 (JE) | KF954161 | KF954151 | – | KF954154 |
| L. convexistipa | Panama (II) | Schäfer‐Verwimp & Verwimp 30937/A (SV) | KX808801 | KX808857 | KY006534 | KX808749 |
| Leptolejeunea dapitana Steph. | Malaysia (I) | Pócs et al. 13160/Q (EGR) | KX808772 | KX808824 | KY006513 | KX808719 |
| L. dapitana | Malaysia (II) | Pócs et al. 13160/L (EGR) | KX808771 | KX808823 | KY006512 | KX808718 |
| L. dapitana | Vietnam | Luong TP211‐004b (EGR) | KX808770 | KX808822 | KY006511 | KX808717 |
| Leptolejeunea elliptica (Lehm. & Lindenb.) Schiffn. | Dominican Republic | Pócs & Pócs 03157/AB (GOET) | KX808795 | KX808854 | KY006532 | KX808744 |
| L. elliptica | Ecuador (I) | Schäfer‐Verwimp & Nebel 32794 (SV) | KX808794 | KX808853 | KY006531 | KX808743 |
| L. elliptica | Ecuador (II) | Schäfer‐Verwimp & Nebel 32834/A (SV) | KX808797 | – | KY006552 | KX808746 |
| L. elliptica | Guadeloupe | Schäfer‐Verwimp & Verwimp 22518 (SV) | KX808793 | KX808852 | KY006541 | – |
| L. elliptica | Jamaica | Schäfer‐Verwimp 34834/E (SV) | KX808796 | KX808855 | KY006549 | KX808745 |
| Leptolejeunea epiphylla (Mitt.) Steph. | Cambodia | Pócs s.n. (SV) | KX808765 | KX808817 | KY006546 | KX808703 |
| L. epiphylla | Malaysia (I) | Pócs et al. 13172/F (EGR) | KX808764 | KX808816 | KY006550 | KX808713 |
| L. epiphylla | Malaysia (II) | Schäfer‐Verwimp & Verwimp 19081 (JE) | KF954163 | – | – | KF954156 |
| L. epiphylla | Mayotte | Pócs et al. 9288/AA (EGR) | – | KX808818 | KY006508 | KX808714 |
| L. epiphylla | Príncipe Island (I) | Shevock 40133 (SV) | KX808767 | KX808819 | KY006509 | KX808715 |
| L. epiphylla | Príncipe Island (II) | Shevock 42132 (EGR) | KX808768 | KX808820 | KY006545 | KX808702 |
| L. epiphylla | Indonesia, Sumatra | Schäfer‐Verwimp & Verwimp 24962/A (SV) | KX808769 | KX808821 | KY006510 | KX808716 |
| L. epiphylla | Thailand | Schäfer‐Verwimp 16245 (SV) | KX808766 | – | – | KX808701 |
| Leptolejeunea exocellata (Spruce) A.Evans | Argentina | Schäfer‐Verwimp & Verwimp 9330 (GOET) | KX808760 | KX808812 | KY006504 | KX808700 |
| L. exocellata | Dominican Republic (I) | Schäfer‐Verwimp & Verwimp 27018/A (SV) | KX808763 | KX808815 | KY006507 | KX808712 |
| L. exocellata | Dominican Republic (II) | Schäfer‐Verwimp & Verwimp 27197/A (SV) | KX808761 | KX808813 | KY006505 | KX808710 |
| L. exocellata | Dominican Republic (III) | Schäfer‐Verwimp & Verwimp 27215/C (SV) | KX808762 | KX808814 | KY006506 | KX808711 |
| L. exocellata | Ecuador | Schäfer‐Verwimp et al. 24407/C (SV) | KX808759 | KX808811 | KY006503 | KX808709 |
| Leptolejeunea foliicola Steph. | Indonesia, Bali | Schäfer‐Verwimp & Verwimp 16689/E (SV) | – | KX808843 | – | KX808734 |
| L. foliicola | Malaysia (I) | Schäfer‐Verwimp & Verwimp 18903/C (SV) | KX808785 | KX808842 | KY006525 | KX808733 |
| L. foliicola | Malaysia (II) | Schäfer‐Verwimp & Verwimp 18976 (SV) | KX808786 | KX808844 | KY006526 | KX808735 |
| Leptolejeunea maculata (Mitt.) Schiffn. | Malaysia (I) | Pócs et al. 13171/G (EGR) | KX808782 | KX808839 | KY006523 | KX808731 |
| L. maculata | Malaysia (II) | Pócs et al. 13168/AE (EGR) | KX808783 | KX808840 | KY006542 | – |
| L. maculata | Malaysia (III) | Pócs et al. 13167/AM (EGR) | – | KX808837 | KY006521 | KX808729 |
| L. maculata | Malaysia (IV) | Schäfer‐Verwimp & Verwimp 18599/A (SV) | KX808781 | KX808838 | KY006522 | KX808730 |
| Leptolejeunea moniliata Steph. | Guadeloupe | Schäfer‐Verwimp & Verwimp 22117/A (SV) | KX808755 | KX808807 | KY006499 | KX808705 |
| Leptolejeunea radicosa (Nees ex Mont.) Grolle | Dominica | Schäfer‐Verwimp & Verwimp 17723/C (JE) | KF954165 | – | – | KF954158 |
| L. radicosa | Guadeloupe (I) | Schäfer‐Verwimp & Verwimp 22305/A (SV) | KX808804 | KX808860 | KY006537 | KX808753 |
| L. radicosa | Guadeloupe (II) | Schäfer‐Verwimp & Verwimp 22417/E (SV) | KX808803 | KX808859 | KY006536 | KX808751 |
| L. radicosa | Guadeloupe (III) | Schäfer‐Verwimp & Verwimp 22414/D (SV) | KX808805 | – | – | KX808752 |
| L. radicosa | Panama | Schäfer‐Verwimp & Verwimp 30795 (SV) | KX808802 | KX808858 | KY006535 | KX808750 |
| Leptolejeunea schiffneri Steph. | Malaysia | Schäfer‐Verwimp & Verwimp 18619/A (SV) | KX808773 | KX808826 | KY006548 | – |
| L. schiffneri | Mayotte (I) | Pócs et al. 05106/BK (SV) | KX808776 | KX808829 | KY006516 | KX808722 |
| L. schiffneri | Mayotte (II) | Pócs et al. 05105/E (EGR) | – | KX808830 | KY006544 | – |
| L. schiffneri | Indonesia, Sumatra (I) | Schäfer‐Verwimp & Verwimp 25233/B (SV) | KX808775 | KX808828 | KY006515 | KX808723 |
| L. schiffneri | Indonesia, Sumatra (II) | Schäfer‐Verwimp & Verwimp 25233/B1 (SV) | KX808774 | KX808827 | KY006547 | KX808721 |
| L. schiffneri | Indonesia, Sumatra (III) | Schäfer‐Verwimp & Verwimp 25228 (SV) | – | KX808825 | KY006514 | KX808720 |
| Leptolejeunea spec. | Thailand (I) | Chantanaorrapint 1352 (EGR) | – | KX808831 | KY006517 | KX808724 |
| Leptolejeunea spec. | Thailand (II) | Schäfer‐Verwimp & Verwimp 16177 (SV) | KX808779 | KX808835 | KY006520 | KX808728 |
| Leptolejeunea subacuta Steph. ex A.Evans published as elliptica (Lehm. & Lindenb.) Schiffn. | China | Koponen et al. 50179 (H) | AY125939 | AY144480 | – | – |
| L. subacuta | Laos | Peregovits NoLaos/8 (EGR) | KX808789 | KX808847 | KY006498 | KX808737 |
| L. subacuta | Japan, Ryukyu Islands | Yamaguchi 15722 (GOET) | KX808787 | KX808845 | KY006527 | KX808736 |
| L. subacuta | Thailand (I) | Schäfer‐Verwimp & Verwimp 23785/C (SV) | – | KX808848 | KY006529 | KX808739 |
| L. subacuta | Thailand (II) | Schäfer‐Verwimp & Verwimp 23791/B (SV) | KX808790 | KX808849 | KY006530 | KX808740 |
| L. subacuta | Thailand (III) | Schäfer‐Verwimp & Verwimp 23834/A (SV) | KX808788 | KX808846 | KY006528 | KX808738 |
| Leptolejeunea cf. subrotundifolia Herzog | Madagascar | Pócs & Szabo 9875/AZ (EGR) | KX808780 | KX808836 | KY006543 | – |
| Leptolejeunea cf. subrotundifolia | Thailand | Pócs & Somadee 1228/C (EGR) | KX808784 | KX808841 | KY006524 | KX808732 |
| Leptolejeunea vitrea (Nees) Schiffn. | Malaysia (I) | Dürhammer D148 (JE) | KF954164 | KF954152 | – | KF954157 |
| L. vitrea | Malaysia (II) | Pócs et al. 13175/O (EGR) | – | KX808833 | KY006518 | KX808726 |
| L. vitrea | Philippines | Schumm & Schwarz 6425 (SV) | KX808778 | KX808834 | KY006519 | KX808727 |
| Pycnolejeunea densistipula (Lehm. & Lindenb.) Steph. | Ecuador | Schäfer‐Verwimp & Preussing 23368 (GOET) | AY548075 | DQ987400 | EF011774 | DQ987294 |
| Pycnolejeunea macroloba (Nees & Mont.) Schiffn. | Brazil | Yano 32740 (M) | KJ408354 | KJ408378 | – | KJ408329 |
| Pycnolejeunea sphaeroides (Sande Lac.) J.B.Jack & Steph. | Malaysia | Schäfer‐Verwimp & Verwimp 18615/B (M) | KJ408355 | KJ408379 | – | KJ408330 |
| Xylolejeunea crenata (Nees & Mont.) X.L.He & Grolle | Brazil | Schäfer‐Verwimp 11225 (GOET) | DQ983740 | DQ987443 | EF011822 | DQ987341 |
| X. crenata | Ecuador | Schäfer‐Verwimp & Nebel 32827/A (M) | KJ408356 | KJ408382 | – | KJ408333 |
| Xylolejeunea grolleana (Pócs) X.L.He & Grolle | Madagascar | Pócs & Szabó 9878/EM (EGR) | KT626911 | KT626928 | – | KT626892 |
2.2. Phylogenetic analyses
Maximum‐likelihood (ML) analyses were conducted using RAxML 8.2.4 (Stamatakis, 2014). The best fit models of evolution selected by jmodeltest 2 (Darriba, Taboada, Doallo, & Posada, 2012) under the Akaike information criterion (AIC; Akaike, 1973) were as follows: TIM3+I+G for rbcL, TPM1uf+G for trnL‐trnF, TIM3+I+G for psbA/psbA‐trnH, and TIM3+I+G for nrITS1‐5.8S‐ITS2. These were not available in RaxML so the best fitting overparameterized model, GTR+G, was used for all markers (Posada, 2008). First, all markers were analyzed separately on the CIPRES Science Gateway (Miller, Pfeiffer, & Schwartz, 2010) using the “thorough ML” option, and an additional analysis was carried out for a combined chloroplast DNA dataset. Clades with bootstrap values (BP) of 70%–94% were regarded as moderately supported and those with BP ≥95% as strongly supported (Erixon, Svennblad, Britton, & Oxelman, 2003). No strongly supported topological contradictions between single markers or the nuclear and plastid datasets were detected. Accordingly, all matrices were concatenated, resulting in an alignment of 3,694 nucleotide positions. Ten thorough ML searches in combination with multiparametric bootstrapping using the autoMRE function (Pattengale, Alipour, Bininda‐Emonds, Moret, & Stamatakis, 2010) were conducted.
Bayesian inference was undertaken with mrbayes 3.2.6 (Ronquist & Huelsenbeck, 2003) using a partition for each marker and a GTR substitution model with rate of invariable sites and gamma rate heterogeneity as recommended by jmodeltest 2. Two metropolis‐coupled Markov chain Monte Carlo (MCMC) analyses, including three heated chains and one cold chain, were run for 10 million generations, sampled every 1,000 generations. TRACER 1.6 (http://tree.bio.ed.ac.uk/software/tracer/) was used to check for convergence and stationarity, and an average standard deviation (SD) of split frequency below 0.01 indicated a sufficiently long run. The initial 25% of sampled trees were discarded as burn‐in. The remainder were summarized with treeannotator 1.8.2 (Drummond, Suchard, Xie, & Rambaut, 2012), and the resulting maximum clade credibility (MCC) tree was visualized using figtree 1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/). BPP values ≥0.95 were regarded as good support (Larget & Simon, 1999).
2.3. Divergence time estimates and biogeography
Dating analyses were performed using BEAST 1.8.2 (Drummond et al., 2012) using the same partitioning scheme and substitution models as the mrbayes analyses. An ultrametric starting tree without time scale was generated by setting the ingroup monophyletic, using linked trees over all partitions, 60 million generations and sampling every 6,000 generations. An uncorrelated log‐normal (UCLN) relaxed clock and a birth–death prior accounting for incomplete sampling (Stadler, 2009) were used. The result was inspected in TRACER, and ESS values >200 indicated good mixing of the MCMC and a sufficient number of generations. A MCC tree was generated with treeannotator 1.8.2 after discarding the first 10% of trees as burn‐in and visualized in figtree. This tree was used as a starting tree for subsequent divergence time estimates. Again, the ingroup was constrained as monophyletic, trees were linked over all partitions, and this analysis ran for 100 million generations sampling every 10,000 generations. As no Leptolejeunea fossils are known, a plastid genome substitution rate of 5 × 10−4 subst./sites/my (Palmer, 1991; Villarreal & Renner, 2012) was used for the three chloroplast markers with a SD of 1 × 10−4 and a normal prior distribution. For the nrITS region, a substitution rate of 1.35 × 10−3 subst./sites/my was adopted from Les, Crawford, Kimball, Moody, & Landolt (2003). A normal prior distribution in combination with the truncate option and upper and lower bounds of 0.4–8.3 × 10−3 subst./sites/my was implemented to allow the rate to vary over the large spectrum of reported nrITS rates (Kay, Whittall, & Hodges, 2006; Villarreal & Renner, 2014). The stepping‐stone sampling in BEAST (Baele, Li, Drummond, Suchard, & Lemey, 2013; Baele et al., 2012; Xie, Lewis, Fan, Kuo, & Chen, 2011) and the Bayes factor (Kass & Raftery, 1995) were used to compare between pure‐birth (Yule), birth–death, and birth–death incomplete sampling tree priors, as well as an UCLN relaxed clock and a strict clock. This resulted in choosing a birth–death incomplete sampling prior in combination with a UCLN relaxed clock model. Log marginal‐likelihood values and Bayes factor values are shown in Table 2. Results of the BEAST run were examined in TRACER, summarized in treeannotator by median branch lengths, and visualized in figtree.
Table 2.
Marginal‐likelihood estimations using stepping‐stone sampling in BEAST and ln Bayes factor calculation resulting in an uncorrelated log‐normal (UCLN) relaxed clock model and a birth–death tree prior accounting for incomplete sampling (BDincompl.) for the Leptolejeunea dataset
| Model 1 | BDincompl., UCLN | BD, UCLN | Yule, UCLN | BDincompl., strict clock | |
|---|---|---|---|---|---|
| Model 2 | Log marginal likelihood | −17,905.64 | −17,911.82 | −17,940.59 | −17,945.11 |
| BDincompl., UCLN | −17,905.64 | 0.00 | −6.17 | −34.95 | −39.46 |
| BD, UCLN | −17,911.82 | 6.17 | 0.00 | −28.78 | −33.29 |
| Yule, UCLN | −17,940.59 | 34.95 | 28.78 | 0.00 | −4.51 |
| BDincompl., strict clock | −17,945.11 | 39.46 | 33.29 | 4.51 | 0.00 |
Afromadagascar, Asia–Australasia, and tropical–subtropical America were chosen as putative areas of endemism, and each specimen was assigned to one of these regions according to the label information. Ancestral areas of distribution were reconstructed using maximum parsimony criteria as implemented in mesquite 3.1 (Maddison & Maddison, 2016) based on the MCC topology from the divergence time analysis. In addition, the R‐package biogeoBEARS (Matzke, 2013a, 2013b, 2014) was employed to infer the ancestral history of Leptolejeunea. This likelihood‐based method implements the LAGRANGE DEC model (Ree & Smith, 2008), DIVA (dispersal‐vicariance analysis; Ronquist, 1997), and BayArea (Landis, Matzke, Moore, & Huelsenbeck, 2013), each of which can be extended with an additional free parameter j accounting for founder‐event speciation. To obtain the recommended operational taxonomic units consisting of monophyletic populations and not individual specimens, specimens of one species with the same putative area of endemism were merged together into a single terminal using the R‐script provided on the biogeoBEARS webpage (http://phylo.wikidot.com/example-biogeobears-scripts#pruning_a_tree). All six models were compared using likelihood values, the AIC, and the AIC corrected for small sample size (AICc) (Matzke, 2014). The maximum number of areas was set to three to account for the assumed pantropical ranges of Leptolejeunea species (Grolle, 1976; Pócs, 2012; Pócs & Lye, 1999; Schuster, 1983).
2.4. Morphological investigation
Specimens were studied under a Carl Zeiss AxioScope A1 compound microscope equipped with a Canon 60D digital camera using transmitted or incident light. The Leptolejeunea convexistipa voucher Schäfer‐Verwimp 35198/A (M) and the L. epiphylla voucher Schäfer‐Verwimp 16245 (M) were digitized (Figure 1). All presented images are digitally stacked photomicrographic composites of up to 20 individual focal planes obtained using the software package HeliconFocus 6.7.1.
3. Results
3.1. Phylogeny
Leptolejeunea splits into three main clades (labeled I, II, III) with clade I placed sister to the remainder of the genus (Figure 2). Clade I includes a Malaysian accession of L. amphiophthalma in an unsupported sister relationship to a robust Neotropical clade consisting of L. moniliata, L. brasiliensis, and L. exocellata. Three accessions of L. brasiliensis were placed sister to a clade with five accessions of L. exocellata. A clade with three accessions of L. exocellata from the Dominican Republic was placed sister to a clade with L. exocellata accessions from Argentina and Ecuador. Clade II achieved a BPP of 1.00 and a BP of 77 and included a lineage with accessions of L. astroidea from Uganda and Príncipe Island, a lineage with Asian accessions of L. subacuta, L. cf. subrotundifolia and L. foliicola, and a Neotropical lineage with accessions of L. convexistipa, L. elliptica, and L. radicosa. An accession of L. radicosa from Panama was placed sister to a clade with accessions from Dominica and Guadeloupe. Clade III comprised Paleotropical accessions (BPP 1.00, BP 99). Leptolejeunea epiphylla split into a clade with two accessions from Sumatra and Malaysia, and a clade with accessions from Cambodia, Malaysia, and Thailand in a sister relationship with accessions from Mayotte and Príncipe Island. The L. epiphylla clade was sister to a clade with accessions assigned to L. dapitana, L. maculata, L. schiffneri, L. vitrea, L. balansae, L. cf. subrotundifolia, and L. spec. indet. The L. schiffneri clade included an Asian lineage and a lineage with accessions from Mayotte. Representatives of other clade III species originated exclusively from Asia. Most species represented by multiple accessions achieved BPPs of 1.00 and BPs >98; L. maculata achieved a BPP of 0.99 and a BP of 62; the monophyly of L. subacuta was unsupported.
Figure 2.
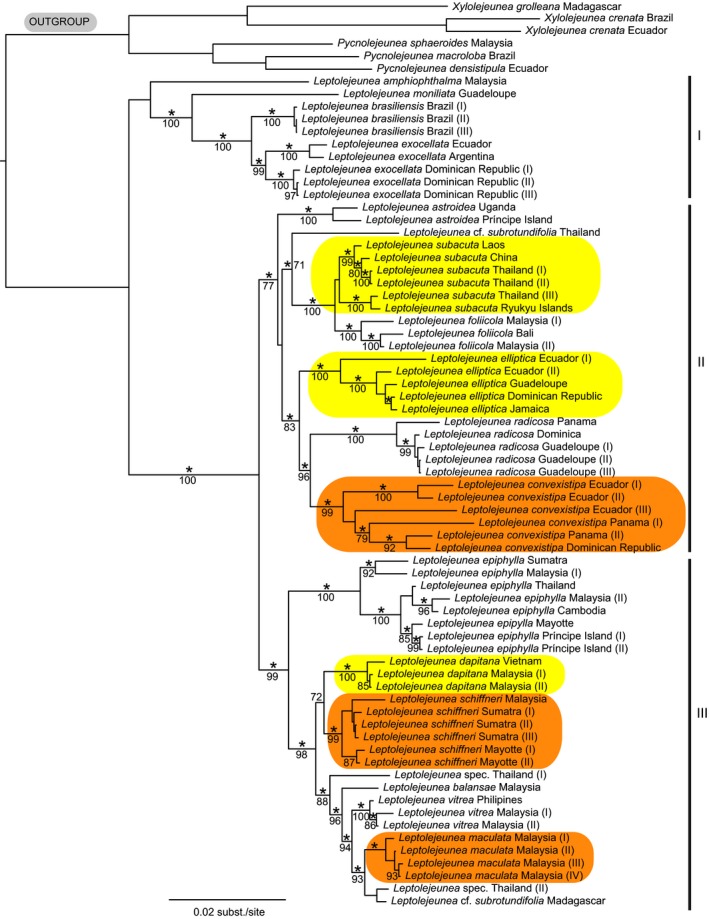
Majority rule consensus tree of trees recovered in stationary phase of Bayesian search. A star indicates a Bayesian Posterior probability >.97. Maximum‐likelihood bootstrap percentage values >70 are also shown at branches. Orange highlighted accessions were earlier considered to belong to Leptolejeunea maculata, and yellow highlighted accessions were earlier considered to belong to L. elliptica
3.2. Divergence time estimates and biogeography
The divergence time analyses (Figure 3) provided evidence for a split between the outgroup and Leptolejeunea in a time interval from the Early Eocene to the Late Cretaceous (67.9 Ma, 95% HPD: 47.9–93.7) and an Oligocene to Eocene (38.4 Ma, 95% HPD: 27.2–52.6) age of the Leptolejeunea crown group. Most of the species clades were established in the Miocene. The biogeoBEARS analyses favored a DIVALIKE+J model for the estimation of ancestral areas (Table 3), and results obtained with this model are shown in Figure 4 in combination with the modified BEAST chronogram. Estimated ancestral area probabilities for selected nodes are given in Table 4. The origin of Leptolejeunea is ambiguous, with the highest probability of an origin in Asia or the Neotropics. Similar results were achieved using maximum parsimony criteria (Figure 3). Neotropical–Paleotropical disjunctions occurred during the Miocene to Eocene. Leptolejeunea epiphylla and L. schiffneri originated in Asia and colonized African islands during the Plio‐Pleistocene.
Figure 3.
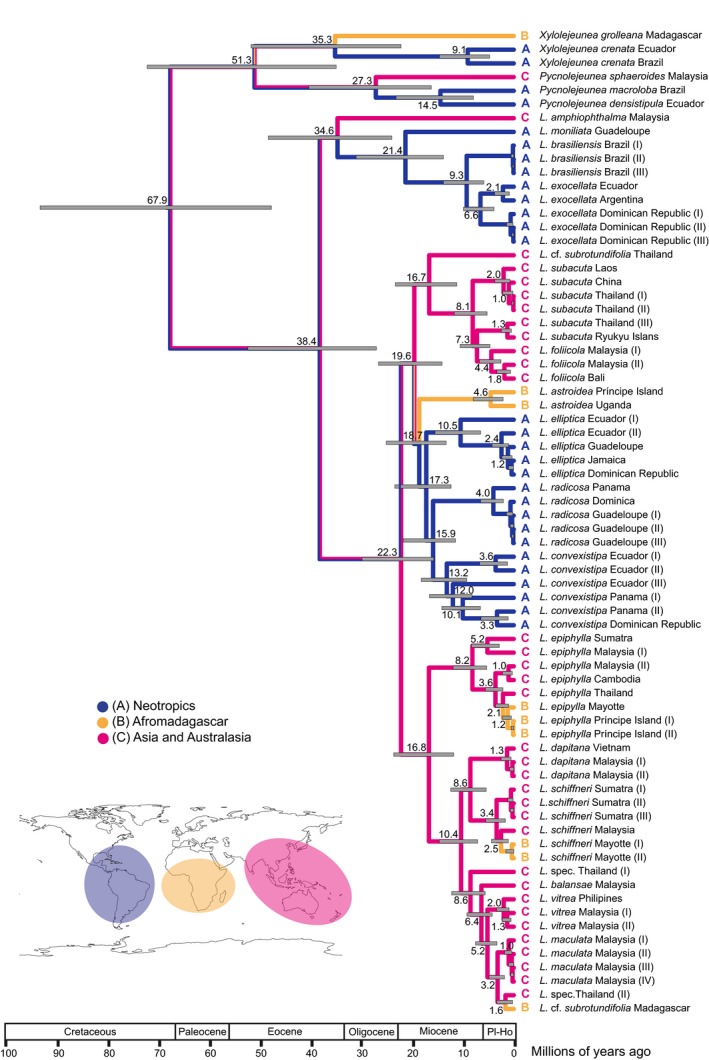
BEAST chronogram with 95% highest posterior density (HPD) intervals and branches colored according to the most parsimonious reconstruction of distributions of Leptolejeunea. Putative areas of endemism are indicated for every accession rather than morphospecies. Node ages ≥1 Ma are reported
Table 3.
Results of the biogeoBEARS analyses favoring a DIVALIKE+J model, as shown in bold, according to model selection by log‐likelihood values (lnL), Akaike information criterion (AIC), and AIC corrected for small sample size (AIC c)
| lnL | n | d | e | j | AIC | AICc | |
|---|---|---|---|---|---|---|---|
| DEC | −42.11 | 2 | 0.008 | 0.003 | 0 | 88.22 | 88.72 |
| DEC+J | −24.95 | 3 | 10−12 | 10−12 | 0.15 | 55.89 | 56.94 |
| DIVALIKE | −38.08 | 2 | 0.009 | 10−12 | 0 | 80.16 | 80.66 |
| DIVALIKE+J | −24.79 | 3 | 10 −12 | 10 −12 | 0.14 | 55.59 | 56.63 |
| BAYAREALIKE | −54.58 | 2 | 0.009 | 0.03 | 0 | 113.2 | 113.7 |
| BAYAREALIKE+J | −25.77 | 3 | 10−7 | 10−7 | 0.14 | 57.54 | 58.58 |
n, number of parameters; d, rate of dispersal; e, rate of extinction; j, relative probability of founder‐event speciation.
Figure 4.
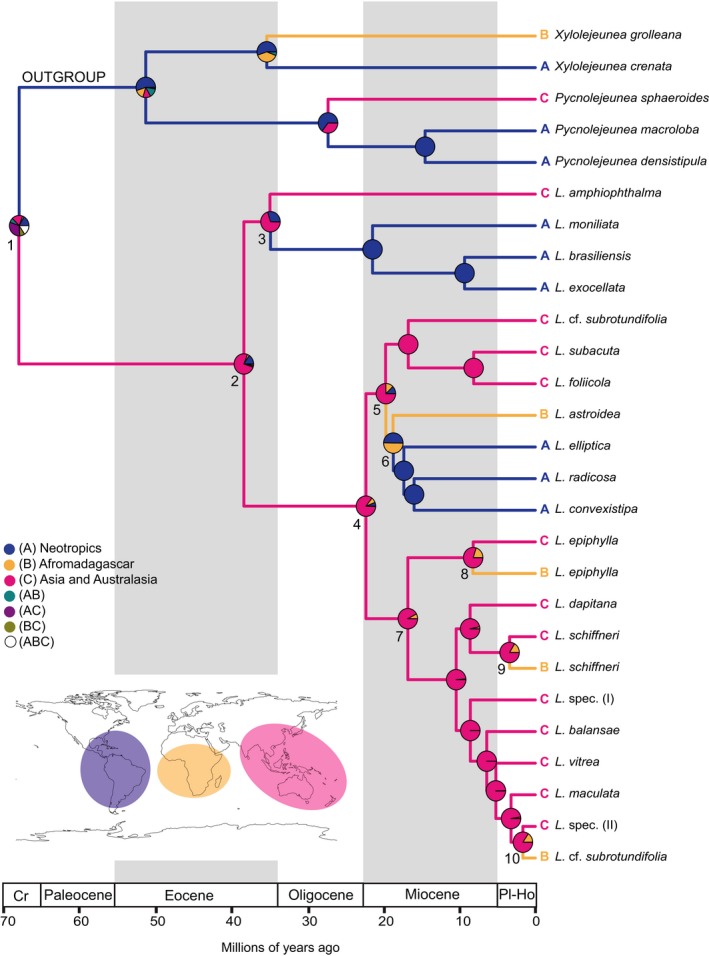
Result of the biogeo BEARS analysis of Leptolejeunea in combination with the modified BEAST chronogram. Circles at nodes represent probabilities for ancestral areas resulting from DIVALIKE analysis accounting for founder‐event speciation. See Table 4 for percent values. Branches are colored according to the most probable area for splits as indicated by biogeo BEARS
Table 4.
Estimated ancestral area probabilities for selected nodes obtained from the biogeoBEARS analysis of Leptolejeunea rounded in percent. Node numbers are displayed in Figure 4. Areas are coded as follows: A, Neotropics; B, Afromadagascar; C, Australasia; AB, AC, BC, ABC are combinations of these areas
| Node | Estimated ancestral area (DIVALIKE+J) |
|---|---|
| 1 | A 17, B 3, C 18, AB 7, AC 31, BC 8, ABC 16 |
| 2 | A 16, B 3, C 76, AC 3, BC 3 |
| 3 | A 30, C 70 |
| 4 | A 6, B 8, C 85, BC 1 |
| 5 | A 12, B 13, C 75 |
| 6 | A 50, B 50 |
| 7 | B 8, C 92 |
| 8 | B 20, C 80 |
| 9 | B 16, C 84 |
| 10 | B 16, C 84 |
4. Discussion
4.1. Bryophyte species in the molecular age
Although intercontinentally disjunct bryophyte species often form monophyla (Heinrichs et al., 2010; Vigalondo et al., 2016), accessions from different continents are often resolved in sister clades (Heinrichs et al., 2011). This pattern of geographically structured phylogenetic relationships suggests gene flow and interbreeding between populations on different continents has ceased, and this may be confirmed by detailed study (Medina, Lara, Goffinet, Garilleti, & Mazimpaka, 2013). Other studies point to the polyphyly of supposedly intercontinentally distributed species (Huttunen & Ignatov, 2010; Renner, 2014) and indicate that monophyletic bryophyte species often have restricted ranges (Medina, Lara, Goffinet, Garilleti, & Mazimpaka, 2012; Medina et al., 2013; Renner et al., 2013). That patterns of phylogenetic and morphological diversification are often decoupled in bryophytes is now well recognized, and many instances of morphologically cryptic species complexes have been documented (Baczkiewicz & Buczkowska, 2016; Kyrkjeeide, Hassel, Flatberg, Shaw, Yousefi, et al., 2016; Odrzykoski & Szweykowski, 1991; Ramaiya et al., 2010; Shaw, Boles, & Shaw, 2008). However, the prevalence of morphologically cryptic divergence, and the number of species resulting from such events, remains unknown. Species circumscription based on morphology may overlook two important features: firstly, the existence of higher phylogenetic diversity than suggested by patterns of morphological variation and secondly, higher geographic structuring than suggested by the distribution of morphological variation (Medina et al., 2013; Ramaiya et al., 2010; Renner, Brown, & Wardle, 2011; Renner et al., 2013).
4.2. Leptolejeunea species ranges and taxonomy
Our study contradicts hypothesized pantropical ranges for two Leptolejeunea species (Figure 2, note highlighted specimens) and supports the hypothesis of Shaw (2001) that morphological uniformity of bryophytes often belies a complex genetic structure. According to our sampling, L. elliptica is restricted to the Neotropics rather than representing a pantropical species (Pócs, 2012; Schuster, 1980). Paleotropical accessions that were earlier assigned to L. elliptica are placed in separate lineages and have been revised to L. dapitana and L. subacuta (Figure 2). The supposedly pantropical L. maculata (Grolle, 1976; Pócs & Lye, 1999) forms three independent lineages (Figure 2). Asian L. maculata s.str. is placed in main clade III, together with a Paleotropical lineage here identified as L. schiffneri. Neotropical accessions of L. maculata belong to main clade II and have been identified as L. convexistipa. Such findings have frequently been explained as instances of cryptic or near cryptic speciation (Shaw, 2001); however, molecular topologies may allow revision of morphological evidence and the identification of morphological character states supporting the different lineages (Forrest et al., 2011; Heinrichs et al., 2015; Renner et al., 2013). Revision of Leptolejeunea specimens is challenging as the taxonomy of this genus relies heavily on the number and distribution of ocelli in the leaves, that is, specialized cells containing only a single large rather than several small oil bodies (He & Piippo, 1999). These often disappear from herbarium specimens. Exceptionally large or small leaf cells in herbarium specimens may be indicative of ocelli; however, ocelli sharing the size of the surrounding leaf cells may not be recognizable in dried materials. A thorough revision of Leptolejeunea thus needs to be based on the investigation of living plants from all parts of the range and sequencing of a comprehensive number of specimens including types or topotypes. New sources of species circumscribing characters also need to be sought. Such work is beyond the scope of this study; however, our data facilitate discrimination between alternative interpretations of species circumscription and to reconstruct the distribution of the main clades. Our data also support the finding of Renner (2015) that morphologically similar leafy liverworts may be placed in different main lineages, despite considerable morphological overlap. Accessions originally assigned to the same species were resolved in different main clades, and the supposedly closely related species L. brasiliensis and L. elliptica (Schuster, 1980) were resolved in main clade I or II (Figure 2). Phylogenies of Lejeuneaceae genera often show a geographical pattern related to the distribution of lineages rather than a morphological pattern. Examples include the genera Lejeunea (Heinrichs et al., 2013) and Diplasiolejeunea (Dong et al., 2012) which exhibit separation into predominantly Neotropical and predominantly Paleotropical lineages. A similar situation manifests in Leptolejeunea.
4.3. Divergence time estimates, biogeography, and infraspecific variation
Our divergence time estimates suggest Cenozoic diversification of Leptolejeunea and contradict Gondwanan vicariance (Raven & Axelrod, 1974) as an explanation for the observed disjunctions. Establishment of the Leptolejeunea crown group in the Eocene accordances well with the appearance of humid megathermal angiosperm forests (Morley, 2011) which provided the preferred epiphyllous habitat of extant Leptolejeunea representatives. Cretaceous gymnosperm forests differed in structure and evaporated less water than tropical angiosperm forests (Boyce & Lee, 2010). Thus, they may not have hosted as diverse epiphyll communities or supported Lejeuneaceae representatives adapted to other niches than modern species (Feldberg et al., 2014). Similar evolutionary processes have been reconstructed for the genera Lejeunea, Harpalejeunea, and Microlejeunea based on molecular and fossil evidence (Heinrichs et al., 2016).
Our reconstruction failed to unambiguously identify the area of origin of Leptolejeunea; however, we need to consider the wide distribution of humid angiosperm forests in the Eocene including the northern “boreotropical” region (Morley, 2011). Lack of fossils and extant species precludes inference of a northern range for Leptolejeunea; however, the Eocene range of Leptolejeunea likely differed from the current distribution. Cooling during the Neogene (Zachos, Pagani, Sloan, Thomas, & Billups, 2001) may have resulted in range contraction and extinction in the north, and possibly the extinction of some early lineages. Caution interpreting biogeographical reconstructions utilizing standard substitution rates is always required; however, our chronogram suggests either lower speciation or higher extinction rates during the early Oligocene cooling phase (Liu et al., 2009), and the establishment of extant Leptolejeunea species predominantly in the Miocene. This pattern could relate to a Miocene reorganization of tropical forests. Miocene origins for extant diversity have also been observed in mosses (Lewis, Rozzi, & Goffinet, 2014; Shaw et al., 2010) and leptosporangiate ferns (Schneider et al., 2010; Wei et al., 2015). The age of the oldest Neotropical–Paleotropical disjunctions could relate to boreotropical migration (Davis, Bell, Matthews, & Donoghue, 2002; Le Péchon et al., 2016) although a thorough reconstruction is precluded by the lack of fossils. Miocene disjunctions are better explained by LDD, as are the island occurrences of several species. Liverworts have dispersed to the African continent and associated islands from both the Neotropics and Asia (Feldberg et al., 2007; Heinrichs et al., 2005). Both biogeographical analyses (Figures 3 and 4) provide evidence for an Asian origin of the African accessions of L. epiphylla, L. schiffneri, and L. cf. subrotundifolia, whereas the origin of the African L. astroidea remains unclear. African taxa nesting in Asian clades have also been described for ferns (Hennequin, Hovenkamp, Christenhusz, & Schneider, 2010; Janssen, Kreier, & Schneider, 2007) and angiosperms (Kulju, Sierra, Draisma, Samuel, & van Welzen, 2007; Li, Dressler, Zhang, & Renner, 2009; Richardson, Chatrou, Mols, Erkens, & Pirie, 2004). Monsoon trade winds were proposed as dispersal agent from Asia to Africa (Li et al., 2009) and could also be responsible for the observed pattern in Leptolejeunea. Alternatively, animal‐mediated dispersal may contribute to current disjunctions. At small spatial scales, millipedes have been demonstrated to move gemmae of species of the moss genus Calymperes (Zona, 2013). Larger animals that move over correspondingly larger spatial scales may also transport propagules and plant fragments (Lewis et al., 2014). In New Zealand, the isolated occurrences of the tropical Calymperes tenerum are congruent with known visitation sites of the predominantly tropical black‐winged petrel (P. J. de Lange, personal communication). Seabirds are known to visit potential or actual breeding sites, even though visiting individuals may not nest there. To visit these sites, which are often forested, birds literally crash through the canopy to the ground, thus coming into close, vigorous contact with leaf and twig surfaces, providing ample opportunity for plant fragments to become deeply embedded within the bird's feather matrix. Seabirds roam widely during their nonbreeding season and routinely traverse oceans and have been known to traverse land masses bridging oceanic waterways.
The island occurrences provide evidence for the ability of Leptolejeunea species to disperse over long distances either by vegetative propagules (Laenen et al., 2016) or by spores (Van Zanten & Gradstein, 1988). However, successful LDD seems rare in Leptolejeunea, as indicated by the plurispecies clades being restricted to either the Neotropics or the Paleotropics, but also by the genetic variation within single morphospecies. Although our data support a narrower species concept and reinstatement of several putative synonyms, some species clades still have a considerable molecular variation, with initial splits in the late Miocene (Figure 3). Examples include a split between mainland South American L. exocellata and accessions from the West Indian Islands, splits within Asian L. epiphylla, and splits within Neotropical L. convexistipa. Considerable molecular variation related to a geographical rather than a morphological pattern has been observed for a larger number of liverworts (Fuselier et al., 2009; Heinrichs et al., 2015; Ramaiya et al., 2010) although it is still somewhat unclear whether this variation is in general indicative of genetically independent entities. Follow‐up studies should thus involve denser sampling and additional markers including microsatellites. Intercontinental gene flow has already been demonstrated for bryophytes, especially for holarctic species of the moss genus Sphagnum (Kyrkjeeide, Hassel, Flatberg, Shaw, Brochmann, et al., 2016; Shaw et al., 2014); however, the epiphyllous habitat of Leptolejeunea species in the understory of tropical forests may lower the LDD success rate compared to Sphagnum species which occur in open wetland systems.
4.4. Perspectives
Every disjunction has its first day; hence, we cannot generally reject intercontinental or even pantropical species ranges (Lewis et al., 2014). On the other hand, a growing body of evidence indicates that LDD occurs only infrequently in bryophytes and that it is thus often associated with speciation. The accumulation of genetic disparity in bryophytes is often not associated with the accumulation of a similar amount of morphological disparity (Baczkiewicz & Buczkowska, 2016; Ramaiya et al., 2010), although there are exceptions (Heinrichs, Gradstein, Groth, & Lindner, 2003). Lack of molecular support for morphology‐based supraspecific taxa such as sections and subgenera (Devos, Renner, Gradstein, Shaw, & Vanderpoorten, 2011) further complicates the understanding of bryophyte evolution and appropriate choice of ingroup representatives. A reliable reconstruction of the evolutionary history and biogeography of bryophytes thus needs to be based on comprehensive molecular phylogenies with complete population‐level sampling. Only such phylogenies will facilitate species identification and refined estimation of bryophyte global diversity and origins.
Conflict of Interest
None declared.
Acknowledgments
We thank the directors and curators of the Herbaria Eger and Göttingen for the loan of specimens and the permission to extract DNA, and Peter de Lange (Auckland) for comments. Financial support by the German Research Foundation (grant HE 3584/6 to JH) and the Alexander von Humboldt Foundation (grant to GEL) is gratefully acknowledged.
Bechteler, J. , Schäfer‐Verwimp, A. , Lee, G. E. , Feldberg, K. , Pérez‐Escobar, O. A. , Pócs, T. , Peralta, D. F. , Renner, M. A. M. and Heinrichs, J. (2017), Geographical structure, narrow species ranges, and Cenozoic diversification in a pantropical clade of epiphyllous leafy liverworts. Ecology and Evolution, 7: 638–653. doi: 10.1002/ece3.2656
References
- Ahonen, I. , Muonen, J. , & Piippo, S. (2003). Inferring the phylogeny of Lejeuneaceae: A first appraisal of molecular data. The Bryologist, 106, 297–308. [Google Scholar]
- Akaike, H. (1973). Information theory as an extension of the maximum likelihood principle In Petrov B. N., & Csâki F. (Eds.), Second International Symposium on Information Theory (pp. 267–281). Budapest: Akadémia Kiado. [Google Scholar]
- Baczkiewicz, A. , & Buczkowska, J. (2016). Differentiation and genetic variability of three cryptic species within the Aneura pinguis complex (Jungermanniidae, Marchantiophyta). Cryptogamie, Bryologie, 37, 3–18. [Google Scholar]
- Baele, G. , Lemey, P. , Bedford, T. , Rambaut, A. , Suchard, M. A. , & Alekseyenko, A. V. (2012). Improving the accuracy of demographic and molecular clock model comparison while accommodating phylogenetic uncertainty. Molecular Biology and Evolution, 29, 2157–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baele, G. , Li, W. L. S. , Drummond, A. J. , Suchard, M. A. , & Lemey, P. (2013). Accurate model selection of relaxed molecular clocks in Bayesian phylogenetics. Molecular Biology and Evolution, 30, 239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechteler, J. , Lee, G. E. , Schäfer‐Verwimp, A. , Pócs, T. , Peralta, D. F. , Renner, M. A. M. , , … Heinrichs, J. (2016). Towards a monophyletic classification of Lejeuneaceae IV: Reinstatement of Allorgella, transfer of Microlejeunea aphanella to Vitalianthus and refinements of the subtribal classification. Plant Systematics and Evolution, 302, 187–201. [Google Scholar]
- Bechteler, J. , Lee, G. E. , Schäfer‐Verwimp, A. , Renner, M. A. M. , Peralta, D. F. , & Heinrichs, J. (2016). Towards a monophyletic classification of Lejeuneaceae V: The systematic position of Pictolejeunea . Phytotaxa, 280, 259–270. [Google Scholar]
- Bischler, H. (1969). Le genre Leptolejeunea (Spruce) Steph. en Amérique. Nova Hedwigia, 17, 265–350. [Google Scholar]
- Boyce, C. K. , & Lee, J.‐E. (2010). An exceptional role for flowering plant physiology in the expansion of tropical rainforests and biodiversity. Proceedings of the Royal Society of London B: Biological Sciences, 277, 3437–3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper, E. D. , Henwood, M. J. , & Brown, E. A. (2012). Are the liverworts really that old? Cretaceous origins and Cenozoic diversifications in Lepidoziaceae reflect a recurrent theme in liverwort evolution. Biological Journal of the Linnean Society, 107, 425–441. [Google Scholar]
- Darriba, D. , Taboada, G. L. , Doallo, R. , & Posada, D. (2012). JModelTest 2: More models, new heuristics and parallel computing. Nature Methods, 9, 772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, C. C. , Bell, C. D. , Matthews, S. , & Donoghue, M. J. (2002). Laurasian migration explains Gondwanan disjunctions: Evidence from Malpighiaceae. Proceedings of the National Academy of Sciences of the United States of America, 99, 6833–6837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos, N. , Renner, M. A. M. , Gradstein, S. R. , Shaw, J. , & Vanderpoorten, A. (2011). Molecular data challenge traditional subgeneric divisions in the leafy liverwort Radula . Taxon, 60, 1623–1632. [Google Scholar]
- Devos, N. , & Vanderpoorten, A. (2009). Range disjunctions, speciation, and morphological transformation rates in the liverwort genus Leptoscyphus . Evolution, 63, 779–792. [DOI] [PubMed] [Google Scholar]
- Dong, S. , Schäfer‐Verwimp, A. , Meinecke, P. , Feldberg, K. , Bombosch, A. , Pócs, T. , … Heinrichs, J. (2012). Tramps, narrow endemics and morphologically cryptic species in the epiphyllous liverwort Diplasiolejeunea . Molecular Phylogenetics and Evolution, 65, 582–594. [DOI] [PubMed] [Google Scholar]
- Drummond, A. J. , Suchard, M. A. , Xie, D. , & Rambaut, A. (2012). Bayesian phylogenetics with BEAUti and the BEAST 1.7. Molecular Biology and Evolution, 29, 1969–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erixon, P. , Svennblad, B. , Britton, T. , & Oxelman, B. (2003). Reliability of Bayesian posterior probabilities and bootstrap frequencies in phylogenetics. Systematic Biology, 52, 665–673. [DOI] [PubMed] [Google Scholar]
- Feldberg, K. , Hentschel, J. , Wilson, R. , Rycroft, D. S. , Glenny, D. , & Heinrichs, J. (2007). Phylogenetic biogeography of the leafy liverwort Herbertus (Jungermanniales, Herbertaceae) based on nuclear and chloroplast DNA sequence data: Correlation between genetic variation and geographical distribution. Journal of Biogeography, 34, 688–698. [Google Scholar]
- Feldberg, K. , Schneider, H. , Stadler, T. , Schäfer‐Verwimp, A. , Schmidt, A. R. , & Heinrichs, J. (2014). Epiphytic leafy liverworts diversified in angiosperm‐dominated forests. Scientific Reports, 4, 5974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrest, L. L. , & Crandall‐Stotler, B. J. (2004). A phylogeny of the simple thalloid liverworts (Jungermanniopsida, subclass Metzgeriidae) as inferred from five chloroplast genes. Monographs in Systematic Botany from the Missouri Botanical Garden, 98, 119–140. [Google Scholar]
- Forrest, L. L. , Salazar‐Allen, N. , Gudiño, J. A. , Korpelainen, H. , & Long, D. G. (2011). Molecular and morphological evidence for distinct species in Dumortiera (Dumortieraceae). The Bryologist, 114, 102–115. [Google Scholar]
- Fuselier, L. , Davison, P. G. , Clements, M. , Shaw, B. , Devos, N. , Heinrichs, J. , … Shaw, A. J. (2009). Phylogeographic analyses reveal distinct lineages of Metzgeria furcata and M. conjugata (Metzgeriaceae) in Europe and North America. Biological Journal of the Linnean Society, 98, 745–756. [Google Scholar]
- Gradstein, S. R. (2013). A classification of Lejeuneaceae based on molecular and morphological evidence. Phytotaxa, 100, 6–20. [Google Scholar]
- Gradstein, S. R. , Churchill, S. P. , & Salazar‐Allen, N. (2001). Guide to the bryophytes of tropical America. Memoirs of the New York Botanical Garden, 86, 1–577. [Google Scholar]
- Grolle, R. (1976). Drepanolejeunea subgen. Kolpolejeunea—eine neue Untergattung aus der Paläotropis. Journal of the Hattori Botanical Laboratory, 40, 191–216. [Google Scholar]
- Hall, T. A. (1999). BioEdit: A user‐friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series, 41, 95–98, Oxford University Press. [Google Scholar]
- Hartmann, F. A. , Wilson, R. , Gradstein, S. R. , Schneider, H. , & Heinrichs, J. (2006). Testing hypotheses on species delimitations and disjunctions in the liverwort Bryopteris (Jungermanniopsida: Lejeuneaceae). International Journal of Plant Sciences, 167, 1205–1214. [Google Scholar]
- He, X. L. , & Piippo, S. (1999). On the taxonomic significance and classification of ocelli characters in the hepatic family Lejeuneaceae. Bryobrothera, 5, 93–97. [Google Scholar]
- Hedenäs, L. , Désamoré, A. , Laenen, B. , Papp, B. , Quandt, D. , Gonzáles‐Mancebo, J. M. , … Stech, M. (2014). Three species for the price of one within the moss Homalothecium sericeum s.l. Taxon, 63, 249–257. [Google Scholar]
- Heinrichs, J. , Dong, S. , Schäfer‐Verwimp, A. , Pócs, T. , Feldberg, K. , Czumaj, A. , … Schneider, H. (2013). Molecular phylogeny of the leafy liverwort Lejeunea (Porellales): Evidence for a Neotropical origin, uneven distribution of sexual systems and insufficient taxonomy. PLoS One, 8, e82547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrichs, J. , Feldberg, K. , Bechteler, J. , Scheben, A. , Czumay, A. , Pócs, T. , … Schäfer‐Verwimp, A. (2015). Integrative taxonomy of Lepidolejeunea (Porellales, Jungermanniopsida): Ocelli allow the recognition of two neglected species. Taxon, 64, 216–228. [Google Scholar]
- Heinrichs, J. , Gradstein, S. R. , Groth, H. , & Lindner, M. (2003). Plagiochila cucullifolia var. anomala var. nov. from Ecuador, with notes on discordant molecular and morphological variation in Plagiochila . Plant Systematics and Evolution, 242, 205–216. [Google Scholar]
- Heinrichs, J. , Hentschel, J. , Bombosch, A. , Fiebig, A. , Reise, J. , Edelmann, M. , … Shaw, A. J. (2010). One species or at least eight? Delimitation and distribution of Frullania tamarisci (L.) Dumort. (Jungermanniopsida, Porellales) inferred from nuclear and chloroplast DNA markers. Molecular Phylogenetics and Evolution, 56, 1105–1114. [DOI] [PubMed] [Google Scholar]
- Heinrichs, J. , Kreier, H.‐P. , Feldberg, K. , Schmidt, A. R. , Zhu, R. L. , Shaw, B. , … Wissemann, V. (2011). Formalizing morphologically cryptic biological entities: New insights from DNA taxonomy, hybridization, and biogeography in the leafy liverwort Porella platyphylla (Jungermanniopsida, Porellales). American Journal of Botany, 98, 1252–1262. [DOI] [PubMed] [Google Scholar]
- Heinrichs, J. , Lindner, M. , Gradstein, S. R. , Groth, H. , Buchbender, V. , Solga, A. , & Fischer, E. (2005). Origin and subdivision of Plagiochila (Jungermanniidae: Plagiochilaceae) in tropical Africa based on evidence from nuclear and chloroplast DNA sequences and morphology. Taxon, 54, 317–333. [Google Scholar]
- Heinrichs, J. , Schäfer‐Verwimp, A. , Czumay, A. , Dong, S. , Scheben, A. , Feldberg, K. , & Schneider, H. (2014). Towards a monophyletic classification of Lejeuneaceae I: Subtribe Leptolejeuneinae subtr. nov. Phytotaxa, 156, 165–174. [Google Scholar]
- Heinrichs, J. , Scheben, A. , Bechteler, J. , Lee, G. E. , Schäfer‐Verwimp, A. , Hedenäs, L. , … Schmidt, A. R. (2016). Crown group Lejeuneaceae and pleurocarpous mosses in Early Eocene (Ypresian) Indian amber. PLoS One, 11, e0156301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennequin, S. , Hovenkamp, P. , Christenhusz, M. , & Schneider, H. (2010). Phylogenetics and biogeography of Nephrolepis—A tale of old settlers and young tramps. Botanical Journal of the Linnean Society, 164, 113–127. [Google Scholar]
- Huttunen, S. , & Ignatov, M. S. (2010). Evolution and taxonomy of aquatic species in the genus Rhynchostegium (Brachytheciaceae, Bryophyta). Taxon, 59, 791–808. [Google Scholar]
- Janssen, T. , Kreier, H.‐P. , & Schneider, H. (2007). Origin and diversification of African ferns with special emphasis on Polypodiaceae. Brittonia, 59, 159–181. [Google Scholar]
- Kass, R. E. , & Raftery, A. E. (1995). Bayes factor. Journal of the American Statistical Association, 90, 773–795. [Google Scholar]
- Kay, K. M. , Whittall, J. B. , & Hodges, S. A. (2006). A survey of nuclear ribosomal internal transcribed spacer substitution rates across angiosperms: An approximate molecular clock with life history effects. BMC Evolutionary Biology, 6, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulju, K. K. M. , Sierra, S. E. C. , Draisma, S. G. A. , Samuel, R. , & van Welzen, P. C. (2007). Molecular phylogeny of Macaranga, Mallotus, and related genera (Euphorbiaceae s.s.): Insights from plastid and nuclear DNA sequence data. American Journal of Botany, 94, 1726–1743. [DOI] [PubMed] [Google Scholar]
- Kyrkjeeide, M. O. , Hassel, K. , Flatberg, K. I. , Shaw, A. J. , Brochmann, C. , & Stenøien, H. K. (2016). Long‐distance dispersal and barriers shape genetic structure of peatmosses (Sphagnum) across the Northern Hemisphere. Journal of Biogeography, 43, 1215–1226. [Google Scholar]
- Kyrkjeeide, M. O. , Hassel, K. , Flatberg, K. I. , Shaw, A. J. , Yousefi, N. , & Stenøien, H. K. (2016). Spatial genetic structure of the abundant and widespread peatmoss Sphagnum magellanicum Brid. PLoS One, 11, e0148447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laenen, B. , Machac, A. , Gradstein, S. R. , Shaw, B. , Patiño, J. , Désamoré, A. , … Vanderpoorten, A. (2016). Geographical range in liverworts: Does sex really matter? Journal of Biogeography, 43, 627–635. [Google Scholar]
- Laenen, B. , Shaw, B. , Schneider, H. , Goffinet, B. , Paradis, E. , Désamoré, A. , … Shaw, A. J. (2014). Extant diversity of bryophytes emerged from successive post‐Mesozoic diversification bursts. Nature Communications, 5, 6134. [DOI] [PubMed] [Google Scholar]
- Landis, M. , Matzke, N. J. , Moore, B. R. , & Huelsenbeck, J. P. (2013). Bayesian analysis of biogeography when the number of areas is large. Systematic Biology, 62, 789–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larget, B. , & Simon, D. L. (1999). Markov chain Monte Carlo algorithms for the Bayesian analysis of phylogenetic trees. Molecular Biology and Evolution, 16, 750–759. [Google Scholar]
- Le Péchon, T. , Zhang, L. , He, H. , Zhou, X.‐M. , Bytebier, B. , Gao, X.‐F. , & Zhang, L.‐B. (2016). A well‐sampled phylogenetic analysis of the polystichoid ferns (Dryopteridaceae) suggests a complex biogeographical history involving both boreotropical migrations and recent transoceanic dispersals. Molecular Phylogenetics and Evolution, 98, 324–336. [DOI] [PubMed] [Google Scholar]
- Les, D. H. , Crawford, D. J. , Kimball, R. T. , Moody, M. L. , & Landolt, E. (2003). Biogeography of discontinuously distributed hydrophytes, a molecular appraisal of intercontinental disjunctions. International Journal of Plant Sciences, 164, 917–932. [Google Scholar]
- Lewis, L. R. , Rozzi, R. , & Goffinet, B. (2014). Direct long‐distance dispersal shapes a New World amphitropical disjunction in the dispersal‐limited dung moss Tetraplodon (Bryopsida: Splachnaceae). Journal of Biogeography, 41, 2385–2395. [Google Scholar]
- Li, Y. , Dressler, S. , Zhang, D. , & Renner, S. (2009). More Miocene dispersal between Africa and Asia—The case of Bridelia (Phyllanthaceae). Systematic Botany, 34, 521–529. [Google Scholar]
- Liu, Z. , Pagani, M. , Zinniker, D. , DeConto, R. , Huber, M. , Brinkhuis, H. , … Pearson, A. (2009). Global cooling during the Eocene‐Oligocene climate transition. Science, 323, 1187–1190. [DOI] [PubMed] [Google Scholar]
- Maddison, D. R. , & Maddison, W. P. (2016) Mesquite: A modular system for evolutionary analysis. Version 3.10. Retrieved from http://mesquiteproject.org. [Google Scholar]
- Matzke, N. J. (2013a). BioGeoBEARS: BioGeography with Bayesian (and likelihood) evolutionary analysis in R Scripts. Vienna, Austria: CRAN: The Comprehensive R Archive Network; Retrieved from http://cran.r-project.org/package=BioGeoBEARS. [Google Scholar]
- Matzke, N. J. (2013b). Probabilistic historical biogeography: New models for founder‐event speciation, imperfect detection, and fossils allow improved accuracy and model‐testing. Frontiers of Biogeography, 5, 242–248. [Google Scholar]
- Matzke, N. J. (2014). Model selection in historical biogeography reveals that founder‐event speciation is a crucial process in island clades. Systematic Biology, 63, 951–970. [DOI] [PubMed] [Google Scholar]
- Medina, R. , Lara, F. , Goffinet, B. , Garilleti, R. , & Mazimpaka, V. (2012). Integrative taxonomy successfully resolves the pseudo‐cryptic complex of the disjunct epiphytic moss Orthotrichum consimile s.l. (Orthotrichaceae). Taxon, 61, 1180–1198. [Google Scholar]
- Medina, R. , Lara, F. , Goffinet, B. , Garilleti, R. , & Mazimpaka, V. (2013). Unnoticed diversity within the disjunct moss Orthotrichum tenellum s.l. validated by morphological and molecular approaches. Taxon, 62, 1133–1152. [Google Scholar]
- Miller, M. A. , Pfeiffer, W. , & Schwartz, T. (2010) Creating the CIPRES Science Gateway for Inference of Large Phylogenetic Trees. Proceedings of the Gateway Computing Environments Workshop (GCE), 14 November 2010, New Orleans, LA, pp. 1–8.
- Morley, R. J. (2011). Cretaceous and tertiary climate change and the past distribution of megathermal rainforests In Bush M. B., Flenley J. R., & Gosling W. D. (Eds.), Tropical Rainforest Responses to Climatic Change (pp. 1–34). Berlin: Springer. [Google Scholar]
- Odrzykoski, I. J. , & Szweykowski, J. (1991). Genetic differentiation without concordant morphological divergence in the thallose liverwort Conocephalum conicum . Plant Systematics and Evolution, 178, 135–151. [Google Scholar]
- Palmer, J. D. (1991). Plastid chromosome, structure and evolution In Bogorad L., & Vasil I. K. (Eds.), The Molecular Biology of Plastids (pp. 5–53). San Diego, CA: Academic Press. [Google Scholar]
- Pattengale, N. D. , Alipour, M. , Bininda‐Emonds, O. R. , Moret, B. M. , & Stamatakis, A. (2010). How many bootstrap replicates are necessary? Journal of Computational Biology, 17, 337–354. [DOI] [PubMed] [Google Scholar]
- Pócs, T. (2012). New or little known epiphyllous liverworts, XVI. A small collection from Laos. Acta Biologica Plantarum Agriensis, 2, 5–10. [Google Scholar]
- Pócs, T. , & Lye, K. A. (1999). New records and additions to the hepatic flora of Uganda. 2. Tropical Bryology, 17, 23–33. [Google Scholar]
- Posada, D. (2008). jModeltest: Phylogenetic model averaging. Molecular Biology and Evolution, 25, 415–430. [DOI] [PubMed] [Google Scholar]
- Ramaiya, M. , Johnson, M. G. , Shaw, B. , Heinrichs, J. , Hentschel, J. , von Konrat, M. , … Shaw, A. J. (2010). Morphologically cryptic biological species within the liverwort Frullania asagrayana . American Journal of Botany, 97, 1707–1718. [DOI] [PubMed] [Google Scholar]
- Raven, P. H. , & Axelrod, D. I. (1974). Angiosperm biogeography and past continental movements. Annals of the Missouri Botanical Garden, 61, 539–673. [Google Scholar]
- Ree, R. H. , & Smith, S. A. (2008). Maximum likelihood inference of geographic range evolution by dispersal, local extinction, and cladogenesis. Systematic Biology, 57, 4–14. [DOI] [PubMed] [Google Scholar]
- Renner, M. A. M. (2014). Radula subg. Radula in Australasia and the Pacific (Jungermanniopsida). Telopea, 17, 107–167. [Google Scholar]
- Renner, M. A. M. (2015). Lobule shape evolution in Radula (Jungermanniopsida): One rate fits all? Botanical Journal of the Linneam Society, 178, 222–242. [Google Scholar]
- Renner, M. A. M. , Brown, E. A. , & Wardle, G. M. (2011). The Lejeunea tumida species group is positively polyphyletic (Lejeuneaceae: Jungermanniopsida). Australian Systematic Botany, 23, 443–462. [Google Scholar]
- Renner, M. A. M. , Devos, N. , Patiño, J. , Brown, E. A. , Orme, A. , Elgey, M. , … von Konrat, M. J. (2013). Integrative taxonomy resolves the cryptic and pseudo‐cryptic Radula buccinifera complex (Porellales, Jungermanniopsida), including two reinstated and five new species. PhytoKeys, 27, 1–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson, J. E. , Chatrou, L. W. , Mols, J. B. , Erkens, R. H. J. , & Pirie, M. D. (2004). Historical biogeography of two cosmopolitan families of flowering plants: Annonaceae and Rhamnaceae. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences, 359, 1495–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist, F. (1997). Dispersal‐vicariance analysis: A new approach to the quantification of historical biogeography. Systematic Biology, 46, 195–203. [Google Scholar]
- Ronquist, F. , & Huelsenbeck, J. P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics, 19, 1572–1574. [DOI] [PubMed] [Google Scholar]
- Scheben, A. , Bechteler, J. , Lee, G. E. , Pócs, T. , Schäfer‐Verwimp, A. , & Heinrichs, J. (2016). Multiple transoceanic dispersals and geographical structure in the pantropical leafy liverwort Ceratolejeunea (Lejeuneaceae, Porellales). Journal of Biogeography, 43, 1739–1749. [Google Scholar]
- Schneider, H. , Kreier, H.‐P. , Janssen, T. , Otto, E. , Muth, H. , & Heinrichs, J. (2010). Key innovations versus key opportunities: Identifying causes of rapid radiations in derived ferns In Glaubrecht M. (Ed.), Evolution in Action (pp. 61–75). Berlin: Springer. [Google Scholar]
- Schuster, R. M. (1980). The Hepaticae and Anthocerotae of North America east of the Hundredth Meridian, Vol. 4 New York, NY: Columbia University Press. [Google Scholar]
- Schuster, R. M. (1983). Phytogeography of the Bryophyta In Schuster R. M. (Ed.), New Manual of Bryology (pp. 463–626). Nichinan: The Hattori Botanical Laboratory. [Google Scholar]
- Shaw, A. J. (2001). Biogeographic patterns and cryptic speciation in bryophytes. Journal of Biogeography, 28, 253–261. [Google Scholar]
- Shaw, A. J. , Boles, S. , & Shaw, B. (2008). A phylogenetic delimitation of the Sphagnum subsecundum complex. American Journal of Botany, 95, 731–744. [DOI] [PubMed] [Google Scholar]
- Shaw, A. J. , Devos, N. , Cox, C. J. , Boles, S. B. , Shaw, B. , Buchanan, A. M. , … Seppelt, R. (2010). Peatmoss (Sphagnum) diversification associated with Miocene Northern Hemisphere climatic cooling? Molecular Phylogenetics and Evolution, 55, 1139–1145. [DOI] [PubMed] [Google Scholar]
- Shaw, A. J. , Golinski, G. K. , Clark, E. G. , Shaw, B. , Stenøien, H. K. , & Flatberg, K. I. (2014). Intercontinental genetic structure in the amphi‐Pacific peatmoss Sphagnum miyabearum (Bryophyta: Sphagnaceae). Biological Journal of the Linnaean Society, 11, 17–37. [Google Scholar]
- Shu, L. , Zhu, R.‐L. , & Pócs, T. (2016). A new species of Leptolejeunea (Lejeuneaceae, Marchantiophyta) from Fiji with special reference to Leptolejeunea tripuncta . Cryptogamie, Bryologie, 37, 157–165. [Google Scholar]
- Söderström, L. , Hagborg, A. , von Konrat, M. , Bartolomew‐Began, S. , Bell, D. , Briscoe, L. , … Zhu, R. L. , (2016). World checklist of hornworts and liverworts. PhytoKeys, 59, 1–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler, T. (2009). On incomplete sampling under birth‐death models and connections to the sampling‐based coalescent. Journal of Theoretical Biology, 261, 58–66. [DOI] [PubMed] [Google Scholar]
- Stamatakis, A. (2014). RAxML version 8: A tool for phylogenetic analysis and post‐analysis of large phylogenies. Bioinformatics, 30, 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stotler, R. E. , & Crandall‐Stotler, B. (1974). A monograph of the genus Bryopteris (Swartz) Nees von Esenbeck. Bryophytorum Bibliotheca, 3, 1–159, + 219 figures on 32 plates. [Google Scholar]
- Sun, Y. , He, X. , & Glenny, D. (2014). Transantarctic disjunctions in Schistochilaceae (Marchantiophyta) explained by early extinction events, post‐Gondwanan radiations and palaeoclimatic changes. Molecular Phylogenetics and Evolution, 76, 189–201. [DOI] [PubMed] [Google Scholar]
- Van Zanten, B. O. , & Gradstein, S. R. (1988). Experimental dispersal geography of neotropical liverworts. Nova Hedwigia Beihefte, 90, 41–94. [Google Scholar]
- Vanderpoorten, A. , Gradstein, S. R. , Carine, M. A. , & Devos, N. (2010). The ghosts of Gondwana and Laurasia in modern liverwort distributions. Biological Reviews, 85, 471–487. [DOI] [PubMed] [Google Scholar]
- Vanderpoorten, A. , Patiño, J. , Dirkse, G. , Blockeel, T. , & Hedenäs, L. (2015). Early divergence of an Azorean endemic species in the moss genus Rhynchostegiella (Brachytheciaceae). Phytotaxa, 210, 60–69. [Google Scholar]
- Vigalondo, B. , Lara, F. , Draper, I. , Valcarel, V. , Garilleti, R. , & Mazimpaka, V. (2016). Is it really you, Orthotrichum acuminatum? Ascertaining a new case of intercontinental disjunction in mosses. Botanical Journal of the Linnean Society, 180, 30–49. [Google Scholar]
- Villarreal, J. C. , & Renner, S. S. (2012). Hornwort pyrenoids, a carbon‐concentrating mechanism, evolved and were lost at least five times during the last 100 million years. Proceedings of the National Academy of Sciences of the United States of America, 109, 18873–18878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarreal, J. C. , & Renner, S. S. (2014). A review of molecular‐clock calibrations and substitution rates in liverworts, mosses, and hornworts, and a timeframe for a taxonomically cleaned‐up genus Nothoceros . Molecular Phylogenetics and Evolution, 78, 25–35. [DOI] [PubMed] [Google Scholar]
- Wei, R. , Xiang, Q. , Schneider, H. , Sundue, M. A. , Kessler, M. , Kamau, P. W. , … Zhang, X. (2015). Eurasian origin, boreotropical migration and transoceanic dispersal in the pantropical fern genus Diplazium (Athyriaceae). Journal of Biogeography, 42, 1809–1819. [Google Scholar]
- Wilson, R. , Gradstein, S. R. , Schneider, H. , & Heinrichs, J. (2007). Unravelling the phylogeny of Lejeuneaceae (Jungermanniopsida): Evidence for four main lineages. Molecular Phylogenetics and Evolution, 43, 270–282. [DOI] [PubMed] [Google Scholar]
- Wilson, R. , Heinrichs, J. , Hentschel, J. , Gradstein, S. R. , & Schneider, H. (2007). Steady diversification of derived liverworts under Tertiary climatic fluctuations. Biology Letters, 3, 566–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, W. , Lewis, P. O. , Fan, Y. , Kuo, L. , & Chen, M.‐H. (2011). Improving marginal likelihood estimation for Bayesian phylogenetic model selection. Systematic Biology, 60, 150–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachos, J. , Pagani, M. , Sloan, L. , Thomas, E. , & Billups, K. (2001). Trends, rhythms, and aberrations in global climate 65 ma to present. Science, 292, 686–693. [DOI] [PubMed] [Google Scholar]
- Zhu, R. L. , & So, M. L. (2001). Epiphyllous liverworts of China. Nova Hedwigia Beiheft, 121, 1–418. [Google Scholar]
- Zona, S. (2013). Millipedes transport gemmae of Calymperes palisotii (Bryophyta: Calymperaceae). Nova Hedwigia, 97, 477–483. [Google Scholar]


