Abstract
Polyhydroxyalkanoate (PHAs) are natural, biodegradable biopolymers, which can be produced from renewable materials. PHAs have potential to replace petroleum derived plastics. Quite a few bacteria can produce PHA under nutritional stress. They generally produce homopolymers of butyrate i.e., polyhydroxybutyrate (PHB), as a storage material. The biochemical characteristics of PHB such as brittleness, low strength, low elasticity, etc. make these unsuitable for commercial applications. Co-polymers of PHA, have high commercial value as they overcome the limitations of PHBs. Co-polymers can be produced by supplementing the feed with volatile fatty acids or through hydrolysates of different biowastes. In this review, we have listed the potential bacterial candidates and the substrates, which can be co-metabolized to produce PHA co-polymers.
Keywords: Bacillus, Biowastes, Co-metabolism, Co-polymers, Polyhydroxyalkanoate, Gram-positive, Gram-negative
Introduction
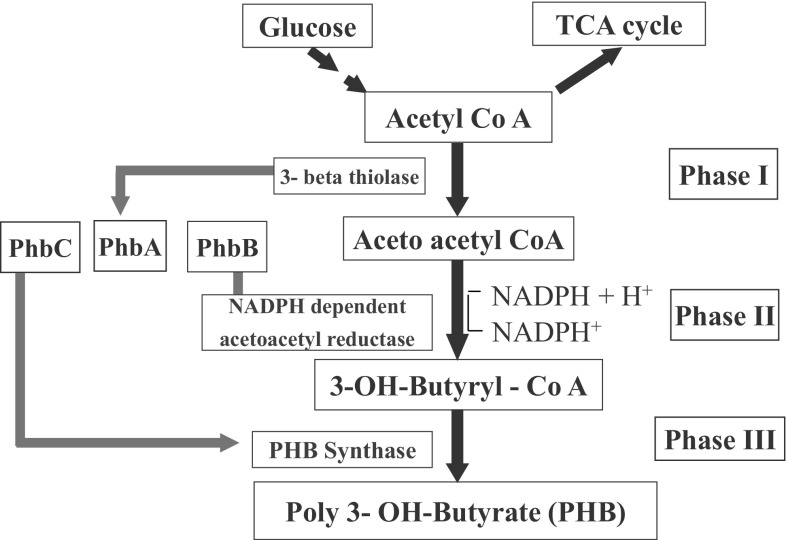
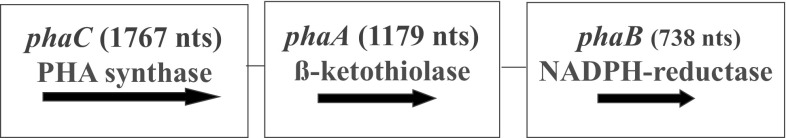
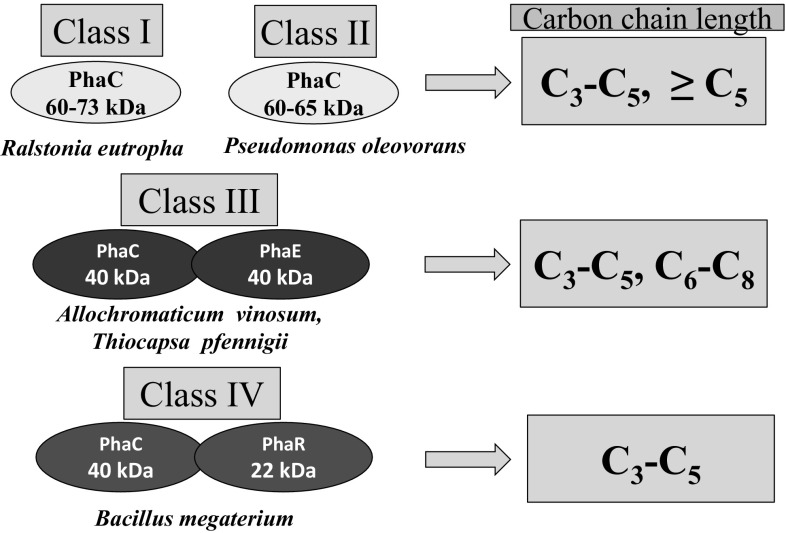
Biopolymers like polyhydroxyalkanoates (PHAs) have gained importance as these can be produced from natural and renewable substrates. Another important characteristic is their biodegradable nature. Their physical and chemical characteristics are very similar to synthetic plastics derived from petroleum products [1, 2]. The basic advantage of this biodegradable plastic is their non-polluting nature and potential to save fossil fuels. Diverse bacteria produce PHAs under nutritionally imbalanced conditions. The PHA biosynthetic pathway operates at high Carbon (C) concentrations and limitations of other nutrients (N, P, K, O, Mg, etc.) in the environment [3]. Here, instead of operating the tri-carboxylic acid cycle for generating energy, the metabolic pathway shifts towards PHA biosynthesis to produce granules, which act as C storage material [4]. Under normal physiological conditions, especially when the C:N ratio is low, i.e. N is present in sufficient quantities, NAD(P)H/NAD(P) ratio decreases and acetyl-CoA goes into the TCA cycle, releasing CoA for the next round of utilization. Accumulation of CoA inhibits the activity of β-ketothiolase, which blocks the PHA synthesis route. β-ketothiolase is the first enzyme of the PHA biosynthetic pathway. On the other hand, PHA production progresses when cell growth is reduced under N-limiting conditions. Here, NAD(P)H/NAD(P) ratio increases, which inhibits citrate synthase and isocitrate dehydrogenase activity resulting in the blockage of the TCA cycle. It leads to high acetyl-CoA concentration and lowers CoA, resulting in the activation of the enzyme—β-ketothiolase. The Phase I of PHA biosynthetic pathway become operative leading to the generation of acetoacetyl-CoA. It then gets transformed to 3-OH-butyryl-CoA, with the aid of NADPH–dependent acetoacetyl reductase in the Phase II. The whole process terminates with the production of polyhydroxybutyrate (PHB), by polymerization of 3OH-butyrate monomers with the help of PHB synthase, i.e. Phase III [5, 6] (Fig. 1). The three enzymes of the PHB biosynthetic pathway are coded by genes: phaA (1179 nucleotides, nts), phaB (738 nts), and phaC (1767 nts), which are organized as CAB operon in Ralstonia eutropha (Fig. 2). The diversity of PHA synthases can be seen in organisms like: (1) R. eutropha, which has class I type, single subunit of PhaC (60–73 kDa), (2) Pseudomonas oleovorans having class II type—single PhaC subunit (60–65 kDa), (3) Allochromaticum vinosum and Thiocapsa pfennigii having class III, composed of two subunits PhaC (40 kDa) and PhaE (40 kDa), and (4) Bacillus megaterium representing class IV composed of subunits PhaC (40 kDa) and PhaR (22 kDa) (Fig. 3). Class I, II and IV type PHA synthases result in C3-C5 PHAs, whereas class III can result in more variable chain length PHAs (Fig. 3).
Fig. 1.
Polyhydroxyalkanoate biosynthetic pathway. PHA is a synthesized by the action of enzymes: PhbA (β-keto thiolase), PhbB (acetoacetyl-CoA reductase) and PhbC (PHA plymerase). TCA tricarboxylic acid cycle
Fig. 2.
phaCAB operon organization in Ralstonia eutropha
Fig. 3.
Diversity of Polyhydroxyalkanoate (PHA) synthases
Polyhydroxyalkanoate (PHAs)
Bacteria have the potential to gather C in the form of PHAs to the extent of 90 % of the total dry cell mass (DCM). The composition of the PHAs depends upon the C chain length, which varies from: (1) C3–C5 i.e., short chain length PHA e.g., in R. eutropha, and (2) C6–C14 i.e., medium chain length PHA e.g., in Pseudomonas oleovorans [3]. The nature of the biopolymers depends upon the growth medium, type and quantity of C source, bacterium, supplements, etc. Most bacteria produce homopolymers as PHB, however, a few have the potential to produce co-polymers, but need specific co-substrate to be present in the medium [1]. R. eutropha and Chromobacterium violaceum grown in the presence of valeric acid (VA) as supplemented material results in PHA co-polymers. The commercial value of PHBs is lower as compared to co-polymers because of the following reasons: (1) brittle nature, (2) low strength, (3) high cost of production, (4) low elasticity, (5) low mechanical resistance, etc. [1]. PHA copolymers have characteristics, which can be compared to petroleum plastics. Here, the improvement in PHA strength is because of high molecular weight and variation in monomeric compositions. These changes can be achieved through variation in: (1) co-substrate, (2) feeding, (3) physiological conditions, (4) genetic modifications, (5) heterologous gene expressions (6) metabolic pathway modification [1, 7, 8]. 3HV monomers when incorporate into a PHA polymer chain, increase material characteristics of PHA co-polymer, such as: (1) melting point, (2) crystallinity, (3) stiffness, and (4) toughness. Co-polymers are thermoplastics, which have a melting temperature of 140 °C, which is close to that of polylactic acids [9].
PHA Co-polymers by Co-metabolism of Diverse Substrates
Gram-Negative Bacteria
Most PHA producers generally belong to gram-negative group of bacteria (Table 1) [10–35]. Ralstonia species are among the most widely studied PHA producers. They have an ability to produce homopolymers and co-polymers. R. eutropha could utilize mixtures of: (1) gluconoate + octanoate, and (2) glycerol + casein hydrolysate (CH) to produce PHB homopolymers, where PHA yield varied from 40 to 50 % of DCM [13, 18]. Different strains of Cupriavidus necator could produce P(3HB-3HV-3HHx) from vegetable oils, and glycerol supplemented with VA or levulinic acid, where 3HV and 3HHx components varied from 6 to 7 mol%. C. necator DSM545 produced homopolymers of PHB from glucose and VA, but co-polymers from FAME + VA [25, 33]. Similarly, Pseudomonas spp. could metabolize mixtures of sugars to PHB homopolymers. However, switching over to substrates such as dodecanoate and gluconaote mixtures resulted in PHA co-polymers—P(HD-HDD-HO-HHx) with P. putida and P(3HB-3HHx) with Aeromonas hyrophila CQ4. A variation in feed to dodecanoate + PA allowed A. hydrophila GAK4 to produce P(3HB-3HV-3HHx) [10, 19–22]. Mixed cultures of Burkholderia and Acidobacteria and other bacteria proved instrumental in transforming acetate and PA combination to P(3HB-3HV) [34]. A few other organisms, which produce PHA copolymers through co-metabolism of substrates are Comamonas and Escherichia coli, whereas others like Azotobacter, Haloferax, Methylobacterium and Azohydromonas did not produce co-polymers inspite of being provided with mixed substrates as feed [11, 13, 15, 17, 27].
Table 1.
Polyhydroxyalkanoate co-polymer production by co-metabolism of diverse substrates by gram-negative microorganism
| Organism | Substrate | Homo-polymers | Co-polymer | References | |||
|---|---|---|---|---|---|---|---|
| PHB | Type | Ratio mol (%) |
Yield (% DCM) | ||||
| mol (%) | Yield (% DCM) | ||||||
| Alcaligenes eutrophus | Glucose + (NH4)2SO4 | 100 | 78 | – | – | – | [16] |
| Ralstonia eutropha PHB-4 | Gluconate + octanoate | 100 | 40.89 | – | – | – | [18] |
| R. eutropha | Glycerol + caesin hydrolysate (CH) | 100 | 50 | – | – | – | [13] |
| Cupriavidus necator H16 | Palm kernel oil + propionic acid (PA) | – | – | P(3HB-3HV-3HHx) | 93:0:7 | 55.5 | [25] |
| Palm kernel oil + valeric acid (VA) | – | – | P(3HB-3HV-3HHx) | 89:6:5 | 52.3 | ||
| C. necator DSM545 | Glucose + VA | 100 | 64.5 | – | – | – | [33] |
| FAME + VA | – | – | P(3HB-3HV) | 4.3 | 63.4 | ||
| C. necator DSM7237 | Glycerol + sunflower meal + levulinic acid | 27 g/L | 72.9 | P(3HB-3HV) | 22.5 | 66.4 | [35] |
| C. necator | Crude glycerol + rapeseed meal | – | – | P(3HB-3HV) | 2.8−8:55.6 | NA | [32] |
| Cupriavidus sp. USMAA1020 | γ-butyrolactone | – | – | P(3HB-4HB) | NA | 52.4 | [23] |
| Pseudomonas pseudoflava | Glucose + xylose | 100 | 22 | – | – | – | [10] |
| P. putida KTOY06 | Dodecanoate + gluconate | – | – | P(3HD-3HDD-3HO- 3HHx) | NA | 84.3 | [19] |
| P. putida KT2440 | Glucose + nonanoic acid | 100 | 75 | – | NA | NA | [21] |
| Burkholderia + Acidobacteria | Acetic acid (AA) + PA | 100 | NA | P(3HB-3HV) | 0–74 | NA | [34] |
| Aeromonas hydrophila CQ4 | Dodecanoate + gluconoate | – | – | P(3HB-3HHx) | 44.67 | [20] | |
| A. hydrophila 4AK4 | Dodecanoate + PA | – | – | P(3HB-3HV-3HHx) | NA | 37.2 | [22] |
| Lauric acid + 1,4-butanediol | – | – | P(3HB-4HB—3HHx) | NA | 23.6 | [28] | |
| Azotobacter sp. | Glucose (5 % w/v) + FP (Fish peptone) | 100 | 85 | – | – | – | [11] |
| Glucose (3 % w/v) + FP + NH4Cl | 100 | 74 | – | – | – | ||
| Glucose (3 % w/v) + FP | 100 | 79 | – | – | – | ||
| A. vinelandii UWD | – | – | P(3HB-3HV) | 4.3 | 58.3 | [12] | |
| Comamonas acidovorans | Glucose + 1,4-butanediol | 100 | 53 | P(4HB) | 0–96 | 40 | [15] |
| Haloferax mediterranei | Rice bran + corn starch (1:8) | 100 | 55.6 | – | – | – | [17] |
| Wheat bran + Corn Starch (1:2) | 100 | 40.2 | – | – | – | ||
| Methylobacterium rhodesianum | Glycerol + CH + Casamino acids | 100 | 65 | – | – | – | [13] |
| E. coli JM109 | Glucose | – | – | P(3HHx-3HO) | NA | 54.0 | [27] |
| Azohydromonas australica | Sucrose + Nitrogen | 100 | 77.0 | – | – | – | [31] |
| Mixed culture | AA + PA + Lactic acid | – | – | P(3HB-3HV) | 31 | NA | [14] |
| Mixed culture | Fermented molasses (VFAs) | 100 | 65 | P(3HB-3HV) | 13 | 30 | [30] |
| Mixed culture | AA + PA | 100 | 78 | P(3HB-3HV) | 15–20 | NA | [26] |
| Mixed culture (Waste activated sludge) | AA + Glucose | 100 | 30 | P(3HB-3HV) | 3.1 | NA | [27] |
| AA + Bovine serum albumin (BSA) | 100 | 29.1 | P(3HB-3HV) | 2.7 | NA | ||
| AA + Glucose + BSA | 100 | 30.8 | P(3HB-3HV) | 3.7 | NA | ||
| Mixed culture (Activated sludge) | VFAs (AA + PA + VA + butyrate) | 100 | 31–47 | P(3HB-3HV) | 53–69 | 48 | [24] |
a Not applicable NA Not available
PHA Polyhydroxyalkanoae
PHB Polyhydroxybutyrate
3HB 3-Hydroxybutyric acid
3HV 3-Hydroxyvaleric acid
4HB 4-Hydroxybutyric acid
3HO 3-Hydroxyoctanoate
3HHx 3-Hydroxyhexenoate
6HHx 6Hydroxyhexanoate
3HD 3-Hydroxydecanoate
3HDD 3Hydroxydodecanoate
Gram-Positive Bacteria
Among gram-positive bacteria, Streptomyces, Corynebacteria, Clostridium, Nocardia, Rhodococcus, Staphylococcus are capable of producing PHA co-polymers [4]. Bacillus spp. are among those few gram-positive bacteria, which have been gaining importance as PHA producers because of their unique metabolic characterstics. These are perhaps the only bacteria in this category, which can produce homopolymers and co-polymers of PHA from sugars and complex biowastes (Table 2) [1, 2, 7, 36–49]. Bacillus species are generally regarded as safe (GRAS) organisms [3, 7]. Bacillus megaterium OU303A and Bacillus sp. 88D utilized glucose, glycerol and acetate to produce PHB homopolymers, whereas addition of PA (<2.5 ml/L) allowed them to convert these mixtures into P(3HB-3HV). Here, 3HV content varied from 2.5 to 6.3 mol% [42, 43]. Bacillus sp. INT005 utilized butyrate to produce PHB, however, glucose in combination with different fatty acids (1 % v/v) resulted in PHA co-polymers with HV content varying from 1.5 to 29 mol% and total PHA yield ranging from 13 to 64.5 % DCM [38].
Table 2.
Polyhydroxyalkanoate Co-polymer production by co-metabolism of diverse substrates by Bacillus sp
| Organism | Substrate | Homo-polymers | Co-polymer | References | |||
|---|---|---|---|---|---|---|---|
| PHB | Type | Ratio mol (%) |
Yield (% DCM) | ||||
| mol (%) | Yield (% DCM) | ||||||
| Bacillus megaterium OU303A | Glucose (2 % w/v) | 100 | 62 | – | – | – | [42] |
| Glucose (2 % w/v) + PA (<2.5 mL/L) | – | – | P(3HB-3HV) | 97.5:2.5 | 58.6 | ||
| Glycerol (2 % w/v) | – | – | P(3HB-3HV) | 95:5 | 52 | ||
| Glycerol (2 % w/v) + PA (<2.5 mL/L) | – | – | P(3HB-3HV) | 86:14 | 57 | ||
| Acetate (2 % w/v) | 100 | 49 | – | – | – | ||
| Acetate (2 % w/v) + PA (<2.5 mL/L) | – | – | P(3HB-3HV) | 96.5:3.5 | 59 | ||
| B. megaterium DSM90 | Glycerol | 100 | 62.4 | – | – | – | [46] |
| B. cereus ATCC14579 | Caprolactone + octanoate | – | – | 3HHx P(3HHx-3HO) |
NA | 2–4 | [36] |
| B. cereus UW85 | γ- caprolactone | – | – | P(3HB-3HV-6HHx) | NA | NA | [37] |
|
Bacillus sp. INT005 |
Butyrate | 100 | NA | – | – | – | [38] |
| Glucose (0.1 % w/v) + Butyrate (1 % v/v) | – | – | P(3HB-3HHx) P(3HB-4HB-3HHx) |
98.5:1.5 | 32.9 | ||
| Glucose (0.1 % w/v) + valerate (1 % v/v) | – | – | P(3HB-3HV) | 51.5:48.5 | 18.8 | ||
| Glucose (0.1 % w/v) + hexanoate (1 % v/v) | – | – | P(3HB-3HHx) | 97.7:2.3 | 13.0 | ||
| Glucose (0.1 % w/v) + octanoate (1 % v/v) | – | – | P(3HB-3HHx) | 97.1:2.9 | 64.5 | ||
| Glucose (0.1 % w/v) + decanoate (1 % v/v) | – | – | P(3HB-3HHx) | 97.1:2.9 | 23.5 | ||
| Glucose (0.1 % w/v) + γ- caprolactone (1 % v/v) | – | – | P(3HB-6HHx-3HHx) | 97.3:2.7 | 23.2 | ||
| Bacillus sp. 88D | Glucose (2 % w/v) | – | – | P(3HB-3HV) | 96:4 | 64.6 | [43] |
| Glucose (2 % w/v) + PA (<2.5 mL/L) | – | – | P(3HB-3HV) | 87:13 | 59.8 | ||
| Glycerol (2 % w/v) | – | – | P(3HB-3HV) | 85:15 | 60.5 | ||
| Glycerol (2 % w/v) + PA (<2.5 mL/L) | – | – | P(3HB-3HV) | 96:4 | 60 | ||
| Acetate (2 %w/v) | 100:0 | 48 | – | – | – | ||
| Acetate (2 %w/v) + PA (<2.5 mL/L) | – | – | P(3HB-3HV) | 93.7:6.3 | 42 | ||
|
Bacillus (Defined mixed strains: B. cereus strains EGU3, EGU43 + EGU44 + EGU520 + B. thuringiensis EGU45 |
Pea-shell slurry (PSS) + glucose | 100 | 18.8 | P(3HB-3HV) | 87:13 | 16.9 | [47] |
| PSS + glucose + PA | – | – | P(3HB-3HV) | 89:11 | 21.6 | ||
| PSS + glucose + VA | – | – | P(3HB-3HV) | 90:10, 93:7 | 16–23 | ||
| B. cereus EGU44 | PSS + glucose | 100 | 30.0 | [47] | |||
| PSS + glucose + PA (0.5–2 % v/v) | – | – | P(3HB-3HV) | 89:11, 84:16, 85:15 | 16 -22 | ||
| PSS + glucose + VA (0.5–2 % v/v) | – | – | P(3HB-3HV) | 83:17, 90:10 | 16 -24 | ||
| B. thuringiensis EGU45 | Effluent from H2-stage + glucose (1 % w/v) + | [49] | |||||
| 1. M9 + GM2 media: 1X + 0.25X | NA | NA | P(3HB-3HV) | 61:39 | 10 | ||
| 2. M9 + GM2 media: 1X + 0.5X | NA | NA | P(3HB-3HV) | 62:38 | 7.6 | ||
| 3. M9 + GM2 media: 1X + 1X | NA | NA | P(3HB-3HV) | 77:23 | 18 | ||
| 4. M9 + GM2 media: 1X + 2X | NA | NA | P(3HB-3HV) | 95:5 | 21 | ||
| B. thuringiensis EGU45 | Crude glycerol (CG) + Peptone (PE) + Yeast extract (YE) + | [48] | |||||
| 1. PA (0.5 % v/v) | – | – | P(3HB-3HV) | 89:11 | 53.9 | ||
| 2. PA (1.0 % v/v) | – | – | P(3HB-3HV) | 94.7:5.3 | 37.3 | ||
| 3. PA (2.0 % v/v) | – | – | P(3HB-3HV) | 98.2:1.8 | 44.2 | ||
| 4. VA (0.5 % v/v) | – | – | P(3HB-3HV) | 95.7:4.3 | 37.8 | ||
| 5. VA (1.0 % v/v) | – | – | P(3HB-3HV) | 98.2:1.8 | 48.5 | ||
| 6. VA (2.0 % v/v) | – | – | P(3HB-3HV) | 99:1.0 | 56.3 | ||
| CG + nutrient broth + | |||||||
| 1. PA (0.5 % v/v) | – | – | P(3HB-3HV) | 86.6:13.4 | 55 | ||
| 2. PA (1.0 % v/v) | – | – | P(3HB-3HV) | 95.7:4.3 | 29 | ||
| 3. PA (2.0 % v/v) | – | – | P(3HB-3HV) | 98.3:1.7 | 36 | ||
| 4. VA (0.5 % v/v) | – | – | P(3HB-3HV) | 96.3:3.7 | 29.7 | ||
| 5. VA (1.0 % v/v) | – | – | P(3HB-3HV) | 98.7:1.3 | 53.1 | ||
| 6. VA (2.0 % v/v) | – | – | P(3HB-3HV) | 98.9:1.1 | 52.2 | ||
| PSS | 100 | 5.8 | – | – | – | [2] | |
| PSS + glucose (1 % w/v) | 100 | 7.7 | – | – | – | ||
| Apple pomace (AP) | – | – | P(3HB-3HV) | 64.3:35.7 | 3.8 | ||
| AP + glucose (1 % w/v) | – | – | P(3HB-3HV) | 75.9:24.1 | 7.5 | ||
| Onion peels (OP) | – | – | P(3HB-3HV) | 80:20 | 8.4 | ||
| OP + glucose (1 % w/v) | – | – | P(3HB-3HV) | 97.5:2.5 | 11.7 | ||
| Potato peels (PP) | – | – | P(3HB-3HV) | 33:67 | 2.6 | ||
| PP + glucose (1 % w/v) | – | – | P(3HB-3HV) | 90.9:9.1 | 38.2 | ||
| PS:AP:2:1 + glucose (1 % w/v) | – | – | P(3HB-3HV) | 78.8:21.2 | 16.4 | ||
| PS:OP:1:2 + glucose (1 % w/v) | – | – | P(3HB-3HV) | 63.4:36.6 | 20.5 | ||
| PS:PP:2:1 + glucose (1 % w/v) | – | – | P(3HB-3HV) | 77:23 | 27.1 | ||
| Bacillus sp. | Glycerol | – | – | NA | NA | 25–52 | [39] |
| Bacillus sp. | Madhuca sp. Flowers (Sugars + malic acid) | – | – | P(3HB-3HV) | 90:10 | 51 | [41] |
| B. licheniformis PHA007 | Glycerol | 100 | 68.8 | – | – | – | [45] |
| B. licheniformis DSM394 | Glycerol | 100 | 17 | – | – | – | [45] |
| B. subtilis DSM10 | Glycerol | 100 | 18.9 | – | – | – | [45] |
| B. cereus PHA037 | Glucose | 100 | 60.7 | – | – | – | |
| B. thuringiensis R1 | Glycerol | 100 | 64.1 | – | – | – | [40] |
| B. sphaericus NII0838 | Glycerol | 100 | 31.0 | – | – | – | [44] |
a Not applicable
NA Not available
PHA Polyhydroxyalkanoate
PHB Polyhydroxybutyrate
3HB 3-Hydroxybutyric acid
3HV 3-Hydroxyvaleric acid
4HB 4-Hydroxybutyric acid
3HO 3-Hydroxyoctanoate
3HHx 3-Hydroxyhexenoate
6HHx 6-Hydroxyhexanoate
3HD 3-Hydroxydecanoate
3HDD 3Hydroxydodecanoate
Bacillus licheniformis, B. cereus, B. subtilis and other Bacillus spp. could not produce PHA co-polymers from glucose or glycerol. However, use of defined mixed cultures of B. cereus and B. thuringiensis produced interesting results: (1) on pea-shell slurry (PSS) + glucose—only PHB 18.8 % of DCM was recorded, whereas (2) PSS + glucose + PA resulted in P(3HB-3HV::87:13), with a yield of 16.9 % of DCM. Addition of VA to PSS + glucose was also quite effective in producing co-polymer having 7–10 mol% of 3HV. In contrast, B. cereus EGU44 was also reported to show results which are quite similar to those recorded with defined mixed cultures of Bacillus [47]. Bacillus thuringiensis EGU45 was able to metabolize effluent from hydrogen production stage and yielded co-polymers of PHA with a 3HV content of 5–39 mol% [48].
Bacillus thuringiensis EGU45 could metabolize CG to PHA co-polymers. The composition of these co-polymers varied with the amount of PA or VA used as a supplement. With PA in Peptone + Yeast extract (PE + YE) medium, PHA co-polymer had 3HV content in the range of 1.8–11 mol%. However, with VA in PE + YE medium, 3HV content varied from 1 to 4.3 mol%. On the other hand, CG + Nutrient broth (NB) supplemented with (1) PA resulted in 1.7–13.4 mol% of 3HV, and (2) VA resulted in 1.1–3.7 mol% of 3HV [49]. A very interesting result was recorded in an effort to provide supplemental fatty acids by hydrolysing different biowastes as mixtures in a wide range of ratios. Hydrolysates of PS was found to produce only AA, whereas apple poamace (AP) hydrolysates had only isovaleric acid. Hydrolysates of potato peels (PP) and onion peels (OP) produced mixtures of AA, butyric acid, and PA. This initial information was found to prove helpful in producing PHA co-polymers by co-metabolizing these biowatses by B. thuringiensis EGU45. PS alone was able to produce only homopolymers i.e., PHB, however, mixtures: (1) PS + AP, (2) PS + OP, and (3) PS + PP resulted in P(3HB-3HV), where HV content varied i.e., 21.2, 36.6, and 23.4 mol%, respectively. It implied that by co-metabolism, it is possible to divert PHA biosynthetic pathway from producing only homopolymers to different co-polymers [2].
Opinion
In order to produce co-polymers of PHA, it seems that in addition to bacterial genetic potential, we also need to choose a right combination of substrates and supplements. Thus co-metabolism is an important approach for producing PHA co-polymers of desired compositions. Among the PHA producers, Bacillus spp. are perhaps the most persistent. They have the ability to produce homopolymers and co-polymers as well from the cometabolizing substrates. It implies how Bacillus can engineer its metabolic pathway to produce PHA co-polymer. This property enables it to be a strong competitor as an industrial PHA producer in future.
Acknowledgments
We are thankful to the Director of CSIR-Institute of Genomics and Integrative Biology (IGIB), and CSIR Project INDEPTH (BSC0111) for providing the necessary funds, facilities and moral support. Authors are also thankful to Academy of Scientific and Innovative Research (AcSIR), New Delhi.
Compliance with Ethical Standards
Conflict of interest
Authors declare no conflict of interests.
References
- 1.Singh M, Kumar P, Ray S, Kalia VC. Challenges and opportunities for customizing polyhydroxyalkanoates. Indian J Microbiol. 2015;55:235–249. doi: 10.1007/s12088-015-0528-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kumar P, Ray S, Kalia VC. Production of co-polymers of polyhydroxyalkanoates by regulating the hydrolysis of biowastes. Bioresour Technol. 2016;200:413–419. doi: 10.1016/j.biortech.2015.10.045. [DOI] [PubMed] [Google Scholar]
- 3.Singh M, Patel SKS, Kalia VC. Bacillus subtilis as potential producer for polyhydroxyalkanoates. Microb Cell Fact. 2009;8:38. doi: 10.1186/1475-2859-8-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Valappil SP, Peiris D, Langley GJ, Herniman JM, Boccaccini AR, Bucke C, Roy I. Polyhydroxyalkanoate (PHA) biosynthesis from structurally unrelated carbon sources by a newly characterized Bacillus spp. J Biotechnol. 2007;127:475–487. doi: 10.1016/j.jbiotec.2006.07.015. [DOI] [PubMed] [Google Scholar]
- 5.Reddy CSK, Ghai R, Kalia VC. Polyhydroxyalkanoates: an overview. Bioresour Technol. 2003;87:137–146. doi: 10.1016/S0960-8524(02)00212-2. [DOI] [PubMed] [Google Scholar]
- 6.Kalia VC, Lal S, Cheema S. Insight into the phylogeny of polyhydroxyalkanoate biosynthesis: horizontal gene transfer. Gene. 2007;389:19–26. doi: 10.1016/j.gene.2006.09.010. [DOI] [PubMed] [Google Scholar]
- 7.Kumar P, Patel SKS, Lee JK, Kalia VC. Extending the limits of Bacillus for novel biotechnological applications. Biotechnol Adv. 2013;31:1543–1561. doi: 10.1016/j.biotechadv.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 8.Opgenorth PH, Korman TP, Bowie JU. A synthetic biochemistry module for production of bio-based chemicals from glucose. Nat Chem Biol. 2016;12:393–395. doi: 10.1038/nchembio.2062. [DOI] [PubMed] [Google Scholar]
- 9.Anjum A, Zuber M, Zia KM, Noreen A, Anjum MN, Tabasum S. Microbial production of polyhydroxyalkanoates (PHAs) and its copolymers: a review of recent advancements. Int J Biol Macromol. 2016;89:161–174. doi: 10.1016/j.ijbiomac.2016.04.069. [DOI] [PubMed] [Google Scholar]
- 10.Bertrand JL, Ramsay BA, Ramsay JA, Chavarie C. Biosynthesis of poly-β-hydroxyalkanoates from pentoses by Pseudomonas pseudovora. Appl Environ Microbiol. 1990;56:3133–3138. doi: 10.1128/aem.56.10.3133-3138.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Page WJ, Cornish A. Growth of Azotobacter vinelandii UWD in fish peptone medium and simplified extraction of poly-β-hydroxybutyrate. Appl Environ Microbiol. 1993;59:4236–4244. doi: 10.1128/aem.59.12.4236-4244.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cho KS, Ryu HW, Park CH, Goodrich PR. Poly(hydroxybutyrate-co-hydroxyvalerate) from swine waste liquor by Azotobacter vinelandii UWD. Biotechnol Lett. 1997;19:7–10. doi: 10.1023/A:1018342332141. [DOI] [Google Scholar]
- 13.Bormann EJ, Roth M. The production of polyhydroxybutyrate by Methylobacterium rhodesianum and Ralstonia eutropha in media containing glycerol and casein hydrolysates. Biotechnol Lett. 1999;21:1059–1063. doi: 10.1023/A:1005640712329. [DOI] [Google Scholar]
- 14.Dionisi D, Majone M, Papa V, Beccari M. Biodegradable polymers from organic acids by using activated sludge enriched by aerobic periodic feeding. Biotechnol Bioeng. 2004;85:569–579. doi: 10.1002/bit.10910. [DOI] [PubMed] [Google Scholar]
- 15.Lee WH, Azizan MNM, Sudesh K. Effects of culture conditions on the composition of poly(3-hydroxybutyrate-co-4-hydroxybutyrate) synthesized by Comamonas acidovorans. Polym Degrad Stab. 2004;84:129–134. doi: 10.1016/j.polymdegradstab.2003.10.003. [DOI] [Google Scholar]
- 16.Zhang S, Norrlow O, Wawrzynczyk J, Dey ES. Poly(3-hydroxybutyrate) biosynthesis in the biofilm of Alcaligenes eutrophus, using glucose enzymatically released from pulp fiber sludge. Appl Environ Microbiol. 2004;70:6776–6782. doi: 10.1128/AEM.70.11.6776-6782.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang TY, Duan KJ, Huang SY, Chen CW. Production of polyhydroxybutyrates from inexpensive extruded rice bran and starch by Haloferax mediterranei. J Ind Microbiol Biotechnol. 2006;33:701–706. doi: 10.1007/s10295-006-0098-z. [DOI] [PubMed] [Google Scholar]
- 18.Chien CC, Chen CC, Choi MH, Kung SS, Wei YHC. Production of poly-β-hydroxybutyrate (PHB) by Vibrio spp. isolated from marine environment. J Biotechnol. 2007;132:259–263. doi: 10.1016/j.jbiotec.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 19.Ouyang SP, Rong CL, Chen SS, Liu Q, Chung A, Wu Q, Chen GQ. Production of polyhydroxyalkanoates with high 3-hydroxydodecanoate monomer content by fadB and fadA knockout mutant of Pseudomonas putida KT2442. Biomacromolecules. 2007;8:2504–2511. doi: 10.1021/bm0702307. [DOI] [PubMed] [Google Scholar]
- 20.Qin LF, Gao X, Liu Q, Wu Q, Chen GQ. Biosynthesis of polyhydroxyalkanoate copolyesters by Aeromonas hydrophila mutant expressing a low-substrate-specificity PHA synthase PhaC2 Ps. Biochem Eng J. 2007;37:144–150. doi: 10.1016/j.bej.2007.04.006. [DOI] [Google Scholar]
- 21.Sun Z, Ramsay JA, Guay M, Ramsay B. Increasing the yield of MCL-PHA from nonanoic acid by co-feeding glucose during the PHA accumulation stage in two-stage fed-batch fermentations of Pseudomonas putida KT2440. J Biotechnol. 2007;132:280–282. doi: 10.1016/j.jbiotec.2007.02.023. [DOI] [PubMed] [Google Scholar]
- 22.Zhao W, Chen GQ. Production and characterization of terpolyester poly (3-hydroxybutyrate-co-3-hydroxyvalerate-co-3-hydroxyhexanoate) by recombinant Aeromonas hydrophila 4AK4 harboring genes phaAB. Process Biochem. 2007;42:1342–1347. doi: 10.1016/j.procbio.2007.07.006. [DOI] [Google Scholar]
- 23.Amirul AA, Yahya ARM, Sudesh K, Azizan MNM, Majid MIA. Biosynthesis of poly (3-hydroxybutyrate-co-4-hydroxybutyrate) copolymer by Cupriavidus sp. USMAA1020 isolated from Lake Kulim, Malaysia. Bioresour Technol. 2008;99:4903–4909. doi: 10.1016/j.biortech.2007.09.040. [DOI] [PubMed] [Google Scholar]
- 24.Bengtsson S, Werker A, Christensson M, Welander T. Production of polyhydroxyalkanoates by activated sludge treating a paper mill wastewater. Bioresour Technol. 2008;99:509–516. doi: 10.1016/j.biortech.2007.01.020. [DOI] [PubMed] [Google Scholar]
- 25.Bhubalan K, Lee WH, Loo CY, Yamamoto T, Tsuge T, Doi Y, Sudesh K. Controlled biosynthesis and characterization of poly(3-hydroxybutyrate-co-3-hydroxyvalerate-co-3-hydroxyhexanoate) from mixtures of palm kernel oil and 3HV-precursors. Polym Degrad Stab. 2008;93:17–23. doi: 10.1016/j.polymdegradstab.2007.11.004. [DOI] [Google Scholar]
- 26.Dias JM, Oehmen A, Serafim LS, Lemos PC, Reis MA, Oliveira R. Metabolic modelling of polyhydroxyalkanoate copolymers production by mixed microbial cultures. BMC Syst Biol. 2008;2:1. doi: 10.1186/1752-0509-2-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tao J, Zheng CG, Zhang B (2008) Synthesis of medium-chain-length Polyhydroxyalkanoate (PHAMCL) by Pseudomonas mendocina NK-01. J Microbiol http://www.oalib.com/paper/1428248&prev=search
- 28.Xie WP, Chen GQ. Production and characterization of terpolyester poly (3-hydroxybutyrate-co-4-hydroxybutyrate-co-3-hydroxyhexanoate) by recombinant Aeromonas hydrophila 4AK4 harboring genes phaPCJ. Biochem Eng J. 2008;38:384–389. doi: 10.1016/j.bej.2007.08.002. [DOI] [Google Scholar]
- 29.Jiang Y, Chen Y, Zheng X. Efficient polyhydroxyalkanoates production from a waste-activated sludge alkaline fermentation liquid by activated sludge submitted to the aerobic feeding and discharge process. Environ Sci Technol. 2009;43:7734–7741. doi: 10.1021/es9014458. [DOI] [PubMed] [Google Scholar]
- 30.Albuquerque MGE, Martino V, Pollet E, Averous L, Reis MAM. Mixed culture polyhydroxyalkanoate (PHA) production from volatile fatty acid (VFA)-rich streams: effect of substrate composition and feeding regime on PHA productivity, composition and properties. J Biotechnol. 2011;151:66–76. doi: 10.1016/j.jbiotec.2010.10.070. [DOI] [PubMed] [Google Scholar]
- 31.Gahlawat G, Srivastava AK. Development of a mathematical model for the growth associated polyhydroxybutyrate fermentation by Azohydromonas australica and its use for the design of fed-batch cultivation strategies. Bioresour Technol. 2013;137:98–105. doi: 10.1016/j.biortech.2013.03.023. [DOI] [PubMed] [Google Scholar]
- 32.Garcia IL, Lopez JA, Dorado MP, Kopsahelis N, Alexandri M, Papanikolaou S, Villar MA, Koutinas AA. Evaluation of by-products from the biodiesel industry as fermentation feedstock for poly (3-hydroxybutyrate-co-3-hydroxyvalerate) production by Cupriavidus necator. Bioresour Technol. 2013;130:16–22. doi: 10.1016/j.biortech.2012.11.088. [DOI] [PubMed] [Google Scholar]
- 33.Spoljaric IV, Lopar M, Koller M, Muhr A, Salerno A, Reiterer A, Malli K, Angerer H, Strohmeier K, Schober S, Mittelbach M. Mathematical modeling of poly [(R)-3-hydroxyalkanoate] synthesis by Cupriavidus necator DSM545 on substrates stemming from biodiesel production. Bioresour Technol. 2013;133:482–494. doi: 10.1016/j.biortech.2013.01.126. [DOI] [PubMed] [Google Scholar]
- 34.Wang Y, Cai J, Lan J, Liu Z, He N, Shen L, Li Q. Biosynthesis of poly (hydroxybutyrate-hydroxyvalerate) from the acclimated activated sludge and microbial characterization in this process. Bioresour Technol. 2013;148:61–69. doi: 10.1016/j.biortech.2013.08.102. [DOI] [PubMed] [Google Scholar]
- 35.Kachrimanidou V, Kopsahelis N, Papanikolaou S, Kookos IK, De Bruyn M, Clark JH, Koutinas AA. Sunflower-based biorefinery: poly (3-hydroxybutyrate) and poly (3-hydroxybutyrate-co-3-hydroxyvalerate) production from crude glycerol, sunflower meal and levulinic acid. Bioresour Technol. 2014;172:121–130. doi: 10.1016/j.biortech.2014.08.044. [DOI] [PubMed] [Google Scholar]
- 36.Caballero KP, Karel SF, Register RA. Biosynthesis and characterization of hydroxybutyrate-hydroxycaproate copolymers. Int J Biol Macromol. 1995;17:86–92. doi: 10.1016/0141-8130(95)93522-Y. [DOI] [PubMed] [Google Scholar]
- 37.Labuzek S, Radecka I. Biosynthesis of PHB tercopolymer by Bacillus cereus UW85. J Appl Microbiol. 2001;90:353–357. doi: 10.1046/j.1365-2672.2001.01253.x. [DOI] [PubMed] [Google Scholar]
- 38.Tajima K, Igari T, Nishimura D, Nakamura M, Satoh Y, Munekata M. Isolation and characterization of Bacillus sp. INT005 accumulating polyhydroxyalkanoate (PHA) from gas field soil. J Biosci Bioeng. 2003;95:77–81. doi: 10.1016/S1389-1723(03)80152-4. [DOI] [PubMed] [Google Scholar]
- 39.Full TD, Jung DO, Madigan MT. Production of poly-β-hydroxyalkanoates from soy molasses oligosaccharides by new, rapidly growing Bacillus species. Lett App Microbiol. 2006;43:377–384. doi: 10.1111/j.1472-765X.2006.01981.x. [DOI] [PubMed] [Google Scholar]
- 40.Rohini D, Phadnis S, Rawal SK. Synthesis and characterization of poly- beta-hydroxybutyrate from Bacillus thuringiensis R1. Indian J Biotechnol. 2006;5:276–283. [Google Scholar]
- 41.Anil-Kumar PK, Shamla TR, Kshama L, Prakash MH, Joshi GJ, Chandrashekar A, Kumari KSL, Divyashree MS. Bacterial synthesis of poly(hydroxybutyrate-co-hydroxyvalerate) using carbohydrate-rich mahua (Madhuca sp.) flowers. J Appl Microbiol. 2007;103:204–209. doi: 10.1111/j.1365-2672.2006.03221.x. [DOI] [PubMed] [Google Scholar]
- 42.Reddy SV, Thirumala M, Mahmood SK. Production of PHA and P (3HB-co-3HV) biopolymers by Bacillus megaterium strain OU303A isolated from municipal sewage sludge. World J Microbiol Biotechnol. 2009;25:391–397. doi: 10.1007/s11274-008-9903-3. [DOI] [Google Scholar]
- 43.Reddy SV, Thirumala M, Mahmood SK. A novel Bacillus sp. accumulating poly (3-hydroxybutyrate-co-3-hydroxyvalerate) from a single carbon substrate. J Ind Microbiol Biotechnol. 2009;36:837–843. doi: 10.1007/s10295-009-0561-8. [DOI] [PubMed] [Google Scholar]
- 44.Sindhu R, Ammu B, Binod P, Deepthi SK, Ramachandran KB, Soccol CR, Pandey A. Production and characterization of poly-3-hydroxybutyrate from crude glycerol by Bacillus sphaericus NII 0838 and improving its thermal properties by blending with other polymers. Braz Arch Biol Technol. 2011;54:783–794. doi: 10.1590/S1516-89132011000400019. [DOI] [Google Scholar]
- 45.Sangkharak K, Prasertsan P. Screening and identification of polyhydroxyalkanoates producing bacteria and biochemical characterization of their possible application. J Gen Appl Microbiol. 2012;58:173–182. doi: 10.2323/jgam.58.173. [DOI] [PubMed] [Google Scholar]
- 46.Naranjo JM, Posada JA, Higuita JC, Cardona CA. Valorization of glycerol through the production of biopolymers: the PHB case using Bacillus megaterium. Bioresour Technol. 2013;133:38–44. doi: 10.1016/j.biortech.2013.01.129. [DOI] [PubMed] [Google Scholar]
- 47.Kumar P, Singh M, Mehariya S, Patel SKS, Lee JK, Kalia VC. Ecobiotechnological approach for exploiting the abilities of Bacillus to produce co-polymer of polyhydroxyalkanoate. Indian J Microbiol. 2014;54:151–157. doi: 10.1007/s12088-014-0457-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Singh M, Kumar P, Patel SKS, Kalia VC. Production of polyhydroxyalkanoate co-polymer by Bacillus thuringiensis. Indian J Microbiol. 2013;53:77–83. doi: 10.1007/s12088-012-0294-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kumar P, Ray S, Patel SK, Lee JK, Kalia VC. Bioconversion of crude glycerol to polyhydroxyalkanoate by Bacillus thuringiensis under non-limiting nitrogen conditions. Int J Biol Macromol. 2015;78:9–16. doi: 10.1016/j.ijbiomac.2015.03.046. [DOI] [PubMed] [Google Scholar]