Abstract
The nucleosome remodelling ATPase ISWI resides in several distinct protein complexes whose subunit composition reflects their functional specialization. Association of ISWI with ACF1, the largest subunit of CHRAC and ACF complexes, improves the efficiency of ISWI-induced nucleosome mobilization by an order of magnitude and also modulates the reaction qualitatively. In order to understand the principle by which ACF1 improves the efficiency of ISWI, we mapped their mutual interaction requirements and generated a series of ACF complexes lacking conserved ACF1 domains. Deletion of the C-terminal PHD finger modules of ACF1 or their disruption by zinc chelation profoundly affected the nucleosome mobilization capability of associated ISWI in trans. Interactions of the PHD fingers with the central domains of core histones contribute significantly to the binding of ACF to the nucleosome substrate, suggesting a novel role for PHD modules as nucleosome interaction determinants. Connecting ACF to histones may be prerequisite for efficient conversion of ATP-dependent conformational changes of ISWI into translocation of DNA relative to the histones during nucleosome mobilization.
Keywords: ATPase, chromatin, remodelling, SNF2H, zinc finger
Introduction
Nucleosomal arrays are rendered dynamic through the action of nucleosome remodelling factors, which use the energy freed by ATP hydrolysis to disrupt histone–DNA contacts. Nucleosome remodelling increases the accessibility of nucleosomal DNA and induces histone octamer translocation on DNA (Becker and Hörz, 2002; Narlikar et al, 2002; Lusser and Kadonaga, 2003). The enzymes dedicated to nucleosome remodelling belong to the SNF2/SWI2 family of ATPases (Eisen et al, 1995). These enzymes typically reside in multiprotein complexes of variable composition. Proposed roles for associated subunits include the modulation of remodelling activity, the targeting of the enzymes to particular sites of action as well as the integration of the remodelling process into more complex regulatory programmes. The ATPase ISWI serves as a paradigm for understanding the mechanism of ATP-dependent nucleosome remodelling and its regulation (Längst and Becker, 2001b). Currently, a number of ISWI-containing complexes have been identified in various species from yeast to man (Längst and Becker, 2001b; Corona and Tamkun, 2004; Mellor and Morillon, 2004). Although their overall subunit composition may vary, ISWI is frequently associated with a larger subunit belonging to a family of related proteins. Members of this family, such as ACF1, WSTF, WCRF, TIP5 and NURF301, share C-terminal plant homeo domain (PHD) finger and bromodomain (Brd) modules besides several conserved sequence motifs of unknown structure and function. Brds and PHD fingers are found in other chromatin regulators as well. Brds can serve to interact with acetylated histone H4 and H3 N-termini and may possibly sensitize a nucleosome remodelling machine for histone acetylation marks (Jenuwein and Allis, 2001). PHD fingers (Aasland et al, 1995) are found in a large number of nuclear proteins involved in chromatin structure modulation. Their mutation or deletion in the context of various regulators, such as the ATRX, ING1 and AF10 proteins, leads to various disease syndromes (references cited in Capili et al, 2001). PHD fingers are supposed to mediate contacts with other proteins and sometimes this function requires coordination with a neighbouring Brd (Aasland et al, 1995; Hsu et al, 2001; Schultz et al, 2001).
The first ISWI interaction partner from this family of PHD–Brd proteins to be identified was Drosophila ACF1, which associates with ISWI to form ACF (Ito et al, 1999) and CHRAC complexes (Poot et al, 2000; Eberharter et al, 2001). These complexes potentially have dual functions as in vitro they may contribute to the assembly of nucleosomal arrays (Ito et al, 1999; Clapier et al, 2002; Vary et al, 2004) and also render these arrays dynamic by mobilizing histone octamers to slide on DNA (Corona et al, 1999; Längst et al, 1999; Eberharter et al, 2001). In certain chromatin assembly protocols, ISWI will not function without ACF1 (Ito et al, 1999; Fyodorov and Kadonaga, 2002b). When it comes to nucleosome sliding, ISWI is able to carry out a basal reaction, but its performance can be boosted by an order of magnitude upon association of ACF1 (Eberharter et al, 2001). Interestingly, ACF1 improves the effectiveness (‘energy-efficiency') of ISWI in trans, that is, although the ATP hydrolysis rate of ISWI does not change upon contact with ACF1, nucleosomes are mobilized much more efficiently. In addition, association of ACF1 modulates the remodelling activity of ISWI qualitatively by altering the directionality of nucleosome movement on small DNA fragments. While ISWI alone catalyses the movement of histone octamers from central positions to the ends of small DNA fragments, ACF triggers movement of end-positioned nucleosomes to more internal positions (Eberharter et al, 2001). The relevance of ‘directionality' of histone octamer translocation on nonphysiological mononucleosome substrates should not be overemphasized, but the phenomenon reveals a regulatory effect of ACF1 on ISWI function in trans, whose explanation in molecular terms will undoubtedly contribute to describing the mechanics of nucleosome remodelling.
The effect of ACF1 on ISWI may be due to an allosteric effect upon interaction of the two proteins, or alternatively be mediated by contacts of ACF1 with the nucleosome substrate. So far, interactions of ISWI and ACF1 with nucleosomal DNA have been characterized (Grüne et al, 2003), but interactions with the histone moiety, although suspected from the substrate requirements of nucleosome remodelling (Clapier et al, 2001), have not been described. In order to distinguish between the two possibilities, we reconstituted ACF complexes by coexpression of both subunits in insect cells from baculovirus vectors and mapped the interaction domains on either factor. Through construction of variant ACF complexes, in which conserved ACF1 domains were deleted, we were able to separate the effect of ACF1 on the directionality of nucleosome mobilization from its contribution to the energy efficiency of the remodelling process. Intriguingly, deletion of the PHD modules largely abolishes the positive effect of ACF1 on nucleosome mobilization. The PHD fingers of ACF1 make crucial contacts with histones and nucleosome particles, a property that has not been previously noted. Our results are consistent with a scenario, according to which contacts of ACF1 with the histone component of the nucleosome substrate improve the effectiveness, with which ISWI can translocate DNA relative to the histone octamer.
Results
Mapping the ACF1 interaction determinant on ISWI
ISWI can be roughly subdivided into an N-terminal ATPase domain and a C-terminal substrate recognition function consisting of HAND, SANT and SLIDE domains (Grüne et al, 2003; Figure 1A). In order determine, which part of ISWI would interact with ACF1, we expressed a series of ISWI fragments (Figure 1A) as glutathione-S-transferase (GST) fusion proteins in Escherichia coli and purified them by chromatography on glutathione-Sepharose. As a binding partner, we expressed FLAG-tagged ACF1 in Sf9 cells (Eberharter et al, 2001) using baculovirus vectors and immobilized it on anti-FLAG (M2) beads. The ISWI fragments were allowed to interact with immobilized ACF1 under stringent conditions and bound protein was resolved by PAGE and detected by Western blotting (Figure 1B). The ACF1 binding determinant localized to the C-terminal part of ISWI (Figure 1B) and could be narrowed down to the very C-terminus of ISWI containing amino acids (aa) 962–991 (Figure 1C). A summary of the interactions is shown in Figure 1A. We will refer to the ISWI domain required for ACF1 binding as AID (for ACF1 Interaction Determinant).
Figure 1.
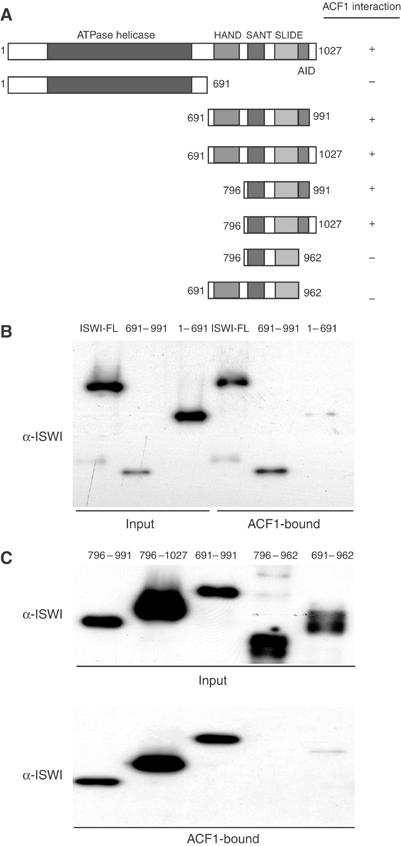
ACF1 interacts with the C-terminus of ISWI. (A) Summary of the domain organization of ISWI known to date. HAND (aa 697–795), SANT (aa 796–850) and SLIDE (aa 886–977) domains were recently defined by Grüne et al (2003). ‘+' and ‘–' behind each derivative indicate the ability to bind as determined in (B) and (C). The AID (between aa 962 and 991) was defined here. (B) FLAG-tagged ACF1 was immobilized on M2 anti-FLAG agarose. The resulting affinity resin was extensively washed and then used in pull-down experiments to monitor the interaction of ISWI. Bacterially expressed ISWI derivatives as indicated were incubated with the ACF1 beads. After extensive washes 30% of bound material was separated by SDS–8% PAGE and detected by Western blotting with an ISWI antibody (‘ACF1-bound'). As a control for interaction with full-length ISWI, we used a whole-cell extract of baculoviral-expressed, untagged ISWI. As reference, 10% of the input was loaded. (C) Smaller parts of ISWI (numbers above lanes correspond to first and last amino acids) were expressed in E. coli and tested for interaction with ACF1, as in (B). The upper panel displays the input of ISWI derivatives, and the lower panel reveals the bound protein.
Determinants on ACF1 for ISWI interaction
To establish the requirements for ISWI interaction in ACF1, we coexpressed a series of myc-tagged ACF1 derivatives with FLAG-tagged full-length ISWI in Sf9 cells (see Figure 2A). Complexes were purified from the cell lysate by affinity chromatography over an anti-FLAG resin and elution by a competing FLAG peptide. The interacting ACF1 derivatives or unbound ACF1 in the supernatant were detected by Western blotting. Deleting the C-terminal Brd and PHD fingers of ACF1 did not affect complex formation (Figure 2B). Deletions within a rather broad region within the N-terminus effectively removing the DDT and BAZ motives (Jones et al, 2000) abolished coelution with ISWI and led to the accumulation of the corresponding ACF1 derivatives in the supernatant (Figure 2B). Similar results were obtained in experiments where in vitro-translated ACF1 fragments were tested in ‘pull-down assays' with immobilized ISWI beads (data not shown). Our mapping of the ISWI interaction domain to a broad, central part of ACF1 is in agreement with previous results from the Kadonaga laboratory (Fyodorov and Kadonaga, 2002a).
Figure 2.
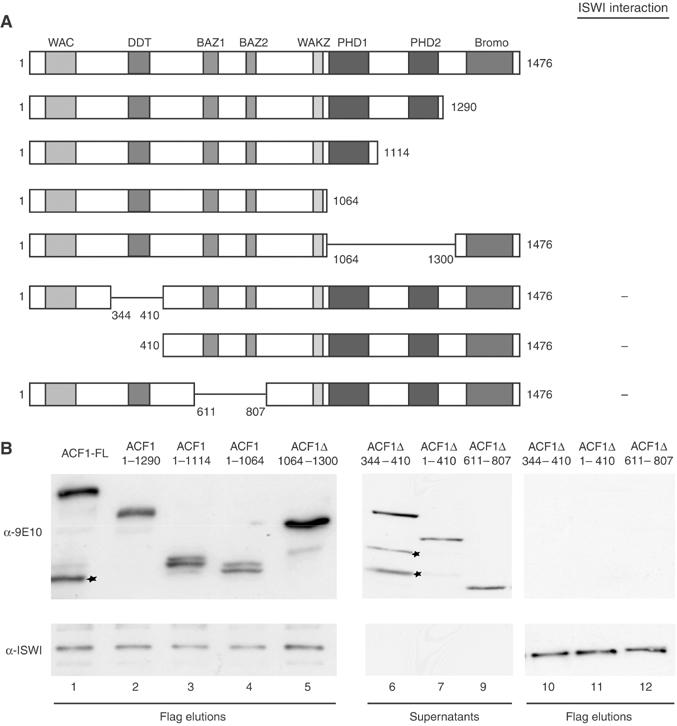
Mapping the ISWI interaction determinants on ACF1. (A) Summary of the known domain organization of ACF1 (Ito et al, 1999). The C-terminal domains extend between the following coordinates: PHD1: aa 1064–1114; PHD2: aa 1240–1300; Brd: aa 1361–1463. ‘+' and ‘−' indicate the ability to bind as determined in (B). (B) Full-length (FL) myc-tagged ACF1 and derivatives bearing various deletions as indicated were coexpressed with FLAG-tagged ISWI in Sf9 cells and resulting complexes were affinity-purified via the FLAG tag. Equal amounts of FLAG-eluted ACF complexes (i.e. ACF1 bound to ISWI) and the corresponding supernatants (i.e. proteins not interacting with ISWI) were separated by SDS–PAGE. ACF1 and ISWI were detected by Western blotting and probing with either monoclonal anti-myc antibody 9E10 for ACF1 detection or with antibody directed against ISWI (provided by J Tamkun).
Functional impact of C-terminal ACF1 deletions
Since the PHD fingers and the Brd motif were dispensable for ACF complex formation (see Figure 2), we were able to investigate whether these domains contributed to its nucleosome remodelling activity. Wild-type (wt) ACF and complexes bearing deletions within the C-terminus of ACF1 exhibited comparable nucleosome-stimulated ATPase activity, when the enzyme input was standardized by the amount of ISWI (data not shown). As a measure of remodelling activity, we monitored the ability of wt and mutated ACF complexes to catalyse the movement of a mononucleosome positioned at the end of a short DNA fragment to more central positions on the DNA (Längst et al, 1999; Eberharter et al, 2001) by a gel retardation assay (Figure 3A). Titrating substoichiometric amounts of wt ACF into a sliding reaction led to progressively more nucleosome movement (Figure 3A, lanes 2–6). Deleting the Brd of ACF1 (ΔBrd) did not impair the nucleosome sliding activity, but even improved the reaction efficiency somewhat (Figure 3A, lanes 7–11). However, further deletion removing the C-terminal PHD finger (ΔPHD2-Brd) clearly reduced the sliding efficiency about four-fold when compared to wt activity and about 10-fold when compared to the ΔBrd mutant (Figure 3A, lanes 12–16). Further deletion of PHD1 also resulted in an impaired enzyme (Figure 3A, lanes 17–21).
Figure 3.
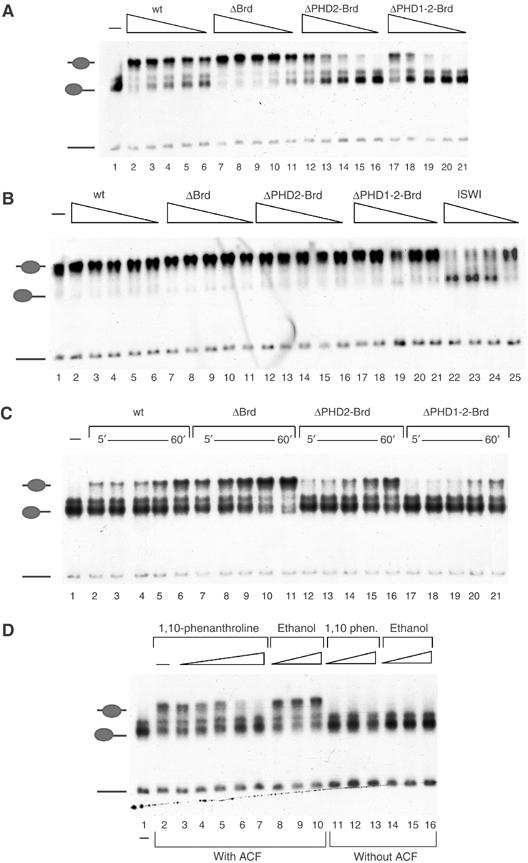
The PHD fingers of ACF1 are important for nucleosome mobilization. (A) 60 fmol of mononucleosomes positioned at the end of a 248 bp DNA fragment were incubated for 1 h with affinity-purified ACF or variant complexes bearing the indicated deletions of ACF1. In order to visualize nucleosome movement, samples were separated on a native 4.5% polyacrylamide gel. Dried gels were exposed to film overnight at −80°C. The reactions contained the following amounts of ACF complexes: 3 fmol (lanes 2, 7, 12 and 17); 1.5 fmol (lanes 3, 8, 13 and 18); 1 fmol (lanes 4, 9, 14 and 19); 0.75 fmol (lanes 5, 10, 15 and 20) and 0.375 fmol (6, 11, 16 and 21). Lane 1 shows the band corresponding to the mononucleosome in the absence of enzyme. The positions of free DNA, end- and centre-positioned nucleosomes are indicated to the left. (B) Nucleosome sliding reaction as in (A), but starting with 60 fmol of a centrally positioned mononucleosome. Lanes 22–25 show reactions driven by 6, 3, 1.5 and 0.75 fmol of ISWI, respectively. The untreated centre-positioned nucleosome is shown in lane 1. (C) Time course of nucleosome sliding. Reactions as in (A) contained 0.5 fmol of each of the indicated ACF complexes and 60 fmol of nucleosomes. Reactions were stopped at different times (5, 10, 15, 30 and 60 min as indicated) by the addition of 200 ng of unlabelled competitor DNA. (D) Nucleosomal sliding reactions as in (A) in the absence (lane 2) or presence of either 1,10 phenanthroline (0.1, 0.5, 1, 2.5 and 5 mM in lanes 3–7, respectively) or corresponding concentrations of the solvent ethanol (0.5, 1.25 or 2.5% in lanes 8–10, respectively). For controls, mononucleosomes were incubated with either 1, 2.5 or 5 mM of 1,10 phenanthroline or 0.5, 1.25 or 25% ethanol and resolved on the gel in lanes 11–13 and 14–16.
Previously, we had seen that the stimulatory effect of ACF1 on ISWI activity correlated with a changed ‘directionality' of nucleosome movement by ACF (Eberharter et al, 2001). It was therefore possible that the reduced sliding of end-positioned nucleosomes upon deletion of the PHD fingers would be accompanied by a corresponding increase of nucleosome movement from a central position. We therefore analysed the ACF complexes deleted in the C-terminus of ACF1 in reactions geared at sliding nucleosomes off central positions (Figure 3B). Recombinant ISWI, devoid of ACF1, was able to catalyse this reaction efficiently (Figure 3B, lanes 22–25). None of the ACF complexes was able to mobilize nucleosomes off the centre of the fragment. Evidently, we have separated the improvement of energy efficiency of ISWI from the qualitative modulation of directionality: whereas the PHD fingers appear crucial for improved energy efficiency, the determinant of ACF-type directionality (end to centre) of nucleosome sliding may reside in the N-terminus of ACF1 or correlate with the mere binding of ACF1 to ISWI.
We further explored the requirement of the ACF1 PHD fingers for efficient nucleosome mobilization by time courses under stringent, low enzyme conditions (60 fmol of nucleosomes versus 0.5 fmol of ACF complex; Figure 3C). The reaction was tuned such that about 40% of nucleosomes were moved by wt ACF during the course of the 60 min incubation (Figure 3C, lanes 2–6). Deletion of Brd improved the sliding efficiency significantly, as before (Figure 3C, lanes 7–11). Further deletion of PHD2 abolished this stimulatory effect and the ΔPHD1-2-Brd derivative was considerably less active than wt ACF (compare 15 min time points, lanes 4, 9, 14 and 19).
PHD domains require zinc for proper folding (Pascual et al, 2000; Capili et al, 2001). Removal of zinc is therefore expected to lead to local unfolding of the domain. To determine whether the activity of ACF1 indeed depended on proper folding, we thought of removing zinc from the protein by inclusion of 1,10-phenanthroline in nucleosome sliding reactions. 1,10-phenanthroline had been used in analogous experiments to evaluate the contribution of zinc-finger structures to the functions of the acetyltransferase MOF (Akhtar and Becker, 2001), the GAGA factor (Pedone et al, 1996), CBP (Kalkhoven et al, 2002) and p300 (Bordoli et al, 2001). Increasing amounts of 1,10-phenanthroline dramatically reduced the sliding ability of ACF (Figure 3D, lanes 3–7), whereas corresponding concentrations of the solvent, ethanol, did not affect the reaction (Figure 3D, lanes 8–10). Neither the drug nor the solvent affected sliding by ISWI (data not shown) or the mobility of nucleosomes alone (Figure 3D, lanes 11–16).
To explore the role of the ACF1 PHD fingers in nucleosome mobilization by ACF, we further expressed an ACF mutant complex lacking both PHD fingers (ACFΔPHD1-2) and analysed it in parallel with the wt ACF complex (Figure 4). wt ACF and ACFΔPHD1-2 were expressed and purified in parallel and carefully standardized with respect to subunit concentrations (Figure 4A). As we have observed for the different ACF complexes (Figure 3), equal amounts of wt and ACFΔPHD1-2 exhibited similar ATPase activity, both in the presence of DNA and nucleosomal substrates (Figure 4B). Moreover, the ATPase activity of these complexes was not affected by 1,10-phenanthroline, under conditions where the zinc chelator inhibits sliding (Figure 4B). 1,7-phenanthroline, which does not chelate zinc (Bird et al, 2003), also had no effect on the ATPase activities. Despite its ability to hydrolyse ATP in response to the nucleosome substrate, ACFΔPHD1-2 was essentially unable to mobilize nucleosomes under conditions where intact ACF functioned efficiently (Figure 4C, compare lanes 2 and 3 with 6 and 7). The addition of 1,10-phenanthroline to the reaction inhibited nucleosome sliding, whereas the nonchelating 1,7-phenanthroline isomer had only a minor effect (Figure 4D, lanes 2–8). The weak sliding activity of ACFΔPHD1-2 (Figure 4D, lane 9) was slightly affected by both 1,10- and 1,7-phenanthroline, pointing to a nonspecific effect (see lanes 10–15).
Figure 4.
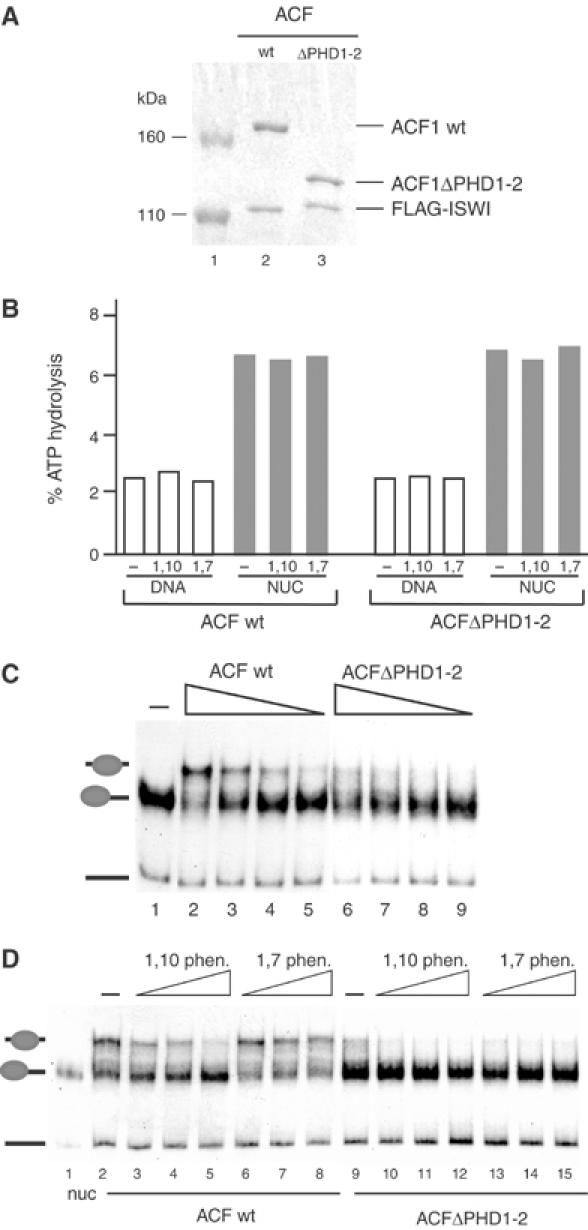
Requirement of the ACF1 PHD fingers for nucleosomal mobilization. (A) Normalization of proteins in ACF wt and ACFΔPHD1-2 complexes to be compared in the following experiments by 6% denaturing PAGE and Coomassie staining. Lane 1: size marker. (B) ATPase assays with 120 ng of naked DNA or 120 ng of nucleosomal DNA using 2 fmol of either wt ACF or the indicated mutant ACF complex. The reactions were performed in the absence (−) or presence of 3 mM of either 1,10-phenanthroline or 1,7-phenanthroline. (C) Nucleosome sliding reactions as in Figure 3A comparing ACF (lanes 2–5) and ACFΔPHD1-2 (lanes 6–9). Protein concentrations were 1 fmol (lanes 2 and 6), 0.5 fmol (lanes 3 and 7), 0.25 fmol (lanes 4 and 8) or 0.125 fmol (lanes 5 and 9). The untreated nucleosome is indicated in lane 1. (D) Nucleosome sliding reactions as in (C) with 1 fmol of either ACF wt (lanes 2–8) or ACFΔPHD1-2 (lanes 9–15). Reactions were carried out in the absence (lanes 2 and 9) or presence of either 1,10-phenanthroline at 2 mM (lanes 3 and 10), 3 mM (lanes 4 and 11) and 4 mM (lanes 5 and 12) or 1,7-phenanthroline at 2 mM (lanes 6 and 13), 3 mM (lanes 7 and 14) and 4 mM (lanes 8 and 15). The untreated nucleosome is indicated in lane 1 (nuc).
This suggests that disruption of the ACF1 PHD fingers neither affects substrate recognition nor the ATPase activity of ISWI, but rather the coupling of ATPase to DNA relocation. This is an example where the activity of a remodelling ATPase is affected in trans by a domain of an associated subunit.
ACF1 binds nucleosomes
Since the PHD fingers of ACF1 are not involved in binding ISWI, we considered interactions with the nucleosome substrate. ACF forms a single, well-defined complex with the mononucleosome sliding substrate in electrophoretic mobility shift assays (EMSAs) (Figure 5A). Deletion of the ACF1 C-terminus including Brd and both PHD fingers did not change this interaction significantly (Figure 5A, lanes 2–13). This overall binding activity is the result of multiple contacts between both subunits of the remodelling factor and the substrate. ISWI alone has a strong preference to interact with nucleosomal DNA (Figure 5A, lanes 14–16; Längst and Becker, 2001a; Grüne et al, 2003), which is likely to dominate the EMSA. A further DNA binding function has been suggested to reside in the N-terminus of ACF1 (Fyodorov and Kadonaga, 2002a). In agreement with dominant DNA binding activities of ACF that are not mediated by the PHD fingers, deletion of the PHD fingers did not abolish the band-shift and 1,10-phenanthroline did not affect this nucleosomal interaction (Figure 5B). The nucleosome binding properties of ACF1 alone have never been analysed since the protein could not be expressed in a soluble form previously (Fyodorov and Kadonaga, 2002a). We optimized baculovirus expression to obtain soluble ACF1 without ISWI, and demonstrate that ACF1 alone binds nucleosomes well (Figure 5A, lanes 17–19; Figure 5C, lanes 2–11). Deletion of the PHD finger did not diminish the affinity to the nucleosome; however, the complexes that were formed were less distinct (the band-shift smeary), possibly because the complex formed under these conditions was less rigid.
Figure 5.
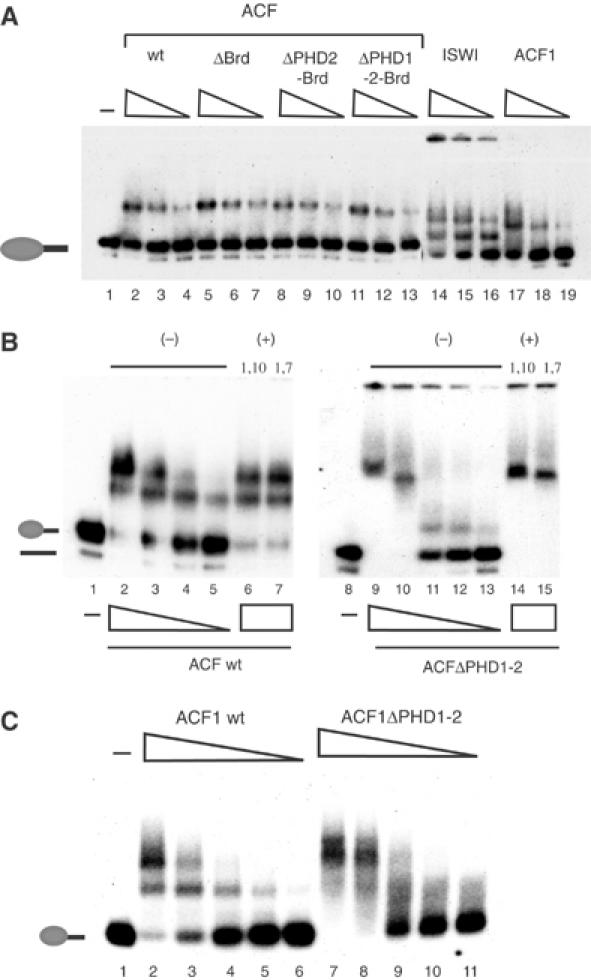
The PHD fingers of ACF1 are important for nucleosome recognition. (A) EMSA monitoring the interaction of various remodelling factors with mononucleosome substrates. Affinity-purified ACF complexes bearing the indicated ACF1 deletions, ISWI and ACF1 were incubated with 60 fmol of end-positioned nucleosome for 15 min. The reactions were then separated on a 1.4% agarose gel, which was dried and exposed to film overnight at −80°C. Protein concentrations were 60 fmol (lanes 2, 5, 8, 11, 14 and 17), 30 fmol (lanes 3, 6, 9, 12, 15 and 18) or 15 fmol (4, 7, 10, 13, 16 and 19). Lane 1 shows the migration of the nucleosome alone. (B) EMSA as in (A) using wt ACF complex (lanes 2–7) and ACFΔPHD1-2 (lanes 9–15). Protein concentrations were 120 fmol (lanes 2, 6, 7, 9, 14 and 15), 90 fmol (lanes 3 and 10), 60 fmol (4 and 11), 30 fmol (lanes 5 and 12) and 15 fmol (lane 13). Where indicated, either 3 mM 1,10-phenanthroline (lanes 6 and 14) or 3 mM 1,7-phenanthroline was added to the reaction. Lanes 1 and 8 show the migration of the nucleosome alone. (C) EMSA as in (A) analysing ACF1 full length (lanes 2–6) and ACFΔPHD1-2 (lanes 7–11). Protein concentrations were 90 fmol (lanes 2 and 7), 60 fmol (lanes 3 and 8), 30 fmol (lanes 4 and 9), 15 fmol (lanes 5 and 10) or 7.5 fmol (lanes 6 and 11). The migration of mononucleosomes in the absence of protein is shown in lane 1.
PHD fingers of ACF1 bind histones
PHD finger structures are known to interact with proteins in other contexts (Aasland et al, 1995; Zhang et al, 1998). In order to address their function directly without interference of the rest of ACF1, we expressed a region of ACF1 containing both fingers (aa 1039–1290) as a C-terminal fusion to a GST (GST-PHD1-2) in bacteria and bound them to glutathione-Sepharose beads, thereby generating an affinity resin. Since we were able to detect significant binding of nucleosomes but not of free DNA to this resin (data not shown), we explored whether the PHD region was able to interact with histones. For reference, we also expressed a GST-Brd protein and coupled it to Sepharose for pull-down studies (GST-Brd). Equivalent volumes of affinity beads were incubated with 1 μg of either purified embryonic Drosophila histones (Figure 6A, upper panel) or recombinant Drosophila histones (middle panel), bound protein was washed stringently, resolved by PAGE and detected by Coomassie blue staining. Both fusion proteins were able to bind a major fraction of the input histones at low and physiological salt, but considering the higher input of GST-Brd over GST-PHD1-2 (Figure 6A, ‘input' lower panel), the latter was more effective. GST beads alone did not bind any histones (not shown). When the stringency of the binding reaction was raised by increasing the ionic strength during the binding reaction, two phenomena were observable. First, the interaction of PHD1-2 appeared considerably stronger (binding at 500 mM salt) than the Brd interaction, since the latter faded as the ionic strength was increased and did not resist to 500 mM salt washes. Second, Brd showed a binding preference for histones H3 and H4 over H2A/H2B, whereas the PHD finger domains contacted all histones equally well. In order to determine whether the PHD finger functioned in the context of a larger structure, we generated additional GST fusion proteins (Figure 6B). As before, our reference was a weak interaction of the GST-Brd protein with H3 and H4. Addition of PHD2 improved the interaction with all four histones somewhat (Figure 6B, lane 5), but inclusion of PHD1 led to profound histone binding (Figure 6B, lane 2). The two PHD fingers alone bound the histones well and this binding was not improved by the presence of the WAKZ domain (lanes 4 and 3). We conclude that the PHD fingers are major determinants of histone interaction. The finding that all four core histones were bound equally well by the PHD finger domain led us to consider that the interaction may not be due to the flexible histone tails but to the central parts of the histones, which all share a common histone fold. Indeed, deletion of the N-termini in the context of histone pairs (labelled ‘tailless' in Figure 6B) did not abolish the interaction (Figure 6B, middle panels). No interaction, however, was observed when we used a dimer of the two histone-fold CHRAC components, CHRAC14/16, in the pull-down assays (data not shown).
Figure 6.
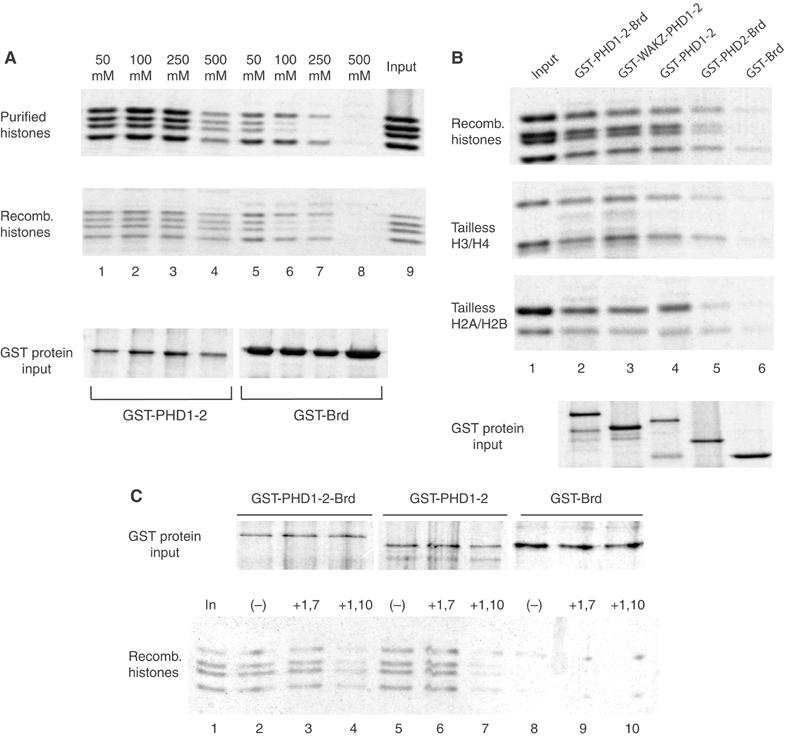
The ACF1 PHD fingers are required for interaction with histones. (A) GST-PHD1-2 and GST-Brd fusion proteins were expressed in bacteria, bound to glutathione-Sepharose 4B and used in ‘pull-down' experiments. About 500 ng of immobilized protein was incubated with 1 μg of Drosophila histone mixtures either purified from embryos (purified histones) or expressed in bacteria (recombinant histones) at the indicated salt concentrations. Histones that remained bound through excessive washes were separated by 15% SDS–PAGE and visualized by Coomassie blue staining. Lane 9 shows the histone input. (B) Indicated domains of ACF1 were expressed as GST fusion proteins and used in ‘pull-down' experiments as in (A). A 1 μg portion of all four recombinant histones (recomb histones) or of recombinant H3/H4 tetramers (tailless H3/H4) or recombinant H2A/H2B dimers (tailless H2A/2B), lacking their N-terminal tail domains, was tested in ‘pull-down' experiments. Bound material (50%) was separated by 15% denaturing PAGE and stained with Coomassie blue. The relative amount of each GST construct is shown in the bottom panel. (C) The indicated domains of ACF1 (upper panel) were expressed and used in pull-downs as described in (A). Before incubation with histones, the indicated GST beads were pretreated for 4 h at RT with buffer alone (−), 3 mM of 1,7-phenanthroline (+1,7) or 3 mM of 1,10-phenanthroline (+1,10). Lane 1 shows the histone input.
In a final experiment, we wished to determine whether the PHD–histone interactions were sensitive to 1,10-phenanthroline. We chelated zinc from the GST fusion proteins by treatment with 3 mM 1,10-phenanthroline before performing pull-down assays. The nonchelating 1,7-phenanthroline served as a control as before (Figure 6C). Interaction of the PHD fingers with histones was sensitive to zinc chelation (lanes 7 and 4), while the 1,7-phenanthroline did not have an effect (lanes 6 and 3). The weak interaction of Brd with H3 and H4 was insensitive against phenanthroline treatment (Figure 6C, lanes 8–10). Taken together, our data suggest that the PHD finger domains of ACF1 contribute to nucleosome mobilization by making crucial contacts with the central histone moiety of the nucleosome substrate.
Discussion
PHD–histone contacts are required for efficient nucleosome remodelling by ISWI
Binding of ACF1 to ISWI leads to a remarkable increase in energy efficiency of the nucleosome remodelling reaction catalysed by ISWI: while the amount of ATP hydrolysed in response to the nucleosome substrate remains unchanged upon ACF1 interaction, the complex moves nucleosomes an order of magnitude more efficiently than ISWI alone (Eberharter et al, 2001). Now, we have found that the PHD modules in the C-terminus of ACF1 are crucially involved in this activation. Previously, we observed this boost of activity only in conjunction with a change of nucleosome sliding directionality (Eberharter et al, 2001). Deletion of the PHD domain selectively affects the efficiency of the sliding reaction, but does not alter the type of nucleosome movement (this study). It appears, therefore, that structures in the N-terminus of ACF1, or simply the fact that ACF1 interacts with ISWI, determine the qualitative outcome of nucleosome mobilization.
PHD fingers have been proposed to serve as protein interaction surfaces (Aasland et al, 1995; Zhang et al, 1998), but our demonstration of contacts with the central parts of core histones in the context of a nucleosome remodelling factor is novel. Disruption of the PHD structure through zinc chelation destroys the interaction of ACF1 with histones and at the same time abolishes the stimulatory effect of ACF1 on the efficiency of nucleosome sliding. Evidently, ACF1–histone contacts are crucial for efficient nucleosome remodelling. Our results are in conflict with those obtained earlier by Fyodorov and Kadonaga (2002a), who did not observe a detrimental effect of deleting the PHD or Brd modules in an ACF-dependent chromatin assembly system. We think that this discrepancy may be due to the different functional assays employed (chromatin assembly versus nucleosome sliding) and perhaps also to differences in the protein expression protocol: whereas Fyodorov and Kadonaga (2002a) were unable to express soluble ACF1, our procedure yielded soluble protein that could be functionally characterized.
According to our favourite model, ISWI-containing remodelling factors lift DNA off the histone surface at the edge of the nucleosome and distort it into a bulge or loop. Propagation of this distortion over the surface of the histone octamer leads to nucleosome relocation (Längst and Becker, 2001a; Becker and Hörz, 2002; Figure 7). We consider that efficient displacement of nucleosomal DNA relative to the histone octamer requires contacts of the remodelling enzyme with both the DNA and histone moieties. ISWI alone binds nucleosomes mainly through interactions of its C-terminal SLIDE domain with nucleosomal DNA (Grüne et al, 2003). Although a segment of the H4 N-terminus is absolutely required for ISWI function (Clapier et al, 2001), we are so far not aware of any stable interactions of ISWI with histones. ISWI may be a relatively inefficient remodelling enzyme because it lacks a stable anchoring point on the histone body (Figure 7A). The PHD–histone contacts documented in our study may provide such an anchoring point for the enzyme, assuring that the presumed conformational changes triggered by ATP binding and hydrolysis are efficiently converted into positional shifts of DNA relative to histones (Figure 7B). According to a variation of this model, ACF plays an active role in the propagation of the DNA distortion (‘the loop') around the histone octamer (Lusser and Kadonaga, 2003; Figure 7C), which would necessitate changing contacts of the remodelling machinery with the histone octamer surface as it traverses around the particle. The observation that the PHD fingers of ACF1 interact with all four histones suggests that they recognize a common structural feature on the histone pairs, and is compatible with models involving multiple, different contacts of the remodeller on the histone octamer.
Figure 7.
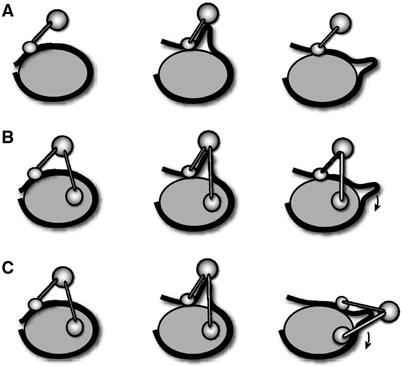
Models explaining the increased effectiveness of nucleosome sliding upon interaction of ACF1 with ISWI. (A) ISWI mainly interacts with linker DNA. (B, C) Additional contact of ACF1 with histones provides an anchor on the histone moiety of the nucleosome that allows efficient conversion of ATP-dependent conformational changes of ISWI into translocation of DNA relative to the histones during nucleosome mobilization.
Widespread occurrence of PHD modules in chromatin modifiers
ACF1 is a prominent member of a family of related proteins in various species, which interact with ISWI to form several distinct nucleosome remodelling complexes such as ACF (Ito et al, 1999), CHRAC (Eberharter et al, 2001), NURF (Xiao et al, 2001), WICH (Bozhenok et al, 2002), WCRF (Bochar et al, 2000) and NoRC (Strohner et al, 2001). These proteins share with ACF1 a similar domain organization including C-terminal PHD and Brds. A region of NURF301, the largest subunit of NURF, containing two PHD fingers and the adjacent Brd binds to all four core histones, but the functional consequences of these interactions have not been determined (Xiao et al, 2001). Interestingly, human ACF1 (alias WCRF180) and the related human WSTF, which associate with the human homologue of ISWI, SNF2H (Bochar et al, 2000; Poot et al, 2000; Bozhenok et al, 2002), function with only one PHD module and the hSNF2H-interacting protein RSF1 does not share any sequence similarity with ACF1, except for one PHD module (Loyola et al, 2003). It is therefore likely that one PHD module may be sufficient for function.
PHD fingers are diverse in sequence and may connect different proteins in various contexts. Our observation that PHD modules of ACF1 serve to tether a nucleosome remodelling enzyme to its substrate adds a new function to the list that should be tested for the known or suspected modifiers of chromatin structure, such as the remodelling ATPase Mi-2 (Zhang et al, 1998), the epigenetic regulators Trithorax, Polycomb-like (Aasland et al, 1995), Ash1 (Tripoulas et al, 1996), Ash2 (Adamson and Shearn, 1996) and Lid (Gildea et al, 2000).
PHD–Brd—an integrated nucleosome recognition module?
PHD modules are frequently found in the direct neighbourhood of a Brd. Examples include the ACF1-related proteins discussed here, and also the histone acetyltransferases CBP and p300 (Bordoli et al, 2001; Kalkhoven et al, 2002) and the KAP1 repressor (Schultz et al, 2001). From their functional analysis of domains involved in KAP-1 repression, Schultz et al concluded that both domains form an integrated, cooperative unit involved in binding the Mi2α subunit of the NuRD complex. In the context of histone binding, the idea of cooperativity between PHD fingers and Brds is attractive. While it is well established that Brds interact preferentially with acetylated N-termini of histones H3 and H4 (Jacobson et al, 2000; Owen et al, 2000), we showed that the PHD modules of ACF1 interact with the central domains of the core histones. This interaction may thus be further modulated by additional contacts of Brd with appropriately modified N-termini, as a means of fine-tuning nucleosome remodelling activity in response to the histone modification status. Whether this principle applies to ACF remains to be seen. However, Aasland and co-workers (Ragvin et al, 2004) have recently functionally characterized the Bromo–PHD modules of the histone acetyltransferase p300 and found that both domains cooperated for preferential binding to highly acetylated nucleosomes in a stringent assay.
Interestingly, deletion of the ACF1 Brd alone did not diminish nucleosome sliding, but consistently improved it. Given our limited knowledge on the structure of the ACF1 C-terminus as an entity, it is difficult to interpret this observation. Conceivably, the interaction of Brd with another domain of the remodelling machinery dampens its activity until a conformational change triggered by its interaction with an acetylated histone N-terminus unleashes full remodelling potential. This scenario suggests a strategy by which histone acetylation could regulate remodelling activity other than by simply increasing the affinity of the remodelling machinery for the substrate.
Apposition of functional domains through ACF1–ISWI interaction
We mapped AID to the very C-terminus of ISWI, directly adjacent to the SANT/SLIDE module, the nucleosome interaction determinant of ISWI (Grüne et al, 2003). Through this interaction, ACF1 will be brought close to the nucleosome surface, but this interaction will also lead to the apposition of several domains on both ACF subunits: the SANT domain of ISWI, which may be involved in contacting histone tails (Peterson and Logie, 2000; Boyer et al, 2002; Grüne et al, 2003); the SLIDE domain of ISWI, which contacts nucleosomal DNA (Grüne et al, 2003); the PHD fingers of ACF1, which binds the histone octamer surface (this study); and finally Brd with its potential for interactions with appropriately modified histone N-termini. Unravelling the sequence and dynamics of enzyme–substrate contacts during the remodelling process remains a major challenge for future research.
Materials and methods
Expression constructs
Baculovirus expression clones. All ACF1 deletions were generated by PCR from the full-length ACF1 cDNA in the pSport1 vector (Gibco-BRL). Each construct was created with a C-terminal myc tag and cloned with appropriate restriction sites into the vector pFastBac1 (Life Technologies Inc.). The clones were verified by sequencing. Details are available upon request. After recombination according to the manufacturer's protocol (Gibco-BRL) each bacmid was transfected into freshly diluted Sf9 cells together with Cellfectin (Life Technologies Inc.). Baculoviruses were amplified three times to obtain high-titer stocks before they were used in protein expression.
ACF1 constructs for expression in E. coli. ACF1 fragments were generated by PCR from full-length ACF1 in pSport1 vector. The GST fusion constructs PHD1-2-Brd (aa 1059–1476) and PHD2-Brd (aa 1237–1476) were subcloned into the EcoRI/XhoI sites of pGEX-4T-3 vector (Amersham Pharmacia Biotech), whereas the ACF1 fragments PHD1-2 (aa 1039–1290) and Brd (1300–1476) were subcloned into pET41c vector (Novagen) using the restriction sites HinDIII/XhoI and Spe/XhoI, respectively. Clones were verified by DNA sequencing.
Purification of ISWI, ACF1 and ACF complexes from Sf9 cells
ACF1-FLAG and FLAG-ISWI were synthesized in Sf9 cells as described previously (Eberharter et al, 2001). The different ACF complexes were generated by coexpression of FLAG-ISWI and variants of ACF1. The proteins were bound to M2 anti-FLAG agarose beads (SIGMA) or, in the case of myc-tagged ACF1 Δ-PHD1-2, to anti-c-myc agarose (SIGMA), extensively washed and purified as a complex or individual proteins as described (Eberharter et al, 2001). In short, Sf9 cells were suspended in HEMG-500 (25 mM HEPES (pH 7.6), 500 mM NaCl, 0.5 mM EDTA, 1.5 mM MgCl2, 10% glycerol, 0.05% NP-40 and protease inhibitors), frozen in liquid nitrogen and sonicated. Elution of the ACF1 Δ-PHD1-2-myc protein was achieved by adding 500 ng/μl MYC peptide (SIGMA) to HEMG-200 containing 1% NP-40 and constant rotation for 4 h at 4°C. To generate ACF1-FLAG beads for ISWI interaction studies, the FLAG peptide elution was omitted and the ACF1 beads were kept in buffer HEMG-250 as a 50% slurry (beads:buffer) at 4°C. A 10 μl volume of slurry of the ACF1-FLAG beads was routinely analysed by SDS–PAGE and Coomassie blue staining.
Synthesis and purification of bacterially expressed proteins
All subfragments of ACF1 were expressed as GST fusion proteins in E. coli. For a typical protein expression, 1 l culture was incubated at 37°C until an OD600 of 0.6, 0.3 mM IPTG was added and incubation proceeded for an additional 3 h. After collecting the cells by centrifugation, 40 ml of resuspension buffer (25 mM Tris–HCl (pH 8.0), 500 mM NaCl, 1% Triton X-100 and protease inhibitors) was added and the suspension was sonicated (Branson 250-D; amplitude 50%) on ice for 30 s with three repetitions. The material was clarified by centrifugation at 4°C for 20 min at 10 000 g. Soluble material was then incubated with pre-equilibrated glutathione-Sepharose 4B beads (Amersham Pharmacia Biotech) for at least 1 h at 4°C on a rotating wheel and washed five times with an excess of HEMG-200 buffer. The GST subfragments of ACF1 were finally resuspended in HEMG-200 and stored as a 50% (beads:buffer) slurry at 4°C.
ISWI subfragments were cloned into the BamHI/EcoRI sites of vector pGEX-4T-3 and expressed in 450 ml of E. coli as GST fusion proteins by induction with 0.15 mM IPTG for 3 h at 37°C. For ISWI 691–991, 796–991, 691–1027 and 796–1027, cells were resuspended in 25 ml of TBS (50 mM Tris–HCl (pH 8.0) and 150 mM NaCl)+10% Triton X-100 and protease inhibitors, sonicated (2 × 20 s, Branson 250-D; amplitude 60%) and cleared by centrifugation (4°C, 20 min, 10 000 g). The supernatant was incubated with 400 μl glutathione-Sepharose 4B beads for 1 h at 4°C. After extensive washes with TBS, proteins were eluted with 5 mM reduced glutathione for 15 min at 4°C under constant mixing. Glycerol was added to a final concentration of 25% and GST subfragments were kept at −20°C. In the case of ISWI 691–962 and 796–962, proteins were isolated from inclusion bodies by resuspending the cells in SAU200 (8 M urea, 20 mM NaAc (pH 5.2), 200 mM NaCl, 1 mM EDTA and 5 mM β-mercaptoethanol) overnight at RT under constant mixing. The material was cleared by centrifugation at 10 000 g for 20 min at 4°C and the supernatant was dialysed three times for 2 h and then overnight against refolding buffer (25 mM HEPES (pH 7.6), 500 mM KCl, 1.25 mM MgCl2, 0.1 mM EDTA, 25% glycerol, 0.05% NP-40 and 1 mM DTT). After dialysis, the material was cleared again by centrifugation and the supernatant was incubated with 300 μl glutathione-Sepharose 4B beads for 1 h at 4°C. Washes, elution and storage of the ISWI subfragments were as above.
ATPase assays
The ATPase assays were performed as described previously (Eberharter et al, 2001).
Nucleosome mobility and binding assays
The generation of mononucleosome particles and the nucleosome sliding reactions were as described (Eberharter et al, 2004). Reactions were stopped by the addition of 200 ng of competitor DNA. The samples were kept on ice before electrophoresis on native 4.5% PAG in 0.5 × TBE. Incubations for the EMSAs were performed at 26°C for 15 min using 60 fmol of nucleosomal substrate and different amounts of the affinity-purified proteins. Samples were electrophoresed on 1.4% agarose gels in 0.3 × TBE at 4°C for 75 min at 150 V. Gels were dried and exposed to film overnight at −80°C. 1,10 phenanthroline or 1,7 phenanthroline (SIGMA-Aldrich) were dissolved as 200 mM stock solutions in ethanol and stored at −20°C. Prior to the experiment, the phenanthroline solutions were diluted into the nucleosome mobility/band-shift buffer.
GST pull-down experiments
Histone octamers were either isolated from Drosophila embryo extract (Simon and Felsenfeld, 1979; Eberharter et al, 2004) or individual recombinant histones were expressed in E. coli (Luger et al, 1999; Clapier et al, 2001). In a typical GST pull-down assay, equal amounts of ACF1-fragment-GST beads were incubated with 1 μg of histones in a final volume of 150 μl of HEMG-X (X for variable salt concentrations) for 4 h at 4°C under constant mixing. Beads were then washed five times with an excess of HEMG-200 or indicated buffers and finally resuspended in 20 μl of loading buffer for SDS–PAGE. Bound material was separated on 15% polyacrylamide gels and stained with Coomassie blue.
Acknowledgments
This research was supported by Deutsche Forschungsgemeinschaft through SFB 594 and the European Commission through network grant HPRN-CT-2000–00078 within the Improvement of Human Potential Programme. We thank D Fyodorov and J Kadonaga for the ACF1-FLAG virus and P Verrijzer and J Tamkun for ISWI antibodies, C Clapier for histones and Ch Schwarzlose for technical support. We also thank Rein Aasland and members of the Becker lab for stimulating discussions and A Ragvin, R Aasland and A Brehm for critical comments on the manuscript.
References
- Aasland R, Gibson TJ, Stewart AF (1995) The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem Sci 20: 56–59 [DOI] [PubMed] [Google Scholar]
- Adamson AL, Shearn A (1996) Molecular genetic analysis of Drosophila ash2, a member of the trithorax group required for imaginal disc pattern formation. Genetics 144: 621–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar A, Becker PB (2001) The histone H4 acetyltransferase MOF uses a C2HC zinc finger for substrate recognition. EMBO Rep 2: 113–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker PB, Hörz W (2002) ATP-dependent nucleosome remodeling. Annu Rev Biochem 71: 247–273 [DOI] [PubMed] [Google Scholar]
- Bird AJ, McCall K, Kramer M, Blankman E, Winge DR, Eide DJ (2003) Zinc fingers can act as Zn2+ sensors to regulate transcriptional activation domain function. EMBO J 22: 5137–5146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochar DA, Savard J, Wang W, Lafleur DW, Moore P, Cote J, Shiekhattar R (2000) A family of chromatin remodeling factors related to Williams syndrome transcription factor. Proc Natl Acad Sci USA 97: 1038–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordoli L, Husser S, Luthi U, Netsch M, Osmani H, Eckner R (2001) Functional analysis of the p300 acetyltransferase domain: the PHD finger of p300 but not of CBP is dispensable for enzymatic activity. Nucleic Acids Res 29: 4462–4471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Langer MR, Crowley KA, Tan S, Denu JM, Peterson CL (2002) Essential role for the SANT domain in the functioning of multiple chromatin remodeling enzymes. Mol Cell 10: 935–942 [DOI] [PubMed] [Google Scholar]
- Bozhenok L, Wade PA, Varga-Weisz P (2002) WSTF–ISWI chromatin remodeling complex targets heterochromatic replication foci. EMBO J 21: 2231–2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capili AD, Schultz DC, Rauscher IF, Borden KL (2001) Solution structure of the PHD domain from the KAP-1 corepressor: structural determinants for PHD, RING and LIM zinc-binding domains. EMBO J 20: 165–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier CR, Längst G, Corona DF, Becker PB, Nightingale KP (2001) Critical role for the histone H4 N-terminus in nucleosome remodeling by ISWI. Mol Cell Biol 21: 875–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier CR, Nightingale KP, Becker PB (2002) A critical epitope for substrate recognition by the nucleosome remodeling ATPase ISWI. Nucleic Acids Res 30: 649–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corona DF, Langst G, Clapier CR, Bonte EJ, Ferrari S, Tamkun JW, Becker PB (1999) ISWI is an ATP-dependent nucleosome remodeling factor. Mol Cell 3: 239–245 [DOI] [PubMed] [Google Scholar]
- Corona DF, Tamkun JW (2004) Multiple roles for ISWI in transcription, chromosome organization and DNA replication. Biochim Biophys Acta 1677: 113–119 [DOI] [PubMed] [Google Scholar]
- Eberharter A, Ferrari S, Langst G, Straub T, Imhof A, Varga-Weisz P, Wilm M, Becker PB (2001) Acf1, the largest subunit of CHRAC, regulates ISWI-induced nucleosome remodelling. EMBO J 20: 3781–3788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberharter A, Längst G, Becker PB (2004) A nucleosome sliding assay for chromatin remodelling factors. Methods Enzymol 377: 344–353 [DOI] [PubMed] [Google Scholar]
- Eisen JA, Sweder KS, Hanawalt PC (1995) Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res 23: 2715–2723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fyodorov DV, Kadonaga JT (2002a) Binding of Acf1 to DNA involves a WAC motif and is important for ACF-mediated chromatin assembly. Mol Cell Biol 22: 6344–6353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fyodorov DV, Kadonaga JT (2002b) Dynamics of ATP-dependent chromatin assembly by ACF. Nature 418: 897–900 [DOI] [PubMed] [Google Scholar]
- Gildea JJ, Lopez R, Shearn A (2000) A screen for new trithorax group genes identified little imaginal discs, the Drosophila melanogaster homologue of human retinoblastoma binding protein 2. Genetics 156: 645–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grüne T, Brzeski J, Eberharter A, Clapier CR, Corona DF, Becker PB, Müller CW (2003) Crystal structure and functional analysis of the nucleosome recognition module of the remodeling factor ISWI. Mol Cell 12: 449–460 [DOI] [PubMed] [Google Scholar]
- Hsu SI, Yang CM, Sim KG, Hentschel DM, O'Leary E, Bonventre JV (2001) TRIP-Br: a novel family of PHD zinc finger- and bromodomain-interacting proteins that regulate the transcriptional activity of E2F-1/DP-1. EMBO J 20: 2273–2285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Levenstein ME, Fyodorov DV, Kutach AK, Kobayashi R, Kadonaga JT (1999) ACF consists of two subunits, Acf1 and ISWI, that function cooperatively in the ATP-dependent catalysis of chromatin assembly. Genes Dev 13: 1529–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson RH, Ladurner AG, King DS, Tjian R (2000) Structure and function of a human TAFII250 double bromodomain module. Science 288: 1422–1425 [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD (2001) Translating the histone code. Science 293: 1074–1080 [DOI] [PubMed] [Google Scholar]
- Jones MH, Hamana N, Nezu J, Shimane M (2000) A novel family of bromodomain genes. Genomics 63: 40–45 [DOI] [PubMed] [Google Scholar]
- Kalkhoven E, Teunissen H, Houweling A, Verrijzer CP, Zantema A (2002) The PHD type zinc finger is an integral part of the CBP acetyltransferase domain. Mol Cell Biol 22: 1961–1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Längst G, Becker PB (2001a) ISWI induces nucleosome sliding on nicked DNA. Mol Cell 8: 1085–1092 [DOI] [PubMed] [Google Scholar]
- Längst G, Becker PB (2001b) Nucleosome mobilization and positioning by ISWI-containing chromatin-remodeling factors. J Cell Sci 114: 2561–2568 [DOI] [PubMed] [Google Scholar]
- Längst G, Bonte EJ, Corona DF, Becker PB (1999) Nucleosome movement by CHRAC and ISWI without disruption or trans-displacement of the histone octamer. Cell 97: 843–852 [DOI] [PubMed] [Google Scholar]
- Loyola A, Huang JY, LeRoy G, Hu S, Wang YH, Donnelly RJ, Lane WS, Lee SC, Reinberg D (2003) Functional analysis of the subunits of the chromatin assembly factor RSF. Mol Cell Biol 23: 6759–6768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Rechsteiner TJ, Richmond TJ (1999) Preparation of nucleosome core particle from recombinant histones. Methods Enzymol 304: 3–19 [DOI] [PubMed] [Google Scholar]
- Lusser A, Kadonaga JT (2003) Chromatin remodeling by ATP-dependent molecular machines. BioEssays 25: 1192–1200 [DOI] [PubMed] [Google Scholar]
- Mellor J, Morillon A (2004) ISWI complexes in Saccharomyces cerevisiae. Biochim Biophys Acta 1677: 100–112 [DOI] [PubMed] [Google Scholar]
- Narlikar GJ, Fan HY, Kingston RE (2002) Cooperation between complexes that regulate chromatin structure and transcription. Cell 108: 475–487 [DOI] [PubMed] [Google Scholar]
- Owen DJ, Ornaghi P, Yang JC, Lowe N, Evans PR, Ballario P, Neuhaus D, Filetici P, Travers AA (2000) The structural basis for the recognition of acetylated histone H4 by the bromodomain of histone acetyltransferase gcn5p. EMBO J 19: 6141–6149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual J, Martinez-Yamout M, Dyson HJ, Wright PE (2000) Structure of the PHD zinc finger from human Williams–Beuren syndrome transcription factor. J Mol Biol 304: 723–729 [DOI] [PubMed] [Google Scholar]
- Pedone PV, Ghirlando R, Clore GM, Gronenborn AM, Felsenfeld G, Omichinski JG (1996) The single Cys2-His2 zinc finger domain of the GAGA protein flanked by basic residues is sufficient for high-affinity specific DNA binding. Proc Natl Acad Sci USA 93: 2822–2826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson CL, Logie C (2000) Recruitment of chromatin remodeling machines. J Cell Biochem 78: 179–185 [DOI] [PubMed] [Google Scholar]
- Poot RA, Dellaire G, Hulsmann BB, Grimaldi MA, Corona DF, Becker PB, Bickmore WA, Varga-Weisz PD (2000) HuCHRAC, a human ISWI chromatin remodelling complex contains hACF1 and two novel histone-fold proteins. EMBO J 19: 3377–3387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragvin A, Valvatne H, Erdal S, Arskog V, Tufteland KR, Breen K, AM OY, Eberharter A, Gibson TJ, Becker PB, Aasland R (2004) Nucleosome binding by the bromodomain and PHD finger of the transcriptional cofactor p300. J Mol Biol 337: 773–788 [DOI] [PubMed] [Google Scholar]
- Schultz DC, Friedman JR, Rauscher FJ III (2001) Targeting histone deacetylase complexes via KRAB-zinc finger proteins: the PHD and bromodomains of KAP-1 form a cooperative unit that recruits a novel isoform of the Mi-2alpha subunit of NuRD. Genes Dev 15: 428–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon RH, Felsenfeld G (1979) A new procedure for purifying histone pairs H2A+H2B and H3+H4 from chromatin using hydroxylapatite. Nucleic Acids Res 6: 689–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strohner R, Nemeth A, Jansa P, Hofmann-Rohrer U, Santoro R, Langst G, Grummt I (2001) NoRC—a novel member of mammalian ISWI-containing chromatin remodeling machines. EMBO J 20: 4892–4900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripoulas N, LaJeunesse D, Gildea J, Shearn A (1996) The Drosophila ash1 gene product, which is localized at specific sites on polytene chromosomes, contains a SET domain and a PHD finger. Genetics 143: 913–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vary JC Jr, Fazzio TG, Tsukiyama T (2004) Assembly of yeast chromatin using ISWI complexes. Methods Enzymol 375: 88–102 [DOI] [PubMed] [Google Scholar]
- Xiao H, Sandaltzopoulos R, Wang HM, Hamiche A, Ranallo R, Lee KM, Fu D, Wu C (2001) Dual functions of largest NURF subunit NURF301 in nucleosome sliding and transcription factor interactions. Mol Cell 8: 531–543 [DOI] [PubMed] [Google Scholar]
- Zhang Y, LeRoy G, Seelig HP, Lane WS, Reinberg D (1998) The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell 95: 279–289 [DOI] [PubMed] [Google Scholar]


