Abstract
Rotavirus, a cause of severe gastroenteritis, contains a segmented double-stranded (ds)RNA genome that replicates using viral mRNAs as templates. The highly conserved 3′-consensus sequence (3′CS), UGUGACC, of the mRNAs promotes dsRNA synthesis and enhances translation. We have found that the 3′CS of the gene (g5) encoding NSP1, an antagonist of interferon signaling, undergoes rapid mutation when rhesus rotavirus (RRV) is serially passaged at high multiplicity of infection (MOI) in cells permitting high titer growth. These mutations increase the promoter activity of the g5 3′-sequence, but decrease its activity as a translation enhancer. The location of the mutations defines the minimal essential promoter for dsRNA synthesis as URN0–5CC. Under passage conditions where cell-to-cell spread of the virus is required to complete infection (low MOI), the 3′CS is retained due to the need for NSP1 to be expressed at levels sufficient to prevent establishment of the antiviral state. These data demonstrate that host cell type and propagation conditions affect the capacity of RRV to produce the virulence gene product NSP1, an important consideration in producing RRV-based vaccines.
Keywords: gene expression, interferon regulatory factor 3, RNA replication, rotavirus, vaccine production
Introduction
Rotavirus, a member of the family Reoviridae, is the primary cause of acute dehydrating diarrhea in infants and young children worldwide (Parashar et al, 2003). The icosahedral core of the triple-layered rotavirus particle contains the 11 segments of double-stranded (ds)RNA that make up the viral genome (Prasad et al, 1988). The segmented nature of the genome demands that the viral RNA-dependent RNA polymerase (RdRP) recognize and employ multiple viral mRNAs as templates for dsRNA synthesis. The synthesis of the dsRNA genome is a process that is coordinated with the packaging of the template mRNAs into previrion cores (Patton and Spencer, 2000). The genome segments are distributed in an orderly manner within the core, each associated with one of the 12 vertices (Prasad et al, 1996). Along with a genome segment, a copy of the viral RdRP (VP1) and mRNA-capping enzyme (VP3) are also associated with each vertex (Estes, 2001). The RdRPs transcribe the genome segments to produce mRNAs that are extruded from the core through channels present at the vertices (Figure 1) (Lawton et al, 1997).
Figure 1.
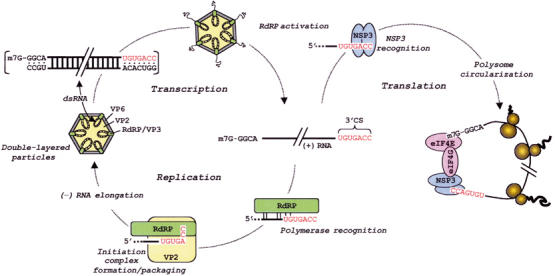
Importance of the 3′CS to rotavirus RNA replication and gene expression. The 11 dsRNA genome segments (upper left) contain highly conserved sequences at their termini, including the 3′CS (red) at the 3′-end of the (+) strand. Activation of the RdRPs within double-layered particles yields (+) RNAs, which are extruded through channels located at the vertices. During gene expression, NSP3 dimers simultaneously interact with the last four to five nucleotides of the 3′CS and with eukaryotic initiation factors, causing polysome circularization. During RNA replication, the RdRP recognizes signals in the 3′CS and upstream region of the (+) RNA. Recognition is followed by the formation a (−) strand initiation complex, an event that requires the 3′CC of the (+) strand template, the RdRP and core scaffold protein VP2, and GTP. Initiation and (−) strand synthesis are associated with the packaging of (+) RNAs into cores. The assembly of a VP6 layer around the core produces a double-layered particle.
Transcription of the rotavirus dsRNA genome produces 11 mostly monocistronic mRNAs that contain 5′-methylated caps but lack 3′-poly(A) tails (Estes, 2001). Sequencing has indicated that the viral mRNAs of the most common group of rotaviruses (group A) begin with 5′-GGC(U/A)7-3′ and, with only rare exception, end with the 3′-consensus sequence (3′CS), 5′-UGUGACC-3′ (Figure 1) (Mitchell and Both, 1990a; Desselberger and McCrae, 1994). The conservation of the 3′CS suggests that it constitutes part of one or more cis-acting signals for processes that all the viral mRNAs undergo during virus replication. Studies on the function of the 3′CS bear this out, having indicated that the element contains information for at least three functions—(i) polymerase recognition: Electrophoretic mobility shift assays (EMSAs) have shown that the 3′CS contains sufficient information to allow the rotavirus RdRP to specifically recognize viral mRNAs. However, the 3′CS represents only a part of the recognition signal since the RdRP can bind specifically to viral mRNAs even upon deletion of the 3′CS (Tortorici et al, 2003); (ii) promoter activity: Template-dependent replication assays have demonstrated that deletion of the 3′CS prevents viral mRNAs from serving as templates for (−) strand synthesis (Patton et al, 1996). Such assays have also indicated that the 3′CC of the mRNAs is of critical importance for dsRNA synthesis, while the remaining residues of the 3′CS are of variable importance (Chen et al, 2001); (iii) efficient translation: The last four to five nucleotides of the 3′CS, (U)GACC, are specifically recognized by dimers of NSP3, a viral nonstructural protein which also has affinity for the cap-associated eukaryotic initiation factor, eIF4GI (Poncet et al, 1994; Piron et al, 1998; Deo et al, 2002; Groft and Burley, 2002). By binding simultaneously to both eIF4GI and the 3′-terminal (U)GACC sequence, NSP3 acts as a translation enhancer that upregulates viral gene expression by promoting the circularization of viral mRNAs in polysomes (Figure 1) (Chizhikov and Patton, 2000; Vende et al, 2000). In this regard, NSP3 operates as a rotavirus-specific surrogate of the eukaryotic poly-A-binding protein (PABP; Kahvejian et al, 2001).
Given the multiple functions for the 3′CS, any mutation of the element could be predicted to lead to a loss of virus viability. However, our analysis of a genome segment of rhesus rotavirus (RRV), a strain which forms the basis of the tetravalent live-attenuated human RotaShield vaccine (Wyeth-Lederle Vaccines and Pediatrics) (Kapikian et al, 1996), reveals that this is not the case. Specifically, we have found that the 3′CS of the gene 5 mRNA (g5(+) RNA) of RRV undergoes rapid and radical change, but only upon serial passage of the virus in highly permissive cells at high multiplicity of infection (MOI). Such changes were not observed upon serial passage in highly permissive cells at low MOI or in less permissive cells. The product of the rotavirus g5(+) RNA is NSP1, a nonstructural protein whose interaction with the cytoplasmic resident protein, interferon (IFN) regulatory factor 3 (IRF3) (Graff et al, 2002), promotes cell-to-cell spread of the virus (Barro and Patton, 2004). This interaction suppresses the translocation of IRF3 to the nucleus, where otherwise the factor activates genes expressing IFN and induces the establishment of the antiviral state (Hiscott, 1999; Barro and Patton, 2004). Consistent with the importance of NSP1 in promoting virus spread is the observation that genetic changes to the rotavirus g5(+) RNA which truncate the NSP1 open reading frame (ORF) yield viruses with small plaque phenotype (Patton et al, 2001).
We determined that mutation of the 3′CS has two overall effects on the RRV g5(+) RNA, increasing its efficiency as a replication template for g5 dsRNA and decreasing its efficiency as a translation template for NSP1. The latter appears to be tolerated under conditions of high MOI passage in highly permissive cells because of the diminished need for the viral protein (NSP1) promoting cell-to-cell spread. Our analysis redefines the essential 3′-terminal sequence required for dsRNA synthesis as URN0–5CC and indicates that the prototypic 3′CS represents a suboptimal promoter for RNA synthesis whose sequence is largely retained due to the role of the element as a translation enhancer. This study illustrates the importance of host cell line and passage conditions as variables affecting the phenotype of progeny viruses. These results also suggest that the efficacy of live-attenuated rotavirus vaccines may be impacted by the methods used in propagating the virus strains included in the vaccines.
Results
3′-Sequence heterogeneity of g5(+) RNA of RRV
Previous studies have shown that the g5(+) RNAs of simian SA11 and SA11-4F rotaviruses grown in the highly permissive MA104 cell line end with the mutant sequence, UGUGAaCC, due to the insertion of an A residue (lower case) into the 3′CS, UGUGACC (Mitchell and Both, 1990b; Patton et al, 2001). Sequencing of a large number of cDNAs generated by RACE confirmed the invariant nature of the mutant g5(+) 3′CS in laboratory stocks of SA11-4F (Table I).
Table 1.
3′-Terminal sequences of gene 5 (+)RNAs of rotavirus propagated in MA104 cells
| Strain | 3′-sequence | Clones |
|---|---|---|
| RRV (rhesus) | UGccuCCa | 6 |
| RRV (rhesus) | UGUuuCC | 5 |
| RRV (rhesus) | UGcuuCC | 3 |
| RRV (rhesus) | UGUuauCC | 3 |
| RRV (rhesus) | UGauuCC | 2 |
| RRV (rhesus) | UGUAaCC | 1 |
| RRV (rhesus) | UGUugCC | 1 |
| RRV (rhesus) | UGUcuCC | 1 |
| SA11-4F (simianb) | UGUGAaCC | 30 |
| UK (bovine) | UGUGACC | 20, 37c |
aFor this and subsequent tables, wild-type and mutant sequences are shown in upper and underlined lower case, respectively.
bGene 4 of SA11-4F is bovine rotavirus-like in origin, other genes are of simian SA11 origin.
cPools originated from separate sources.
To examine whether the mutant g5(+) 3′CS characteristic of SA11 virus was common to other primate strains of rotavirus, cDNA clones of the g5 dsRNA recovered from laboratory stocks of RRV grown in MA104 cells were generated by RACE. Sequencing showed that the RRV g5(+) RNAs ended with neither the prototypic 3′CS nor the mutant g5(+) 3′CS of SA11 viruses (Table I). Instead, the RRV g5(+) 3′-sequences were heterogenous in nature, most differing by two or three residues from the 3′CS due to base substitutions at the −3 (A → U or G), −4 (G → U, C or A), and, in some cases, −5 (U → C or A) positions. Although the majority of the g5(+) 3′-sequences retained the heptamer length 3′CS, an insertional mutation caused the 3′CS to become octameric in some cases, that is, UGUuauCC. Notably, the terminal CC (−1, −2) and upstream UG (−6, −5) of the 3′CS were retained in all the RRV g5(+) RNAs. Unlike the g5(+) RNAs of SA11, SA11-4F and RRV, RACE analysis showed that all g5(+) RNAs contained in two independent lots of UK bovine rotavirus grown in MA104 cells ended with the 3′CS (Table I).
Plaque isolation of RRV g5 variants
To determine whether RRV variants with mutant g5(+) 3′CS could be clonally isolated, RRV grown in MA104 cells was subjected to triple-plaque-to-plaque purification. Afterwards, the plaque-purified viruses from the second and third cycles were amplified once at low MOI to generate dsRNAs for RACE analysis. Sequencing led to the identification of five unique variants, characterized largely by the substitution of residues at the −3, −4, and, less frequently, −5 position of the 3′CS with U (Table II). Little or no heterogeneity was detected for the g5(+) 3′-sequences of the variants RRVg5-auu, -cuu, -u, and -uu, recovered after the second and third rounds of plaque purification. The predominant g5(+) 3′-sequence of the variant RRVg5-u contained not only a base substitution but also a deletion. Thus, virus with g5 dsRNAs ending with a hexameric sequence instead of the heptameric 3′CS is viable. Analysis of the triple-plaque-purified variant RRVg5-ccuu gave results suggesting that its g5(+) 3′-sequence was not genetically stable, shifting between the octamer UGccuuCC and the tetramer UGCC (Table II). The UGCC sequence is significant as it is similar to the conserved sequence, (A/U)GCC, found at the 3′-end of the (−) strand of all rotavirus RNAs (3,16). The conservation of the (A/U)GCC sequence and its location at the 3′-end of the (−) strand of rotavirus RNAs indicates that it forms all or part of the transcription promoter for (+) strand synthesis. RACE analysis of the plaque-purified variant RRV-uu revealed that its g9(+) RNA ended with the prototypic 3′CS (data not shown). This finding indicates that mutation of the g5(+) 3′CS can occur in the absence of changes to the 3′-ends of the other viral (+) RNAs.
Table 2.
3′-Sequence of g5(+) RNA of RRV variants isolated by triple plaque-to-plaque purification on MA014 cellsa
| Cycle two |
Cycle three |
|||
|---|---|---|---|---|
| Variant | 3′-end | Clones | 3′-end | Clones |
| RRVg5-u | UGUuCC | 14 | UGUuCC | 12 |
| UGUuuCC | 1 | |||
| RRVg5-uu | UGUuuCC | 6 | UGUuuCC | 9 |
| UGUuCC | 1 | UGUuCC | 1 | |
| UGUuuuCC | 1 | |||
| RRVg5-auu | UGauuCC | 9 | UGauuCC | 17 |
| UGauCC | 1 | |||
| RRVg5-cuu | UGcuuCC | 13 | UGcuuCC | 14 |
| RRVg5-ccuu | UGccuuCC | 7 | UGccuuCC | 7 |
| UG–––CC | 3 | UG–––CC | 5 | |
| UGccuCC | 1 | |||
aGenBank accession numbers for g5(+) RNAs: RRVg5-cs, AY117048; RRVg5-u, AY117052; RRVg5-uu, AY117053; RRVg5-auu, AY117049; RRVg5-cuu, AY117051; RRVg5-ccuu, AY117050.
Plaque isolation of a variant from a pool of DS1xRRV grown in rhesus fetal lung FRhL2 cells revealed that viruses with mutant g5(+) 3′-ends could also be isolated, which contained other than just RRV-specific genes. The reassortant virus DS1xRRV contains 10 genome segments from RRV and one segment (gene 9) from the human strain DS1 (Midthun and Kapkian, 1996). The g5(+) RNA of the variant, DS1xRRVg5-uu, was like that of RRVg5-uu, ending with the sequence UGUuuCC (Table II). One-step growth curves revealed no differences in virus production by DS1xRRVg5-uu and DS1xRRV containing g5(+) RNA with the prototypic 3′CS (DS1xRRVg5-cs) in MA104 cells (data not shown).
Full-length cDNA clones were prepared from the g5 dsRNAs of DS1xRRV and RRV variants and their corresponding wild-type viruses, that is, DS1xRRVg5-cs and RRVg5-cs, respectively. Sequencing revealed that the g5(+) RNAs of DS1xRRVg5-uu and DS1xRRVg5-cs were identical, except for the expected 3′-terminal variations. Similarly, the sequences of the g5(+) RNAs of the RRV variants were identical to that of RRVg5-cs, except for the expected differences in their 3′-ends, and in some cases the replacement of nucleotide G178 in the RRVg5-cs sequence with a T in the variant sequences. When present, the G178>T change resulted in the conservative amino-acid substitution V50>L in the g5 product, NSP1. RACE analysis indicated that the sequence 5′-A8CGCC-3′ was present at the 3′-ends of the (−) strand of the g5 dsRNAs recovered from the RRV variants and from RRVg5-cs. Hence, the portions of the g5 dsRNAs predicted to be involved in initiation of transcription (i.e., (+) strand synthesis) were the same for the variant and wild-type viruses (Figure 1). Electrophoretic analysis revealed no differences in the sizes of any of the dsRNA genome segments among the RRV and DS1xRRV isolates (data not shown). Although their g5 dsRNAs lacked the 3′CS, the variant and wild-type viruses were able to grow to similar high titers (⩾108 plaque-forming units (PFU)/ml) in MA104 cells.
Impact of cell passage on 3′-sequence variation
Although the g5(+) 3′-ends of RRV grown in MA104 cells were variable and atypical in sequence (Table I), RACE analysis revealed that g5(+) RNAs of plaque-purified RRV serially passaged seven times in FRhL2 cells ended exclusively with the 3′CS (Table III). To explore the possibility that growth of RRV in MA104 cells promoted deviation of the g5(+) 3′CS, FRhL2-derived RRV was passaged serially in MA104 cells using low dilutions of infected cell lysates as inocula (initial MOI >10). RACE revealed that, even after a single passage in MA104 cells, g5(+) RNAs which lacked the 3′CS were present (Table III). By the third passage, 50% of the g5(+) RNAs lacked the 3′CS and, by the fifth passage, no g5(+) RNAs were detected that contained the 3′CS. Thus, serial passage of RRV at high MOI in MA104 cells strongly favored the production of virus lacking the g5(+) 3′CS.
Table 3.
Generation of RRV gene 5 variants by serial passage in MA104 cells
| MA104 cells (clones) |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| High MOI |
Low moi |
||||||||||
| FRhL2 cells | (48) | Pass 1 | (27) | Pass 2 | (28) | Pass 3 | (40) | Pass 5 | (36) | Pass 5 | (31) |
| UGUGACC | 48 | UGUGACC | 23 | UGUGACC | 18 | UGUGACC | 19 | UGUuauCC | 24 | UGUGACC | 29 |
| UGUuuCC | 2 | UGUu-CC | 4 | UGUu-CC | 8 | UGUu-CC | 6 | UGUuuCC | 1 | ||
| UGUu-CC | 2 | UGUuuCC | 3 | UGUuuCC | 7 | UGUuuCC | 4 | UGUugCC | 1 | ||
| UGUuACC | 1 | UGUuauCC | 2 | UGcu-CC | 1 | ||||||
| UGUuauCC | 1 | UGccuCC | 2 | UGU–CC | 1 | ||||||
| UGUuuuuCC | 1 | UGUauuCC | 1 | ||||||||
| UGUGAaCC | 1 | ||||||||||
Inspection of RRV passaged serially at high MOI revealed that the initial base substitutions at the g5(+) 3′-end were not random, but mostly involved replacement of A(−3) and/or G(−4) with U (Table III, pass 1 and 2). Both deletion and insertion of residues between −3 and −4 were also events observed early in the passage of RRV. Interestingly, the g5(+) 3′-end UGUuauCC that predominated in the pass-5 RRV pool (24/36) was not the predominant 3′-end of any earlier virus passage. This suggests that, although other variant 3′-ends (e.g., UGUGuuCC) may have evolved more readily during serial passage, stronger selective pressure existed favoring utilization and retention of the UGUuauCC end.
Analysis of RRV serially passaged 50 times at high MOI in MA104 cells showed that the g5(+) 3′-ends were still somewhat heterogenous in nature (Table IV). However, the predominant g5(+) 3′-end UGUugCC of the pass 50 pool was unlike the predominant 3′-end of the pass 5 pool (Table III), instead showing similarity to the transcription-promoter sequence (A/U)GCC found at the 3′-end of rotavirus (−) sense RNAs (Figure 1). The UGUugCC end was not detected in any of the pass 1–5 pools of RRV (Table III), suggesting that it may have evolved by a multi-step process whereby variant g5(+) 3′-ends underwent successive rounds of mutation. The second most prevalent g5(+) 3′-end in the pass 50 pool was UGUGAaCC, and thus identical to the g5(+) 3′-ends of virus strains SA11, K9, and KU (Table VI).
Table 4.
Effect of cell line on the variation of the 3′-end of RRV g5(+) RNA
| Virus | Cell type | Passagea | 3′-sequence | Clones |
|---|---|---|---|---|
| RRVb | AGMK | 50 | UGcugCC | 46 |
| UGUGAaCC | 5 | |||
| UGUugCC | 2 | |||
| MA104 | 50 | UGUugCC | 32 | |
| UGUGAaCC | 8 | |||
| UGUG-CC | 2 | |||
| HCT | 15 | UGUGACC | 7 | |
| Vero | 10 | UGUauCC | 5 | |
| UGUuauCC | 3 | |||
| UGUuuCC | 1 | |||
| UGUu-CC | 1 | |||
| UGcuuCC | 1 | |||
| DS1xRRVg5-cs | MA104 | 10 | UGUuCC | 11 |
| UGUGACC | 10 | |||
| UGUGAuCC | 8 | |||
| UGUuuCC | 5 | |||
| UGcuuCC | 1 | |||
| aGcuuCC | 1 | |||
| SA11-4F | MA104 | 10 | UGUGACC | 17 |
aVirus was serially passaged the indicated times using 1:3 dilutions of infected cell lysate to fresh media as inoculum.
bRRV prepared in FRhL2 cells (see Table III).
In contrast to the extensive mutation of the 3′CS that occurred upon serial passage of the virus at high MOI, the g5(+) RNAs of RRV recovered after five cycles of serial passage at high dilution (1:4200) in MA104 cells (initial MOI <0.1) terminated almost exclusively with the 3′CS (29 3′CS/32 total ends) (Table III). The differences in results obtained upon serial passage of RRV at low and high dilutions in MA104 cells indicates that MOI is a factor affecting the extent of change to the g5(+) 3′-end. Notably, at lower MOI where complete infection of the MA104 culture requires cell-to-cell spread, the g5(+) RNA retains the 3′CS. At higher MOI, where all cells in the culture are initially infected, negating the need for cell-to-cell spread, the g5(+) 3′-end undergoes rapid and extensive change.
Genetic instability was also seen for the g5(+) 3′-end of DS1xRRV upon serial passage at low dilution (initial MOI >10) in MA104 cells. Specifically, 10 rounds of such passage yielded virus with g5(+) RNAs that ended predominantly with other than the 3′CS (10 3′CS/46 total ends) (Table IV). However, similar high MOI serial passage of SA11-4F in MA104 cells did not result in changes to its g5(+) 3′ sequence (Table IV). Hence, the potential for hypervariability of the g5(+) 3′-end is a characteristic of some but not all strains of rotavirus, and only some genes of RRV are required for the hypervariable phenotype.
Impact of cell line on 3′-sequence variation
The impact of cell line on mutation of the g5(+) 3′CS was further examined by serial passage of FRhL2-derived RRV at low dilution in primary or secondary African green monkey AGMK cells, human ileocecal adenocarcinoma HCT-8, and African green monkey Vero cells. A number of these (i.e., AGMK, FRhL2 and Vero) have been employed in the production of live-attenuated human rotavirus vaccines (Midthun and Kapikian, 1996; Bernstein et al, 1998). The analysis showed that the g5(+) RNAs of RRV passaged 50 times in AGMK cells ended not with the 3′CS, but predominantly with the 3′-sequence UGcugCC (Table IV). This end differs by only a single pyrimidine substitution at the −5 position from the predominant 3′-end detected upon passage of RRV 50 times in MA104 cells (UGcugCC versus UGUugCC), suggesting similarities in mutation rates, types of mutations, and selective pressures for the 3′-end of the RRV g5 RNA in both cell types. This suggestion is further supported by the observation that the second most common g5(+) 3′-ends detected in the 50-pass RRV pools from MA104 and AGMK cells were identical: UGUGAaCC (Table IV).
Analysis of FRhL2-derived RRV passed 10 times in Vero cells at low dilution showed that this cell line, like the AGMK and MA104 lines, promoted deviation of the g5 3′CS (Table IV). In contrast, the g5 RNAs of RRV passaged even 15 times in the HCT-8 cells retained the prototypic 3′CS, a result reminiscent of passage of RRV in FRhL2 cells (Table III), which caused little or no deviation of the 3′CS. Importantly, AGMK, MA104, and Vero cells support the high titer growth of RRV, yielding infected cell lysates that are typically ⩾108 PFU/ml. In contrast, RRV titers achieved in FRhL2 cells are 1–2 logs lower. Likewise, HCT-8 cells are less efficient in supporting RRV growth, and, in fact, the virus is lost upon multiple rounds of passage in this cell line. Thus, hypervariability of the g5(+) 3′-end of RRV is a characteristic that can be correlated with serial passage of the virus in cell lines supporting high titer growth. Owing to their ability to support high titer growth, serial passage of RRV in AGMK, MA104, and Vero cells even at a the same low dilution (1:3) used in serial passage of the virus in FRhL2 and HCT-8 cells means that the AGMK, MA104, and Vero cells are likely infected at much higher MOI at each cycle than the FRhL2 and HCT-8 cells. As a consequence, complete infection of the FRhL2 and HCT-8 cell cultures is likely to be more dependent upon the efficient cell-to-cell spread of the serially passaged virus than is required for complete infection of the AGMK, MA104, and Vero cell cultures.
3′-Sequence of g5 RNAs of RRV vaccine strains
Rotashield, the RRV-based tetravalent vaccine, was found to be highly efficacious in protecting against the four major rotavirus serotypes (G1–G4) that cause severe diarrhea in humans (Kapikian et al, 1996). The G1, G2, and G4 components of the vaccine were reassortant viruses containing a single gene (VP7) from human rotaviruses and 10 genes including g5 from RRV. RRV itself served as the G3 component (Kapikian et al, 1985; Midthun and Kapikian, 1996). VP7 is an outer capsid protein of rotavirus which stimulates protection in vaccines by inducing anti-VP7-neutralizing antibodies.
The virus components of the vaccine were prepared by passage six to seven times in AGMK cells, followed by passage seven (RRV), 11 (DS1xRRV), 13 (ST3xRRV), or 16 (DxRRV) times in FRhL2 cells (Midthun and Kapikian, 1996). To gain further insight into the impact of the FRhL2 cell line on RRV g5(+) 3′CS variation, we analyzed the g5(+) 3′-ends of viruses contained in the vaccine. The analysis showed that, despite passage of the DxRRV, DS1xRRV, and RRV vaccine components up to 16 times in FRhL2 cells, these viruses contained few if any RRV g5(+) RNAs that lacked the 3′CS (Table V). Although significantly greater numbers of g5(+) RNAs lacking the 3′CS were present for the ST3xRRV component, the majority of the RNAs possessed the 3′CS. Overall, these results suggested that serial passage of the vaccine components in FRhL2 cells favored retention of the RRV g5(+) 3′CS.
Table 5.
3′-Sequences of g5(+) RNAs of rotavirus vaccine strains propagated in FRhL2 cells
| RRV-based vaccine component (passage cycles in FRhL2 cells) | |||||||
|---|---|---|---|---|---|---|---|
| DxRRV-G1 | (16) | DS1xRRV-G2 | (11) | RRV-G3 | (7) | ST3xRRV-G4 | (13) |
| UGUGACC | 22 | UGUGACC | 21 | UGUGACC | 23 | UGUGACC | 14 |
| UGUuuCC | 1 | UGUu-CC | 1 | UGUauCC | 4 | ||
| UGacuCC | 1 | UGUuauCC | 1 | UGUu-CC | 3 | ||
| UGUuuCC | 1 | ||||||
| UGUGAuCC | 1 | ||||||
| UK-based vaccine component | |||||||
| UK-G3 |
(7) |
|
|
|
|
|
|
| UGUGACC | 48 | ||||||
3′-Motif necessary for genome replication
The results presented above indicate that RRV remains viable even upon significant deviation of the g5(+) 3′CS. Considered en toto (see Tables I, II, III, IV and V), the data reveal that any g5(+) RNA can function as a template for dsRNA synthesis in vivo as long as it ends with the motif, UGN0–5CC (N=any nucleotide) (Table VI). Although there are several other rotaviruses with g5(+) 3′-ends that deviate from the 3′CS (e.g., K9, KU; Okada et al, 1999), their atypical g5(+) 3′-ends do not exhibit the extreme sequence hypervariability detected for RRV (Table VI).
Table 6.
Common 3′-motif for group A rotavirus (+) RNAs
| 3′-consensus sequence for RRV gene 5 | UGN0–5CC |
| 3′-ends of other group A viruses: | |
| SA11 gene 2 (VP2) | UaUGACC |
| K9 gene 5 (NSP1) | UGUGAaCC |
| KU gene 5 (NSP1) | UGUGAaCC |
| SA11 gene 5 (NSP1) | UGUGAaCC |
| SA11 gene 7 (NSP3) | UGUGgCC |
| Common 3′-motif | URN0–5CC |
| Common 3′-motif excluding gene 5 | URUGRCC |
R=ATP, GTP; N=ATP, CTP, GTP, UTP.
Deviations in the 3′CS have also been noted for rotavirus genes, which, unlike g5, encode proteins that are essential for virus replication (Table VI). In particular, we confirmed by RACE the earlier finding that SA11 g2(+) (Mitchell and Both, 1990a) and g7(+) (Both et al, 1984) RNAs have atypical 3′-ends, resulting from the purine (R) substitutions G(−6) → A and A(−3) → G, respectively. The G(−6) → A deviation in the SA11 g2(+) 3′-end corresponds to one of the few residues of the 3′-end of the RRV g5(+) RNA noted for its absolute degree of conservation (UGN0–5CC). Though usually a G, the presence of the A at the −6 position of SA11 g2(+) RNA suggests that any purine at this position is sufficient to satisfy the requirements for virus viability (Table VI). The A(−3) → G deviation in the SA11 g7(+) 3′CS falls within the NSP3-recognition signal (U)GACC (Poncet et al, 1994) and, hence, would presumably interfere with the ability of NSP3 to enhance the translation efficiency of the g7(+) RNA. The G(−6) → A and A(−3) → G variations in the SA11 g2 and g7(+) RNAs have yet to be found in the g2 and g7 RNAs of other strains of rotavirus. Collectively, the available sequence data for the 3′-ends of all group A rotavirus (+) strand RNAs indicate that the motif required for virus viability is URN0–5CC (Table VI). Excluding the data for 3′-ends of g5 RNAs, the common 3′-motif for the rotavirus genes becomes URUGRCC, a sequence which shows much more similarity to the 3′CS (UGUGACC).
Effect of 3′-sequence variations on RNA replication
Previous analyses of rotavirus (+) RNAs in open-core replication assays have shown that changes to the 3′CS can alter the ability of the RNAs to serve as templates for dsRNA synthesis (Patton et al, 1996; Chen et al, 2001). To understand how the changes identified at the 3′-end of the RRV g5(+) RNAs affected their template activity, such RRV g5(+) RNAs were prepared by T7 transcription (Figure 2A) and incubated with open cores. Electrophoretic analysis of the assay products showed that all the RNAs functioned as templates for dsRNA synthesis (Figure 2B). Quantitation of the dsRNA product indicated that, in most cases, changes to the 3′CS caused the RRV g5(+) RNAs to support levels of replication greater than g5(+) RNAs with the 3′CS (Figure 2C). This included the g5(+) RNAs of the triple-plaque-purified isolates RRVg5-u, -uu, -auu, and -cuu, which were replicated by open cores with roughly twice the efficiency of g5(+) RNAs with the 3′CS. The fact that most mutations to the 3′CS caused the g5(+) RNA to replicate with greater efficiency suggests that the 3′CS has suboptimal activity in promoting (−) strand synthesis.
Figure 2.
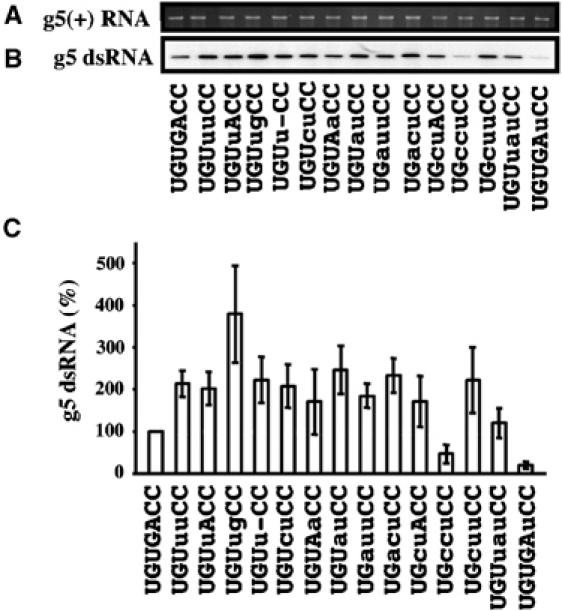
Impact of 3′-sequence variation on replication of RRV g5(+) RNAs. g5(+) RNAs, prepared by T7 transcription, were analyzed for quality by electrophoresis (A). The RNAs were assayed for template activity using DS1xRRV open cores. 32P-labeled dsRNA products were resolved by SDS–PAGE, detected by autoradiography (B), and quantified with a phosphorimager. (C) The averaged values of three independent experiments normalized to 100% for the 3′CS (UGUGACC), and standard errors are shown. Mutations in the 3′CS are indicated in lower case letters.
Of all the g5(+) RNAs evaluated, those containing the 3′-sequence, UGUugCC, were replicated with the greatest efficiency in vitro (Figure 2). As noted above, this sequence is remarkable because of its similarity to the transcription-promoter sequence present at the 3′-end of all rotavirus (−) strand RNAs (Figure 1). Furthermore, this sequence is interesting because nearly all g5(+) RNAs end with this sequence upon 50 cycles of serial passage of RRV in MA104 cells (Table IV). In combination, these observations suggest that, during serial passage of RRV, a strong selective pressure is exerted on the g5(+) 3′CS to undergo mutation to a sequence with greater promoter activity for (−) strand synthesis.
Although most of the g5(+) RNAs were replicated more efficiently in vitro upon mutation of the 3′CS, there were exceptions. In particular, RNAs ending with UGUGAuCC or UGccuCC were found to replicate less efficiently than those ending with the 3′CS (Figure 2). The basis for the occurrence of these weak promoter sequences is not clear, but may simply reflect the randomness associated with the introduction of base substitutions at the 3′-end of the RRV g5(+) RNA that, occasionally, may generate sequences that have decreased promoter activity. The fact that neither of these promoters were detected on RRV g5(+) RNAs following 50 rounds of serial passage at high MOI in highly permissive cells indicates that these 3′-ends were not evolutionarily favored. This would be consistent with the poor behavior of these RNAs as templates for replication.
Similar results were obtained when replication assays were performed with open cores from SA11-4F instead of DxRRV (data not shown). Thus, the source of the RdRP appears not to be a factor affecting the template activities of the RRV g5(+) RNAs.
Effect of 3′-sequence variations on NSP1 expression
To examine how changes in the NSP3-recognition signal at the 3′-ends of RRV g5(+) RNAs affected the expression of the g5 product NSP1, MA104 cells were infected with the plaque-purified viruses RRVg5-cs, -auu, -u, -uu, -cuu, and -ccuu. The cells were labeled with 35S-amino acids and then analyzed by polyacrylamide gels containing sodium dodecyl sulfate (SDS–PAGE) and autoradiography (Figure 3A). The analysis revealed no obvious differences in the overall expression of viral proteins, a result consistent with the observation that the viruses grew to similar titers. Western blot assay was used to examine the levels of NSP1 and NSP2 in the infected-cell lysates (Figure 3B). Based on a comparison of NSP1:NSP2 chemiluminescence signals, the amount of NSP1 produced in cells infected with RRVg5-cs was at least four-fold higher than that produced in cells infected with any of the RRV variants. Thus, the loss of the prototypic 3′CS from the g5(+) RNAs of RRV was correlated with a decrease in NSP1 expression.
Figure 3.
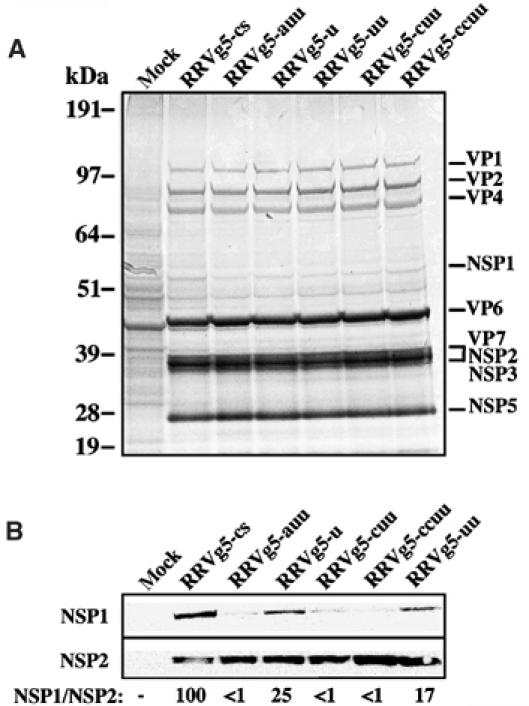
Expression of NSP1 by RRV variants. (A) Lysates prepared from MA104 mock- or RRV-infected MA104 cells, maintained in 35S-amino acids, were analyzed by electrophoresis and autoradiography. NSP1 detection is obscured by a co-migrating host protein. (B) NSP1 and NSP2 in the lysates were detected by Western blot assay with NSP2–NSP1(RRV-C19) antisera. The chemiluminescence exposure shown for NSP2 is for a shorter time than that for NSP1. Intensity values obtained with a phosphorimager were used to calculate the relative ratio of NSP1:NSP2, with the value determined for the RRVg5-cs-infected cells arbitrarily defined as 100. The g5 3′-sequences of the variants are as presented in Table II.
To address the possibility that the decrease in NSP1 expression resulted from a reduction in the binding activities of NSP3 for the mutant g5(+) 3′-ends of the RRV variants, His-tagged recombinant (r)NSP3 was expressed in bacteria and purified by Ni2+-affinity chromatography (Patton et al, 2001). EMSAs were then performed using rNSP3 and 32P-labeled RNA probes representing wild-type and mutant g5(+) RNAs. The analysis showed that rNSP3 interacted with the 3′CS probe to form a ribonucleoprotein complex (major RNP) (Figure 4). Little, if any, of this complex was formed when rNSP3 was incubated with probes representing the mutant g5(+) 3′-ends of the variant viruses. Instead, the probes of the atypical g5(+) 3′-ends interacted with rNSP3 to form a faster migrating RNP complex (minor RNP). The major RNP complex is formed by interaction of the NSP3 dimer with the 3′CS, a complex in which one protein monomer binds to the 3′CC and the other monomer binds to the immediately upstream GA (−4, −3) (Deo et al, 2002). The minor RNP is likely the result of the binding of only one monomer to the g5(+) 3′-end, an outcome that is expected if residues at the −3 or −4 positions are mutated. The results of the EMSA are consistent with earlier studies which showed that changes to the 3′CS interfere with NSP3 binding and decrease the translation efficiency of the (+) RNA (Vende et al, 2000; Patton et al, 2001; Deo et al, 2002). In combination, these data provide compelling evidence that mutations to the RRV g5(+) 3′CS can give rise to viruses that differ in their ability to express NSP1, and, as a consequence, may differ in their ability to modulate innate antiviral responses.
Figure 4.
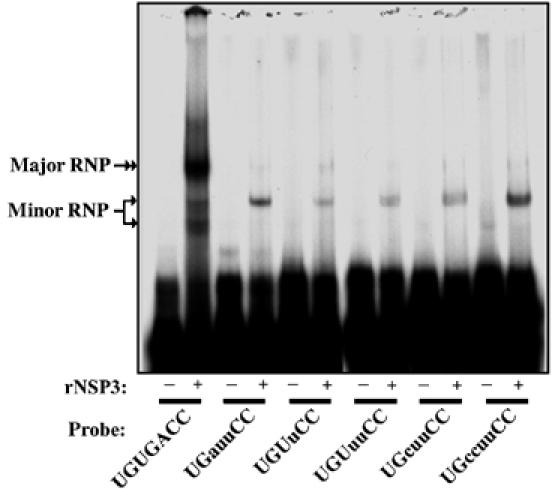
Effect of variations at the 3′-end of RRV g5(+) RNAs on the RNA-binding activity of NSP3. RNPs, formed by incubating purified rNSP3 with 32P-labeled g5-specific riboprobes ending with the 3′CS or variant sequences, were resolved by electrophoresis under nondenaturing conditions and detected by autoradiography.
Discussion
The lack of a reverse genetics system has made it impossible to generate the appropriate mutant rotaviruses required to evaluate the importance of the individual residues of the 3′CS on the replication and translation of viral (+) RNAs in the infected cell. In the absence of such a system, the discovery that the RRV g5(+) 3′CS can undergo extensive change as a function of host cell type and passage conditions has been particularly useful in providing insight into the role of the 3′CS. Notably, our analysis of the g5(+) 3′-ends of RRV passed at high MOI in cell lines permissive for high titer growth show that the 3′CS, or even minor variants of it, are not needed for viral (+) RNAs to replicate efficiently. Instead, any viral (+) RNA ending with the sequence URN0–5CC appears to be a suitable template for dsRNA synthesis. Thus, it is URN0–5CC that represents the minimal essential promoter for (−) strand synthesis. Our studies indicate that the high degree of conservation associated with the 3′CS stems largely from its role as a translation enhancer and not from its role as a (−) strand promoter.
The strict conservation of the CC (−2,−1) within the minimal essential promoter is expected, given that these residues are necessary for the interaction of viral (+) RNA with the RdRP to form an initiation complex for (−) strand synthesis (Chen and Patton, 2000). Likewise, the conservation of the UR (−7,−6) suggests that these residues play a critical role in genome replication. Based on EMSA, the UR and upstream regions of the viral (+) RNA form recognition signals for the RdRP (Tortorici et al, 2003). The fact that viral (+) RNAs with base substitutions, deletions, and/or insertions in the UGA (−5>−3) portion of the 3′CS can be replicated in vivo indicates that residues between the UR and CC do not have a critical role in genome replication and that the UR and CC can function independently from one another, at least with respect to distance.
During rotavirus replication, the RdRP is believed to bind initially to the viral (+) RNA via recognition signals formed by the UR (−7,−6) and upstream sequences. This is followed by repositioning of the RdRP on the (+) strand template such that the 3′CC becomes situated at the site governing (−) strand initiation. This complex, in the presence of GTP, divalent cations, and the core shell protein VP2, gives rise to a salt-stable (−) strand initiation complex (Tortorici et al, 2003). The switch from (−) strand initiation to elongation is accompanied by promoter escape and movement of the 3′CC from the catalytic site of the RdRP. The location of sequence variability at the RRV g5(+) 3′-end suggests that the RdRP has a tendency to add nontemplated nucleotides to the nascent CC as it undergoes transitions into its elongation mode. Since the g5(+) 3′CS of simian SA11 and bovine UK do not exhibit the same sequence hypervariability as that of RRV, the mechanism driving the hypervariability of the RRV g5(+) 3′-end appears not to be a universal phenomena among rotaviruses.
An intriguing question from our studies is why the g5(+) 3′CS changes so extensively when RRV is passed at high MOI (low dilution) in cell lines permissive for high titer growth, but not at low MOI or when passed in less permissive cell lines. The answer seems to be connected to whether the g5 product, NSP1, is necessary for virus replication under the particular propagation condition. When passed serially at high MOI in highly permissive cells (AGMK, Vero, MA104), all cells in the culture are infected at the time of virus adsorption, negating any need for the virus to spread subsequently from infected to uninfected cells. As a consequence, the role for NSP1 in the viral life cycle is decreased, since there is no demand for the protein to suppress establishment of the antiviral state in uninfected cells. From this, it can be predicted that the selective pressure exerted on the g5(+) 3′CS will shift favoring its evolution towards a stronger promoter for (−) strand synthesis and away from its function as a translation enhancer for NSP1 expression. Multiple findings support the idea that the RRV g5(+) 3′CS comes under such a change in selective pressure during high MOI serial passage: (i) the vast majority of mutations detected in the 3′CS caused RRV g5(+) RNAs to function more efficiently as templates for dsRNA in vitro; (ii) the g5(+) 3′-end of RRV serially passaged for 50 cycles in highly permissive cells evolved such that it was similar to the highly active promoter for viral transcription (+) strand synthesis; (iii) RRV variants with g5(+) RNAs lacking the 3′CS expressed less NSP1 than wild-type RRV and (iv) mutations introduced into the g5(+) 3′CS due to serial passage of RRV decreased the affinity of NSP3 for the translation enhancer.
Our analyses indicates that the RRV g5(+) 3′CS is relatively stable when the virus is passed serially at high dilution (low MOI) in cell lines that are permissive for high titer growth (e.g., MA104). Under these conditions, the virus must spread from infected cells to uninfected cells to achieve complete infection of the culture, and thus there exists a need for NSP1 to suppress establishment of the antiviral state. This need can be expected to provide a selective pressure that causes retention of the g5(+) 3′CS, since this element includes the translation enhancer for NSP1 expression.
RRV titers produced by infection of the less permissive cell lines (e.g., FRhL2 and HCT-8) are significantly less than those produced by infection of more permissive cell lines (e.g., AGMK, MA104, and Vero). Thus, when RRV is passaged serially at low dilution in the less permissive cells, the MOI at which the cultures are infected is lower than that when the virus is passaged serially at the same dilution in the more permissive cells. Hence, complete infection of less permissive cell cultures is more likely to require cell-to-cell spread of the virus even upon low-dilution passage, and therefore, more likely to require efficient expression of NSP1. This requirement would favor retention of the g5(+) 3′CS, a suggestion supported by the observation that this element is conserved upon serial passage of RRV at low dilution in FRhL2 and HCT-8 cells. However, other factors may also be connected to stability of the 3′CS in these cells. These include the fact that, since fewer dsRNA genome segments are made in less permissive cells, the opportunity for random mutation events to alter the g5(+) 3′CS is reduced. Also, since the replication cycle takes longer to complete in the FRhL2 cells, there may be a greater need for NSP1, through its targeted degradation of IRF3, to prevent IRF7 from inducing expression of antiviral factors (e.g., PKR, RNase L) in the infected cell (Hiscott, 1999). Finally, the possibility exists that due to variations either in the cytoplasmic levels or sequence of IRF3, more NSP1 is required in FRhL2 cells to prevent IRF3-dependent activation of IFN than in more permissive cells.
Given the role of NSP1 as an antagonist of the IFN-signaling pathway, our results show that the methods selected for propagating vaccine strains of rotavirus may alter the virulence of the progeny virus. Currently, FRhL2 and Vero cell lines are used to produce the pools of live-attenuated viruses that make up candidate rotavirus vaccines. In the absence of other factors, the ability of Vero cells to support rotavirus growth to titers that are higher than in FRhL2 cells makes it the obvious cost-effective choice for producing vaccines. On the other hand, FRhL2 cells seemingly provide a growth environment favoring genetic stability of some rotaviruses, a characteristic that may allow for better reproducibility in the production and behavior of vaccine lots. Although we detected variability in the 3′-sequences of RRV g5(+) RNAs, this was not observed for the UK bovine and SA11 g5(+) RNAs. As a consequence, this variability may be unique to RRV and therefore not of concern in preparing other types of rotavirus vaccines in Vero cells (e.g., UK bovine). However, given the propensity of Vero cells to promote mutation of the multifunctional RRV g5(+) 3′CS, it may be prudent to continue the current practice of preparing the RRV tetravalent vaccine in FRhL2 cells until such time that we have a fuller appreciation of the impact of these mutations on RRV viability, virulence, and immunogenicity in the vaccinee.
In summary, this study reinforces the concept that cell lines and propagation conditions should be considered as significant variables influencing the biological behavior of viruses. In the case of RRV, changes in cell line and propagation conditions led to changes in the selective pressures exerted on a multifunctional determinant affecting replication and expression of the g5(+) RNA. These changes in selective pressure revealed that the highly conserved nature of the determinant was the product of the balanced need for its sequence to function efficiently in two exclusive processes.
Materials and methods
Virus and cells
RRV strain MMU18006, isolated from the diarrheal stool of a rhesus monkey, was passaged twice in cynomologous monkey kidney cells (CMK-2), seven times in primary or secondary African green monkey kidney cells (AGMK) cells including triple-plaque purification, and six times in (FRhL2) cells, yielding virus pool RRV-1 (Kapikian et al, 1985). RRVg5-cs was isolated from RRV-1 by triple-plaque purification in MA104 cells. The variants RRVg5-u, -uu, -auu, -cuu, and -ccuu were isolated by triple-plaque purification from RRV passaged 30 times in MA104 cells. RRV used in serial passage experiments was triple-plaque purified in MA104 cells from the RRV-1 pool, amplified once in AGMK cells, and then serially passaged for the indicated times in MA104, AGMK, human ileocecal adenocarcinoma HCT-8, or African green monkey kidney Vero cells.
The RRV tetravalent vaccine components DxRRV, DS-1xRRV, and ST3xRRV were made by co-infection of AGMK cells with RRV and the human rotaviruses D, DS1, or ST3, respectively. RRV MMU18006 was the fourth component of the vaccine (Midthun and Kapikian, 1996). The appropriate reassortants were triple-plaque purified in AGMK cells and passaged multiple times in AGMK and FRhL2 cells: DxRRV, AGMK-6 (passed six times in AGMK cells), FRhL2-16; DS1xRRV, AGMK-6, FRhL2-11; RRV-1 (as noted above); ST3xRRV, AGMK-7, FRhL2-13.
UK (Compton) bovine rotavirus was isolated from the diarrheal stool of a calf, serially passed eight times in primary calf kidney cells, four times in primary embryonic kidney cells, and four times in AGMK cells. Clone 22 was isolated from this pool by triple-plaque purification in AGMK cells, then serially passaged five times in primary calf kidney cells and subsequently amplified in the same cell type, yielding the virus pool Lot BR-3. The Lot BR-4 pool was generated from the Lot BR-3 pool by serial passage seven times in FRhL2 cells (Wyatt et al, 1985).
RACE analysis
Genomic dsRNAs were prepared by phenol–chloroform extraction from virus recovered from infected cell lysates. Terminal sequences of denatured dsRNAs were determined with a 5′-RACE kit (InVitrogen) or an RLM-RACE kit (Ambion). For 5′-RACE, cDNAs of the RRV g5(−) 5′-end were made by annealing the first-strand primer 5′-GAGACATGGATTGGAAGATGG-3′ (g5(+) 1032–1052) to denatured dsRNAs and incubating the hybrids with Superscript II reverse transcriptase (Patton et al, 2001). After RNase treatment, cDNAs were recovered with GlassMAX cartridges and dA-tailed using terminal deoxynucleotide transferase. The tailed cDNAs were amplified by polymerase chain reaction (PCR) in mixtures containing Taq DNA polymerase, oligo dT, and the nested g5(+) primer, 5′-ATTGGATCCAATCTTGCTATAAAAGTAATG-3′ (g5(+) 1086–1115). The PCR products were ligated into the pCR2.1 vector (InVitrogen) and transformed into DH5α bacteria (InVitrogen). To obtain cDNAs of the RRV g5(+) 5′-end, the first strand primer, 5′-TTTACTTGATCTAACAATAG-3′ (g5(−) 608–586), nested primer, 5′-TATGCATGGTGAGGATCCTTCATA-3′ (g5(−) 540–517), and oligo dC primer, 5′-GGGCCCGAGCTCCGCCGGCGGCCIICCCIICCCIIC-3′ were used instead, and the cDNAs were tailed with dGTP. The cDNAs of the g5(+) 5′-end were cloned into the SP72 vector.
For RLM-RACE, cDNAs of the RRV g5(−) 5′-end were made by ligating the 5′-RACE adaptor 5′-GCUGAUGGCGAUGAAUGAACACUGCUUUGCUGGCU UUGAUGAAA-3′ to the 5′-end of decapped denatured RRV dsRNAs. Afterwards, first-strand cDNAs were produced by incubation of the RNA with first-strand primer g5(+) 608–586 (see above) and M-MuLV reverse transcriptase. The cDNAs were amplified by PCR in mixtures containing the nested primer g5(+) 540–517 and a primer complementary to the adaptor, 5′-CAGTGTTCATTCATCGCCATCAGC-3′. The cDNA products were cloned into pCR2.1 and sequenced with an Applied Biosystems 310 automated sequencer.
g5 plasmids and sequences
cDNAs of the (+) strand of RRV g5 dsRNAs were prepared with the Titan One Tube RT–PCR System (Roche) and the oligonucleotide primers 5′-AGTCTTGTGTTAGCCATGGC-3′ (g5(+) 16–35) and 5′-AATTCAATCTTAGTCGTCATC-3′ (g5(−) 1525–1504). The cDNAs were ligated into pCR2.1 and two independently derived cDNAs were analyzed to determine g5 sequences. g5 sequences of the virus isolates RRVg5-cs, AY117048; RRVg5-auu, AY117049; RRVg5-ccuu, AY117050; RRVg5-cuu, AY117051; RRVg5-u, AY117052; and RRVg5-uu, AY117053 were submitted to GenBank.
The plasmid SP65/RRVg5-cs was made similarly, except that the forward and reverse primers were 5′-TAATACGACTCACTATAGGCTTTTTTTATGAAAAG TCTTGTGTT-3′ (T7-RRVg5P) and 5′-GGTCACATTTTTTGCCAGCTAGGCGCTACT-3′ (RRVg5M-wt), respectively.
In vitro replication assays
PCR was used to link T7 promoters to RRVg5 cDNAs with variable 3′-sequences (Patton et al, 1996). The amplification reactions contained Platinum Pfx DNA polymerase (InVitrogen), SP65/RRVg5-cs, the T7g5/5′ forward primer, 5′-TAATACGACTCACTATAGGCTTTTTTTATGAAAAG TCTTGTGTT-3′, and the g5/3′cs reverse primer 5′-GGTCACATTTTTTGCCAGCTAGGCGCTACT-3′, or the appropriate derivative of it. The PCR products were transcribed with an Ambion MEGAscript T7 transcription kit.
In vitro replication assays were carried out using DS1xRRV open cores as described earlier (Chen et al, 2001). The 32P-dsRNA products were analyzed by electrophoresis on 12% SDS–PAGE.
Production of NSP2–NSP1(RRV-C19) antisera
The bacterial expression vector pQE60g8-RRVNSP1-C19 produces a protein in which the C-terminal 19 amino acids of RRV NSP1 are linked to SA11 NSP2 protein via a GPGP hinge and a 6X-His affinity tag. The pQE60g8-RRVNSP1-C19 was made by PCR-mediated modification of pQE60g8 (Taraporewala et al, 1999), a vector that encodes NSP2 with a C-terminal 6X-His tag. The PCR reaction mixture included the forward primer: 5′-CTTCTGATCTCGGACTCTGAAGATGACGATTAAGC TTAATTAGCTGAGCTTGGACTCCTG-3′ and the reverse primer: 5′ CTCATATTCTTCAGATAGTTTTCCTTCCGGGCCTG GTCCGTGATGGTGATGGTGATGAGATCTAACGCC-3′, pQE60g8, and a mixture of Taq DNA polymerase and Pwo DNA polymerase mixture (Roche). The amplified DNA was blunt-ended with T4 DNA polymerase, treated with T4 DNA polynucleotide kinase and self-ligated with T4 DNA ligase. Following transformation into Escherichia coli DH5α, positive clones were selected based on antibiotic resistance and sequencing.
NSP2–NSP1(RRV-C19) was expressed and purified under the same conditions as described for NSP2 (Taraporewala et al, 1999). Polyclonal anti-NSP2–NSP1(RRV-C19) was produced in guinea-pigs immunized initially with 0.2 mg of the protein in Freunds complete adjuvant and then at weeks 2, 4, and 5, each with 0.2 mg of the protein in Freunds incomplete adjuvant.
Preparation of infected cell extracts
MA104 cells were infected with trypsin-activated RRV at an MOI of 20 in serum-free medium and maintained, from 3 to 7 h p.i., in 80% met-cys free minimal essential medium containing 20 μCi/ml of 35S-Express (1.175 mCi/mmol; NEN). At 7 h p.i., the cells were lysed in buffer containing 3 mM Tris–HCl, pH 8.5, 0.5 mM MgCl2, 3 mM NaCl, 0.5% Triton X-100, and protease inhibitor cocktail (Roche). Proteins were analyzed by electrophoresis on NuPAGE 10% polyacrylamide gels (InVitrogen) and by autoradiography. NSP1 and NSP2 were detected by Western blot assay using guinea-pig anti-NSP2–NSP1(RRV-C19) antisera (1:100) and peroxidase-labeled goat anti-guinea-pig IgG (KPL) (1:10 000) and a SuperSignal West Pico chemiluminescence detection kit (Pierce).
EMSA
DNA templates for synthesis of RNA probes were generated by PCR. Amplification reactions contained the plasmid pSP72-(T3)RRVg5-3′72, Elongase (Invitrogen), the forward primer 5′-GGAAGGCAGTTTTTGCTGGCTAGGCGCTAC-3′, and an appropriate reverse primer complimentary to 3′-terminal 18 residues of RRV g5(+) RNA. PCR products were transcribed with an MEGASHORTscript T3 transcription kit (Ambion) in the presence of 32P-UTP (Patton et al, 2001). Reaction mixtures containing 3 pmol of 32P-labeled RNA probe, 3 pmol of rNSP3 (Patton et al, 2001), and 0.1 μg of luciferase RNA were incubated for 30 min at room temperature. rNSP3–probe complexes in the mixtures were resolved by electrophoresis on nondenaturing 8% polyacrylamide gels (Patton et al, 2001).
References
- Barro M, Patton JT (2004) Rotavirus NSP1 subverts innate immunity by preventing nuclear translocation and inducing proteasomal-mediated degradation of the transcription factor IRF-3 (in review)
- Bernstein DI, Smith VE, Sherwood JR, Schiff GM, Sander DS, DeFeudist D, Spriggs DR, Ward RL (1998) Safety and immunogenicity of live, attenuated human rotavirus vaccine 89–12. Vaccine 16: 381–387 [DOI] [PubMed] [Google Scholar]
- Both GW, Bellamy AR, Siegman LJ (1984) Nucleotide sequence of the dsRNA genomic segment 7 of simian 11 rotavirus. Nucleic Acids Res 12: 1621–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Barro M, Spencer E, Patton JT (2001) Features of the 3′-consensus sequence of rotavirus mRNAs critical to minus strand synthesis. Virology 281: 221–229 [DOI] [PubMed] [Google Scholar]
- Chen D, Patton JT (2000) De novo synthesis of minus strand RNA by the rotavirus RNA polymerase in a cell-free system involves a novel mechanism of initiation. RNA 6: 1455–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chizhikov V, Patton JT (2000) A four-nucleotide translation enhancer in the 3′-terminal consensus sequence of the nonpolyadenylated mRNAs of rotavirus. RNA 6: 814–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deo RC, Groft CM, Rajashankar KR, Burley SK (2002) Recognition of the rotavirus mRNA 3′ consensus by an asymmetric NSP3 homodimer. Cell 108: 71–81 [DOI] [PubMed] [Google Scholar]
- Desselberger U, McCrae MA (1994) The rotavirus genome. Curr Top Microbiol Immunol 185: 31–66 [DOI] [PubMed] [Google Scholar]
- Estes MK (2001) Rotaviruses and their replication. In Fields Virology, Knipe DM, Howley PM, Griffin DE, Martin MA, Lamb RA, Roizman B, Straus SE (eds), 4th edn, pp 1747–1785. Philadelphia, PA: Lippincott Williams & Wilkins [Google Scholar]
- Graff JW, Mitzel DN, Weisend CM, Flenniken ML, Hardy ME (2002) Interferon regulatory factor 3 is a cellular partner of rotavirus NSP1. J Virol 76: 9545–9550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groft CM, Burley SK (2002) Recognition of eIF4G by rotavirus NSP3 reveals a basis for mRNA circularization. Mol Cell 9: 1273–1283 [DOI] [PubMed] [Google Scholar]
- Hiscott J (1999) Triggering the interferon response: the role of IRF-3 transcription factor. J Interferon Cytokine Res 19: 1–13 [DOI] [PubMed] [Google Scholar]
- Kahvejian A, Roy G, Sonenberg N (2001) The mRNA closed-loop model: the function of PABP and PABP-interacting proteins in mRNA translation. Cold Spring Harb Symp Quant Biol 66: 293–300 [DOI] [PubMed] [Google Scholar]
- Kapikian AZ, Hoshino Y, Chanock RM, Perez-Schael I (1996) Efficacy of a quadrivalent rhesus rotavirus-based human rotavirus vaccine aimed at preventing severe rotavirus diarrhea in infants and young children. J Infect Dis 174 (Suppl 1): S65–S72 [DOI] [PubMed] [Google Scholar]
- Kapikian AZ, Midthun K, Hoshino Y, Flores J, Wyatt RG, Glass RI, Askaa J, Nakagomi O, Nakagomi T, Chanock RM, Levine MM, Clements ML, Dolin R, Wright PF, Belshe RB, Anderson EL, Potash L (1985), In Vaccines 85. Modern Approaches to Vaccines: Molecular and Chemical Basis for Resistance to Viral, Bacterial, and Parasitic Diseases, Lerner L, Brown F, Chanock RM (eds), pp 357–367. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory [Google Scholar]
- Lawton JA, Estes MK, Prasad BV (1997) Three-dimensional visualization of mRNA release from actively transcribing rotavirus particles. Nat Struct Biol 4: 118–121 [DOI] [PubMed] [Google Scholar]
- Midthun K, Kapikian AZ (1996) Rotavirus vaccines: an overview. Clin Microbiol Rev 9: 423–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell DB, Both GW (1990a) Completion of the genomic sequence of the simian rotavirus SA11: nucleotide sequences of segments 1, 2 and 3. Virology 177: 324–331 [DOI] [PubMed] [Google Scholar]
- Mitchell DB, Both GW (1990b) Conservation of a potential metal binding motif despite extensive sequence diversity in the rotavirus nonstructural protein NS53. Virology 174: 618–621 [DOI] [PubMed] [Google Scholar]
- Okada J, Kobayashi N, Taniguchi K, Shiomi H (1999) Analysis on reassortment of rotavirus NSP1 genes lacking coding region for cysteine-rich zinc finger motif. Arch Virol 144: 1439–1449 [DOI] [PubMed] [Google Scholar]
- Parashar UD, Hummelman EG, Bresee JS, Miller MA, Glass RI (2003) Global illness and deaths caused by rotavirus disease in children. Emerg Infect Dis 9: 565–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton JT, Spencer E (2000) Genome replication and packaging of segmented double-stranded RNA viruses. Virology 277: 217–225 [DOI] [PubMed] [Google Scholar]
- Patton JT, Taraporewala Z, Chen D, Chizhikov V, Jones M, Elhelu A, Collins M, Kearney K, Wagner M, Hoshino Y, Gouvea V (2001) Effect of intragenic rearrangement and changes in the 3′ consensus sequence on NSP1 expression and rotavirus replication. J Virol 75: 2076–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton JT, Wentz M, Xiaobo J, Ramig RF (1996) cis-Acting signals that promote genome replication in rotavirus mRNA. J Virol 70: 3961–3971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piron M, Vende P, Cohen J, Poncet D (1998) Rotavirus RNA-binding protein NSP3 interacts with eIF4GI and evicts the poly(A) binding protein from eIF4F. EMBO J 17: 5811–5821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poncet D, Laurent S, Cohen J (1994) Four nucleotides are the minimal requirement for RNA recognition by rotavirus non-structural protein NSP3. EMBO J 13: 4165–4173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad BVV, Rothnagel R, Zeng CQ-Y, Jakana J, Lawton JA, Chui W, Estes MK (1996) Visualization of ordered genomic RNA and localization of transcriptional complexes in rotavirus. Nature 382: 471–473 [DOI] [PubMed] [Google Scholar]
- Prasad BVV, Wang GJ, Clerx JPM, Chiu W (1988) Three-dimensional structure of rotavirus. J Mol Biol 199: 269–275 [DOI] [PubMed] [Google Scholar]
- Taraporewala ZF, Chen D, Patton JT (1999) Multimers formed by the rotavirus nonstructural protein NSP2 bind to RNA and have nucleoside triphosphatase activity. J Virol 73: 9934–9943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortorici MA, Broering TJ, Nibert ML, Patton JT (2003) Template recognition and formation of initiation complexes by the replicase of a segmented double-stranded RNA virus. J Biol Chem 278: 32673–32682 [DOI] [PubMed] [Google Scholar]
- Vende P, Piron M, Castagne N, Poncet D (2000) Efficient translation of rotavirus mRNA requires simultaneous interaction of NSP3 with the eukaryotic translation initiation factor eIF4G and the mRNA 3′ end. J Virol 74: 7064–7071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt RG, Kapikian AZ, Hoshino Y, Flores J, Midthun K, Greenberg HB, Glass RI, Askaa J, Levine M, Black RE, Clements ML, Potash L, London WT (1985) Development of rotavirus vaccines. In Control and Eradication of Infectious Diseases. An International Symposium, PAHO Copublication Series No. 1, pp 17–28. Washington, DC: Pan American Health Organization [Google Scholar]


