Abstract
GGA proteins coordinate the intracellular trafficking of clathrin-coated vesicles through their interaction with several other proteins. The GAT domain of GGA proteins interacts with ARF, ubiquitin, and Rabaptin5. The GGA–Rabaptin5 interaction is believed to function in the fusion of trans-Golgi-derived vesicles to endosomes. We determined the crystal structure of a human GGA1 GAT domain fragment in complex with the Rabaptin5 GAT-binding domain. In this structure, the Rabaptin5 domain is a 90-residue-long helix. At the N-terminal end, it forms a parallel coiled-coil homodimer, which binds one GAT domain of GGA1. In the C-terminal region, it further assembles into a four-helix bundle tetramer. The Rabaptin5-binding motif of the GGA1 GAT domain consists of a three-helix bundle. Thus, the binding between Rabaptin5 and GGA1 GAT domain is based on a helix bundle–helix bundle interaction. The current structural observation is consistent with previously reported mutagenesis data, and its biological relevance is further confirmed by new mutagenesis studies and affinity analysis. The four-helix bundle structure of Rabaptin5 suggests a functional role in tethering organelles.
Keywords: crystal structure, GAT domain, GGA proteins, intracellular trafficking, Rabaptin5
Introduction
Clathrin-coated vesicles (CCVs) represent a major type of vesicular carrier that traffics between trans-Golgi network (TGN) and endosomes, as in the case of lysosomal enzyme transport, and from the plasma membrane to early endosomes, as in the case of receptor-mediated endocytosis (Kirchhausen, 2000). The clathrin lattice interacts with membrane or membrane-bound proteins through adaptor proteins. The GGA family of adaptor proteins (Golgi-localized, γ-ear-containing, and ARF-binding) plays important roles in the CCV trafficking between TGN and endosome compartments (Boman et al, 2000; Dell'Angelica et al, 2000; Hirst et al, 2000; Poussu et al, 2000) and probably also in endocytosis (Puertollano and Bonifacino, 2004), including cargo selection and recruitment of clathrin and accessory proteins to the budding site. GGAs are monomeric proteins present in a variety of organisms. There are three isoforms, GGA1–3, in higher eukaryotic cells. The cellular function of GGA proteins in vesicular trafficking is manifested largely through the interactions of its four domains with other protein partners. In the order of N- to C-terminus, the four domains are VHS (VPS27, Hrs, and STAM homologous), GAT (GGA and TOM1 homologous), hinge region, and GAE (γ-adaptin ear homologous) (Dell'Angelica et al, 2000; Hirst et al, 2000; Takatsu et al, 2000, 2002; Puertollano et al, 2001). The GAT domain has been shown to interact with the membrane-bound form of ARF, a GTPase molecular switch initiating CCV assembly (Moss and Vaughan, 1998). The interaction between GAT domain and ARF (Dell'Angelica et al, 2000; Boman, 2001; Takatsu et al, 2002) defines the vesicle-assembling site on the membrane and facilitates subsequent interactions with other proteins in the assembly complex (Dell'Angelica et al, 2000). Recently, the GAT domain is shown to interact with Rabaptin5 and ubiquitin using overlapped binding sites (Zhai et al, 2003; Mattera et al, 2004; Shiba et al, 2004). The GAE domain has an immunoglobulin fold and interacts with an array of accessory ligands, including an extended peptide strand from Rabaptin5 (Hirst et al, 2000; Takatsu et al, 2000; Miller et al, 2003). Since the GAT domain also interacts with Rabaptin5, the interaction between GGA and Rabaptin5 is divalent in nature. The structure of GGA1 GAT domain comprises four helices in an overall elongated shape, in which the longest helix participates in two structural motifs: an N-terminal helix–loop–helix motif and a C-terminal three-helix bundle (Collins et al, 2003; Shiba et al, 2003; Suer et al, 2003; Zhu et al, 2003). The N-terminal motif contains a hydrophobic surface patch, which is found to interact directly with ARF in a crystal structure (Shiba et al, 2003). The C-terminal three-helix bundle is involved in the binding with Rabaptin5 (Zhai et al, 2003; Mattera et al, 2004) and ubiquitin (Scott et al, 2004; Shiba et al, 2004).
Rabaptin5, a 99-kDa protein, was initially identified as an effector of Rab5 in the fusion of early endosomes (Stenmark et al, 1995). The involvement of Rabaptin5 in this process is an essential and rate-limiting step. While located mainly in the cytosol, Rabaptin5 is also present at low levels on the cell membrane (Gournier et al, 1998). Secondary structure prediction from the amino-acid sequence suggests that Rabaptin5 contains multiple coiled-coil regions, each of which is capable of forming homodimers in vitro (Stenmark et al, 1995). It has been shown that one of the predicted coiled-coil regions in human Rabaptin5, consisting of residues 551–661, binds the GAT domain of GGA1 and GGA2 (Mattera et al, 2003). Besides GGAs, Rabaptin5 divalently interacts with the γ-adaptin subunit of AP-1 complex (Shiba et al, 2002; Mattera et al, 2003), which is a major component of the CCV coating complex at the TGN. Rabaptin5 is also involved in the endosome-recycling pathway through its interaction with Rab4 (Vitale et al, 1998). Members of the Rabaptin5 family from different species are highly homologous in their primary sequences. In human cells, there exist a number of Rabaptin5 splicing variants (Korobko et al, 2002; Deneka et al, 2003), but their functional roles are yet to be determined. Among these splicing variants, there is no deletion/insertion observed around the GAT-binding region.
While it is known that a short peptide motif in Rabaptin5 is responsible for interacting with the GAE domain of GGAs, the structural motif of Rabaptin5 mediating the GAT binding remains to be identified. Here, we present the crystal structure of the three-helix bundle motif of human GGA1 GAT domain in complex with the GAT-binding domain of Rabaptin5. The GAT–Rabaptin5 interaction has a helix bundle–helix bundle-binding mode, in which one GAT domain binds to a pseudosymmetric Rabaptin5 dimer. Furthermore, a four-helix bundle formation is observed for the Rabaptin5551–661 fragment in both crystal and solution, suggesting a direct functional role of Rabaptin5 in tethering organelles.
Results
Overall structure of the GAT–Rabaptin5 complex
To determine the complex structure of GGA1 GAT and Rabaptin5 fragments, we expressed and purified recombinant proteins of GGA1210–302 (i.e. residues 210–302 of GGA1, the three-helix bundle motif of the GAT domain) and Rabaptin5551–661 (Figure 1A) from Escherichia coli and cocrystallized them. The crystal form belongs to P3221 space group with one GGA1210–302 and two Rabaptin5551–661 molecules per asymmetric unit (asu) (Table I, Figure 1B). The molecular surface of this GAT–Rabaptin52 complex is dominated by negatively charged electrostatic potential (Figure 1C), consistent with the calculated pI values of 4.7 and 5.2 for the GGA1210–302 and Rabaptin5551–661 recombinant proteins, respectively. The structure of the three-helix bundle (i.e. helices α2–α4) of GAT is essentially the same as that from the full-length GAT (Protein Data Bank (PDB) file 1OXZ) (Zhu et al, 2003), with a Cα-atom root mean square deviation (r.m.s.d.) of 0.92 Å between them. The Rabaptin5-binding site has an even better agreement between the two structures (a 0.47 Å Cα-r.m.s.d. for residues 257–270 and 274–288). A few regions of high mobility in GGA1210–302, including the connecting loop between helices α2 and α3 and a couple of residues from both termini, are omitted from the final refined model. Similarly omitted are mobile residues in Rabaptin5551–661 that are either at the N-terminus or C-terminal to Glu640R (the superscript R stands for Rabaptin5). Each of the two Rabaptin5551–661 chains contains one long helix of 24 turns (∼90 residues) with an overall length of 135 Å. The two Rabaptin5 chains in one asu form a parallel coiled-coil homodimer at their N-terminal region, which binds to helices α3 and α4 of the three-helix bundle of GGA1210–302. The helix–helix distance (9.3 Å) and crossing angle (20°) in this Rabaptin5 N-terminal region are typical for a coiled-coil dimer (Lupas, 1996). Furthermore, this homodimer region bends by about 10° from the dyad axis of the remaining homodimer (Figure 1B), where the helix–helix distance and crossing angle become 13.5 Å and 28°, respectively (calculated from residues 611–620). The bending of the Rabaptin5 dimer in the GAT-binding region toward its ligand could be an artifact of crystallization, but it is more likely that the bending is induced by the intimate interaction with GAT. Thus, the conformations of two Rabaptin5551–661 chains in one asu (i.e. the green and blue chains in Figure 1B) are similar but not identical to each other. Their overall Cα-r.m.s.d. is 2.3 Å, as compared to that of 0.93 Å for the N-terminal coiled-coil homodimer (i.e. residues 555–576) alone and 0.75 Å for the rest of the long helix. Furthermore, we found unexpectedly in the crystal that two crystallographic symmetry-related Rabaptin5 dimers form a homotetramer using their C-terminal helical fragments, which is organized into a left-hand twisted four-helix bundle (see below).
Figure 1.
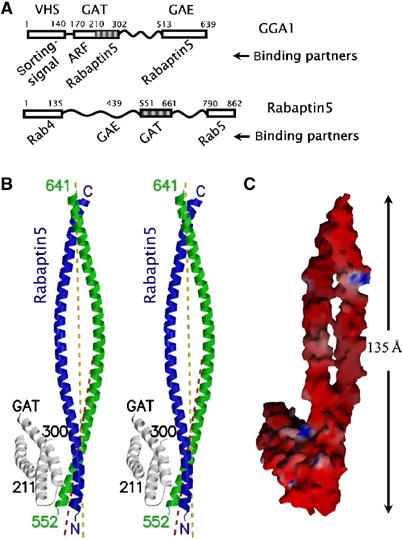
GAT–Rabaptin5 complex. (A) Schematic diagram of the constructs of GAT and Rabatin5 fragments. Known domains are shown as boxes with the residue numbers and names (if existing) marked on top. Relevant binding partners are listed below. Regions included in the crystal are shaded. (B) Stereo view of a ribbon diagram of the complex of GGA1 GAT three-helix bundle domain (white) and Rabaptin5551–661 (blue and green). The crystallographically visible peptide termini are labeled either as N/C or with residue numbers. The major local two-fold axis of the Rabaptin5551–661 homodimer is depicted as a yellow dash line, and the two-fold axis of the GAT-binding region is shown as a red dash line. (C) Molecular surface model of the complex. The color intensity corresponds to the electrostatic potential calculated with the program GRASP and its default parameters, from −8 kT e−1 (intense red) to +8 kT e−1 (intense blue). Figures 1B, 2A, 4A, and 6 were drawn with the programs MolScript and Raster3D. Figures 1C and 2B were drawn with the program GRASP.
Table 1.
Statistics of crystallographic data, phasing, and refinement
| Data collection | |||||
| Space group | P3221 | ||||
| Cell dimensions | a=b=155.2 Å, c=53.1 Å | ||||
| Se-Met | |||||
| Native | λ1 (remote) | λ2 (peak) | λ3 (inf.) | ||
| Wavelength used (Å) | 1.5418 | 0.95667 | 0.97938 | 0.97952 | |
| Resolution range (Å) | 50 (2.49)a–2.40 | 20 (2.90)–2.80 | 20 (2.90)–2.80 | 20 (2.90)–2.80 | |
| No. of measurements | 148 886 | 126 001 | 122 613 | 122 550 | |
| No. of unique reflections | 28 214 (2217) | 18 160 | 18 008 | 17 972 | |
| Completeness (%) | 97.2 (77.3) | 98.6 (99.5) | 98.0 (95.6) | 97.9 (95.6) | |
| I/σ(I) | 30.1 (2.3) | 19.2 (4.3) | 20.6 (4.7) | 20.6 (4.9) | |
| Rmerge b (%) | 5.4 (40.5) | 8.7 (39.8) | 8.3 (34.2) | 8.0 (33.1) | |
| Phase powerc and figure of merit (FOM) | Observed diffraction ratiosd | ||||
| Resolutions (Å) | 20(2.92)–2.79 | λ1 | λ2 | λ3 | |
| Phase power | λ1 | 0.085 | 0.082 | 0.105 | |
| λ1–λ1− | 0.91 (0.55) | λ2 | 0.077 | 0.096 | |
| λ1–λ2+ | 0.80 (0.58) | λ3 | 0.088 | ||
| λ1–λ2− | 1.32 (0.68) | ||||
| λ1–λ3+ | 1.04 (0.51) | ||||
| λ1–λ3− | 1.38 (0.69) | ||||
| FOM | 0.50 (0.28) | ||||
| Refinement | |||||
| Resolution (Å) | 55–2.4 | ||||
| Rworking e (%)/no. of reflections | 23.3/22 743 | ||||
| Rfree e (%)/no. of reflections | 28.3/910 | ||||
| No. of non-hydrogen atoms | 2246 | ||||
| R.m.s.d. from ideal values | |||||
| Bond length (Å) | 0.013 | ||||
| Bond angle (deg) | 1.4 | ||||
| Improper angle (deg) | 0.91 | ||||
| Dihedral angle (deg) | 16.9 | ||||
| Average B-factor (Å2) | 58.3 (45.9)f | ||||
| aValues in parentheses are for the highest resolution bin. | |||||
| bRmerge=∑h∑i∣Ii(h)–I(h)〉∣/∑h∑iIi(h), where Ii(h) is the ith measurement and 〈I(h)〉 is the mean of all measurements of I(h) for Miller indices h. | |||||
| cMAD phasing power is defined as (〈∣FD–FN∣2〉/∫ϕP(ϕ)(∣FN exp(iϕ)+ΔFH∣–FD)2 dϕ)1/2, where P(ϕ) is the experimental phase probability distribution, FN is the amplitude of the structure factor at the reference wavelength λ1, FD corresponds to the structure factor at wavelength λi (indicated by the superscript ‘+') or its Friedel mate (indicated by the superscript ‘−'), and ΔFH is the difference in heavy atom structure factors between the two wavelengths. Similarly, the phasing power of a derivative is defined between the derivative and native data. | |||||
| dValues are 〈(ΔF)2)〉1/2/〈F2〉1/2, where ΔF is the dispersive (off-diagonal elements) or Bijvoet difference (diagonal elements), computed between 30 and 2.8 Å resolution. | |||||
| eR=∑(Fobs−kFcalc)/∑Fobs; all reflections of F>0 in the resolution range were included. | |||||
| fWilson thermal factor of the native dataset. | |||||
GAT–Rabaptin5 interaction
The N-terminal homodimer and the C-terminal four-helix bundle regions of Rabaptin5551–661 have individual local symmetries. However, only one GGA1210–302 domain binds with the N-terminal symmetric homodimer, although it interacts with both monomers. The solvent-accessible surface (SAS) buried upon complex formation between one GAT molecule and Rabaptin5 homodimer is 1770 Å2, with 52% from hydrophobic atoms. The only other interaction between GAT and Rabaptin5 observed in the crystal packing is significantly smaller, with buried SAS less than 600 Å2. It unlikely bears any biological function.
In the crystal structure, the Rabaptin5-binding residues from GGA1210–302 form a central hydrophobic strip that is surrounded by charged or polar residues (Figure 2 and Supplementary Figure S1). The interacting residues in Rabaptin5551–661 include Gln556R, Lys559R, Met563R, Gln566R, and Lys573R from one monomer, and Arg554R, Gln561R, Leu564R, Arg565R, Asn568R, and Met575R from the other. They also form a hydrophobic strip in the center of the binding patch (Figure 2B). The interaction between the two proteins is extensive and fairly complimentary. For example, a surface socket formed by residues Phe264, Ser268, Leu277, and Leu281 of GGA1 is complementary with the side chain of Met563R. Similarly, the GGA1 Phe264 plugs into a socket in the groove between the two Rabaptin5 helices. The ARF-binding site of the GAT domain, which is not included in the construct used for crystallization, would be on the opposite side and located in the helix–loop–helix motif ∼35 Å from the Rabaptin5-binding site (Zhu et al, 2003). The predicted ubiquitin-binding site (Shiba et al, 2004), however, overlaps with the Rabaptin5-binding area. No steric inhibition is observed in the symmetric potential GAT-binding site that would require major conformational changes for a second GAT–Rabaptin5 interaction to occur. An electrostatic potential change induced by the first GAT binding may provide an explanation for the affinity reduction in the symmetric binding site.
Figure 2.
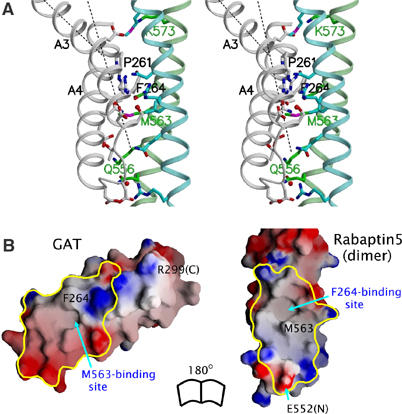
GAT–Rabaptin5 interaction. (A) Stereo view of the interface. Helix backbones are shown in a ribbon representation and colored as white, cyan, and green for GAT and the two Rabaptin5 monomers, respectively. Side chains of residues directly involved in the interface are shown in stick models and colored in blue (for nitrogen atoms), red (oxygen), and magenta (sulfur), or according to their backbone colors (carbon). Helices α3 and α4 of GAT are labeled as A3 and A4 (Zhu et al, 2003). The kink of helix α3 is depicted by the axes of the two portions of the helix (black dash lines). (B) Molecular surface models of the GAT three-helix bundle (left) and N-terminal part of Rabaptin5551–661 dimer (right). The color scheme is the same as Figure 1C. The interface regions, based on a calculation of buried SAS, are enclosed with yellow lines. Selected residues and (N- or C-) termini are labeled.
Binding assay on the GAT–Rabaptin5 interaction
Results from our previous mutagenesis studies on GGA1 GAT (Zhai et al, 2003) are consistent with the notion that the complex observed in the current crystal structure also exists in solution. The reduction of binding affinity to Rabaptin5 by any one of the point mutations P261E (i.e. Pro261 to Glu substitution), F264R, R265E, L277R, N284S, or D285R in GGA1 GAT (Zhai et al, 2003) can be explained based on the current complex structure: P261E introduces an acidic residue that interferes with Glu572R, F264R disrupts a hydrophobic interaction, R265E increases the negative charge of GAT in the vicinity of Glu572R, L277R interferes with Arg554R and/or destabilizes hydrophobic interaction, N284S causes a loss of one critical hydrogen bond with Gln566R, and D285R introduces a repulsive interaction with Lys559R (Figure 2A and Supplementary Figure S2). Meanwhile, our GAT surface mutations not part of the structurally observed Rabaptin5-binding site show no effect on the binding (Zhai et al, 2003), suggesting that the GAT domain uses only one surface patch to interact specifically with Rabaptin5.
The current complex structure further allows us to test key surface residues from both monomers of the Rabaptin5551–661 dimer through mutagenesis studies by making point mutations Q561RA, M563RR, Q566RA, Q566RR, N568RA, and N568RR in the Rabaptin5551–661 parental construct. In the pull-down experiments, GGA1141–326 (i.e. full-length GAT domain) was fused to glutathione S-transferase (GST), immobilized to resin beads, and incubated with the cell lysate containing individually expressed Rabaptin5551–661 variants. The bound Rabaptin5551–661 variants were further analyzed with SDS–PAGE. Alanine or arginine replacements at the selected positions of Rabaptin5551–661 diminished the binding to GAT (Figure 3A), indicating that important components for Rabaptin5–GAT interactions seen in the complex structure are also operative in solution. To address the biological relevance of this interaction further, we employed a competition assay to determine if the full-length GAT domain–full-length Rabaptin5 interaction can be disrupted by the presence of the Rabaptin5551–661 fragment (Figure 3B). Full-length Rabaptin5 was expressed in mammalian cells, and the cell lysate was pulled down with GST-GGA1141–326 in the presence of varied concentrations of Rabaptin5551–661. As the Rabaptin5551–661 concentration increased, the binding of full-length Rabaptin5 to GAT diminished. This result suggests that in vivo Rabaptin5–GAT binding uses the same interface that is observed in the crystal structure.
Figure 3.
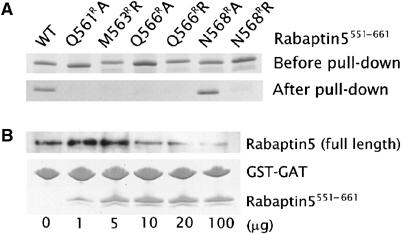
Binding affinity between Rabaptin5 fragment and GGA1 GAT domain. (A) GST-mediated pull-down assay. Purified recombinant GST-GGA1141–326 is immobilized to GSH resin. Rabaptin5551–661 variants from cell lysate (i.e. samples labeled as ‘before pull-down') were pulled down by the GAT fusion protein and visualized with CBB stain (labeled as ‘after pull-down'). (B) Competition of Rabaptin5551–661 with full-length Rabaptin5 for GGA1 GAT. GST-GGA1141–326 (10 μg) were preincubated with GSH-Sepharose beads and cell lysate containing full-length Rabaptin5 in the presence of different concentrations of Rabaptin5551–661 (0–100 μg) to a final volume of 400 μl. The material bound to the beads was subjected to SDS–PAGE and CBB stain (for GST-GGA1141–326 and Rabaptin5551–661) or anti-Rabaptin5 Western blot (for full-length Rabaptin5). The experiment was repeated multiple times and was reproducible.
Four-helix bundle of Rabaptin5
In the crystal structure, two Rabaptin5551–661 homodimers from neighboring asymmetric units form a 190-Å-long tetramer related by the crystallographic two-fold symmetry. A single homo-four-helix bundle constitutes the entire tetramer interaction and possesses a pseudo-222-point symmetry (Figure 4A). Each helix of the bundle is about 50 residues long (i.e. residues 590–640) and is antiparallel to its nearest neighbors. Hydrophobic residues are packed in the core of the bundle. Charged and polar residues form extensive salt-bridge and hydrogen bonds on the surface. The buried SAS between the two symmetric dimers is 8300 Å2, with 67% contributed by hydrophobic atoms. In comparison, the SAS buried between the two monomers in an isolated parallel Rabaptin5551–661 homodimer is 2050 Å2 and is mainly contributed by the N-terminal regions of the long helices.
Figure 4.
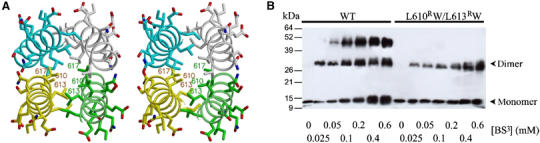
Rabaptin5 tetramerization. (A) Stereo view of the central section of the four-helix bundle of Rabaptin5551–661. Backbones of the four helices are shown in ribbon representation and colored in yellow, green, cyan, and white, respectively. Side chains are shown in stick models and their carbon atoms are colored corresponding to their respective backbones. Other atoms are colored as in Figure 2A. Selected residues from the hydrophobic core are labeled. The yellow and white molecules belong to one parallel dimer, as do the cyan and green molecules, whereas the two dimers are antiparallel to each other. (B) Chemical crosslink. Tubes containing equal amounts of His6-Rabaptin5551–661 WT (left) or L610RW/L613RW mutant (right) were incubated with varied concentrations of BS3 (0–0.6 mM) for 30 min at 22°C. The final crosslinked products were then subjected to 12% homogenous SDS–PAGE and analyzed with anti-His Western blot.
The existence of this tetramerization structure in solution was detected with chemical crosslinking (Figure 4B), using a His-tagged recombinant protein sample directly purified from the soluble fraction of E. coli cell lysate. It showed that a significant fraction of Rabaptin5551–661 can be crosslinked in forms of a molecular weight range higher than a dimer. This result was reproducible in a 300-fold range of protein concentration (data not shown), indicating that the oligomerization is not an artifact of high protein concentration. Furthermore, we constructed a Rabaptin5551–661 variant of a double mutation L610RW/L613RW, which presumably disrupts the tetramer formation (see Figure 4A). Repeating the chemical crosslinking experiment with this Rabaptin5 mutant essentially eliminated the trimer/tetramer species but retained the dimer in the crosslinking product (Figure 4B). This result further supports that the crystallographically observed Rabaptin5 tetramer exists in solution. In addition, this L610RW/L613RW double mutant retained full GAT-binding ability as the wild type in a GST-mediated pull-down assay (data not shown), suggesting that the GAT-binding ability of Rabaptin5 is structurally independent of its tetramerization.
Discussion
GGA adaptors function in the vesicle transport pathway between TGN and endosomes in concert with an array of protein partners. Among them, Rabaptin5 binds with all three isoforms of human GGAs. Two Rabaptin5-binding sites have been identified on the GGA proteins (Mattera et al, 2003) (Supplementary Figure S3). One is located in the GAE domain of GGA1–3 (Hirst et al, 2000; Mattera et al, 2003). A crystal structure of GGA3 GAE in complex with a peptide of Rabaptin5435–447 has been reported (Miller et al, 2003). This interaction occurs between an extended peptide of Rabaptin5 containing a sequence motif of FxxΦ (where Φ stands for a hydrophobic amino-acid residue) and the surface depression formed along the edges of two β-sheets present in the GAE domain. The second binding site occurs in the GAT domain of GGA1 and GGA2, but not in GGA3. Here, we demonstrate that this second Rabaptin5–GGA interaction is between the three-helix bundle of GGA GAT domain and a Rabaptin5551–661 parallel coiled-coil homodimer. This GAT-binding region of Rabaptin5551–661 is locally symmetrical; however, only one GAT domain is found to interact with the dimer in the current crystal structure (Figure 1B). Such an interaction may also occur in vivo for the following reasons. First, if two GAT domains could bind simultaneously to the Rabaptin5 dimer in a symmetrical way while both also attach to the membrane, it would require the two-fold axis of Rabaptin5 dimer to be more or less perpendicular to the membrane. Such a scenario is unlikely because the GAT-binding site is located in the middle of the elongated Rabaptin5 molecule. Second, the surfaces of both GGA1210–302 and Rabaptin5551–661 structures are highly negatively charged (Figure 1C) except for the binding interface (Figure 2B) where hydrophobic interaction contributes significantly. Therefore, electrostatic repulsion may prevent two GAT domains binding simultaneously to the Rabaptin5 dimer.
Although Rabaptin5 interacts equally well with the GAE domains from all three isoforms of human GGAs (Hirst et al, 2000; Mattera et al, 2003), it shows different binding affinities toward their GAT domains (Mattera et al, 2003; Zhai et al, 2003), that is, preferring the GAT domain of GGA1 and GGA2 over GGA3. The functional consequence of this difference is unclear at present. Among the residues of GGA1 GAT domain that directly interact with Rabaptin5551–661, only residues 261 (Pro/Arg in GGA1 and GGA3, respectively), 265 (Arg/Lys), 284 (Asn/Ser), and 288 (Thr/Ser) are different between GGA1 and GGA3. Residue 261 is located at the kink position of helix α3, which appears critical to the Rabaptin5 binding (Figure 2A). Its importance is supported by the deleterious effect of the P261E mutant (Zhai et al, 2003). Nevertheless, the GGA3-like P261R mutant interacts well with Rabaptin5, arguing that Pro261 may not be essential for the Rabaptin5-binding preference of GGA1. The two conserved mutations between GGA1 and GGA3, R265K and T288S, are expected to maintain similar interactions with the ligand; thus, they are unlikely to be the structural determinants causing the difference in preference. A more serious change is the N284S substitution that results in the loss of one hydrogen bond with Gln566R and has been shown to diminish Rabaptin5 binding (Zhai et al, 2003). Moreover, it is interesting to see that a reversed mutant, S283N, can instill a significant Rabaptin5-binding ability in GGA3 (Zhai et al, 2003; Mattera et al, 2004). Furthermore, nonconserved residues that do not directly interact with Rabaptin5 may also influence the binding ability through subtle environmental changes (Mattera et al, 2004). For example, the region of sequence difference N-terminal to the ‘kink' region of helix α3, which does not directly contact Rabaptin5, may result in a local conformational change in the ‘kink' region on the complex interface and subsequently disturb a salt-bridge bond between Lys573R and the position cognate to Glu257 of GGA1. In addition, the current crystal structure is consistent with the observation that Rabaptin5 does not interact with the GAT domain from TOM1 or TOM1-L1 (Mattera et al, 2004), since 11 out of 14 GGA1 residues that interact with Rabaptin5 in the current structure differ from those in TOM1 (Dell'Angelica et al, 2000). Interestingly, the same ‘Rabaptin5-binding' region in the GAT domain from either GGA1 or GGA3, but not GGA2, is shown to bind with ubiquitin (Shiba et al, 2004). This interaction appears to be strengthened by ARF binding in the presence of N-terminal VHS domain of GGA, although structural modeling suggests that a ubiquitin monomer and ARF would bind to distinct structural motifs in the GAT domain that are over 30 Å away from each other. Similar ubiquitin binding has also been observed in the GAT domain of human TOM1 despite its relatively low sequence homology with GGAs (e.g. lower than that between GGA1 and GGA2). It appears that Rabaptin5 and ubiquitin would compete for the same binding site on the GAT domain three-helix bundle region, particularly the surface hydrophobic patch formed by helices α3 and α4 (Mattera et al, 2004; Shiba et al, 2004). Such a potential interference between the two interactions could form an additional layer of regulation in vesicle trafficking, that is, Rabaptin5–GAT interaction could be regulated by ubiquitin binding, or vice versa.
While there has been evidence suggesting that purified Rabaptin5 exists in solution as high-molecular-weight oligomers, the complexes are often interpreted as dimers (Vitale et al, 1998). Besides the GAT–(Rabaptin5)2 interaction, to our surprise, our crystal structure reveals an unexpected four-helix bundle mediated homotetramerization of Rabaptin5. The interactions between the helices appear to be also stable in solution (Figure 4B). The Rabaptin5551–661 dimer dimerization buries more than 8000 Å2 SAS, and two-thirds of the buried SAS are from hydrophobic atoms. At present, we could not rule out the possibility that the tetramerization is an artifact of the recombinant protein fragment. For example, in full-length Rabaptin5, intramolecular interactions could replace the structurally observed tetramer interactions. Nevertheless, the extensive and complementary tetramer interface seen in the crystal structure favors an alternative interpretation. Although we have not determined the affinity for the tetramerization, structural comparison strongly suggests that the strength of the dimer–dimer interaction between Rabaptin5551–661 is significantly higher than that of the GGA1 GAT–Rabaptin5551–661 interaction. Unless drastically reorganized, an isolated Rabaptin5551–661 dimer from the current structure would expose a large hydrophobic surface to solvent. Thus, a permanent tetramerization appears a way to stabilize this portion of the Rabaptin5 protein in solution (Nooren and Thornton, 2003). Since the surface of the four-helix bundle is dominated by negative charges, it seems unlikely that this Rabaptin5 fragment would form higher orders of homo-oligomers in solution once tetramerized. Because regions of Rabaptin5 on both sides of the GAT-binding domain also form homodimers (Vitale et al, 1998), the detailed dimerization scheme in the Rabaptin5 tetramer could be topologically complicated. A similar antiparallel tetramerization of two parallel dimers was proposed for another trafficking accessory protein, the Rab5 activator Rin2 (Saito et al, 2002), although it is not known whether the two cases of tetramerization are the same because of lack of detailed structural information on Rin2. The GAT-binding and tetramer-forming region of Rabaptin5 is highly conserved in evolution, with a better than 90% sequence identity in this region in general among species (in current GenBank). Compared with human Rabaptin5, the chicken Rabaptin5 has the lowest homology, being 84% identical and 91% conserved in this region (Figure 5). Nevertheless, in a modeling test, the current crystal structure can accommodate the chicken Rabaptin5 sequence without significant conformational adjustment (data not shown). In addition, the human Rabaptin5 homolog, Rabaptin5β (Gournier et al, 1998), is conserved in the four-helix bundle region but contains a deletion in the GAT-binding region compared to Rabaptin5 (Figure 5). It suggests that the four-helix bundle and the GAT-binding dimer region of Rabaptin5 are modular structural motifs, each bearing a distinct function. The coiled-coil four-helix bundle may hold multiple functions. For example, it could provide mechanical support and scaffold for other protein domains and/or bind specifically to some yet to be recognized partners.
Figure 5.
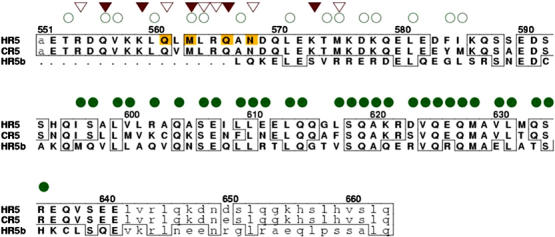
Sequence alignment of Rabaptin5 homolog proteins in the ‘GAT-binding' region. The order from top to bottom is human Rabaptin5 (HR5, residues 551–663), chicken Rabaptin5 (CR5, residues 549–661, GenBank ID BAA21785), and human Rabaptin5β (HR5b, residues 287–383, GenBank ID RNU34932). The region of residues visible in the GAT–Rabaptin5 complex crystal is represented with capital letters, and mobile residues are in lowercase letters. HR5 residue numbers are labeled above the sequence. Residues identical to that of HR5 are boxed. Positions involved in the GAT binding from the two monomers of the Rabaptin5 dimer are marked with open and filled triangles, respectively. Positions involved in the N-terminal dimerization and C-terminal tetramerization (4 Å cutoff) are marked with open and filled circles, respectively. Residues of HR5 subjected to mutagenesis studies are highlighted.
Because Rabaptin5 plays important roles in early endosome fusion (Stenmark et al, 1995), it is hypothesized that the GGA–Rabaptin5 interaction enables fusion of TGN-derived vesicles with endosomes (Mattera et al, 2003), allowing the unloading of cargos from the TGN to endosomal system (Puertollano et al, 2003). Rabaptin5 interfering with the ARF–GGA–clathrin binding through steric and/or electrostatic mechanism may mediate this function. Currently available complex crystal structures allow us to assemble a model of the ARF–GAT–Rabaptin5551–661 interaction with high confidence in its geometry (Figure 6). In this model, two GGA1 GAT domains independently bind to the opposite two ends of the Rabaptin5551–661 tetramer of the current structure, retaining the 1:2 GAT (monomer) to Rabaptin5 (monomer) ratio. One ARF molecule interacts with each of the two GAT domains, assuming that ARF and Rabaptin5 simultaneously bind to the GAT domain (thus the GGA protein) (Zhai et al, 2003). Should such a complex exist in vivo, it could provide a linkage between two ARF molecules 200 Å apart.
Figure 6.
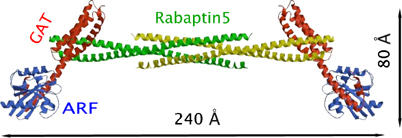
Ribbon diagram of a hypothetical ARF–GAT–(Rabaptin5)4–GAT–ARF complex. The ARF molecule and ARF–GAT interaction are modeled according to the crystal structure of ARF–GAT peptide complex (PDB file 1J2J). The GAT domain is copied from the crystal structure of GGA1 GAT domain (1OXZ). The GAT–Rabaptin5 interaction and Rabaptin5 tetramer are based on the current crystal structure. Since the two ARF-binding sites are independent of each other, in solution an alterative arrangement is also possible, that is, the right side GAT–ARF complex could flip 180° about the dyad axis of the yellow Rabaptin5 dimer.
Among the proteins of the current discussion, both GGA and Rabaptin5 are multidomain proteins, which dynamically interact with an array of partners in vesicular transport. With regard to the divalent interaction between GGA and Rabaptin5, it is interesting that GAT–Rabaptin5 interaction has a stoichiometry of 1:2 but the GAE–Rabaptin5 interaction has a 1:1 ratio (Supplementary Figure S3). Possibly, this would free up one extended peptide containing the FxxΦ motif from the Rabaptin5 dimer not bound to GAE to interact with another protein, and thereby expand the Rabaptin5 interacting network. Furthermore, the GAT and GAE domains interacting with one Rabaptin5 molecule may come from different GGA molecules. With the tetramerization, Rabaptin5 potentially assumes an even greater role in network formation. For example, its Rab5-binding motifs at the C-termini of the four monomers could reach the endosome membrane and regulate the tethering and fusion process.
Materials and methods
Protein expression and purification
Fusion constructs of GST-GGA1210–302 (i.e. the three-helix bundle region of the GGA1 GAT domain) and GST-GGA1141–326 (i.e. the full-length GAT domain of GGA1) were inserted into pGEX6P1 and pGEX2T (Amersham-Biosciences), respectively, as described previously (Zhai et al, 2003). Each was expressed as soluble recombinant protein in E. coli B21 (DE3) cells and purified with GSH-Sepharose 4B affinity chromatography (Amersham-Biosciences). The GGA1210–302 protein was separated from gel-immobilized GST after cleavage with the PreScission protease (Amersham-Biosciences) and was further purified with Resource-Q anion-exchange chromatography (Amersham-Biosciences). The protein sample was concentrated to 20 mg ml−1 (based on ɛ280=2.7 AU mM−1, estimated using the program VectorNTI from InforMax Inc.). The GST-GGA1141–326 fusion protein was left intact for the pull-down assay, purified as previously described (Zhai et al, 2003), and concentrated to 12 mg ml−1 (ɛ280=1.0 AU mM−1, VectorNTI). Se-Met-substituted GGA1210–302 was prepared by using the same construct as the native protein. It was expressed in E. coli B834 (DE3) pLysS cells (Novagen) grown in minimal media plus 40 mg l−1 Se-Met (Sigma). The construct encoding Rabaptin5551–661 was subcloned into pET11a (Novagen), expressed as inclusion bodies, refolded using the rapid dilution method (Lin et al, 1989) with dithiothreitol (DTT) as the reducing agent, and purified as described before (Zhai et al, 2003). For crystallization and pull-down assay, the Rabaptin5 protein was concentrated to 8 mg ml−1 (based on the Bradford protein assays, Bio-Rad Laboratories) in 10 mM HCl–Tris (pH 8.0) and stored on ice. Mass spectroscopic and N-terminal analyses showed that this sample contained two species of residues 551–642 and 551–648, respectively, both of which had an extra N-terminal methionine derived from the starting codon. The ratio of the two species (∼1:1) remained unchanged during a 2-month period according to SDS–PAGE analyses. For the crosslink experiment, a His6-Rabaptin5551–661 fusion construct and its L610RW/L613RW double mutant were inserted separately into the pET15b vector (Novagen) and expressed in E. coli BL21 (DE3) cells. The cell culture of each variant was lysed using the CelLytic bacterial lysis buffer (Sigma) containing protease inhibitors and DNAse (Roche), incubated for 15 min at 22°C, and clarified by centrifugation for 15 min at 60 000 g. The supernatant was then applied to His-select affinity resin (Sigma), washed, and eluted. The sample was dialyzed against 1 × phosphate-buffered saline (PBS, pH 7.5), and its concentration was estimated by comparison to known concentrations of bovine serum albumin since the 280-nm extinction coefficient of the Rabaptin5 sample is extremely low to nonexistent.
Crystallization and data collection
The initial crystallization condition of the GGA1210–302–Rabaptin5551–661 complex was obtained using Hampton Research crystallization screening kits, by varying the concentration ratio of the two protein components and cross-seeding with a tetragonal form of GGA1210–302 crystal (G Zhu and XC Zhang, unpublished data). Crystals of good diffraction quality were obtained under the following condition: protein samples of GGA1210–302 (20 mg ml−1) and Rabaptin5551–661 (8 mg ml−1) were first mixed 1:2 (v/v), then this sample was mixed 2:1 (v/v) with the reservoir solution containing 1.1 M (NH4)2SO4, 0.1 M Tris (pH 9.0), and 0.1% (v/v) β-mercaptoethanol. The complex crystals were grown at 20°C using the hanging drop method. Native crystals grew to 0.8 × 0.4 × 0.3 mm from a 4.5 μl drop over a 500 μl reservoir in 10 days, and Se-Met derivative crystals to 0.5 × 0.2 × 0.15 mm under similar conditions. The crystals were soaked in a mixture of saturated Li2SO4 and the reservoir solution (4:1) for cryoprotection before transference to a 100 K nitrogen gas stream for data collection.
X-ray diffraction data from the crystal of native GGA1210–302–Rabaptin5551–661 complex were collected up to 2.4 Å resolution from an in-house MAR345 image plate (Mar Research Inc.) using a single crystal. For the Se-Met derivative crystal, a three-wavelength multiple anomalous dispersion (MAD) dataset was collected to 2.8 Å resolution at the F2 beam line of the Cornell High Energy Synchrotron Source (CHESS) using a Quantum-210 CCD detector system. Data were processed with the HKL program suite (Otwinowski and Minor, 1997). Both the native and Se-Met derivative crystals were shown to be hemihedrally twinned (Yeates, 1997), with twinning fractions of 0.41 and 0.13, respectively. The data were detwinned correspondingly as the last step of data processing.
Structural determination and refinement
Most of the crystallographic calculations, including detwinning the data, were performed with the program suite CNS (Brunger et al, 1998). The MAD dataset yielded four selenium sites, which correspond to the four Met residues positioned at 231, 233, 249, and 259 previously found in one GGA1 GAT domain (Zhu et al, 2003). The MAD phases of the Se-Met derivative yielded an initial experimental electron density map at 2.8 Å resolution. The phases were further improved and extended to 2.4 Å resolution by density modification and switching to the native dataset. The resulting density map was interpretable for the most part. The crystal form was found to belong to the P3221 space group, with a 5.4 Å3 Da−1 VM, assuming one GAT and two Rabaptin5 fragments per asu. Manual model building was carried out with the program Turbo-Frodo (Roussel and Cambillau, 1989), with the three-helix bundle model from our previously reported GGA1 GAT crystal structure (PDB file 1OXZ) as a partial initial model. Structural refinement consisted of several rounds of torsion angle simulated-annealing and restrained individual B-factor refinements, using a maximum likelihood target function against native data up to the highest resolution available. The final Rworking was 0.235 for the detwinned dataset, and 0.181 for the twinned dataset using a corresponding target function in CNS. SAS was calculated using a 1.4 Å probe radius.
GST-mediated pull-down assay for studying GAT–Rabaptin5 interaction
To verify Rabaptin5 residues involved in GAT binding, point mutations were introduced into the Rabaptin5551–661 parental construct and expressed in E. coli. The soluble form of variant protein was used directly from cell lysate without further purification. GST-GGA1141–326 was used for its stronger binding with Rabaptin5551–661 than that of the GST-GGA1210–302 fusion construct. In the pull-down experiment, recombinant fusion protein GST-GGA1141–326 (30 μg) was incubated for 30 min at 22°C with 40 μl of a 50% slurry of GSH-Sepharose 4B in a final volume of 500 μl binding buffer containing 1 × PBS (pH 7.4), 0.2% (v/v) Triton X-100, and 0.2% (v/v) β-mercaptoethanol, supplemented with protease inhibitor cocktail. Beads containing immobilized GST fusion protein were washed three times by resuspension in 1 ml of 1 × PBS and centrifugation (3 min, 2000 g). Washed beads were incubated for 2 h at 22°C with equal volumes of cell lysate containing Rabaptin5551–661 variants (∼100 μg total protein) in 1 ml binding buffer, washed three times with 1 ml of binding buffer, and resuspended in 30 μl of 2 × SDS sample buffer. The sample was subjected to 20% SDS–PAGE analysis and visualized by Coomassie brilliant blue (CBB) stain.
Competition assay testing GAT–full-length Rabaptin5 interaction
A pcDNA3.1 expression vector containing the full-length human Rabaptin5 was transfected into HEK293 cells using the Lipofectamin 2000 transfection kit (Invitrogen). The transfected cells were grown in a T-25 culture flask for 36 h, resuspended in buffer A containing 1 × PBS, 1 mM EDTA, 1 mM MgCl2, 1 mM DTT, 1% (v/v) NP-40 detergent, and protease inhibitor cocktail, and then lysed by repeated ejection through a 21-gauge needle. The lysate was clarified by 10 000 g centrifugation for 10 min at 4°C followed by 120 000 g for 1 h at 4°C. An aliquot of clarified supernatant (200 μl containing ∼100 μg total protein) was incubated in buffer A containing 20 μg of GST-GGA1141–326 (prebound to 40 μl of 50% slurry GSH-Sepharose 4B beads) in the presence of varied concentrations of Rabaptin5551–661 (400 μl final volume) for 2 h at 22°C with gentle shaking. The beads were washed three times with 500 μl of buffer A, 25 μl of 2 × SDS sample buffer was added, and then the sample was subjected to 10% SDS–PAGE and blotted onto PVDF membrane (Millipore Co.). The blot was incubated sequentially with monoclonal mouse anti-Rabaptin5 antibody (BD Biosciences), which does not crossreact with the Rabaptin5551–661 fragment, then peroxidase-conjugated anti-mouse IgG (Sigma) and detected on film using Renaissance Chemiluminescence Reagent plus (Amersham-Biosciences). The presence of Rabaptin5551–661 in the complex was confirmed by CBB stain.
Chemical crosslink of Rabaptin5551–661
To test for oligomer formation, recombinant protein of His6-Rabaptin5551–661 purified from the soluble fraction of E. coli cell lysate was crosslinked using bis-(sulfosuccinimidyl suberate) (BS3, Pierce) reagent following the manufacturer's recommended protocol. Aliquots of His6-Rabaptin5551–661 (4 μg ml−1) in 1 × PBS (pH 7.5) were added to each tube and incubated with varying concentrations of BS3 (0–0.6 mM) for 30 min at 22°C. The reaction was quenched by adding 60 μl of 1 M Tris–HCl (pH 7.5) to each tube and incubated for 15 min at 22°C. For final analysis, the samples were applied to a 12% SDS mini-gel (Bio-Rad), transferred to PVDF membrane using a semidry transfer cell (Bio-Rad), and immunolabeled with mouse anti-His antibodies (Amersham-Biosciences). The anti-His-positive proteins were detected on film after a 4-min exposure following treatment of the membrane with ECL™ Western Blotting reagents (Amersham-Biosciences). To test for concentration dependence of crosslink oligomer formation, the same experiment was repeated with varied protein concentrations (0.7–220 μg ml−1), analyzed with SDS–PAGE, and stained with anti-His Western blot or CBB. To confirm the importance of the hydrophobic core in tetramer formation in solution, we introduced a double mutant, L610RW/L613RW, into the His6-Rabaptin5551–661 construct and repeated the chemical crosslink experiment against that of the wild type.
Structural data deposition
Coordinates and the experimental structural factors of the GGA1210–302–Rabaptin5551–661 complex crystal structure have been deposited in PDB under code 1X79.
Supplementary Material
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure Legends
Acknowledgments
We thank W Radosevich for excellent technical assistance, Dr Q Hao and the staff of the F2 beam line at the CHESS synchrotron facility for help in data collection, and Dr T Mather for critical reading of the manuscript. We are grateful to Dr M Robinson (University of Cambridge) for providing the cDNA of human GGAs, and Dr G Grenningloh (University of Lausanne) for the Rabaptin5 cDNA. The Molecular Biology Resource Facility of University of Oklahoma Health Sciences Center performed DNA oligomer synthesis, total amino-acid analysis, and mass spectroscopic analysis. This work was supported in part by a grant from Oklahoma Center for the Advancement of Science and Technology to XZ, an NIH grant (AG-18933) and Alzheimer's Association Pioneer Award to JT, and an NSF CAREER award to GL.
References
- Boman AL (2001) GGA proteins: new players in the sorting game. J Cell Sci 114: 3413–3418 [DOI] [PubMed] [Google Scholar]
- Boman AL, Zhang C, Zhu X, Kahn RA (2000) A family of ADP-ribosylation factor effectors that can alter membrane transport through the trans-Golgi. Mol Biol Cell 11: 1241–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Collins BM, Watson PJ, Owen DJ (2003) The structure of the GGA1-GAT domain reveals the molecular basis for ARF binding and membrane association of GGAs. Dev Cell 4: 321–332 [DOI] [PubMed] [Google Scholar]
- Dell'Angelica EC, Puertollano R, Mullins C, Aguilar RC, Vargas JD, Hartnell LM, Bonifacino JS (2000) GGAs: a family of ADP ribosylation factor-binding proteins related to adaptors and associated with the Golgi complex. J Cell Biol 149: 81–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deneka M, Neeft M, Popa I, van Oort M, Sprong H, Oorschot V, Klumperman J, Schu P, van der Sluijs P (2003) Rabaptin-5alpha/rabaptin-4 serves as a linker between rab4 and gamma(1)-adaptin in membrane recycling from endosomes. EMBO J 22: 2645–2657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gournier H, Stenmark H, Rybin V, Lippe R, Zerial M (1998) Two distinct effectors of the small GTPase Rab5 cooperate in endocytic membrane fusion. EMBO J 17: 1930–1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst J, Lui WW, Bright NA, Totty N, Seaman MN, Robinson MS (2000) A family of proteins with gamma-adaptin and VHS domains that facilitate trafficking between the trans-Golgi network and the vacuole/lysosome. J Cell Biol 149: 67–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhausen T (2000) Clathrin. Annu Rev Biochem 69: 699–727 [DOI] [PubMed] [Google Scholar]
- Korobko EV, Kiselev SL, Korobko IV (2002) Multiple Rabaptin-5-like transcripts. Gene 292: 191–197 [DOI] [PubMed] [Google Scholar]
- Lin XL, Wong RN, Tang J (1989) Synthesis, purification, and active site mutagenesis of recombinant porcine pepsinogen. J Biol Chem 264: 4482–4489 [PubMed] [Google Scholar]
- Lupas A (1996) Coiled coils: new structures and new functions. Trends Biochem Sci 21: 375–382 [PubMed] [Google Scholar]
- Mattera R, Arighi CN, Lodge R, Zerial M, Bonifacino JS (2003) Divalent interaction of the GGAs with the Rabaptin-5–Rabex-5 complex. EMBO J 22: 78–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattera R, Puertollano R, Smith WJ, Bonifacino JS (2004) The tri-helical bundle subdomain of the GGA proteins interacts with multiple partners through overlapping but distinct sites. J Biol Chem 279: 31409–31418 [DOI] [PubMed] [Google Scholar]
- Miller GJ, Mattera R, Bonifacino JS, Hurley JH (2003) Recognition of accessory protein motifs by the gamma-adaptin ear domain of GGA3. Nat Struct Biol 10: 599–606 [DOI] [PubMed] [Google Scholar]
- Moss J, Vaughan M (1998) Molecules in the ARF orbit. J Biol Chem 273: 21431–21434 [DOI] [PubMed] [Google Scholar]
- Nooren IM, Thornton JM (2003) Diversity of protein–protein interactions. EMBO J 22: 3486–3492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Poussu A, Lohi O, Lehto VP (2000) Vear, a novel Golgi-associated protein with VHS and gamma-adaptin ‘ear' domains. J Biol Chem 275: 7176–7183 [DOI] [PubMed] [Google Scholar]
- Puertollano R, Aguilar RC, Gorshkova I, Crouch RJ, Bonifacino JS (2001) Sorting of mannose 6-phosphate receptors mediated by the GGAs. Science 292: 1712–1716 [DOI] [PubMed] [Google Scholar]
- Puertollano R, Bonifacino JS (2004) Interactions of GGA3 with the ubiquitin sorting machinery. Nat Cell Biol 6: 244–251 [DOI] [PubMed] [Google Scholar]
- Puertollano R, van der Wel NN, Greene LE, Eisenberg E, Peters PJ, Bonifacino JS (2003) Morphology and dynamics of clathrin/GGA1-coated carriers budding from the trans-Golgi network. Mol Biol Cell 14: 1545–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussel A, Cambillau C (1989) TURBO-FRODO. In Sillicon Graphics Geometry Partners Directory, pp 77–79. Mountain View, CA: Silicon Graphics [Google Scholar]
- Saito K, Murai J, Kajiho H, Kontani K, Kurosu H, Katada T (2002) A novel binding protein composed of homophilic tetramer exhibits unique properties for the small GTPase Rab5. J Biol Chem 277: 3412–3418 [DOI] [PubMed] [Google Scholar]
- Scott PM, Bilodeau PS, Zhdankina O, Winistorfer SC, Hauglund MJ, Allaman MM, Kearney WR, Robertson AD, Boman AL, Piper RC (2004) GGA proteins bind ubiquitin to facilitate sorting at the trans-Golgi network. Nat Cell Biol 6: 252–259 [DOI] [PubMed] [Google Scholar]
- Shiba T, Kawasaki M, Takatsu H, Nogi T, Matsugaki N, Igarashi N, Suzuki M, Kato R, Nakayama K, Wakatsuki S (2003) Molecular mechanism of membrane recruitment of GGA by ARF in lysosomal protein transport. Nat Struct Biol 10: 386–393 [DOI] [PubMed] [Google Scholar]
- Shiba Y, Katoh Y, Shiba T, Yoshino K, Takatsu H, Kobayashi H, Shin HW, Wakatsuki S, Nakayama K (2004) GAT (GGA and Tom1) domain responsible for ubiquitin binding and ubiquitination. J Biol Chem 279: 7105–7111 [DOI] [PubMed] [Google Scholar]
- Shiba Y, Takatsu H, Shin HW, Nakayama K (2002) gamma-Adaptin interacts directly with Rabaptin-5 through its ear domain. J Biochem (Tokyo) 131: 327–336 [DOI] [PubMed] [Google Scholar]
- Stenmark H, Vitale G, Ullrich O, Zerial M (1995) Rabaptin-5 is a direct effector of the small GTPase Rab5 in endocytic membrane fusion. Cell 83: 423–432 [DOI] [PubMed] [Google Scholar]
- Suer S, Misra S, Saidi LF, Hurley JH (2003) Structure of the GAT domain of human GGA1: a syntaxin amino-terminal domain fold in an endosomal trafficking adaptor. Proc Natl Acad Sci USA 100: 4451–4456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatsu H, Yoshino K, Nakayama K (2000) Adaptor gamma ear homology domain conserved in gamma-adaptin and GGA proteins that interact with gamma-synergin. Biochem Biophys Res Commun 271: 719–725 [DOI] [PubMed] [Google Scholar]
- Takatsu H, Yoshino K, Toda K, Nakayama K (2002) GGA proteins associate with Golgi membranes through interaction between their GGAH domains and ADP-ribosylation factors. Biochem J 365: 369–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitale G, Rybin V, Christoforidis S, Thornqvist P, McCaffrey M, Stenmark H, Zerial M (1998) Distinct Rab-binding domains mediate the interaction of Rabaptin-5 with GTP-bound Rab4 and Rab5. EMBO J 17: 1941–1951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeates TO (ed) (1997) Detecting and Overcoming Crystal Twinning. New York: Academic Press [PubMed] [Google Scholar]
- Zhai P, He X, Liu J, Wakeham N, Zhu G, Li G, Tang J, Zhang XC (2003) The interaction of the human GGA1 GAT domain with Rabaptin-5 is mediated by residues on its three-helix bundle. Biochemistry 42: 13901–13908 [DOI] [PubMed] [Google Scholar]
- Zhu G, Zhai P, He X, Terzyan S, Zhang R, Joachimiak A, Tang J, Zhang XC (2003) Crystal structure of the human GGA1 GAT domain. Biochemistry 42: 6392–6399 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure Legends


