Abstract
Over the past 5 years, massive accumulations of holopelagic species of the brown macroalga Sargassum in coastal areas of the Caribbean have created “golden tides” that threaten local biodiversity and trigger economic losses associated with beach deterioration and impact on fisheries and tourism. In 2015, the first report identifying the cause of these extreme events implicated a rare form of the holopelagic species Sargassum natans (form VIII). However, since the first mention of S. natans VIII in the 1930s, based solely on morphological characters, no molecular data have confirmed this identification. We generated full‐length mitogenomes and partial chloroplast genomes of all representative holopelagic Sargassum species, S. fluitans III and S. natans I alongside the putatively rare S. natans VIII, to demonstrate small but consistent differences between S. natans I and VIII (7 bp differences out of the 34,727). Our comparative analyses also revealed that both S. natans I and S. natans VIII share a very close phylogenetic relationship with S. fluitans III (94‐ and 96‐bp differences of 34,727). We designed novel primers that amplified regions of the cox2 and cox3 marker genes with consistent polymorphic sites that enabled differentiation between the two S. natans forms (I and VIII) from each other and both from S. fluitans III in over 150 Sargassum samples including those from the 2014 golden tide event. Despite remarkable gene synteny and sequence conservation, the three Sargassum forms differ in morphology, ecology, and distribution patterns, warranting more extensive interrogation of holopelagic Sargassum genomes as a whole.
Keywords: accumulations, chloroplast genome, macroalgae, mitogenome, Sargasso Sea, strandings
1. Introduction
The brown alga Sargassum is one of the most diverse marine macroalgal genera with 351 recognized species worldwide (Guiry & Guiry, 2016). Members of the genus Sargassum are widespread, and most species are benthic with holdfasts, with only two recognized holopelagic species Sargassum natans (Linnaeus) Gaillon and Sargassum fluitans (Boergesen) Boergesen that do not attach to substrates (ibid). These species have gas vesicles and drift and reproduce vegetatively at the surface of the ocean (Dawes & Mathieson, 2008). They are most abundant in the North Atlantic Ocean subtropical gyre, also referred to as the Sargasso Sea (Figure 1), and also occur in the Gulf of Mexico and Caribbean Sea. Holopelagic Sargassum has been called the “golden floating rainforest of the Atlantic Ocean” (Laffoley et al., 2011) and functions as an ecosystem engineer that creates a unique floating pelagic biome in substrate‐poor, low‐nutrient open‐ocean waters. The floating Sargassum supports over 100 species each of invertebrates and fishes, including ten endemic taxa. Sargassum also serves as a nursery habitat for a host of important commercial and threatened species, including large pelagic fish such as tuna and bill fish, and four species of endangered sea turtles (Coston‐Clements, Settle, Hoss, & Cross, 1991).
Figure 1.
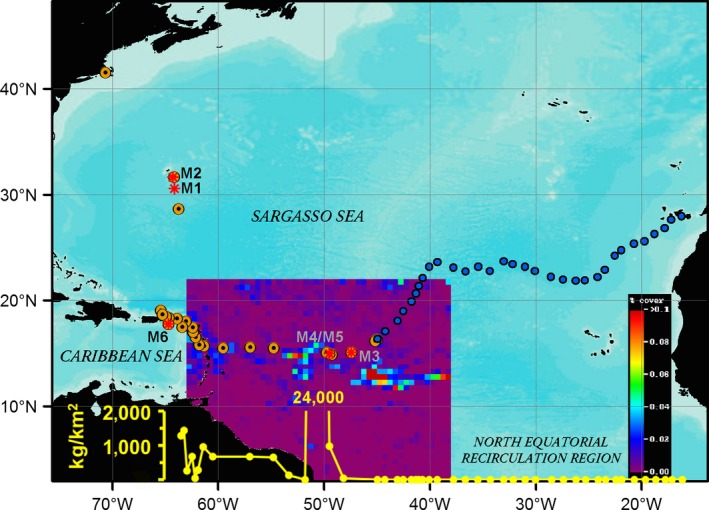
Map of sampling locations where we collected Sargassum (yellow dots with black centers), including open‐ocean Atlantic and Caribbean, as well as specimens collected from the shore (Caribbean and Cape Cod, USA). Stations where Sargassum was absent from net tows are shown as blue dots, and locations of mitogenome samples as red asterisks. The heatmap that overlies from approximately 38–63°W and 0–22°N depicts estimates of Sargassum accumulations from satellite data integrated over the 12‐day period coincident with the timing of the sample collection in that region. The scale of the heatmap at the lower right shows percent of the ocean surface covered by Sargassum from 0% to >0.1% (satellite data courtesy of University of South Florida Oceanography Lab, http://optics.marine.usf.edu). The yellow line graph at the bottom of the figure shows Sargassum quantity from the cruise net tows, showing the peak around 50°W and good correspondence with estimated densities from the satellite data
Although typically found offshore, unusual accumulations of Sargassum dubbed “golden tides” (Smetacek & Zingone, 2013), began washing ashore on islands in the Caribbean during 2011 then again in 2014 and 2015, burying beaches, impacting coastal fisheries, restricting harbors, and smothering sea turtle nests (Maurer et al., 2015). Reports of golden tides like those in the Caribbean have also been reported in western Africa and Brazil (De Széchy, Guedes, Baeta‐Neves, & Oliveira, 2012; Smetacek & Zingone, 2013) impacting tourism, food security, and the limited budgets of coastal towns trying to remove the rotting biomass from their beaches. Whether these golden tides represent changes in distribution of existing biomass or result from unusual accumulations due to higher growth rates (“blooms”) has not been established, but the most popular hypothesis is that nutrients supplied by the Amazon and Congo River basins, and also equatorial and coastal upwelling regions along west Africa are allowing fast‐growing Sargassum to reach very high concentrations in an area known as the North Equatorial Recirculation Region (NERR, Figure 1), with subsequent flushing toward the Caribbean (Johnson, Ko, Franks, Moreno, & Sanchez‐Rubio, 2013). The impacts of potential “bloom” conditions on the structure of the Sargassum populations and the functional diversity of the community of attached and mobile fauna dependent on the Sargassum biome are unknown. We also do not know the impacts of the Sargassum on coastal ecosystems and the potential for associated fauna to become invasive once the rafts of algae wash ashore.
Only two species of holopelagic Sargassum are recognized in contemporary taxonomic guides: S. natans and S. fluitans with a range historically limited to the Sargasso Sea, Gulf of Mexico, and Caribbean (Figure 2a, b). Extensive collections and early taxonomic work on holopelagic Sargassum (Parr, 1939; Winge, 1923) revealed distinct morphological forms of S. natans (I, II, VIII, IX) and S. fluitans (III, X). Two forms, S. natans I and S. fluitans III, proved most abundant in extensive field surveys in the 1930s (Parr, 1939), and despite annual sampling, none of the rarer forms were documented again until the “re‐emergence” of S. natans VIII in 2014 (Schell, Goodwin, & Siuda, 2015; Figure 2c, d). These authors noted that during their November 2014 cruise, Sargassum concentrations were an order of magnitude higher than previously recorded in their 20‐year data set. Although morphological features identified the accumulating Sargassum as the formerly rare form S. natans VIII, no genetic analyses have confirmed this observation or Parr's original morphology‐based descriptions.
Figure 2.

The most common forms of holopelagic Sargassum found in Atlantic: (a) herbarium specimen of Sargassum natans I from Parr's collection showing characteristic spine‐like appendages on vesicles; (b) herbarium specimen of Sargassum fluitans III from Parr's collection showing characteristic axis thorns; (c) herbarium specimen of S. natans VIII from Parr's collection showing neither spine‐like appendages nor thorns; (d) live fragment of S. natans VIII from sample used for one of the sequenced mitogenomes that matches Parr's morphological description of S. natans VIII. Scale bars are 1 cm. Herbarium images a, b, c from the Peabody Museum of Natural History, Yale University (YU); peabody.yale.edu
In order to understand changes in Sargassum population structure and potentially mitigate the impact of the Sargassum golden tides, marine scientists and managers have to be able to identify the algal species involved and where it is coming from. However, recent efforts to characterize members of the Sargassum subgenus Sargassum have encountered challenges due to low genetic variability among some clades (Cheang et al., 2010; Cho, Lee, Ko, Mattio, & Boo, 2012; Dixon et al., 2014; Mattio, Bolton, & Anderson, 2015; Mattio et al., 2013) and a lack of marker genes capable of distinguishing between closely related taxa in general (Camacho, Mattio, Draisma, Fredericq, & Diaz‐Pulido, 2014; Mattio & Payri, 2010). Thus, new molecular ecology resources are urgently needed to aid this endeavor. The use of mitogenomes to differentiate between different species of Sargassum has recently provided a bit of traction in terms of offering new sets of genetic markers for population studies of Sargassum species from the Pacific (Bi & Zhou, 2016; Liu & Pang, 2014; Liu, Pang, & Chen, 2016; Liu, Pang, Li, & Li, 2015; Liu, Pang, & Luo, 2014). In addition to mitochondrial genomes, the first Sargassum chloroplast genome from the benthic species S. horneri recently became available providing another source of potential marker genes (Liu & Pang, 2015).
Here, we explored the potential for mitogenomes and chloroplast coding regions to distinguish between Atlantic Sargassum holopelagic species by generating and comparing complete mitochondrial genomes and partial chloroplast genomes from all of the known holopelagic Sargassum species using next‐generation sequencing approaches. From our reference mitogenomes, we developed two marker gene primer sets that are capable of differentiating between all three holopelagic Sargassum forms encountered in the Atlantic: S. natans I, S. natans VIII, and S. fluitans III. Using all Sargassum mitogenomes available to date including the six new ones we generated, we also provide a phylogenetic placement for the three forms of Sargassum most commonly encountered in the tropical and subtropical seas of the western Atlantic.
2. Materials and Methods
2.1. Sample collection and DNA extraction
We collected samples for our six reference mitochondrial and partial chloroplast genomes and 155 marker gene validations from 71 independent clumps of Sargassum from across the NW Atlantic Ocean and in the Caribbean Sea (see Figure 1 and Table S1 for details). Samples of holopelagic Sargassum were collected in towed neuston nets, as well as in dip nets during four separate cruises in 2012, 2014, 2015, and 2016 in the North Atlantic Ocean. Additional samples of clumps that had stranded on shore were collected by hand in 2014 and 2016 (see Table S1).
Clumps of macroalgae were assigned species identifications using morphological characters defined by Parr (1939) including frond characteristics, the presence or absence of thorns on the axes, and spine‐like appendages on the air bladders (vesicles), as well as by blade (leaf) size. According to Parr's definitions, S. fluitans III can be distinguished from both S. natans I and S. natans VIII based on the presence of thorns on its axes. S. fluitans also does not have spine‐like appendages on its vesicles that are usually present on vesicles of S. natans I and occasionally found on vesicles of S. natans VIII. The two species of S. natans can also be distinguished based on their blade width. S. natans I has blades that are much narrower than S. natans VIII. Figure 2 depicts museum vouchers for Parr's original species morphological descriptions of the different forms sequenced in this study.
After identifying a particular sample, we removed 5 cm of the axis from each individual clump, with all attached blades and vesicles, for processing and preservation. In the laboratory, samples were rinsed in 0.22‐μm‐filtered seawater, shaken, blotted with a paper towel to remove the excess water, and then placed in individual plastic bags containing 15 g of silica gel (Activa Products, Marshall, TX, USA). The fronds became fully desiccated after 24 hr in the silica gel.
Samples for molecular characterizations included those preserved in silica gel and others in RNAlater® (ThermoFisher, Waltham, MA, USA). Voucher material has been archived for all the samples in the Amaral‐Zettler laboratory and is available for viewing upon request. RNAlater® sample extraction followed protocols of Zettler, Mincer, and Amaral‐Zettler (2013). For Sargassum preserved in silica gel, we used a modified Qiagen DNeasy Plant Mini‐kit protocol (Qiagen, Valencia, CA, USA) to extract genomic DNA. The modified protocol followed the manufacturer's instructions with a few changes. In place of step one of the Qiagen protocol, we crushed approximately 0.05 g of preserved Sargassum tissue in an individual 1.5‐ml microfuge tube with a mini‐pestle (USA Scientific, Ocala, FL) to prepare the sample for extraction. In step two, after reagents were added, the samples were vortexed on high speed for 30 s and then shaken in a vortex mixer with a 24‐tube holder for 3 min. In step nine, the columns were spun at 14,000 rpm for 3 min to remove any residual buffer. Finally, the elutions in steps 11 and 12 occurred with 50 μl of buffer AE, for a total elution volume of 100 μl. All extracted genomic DNA was cleaned using the MoBio PowerClean Pro DNA Clean‐Up Kit (MoBio, Carlsbad, CA, USA), following manufacturer's instructions.
2.2. Library construction and sequencing
We constructed genomic libraries for representative samples of each Sargassum form (number of individuals sequenced in parentheses): S. natans VIII (three specimens); S. natans I (one specimen); and S. fluitans III (two specimens). Table S1 and Figure 1 (M1–M6) provide the GPS coordinates and locations where the samples used in metagenomic library constructions originated. We prepared short (175 bp) and/or long (~250–450 bp) insert libraries by shearing the genomic DNAs to the corresponding lengths on the Covaris S220 (Covaris, Inc., Woburn, MA, USA) and constructed genomic libraries using Ovation Ultralow DR Multiplex Systems 9–16 kits (NuGEN Technologies Inc., San Carlos, CA, USA). We sequenced samples on either a HiSeq 1000 or NextSeq 500 Illumina platform according to manufacturer protocols. Table S1 contains specific details on library preparation and sequencing performed for each sample.
2.3. Bioinformatics and phylogenetics
Resulting Illumina raw reads were merged and quality‐checked using a series of Python scripts: “iu‐merge‐pairs” with the “enforce‐Q30‐check” option, to remove low‐quality reads and merge pairs, followed by “iu‐filter‐merged‐reads” to retain only merged pairs with no mismatches in the overlapping region (Minoche, Dohm, & Himmelbauer, 2011). We used CLC genomic workbench to assemble and map reads back to single mitochondrial genomic contigs (http://www.clcbio.com) and Geneious v. 8.0.5 (Biomatters, Ltd, Aukland, New Zealand) for gene annotation and visualization of the mitogenomes by importing annotation tracks from existing sequenced mitogenomes of other Sargassum species in GenBank. We used a similar strategy for mapping chloroplast gene reads back to the S. horneri chloroplast genome in order to access variation of polymorphic sites in the chloroplast coding regions of our representative Sargassum species.
For our phylogenetic analyses of mitogenomes, we removed any positions of uncertain homology in the mitochondrial alignments and inferred phylogenetic trees using 34,290 homologous nucleotide positions in RAxML version 7.2.8 (Stamatakis, 2006) and a general time reversible substitution model with gamma and invariant sites implemented through Geneious. One thousand bootstrap replicates determined the confidence of the branch support in the resulting phylogeny.
2.4. Confirmation of polymorphic sites via PCR
To assess whether the variant sites within mitochondrial genomes that we discovered were fixed, we designed primers that targeted the relevant cytochrome oxidase subunit 2 (cox2) and cytochrome oxidase subunit 3 (cox3) gene regions and obtained sequences for 71 geographically diverse individuals, including representatives of each holopelagic Sargassum form (number of individuals sequenced in parentheses): (S. natans VIII (n = 53); S. natans I (n = 13); and S. fluitans III (n = 5): See Supplementary Online Material for details, different parts of some individuals were extracted and sequenced more than once). Confirmation of polymorphic sites differentiating our three Sargassum mitogenomes was accomplished using primers targeting cox2 (cox2‐370F: 5′‐CAAAGATGGATTCGACGGTTGG‐3′, cox2‐776R: 5′‐CCGGTATCAAACTCGCCCTT‐3′) and cox3 (cox3‐467F: 5′‐GGTTCAACGACACCCATTT‐3′, cox3‐901R: 5′‐TAGCGTGATGAGCCCATG‐3′) gene regions. PCR was carried out in triplicate 25 μl reactions with 1× NEB OneTaq (New England BioLabs, Ipswich, MA, USA), 1 μl of 10 mmol/L forward primer, and 1 μl of 10 mmol/L reverse primer. Cycling conditions for both primer sets were as follows: an initial 94°C denaturation step for 4 min; 30 cycles of 94°C for 1 min, 50°C for 30 s, 72°C for 1 min; and a final 7‐minute extension at 72°C. The triplicate PCRs were pooled after amplification and purified using MinElute PCR Purification spin columns (Qiagen). Purified DNA was eluted in 10 μl of MinElute buffer EB. Cleaned PCR products were amplified for 60 cycles (96°C for 10 s, 50°C for 5 s, 60°C for 4 min) with BigDye at 1/16th concentration and sequenced on an ABI 3730XL (Applied Biosystems, Foster City, CA, USA) capillary sequencer at the W. M. Keck Ecological and Evolutionary Genetics Facility at the Marine Biological Laboratory.
3. Results
Our phylogenetic analyses based on mitochondrial sequence data revealed three genetically distinct forms of holopelagic Sargassum. The holopelagic Sargassum responsible for the golden tides in the Caribbean fell into the S. natans species complex with high bootstrap support (100%) and was not S. fluitans as often reported. Figure 3 depicts a maximum‐likelihood phylogenetic tree based on complete mitochondrial genomes of the three holopelagic Sargassum forms and six additional Sargassum species (representing all the available Sargassum mitogenomes in GenBank). All three holopelagic Sargassum mitogenomes are 34,727 bp in length and share identical gene synteny and content over the entire mitogenome (65 genes), as well as a similar AT content (63.8%). At the nucleotide level, the two S. natans forms differed at only seven sites of 34,727 bp (Table 1; differences are highlighted between the two S. natans forms), and S. natans I and S. natans VIII differed from S. fluitans III at 93 and 96 sites, respectively (Table 1). For the two S. natans mitogenomes, differences occurring in coding regions resulted in amino acid differences in five protein‐coding genes (ribosomal protein L5 (rpl5), ribosomal protein S19 (rps19), ribosomal protein S13 (rps13), cytochrome oxidase 3 (cox3), and NADH dehydrogenase subunit 6 (nad6)). We also found a single polymorphism across the two S. natans forms in both the 23S ribosomal RNA (23S rRNA) and 16S rRNA genes. The three holopelagic Sargassum differed at 370, 372, and 383 sites from the closest species with a sequenced mitogenome—S. vachellianum, a benthic Sargassum species endemic to China.
Figure 3.
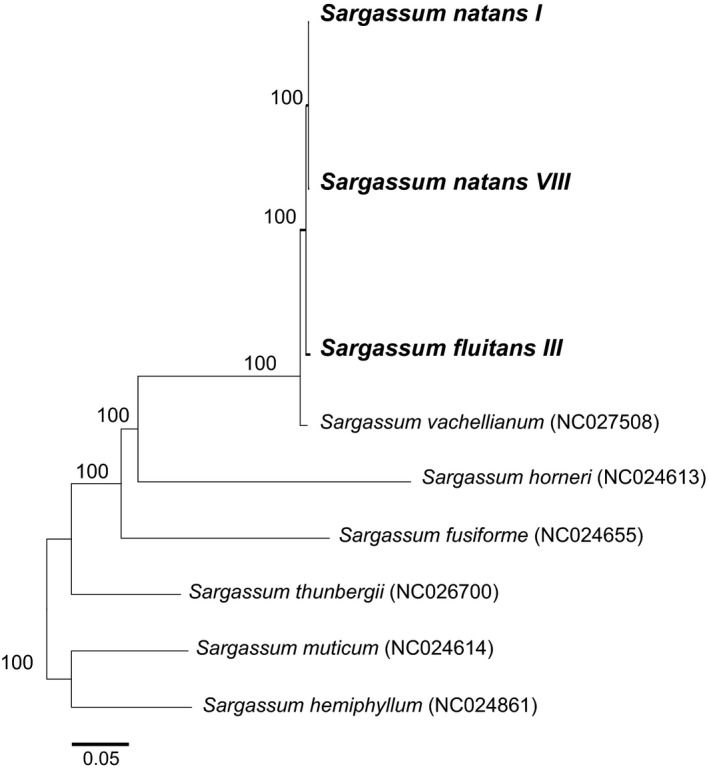
A maximum‐likelihood‐inferred phylogeny of mitogenomes from all available Sargassum species alongside the holopelagic species sequenced in this study (in bold). Scale bar represents evolutionary distance, and numbers at the nodes represent bootstrap confidence values. GenBank numbers follow the names of published mitogenomes
Table 1.
Nucleotide differences and corresponding amino acid changes across the complete mitogenomes of the three holopelagic Sargassum forms
| Genomic position | Locus | Nucleotide | Amino acid | ||
|---|---|---|---|---|---|
| Sargassum fluitans III | Sargassum natans I | S. natans VIII | |||
| 1279 | 23S rRNA | C | T | T | – |
| 1883 | 23S rRNA | C | T | C | – |
| 2658 | 23S rRNA | A | G | G | – |
| 2662 | Intergenic Region | C | T | T | – |
| 2688 | Intergenic Region | C | T | T | – |
| 2691 | Intergenic Region | G | T | T | – |
| 2714 | Intergenic Region | G | T | T | – |
| 2715 | Intergenic Region | A | T | T | – |
| 2716 | Intergenic Region | T | A | A | – |
| 2717 | Intergenic Region | T | A | A | – |
| 2718 | Intergenic Region | A | T | T | – |
| 2719 | Intergenic Region | A | C | C | – |
| 4629 | rpl6 | C | T | T | Ala > Ala |
| 4928 | rps2 | C | T | T | Pro > Pro |
| 5147 | rps2 | C | T | T | Thr > Thr |
| 5424 | rps4 | G | A | A | Ala > Ala |
| 5470 | rps4 | T | C | C | Leu > Leu |
| 5491 | rps4 | A | G | G | Ile > Val |
| 5832 | rps4 | C | T | T | Pro > Pro |
| 6097 | Intergenic Region | T | C | C | – |
| 6805 | nad1 | T | C | C | Val > Val |
| 7033 | nad1 | C | T | T | Arg > Arg |
| 7237 | tatC | T | A | A | Ile > Phe |
| 7720 | tatC | G | A | A | Leu > Leu |
| 7837 | Intergenic Region | A | T | T | – |
| 7903 | trnW(cca) | A | G | G | – |
| 8090 | orf39 | T | C | C | Phe > Ser |
| 8250 | trnQ(uug) | G | A | A | – |
| 8438 | rps12 | G | A | A | Ala > Thr |
| 8857 | rps7 | A | G | G | Ser > Gly |
| 9030 | rps7 | A | T | T | Ile > Ile |
| 9153 | rps7 | A | G | G | Ala > Ala |
| 9863 | rpl14 | C | A | A | Val > Val |
| 10018 | rpl14 | C | T | T | Ser > Leu |
| 10313 | rpl5 | C | C | T | Pro > Ser |
| 10783 | orf129 | C | A | A | Glu > Stop |
| 12021 | rps3 | A | T | T | Ser > Ser |
| 12429 | rps19 | G | G | T | Gln > Lys |
| 12821 | rpl2 | T | C | C | Gly > Gly |
| 12878 | rpl2 | G | T | T | Ala > Ala |
| 13502 | rps13 | G | G | T | Ala > Ser |
| 13537 | rps13 | T | C | C | Phe > Phe |
| 13583 | rps13 | G | C | C | Val > Leu |
| 14111 | rps11 | T | C | C | Val > Val |
| 14466 | cox3 | A | G | G | Leu > Leu |
| 14516 | cox3 | G | A | G | Gly > Asp > Gly |
| 15108 | cox3 | G | A | A | Gly > Gly |
| 15249 | Intergenic Spacer | G | C | C | – |
| 17135 | nad2 | C | T | T | Val > Val |
| 18237 | cox1 | T | C | C | Phe > Phe |
| 18645 | cox1 | G | T | T | Thr > Thr |
| 19527 | Intergenic Spacer | T | C | C | – |
| 19698 | nad9 | T | C | C | Tyr > His |
| 20527 | cob | T | C | C | Gly > Gly |
| 21649 | cox2 | A | G | G | Leu > Leu |
| 21693 | cox2 | C | G | G | Thr > Ser |
| 21780 | cox2 | G | C | C | Gly > Ala |
| 21940 | cox2 | A | G | G | Leu > Leu |
| 22037 | cox2 | C | T | T | Leu > Leu |
| 22216 | cox2 | A | T | T | Ser > Ser |
| 22453 | cox2 | C | T | T | Thr > Thr |
| 22469 | cox2 | G | A | A | Glu > Lys |
| 22474 | cox2 | T | C | C | Ser > Ser |
| 23156 | cox2 | C | T | T | Pro > Ser |
| 23290 | cox2 | G | A | A | Val > Val |
| 23294 | cox2 | T | G | G | Tyr > Asp |
| 23577 | cox2 | G | A | A | Arg > Lys |
| 23856 | cox2 | T | C | C | Leu > Pro |
| 24568 | cox2 | T | A | A | Asp > Glu |
| 24665 | nad4 | G | A | A | Gly > Glu |
| 24706 | nad4 | T | G | G | Ser > Ala |
| 24861 | nad4 | C | T | T | Thr > Thr |
| 26136 | trnl(uau) | T | A | A | – |
| 26272 | nad5 | T | C | C | Cys > Cys |
| 26536 | nad5 | C | T | T | Phe > Phe |
| 26926 | nad5 | A | T | T | Val > Val |
| 26962 | nad5 | T | A | A | Ala > Ala |
| 27508 | nad5 | C | T | T | Pro > Pro |
| 28323 | nad6 | G | A | A | Ala > Ala |
| 28444 | nad6 | T | T | A | Phe > Ile |
| 28463 | nad6 | C | G | G | Thr > Ser |
| 28656 | nad6 | C | T | T | Thr > Thr |
| 28890 | nad6 | A | G | G | Leu > Leu |
| 28895 | nad6 | T | G | G | Leu > Trp |
| 29053 | nad11 | T | C | C | Asn > Asn |
| 29299 | nad11 | A | T | T | Leu > Phe |
| 29518 | nad11 | C | T | T | Cys > Cys |
| 29976 | rps14 | G | A | A | Ser > Asn |
| 30082 | rps14 | G | A | A | Arg > Arg |
| 30958 | rps10 | T | A | A | Leu > Leu |
| 31048 | rps10 | C | T | T | Ser > Ser |
| 31120 | rps10 | G | C | C | Lys > Asn |
| 31578 | Intergenic Spacer | T | A | A | – |
| 31920 | 16S rRNA | A | A | C | – |
| 32415 | 16S rRNA | C | T | T | – |
| 33027 | 16S rRNA | G | A | A | – |
| 33828 | nad7 | C | A | A | Leu > Leu |
| 34589 | nad7 | G | A | A | Stop > Stop |
Shaded areas highlight differences between S. natans I and VIII.
Sequencing partial cox2 and cox3 genes for 71 different holopelagic Sargassum individuals revealed no polymorphisms within forms at the nucleotide sites that differentiated the two S. natans from each other or at sites that distinguished both S. natans from S. fluitans III; that is, the differences between forms and species appear to be fixed. Further interrogating our genomic datasets, we also could not detect differences between the three Sargassum forms using barcoding genes often used in phylogenetic or population genetic studies: nuclear genes 18S rRNA, Internal Transcribed Spacer 2 (ITS‐2) and 5.8S rRNA, or chloroplast genes ribulose‐1,5‐bisphosphate carboxylase/oxygenase large subunit—intergenic spacer—ribulose‐1,5‐bisphosphate carboxylase/oxygenase small subunit (rbcL—intergenic spacer—rbcS). Both nuclear 18S and 5.8S rRNA genes were identical across the forms, while the intergenic spacer between rbcL and rbcS and three sites in the rbcL gene distinguished S. fluitans from S. natans, but could not differentiate between the two S. natans forms. ITS‐2 genes for S. natans I and S. fluitans III were identical, while S. natans VIII showed a single bp difference. Across the gold‐standard barcoding gene, cox1, S. fluitans differed from the S. natans forms at only two sites, and the two S. natans forms were identical.
In addition to comparisons between mitogenomes and nuclear marker genes used in other studies targeting Sargassum, we also compared the chloroplast coding regions that we were able to recover from mapping reads from our six representative samples against the full‐length chloroplast genome of S. horneri as a reference (Liu & Pang, 2015). Near‐complete chloroplast genomes were recovered from one of the S. natans VIII samples (C6) and the S. natans I sample (C2) and included all of the coding regions reported in S. horneri. Table S2 lists all the coding regions found in S. horneri and their status (i.e., level of completeness) in our reference Sargassum samples. Polymorphic sites occurred in the following S. natans I and S. natans VIII genes (indicated with a number in Table S2 corresponding to a polymorphic location in a gene): light‐independent protochlorophyllide reductase subunit B gene (chlB), clpC, conserved hypothetical gene (orf 219), ribosomal protein L2 (rpl2), DNA‐directed RNA polymerase subunit beta (rpoB), DNA‐directed RNA polymerase subunit C1 (rpoC1), DNA‐directed RNA polymerase subunit C2 (rpoC2), thiazole synthase (thiG), and conserved hypothetical gene (ycf19) coding regions. Additional polymorphic sites were detected in the 23S rRNA gene, as well as the tRNA coding region for tRNA‐Threonine and one copy of the tRNA‐Methionine. In addition to these variable sites—we discovered candidate polymorphisms that may reveal population‐level differences in the Photosystem I iron–sulfur center (psaC: S. fluitans III), 23S rRNA (S. natans VIII), and tRNA‐Isoleucine (S. fluitans III) genes, but these particular variations occurred in regions of low coverage and thus need to be confirmed with additional sequencing. Overall, we detected 12 genes that contained variable regions between S. natans VIII and S. natans I samples, 17 genes between S. natans VIII and S. fluitans III, and 19 genes between S. natans I and S. fluitans III (Table S2).
For chloroplast genomes that were most complete S. natans VIII (C6) and the S. natans I (C2), gene synteny and content were identical to S. horneri with 173 genes (Table S2), including six coding for rRNA genes (two copies each) and 28 coding for tRNA genes (with multiple copies of some, designated with a “1” or a “2” in Table S2). As described extensively in Liu and Pang (2015), there is a high degree of conservation in chloroplast DNA among the brown algae and our results were consistent with this finding.
4. Discussion
Despite massive strandings of Sargassum on beaches in the Caribbean, Brazil, and Africa during the last several years, it has not been clear what species are involved in these events or where the accumulations originated. We sampled from beaches and waters of the Caribbean Sea, as well as from large accumulations in the open Atlantic, where our net tows revealed unusually high concentrations of over 24,000 kg/km2; satellite data supported the idea that concentrations we sampled were part of a larger accumulation that extended from the Caribbean toward the NERR (Figure 1). Our molecular results confirmed that S. natans VIII is causing the large accumulations and associated strandings (a.k.a. golden tides) and that it is genetically distinct from the S. natans I and S. fluitans III reported from the Atlantic in the past (Schell et al., 2015). Based on morphological features of archived specimens of Parr's original type material, we confirmed that the accumulating form encountered in 2014 and 2015 is the formerly rare S. natans VIII, not the better‐known S. fluitans III that is morphologically similar. S. natans VIII was the dominant form of Sargassum collected in 2014 and 2015 from cruises and beach collections used in our molecular analyses (Figure 1).
Taxonomic problems within the genus Sargassum C. Agardh are broadly reported in the literature due to the morphological variability of the genus and the fact that standard marker genes (ITS2, rbcLS, cox3, mtsp, etc.) fail to resolve relationships between the closely related members of the holopelagic species. While cox3 was previously suggested as a possible marker to delineate Sargassum species (Camacho et al., 2014), existing published primers target a portion of this gene that fails to differentiate between the holopelagic members of the subgenus Sargassum including S. natans VIII responsible for the recent strandings. As limited information is available from public databases for the regions of the cox2 and cox3 genes covered by our new primers, it is difficult to predict their utility to differentiate between other Sargassum subgenera. However, our cox3 primer set has an identical match to S. vachellianum, for example, so may be able to differentiate between other related benthic species. Limited application of our cox3 primers to benthic Sargassum spp. revealed that the primers may also differentiate between benthic Sargassum spp. from the Caribbean (Amaral‐Zettler, unpublished observation). Our cox2 primers, on the other hand, have at least three mismatches in either or both the forward or reverse priming region in benthic Sargassum spp. with available cox2 genes for comparison, so are unlikely to be useful for population studies beyond the holopelagic forms.
Due to limitations in using single marker genes for comparative brown algal phylogenetic studies, investigators are increasingly turning to comparative genomics of organelles as tools to differentiate between closely related taxa. Our study is the first to demonstrate genetic variability between the different morphotypes of Sargassum originally described in the first part of the 20th century. The fact that we were able to confirm that the species involved in the “golden tides” are a genetically distinct and formerly rare form of holopelagic Sargassum is important because it suggests that the absolute and relative abundance of the different forms of Sargassum is shifting in the NW Atlantic and Caribbean Sea. While the reasons for this shift are unknown, our findings support the existence of a formerly overlooked, genetically distinct population from the NERR that is increasing in abundance and expanding into other areas of the tropical Atlantic.
Since 2011, several groups have tried to model potential sources of the emerging Sargassum accumulations based on known distributions of S. natans and S. fluitans in the Sargasso and Caribbean Seas. Johnson et al. (2013) could not trace a Sargasso Sea source for the Caribbean strandings using satellite‐tracked drifters, but using back‐tracking models argued for a source in the NERR. Likewise, modeling work of Gower, Young, and King (2013) suggested a separate origin to the 2011 Sargassum strandings distinct from earlier satellite detections in 2005 and 2003‐2005 where accumulations were localized to the Gulf of Mexico. As satellite observations cannot differentiate between the distinct species of Sargassum, much confusion originated from these original publications regarding the probable source populations responsible for the accumulations. Our results, coupled with the modeling results, as well as visual sightings of large mats of Sargassum in the South Atlantic off of Africa and Brazil (De Széchy et al., 2012) support the hypothesis that a genetically distinct population of S. natans VIII always occurred in the NERR but never experienced the proper set of conditions to form large accumulations like the ones that are causing the recent strandings. Parr (1939) consistently reported S. natans VIII from the Caribbean, but always in small amounts. Our confirmation of a distinct population and the scale of recent strandings pose the questions: What is causing this previously rare taxon to expand its range and dominate areas where it was formerly rare? Are we witnessing a regime shift in the NERR and North Atlantic ecosystems?
The ability to accurately identify the type of Sargassum is important as we start to address these questions, but recent publications disagree on morphological characters that differentiate between Sargassum forms (De Széchy et al., 2012), such as the presence or absence of thorns on the axes as a character differentiating S. fluitans from S. natans (see Schell et al., 2015: Figure 1b vs. De Széchy et al., 2012: Figure 4). Sargassum fluitans III has a range that overlaps S. natans VIII and the two can be readily confused; even with considerable expertise of the scientists identifying our samples using morphological characters alone, our molecular results showed that some specimens had been misclassified, highlighting the importance of genetic analyses to confirm the identity of these closely related species. The primers and methods described in this study address this need by providing accurate and nonsubjective identification of the holopelagic species.
Despite consistent differences at the mitogenome and marker gene levels, our molecular results demonstrate that overall, the holopelagic Sargassum species exhibit remarkable genetic similarity. Likewise, differences in chloroplast coding regions are modest at best. In their comparative chloroplast genomics of four brown algae including S. horneri, Fucus vesiculosus, Saccharina japonica, and Ectocarpus siliculosus, Liu and Pang (2015) found that despite a smaller chloroplast genome size, S. horneri shared all the same genes and gene order (synteny) as F. vesiculosus. A list of all the coding regions in the chloroplast genome for S. natans I and VIII can be found in Table S2. Deeper sequencing is required to interrogate the variation in intergenic spacer regions of holopelagic Sargassum chloroplast genomes, as well as nuclear genomes overall and should be the focus of future efforts.
Recent studies report temporal changes in the richness of Sargassum's attached and mobile faunal communities (Huffard, Von Thun, Sherman, Sealey, & Smith, 2014) and in the relative proportions of the different forms of the two dominant Sargassum species S. fluitans and S. natans (Schell et al., 2015). We do not know what is causing these changes, whether it is part of a natural cycle or represents a shift in open‐ocean ecosystems due to anthropogenic impacts such as nutrient input and climate change. Something is leading to increased abundance and dominance of S. natans VIII, a form that was previously rare, and there is evidence that the different forms have different ecologies and associated fauna (Schell et al., 2015). This suggests that the changes in Sargassum populations and distribution will have corresponding impacts on both open‐ocean communities and the coastal regions where golden tides occur. Understanding of the holopelagic Sargassum ecosystem and environmental costs of recent and future golden tide events cannot proceed without proper species identification of the Sargassum involved. Our PCR‐screening approach of cox2 and cox3 genes offers a straight‐forward means of confirming morphological identifications of the holopelagic Sargassum species.
This study confirms that not only the amount, but also the population structure of Sargassum is changing and that recent Sargassum strandings represent a major biological shift in open‐ocean and coastal ecosystems around the Atlantic. Our study identifies the Sargassum responsible as a formerly rare form and generates important questions about the diversification of holopelagic species possessing such limited variability at the genomic scales documented in our study: Do they represent recently diverged lineages? The case of Sargassum is interesting in that, despite clear morphological, ecological, and distribution differences, the evolutionary distances between S. natans and S. fluitans observed in our phylogenetic analysis are very small compared to their benthic counterparts and suggest that they are relatively new species. Current theories argue that human‐mediated environmental changes such as increases in temperature and nutrient loads are leading to blooms of previously “rare” genotypes already existing in a population. The increase in abundance along with asexual reproduction and gas vesicles in the holopelagic forms enhance their ability to be transported long distances and may increase their invasion potential (Paula & Eston, 1987). With the new repertoire of genetic information now available through our efforts, we are poised to expand our understanding of population‐level variation in holopelagic Sargassum and its associated taxa.
Conflict of Interests
We have no competing interests.
Data Accessibility
GenBank accessions are KY084907–KY084912 for Sargassum mitochondrial genomes. KY126998–KY127307 for cox2 and cox3 population sets and KY206015–KY206739 for chloroplast genes from Sargassum references.
Supporting information
Acknowledgments
Students and staff of SEA Semester academic programs (www.sea.edu) helped collect open‐ocean samples aboard the vessel SSV Corwith Cramer, and Dr. Maureen Conte provided samples collected off of Bermuda. We thank Greg Boyd for provided field assistance, A.N.S. Siuda for discussions and sailing as co‐Chief Scientist on cruise C‐241, Chuanmin Hu and Mengqiu Wang for satellite data, and Patrick Sweeney and Eric Lazo‐Wasem at the Yale Peabody Museum for access to Parr's original specimens.
Amaral‐Zettler, L. A. , Dragone, N. B. , Schell, J. , Slikas, B. , Murphy, L. G. , Morrall, C. E. and Zettler, E. R. (2017), Comparative mitochondrial and chloroplast genomics of a genetically distinct form of Sargassum contributing to recent “Golden Tides” in the Western Atlantic. Ecology and Evolution, 7: 516–525. doi: 10.1002/ece3.2630
Funding Information
This work was supported by a US National Science Foundation (NSF) collaborative grant to LAA‐Z (OCE‐1155571) and ERZ (OCE‐1155379), and an NSF TUES grant (DUE‐1043468) to LAA‐Z and ERZ.
References
- Bi, Y. , & Zhou, Z. (2016). Complete mitochondrial genome of the brown alga Sargassum vachellianum (Sargassaceae, Phaeophyceae). Mitochondrial DNA. Part A, 27, 2796–2797. [DOI] [PubMed] [Google Scholar]
- Camacho, O. , Mattio, L. , Draisma, S. , Fredericq, S. , & Diaz‐Pulido, G. (2014). Morphological and molecular assessment of Sargassum (Fucales, Phaeophyceae) from Caribbean Colombia, including the proposal of Sargassum giganteum sp. nov., Sargassum schnetteri comb. nov. and Sargassum section Cladophyllum sect. nov . Systematics and Biodiversity, 13, 105–130. [Google Scholar]
- Cheang, C. C. , Chu, K. H. , Fujita, D. , Yoshida, G. , Hiraoka, M. , Critchley, A. , … Ang, P. O. (2010). Low genetic variability of Sargassum muticum (Phaeophyceae) revealed by a global analysis of native and introduced populations. Journal of Phycology, 46, 1063–1074. [Google Scholar]
- Cho, S. M. , Lee, S. M. , Ko, Y. D. , Mattio, L. , & Boo, S. M. (2012). Molecular systematic reassessment of Sargassum (Fucales, Phaeophyceae) in Korea using four gene regions. Botanica Marina, 55, 473–484. [Google Scholar]
- Coston‐Clements, L. , Settle, L. , Hoss, D. , & Cross, F. (1991) Utilization of the Sargassum habitat by marine invertebrates and vertebrates: A review (32 pp.). Beaufort, NC: National Marine Fisheries Service, NOAA. [Google Scholar]
- Dawes, C. J. , & Mathieson, A. C. (2008). The seaweeds of Florida (592 pp.). Gainesville, FL: University Press of Florida. [Google Scholar]
- De Széchy, M. T. M. , Guedes, P. M. , Baeta‐Neves, M. H. , & Oliveira, E. N. (2012). Verification of Sargassum natans (Linneaus) Gaillon (Heterokontophyta: Phaeophyceae) from the Sargasso Sea off the coast of Brazil, western Atlantic Ocean. Check List, 8, 638–641. [Google Scholar]
- Dixon, R. R. , Mattio, L. , Huisman, J. M. , Payri, C. E. , Bolton, J. J. , & Gurgel, C. F. D. (2014). North meets south—Taxonomic and biogeographic implications of a phylogenetic assessment of Sargassum subgenera Arthrophycus and Bactrophycus (Fucales, Phaeophyceae). Phycologia, 53, 15–22. [Google Scholar]
- Gower, J. , Young, E. , & King, S. (2013). Satellite images suggest a new Sargassum source region in 2011. Remote Sensing Letters, 4, 764–773. [Google Scholar]
- Guiry, M. D. , & Guiry, G. M. (2016). AlgaeBase. World‐wide Electronic Publication, National University of Ireland; Retrieved from http://www.algaebase.org [Google Scholar]
- Huffard, C. L. , Von Thun, S. , Sherman, A. D. , Sealey, K. , & Smith, K. L. (2014). Pelagic Sargassum community change over a 40‐year period: Temporal and spatial variability. Marine Biology, 161, 2735–2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, D. , Ko, D. , Franks, J. , Moreno, P. , & Sanchez‐Rubio, G. (2013) The Sargassum invasion of the eastern Caribbean and dynamics of the equatorial North Atlantic. Proceedings of the 65th Gulf and Caribbean Fisheries Institute, 65, 102–103. [Google Scholar]
- Laffoley, D. D. A. , Roe, H. S. J. , Angel, M. V. , Ardron, J. , Bates, N. R. , Boyd, I. L. , … Vats, V. (2011) The protection and management of the Sargasso Sea: The golden floating rainforest of the Atlantic Ocean. Summary science and supporting evidence case (44 pp.). Sargasso Sea Alliance. [Google Scholar]
- Liu, F. , & Pang, S. (2014). Complete mitochondrial genome of the invasive brown alga Sargassum muticum (Sargassaceae, Phaeophyceae). Mitochondrial DNA, 27, 1129–1130. [DOI] [PubMed] [Google Scholar]
- Liu, F. , & Pang, S. (2015). Chloroplast genome of Sargassum horneri (Sargassaceae, Phaeophyceae): Comparative chloroplast genomics of brown algae. Journal of Applied Phycology, 28, 1419–1426. [Google Scholar]
- Liu, F. , Pang, S. , & Chen, W. (2016). Complete mitochondrial genome of the brown alga Sargassum hemiphyllum (Sargassaceae, Phaeophyceae): Comparative analyses. Mitochondrial DNA, 27, 1468–1470. [DOI] [PubMed] [Google Scholar]
- Liu, F. , Pang, S. , Li, X. , & Li, J. (2015). Complete mitochondrial genome of the brown alga Sargassum horneri (Sargassaceae, Pharophyceae): Genome organization and phylogenetic analyses. Journal of Applied Phycology, 27(1), 469–478. [Google Scholar]
- Liu, F. , Pang, S. , & Luo, M. (2014). Complete mitochondrial genome of the brown alga Sargassum fusiforme (Sargassaceae, Phaeophyceae): Genome architecture and taxonomic consideration. Mitochondrial DNA, 27, 1158–1160. [DOI] [PubMed] [Google Scholar]
- Mattio, L. , Bolton, J. J. , & Anderson, R. J. (2015). Contribution to the revision of the genus Sargassum (Fucales, Phaeophyceae) in Madagascar using morphological and molecular data. Cryptogamie Algologie, 36, 143–169. [Google Scholar]
- Mattio, L. , & Payri, C. E. (2010). Assessment of five markers as potential barcodes for identifying Sargassum subgenus Sargassum species (Phaeophyceae, Fucales). Cryptogamie Algologie, 31, 467–485. [Google Scholar]
- Mattio, L. , Zubia, M. , Loveday, B. , Crochelet, E. , Duong, N. , Payri, C. E. , … Bolton, J. J. (2013). Sargassum (Fucales, Phaeophyceae) in Mauritius and Reunion, western Indian Ocean: Taxonomic revision and biogeography using hydrodynamic dispersal models. Phycologia, 52, 578–594. [Google Scholar]
- Maurer, A. S. , De Neef, E. , & Stapleton, S. (2015). Sargassum accumulation may spell trouble for nesting sea turtles. Frontiers in Ecology and the Environment, 13(7), 394–395. [Google Scholar]
- Minoche, A. E. , Dohm, J. C. , & Himmelbauer, H. (2011). Evaluation of genomic high‐throughput sequencing data generated on Illumina HiSeq and genome analyzer systems. Genome Biology, 12, 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parr, A. E. (1939). Quantitative observations on the pelagic Sargassum vegetation of the western North Atlantic: Bulletin of the Bingham Oceanographic Collection, Yale Univ, 6, p. 52. [Google Scholar]
- Paula, J. , & Eston, V. R. (1987). Are there other Sargassum species potentially as invasive as S. muticum . Botanica Marina, 30, 405–410. [Google Scholar]
- Schell, J. M. , Goodwin, D. S. , & Siuda, A. N. S. (2015). Recent Sargassum inundation events in the Caribbean: Shipboard observations reveal dominance of a previously rare form. Oceanography, 28, 8–10. [Google Scholar]
- Smetacek, V. , & Zingone, A. (2013). Green and golden seaweed tides on the rise. Nature, 504, 84–88. [DOI] [PubMed] [Google Scholar]
- Stamatakis, A. (2006). RAxML‐VI‐HPC: Maximum likelihood‐based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics, 22, 2688–2690. [DOI] [PubMed] [Google Scholar]
- Winge, O. (1923). The Sargasso Sea, its boundaries and vegetation. Report on the Danish Oceanographical Expeditions 1908‐10 to the Mediterranean and Adjacent Seas (33 pp.). Carlsberg Physiological Laboratory, Copenhagen, Denmark. [Google Scholar]
- Zettler, E. R. , Mincer, T. J. , & Amaral‐Zettler, L. A. (2013). Life in the “Plastisphere”: Microbial communities on plastic marine debris. Environmental Science Technology, 47, 7137–7146. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
GenBank accessions are KY084907–KY084912 for Sargassum mitochondrial genomes. KY126998–KY127307 for cox2 and cox3 population sets and KY206015–KY206739 for chloroplast genes from Sargassum references.


