Abstract
Sex differentiation of flower buds is an important developmental process that directly affects fruit yield of cucumber (Cucumis sativus L.). Plant hormones, such as gibberellins (GAs) and ethylene can promote development of male and female flowers, respectively, however, the regulatory mechanisms of GA-induced male flower formation and potential involvement of ethylene in this process still remain unknown. In this study, to unravel the genes and gene networks involved in GA-regulated cucumber sexual development, we performed high throughout RNA-Seq analyses that compared the transcriptomes of shoot tips between GA3 treated and untreated gynoecious cucumber plants. Results showed that GA3 application markedly induced male flowers but decreased ethylene production in shoot tips. Furthermore, the transcript levels of M (CsACS2) gene, ethylene receptor CsETR1 and some ethylene-responsive transcription factors were dramatically changed after GA3 treatment, suggesting a potential involvement of ethylene in GA-regulated sex expression of cucumber. Interestingly, GA3 down-regulated transcript of a C-class floral homeotic gene, CAG2, indicating that GA may also influence cucumber sex determination through an ethylene-independent process. These results suggest a novel model for hormone-mediated sex differentiation and provide a theoretical basis for further dissection of the regulatory mechanism of male flower formation in cucumber.
Statement: We reveal that GA can regulate sex expression of cucumber via an ethylene-dependent manner, and the M (CsACS2), CsETR1, and ERFs are probably involved in this process. Moreover, CAG2, a C-class floral homeotic gene, may also participate in GA-modulated cucumber sex determination, but this pathway is ethylene-independent.
Keywords: cucumber, ethylene, gibberellin, sex expression, transcriptome
Introduction
Cucumber (Cucumis sativus L.) is a typical monoecious plant with distinct male and female flowers, and has been served as a model system for studying physiological and molecular aspects of sex determination in plants (Malepszy and Niemirowicz-Szczytt, 1991; Bai and Xu, 2013). During the early stages of cucumber flower development, both stamen primordia and carpel primordia are initiated, however, sex differentiation occurs just after the hermaphroditic stage, subsequently, female or male flower is formed and developed through the selective developmental arrest of stamen or carpel, respectively (Bai et al., 2004).
Sex differentiation in cucumber is mainly determined by F, M, and A genes. Among them, F (CsACS1G) and M (CsACS2) genes encoding two ACC synthases (key enzymes in ethylene biosynthetic pathway) govern female sex expression in cucumber, and the F gene promotes female flower development (Trebitsh et al., 1997; Mibus and Tatlioglu, 2004; Knopf and Trebitsh, 2006), while the M gene inhibits stamen development in flower buds (Yamasaki et al., 2001, 2003; Saito et al., 2007; Li et al., 2009, 2012). In contrast, the A gene inhibits female flower development and facilitates male flower formation (Pierce and Wehner, 1990). The interaction of F, M, and A genes eventually determines various sexual phenotypes of cucumber.
In addition to genetic control, sex expression of cucumber can be affected by phytohormones, such as ethylene and GAs. Particularly, ethylene is considered as a potent sex hormone in cucumber that can induce formation of female flowers (Malepszy and Niemirowicz-Szczytt, 1991). Ethylene content in shoot tip of gynoecious cucumber is higher than that of monoecious plant (Rudich et al., 1972; Fujita and Fujieda, 1981; Trebitsh et al., 1987). Treatment with exogenous ethylene or ethylene-releasing reagent can increase the numbers of female and bisexual flowers in monoecious and andromonoecious lines, respectively (MacMurray and Miller, 1968; Iwahori et al., 1969). Until now, the molecular mechanism of ethylene-regulated sex determination of cucumber has been well understood. Except for the F and M genes, other ethylene biosynthetic genes, such as CSACO2 and CSACO3, which encode ACC oxidases, are also involved in sex expression of cucumber, but the transcript levels of CSACO2 and CSACO3 in the shoot tips show a negative correlation with femaleness, indicating an existence of a feedback inhibition mechanism underlying such correlation (Kahana et al., 1999). Overexpression of CsACO2, driven by the AP3 promoter, can arrest the stamen development by inducing chromatin condensation in Arabidopsis (Hao et al., 2003; Duan et al., 2008). Moreover, an ethylene receptor, CsETR1, has been demonstrated to play a key role in stamen arrest in female cucumber flowers through induction of DNA damage (Wang et al., 2010).
Gibberellins, one class of tetracyclic diterpenoid phytohormones, can promote the male tendency in cucumber. GA production in andromonoecious cucumber in higher than that in gynoecious and monoecious plants (Hemphill et al., 1972). Exogenous GA3 application can increase the ratio of maleness to femaleness in monoecious cucumber and induce the formation of male flowers in gynoecious plants (Wittwer and Bukovac, 1962; Pike and Peterson, 1969). In addition, GA signaling pathway is involved in stamen and anther development in hermaphroditic plants, such as Arabidopsis and rice (Oryza sativa) (Cheng et al., 2004; Fleet and Sun, 2005; Aya et al., 2009; Sun, 2010, 2011; Plackett et al., 2011; Song et al., 2013). In this pathway, GA first binds with GID1 receptor and promotes the interaction between GID1 and DELLA proteins (repressors of GA signaling), leading to a rapid degradation of DELLA proteins by an ubiquitin-proteasome pathway, and the proteolysis of DELLA proteins releases their inhibitory effect on GA action and allows plant growth and development (Fleet and Sun, 2005; Murase et al., 2008; Harberd et al., 2009; Sun, 2010, 2011; Plackett et al., 2014). GAMYB is a positive regulator in GA signaling pathway and acts as an important downstream gene of DELLA proteins (Olszewski et al., 2002; Achard et al., 2004; Fleet and Sun, 2005). GA can induce GAMYB transcript through degradation of DELLA proteins, resulting in an enhanced flowering and anther development (Achard et al., 2004). In our previous studies, we identified two GA signaling genes, CsGAIP and CsGAMYB1, which belong to DELLA and GAMYB family, respectively. Both of them were predominantly expressed in the male specific organs during cucumber flower development. CsGAIP can inhibit stamen development through transcriptional repression of B class floral homeotic genes APETALA3 (AP3) and PISTILLATA (PI) in Arabidopsis (Zhang et al., 2014a). However, whether CsGAIP is involved in GA-regulated sex determination in cucumber flowers is still unknown. Notably, CsGAMYB1 can also mediate sex expression of cucumber. Knockdown of CsGAMYB1 in cucumber results in decreased ratio of nodes with male to female flowers (Zhang et al., 2014b). Despite the current knowledge of GA-regulated sex expression of cucumber, the precise regulatory pathway in this complex process remains elusive.
Although both ethylene and GA can mediate sex expression of cucumber, their regulatory functions appear to be opposite. Atsmon and Tabbak (1979) interpreted that the GA-regulated sex differentiation has no effect on ethylene production, and there is a balance in the content of ethylene and GA in controlling the sex expression of cucumber. However, Yin and Quinn (1995) proposed a “one-hormone hypothesis” which posited that ethylene plays a dominant role in cucumber sex determination and GA may regulate the maleness through inhibiting ethylene production. However, our previous studies demonstrated that GA-CsGAMYB1 signaling could regulate sex differentiation in cucumber through an ethylene-independent process (Zhang et al., 2014b). Therefore, a potential crosstalk between GA and ethylene pathways that determine sex expression in cucumber still remains unclear.
Besides, members of the MADS-box gene family can also regulate the sexual development in cucumber. AGAMOUS (AG), the C-class floral homeotic gene, specifies stamen and carpel identity (Lohmann and Weigel, 2002). There are three AG homologs in cucumber, CAG1, CAG2, and CAG3, in which CAG1 and CAG3 are expressed in both stamen and carpel, while CAG2 is particularly restricted to the carpel. However, the expression levels of these three genes do not appear to be mediated by ethylene and GA (Kater et al., 1998; Perl-Treves et al., 1998). Moreover, ERAF17, an another MADS-box gene, can be induced by ethylene and may be involved in female flowers formation in cucumber (Ando et al., 2001).
In our study, in order to understand the genes and gene networks that may be involved in GA-modulated cucumber sex determination, we performed RNA-Seq analyses to compare the transcriptomes of shoot apices between GA3-treated and control gynoecious cucumber plants. GA3 application induced male flowers but reduced ethylene production in the shoot apices. Notably, GA-regulated sex differentiation was associated with the changes in transcript levels of M (CsACS2) gene, ethylene receptor ETR1, ethylene-responsive transcription factors, and CAG2 (a C-class floral homeotic gene), suggesting a potential involvement of both ethylene-dependent and -independent processes in GA-mediated cucumber sexual development. Thus, our results built a foundation for dissecting the molecular mechanism of male flower development in cucumber.
Materials and Methods
Plant Materials and Growth Conditions
A gynoecious cucumber (C. sativus L.) line 13-3B was used in this study. The seeds were germinated on wet filter paper in a Petri dish at 28°C in dark overnight. Then the resulting seedlings were grown in a growth chamber under 16 h/8 h with 25°C/18°C in day/night, respectively. Upon two true-leaf stage, plants were transferred to a greenhouse in the experimental field of the Northwest A&F University. Pest control and water management were carried out according to standard practices.
Exogenous GA3 Treatment
For male flowers induction in the gynoecious cucumber 13-3B, 1000 ppm GA3 (dissolved in 0.1% ethanol) or deionized water with 0.1% ethanol (Control) were applied by foliar spray for three times at 7 day intervals, starting when the first true leaf was approximately 2.5 cm in diameter. The sex of the flowers on each node of the main stem was recorded until anthesis of flowers on node 25.
In addition, ethylene production in shoot apices was measured after 7 days of the third GA3 treatment. And the RNA-Seq analyses were performed in shoot apices from the cucumber plants firstly treated with GA3 for 6 h, 12 h and the Control, respectively. GA3 was acquired from Sigma-Aldrich Chemical Co. (Shanghai, China).
Quantification of Ethylene
The ethylene production was measured by gas chromatography as described previously with some modifications (Zhang et al., 2014b). In brief, the excised shoot apices from cucumber plants treated with exogenous GA3 and the Control were enclosed in 10 mL vessels after weighing and sealed with rubber stoppers. After incubation at 25°C for 16 h, 1 mL of head gas was withdrawn from each vessel using a syringe and injected into a gas chromatograph (GC-9A, Shimadzu, Japan) equipped with a hydrogen FID and an activated alumina column for the measurement of ethylene production. Standard ethylene gas was used for calibrating the instrument. Amount of ethylene was calculated per 1 g fresh weight and per hour.
RNA Extraction and Quality Test
The shoot apices from the gynoecious cucumber plants were collected at 6 and 12 h after the first treatment with GA3 and the Control. Samples were immediately frozen in liquid nitrogen and stored at -80°C for RNA-Seq analyses. Total RNA was isolated using the RNA extraction kit (Promega, USA). RNA was checked by RNase-free agarose gel electrophoresis to avoid possible degradation and contamination, and then examined using the NanoPhotometer spectrophotometer (IMPLEN, Westlake Village, CA, USA) for RNA purity. RNA concentration and integrity were measured and assessed using the Qubit RNA Assay Kit in Qubit 2.0 Flurometer (Life Technologies, Carlsbad, CA, USA) and RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, CA, USA), respectively.
Digital Gene Expression (DGE) Library Construction and Sequencing
Digital gene expression libraries were constructed using the NEBNext Ultra Directional RNA Library Prep Kit for Illumina (NEB, Ispawich, USA) following instructions of manufacturer and six index codes were added to attribute sequences to various samples (Wang et al., 2009). Briefly, poly (A) mRNA was isolated from 3 μg total RNA using oligo-dT magnetic beads (Life Technologies, Carlsbad, CA, USA), and then broken into short fragments by adding fragmentation buffer. First-strand cDNA was synthesized using random hexamer-primed reverse transcription, followed by the synthesis of the second-strand cDNA using RNase H and DNA polymerase I. After adenylation of the 3′ ends of cDNA fragments, NEBNext adapter oligonucleotides were ligated to prepare for hybridization, and then the cDNA fragments were purified using AMPure XP system (Beckman Coulter, Beverly, MA, USA) to select the fragments of preferentially 150–200 bp in length. The size-selected, adaptor-ligated cDNA fragments were enriched using Phusion High-Fidelity DNA polymerase, Universal PCR primers and Index Primer in the PCR reaction. PCR products were purified with AMPure XP system and library quality was assessed using the Agilent Bioanalyzer 2100 system. At last, the cDNA libraries were sequenced on an Illumina HiSeq 4000 platform using the paired-end technology by Novogene Co. (Beijing, China).
Bioinformatics Analysis of DGE Data
Raw reads were pre-processed to remove low quality sequences (there were more than 50% bases with quality lower than 20 in one sequence), reads with more than 5% N bases (bases unknown) and reads containing adaptor sequences. Then the clean reads were mapped to the cucumber genome (Chinese long) v21 using TopHat (Huang et al., 2009; Trapnell et al., 2009), allowing up to one mismatch. Unigenes mapped by at least one read, in at least one sample, were identified for further analysis.
In this study, samples from three treatments (GA 6 h, GA 12 h, and Control) were prepared for genome-wide expression analyses. Two biological replicates were performed for each treatment, and thus six DGE libraries were sequenced. 44.28–60.29 million raw reads from each library were generated. After removal of low-quality tags and adapter sequences, 42.31–57.63 million high-quality clean reads with a total of 6.35–8.64G bases were obtained. Among these clean reads, the percentage of Q20 (base quality more than 20) and GC was 97.28–97.52% and 43.46–43.63%, respectively (Table 1). Furthermore, we clustered these clean reads into unique tags, which were mapped to the cucumber genome using TopHat (Huang et al., 2009; Trapnell et al., 2009). About 36.28–49.30 million clean reads (85.21–85.74% of total clean reads) from RNA-Seq data in the six libraries were mapped uniquely to the cucumber genome (Table 1).
Table 1.
Summary of the transcriptome assembly.
| Samples | Control_rep1 | Control _rep2 | GA 6 h_rep1 | GA 6 h_rep2 | GA 12 h_rep1 | GA 12 h_rep2 |
|---|---|---|---|---|---|---|
| Raw reads | 50,991,998 | 56,788,784 | 60,288,014 | 58,393,488 | 44,280,996 | 52,072,848 |
| Clean reads (%) | 48,705,848 (95.52) | 54,275,188 (95.57) | 57,631,340 (95.59) | 55,720,738 (95.42) | 42,311,862 (95.55) | 49,993,570 (96.01) |
| Clean bases | 7.31G | 8.14G | 8.64G | 8.36G | 6.35G | 7.50G |
| Q20 (%) | 97.52 | 97.42 | 97.38 | 97.43 | 97.28 | 97.36 |
| GC (%) | 43.63 | 43.60 | 43.51 | 43.53 | 43.46 | 43.54 |
| Mapped clean reads (%) | 42,119,358 (86.48) | 46,783,558 (86.20) | 49,881,555 (86.55) | 48,281,412 (86.65) | 36,728,637 (86.80) | 43,283,227 (86.58) |
| Unique mapped clean reads (%) | 41,622,535 (85.46) | 46,245,871 (85.21) | 49,302,733 (85.55) | 47,674,994 (85.56) | 36,276,717 (85.74) | 42,766,514 (85.54) |
The R package edgeR used to identify the DGEs (Robinson et al., 2010). The expression level of each gene was calculated and normalized to FPKM. The FDR was used to determine the threshold of the P-value in multiple tests. In our study, the FDR < 0.05 and fold change > 2 were used as significance cut-offs of the expression differences.
Furthermore, GO enrichment analysis of DGEs was performed using the GOseq R package (Young et al., 2010). GO terms with corrected P-value < 0.05 were considered significantly enriched by differential expressed genes.
Quantitative Real-Time PCR (qRT-PCR) Validation
Quantitative Real-Time PCR analyses were performed using the independent cucumber shoot apices in the same time point of GA3 application as those used for RNA-Seq. Total RNA was isolated using the RNA extraction kit (Promega, USA), and cDNA was synthesized using the PrimeScript RT reagent Kit (TaKaRa, China). qRT-PCR was carried out using SYBR Premix Ex Taq (TaKaRa, China) on an ABI 7500 Real-Time PCR System (Applied Biosystems, USA). The cucumber α-TUBULIN (TUA) gene was used as an internal control in analyzing gene expression. Three biological replicates were performed for each experiment. The gene specific primers for qRT-PCR are listed in Supplementary Table S5.
Results
Exogenous GA3 Induces Male Flowers Formation and Inhibits Ethylene Production in Gynoecious Cucumber
To verify the effect of GA on sex expression of cucumber, a gynoecious cucumber line 13-3B was treated with GA3 and the sex of flowers on each node was recorded until anthesis of the flowers on node 25 of the main stems. As shown in Figure 1, there were no male flower nodes in the control plants, however, GA3 treatment induced male flowers in the gynoecious cucumber line (Figure 1A), accounting for 51.2% of nodes with male flowers (Figure 1B). Interestingly, formation of male flowers occurred mainly at the lower node positions as compared with the location of female flowers on the main stems (Figure 1A). These observations suggested that exogenous GA can promote male flowers formation in cucumber.
FIGURE 1.
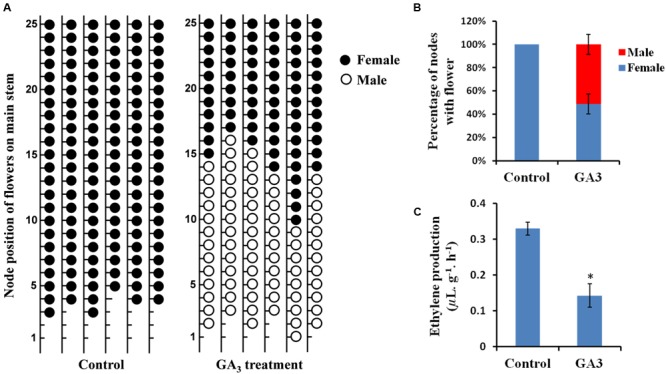
Effects of exogenous GA3 on sex expression and ethylene production in gynoecious cucumber. (A) Sex expression of the flowers on the first 25 nodes of the main stems in gynoecious cucumber plants treated with deionized water (with 0.1% ethanol) (Control) or 1000 ppm GA3. The black and white circles represent female and male flowers, respectively. Lower nodes without circle indicate the vegetative nodes. (B) The percentage of the nodes with female or male flowers up to the 25th node on the main stems of the Control and GA3 treatment lines. Values are the means ± SE from six independent plants. (C) Quantification of ethylene released from shoot tips in gynoecious cucumber plants at 7 days after treatment with GA3 and the Control. Six biological replicates were performed for this experiment. Vertical bars represent the standard errors. The asterisk indicates the significant difference (P < 0.01) between the Control and GA3 treatment lines by Duncan’s test.
It is well known that ethylene can also control sex determination of cucumber (Malepszy and Niemirowicz-Szczytt, 1991; Bai and Xu, 2013). To assess potential involvement of ethylene in GA-regulated sex expression of cucumber, ethylene production in the shoot apices was measured in GA3-treated and control plants. As shown in Figure 1C, ethylene production was significantly decreased after GA3 treatment, suggesting that ethylene might function as a negative factor in GA-regulated male flower formation in cucumber.
Identification of Differentially Expressed Genes (DEGs) in Shoot Apices from the Gynoecious Cucumber Plants Treated with GA3 and the Control
To identify the genes and gene networks that are involved in GA-regulated cucumber sex expression, the genome-wide expression analyses were performed to compare the transcriptome profiles of the shoot apices between the gynoecious cucumber plants treated with GA3 for different time points (6 and 12 h) and the Control through the digital gene expression (DGE) approach (Eveland et al., 2010). Based on deep sequencing, 23,911 unigenes were collected in six libraries. Using fold change > 2 and FDR < 0.05 as the significance cut-offs, we identified 1073 DEGs including 727 up-regulated genes and 346 down-regulated genes after GA3 treatment for 6 h compared with the Control. And we also found that 1590 genes were differentially expressed, in which 765 genes were up-regulated and 825 genes were down-regulated after GA3 treatment for 12 h (Figure 2; Supplementary Tables S1 and S2). Moreover, 594 DEGs containing 303 up-regulated genes (Figure 2A) and 291 down-regulated genes (Figure 2B) were shared in the two sets of transcriptome comparisons.
FIGURE 2.

Venn diagrams of DEGs that were significantly upregulated (A) or downregulated (B) after 6 or 12 h of GA3 treatment.
Verification of RNA-Seq Data by Quantitative Real Time RT-PCR Analyses
To validate the DEGs identified by RNA-Seq, we performed quantitative real time RT-PCR (qRT-PCR) assays using the independent cucumber shoot apices in the same time point after GA3 treatment as those used for RNA-Seq analysis. Twenty DEGs were randomly chosen for qRT-PCR analyses, in which 10 genes including five up-regulated genes and five down-regulated genes were from the set of GA 6 h vs. Control, and the other 10 genes containing five up-regulated genes and five down-regulated genes were from the set of GA 12 h vs. Control. As shown in Figure 3, all the 20 genes showed the similar expression patterns in the qRT-PCR analyses as those in the RNA-Seq data, although the particular values of fold-change were different. The Pearson correlation coefficient between qRT-PCR and RNA-Seq data was 0.975 (P = 3.5E-13), indicating that the RNA-Seq results were highly reliable.
FIGURE 3.
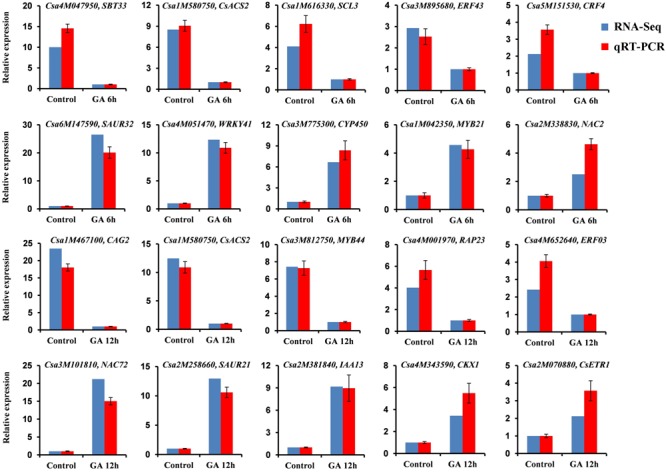
qRT-PCR validation of DEGs identified by RNA-Seq. Twenty DEGs including ten up-regulated genes and ten down-regulated genes from the two sets of transcriptome comparisons (GA 6 h vs. Control and GA 12 h vs. Control) were randomly selected for qRT-PCR confirmation. The blue and red bars represent RNA-Seq and qRT-PCR data, respectively. The cucumber TUA gene was used as an internal control, and these experiments were repeated with three biological samples. Error bars indicate the standard errors.
The M Gene Is Involved in GA-Regulated Sex Differentiation of Cucumber
Given that ethylene production was significantly decreased in the cucumber plants treated with GA3 (Figure 1C), we screened the ethylene biosynthetic genes in the DGEs. We found that the transcript of M (CsACS2) gene encoding an ACC synthase was inhibited in both sets of transcriptome comparisons (GA 6 h vs. Control and GA 12 h vs. Control) (Table 2), and the qRT-PCR verification displayed the same expression pattern (Figure 3). Since the M gene is believed to inhibit stamen development in flower buds (Saito et al., 2007; Li et al., 2009, 2012), we speculated that GA might release the inhibitory effect of the M gene and allow male flowers to develop in cucumber.
Table 2.
List of differentially expressed ethylene biosynthetic genes identified by RNA-Seq in the shoot apices of GA3 treatment plants and the Control.
| Gene ID | Gene annotation | Fold change (GA 6 h/Control) | FDR | Fold change (GA 12 h/Control) | FDR |
|---|---|---|---|---|---|
| Csa1M580750 | M (CsACS2, ACC Synthase 2) | –8.53 | 5.33E-23 | –12.47 | 9.94E-22 |
| Csa6M160180 | CsACO1 (ACC Oxidase 1) | 2.24 | 1.25E-06 | 2.56 | 5.59E-05 |
| Csa6M421630 | CsACO3 (ACC Oxidase 2) | / | / | –2.07 | 6.92E-04 |
The oblique line represents that the gene expression has no change in GA 6 h vs. Control.
In addition, another two ethylene biosynthetic genes, CsACO1 and CsACO3 which encode two ACC oxidases, were also differently expressed in the shoot apices after GA3 treatment. CsACO3 expression was dramatically down-regulated in the set of GA 12 h vs. Control, but not changed in GA 6 h vs. Control. However, there was an increase in the transcript level of CsACO1 in both GA 6 h vs. Control and GA 12 h vs. Control, but the fold change was lower than that of M gene (Table 2). These results suggested that the decreased transcript levels of M and CsACO3 might inhibit ethylene biosynthesis in the cucumber plants treated with GA3. Nonetheless, an increased expression of CsACO1 following GA3 treatment was insufficient to rescue the effect of M and CsACO3 on ethylene production.
The Ethylene-Responsive Transcription Factors and Ethylene Receptor CsETR1 Participate in GA-Modulated Cucumber Sex Expression
To further understand the potential functions of DEGs identified by DGE, GO term enrichment analyses (Corrected P-value < 0.05) were carried out in both sets of RNA-Seq data. We found that the DEGs were markedly enriched in biological process and molecular function (MF) groups. For the biological process category, the most significantly enriched GO terms were “cellular carbohydrate biosynthetic process” (P = 1.1E-02) and “regulation of cellular macromolecule biosynthetic process” (P = 5.8E-04) in GA 6 h vs. Control and GA 12 h vs. Control groups, respectively (Figures 4 and 5). While five GO terms including “oxidoreductase activity, acting on paired donors” (P = 1.1E-02), heme binding (P = 1.1E-02), tetrapyrrole binding (P = 1.5E-02), iron ion binding (P = 2.5E-02), and “sequence-specific DNA binding transcription factor activity” (P = 4.6E-02) in the set of GA 6 h vs. Control (Figure 4) and two terms containing “sequence-specific DNA binding transcription factor activity” (P = 2.2E-03) and “DNA binding” (P = 1.1E-02) in the GA 12 h vs. Control group (Figure 5) were detected in the MF category. Furthermore, the transcription factors were dramatically enriched in the DGEs in both sets of data. Accordingly, many ethylene-responsive transcription factors (ERFs) including four in GA 6 h vs. Control (Table 3 and Supplementary Table S3) and nine in GA 12 h vs. Control (Table 4 and Supplementary Table S4) were identified to be down-regulated, consistent with the reduced ethylene production (Figure 1C). Among them, three genes, ERF43 (Csa3M895680) and CRF2s (Csa5M139630 and Csa4M051360), were shared in the two sets. These observations indicated that ERFs may be implicated in cucumber sex expression. Because the ERFs act as positive regulators in ethylene signal transduction pathway (Wang et al., 2002; Guo and Ecker, 2004; Prescott et al., 2016; Zhang et al., 2016), we speculated that the decreased ethylene production inhibited the expression of ERFs, followed by modulated sexual development in cucumber plants treated with GA3.
FIGURE 4.
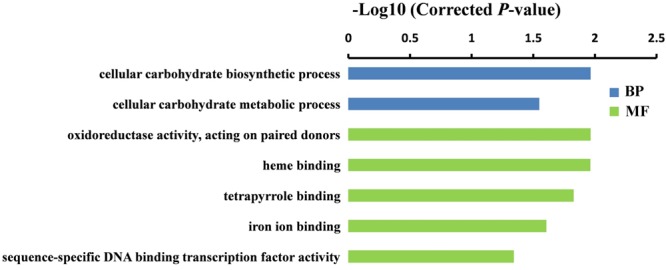
Gene ontology (GO) terms that were significantly enriched in the DEGs after 6 h of GA3 treatment. GO terms belong to biological processes (BP) and molecular functions (MFs) were shown in blue and green, respectively. GO terms were sorted based on corrected P-value, and the corrected P-value < 0.05 was used as the significance cut-off.
FIGURE 5.
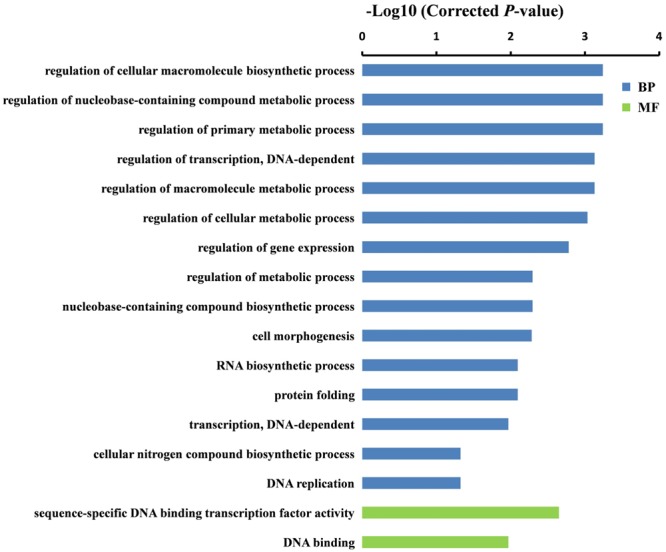
Gene ontology terms that were significantly enriched in the DEGs after 12 h of GA3 treatment. GO terms belong to BP and MFs were shown in blue and green, respectively. GO terms were sorted based on corrected P-value, and the corrected P-value < 0.05 was used as the significance cut-off.
Table 3.
List of selected ethylene signaling factors in the DEGs with enriched GO terms after GA3 treatment for 6 h.
| Gene ID | Gene annotation | Fold change (GA 6 h/Control) | FDR |
|---|---|---|---|
| Sequence-specific DNA binding transcription factor activity | |||
| Csa3M895680 | ERF43 (Ethylene-responsive transcription factor ERF043) | –2.93 | 1.08E-16 |
| Csa5M139630 | CRF2 (Ethylene-responsive transcription factor CRF2) | –2.51 | 1.89E-02 |
| Csa4M051360 | CRF2 (Ethylene-responsive transcription factor CRF2) | –2.23 | 2.66E-02 |
| Csa5M151530 | CRF4 (Ethylene-responsive transcription factor CRF4) | –2.12 | 1.14E-03 |
Table 4.
List of selected ethylene signaling factors and AGAMOUS (AG) homolog in the DEGs with enriched GO terms after GA3 treatment for 12 h.
| Gene ID | Gene annotation | Fold change (GA 12 h/Control) | FDR |
|---|---|---|---|
| equence-specific DNA binding transcription factor activity | |||
| Csa2M092800 | BBM2 (AP2-like ethylene-responsive transcription factor BBM2) | –∞ | 1.86E-02 |
| Csa2M382540 | ERF03 (Ethylene-responsive transcription factor ERF003) | –3.44 | 3.98E-02 |
| Csa2M382550 | ERF03 (Ethylene-responsive transcription factor ERF003) | –2.03 | 8.55E-05 |
| Csa3M895680 | ERF43 (Ethylene-responsive transcription factor ERF043) | –3.06 | 1.93E-13 |
| Csa4M001970 | RAP23 (Ethylene-responsive transcription factor RAP2-3) | –4.03 | 5.42E-10 |
| Csa4M051360 | CRF2 (Ethylene-responsive transcription factor CRF2) | –2.44 | 3.40E-02 |
| Csa4M192030 | RA211 (Ethylene-responsive transcription factor RAP2-11) | –3.87 | 4.03E-02 |
| Csa4M652640 | ERF03 (Ethylene-responsive transcription factor ERF003) | –2.42 | 5.66E-03 |
| Csa5M139630 | CRF2 (Ethylene-responsive transcription factor CRF2) | –3.02 | 9.74E-03 |
| Csa1M467100 | CAG2 (Floral homeotic protein AGAMOUS 2) | –23.48 | 7.84E-03 |
| DNA binding | |||
| Csa2M070880 | CsETR1 (Cucumber Ethylene receptor 1) | 2.12 | 6.99E-06 |
Interestingly, we also noticed that an ethylene receptor, CsETR1, was enriched in the GO term of “DNA binding” (Figure 5), and its expression was increased by 2.12-fold in shoot apices after GA3 treatment for 12 h (Table 4 and Supplementary Table S4), but not changed in GA 6 h vs. Control group. While the qRT-PCR verification revealed the same expression pattern (Figure 3). ETR1 is an important member of ethylene receptors family that acts as negative regulator in the ethylene signaling pathway (Wang et al., 2002; Guo and Ecker, 2004; Light et al., 2016; Prescott et al., 2016). Previous studied have confirmed that CsETR1 plays a negative role in stamen arrest during development of flower buds in cucumber (Wang et al., 2010). In accordance with these findings, we speculated that up-regulated CsETR1 may promote male flowers formation by alleviating stamen arrest in cucumber after GA3 treatment.
GA May Restrain the Female Tendency via Transcriptional Inhibition on CAG2 in Cucumber
Through GO term enrichment analyses, we further identified an AG (C-class floral homeotic gene) homolog CAG2, which was enriched in the “sequence-specific DNA binding transcription factor activity” group. CAG2 is one of the three AG genes in cucumber that controls pistil development due to its specific expression in the carpel (Kater et al., 1998; Perl-Treves et al., 1998). We found that the transcript level of CAG2 was significantly decreased by 23.48-fold in shoot apices after 12 h of GA3 treatment (Table 4 and Supplementary Table S4), and the qRT-PCR assay showed the same expression pattern (Figure 3). Our data implied that GA may restrain the femaleness via inhibiting the CAG2 expression in cucumber.
Discussion
Sex differentiation of flower buds is an important developmental process that directly affects the product yield in cucumber. In addition to genetic control, sex expression can be modified by plant hormones and environmental conditions. (Malepszy and Niemirowicz-Szczytt, 1991). Among various plant hormones, ethylene can induce female flowers formation (MacMurray and Miller, 1968; Iwahori et al., 1969), and the underlying molecular mechanism has been widely documented (Yamasaki et al., 2001, 2003; Mibus and Tatlioglu, 2004; Knopf and Trebitsh, 2006; Saito et al., 2007; Li et al., 2009, 2012; Wang et al., 2010). GA can promote male flowers development (Wittwer and Bukovac, 1962; Pike and Peterson, 1969), but the regulatory pathway remains elusive. In addition, a potential crosstalk between GA and ethylene in controlling sex determination of cucumber still remains disputed. In this study, through genome-wide expression analyses, we showed that GA may promote cucumber maleness via an ethylene-dependent pathway by altering expression of the M (CsACS2) gene, ethylene receptor CsETR1 and ethylene-responsive transcription factors. Nevertheless, we also found that GA may also restrain femaleness through an ethylene-independent pathway regulating CAG2, a C-class floral homeotic gene (Figure 6).
FIGURE 6.

A proposed model showing GA-regulated sex expression in cucumber. GA can promote male flowers formation via ethylene-dependent (right) and ethylene-independent (left) pathways. Arrows and T-bars indicate positive and negative effects, respectively. The solid lines define proved regulations, whereas the dotted lines represent proposed regulations.
GA May Promote Male Tendency via an Ethylene-Dependent Pathway in Cucumber
Upon exogenous GA3 treatment in the gynoecious cucumber line 13-3B, the male flowers were markedly induced, meanwhile, ethylene production in the shoot apices was significantly decreased (Figure 1) due to collaborative regulation by three ethylene biosynthetic genes such as M (CsACS2), CsACO1, and CsACO3. Notably, M and CsACO1 were significantly down-regulated and up-regulated in both sets of transcriptome comparisons, respectively, however, CsACO3 transcript was only decreased in GA 12 h vs. Control group (Figure 3; Table 2). Since both CsACO1 and CsACO3 encode ACC oxidases, and they showed opposite expression patterns and similar fold changes in RNA-Seq data (Table 2), we speculated that interplay between up-regulation of CsACO1 and down-regulation of CsACO3 offset the ACC oxidase activity in shoot apices after 12 h of GA3 treatment. This implied that the reduced M transcript might play major role in inhibiting ethylene biosynthesis. Given that the M gene can directly inhibit stamen development in cucumber flower buds (Saito et al., 2007; Li et al., 2009, 2012), our data revealed that GA might release the inhibitory effect of the M gene on stamen arrest and restrain ethylene production, by down-regulating the M gene expression (Figure 6, right panel).
In ethylene signal transduction pathway, the receptors such as ETRs function as negative regulators, while ERFs, downstream components of receptors, act as positive transcription factors (Wang et al., 2002; Guo and Ecker, 2004; Light et al., 2016; Prescott et al., 2016; Zhang et al., 2016). RNA-Seq data displayed that the ethylene receptor CsETR1 in cucumber, which is thought to be a negative regulatory factor in stamen arrest of flower buds (Wang et al., 2010), was markedly up-regulated after 12 h of GA3 treatment (Figure 3; Table 4 and Supplementary Table S4). Up-regulation of CsETR1 occurred at relatively later time point than down-regulation of the M gene (Table 2), suggesting that the increased CsETR1 transcript was not directly caused by GA3 rather by reduced ethylene production. Moreover, the expression levels of some ERFs were dramatically decreased after GA3 treatment (Tables 3 and 4, Supplementary Tables S3 and S4), and they were probably involved in cucumber sex expression through inhibiting maleness or promoting femaleness. However, this understanding was based on bioinformatics analysis, the precise roles of ERFs on cucumber flower development remained unclear and should be verified in future studies using advanced physiological and molecular techniques. These observations indicated that increased CsETR1 expression may stimulate male tendency through direct inhibition in stamen arrest or down-regulation on the ERFs transcript in cucumber after GA3 treatment (Figure 6, right panel).
Furthermore, despite we have shown that GA can regulate cucumber sex expression in cooperation with ethylene, how GA modulated ethylene and which genes participated in this process were unknown. Given that the DELLA proteins are central repressors of GA responses (Sun, 2010, 2011; Plackett et al., 2014), and accumulating evidence suggested that DELLA proteins play important roles in ethylene-mediated plant growth and development processes through interactions with some regulatory factors in ethylene signaling pathway, such as CTR1 (CONSTITUTIVE TRIPLE RESPONSE1), EIN3/EIL1 (ETHYLENE INSENSITIVE 3/EIN3-LIKE 1), RAP 2.3 (RELATED TO APETALA 2.3), and ERF11 (ETHYLENE RESPONSE FACTOR 11) (Achard et al., 2003, 2007; Pierik et al., 2009; An et al., 2012; Luo et al., 2013; Marín-de la Rosa et al., 2014; Zhou et al., 2016). Therefore, we proposed that DELLA proteins may be involved in the sex differentiation of cucumber coupled with GA and ethylene in a collaborative regulation at the protein level, since the expressions of four DELLA homologs in cucumber, CsGAIP, CsGAI1, CsGAI2, and CsGAI3 (Zhang et al., 2014a), had no change after GA3 treatment (Supplementary Tables S1 and S2).
In addition, GA and ethylene can also cooperatively regulate other aspects of plant growth and development. For example, GA promoted apical hook development of Arabidopsis, in part through transcriptional regulation of several genes in ethylene biosynthetic pathway mediated by DELLA proteins (Gallego-Bartolome et al., 2011). In this process, GA induced ethylene production, that was opposite to our results in cucumber sex expression. We speculated that this distinction may be due to different roles of hormones in various plant developmental processes. In fact, GA and ethylene showed similar functions in hook development, but they may play opposite roles in sex determination, thus, the regulatory mechanisms were different.
GA May Inhibit Femaleness or Induce Maleness of Cucumber via an Ethylene-Independent Pathway
AGAMOUS, the C-class floral homeotic gene, belongs to the MADS-box family. In Arabidopsis, GA can induce the expression of AG gene (Yu et al., 2004). However, our data provided a novel point that an AG homolog CAG2 in cucumber was down-regulated upon GA3 treatment (Figure 3; Table 4 and Supplementary Table S4). This distinction may be due to different abilities of these two genes to induce reproductive organ fate, in which, AG in Arabidopsis controls stamen and carpel development (Lohmann and Weigel, 2002), but CAG2 is particularly restricted to the carpel (Kater et al., 1998; Perl-Treves et al., 1998). Previous studies showed that CAG2 transcripts were not mediated by ethylene (Perl-Treves et al., 1998), hence, we speculated that GA probably suppressed pistil development through inhibiting the CAG2 expression, thereby allowing male flowers to develop, and ethylene was not involved in this process. Moreover, in our previous study, we demonstrated that transcript of CsGAMYB1 was upregulated by GA3 treatment in male flower buds and silencing of CsGAMYB1 could suppress masculinization of cucumber, but the ethylene production and expression of F and M genes were not changed in the CsGAMYB1-RNAi lines (Zhang et al., 2014b). These observations suggested that GA can also regulate sex expression of cucumber via an ethylene-independent pathway (Figure 6, left panel). However, the relationship between CAG2 and CsGAMYB1 regulations remains obscure and that should be verified in further studies.
Besides, DELLA proteins can down-regulate the expression of floral homeotic genes, AP3, PI and AG, subsequently inhibit flower development in Arabidopsis (Yu et al., 2004). CsGAIP, a DELLA homolog in cucumber, may restrain staminate development through transcriptional repression of AP3 and PI in Arabidopsis (Zhang et al., 2014a). However, the expression levels of AP3 and PI were not changed after GA3 treatment (Supplementary Tables S1 and S2), indicating that they did not participate in GA-regulated male tendency. Accordingly, there may be a regulatory relationship between DELLA proteins and floral homeotic gene CAG2 during GA-modulated sexual development in cucumber, however, the mechanism would be possibly different from that of Arabidopsis.
In summary, our data revealed a novel viewpoint that GA might control sex differentiation of cucumber via both ethylene-dependent and ethylene-independent pathways, and DELLA proteins were likely to be involved in both processes. However, this model was proposed by bioinformatics data, therefore, elucidation of the critical roles of DELLA proteins in flower development by cucumber transformation, and identification of the relationships among DELLAs and ethylene regulatory factors, GA-DELLA-CsGAMYB1 signaling and CAG2 gene, will shed new light on the molecular details of GA-regulated sex expression in cucumber.
Evolution of Unisexual Flower in Cucumber and Potential Involvement of Hormones
Generally, typical unisexual flowers have two morphological types. The type I is unisexual by abortion. Initiation of stamen and pistil occurs in all flowers, followed by the developmental arrest in one or another organ. The type II is unisexual from inception. Only stamen or pistil is initiated and it does not go through a hermaphroditic stage (Lebel-Hardenack and Grant, 1997; Ainsworth, 2000; Mitchell and Diggle, 2005). Now, it is believed that the morphology of cucumber flowers belongs to the type I (Bai et al., 2004), but its evolutionary mechanism is largely unknown. Bai and Xu proposed a “miR initiative” hypothesis (Bai and Xu, 2013), where they speculated that unisexual cucumber flowers are evolved from a hermaphrodite ancestor. The first step in the evolutionary process might be the miRNA-regulated arrest of ovary development, and this predication is based on the altered expression of miRNAs, such as miR396a, 156b, 159a, 171b, and 166a, in male flowers (Bai et al., 2004; Sun et al., 2010). And this event leads to environment-dependent andromonoecy which has no progeny. Then, the M gene is recruited. On the one hand, the M gene promotes ethylene biosynthesis, resulting in the rescue of ovary development for seed set, because the ethylene might regulate the miRNA production. On the other hand, the M gene inhibits stamen development to avoid self-pollination and maintain cross-pollination. So, the monoecious genotype is generated through the cooption of the M gene. The andromonoecious genotype is produced by the loss-of-function m gene, which is regarded as a reverted point mutation. Further, the F gene is coopted and generate the gynoecious genotype (Sun et al., 2010).
Until now, a potential role of GA in unisexual flower evolution of cucumber has not been reported. But based on the possible function of M gene on evolutionary development of cucumber flower and the effect of GA on the transcript of M gene (Table 2), we speculated that GA might be involved in the process of cucumber flower evolution through interaction with ethylene. In addition, previous studies showed that GA signaling system can regulate anther development by modulation of miR159/GAMYB (Achard et al., 2004). In this pathway, miR159 acts as a post-transcriptional regulator of GAMYB transcript levels. GA relieves the DELLA repression of GAMYB, which is mediated by the GA activation of miR159. As mentioned above, miR159 is likely to participate in the arrest of ovary development in the evolutionary process of cucumber flower. And the GAMYB homolog CsGAMYB1 can regulate cucumber sex expression via an ethylene-independent pathway (Zhang et al., 2014b). These observations further revealed the possible involvement of GA in unisexual flower evolution of cucumber, but this process might be dependent on miR159 and GAMYB, and have no relationship with ethylene. Finally, it is worth noting that these viewpoints are built on the basis of the “miR initiative” hypothesis and needed to be tested in further work.
Author Contributions
YZ and YaL designed the experiments. YZ, GZ, and NM performed the experiments. YZ, YuL, and JZ analyzed the data. YZ wrote the paper along with YaL. All authors reviewed the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Huazhong Ren (China Agricultural University) for providing the cucumber 13-3B seeds, and members of the Liang Laboratory for helpful discussions and technical assistance.
Abbreviations
- ACC
1-aminocyclopropane-1-carboxylate
- DEG
differentially expressed gene
- DGE
digital gene expression
- FDR
false discovery rate
- FID
flame ionization detector
- FPKM
fragments per kilobase of transcript sequence per millions base pairs sequenced
- GA
gibberellin
- GO
gene ontology
- qRT-PCR
quantitative real-time PCR
Funding. This work was supported by National Natural Science Foundation of China (31601770), Science and Technology Research and Development Program of Shaanxi Province (2015NY098), Fundamental Research Funds for Northwest A&F University (2452015024), and Doctoral Scientific Research Foundation of Northwest A&F University (Z109021504) to YZ.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00010/full#supplementary-material
References
- Achard P., Baghour M., Chapple A., Hedden P., Van Der Straeten D., Genschik P., et al. (2007). The plant stress hormone ethylene controls floral transition via DELLA-dependent regulation of floral meristem-identity genes. Proc. Natl. Acad. Sci. U.S.A. 104 6484–6489. 10.1073/pnas.0610717104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achard P., Herr A., Baulcombe D. C., Harberd N. P. (2004). Modulation of floral development by a gibberellin-regulated microRNA. Development 131 3357–3365. 10.1242/dev.01206 [DOI] [PubMed] [Google Scholar]
- Achard P., Vriezen W. H., Van Der Straeten D., Harberd N. P. (2003). Ethylene regulates arabidopsis development via the modulation of DELLA protein growth repressor function. Plant Cell 15 2816–2825. 10.1105/tpc.015685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An F., Zhang X., Zhu Z., Ji Y., He W., Jiang Z., et al. (2012). Coordinated regulation of apical hook development by gibberellins and ethylene in etiolated Arabidopsis seedlings. Cell Res. 22 915–927. 10.1038/cr.2012.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando S., Sato Y., Kamachi S., Sakai S. (2001). Isolation of a MADS-box gene (ERAF17) and correlation of its expression with the induction of formation of female flowers by ethylene in cucumber plants (Cucumis sativus L.). Planta 213 943–952. 10.1007/s004250100571 [DOI] [PubMed] [Google Scholar]
- Ainsworth C. (2000). Boys and girls come out to play: the molecular biology of dioecious plants. Ann. Bot. 86 211–221. 10.1006/anbo.2000.1201 [DOI] [Google Scholar]
- Atsmon D., Tabbak C. (1979). Comparative effects of gibberellin, silver nitrate and aminoethoxyvinyl glycine on sexual tendency and ethylene evolution in the cucumber plant (Cucumis sativus L.). Plant Cell Physiol. 20 1547–1555. [Google Scholar]
- Aya K., Ueguchi-Tanaka M., Kondo M., Hamada K., Yano K., Nishimura M., et al. (2009). Gibberellin modulates anther development in rice via the transcriptional regulation of GAMYB. Plant Cell 21 1453–1472. 10.1105/tpc.108.062935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai S. L., Peng Y. B., Cui J. X., Gu H. T., Xu L. Y., Li Y. Q., et al. (2004). Developmental analyses reveal early arrests of the spore-bearing parts of reproductive organs in unisexual flowers of cucumber (Cucumis sativus L.). Planta 220 230–240. 10.1007/s00425-004-1342-2 [DOI] [PubMed] [Google Scholar]
- Bai S. N., Xu Z. H. (2013). Unisexual cucumber flowers, sex and sex differentiation. Int. Rev. Cell Mol. Biol. 304 1–55. 10.1016/B978-0-12-407696-9.00001-4 [DOI] [PubMed] [Google Scholar]
- Cheng H., Qin L., Lee S., Fu X., Richards D. E., Cao D., et al. (2004). Gibberellin regulates Arabidopsis floral development via suppression of DELLA protein function. Development 131 1055–1064. 10.1242/dev.00992 [DOI] [PubMed] [Google Scholar]
- Duan Q. H., Wang D. H., Xu Z. H., Bai S. N. (2008). Stamen development in Arabidopsis is arrested by organ-specific overexpression of a cucumber ethylene synthesis gene CsACO2. Planta 228 537–543. 10.1007/s00425-008-0756-7 [DOI] [PubMed] [Google Scholar]
- Eveland A. L., Satoh-Nagasawa N., Goldshmidt A., Meyer S., Beatty M., Sakai H., et al. (2010). Digital gene expression signatures for maize development. Plant Physiol. 154 1024–1039. 10.1104/pp.110.159673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleet C. M., Sun T. P. (2005). A DELLAcate balance: the role of gibberellin in plant morphogenesis. Curr. Opin. Plant Biol. 8 77–85. 10.1016/j.pbi.2004.11.015 [DOI] [PubMed] [Google Scholar]
- Fujita Y., Fujieda K. (1981). Relation between sex expression types and cotyledon etiolation of cucumber in vitro. I. on the role of ethylene evolved from seedlings. Plant Cell Physiol. 22 667–674. [Google Scholar]
- Gallego-Bartolome J., Arana M. V., Vandenbussche F., Zadnikova P., Minguet E. G., Guardiola V., et al. (2011). Hierarchy of hormone action controlling apical hook development in Arabidopsis. Plant J. 67 622–634. 10.1111/j.1365-313X.2011.04621.x [DOI] [PubMed] [Google Scholar]
- Guo H., Ecker J. R. (2004). The ethylene signaling pathway: new insights. Curr. Opin. Plant Biol. 7 40–49. 10.1016/j.pbi.2003.11.011 [DOI] [PubMed] [Google Scholar]
- Hao Y. J., Wang D. H., Peng Y. B., Bai S. L., Xu L. Y., Li Y. Q., et al. (2003). DNA damage in the early primordial anther is closely correlated with stamen arrest in the female flower of cucumber (Cucumis sativus L.). Planta 217 888–895. 10.1007/s00425-003-1064-x [DOI] [PubMed] [Google Scholar]
- Harberd N. P., Belfield E., Yasumura Y. (2009). The angiosperm gibberellin-GID1-DELLA growth regulatory mechanism: how an “inhibitor of an inhibitor” enables flexible response to fluctuating environments. Plant Cell 21 1328–1339. 10.1105/tpc.109.066969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemphill D. D., Baker L. R., Sell H. M. (1972). Different sex phenotypes of Cucumis sativus L. and C. melo L. and their endogenous gibberellin activity. Euphytica 21 285–291. 10.1007/BF00036769 [DOI] [Google Scholar]
- Huang S., Li R., Zhang Z., Li L., Gu X., Fan W., et al. (2009). The genome of the cucumber, Cucumis sativus L. Nat. Genet. 41 1275–1281. 10.1038/ng.475 [DOI] [PubMed] [Google Scholar]
- Iwahori S., Lyons J. M., William L. S. (1969). Induced femaleness in cucumber by 2-chloroethanephosphonic acid. Nature 222 271–272. 10.1038/222271a0 [DOI] [PubMed] [Google Scholar]
- Kahana A., Silberstein L., Kessler N., Goldstein R. S., Perl-Treves R. (1999). Expression of ACC oxidase genes differs among sex genotypes and sex phases in cucumber. Plant Mol. Biol. 41 517–528. 10.1023/A:1006343707567 [DOI] [PubMed] [Google Scholar]
- Kater M. M., Colombo L., Franken J., Busscher M., Masiero S., Van Lookeren Campagne M. M., et al. (1998). Multiple AGAMOUS homologs from cucumber and petunia differ in their ability to induce reproductive organ fate. Plant Cell 10 171–182. 10.1105/tpc.10.2.171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knopf R. R., Trebitsh T. (2006). The female-specific Cs-ACS1G gene of cucumber. A case of gene duplication and recombination between the non-sex-specific 1-aminocyclopropane-1-carboxylate synthase gene and a branched-chain amino acid transaminase gene. Plant Cell Physiol. 47 1217–1228. 10.1093/pcp/pcj092 [DOI] [PubMed] [Google Scholar]
- Lebel-Hardenack S., Grant S. R. (1997). Genetics of sex determination in flowering plants. Trends Plant Sci. 2 130–136. 10.1016/S1360-1385(97)01012-1 [DOI] [Google Scholar]
- Li Z., Huang S., Liu S., Pan J., Zhang Z., Tao Q., et al. (2009). Molecular isolation of the M gene suggests that a conserved-residue conversion induces the formation of bisexual flowers in cucumber plants. Genetics 182 1381–1385. 10.1534/genetics.109.104737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Wang S., Tao Q., Pan J., Si L., Gong Z., et al. (2012). A putative positive feedback regulation mechanism in CsACS2 expression suggests a modified model for sex determination in cucumber (Cucumis sativus L.). J. Exp. Bot. 63 4475–4484. 10.1093/jxb/ers123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Light K. M., Wisniewski J. A., Vinyard W. A., Kieber-Emmons M. T. (2016). Perception of the plant hormone ethylene: known-knowns and known-unknowns. J. Biol. Inorg. Chem. 21 715–728. 10.1007/s00775-016-1378-3 [DOI] [PubMed] [Google Scholar]
- Lohmann J. U., Weigel D. (2002). Building beauty: the genetic control of floral patterning. Dev. Cell 2 135–142. 10.1016/S1534-5807(02)00122-3 [DOI] [PubMed] [Google Scholar]
- Luo J., Ma N., Pei H., Chen J., Li J., Gao J. (2013). A DELLA gene, RhGAI1, is a direct target of EIN3 and mediates ethylene-regulated rose petal cell expansion via repressing the expression of RhCesA2. J. Exp. Bot. 64 5075–5084. 10.1093/jxb/ert296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacMurray A. L., Miller C. M. (1968). Cucumber sex expression modified by 2-chloroethanephosphonic acid. Science 162 1397–1398. 10.1126/science.162.3860.1397 [DOI] [PubMed] [Google Scholar]
- Malepszy S., Niemirowicz-Szczytt K. (1991). Sex determination in cucumber (Cucumis sativus) as a model system for molecular biology. Plant Sci. 80 39–47. 10.1016/0168-9452(91)90271-9 [DOI] [Google Scholar]
- Marín-de la Rosa N., Sotillo B., Miskolczi P., Gibbs D. J., Vicente J., Carbonero P., et al. (2014). Large-scale identification of gibberellin-related transcription factors defines group VII ETHYLENE RESPONSE FACTORS as functional DELLA partners. Plant Physiol. 166 1022–1032. 10.1104/pp.114.244723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mibus H., Tatlioglu T. (2004). Molecular characterization and isolation of the F/f gene for femaleness in cucumber (Cucumis sativus L.). Theor. Appl. Genet. 109 1669–1676. 10.1007/s00122-004-1793-7 [DOI] [PubMed] [Google Scholar]
- Mitchell C. H., Diggle P. K. (2005). The evolution of unisexual flowers: morphological and functional convergence results from diverse developmental transitions. Am. J. Bot. 92 1068–1076. 10.3732/ajb.92.7.1068 [DOI] [PubMed] [Google Scholar]
- Murase K., Hirano Y., Sun T. P., Hakoshima T. (2008). Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature 456 459–463. 10.1038/nature07519 [DOI] [PubMed] [Google Scholar]
- Olszewski N., Sun T. P., Gubler F. (2002). Gibberellin signaling: biosynthesis, catabolism, and response pathways. Plant Cell 14(Suppl.) S61–S80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perl-Treves R., Kahana A., Rosenmann N., Xiang Y., Silberstein L. (1998). Expression of multiple AGAMOUS-like genes in male and female flowers of cucumber (Cucumis sativus L.). Plant Cell Physiol. 39 701–710. 10.1093/oxfordjournals.pcp.a029424 [DOI] [PubMed] [Google Scholar]
- Pierce L. K., Wehner T. C. (1990). Review of genes and linkage groups in cucumber. HortScience 25 605–615. [Google Scholar]
- Pierik R., Djakovic-Petrovic T., Keuskamp D. H., de Wit M., Voesenek L. A. (2009). Auxin and ethylene regulate elongation responses to neighbor proximity signals independent of gibberellin and della proteins in Arabidopsis. Plant Physiol. 149 1701–1712. 10.1104/pp.108.133496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pike L. M., Peterson C. E. (1969). Gibberellin A4/A7, for induction of staminate flowers on the gynoecious cucumber (Cucumis sativus L.). Euphytica 18 106–109. [Google Scholar]
- Plackett A. R., Ferguson A. C., Powers S. J., Wanchoo-Kohli A., Phillips A. L., Wilson Z. A., et al. (2014). DELLA activity is required for successful pollen development in the Columbia ecotype of Arabidopsis. New Phytol. 201 825–836. 10.1111/nph.12571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plackett A. R., Thomas S. G., Wilson Z. A., Hedden P. (2011). Gibberellin control of stamen development: a fertile field. Trends Plant Sci. 16 568–578. 10.1016/j.tplants.2011.06.007 [DOI] [PubMed] [Google Scholar]
- Prescott A. M., McCollough F. W., Eldreth B. L., Binder B. M., Abel S. M. (2016). Analysis of network topologies underlying ethylene growth response kinetics. Front. Plant Sci. 7:1308 10.3389/fpls.2016.01308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M. D., McCarthy D. J., Smyth G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26 139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudich J., Halevy A. H., Kedar N. (1972). The level of phytohormones in monoecious and gynoecious cucumbers as affected by photoperiod and ethephon. Plant Physiol. 50 585–590. 10.1104/pp.50.5.585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito S., Fujii N., Miyazawa Y., Yamasaki S., Matsuura S., Mizusawa H., et al. (2007). Correlation between development of female flower buds and expression of the CS-ACS2 gene in cucumber plants. J. Exp. Bot. 58 2897–2907. 10.1093/jxb/erm141 [DOI] [PubMed] [Google Scholar]
- Song S., Qi T., Huang H., Xie D. (2013). Regulation of stamen development by coordinated actions of jasmonate, auxin, and gibberellin in Arabidopsis. Mol. Plant 6 1065–1073. 10.1093/mp/sst054 [DOI] [PubMed] [Google Scholar]
- Sun J. J., Li F., Li X., Liu X. C., Rao G. Y., Luo J. C., et al. (2010). Why is ethylene involved in selective promotion of female flower development in cucumber? Plant Signal. Behav. 5 1052–1056. 10.4161/psb.5.8.12411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T. P. (2010). Gibberellin-GID1-DELLA: a pivotal regulatory module for plant growth and development. Plant Physiol. 154 567–570. 10.1104/pp.110.161554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T. P. (2011). The molecular mechanism and evolution of the GA-GID1-DELLA signaling module in plants. Curr. Biol. 21 R338–R345. 10.1016/j.cub.2011.02.036 [DOI] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S. L. (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25 1105–1111. 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trebitsh T., Rudich J., Riov J. (1987). Auxin, biosynthesis of ethylene and sex expression in cucumber (Cucumis sativus). Plant Growth Regul. 5 105–113. 10.1007/BF00024738 [DOI] [Google Scholar]
- Trebitsh T., Staub J. E., O’Neill S. D. (1997). Identification of a 1-aminocyclopropane-1-carboxylic acid synthase gene linked to the female (F) locus that enhances female sex expression in cucumber. Plant Physiol. 113 987–995. 10.1104/pp.113.3.987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D. H., Li F., Duan Q. H., Han T., Xu Z. H., Bai S. N. (2010). Ethylene perception is involved in female cucumber flower development. Plant J. 61 862–872. 10.1111/j.1365-313X.2009.04114.x [DOI] [PubMed] [Google Scholar]
- Wang L. C., Li H., Joseph R. E. (2002). Ethylene biosynthesis and signaling networks. Plant Cell 14(Suppl. 14) S131–S151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Gerstein M., Snyder M. (2009). RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10 57–63. 10.1038/nrg2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittwer S. H., Bukovac M. I. (1962). Staminate flower formation on gynoecious cucumber as influenced by the various gibberellins. Naturwissenshaften 49 305–306. 10.1007/BF00622719 [DOI] [Google Scholar]
- Yamasaki S., Fujii N., Matsuura S., Mizusawa H., Takahashi H. (2001). The M locus and ethylene-controlled sex determination in andromonoecious cucumber plants. Plant Cell Physiol. 42 608–619. 10.1093/pcp/pce076 [DOI] [PubMed] [Google Scholar]
- Yamasaki S., Fujii N., Takahashi H. (2003). Characterization of ethylene effects on sex determination in cucumber plants. Sex. Plant Reprod. 16 103–111. 10.1007/s00497-003-0183-7 [DOI] [Google Scholar]
- Yin T., Quinn J. A. (1995). Tests of a mechanistic model of one hormone regulating both sexes in Cucumis sativus (Cucurbitaceae). Am. J. Bot. 82 1537–1546. 10.2307/2446182 [DOI] [Google Scholar]
- Young M. D., Wakefield M. J., Smyth G. K., Oshlack A. (2010). Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11:R14 10.1186/gb-2010-11-2-r14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Ito T., Zhao Y., Peng J., Kumar P., Meyerowitz E. M. (2004). Floral homeotic genes are targets of gibberellin signaling in flower development. Proc. Natl. Acad. Sci. U.S.A. 101 7827–7832. 10.1073/pnas.0402377101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Li A., Zhang Z., Huang Z., Lu P., Zhang D., et al. (2016). Ethylene response factor TERF1, regulated by ETHYLENE-INSENSITIVE3-like factors, functions in reactive oxygen species (ROS) scavenging in tobacco (Nicotiana tabacum L.). Sci. Rep. 6:29948 10.1038/srep29948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu B., Yang S., An J., Chen C., Zhang X., et al. (2014a). A cucumber DELLA homolog CsGAIP may inhibit staminate development through transcriptional repression of B class floral homeotic genes. PLoS ONE 9:e91804 10.1371/journal.pone.0091804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Zhang X., Liu B., Wang W., Liu X., Chen C., et al. (2014b). A GAMYB homologue CsGAMYB1 regulates sex expression of cucumber via an ethylene-independent pathway. J. Exp. Bot. 65 3201–3213. 10.1093/jxb/eru1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Zhang Z. L., Park J., Tyler L., Yusuke J., Qiu K., et al. (2016). The ERF11 transcription factor promotes internode elongation by activating gibberellin biosynthesis and signaling. Plant Physiol. 171 2760–2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


