Abstract
Proteae, and especially Proteus mirabilis, are often the cause of urinary tract infections (UTIs) in humans. They were reported as carriers of extended-spectrum β-lactamase (ESBL) genes, and recently of carbapenemases, mostly carried by the Salmonella genomic island 1 (SGI1) and Proteus genomic island 1 (PGI1). Proteae have also lately become an increasing cause of UTIs in companion animals, but antimicrobial susceptibility data in animals are still scarce. Here, we report the characterization of 468 clinical epidemiologically unrelated Proteae strains from animals collected between 2013 and 2015 in France. Seventeen P. mirabilis strains (3.6%) were positive for SGI1/PGI1 and 18 Proteae (3.8%) were resistant to extended-spectrum cephalosporins (ESC). The 28 isolates carrying SGI1/PGI1 and/or ESC-resistance genes were isolated from cats, dogs, and horses. ESBL genes were detected in six genetically related P. mirabilis harboring blaV EB-6 on the SGI1-V variant, but also independently of the SGI1-V, in 3 P. mirabilis strains (blaVEB-6 and blaCTX-M-15) and 1 Providencia rettgeri strain (blaCTX-M-1). The AmpC resistance genes blaCMY -2 and/or blaDHA-16 were detected in 9 P. mirabilis strains. One strain presented both an ESBL and AmpC gene. Interestingly, the majority of the ESBL/AmpC resistance genes were located on the chromosome. In conclusion, multiple ESC-resistance genetic determinants are circulating in French animals, even though SGI1-V-carrying P. mirabilis seems to be mainly responsible for the spread of the ESBL gene blaVEB-6 in dogs and horses. These results are of public health relevance and show that companion animals in close contact with humans should be regarded as a potential reservoir of ESC-resistant bacteria as well as a reservoir of ESC-resistance genes that could further disseminate to human pathogens.
Keywords: SGI1, PGI1, Proteus, ESBL, AmpC, animal, dog
Introduction
Proteae are specific Enterobacteriaceae comprising bacterial species present in water, soil or in the intestinal tract of humans and animals. In humans, Proteae, and especially Proteus mirabilis, are often the cause of urinary tract infections (UTIs; Schaffer and Pearson, 2015). They are otherwise opportunistic pathogens responsible for various infections ranging from minor to life-threatening issues. In veterinary medicine, P. mirabilis and other Proteae are rarely found as pathogens except as a cause of UTIs in companion animals (Bubenik et al., 2007). This pathology is only rarely treated with extended-spectrum cephalosporins (ESC), and the recommended antibiotics are sulphonamides, aminoglycosides, or fluoroquinolones.
Proteus mirabilis is naturally susceptible to β-lactams and β-lactamases inhibitors (Stock, 2003). In the late 1990s, the emergence of P. mirabilis isolates expressing acquired β-lactamases was first reported in France (Chanal et al., 2000). Extended-spectrum (ESBL) and AmpC β-lactamases are of a critical importance because they both confer resistance to nearly all β-lactams, including ESC. Interestingly, even though the corresponding genes (mostly blaCTX-M-types and blaVEB-6 for ESBL and blaCMY -2 and blaDHA-16 for AmpC) are generally located on plasmids which allow an easy intra- or inter-species dissemination, several studies revealed chromosome-encoded ESBL/AmpC genes in P. mirabilis (Song et al., 2011; Harada et al., 2012). These chromosome-encoded genes are often carried by genomic islands (such as the Salmonella Genomic Island 1, SGI1) or integrating conjugative elements (ICEs) that may also be transmitted (Harada et al., 2010; Mata et al., 2011). While ESBL-producing P. mirabilis are nowadays commonly isolated from humans, the first CTX-M-55-producing P. mirabilis in animals was only reported in 2011 from a macaque imported from Vietnam to France (Dahmen et al., 2013).
Besides, P. mirabilis can also carry various genomic islands conferring multidrug resistance. For example SGI1, the genomic island widely disseminated in Salmonella, was first identified in a clinical P. mirabilis from a diabetic patient from Palestine in 2006 (Ahmed et al., 2007). SGI1 is a site-specific integrative mobilizable element conferring multidrug resistance initially described in Salmonella enterica serovar Typhimurium DT104 (Boyd et al., 2001).
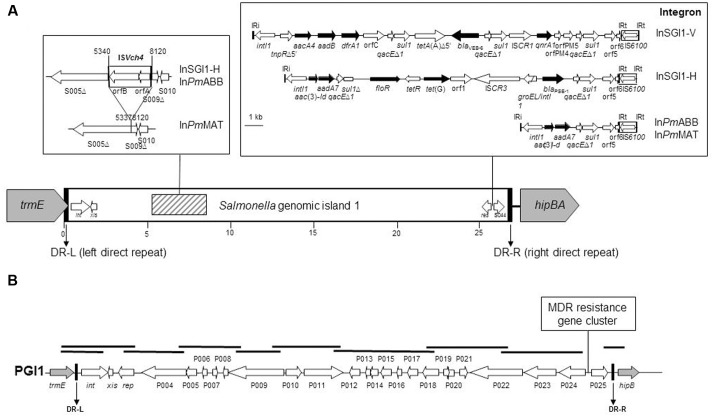
Salmonella genomic island 1 is the first MDR genomic island identified in S. enterica, and contains a complex class 1 integron, named In104 (Hall, 2010). Since the identification of SGI1 in S. Typhimurium DT104, more than 30 different SGI1 variants carrying different combinations of antimicrobial resistance genes were described so far (Hall, 2010). The complex In104 integron variants classically possess one or two cassette attachment sites (attI1) carrying various resistance gene cassette arrays, contain an IS6100 element, may contain additional resistance genes, and are bound by 25-bp inverted repeats IRi and IRt (Boyd et al., 2001) (Figure 1). In the great majority of these variants, the complex In104 integron or its variants are found always at the same position in the SGI1 scaffold, i.e., between the resolvase gene res (also named tnpR) and the open reading frame (ORF) S044 at the 3′ end of SGI1 (Hall, 2010). A few additional variations occurred in the SGI1 backbone of some variants, especially the insertion/deletion created by ISVch4 between ORF S005 and S009 that was found in several SGI1 variants (SGI1-H, -Ls, -Ks, -Ps, Qs, -PmABB, and -PmMAT) in S. enterica and P. mirabilis (Doublet et al., 2008; Siebor and Neuwirth, 2013).
FIGURE 1.
Schematic view of SGI1 in the Proteus mirabilis chromosome. (A) Specific genetic traits of the SGI1 structure, the genetic rearrangement due to ISVch4, and the complex class 1 integrons are represented in frames. Genes and ORFs are shown as arrows, with their orientations of transcription indicated by the arrowheads. Black arrows indicate antibiotic resistance genes. White arrows within boxes represent insertion sequence elements. IRi and IRt are 25-bp imperfect inverted repeats defining the integrase and tni ends of complex class 1 integrons. The complex class 1 integrons were drawn from PCR mapping results and GenBank accession numbers AF261825 (SGI1), HQ888851 (SGI1-V), AY458224 (SGI1-H), and JX121638 (SGI1-PmABB). (B) Schematic view of PGI1 in the P. mirabilis chromosome. PCRs carried out to map the PGI1 backbone and junctions with the chromosome are indicated by thick black bars. Gray and white arrows represent the chromosomal genes and the backbone of the island, respectively.
Salmonella genomic island 1 is found integrated most of the time within the last 18 bp of the well-conserved chromosomal trmE gene (also named thdF). SGI1 is specifically mobilized in trans by conjugative plasmids of the IncA/C family (Doublet et al., 2005). Only this plasmid family has been shown to be able to mobilize SGI1 (Douard et al., 2010). The main reason of this specificity is that the SGI1 excision from the chromosome is triggered by the master activator AcaDC encoded by IncA/C conjugative plasmids (Carraro et al., 2014; Kiss et al., 2015). Then, as an extrachromosomal form, SGI1 is able to hijack the conjugative apparatus encoded by IncA/C plasmids to be conjugally transferred to a recipient cell (Carraro et al., 2014).
Since 2006, P. mirabilis strains carrying different SGI1 variants have only been reported in China and France (Boyd et al., 2008; Siebor and Neuwirth, 2013; Qin et al., 2015). Importantly, the SGI1-V variant, which is specifically found in P. mirabilis, harbors the blaVEB-6 gene and was first reported in a lethal human case in France (Siebor and Neuwirth, 2011).
Recently, a new multidrug resistant genomic island named Proteus Genomic Island 1 (PGI1) was described in human P. mirabilis isolates in France (Siebor and Neuwirth, 2014). A specific PGI1 variant, PGI1-PmPEL, was shown to harbor both the blaVEB-6 gene and the carbapenemase-encoding gene blaNDM-1 (Girlich et al., 2015). P. mirabilis isolates of animal origin were also shown to carry SGI1 or PGI1. Indeed, SGI1-positive P. mirabilis isolates were reported in poultry and swine farms in China (Lei et al., 2014, 2015). In France, we recently described the very first cases of SGI1 (including the VEB-6-producing SGI1-V variant) or PGI1-positive P. mirabilis in dogs (Schultz et al., 2015).
The SGI1/PGI1-positive P. mirabilis isolates reported in animals and humans so far were sporadic cases. Considering the apparent emergence of these genetic determinants in P. mirabilis, our aim was to investigate the prevalence of SGI1 and PGI1 in Proteae of animal origin in France, and to characterize molecularly the collected strains. In line with recent observations that a SGI1-V-carrying P. mirabilis clonal population was shared between humans and animals, we also investigated the genetic relationship of those isolates in order to draw hypotheses on a possible transfer between the two populations. Finally, as these islands in P. mirabilis isolates were also shown to occasionally capture ESBL or carbapenemase genes, we investigated the global prevalence of those genes in the same collection.
Materials and Methods
Bacterial Strains and Antibiotic Susceptibility Testing
Between April, 2013 and February, 2015, a total of 468 clinical non-duplicate Proteae isolates (P. mirabilis, n = 459; P. vulgaris, n = 1; P. penneri, n = 1; Morganella morganii, n = 2; Providencia rettgeri, n = 4; P. stuartii, n = 1) were collected from various animal hosts corresponding to distinct and epidemiological unrelated individuals (dogs, n = 411; cats, n = 25; horses, n = 13; bovine, n = 7; rabbit, n = 3; ovine, n = 3; ferret, n = 2; snake, n = 2; chicken; n = 1; bird, n = 1) and originating from distant geographical areas throughout France. These isolates were collected through the Resapath, the French antimicrobial resistance surveillance network in animal pathogens1. Identification was performed by peripheral veterinary laboratories and then confirmed at the Anses laboratory in Lyon, France, using Api20E strips (bioMérieux, Marcy l’Etoile, France). Isolates were screened for antibiotic resistance by the disk diffusion method according to the guidelines of the CA-SFM with 30 antibiotics of veterinary or human interest2.
SGI1/PGI1 Detection and PCR Mapping
Salmonella genomic island 1 detection was performed by PCR amplification with degenerated primers designed in order to amplify all known SGI1 and PGI1 integrase genes (FwintSGI1HR, 5′ATGTTGCGTCAGGCYGAGGC; RvintSGI1HR, 5′GAGTGYCCAAGAAGSCGAGAG). The chromosomal location was confirmed by PCR amplification of the left and right junctions in the chromosome, as previously described (Schultz et al., 2015). The genetic diversity of SGI1 was assessed by PCR mapping covering the entire island using primers previously described (Siebor and Neuwirth, 2011, 2013). PGI1 detection, chromosomal location, backbone mapping and resistance gene detection were performed by PCR, as previously described (Figure 1) (Siebor and Neuwirth, 2014).
Identification and Genetic Location of ESBL and AmpC Genes
β-lactamase genes detection was performed by PCR, as previously described (Dallenne et al., 2010). For the CTX-M-1 group, an additional PCR was performed using external primers (ISEcp1L1, 5′ CAGCTTTTATGACTCG; P2D, 5′ CAGCGCTTTTGCCGTCTAAG) and the amplicons were sequenced.
The chromosomal location of these genes was assessed with the I-CeuI (New England Biolabs, Hertfordshire, UK) technique (Liu et al., 1993). After digestion of the complete DNA, DNA fragments were separated by Pulsed-Field Gel Electrophoresis (PFGE) in TBE 0.5 X at 14°C using a CHEF Mapper (Bio-Rad Laboratories, Richmond, CA, USA). Running conditions were 6 V/cm with a switch time of 5.3–49.9 s for 19.7 h. Southern blots were performed by transferring the DNA on a Hybond-N+ membrane and hybridizing the membrane with DIG-labeled probes specific for the 23S rDNA, the ESBL (blaVEB-6, blaCTX-M-1, or blaCTX-M-15) and AmpC (blaCMY -2 or blaDHA-16) genes of interest (see detailed primers in Supplementary Table S1). Probes were prepared by PCR using labeled DIG-dUTP (PCR DIG probe synthesis kit; Roche Diagnostics, Indianapolis, IN, USA). Detection was performed using the DIG DNA Labeling and Detection Kit (Roche Diagnostics) according to the manufacturer’s instructions.
The plasmidic location of these genes was assessed by Southern blots on S1-PFGE gels (New England Biolabs, Hertfordshire, UK). Running conditions were 6 V/cm with a switch time of 1–30 s for 20 h. Hybridization was performed as described above for Southern blots on I-CeuI gels, using the same ESBL/AmpC probes and the DIG DNA Labeling and Detection Kit.
Finally, all SGI1/PGI1-positive, ESBL and AmpC strains were analyzed by PCR-based replicon typing (PBRT kit; Diatheva, Fano, Italy) in order to type the plasmids carrying the ESBL/AmpC genes.
Genetic Relationship of the Isolates
The genetic diversity was investigated by PFGE. DNA plugs were digested with SmaI (Promega, Madison, WI, USA) and genomic DNA was separated by CHEF Mapper gel electrophoresis in TBE 0.5 X at 14°C. The running conditions were 6 V/cm with a switch time of 5–20 s for 22 h. XbaI-digested S. enterica serovar Braenderup strain H9812 was used as size ladder. The DNA patterns were analyzed using BioNumerics software version 4.5 (Applied Maths, Sint-Martens-Latem, Belgium) to construct a phylogenetic tree. Analysis was performed using the Dice coefficient with optimization set at 0.5% and tolerance at 1%.
Results and Discussion
Prevalence and Molecular Characterization of the SGI1/PGI1 Genomic Islands
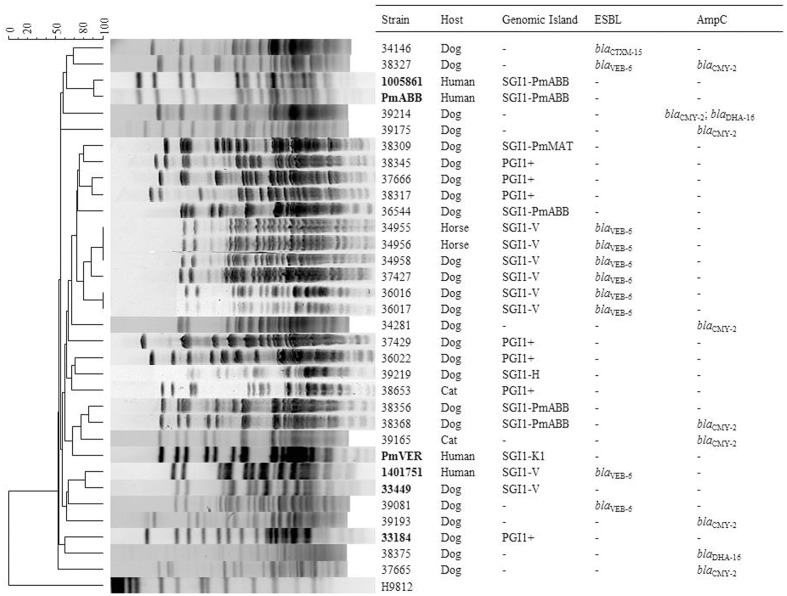
Salmonella genomic island 1/Proteus genomic island 1-carrying P. mirabilis are emerging pathogens in humans and animals. In a recent study, SGI1/PGI1-carrying P. mirabilis were reported in animals in France but no large-scale non-biased data on their prevalence were available (Schultz et al., 2015). Here, we investigated a large collection of 468 Proteae isolates of animal origin in France to estimate the prevalence of the SGI1/PGI1 elements. We also assessed the prevalence of resistance to broad-spectrum cephalosporins in those Proteae isolates considering that the blaVEB-6 gene was recurrently reported on the SGI1-V variant in P. mirabilis (Siebor and Neuwirth, 2011; Schultz et al., 2015). Among the 468 isolates studied, 17 P. mirabilis (17/468, 3.6%) were positive for SGI1 (11 isolates) or PGI1 (6 isolates) by PCR (Figure 2 and Table 1). They were mostly recovered from dogs (n = 13; 13/411, 3.2%), but also from cats (n = 2; 2/25, 8%), and horses (n = 2; 2/13, 15.4%). While Proteae are more frequently isolated from dogs, SGI1/PGI1 elements were more prevalent in cats and horses. This would deserve confirmation due to the low number of isolates collected from these two animal categories.
FIGURE 2.
Analysis of SmaI-Pulsed-Field Gel Electrophoresis (PFGE) patterns obtained from the relevant P. mirabilis isolates. PFGE profiles were compared using BioNumerics software version 4.5 (Applied Maths) with settings of 0.5% optimization and 1.0% tolerance. DNA of S. enterica serovar Braenderup strain H9812 was used as standard size marker in PFGE experiments and as outgroup in analysis (Hunter et al., 2005). The isolates in bold correspond to previously published profiles included for comparison purposes (Schultz et al., 2015).
Table 1.
Antibiotic resistance profiles of Proteus mirabilis strains of interest in this study.
| Strain | Isolation date (yyyy/mm/dd) | Host | Geographic areab | Pathology | Antibiotic resistance profilea |
|---|---|---|---|---|---|
| 34146 | 2013/04/17 | dog | Haute-Savoie | Urinary tract infection | ESBL; StrKanAprTobNetSss |
| 34381 | 2013/10/09 | dog | Alpes-Maritimes | Urinary tract infection | AmpC; Chl |
| 34955 | 2013/11/22 | horse | Oise | Skin infection | ESBL; KanTobNetChlSssTmpNalEnr |
| 34956 | 2013/10/25 | horse | Calvados | Skin infection | ESBL; StrKanTobNetChlSssTmpNalEnr |
| 34958 | 2013/07/30 | dog | Paris | Urinary tract infection | ESBL; StrKanAmkAprGenTobNetChlSssTmpNalEnr |
| 36016 | 2013/11/05 | dog | Val-de-Marne | Abdominal infection | ESBL; StrKanAmkGenTobNetChlSssTmpNalEnr |
| 36017 | 2013/22/04 | dog | Val-de-Marne | Urinary tract infection | ESBL; StrKanGenTobNetChlSssTmpNalEnr |
| 36022 | 2013/11/10 | dog | Alpes-Maritimes | Urinary tract infection | StrSss |
| 36544 | 2014/03/05 | dog | Paris | Urinary tract infection | StrChlSssTmpNalEnr |
| 37427 | 2014/04/04 | dog | Val-de-Marne | Urinary tract infection | ESBL; KanGenTobNetChlSssTmpNalEnr |
| 37429 | 2014/03/14 | dog | Loire | Otitis | StrKanGenTobChlSss |
| 37665 | 2014/04/22 | dog | Paris | Urinary tract infection | AmpC |
| 37666 | 2014/03/21 | dog | Val-de-Marne | Wound | AmxStrKanGenTobSssTmpNalEnr |
| 38309 | 2014/09/09 | dog | Haute-Savoie | Unknown | StrSssTmpNalEnr |
| 38317 | 2014/09/09 | dog | Tarn | Otitis | AmxStrKanGenTobChlSssTmpNalEnr |
| 38327 | 2014/09/29 | dog | Oise | Skin infection | ESBL + AmpC; StrKanChlSssTmp |
| 38346 | 2014/07/09 | dog | Hauts-de-Seine | Urinary tract infection | AmxStrKanGenTobChlSss |
| 38356 | 2014/07/15 | dog | Aisne | Otitis | StrChlSssTmpNalEnr |
| 38368 | 2014/08/21 | dog | Loire-Atlantique | Urinary tract infection | AmpC; ChlSssTmpNalEnr |
| 38375 | 2014/08/21 | dog | Côtes-d’Armor | Otitis | AmpC; Chl |
| 38653 | 2014/11/03 | cat | Seine-St-Denis | Urinary tract infection | StrKanGenTobChlSss |
| 39081 | 2014/12/01 | dog | Val-de-Marne | Urinary tract infection | ESBL; StrKanAmkGenTobNetChlSssTmpNalEnr |
| 39165 | 2014/10/02 | cat | Loire-Atlantique | Urinary tract infection | AmpC; SssTmpNalEnr |
| 39175 | 2014/10/31 | dog | Val-de-Marne | Otitis | AmpC; AprChlNal |
| 39193 | 2014/10/09 | dog | Paris | Otitis | AmpC; StrChlTmpNal |
| 39214 | 2014/11/18 | dog | Val-de-Marne | Skin infection | AmpC; AprChlNal |
| 39219 | 2015/01/26 | dog | Somme | Unknown | StrChlSssTmpNalEnr |
| 39465c | 2014/09/19 | cat | Gironde | Unknown | ESBL; SssTmpNalEnr |
aESBL, extended-spectrum beta-lactamase; Amx, amoxicillin; Chl, chloramphenicol; Ffc, florfenicol; Amk, amikacin; Apr, apramycin; Gen, gentamicin; Tob, tobramycin; Net, netilmicin; Kan, kanamycin; Str, streptomycin; Sss, sulfonamides; Tmp, trimethoprim; Nal, nalidixic acid; Enr, enrofloxacin. bFrench department: The department is the administrative subdivision of France. cP. rettgeri strain.
Among the 11 SGI1-positive P. mirabilis, six showed an ESBL phenotype using the double disk synergy test by antibiogram. PCR mapping of the SGI1 resistance gene cluster and backbone revealed that all six ESBL-positive isolates harbored the SGI1-V variant carrying the blaVEB-6 gene (Figure 1). In the five other SGI1-positive P. mirabilis isolates, PCR mapping of the antimicrobial resistance gene cluster gave positive results for different complex class 1 integrons, i.e., InSGI1-H, InPmABB and InPmMAT (Figures 1, 2 and Table 1). PCR mapping of the SGI1 backbone confirmed that SGI1 elements with InSGI1-H, InPmABB, and InPmMAT harbored the same 2789 bp deletion at position 5340-8120 (GenBank accession number AF261825) in the region spanning from ORF S005 to ORF S009, as previously described. Except for SGI1-InPmMAT, this deletion was replaced by the insertion of ISVch4 (also called IS1359) (Figure 1). Interestingly, this region contains two ORFs S007 and S006 coding for homologs of the master activator AcaDC of IncA/C conjugative plasmids and named SgaDC (SGI1 activator, subunits D and C; Poulin-Laprade et al., 2015; Muranyi et al., 2016). The SgaDC activator (also named FlhDCSGI1) was shown to be active on the same AcaDC-dependent promoter regions, i.e., Pxis, and thus should be also implicated in transfer and/or maintenance of SGI1 (Muranyi et al., 2016). The absence of flhDCSGI1 in these variants and the partial deletion of ORF S005 (traN) may have implications in their spread that needs to be further studied. In addition to the classical penta-resistance of SGI1 to amoxicillin, chloramphenicol, streptomycin and sulphonamides, other SGI1/PGI1-conferred resistances were amikacin, apramycin, gentamicin, tobramycin, netilmicin, kanamycin, nalidixic acid, trimethoprim, and enrofloxacin.
The six PGI1-positive P. mirabilis isolates were characterized by PCR mapping of the whole backbone. PCR products of the expected sizes were obtained for the complete mapping indicating a conserved PGI1 genetic structure in these isolates (Figure 1). All six isolates showed similar antibiotic resistance profiles as the ones previously described (Siebor and Neuwirth, 2014; Schultz et al., 2015). Interestingly PGI1, which was only recently reported in P. mirabilis, was represented in more than one-third of the genomic islands characterized here, thus confirming the proportion observed in a recent study on a much smaller number of isolates (Schultz et al., 2015). This may suggest either a previously undetected situation in the Proteae population or a recent and rapid spread of PGI1 elements, which were first described in 2014 (Siebor and Neuwirth, 2014). Our data also indicate that the dissemination of SGI1/PGI1 in multidrug-resistant P. mirabilis in animals is not a sporadic phenomenon and should be considered with great attention.
Extended-Spectrum and AmpC β-Lactamases
Eighteen strains (18/468, 3.8%) harbored an ESBL profile (n = 9), an AmpC profile (n = 8), or a combination of both (n = 1) after antibiotic susceptibility analysis. Only one AmpC-producing strain did not present any additional resistance (Table 1). Otherwise, proportions of strains resistant to non-β-lactam antibiotics were particularly high for chloramphenicol (14/18, 77.8%), streptomycin (10/18, 55.6%), trimethoprim/sulphonamides (13/18, 72.2%), nalidixic acid (13/18, 72.2%), and enrofloxacin (10/18, 55.6%). Of note, none of the strains were resistant to carbapenems.
In addition to the six SGI1-V positive P. mirabilis described above, the ESBL phenotype was detected in four other strains. Two P. mirabilis isolates (38327 and 39081) harbored the blaVEB-6 gene independently of the SGI1 element (Figure 2), whereas the two last ESBL-producing isolates harbored the blaCTX-M-15 gene (P. mirabilis 34146) and the blaCTX-M-1 gene (P. rettgeri 39465). The prevalence of ESBL producers was lower than the one recently reported in France. However, the previous sampling was based on isolates conserved by the veterinary laboratories and a bias toward non-susceptible isolates cannot be excluded (Schultz et al., 2015). On the contrary, no ESBL-producing isolate was detected in Japan (Harada et al., 2014). In any case, Proteae from animal origin present much less ESC-resistant isolates than those from human origin, since the nowadays ESBL rate in animals is very close to the French situation in human medicine in the late 1990s (Chanal et al., 2000).
Concerning AmpC resistance phenotypes, the blaCMY -2 gene was detected alone in 7 P. mirabilis isolates whereas another P. mirabilis isolate (38375) presented a blaDHA-16 gene (Figure 2). Finally, one P. mirabilis isolate (39214) possessed both blaCMY -2 and blaDHA-16 genes (Figure 2). The presence of multiple ESC-resistance genes in two strains (blaVEB-6/blaCMY -2 in 38327 and blaCMY -2/blaDHA-16 in 39214) shows the capacity of P. mirabilis to accumulate redundant resistance genes and thus acting as a potential reservoir.
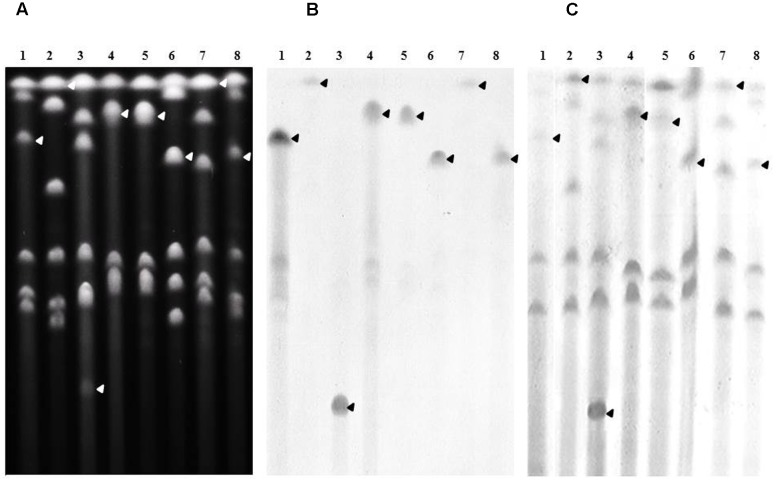
The chromosomal location of genes was assessed by Southern-blots on I-CeuI-PFGE using the probes corresponding to the ESBL/AmpC genes carried by the studied isolates, as well as probes specific for the 23S rDNA. This method revealed that all blaCMY -2 (Figure 3) and blaDHA-16 genes (Supplementary Figure S1) were located on the bacterial chromosome. All blaVEB-6 genes carried by the SGI1-V variant as well as the blaVEB-6 gene identified in isolate 39081 were also proved to be encoded by the chromosome (Supplementary Figure S2). On the contrary, the plasmidic localization of genes was proved by Southern-blots on S1-nuclease-PFGE using adequate probes. The last non-chromosomal blaVEB-6 (38327; Supplementary Figure S3) and blaCTX-M-1 (P. rettgeri 39465; Figure 4) genes were thus shown to be carried on plasmids, which were considered as non-typable because of the total absence of amplification using the PBRT kit. Finally, the P. mirabilis isolate 34146 carried two copies of the ESBL gene blaCTX-M-15, one located on the chromosome and the other on a non-typable plasmid (Figure 4). Whereas ESBL genes were mostly reported on plasmids in Enterobacteriaceae, it is noteworthy to observe that it is seemingly not the case in Proteae. These data strongly suggest that Proteae may be more prone than other Enterobacteriaceae to integrate resistance determinants into the chromosome, mostly on SGI1/PGI1 or other genomic islands.
FIGURE 3.
Chromosomal localization of blaCMY -2 in the relevant P. mirabilis isolates. (A) Whole genomic DNAs of isolates 34381 (lane1), 37665 (lane 2), 38327 (lane 3), 38368 (lane 4), 39165 (lane 5), 39175 (lane 6), 39193 (lane 7), and 39214 (lane 8) were digested with I-CeuI, and the restricted fragments subjected to PFGE. DNA fragments were transferred to a nylon membrane and hybridized with probes specific to blaCMY -2 (B), and the 23S rRNA gene (C). The arrows indicate the bands of interest.
FIGURE 4.
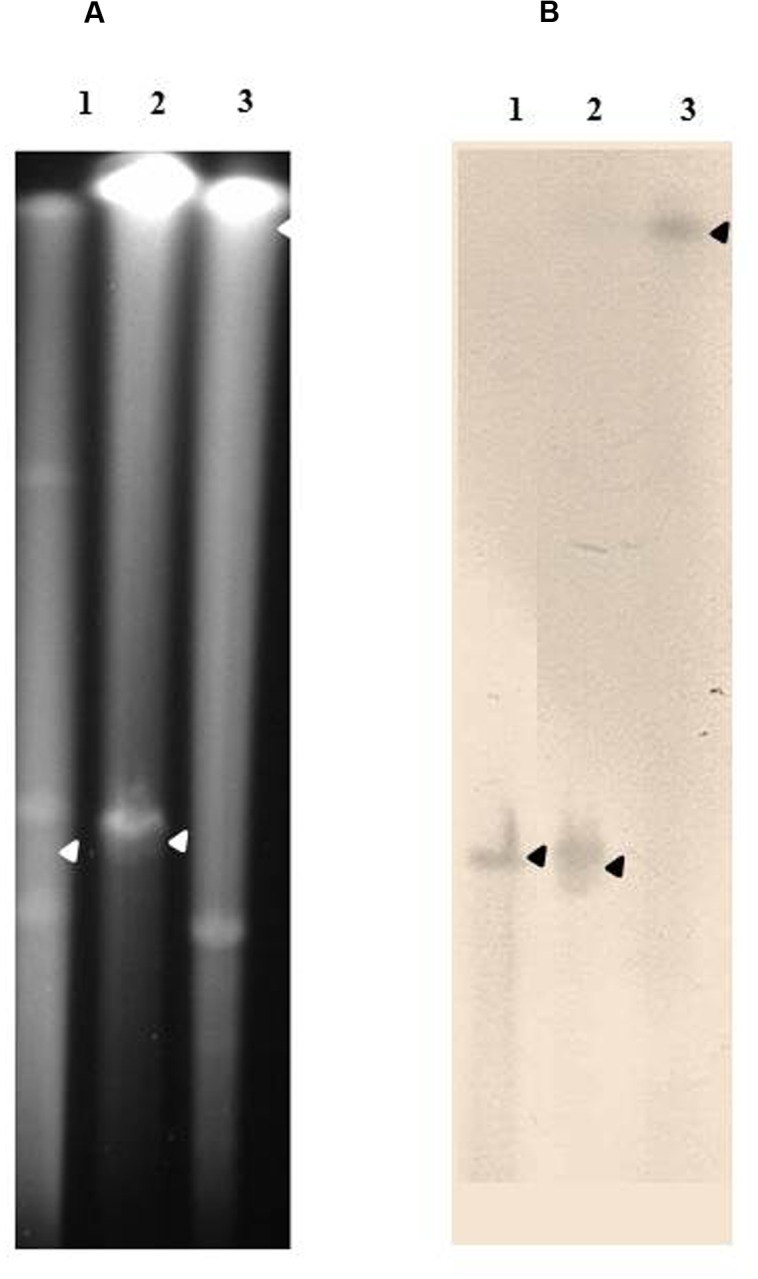
Plasmidic localization of blaCTX-M-1 group genes in the relevant isolates. (A) Whole genomic DNAs of a CTX-M-15-carrying E. coli isolate used as a positive control (lane 1), P. mirabilis isolate 34146 (lane 2), and P. rettgeri isolate 39465 (lane 3), were digested with S1-nuclease, and the restricted fragments subjected to PFGE. DNA fragments were transferred to a nylon membrane and hybridized with a probe specific to blaCTX-M (B). The arrows indicate the bands of interest.
Genetic Diversity of the Isolates
SmaI PFGE revealed that all SGI1/PGI1-positive isolates were genetically unrelated as were the ESBL/AmpC producing isolates (Figure 2), except the six SGI1-V positive isolates that belonged to the same cluster but interestingly differed from the ones previously reported (Schultz et al., 2015). This clustering suggests the spread of a clonal population among different and unrelated individuals. Such a dominance of a clonal population of SGI1-V/blaVEB-6 P. mirabilis strains in humans and companion animals may result from close contacts between these two populations, as also demonstrated for the transfer of other multidrug resistant bacteria or plasmids, such as those carrying ESBL genes. Indeed, the transmission of bacterial clones from animals to humans (or vice-versa) through physical contacts or contact with contaminated saliva or feces has been described (Espinosa-Gongora et al., 2015; Ljungquist et al., 2016).
Conclusion
In this study, we showed a significant prevalence rate (∼4%) of ESBLs/AmpC in P. mirabilis of animal sources. A long-term survey is now needed to decipher whether these are emerging phenotypes or only sporadic cases. The multidrug resistance genomic islands SGI1 and PGI1 play a major role in the dissemination of ESBLs/AmpC genes as well as other non-β-lactam resistance genes. Moreover, we report the spread of the SGI1-V/blaVEB-6-carrying P. mirabilis clonal population to horses. This peculiar ESBL-producing P. mirabilis population was previously recognized in humans and dogs and highly suspected in poultry isolates (Seiffert et al., 2013). Altogether, these data suggest an inter-transmission pathway of public health relevance that needs further investigations to be clarified. Therefore, P. mirabilis should be regarded as a potential reservoir of resistance traits in companion animals making us believe that veterinarians should pay more attention to P. mirabilis as an opportunistic multidrug-resistant pathogen.
Author Contributions
ES, MH, J-YM, BD, and AC designed the experiments. ES did the experiments. ES, MH, and BD analyzed the data. ES and MH drafted the manuscript. BD, J-YM, and AC actively contributed to the manuscript’s writing. All authors approved the final version of this manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Cécile Ponsin for the technical help in the susceptibility testing, Pierre Châtre for performing the BioNumerics analyses, and Karine Praud for the help in the characterization of the genomic islands.
Funding. This work was supported by the French Agency for Food, Environmental and Occupational Health and Safety (ANSES). ES is supported by a Ph.D. fellowship from ANSES and INRA.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.00032/full#supplementary-material
References
- Ahmed A. M., Hussein A. I., Shimamoto T. (2007). Proteus mirabilis clinical isolate harbouring a new variant of Salmonella genomic island 1 containing the multiple antibiotic resistance region. J. Antimicrob. Chemother. 59 184–190. 10.1093/jac/dkl471 [DOI] [PubMed] [Google Scholar]
- Boyd D., Peters G. A., Cloeckaert A., Boumedine K. S., Chaslus-Dancla E., Imberechts H., et al. (2001). Complete nucleotide sequence of a 43-kilobase genomic island associated with the multidrug resistance region of Salmonella enterica serovar Typhimurium DT104 and its identification in phage type DT120 and serovar Agona. J. Bacteriol. 183 5725–5732. 10.1128/JB.183.19.5725-5732.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd D. A., Shi X., Hu Q. H., Ng L. K., Doublet B., Cloeckaert A., et al. (2008). Salmonella genomic island 1 (SGI1), variant SGI1-I, and new variant SGI1-O in Proteus mirabilis clinical and food isolates from China. Antimicrob. Agents Chemother. 52 340–344. 10.1128/AAC.00902-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubenik L. J., Hosgood G. L., Waldron D. R., Snow L. A. (2007). Frequency of urinary tract infection in catheterized dogs and comparison of bacterial culture and susceptibility testing results for catheterized and noncatheterized dogs with urinary tract infections. J. Am. Vet. Med. Assoc. 231 893–899. 10.2460/javma.231.6.893 [DOI] [PubMed] [Google Scholar]
- Carraro N., Matteau D., Luo P., Rodrigue S., Burrus V. (2014). The master activator of IncA/C conjugative plasmids stimulates genomic islands and multidrug resistance dissemination. PLoS Genet. 10:e1004714 10.1371/journal.pgen.1004714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanal C., Bonnet R., De Champs C., Sirot D., Labia R., Sirot J. (2000). Prevalence of beta-lactamases among 1,072 clinical strains of Proteus mirabilis: a 2-year survey in a French hospital. Antimicrob. Agents Chemother. 44k1930–1935. 10.1128/AAC.44.7.1930-1935.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahmen S., Madec J. Y., Haenni M. (2013). F2:A-:B- plasmid carrying the extended-spectrum beta-lactamase blaCTX-M-55/57 gene in Proteus mirabilis isolated from a primate. Int. J. Antimicrob. Agents 41 594–595. 10.1016/j.ijantimicag.2013.02.004 [DOI] [PubMed] [Google Scholar]
- Dallenne C., Da Costa A., Decre D., Favier C., Arlet G. (2010). Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 65 490–495. 10.1093/jac/dkp498 [DOI] [PubMed] [Google Scholar]
- Douard G., Praud K., Cloeckaert A., Doublet B. (2010). The Salmonella genomic island 1 is specifically mobilized in trans by the IncA/C multidrug resistance plasmid family. PLoS ONE 5:e15302 10.1371/journal.pone.0015302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doublet B., Boyd D., Mulvey M. R., Cloeckaert A. (2005). The Salmonella genomic island 1 is an integrative mobilizable element. Mol. Microbiol. 55 1911–1924. 10.1111/j.1365-2958.2005.04520.x [DOI] [PubMed] [Google Scholar]
- Doublet B., Praud K., Bertrand S., Collard J. M., Weill F. X., Cloeckaert A. (2008). Novel insertion sequence- and transposon-mediated genetic rearrangements in genomic island SGI1 of Salmonella enterica serovar Kentucky. Antimicrob. Agents Chemother. 52 3745–3754. 10.1128/AAC.00525-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa-Gongora C., Shah S. Q., Jessen L. R., Bortolaia V., Langebaek R., Bjornvad C. R., et al. (2015). Quantitative assessment of faecal shedding of beta-lactam-resistant Escherichia coli and enterococci in dogs. Vet. Microbiol. 181 298–302. 10.1016/j.vetmic.2015.10.004 [DOI] [PubMed] [Google Scholar]
- Girlich D., Dortet L., Poirel L., Nordmann P. (2015). Integration of the blaNDM-1 carbapenemase gene into Proteus genomic island 1 (PGI1-PmPEL) in a Proteus mirabilis clinical isolate. J. Antimicrob. Chemother. 70 98–102. 10.1093/jac/dku371 [DOI] [PubMed] [Google Scholar]
- Hall R. M. (2010). Salmonella genomic islands and antibiotic resistance in Salmonella enterica. Future Microbiol. 5 1525–1538. 10.2217/fmb.10.122 [DOI] [PubMed] [Google Scholar]
- Harada K., Niina A., Shimizu T., Mukai Y., Kuwajima K., Miyamoto T., et al. (2014). Phenotypic and molecular characterization of antimicrobial resistance in Proteus mirabilis isolates from dogs. J. Med. Microbiol. 63 1561–1567. 10.1099/jmm.0.081539-0 [DOI] [PubMed] [Google Scholar]
- Harada S., Ishii Y., Saga T., Kouyama Y., Tateda K., Yamaguchi K. (2012). Chromosomal integration and location on IncT plasmids of the blaCTX-M-2 gene in Proteus mirabilis clinical isolates. Antimicrob. Agents Chemother. 56 1093–1096. 10.1128/AAC.00258-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada S., Ishii Y., Saga T., Tateda K., Yamaguchi K. (2010). Chromosomally encoded blaCMY-2 located on a novel SXT/R391-related integrating conjugative element in a Proteus mirabilis clinical isolate. Antimicrob. Agents Chemother. 54 3545–3550. 10.1128/AAC.00111-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter S. B., Vauterin P., Lambert-Fair M. A., Van Duyne M. S., Kubota K., Graves L., et al. (2005). Establishment of a universal size standard strain for use with the PulseNet standardized pulsed-field gel electrophoresis protocols: converting the national databases to the new size standard. J. Clin. Microbiol. 43 1045–1050. 10.1128/JCM.43.3.1045-1050.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss J., Papp P. P., Szabo M., Farkas T., Muranyi G., Szakallas E., et al. (2015). The master regulator of IncA/C plasmids is recognized by the Salmonella Genomic island SGI1 as a signal for excision and conjugal transfer. Nucleic Acids Res. 43 8735–8745. 10.1093/nar/gkv758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei C. W., Zhang A. Y., Liu B. H., Wang H. N., Guan Z. B., Xu C. W., et al. (2014). Molecular characteristics of Salmonella genomic island 1 in Proteus mirabilis isolates from poultry farms in China. Antimicrob. Agents Chemother. 58 7570–7572. 10.1128/AAC.03992-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei C. W., Zhang A. Y., Liu B. H., Wang H. N., Yang L. Q., Guan Z. B., et al. (2015). Two novel Salmonella genomic island 1 variants in Proteus mirabilis isolates from swine farms in China. Antimicrob. Agents Chemother. 59 4336–4338. 10.1128/AAC.00120-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S. L., Hessel A., Sanderson K. E. (1993). Genomic mapping with I-Ceu I, an intron-encoded endonuclease specific for genes for ribosomal RNA, in Salmonella spp., Escherichia coli, and other bacteria. Proc. Natl. Acad. Sci. U.S.A. 90 6874–6878. 10.1073/pnas.90.14.6874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljungquist O., Ljungquist D., Myrenas M., Ryden C., Finn M., Bengtsson B. (2016). Evidence of household transfer of ESBL-/pAmpC-producing Enterobacteriaceae between humans and dogs - a pilot study. Infect. Ecol. Epidemiol. 6:31514 10.3402/iee.v6.31514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata C., Navarro F., Miro E., Walsh T. R., Mirelis B., Toleman M. (2011). Prevalence of SXT/R391-like integrative and conjugative elements carrying blaCMY -2 in Proteus mirabilis. J. Antimicrob. Chemother. 66 2266–2270. 10.1093/jac/dkr286 [DOI] [PubMed] [Google Scholar]
- Muranyi G., Szabo M., Olasz F., Kiss J. (2016). Determination and analysis of the putative AcaCD-responsive promoters of Salmonella Genomic Island 1. PLoS ONE 11:e0164561 10.1371/journal.pone.0164561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulin-Laprade D., Carraro N., Burrus V. (2015). The extended regulatory networks of SXT/R391 integrative and conjugative elements and IncA/C conjugative plasmids. Front. Microbiol. 6:837 10.3389/fmicb.2015.00837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin S., Qi H., Zhang Q., Zhao D., Liu Z. Z., Tian H., et al. (2015). Emergence of extensively drug-resistant Proteus mirabilis harboring a conjugative NDM-1 plasmid and a novel Salmonella Genomic Island 1 Variant, SGI1-Z. Antimicrob. Agents Chemother. 59 6601–6604. 10.1128/AAC.00292-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer J. N., Pearson M. M. (2015). Proteus mirabilis and urinary tract infections. Microbiol. Spectr. 3 10.1128/microbiolspec.UTI-0017-2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz E., Haenni M., Mereghetti L., Siebor E., Neuwirth C., Madec J. Y., et al. (2015). Survey of multidrug resistance integrative mobilizable elements SGI1 and PGI1 in Proteus mirabilis in humans and dogs in France, 2010-13. J. Antimicrob. Chemother. 70 2543–2546. 10.1093/jac/dkv154 [DOI] [PubMed] [Google Scholar]
- Seiffert S. N., Tinguely R., Lupo A., Neuwirth C., Perreten V., Endimiani A. (2013). High prevalence of extended-spectrum-cephalosporin-resistant enterobacteriaceae in poultry meat in Switzerland: emergence of CMY-2- and VEB-6-possessing Proteus mirabilis. Antimicrob. Agents Chemother. 57 6406–6408. 10.1128/AAC.01773-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebor E., Neuwirth C. (2011). The new variant of Salmonella genomic island 1 (SGI1-V) from a Proteus mirabilis French clinical isolate harbours blaVEB-6 and qnrA1 in the multiple antibiotic resistance region. J. Antimicrob. Chemother. 66 2513–2520. 10.1093/jac/dkr335 [DOI] [PubMed] [Google Scholar]
- Siebor E., Neuwirth C. (2013). Emergence of Salmonella genomic island 1 (SGI1) among Proteus mirabilis clinical isolates in Dijon, France. J. Antimicrob. Chemother. 68 1750–1756. 10.1093/jac/dkt100 [DOI] [PubMed] [Google Scholar]
- Siebor E., Neuwirth C. (2014). Proteus genomic island 1 (PGI1), a new resistance genomic island from two Proteus mirabilis French clinical isolates. J. Antimicrob. Chemother. 69 3216–3220. 10.1093/jac/dku314 [DOI] [PubMed] [Google Scholar]
- Song W., Kim J., Bae I. K., Jeong S. H., Seo Y. H., Shin J. H., et al. (2011). Chromosome-encoded AmpC and CTX-M extended-spectrum beta-lactamases in clinical isolates of Proteus mirabilis from Korea. Antimicrob. Agents Chemother. 55 1414–1419. 10.1128/AAC.01835-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock I. (2003). Natural antibiotic susceptibility of Proteus spp., with special reference to P. mirabilis and P. penneri strains. J. Chemother. 15 12–26. 10.1179/joc.2003.15.1.12 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.