Abstract
Screening for drug compounds that exhibit therapeutic properties in the treatment of various disease remain a challenge even after considerable advancements in biomedical research. Here we introduce an integrated platform that exploits gene expression compendia generated from drug-treated cell lines and primary tumor tissue to identify therapeutic candidates that can be used in the treatment of acute myeloid leukemia (AML). Our framework combines these data with patient survival information to identify potential candidates that presumably have significant impact on AML patient survival. We use a Drug Regulatory Score (DRS) to measure the similarity between drug-induced cell line and patient tumor gene expression profiles, and show that these computed scores are highly correlated with in vitro metrics of pharmacological activity. Furthermore, we conducted several in vivo validation experiments of our potential candidate drugs in AML mouse models to demonstrate the accuracy of our in silico predictions.
Introduction
Today’s drug discovery pipelines suffer from high rates of drug candidate failure that impose an economic burden on healthcare. Due to the complexity of drug action, identifying drugs that exhibit therapeutic effect for a specific disease is a highly involved process. It has been estimated that US$1.78 billion and 13.5 years are required to take a single therapeutic from its initial identification as a candidate to its availability in the clinic 1. Therefore, there is enormous opportunity to develop more efficient drug identification platforms that integrate in silico approaches with available genomic data. In the case of cancer, there have been several efforts to characterize drug action at the genomic level to understand its functional effect. Large consortia such as the Connectivity Map (CMap) and Genomics of Drug Sensitivity (GDSC) have generated cell-line derived gene expression and drug response data to identify genomic predictors of drug effectiveness 2–4. Here, we chose to utilize data from blood-derived cell lines to develop a novel integrated pre-clinical drug-screening framework to identify drug leads for the treatment of acute myeloid leukemia (AML).
AML is a relatively rare, liquid cancer that is induced upon development of genetic lesions in immature hematocytes that comprise the myeloid lineage. The current model of AML holds that hematopoietic cells contain sequential mutations that propagate through the myeloid lineage during differentiation and self-renewal 5. As the age of an individual increases, mutations accumulate until a key driver mutation occurs that drives uncontrolled cell proliferation 5. Unfortunately, the few therapeutics have been introduced into the market over the past decades 6. To address this issue, we have developed an integrated in silico and in vivo drug-screening framework that utilizes readily available high-throughput genomic data to expedite pre-clinical drug discovery efforts in AML.
Gene expression profiles (GEPs) have been used to computationally search for drug candidates that exhibit potential anticancer activity 7–12. Hassane et al. utilized the gene expression signature of parthenolide, a known AML therapeutic, to search for drugs that could induce a similar signature using a correlation-based procedure 8. Furthermore, Sirota et al. performed a systematic repositioning analysis to identify potential drug candidates for several diseases using a compendia of publicly available gene expression data 9. Similarly, Dudley et al. identified topiramate as a potential candidate to treat inflammatory bowel disease 10. However, these studies did not incorporate patient clinical information into their prediction analyses, which is a key indictor of drug effectiveness.
In this study, we utilize an integrated approach whereby we implement IDEA in AML to derive a set of drug candidate predictions 13. IDEA was previously developed to predict drug candidates by identifying drugs that could induce a GEP associated with breast cancer patient survival 13. This approach differs from previous methods by taking into account time-to-event patient clinical information and by using a more sensitive GEP-matching algorithm. We show that by applying IDEA, drugs associated with AML patient survival correlate with in vitro pharmacological metrics of drug potency and drug-associated molecular features. Finally, we carried out validation experiments in an AML mouse model to show that our predicted survival-associated drugs indeed exhibit pharmacological activity in vivo by substantially slowing tumor growth. Overall, we present a novel integrated pre-clinical drug discovery platform that combines both data-driven and experimental methodologies.
Materials and Methods
Processing of drug treatment profiles
1229 drug treatment profiles (DTPs) corresponding to 1078 different drugs were downloaded from the CMap (Connectivity Map) database 2. Data used in this study were derived from HL-60 (human promyelocytic leukemia cells) cells treated with 1078 different drugs and gene expression was measured by using Affymetrix arrays. The raw .CEL files were processed using the Robust Microarray Analysis method implemented in the R package “affy” 14; all probe sets were represented as absolute expression values. For each drug, relative log2 expression profiles were generated by comparing profiles from treated samples to a common reference profile containing the basal expression values of untreated HL60 cell lines. Probe set expression values were collapsed into gene level expression by taking the average of the probe set log2 ratios, resulting in final DTPs. For some drugs, multiple treatment profiles were available which corresponded to different biological replicates and treatment with different dose concentrations. Each drug’s treatment profile is represented as a vector of log2 ratios, indicating the expression changes of all genes in response to the drug treatment. Each DTP was then split into upregulated gene and downregulated gene groups and z-transformed to derive a p-value. The p-values were then −log10 transformed and used as input into IDEA. Detailed explanation of this process can be found in Ung et al. 13.
AML gene expression data
Normalized AML microarray data containing 562 patient GEPs from Herold et al. were downloaded from GEO under accession number GSE37642 15, 16. 170 AML GEPs published by Wilson et al. was downloaded from the NCI caArray database under the accession number willm-0019 17. The Wouters (n=526) and Valk (n=293) AML datasets were downloaded from the GEO database under the accession numbers GSE14468 and GSE1159, respectively 17–19.
In silico identification of survival-associated drugs
The IDEA computational framework was used to screen for drugs associated with patient survival in AML. Briefly, the drug treatment profiles characterize the effect of drugs on the expression of genes. Genes with larger positive/negative log ratios are those that are regulated more intensively by a drug. We examined the baseline expression levels of the genes in AML patient samples and calculated DRSs for each DTP-tumor sample pair. DRS is a quantitative measure of similarity between a drug treatment profile and an AML sample’s GEP. If genes that are upregulated/downregulated in an AML patient sample also tend to be upregulated/downregulated in HL60 cell lines in response to drug exposure, then the sample will be assigned a large positive DRS corresponding to the drug. Conversely, if an inverse relationship is observed, the corresponding AML sample will be assigned a large negative DRS. When drug regulated genes are randomly distributed in a sorted AML expression profile, the sample will yield a DRS close to 0. The DRS is calculated using a random-walk based algorithm implemented by IDEA. Detailed description of the IDEA framework can be found in Ung et al. 13.
Machine learning analysis
Unsupervised clustering of patients from the Wouters dataset was performed using DRS profiles that were most deviant between good and poor/intermediate cytogenetic risk patients as determined by Wilcoxon rank-sum test 19. This resulted in 94 significant DRS profiles (P<1E-4) that were used as features in the clustering analysis using complete linkage and Euclidean distance. A random forest machine learning model was trained using all DRS profiles as features to classify patients into two (good and poor/intermediate) or three (good, poor, and intermediate) cytogenetic risk groups. 10-fold cross-validation and calculation of the AUC from the ROC curve was performed to evaluate model performance for the two-group classification task. Accuracy (number correct/total) was calculated to evaluate performance in the three-group classification task. Linear discriminant analysis (LDA) was used to validate results from the random forest model. For the two-group classification task, the 94 features identified in the clustering analysis were used. Features for the LDA analysis in the three-group classification task were selected by performing an ANOVA to compare DRS profiles between the three sample groups and an adjusted P<0.01 (Benjamini-Hochberg) was used as the cutoff. This yielded a total of 144 DRS profiles that were used to train the LDA model. R packages “gplots”, “randomForest”, and “MASS” were used to implement clustering, random forest, and LDA analyses, respectively.
Correlation of Trichostatin A DRS with IC50 and mRNA expression
GEPs for 102 blood-derived cell lines along with IC50 values across these cell lines for 125 drugs were downloaded from the GDSC (Genomics of Drug Sensitivity) database 4. Trichostatin A (TSA) DRS was correlated with the IC50 of each drug in the GDSC database across the 102 blood-derived cell lines using Spearman correlation. Correlation of TSA DRS with HDAC2, MEK, and Bcl2 mRNA expression across the same cell lines was performed using the same method.
In vivo validation of survival-associated drugs
The following animal study was approved by the IACUC of National Chung Hsing University. Athymic BALB/c nu/nu nude mice (4–6 weeks of age) were purchased from the National Laboratory Animal Center (Taipei, Taiwan) and mice were maintained in pathogen-free conditions with irradiated chow. HL-60 (ATCC, CCL-240) cells were re-suspended in serum free RPMI-1640 medium mixed with Matrigel (BD Biosciences, San Jose, CA) at a 1:1 ratio. Mice were injected s.c with 5x106 cells in 0.5 ml matrigel into the ventral flank and tumors were allowed to grow for 10 days or until palpable tumors formed (approximately 50 mm3). Tumor-bearing mice were randomly assigned to the following treatment groups: Control (10% DMSO + 90% glyceryl trioctanoate), sulfasalazine (250 mg/kg), Fluoxetine (30 mg/kg), Betulinic acid (20 mg/kg), Clozapine (1 mg/kg). All four drugs were purchased from Sigma-Aldrich (St Louis, MO) and the drug dosages were used by referring to previous reports describing anti-tumor activity of these compounds 20–23. Mice were treated every other day by intraperitoneal injection using 100 μL total volumes. Mean tumors volumes were measured according to the formula: length × width × thickness × 0.5, and expressed as mm3 values before each treatment. Mice were euthanized when tumors reached a size of 200 cm3 or became ulcerated. If individual mice within a group were euthanized, the final measurement was carried over to subsequent time points.
Identification of candidate drugs for AML treatment
We calculated the DRS for 1078 drugs in all samples of an AML expression dataset. To identify the candidate drugs that might be effective for treating AML, we examined the correlation between DRS of these drugs and patient survival. Our rationale is that if the expression of drug-regulated genes is correlated with patient survival, then the drug might be used to modulate these genes in AML to induce a pharmacological effect. For each drug, we fitted a univariate Cox proportional hazards model using the DRS as the independent variable and patient survival as the dependent variable 24. We also fitted multivariate Cox regression models to adjust for potential confounding clinical factors such as age, tumor stage, tumor grade, ER status, etc. Analysis of Schoenfeld residuals was used to evaluate the proportional hazards assumption for all models. The Wald test was used to assess the significance of the model parameters and p-values were adjusted for multiple hypotheses testing using the Benjamini-Hochberg procedure 25. The “survival” R package was used to implement the survival analysis.
GO enrichment analysis
Genes that were up- or downregulated two-fold upon treatment with TSA were used to calculate GO enrichment of BP terms via the DAVID bioinformatics tool (http://david.abcc.ncifcrf.gov/) 26, 27.
Results
Overview of integrated framework
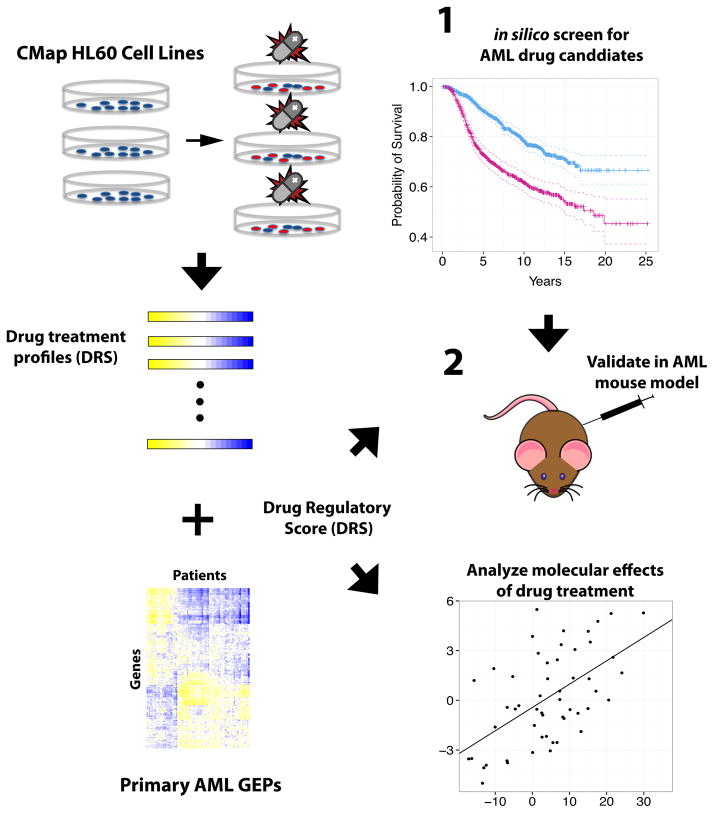
Our integrated pre-clinical drug-screening procedure begins by implementing IDEA (Integrated Drug Expression Analysis), a computational drug prediction framework, to identify drug candidates for AML treatment. IDEA integrates drug treatment profiles from CMap, AML patient tumor GEPs, and AML patient survival information to output drugs that may induce a pharmacological effect that impacts patient survival (Fig 1). Briefly, IDEA calculates a Drug Regulatory Score (DRS) between a drug treatment profile (DTP) and an AML tumor gene expression profile (GEP) which quantitatively measures the level of similarity or dissimilarity between the two profiles (See Methods) 13. DRS are calculated for all DTP and tumor GEP pairs and the DRS profile of each drug (DRS across patient samples) were fitted with a Cox proportional hazards model to identify top candidates that presumably have an effect on patient survival (Fig 1). Several predicted candidates are then selected for experimental validation in an AML mouse model (Fig 1).
Fig. 1. Flowchart of integrated screening pipeline.
Drug treatment profiles from HL60 cell lines were integrated with AML patient gene expression profiles to derive DRS profiles for each drug. Survival analysis of DRS profiles was implemented to identify drugs associated with patient survival. Selected candidates were then validated in an AML mouse model.
Systematic screening of drugs associated with AML patient survival
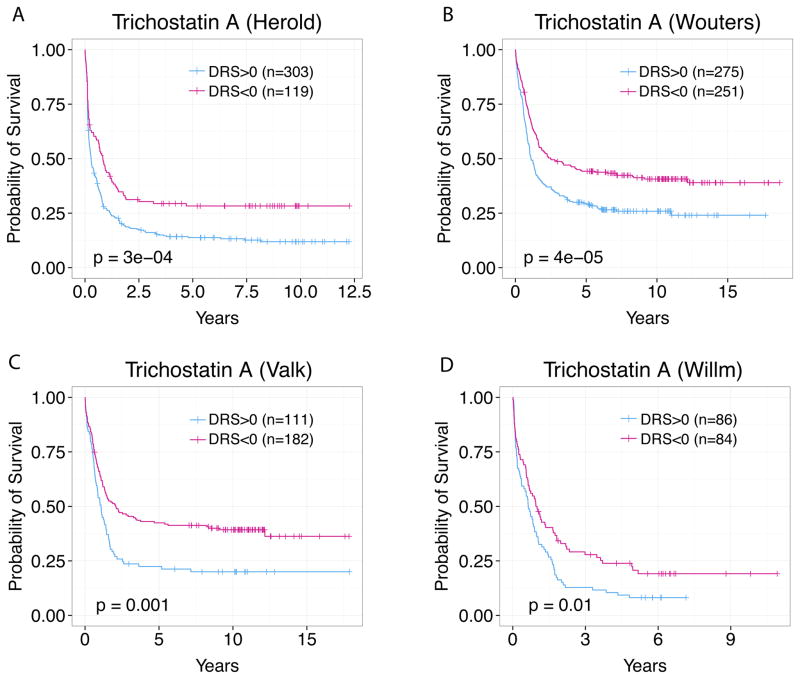
At an adjusted p-value cutoff of 0.01, we identified 66 drugs using the Herold dataset that are significantly associated with AML patient survival to validate in vivo (Suppl. Table S1) 28. We re-implemented this analysis in the Verhaak, Valk, and Wilson AML datasets and achieved similar results 17–19. Figure 2 shows an example where TSA DRS profiles effectively stratify AML patients into favorable and poor prognosis groups in four independent datasets 17–19. This indicates that IDEA’s output is reproducible and that TSA exhibits pharmacological activity in several cancer types, as shown by previous studies 21, 24, 29–31. This in silico screening approach filters out biologically inactive drugs and efficiently identifies the most probable candidates based on gene expression and patient survival.
Fig. 2. TSA was identified as a drug candidate in four datasets.
TSA DRS profile is prognostic in the (A) Herold dataset, (B) Verhaak dataset, (C) Valk dataset, (D) and William dataset with p<0.01. Patients were stratified at DRS = 0
Drug DRS profiles are predictive of cytogenetic risk in AML
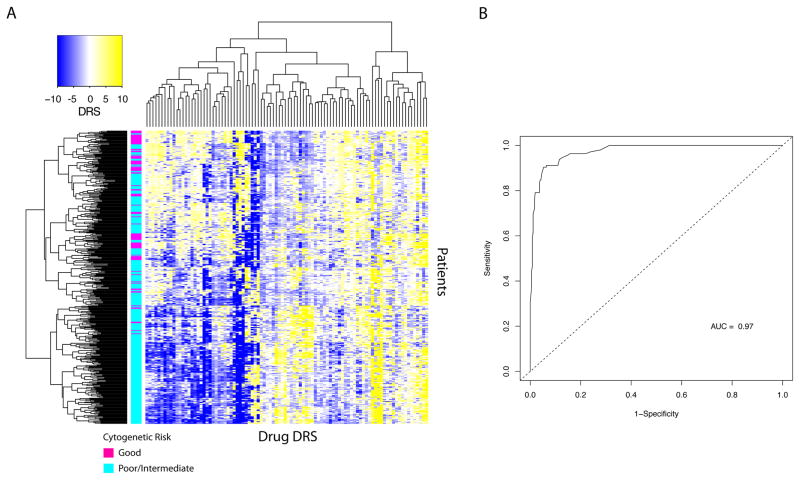
Since drug DRS are prognostic, we hypothesized that DRS can predict cellular phenotypes that have been traditionally known to correlate with patient survival. As such, we attempted to predict cytogenetic risk of patients based on drug DRS profiles 32, 33. Using the Wouters dataset, we selected DRS profiles that differed significantly between patients classified as having “good” cytogenetic risk or “poor/intermediate” cytogenetic risk using an adjusted p-value cutoff of 1E-4 (Wilcoxon rank-sum test) 19. This yielded 94 DRS profiles that were used to cluster patients into subgroups. We found that these DRS profiles were informative such that they were able to cluster patients into cytogenetic risk groups (Fig 3A). In particular, we identified three apparent clusters corresponding to patient cytogenetic risk. The first cluster (top) had 50 samples with good cytogenetic risk (49.4%), the second cluster had 40 samples with good cytogenetic risk (25.3%), and the third cluster had 7 samples with good cytogenetic risk (3.8%) (Fig 3A). This indicates that DRS profiles effectively identified differences in cytogenetic risk between acute myeloid leukemia samples, showing that they reflect prognostic molecular features of tumors.
Fig. 3. DRS profiles are predictive of cytogenetic risk in AML.
(A) Unsupervised clustering of DRS profiles. Magenta sample labels denote good cytogenetic risk and aqua sample labels indicate poor/intermediate cytogenetic risk (B) Receiver operating characteristic curve for random forest classification of patients into good or poor/intermediate cytogenetic risk categories.
To further evaluate our unsupervised results, we trained a random forest model using DRS profiles from all drugs to verify their ability predict patient cytogenetic risk. We evaluated the model’s performance by implementing a 10-fold cross validation procedure, from which the model achieved an AUC of 0.97 calculated from the ROC (Receive Operating Characteristic) curve (Fig 3B). Furthermore, we extended the random forest analysis to categorize patient samples into good, intermediate, or poor cytogenetic risk groups and was able to correctly classify 71% of the samples. As validation, we also implemented a linear discriminant analysis (LDA) model that achieved an AUC of 0.97 for the two-group classification task. Additionally, the LDA model was able to correctly classify 76% of the samples for the three-group task (See methods). Similarly, Zhou et al. reported comparable accuracy when predicting cytogenetic risk using GEPs 34. Ultimately, these results indicate that DRS profiles contain information that reflects phenotypic differences between patient groups.
Correlation of DRS with in vitro pharmacological metrics
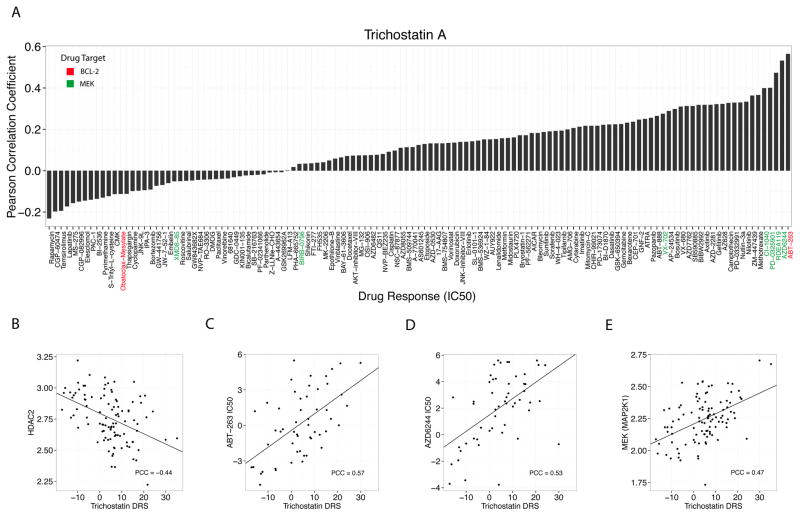
To explore potential mechanisms of action underlying our predicted drugs, we adopted a novel integrated validation procedure whereby we implemented IDEA in 102 GDSC (Genomics of Drug Sensitivity) blood-derived cell lines and correlated the outputted DRS with treatment response metrics. Since the drugs included in the GDSC dataset have known targets, this correlation analysis allowed us to gain insight into the biological mechanisms underlying drug treatment with CMap drugs. We calculated DRS between CMap DTPs and GDSC blood-derived cell line GEPs and correlated the DRS of each CMap drug with the IC50 of each GDSC drug across blood-derived GDSC cell lines (Fig 4A). We repeated this analysis using other pharmacological metrics (Area under the curve and EC50) derived from dose-response curves and achieved consistent results. To note, CMap provides before and after drug treatment GEPs for three cell lines (drug-centric), whereas GDSC provides profiles that measure basal gene expression of several cell lines (cell line-centric).
Fig. 4. Correlation of TSA with drug IC50 and mRNA expression across 102 blood-derived GDSC cell lines.
(A) Correlation of TSA DRS with IC50 of 125 drugs from the GDSC database (B) TSA DRS is anticorrelated with HDAC2 mRNA expression, (C) TSA DRS is correlated with ABT-263 (Bcl2 inhibitor) IC50 (D) TSA DRS is correlated with AZD6244 (MEK inhibitor) IC50 (E) TSA DRS is correlated with MEK mRNA expression.
Interestingly, we found that TSA DRS was most correlated with the IC50 of ABT-263 (navitoclax), a Bcl2 inhibitor, with a Pearson correlation coefficient (PCC) of 0.57 and P=1E-5 (Fig 4A, 4C). Furthermore, we identified MEK inhibitors that were also highly correlated including AZD6244 (PCC=0.53), RDEA119 (PCC=0.47), PD0325901 (PCC=0.4), and CI-1040 (PCC=0.4) (Fig 4A). These results suggest that Bcl2 and MEK pathways are involved in response to TSA treatment. To further evaluate their involvement, we correlated the TSA DRS with HDAC2, Bcl2, and MEK mRNA expression across the 102 GDSC blood-derived cell lines. As expected, the TSA DRS was anti-correlated with HDAC2 expression. (PCC=−0.44, P=4.5E-6) (Fig 4B). However, we found that there was a significant, albeit weak, anti-correlation between TSA DRS and Bcl2 expression (PCC=-0.2, P=0.04) even though there was a strong correlation between TSA DRS and ABT-263 IC50 (Fig S1, Fig 4C). In spite of this, the directionality of the correlation is in accordance with previous studies reporting that increased Bcl2 expression confers sensitivity to Bcl2 inhibitors 35, 36. However, the small effect size suggests a more complicated relationship between HDAC inhibition and BCL-2 expression. In the case of MEK, we observed that TSA DRS was correlated with both MEK expression (PCC=0.47, P=6.2E-7) and AZD6244 IC50 (PCC=0.53, P=4.1E-5) (Fig 4D, 4E). This suggests that high TSA DRS, which indicates decreased HDAC expression, results in an upregulation of MEK that explains why increased dosage concentration of AZD6244, RDEA119, PD0325901, and CI-1040 is required to achieve 50% cellular inhibition in vitro. Indeed, several studies have shown that HDAC inhibitors and MEK inhibitors exhibit synergistic effects in leukemia indicating that our analysis was able to identify molecular mechanisms underlying drug effect 37–40.
To compare, we carried out Gene Ontology enrichment analysis of the top up- and downregulated genes in the TSA drug treatment profile (Suppl. Table S2) 41. Surprisingly, we found no significant cancer-related biological processes enriched in the differentially expressed genes. We speculate that since TSA is a histone deacetylase inhibitor, it will have widespread, yet small downstream effects on genes that may or may not have a functional effect. As a result, standard enrichment analysis may not be sensitive enough to detect key genes involved in apoptosis or cell proliferation. These results suggest that correlating TSA DRS with phenotypic response to targeted inhibitors may be a better strategy for identifying functional pathways that result from TSA treatment. However, we caution that this interpretation is only valid for TSA, and may not be generalizable to other drugs.
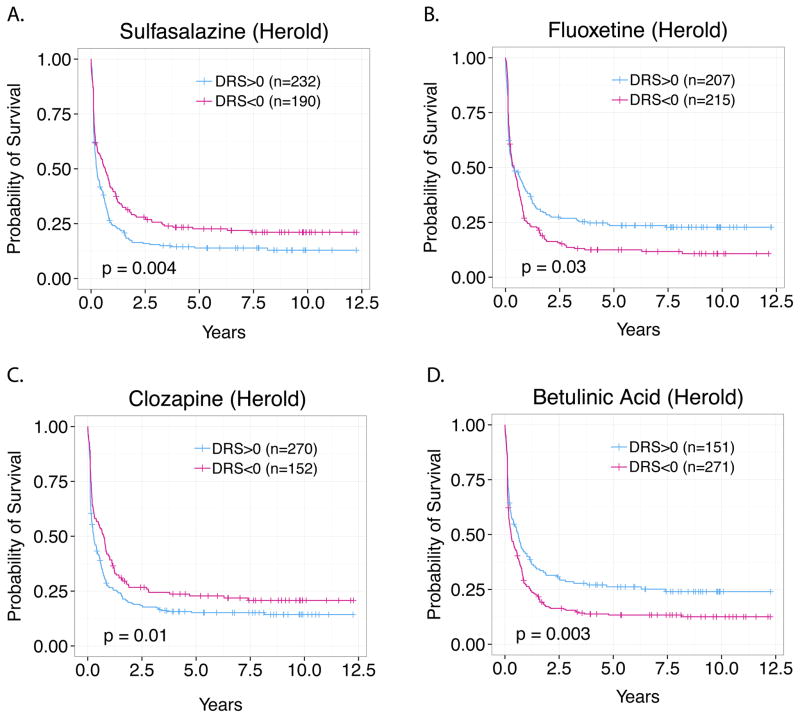
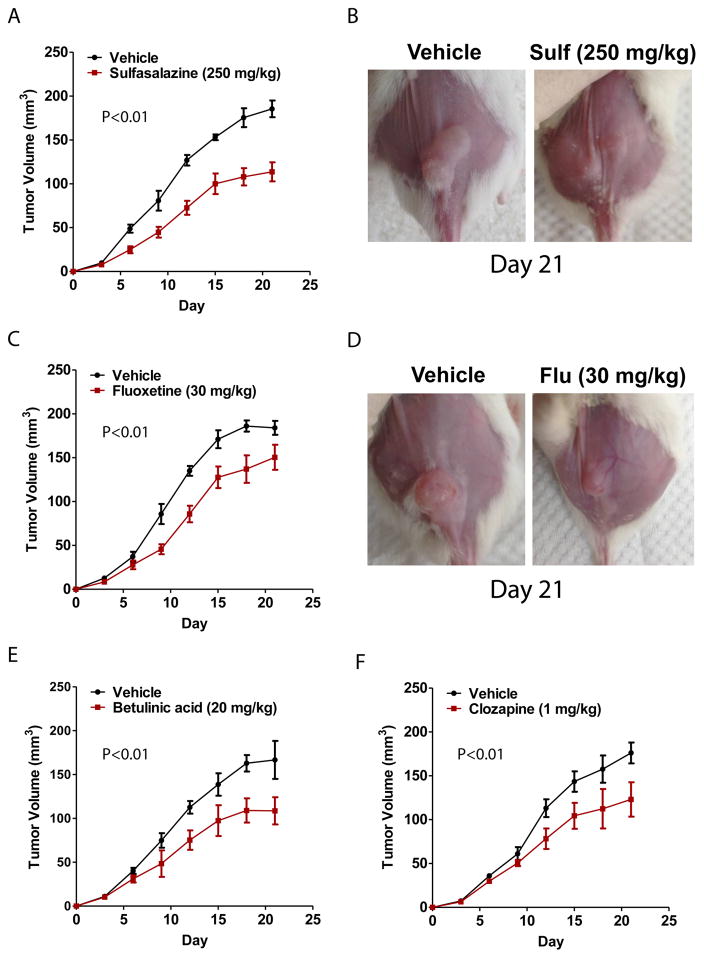
In vivo validation of novel predicted drugs for AML treatment: Sulfasalazine, Fluoxetine, Betulinic Acid, Clozapine
To translate our in silico drug-screening procedure into the pre-clinical testing phase, we identified five novel survival-associated drugs that were predicted by IDEA and experimentally evaluated their effectiveness in an AML mouse model. In particular, we tested sulfasalazine, fluoxetine, clozapine, betulinic acid, and ceforanide, which were originally intended to treat arthritis, depression, schizophrenia, viral infections, and bacterial infections, respectively 42–46. All five of these drugs were predicted to impact patient survival via pharmacological activity as shown by Figs. 5A–5D in the Herold dataset 15. To generate our AML mouse models we engrafted athymic BALB/c nu/nu mice with HL-60 (Human promyelocytic leukemia cells) via subcutaneous xenografts. Each drug was then tested for therapeutic activity by treating mice with either vehicle (10% DMSO + 90% glyceryl trioctanoate) alone or standard doses of the drug candidate. We found that 4 out of 5 (ceforanide showed no substantial effect) of our predicted drugs exhibited significant therapeutic activity. First, we found that treatment with 250 mg/kg of sulfasalazine substantially decelerated tumor growth compared to vehicle over a 21-day period (Figs. 6A, 6B). Second, in the case of fluoxetine, daily treatment with 30 mg/kg also decreased the rate of tumor growth compared to vehicle (Figs. 6C, 6D). Third, we observed that daily treatment with 20 mg/kg of betulinic acid also decelerated tumor growth (Fig. 6E). Lastly, we observed similar antineoplastic activity of clozapine at a dose of 1 mg/kg (Fig. 6F). These results suggest that our initial in silico screen was able to output several survival-associated drugs, the majority of which could be verified in vivo.
Fig. 5. Survival analysis of Sulfasalazine, Fluoxetine, Clozapine, and Betulinic Acid DRS profiles.
(A) Sulfasalazine, (B) Fluoxetine, (C) Clozapine, (D) and Betulinic Acid were identified as drugs associated with AML patient survival and chosen for further experimental validation. DRS profiles of all drugs were associated with AML patient survival with P<0.05 (Logrank test).
Fig. 6. In vivo validation of Sulfasalazine, Fluoxetine, Clozapine, and Betulinic Acid in AML mouse models.
(A) 250 mg/kg Sulfasalazine decreased the rate of tumor growth compared to vehicle-treated control over a course of 21 days. (B) Image of tumor from mouse treated with 250 mg/kg Sulfasalazine compared to control (C) 30 mg/kg Fluoxetine decreased the rate of tumor growth compared to vehicle-treated control over a course of 21 days. (D) Image of tumor from mouse treated with 250 mg/kg Fluoxetine compared to control. (E) 20 mg/kg Betulinic acid decreased the rate of tumor growth compared to vehicle-treated control over a course of 21 days. (F) 1 mg/kg Clozapine decreased the rate of tumor growth compared to vehicle-treated control over a course of 21 days.
Discussion
Drug discovery has long been a focus of intense research due to the constant need for therapeutics that can treat disease and ameliorate symptoms. In the case of cancer, rapid development of acquired resistance to commonly prescribed chemotherapeutics and targeted therapies necessitates the formulation of faster and more efficient drug-screening pipelines. Here, we implemented a computational drug prediction framework in acute myeloid leukemia and were able to experimentally validate several of our drug predictions to identify candidates. We explored the association between IDEA and pharmacological metrics to gain mechanistic insight into drug action and our experimental results show a substantial reduction in tumor growth when mice were treated with 4 different survival-associated drugs over a 21-day period.
We note that there are limitations to our approach. First, the reliability of drug treatment profiles, which were derived by averaging gene expression over several replicate experiments, may exhibit variability. Second, these drug treatment profiles were generated over different concentrations and may not reflect optimal drug activity. Third, we note that the limited time interval over which mice were treated provides a short-term evaluation of drug effectiveness and that anti-tumor activity may not be sustained and/or side effects may present itself after prolonged exposure. Finally, it is difficult to interpret the hazard ratios of the top drugs outputted by IDEA. Since patient tumors from our datasets were collected prior to treatment, the survival results may have been influenced by subsequent therapy. This may be why some known anti-cancer drugs were associated with a hazard ratio >1. Thus, we claim that any significant association that exists between the drug and patient survival indicates pharmacological activity, which merits further experimental investigation.
Despite these obstacles, we maintain that our integrated pipeline is robust and sensitive enough to detect potential drug candidates. In particular, we have shown that we could computationally identify known therapeutics (e.g. TSA) and novel candidates. We further support our predictions by conducting in vivo experiments that show our drug leads exhibit therapeutic activity in an AML mouse model. Our results strongly support the effectiveness of using an integrated in silico/in vivo approach to drug screening in the context of patient survival.
Supplementary Material
Fig. S1. Correlation of TSA DRS with Bcl2 mRNA expression.
Acknowledgments
We would like to thank Tobias Herold, Wolfgang Hiddemann, Thomas Büchner, Karsten Spiekermann, Stephanie Schneider, Maria Cristina Sauerland for providing us with the patient survival information for the Herold dataset. We also thank Roel Veerhak for providing us with the patient survival information for the Wouter’s dataset.
Funding: This work was supported by the American Cancer Society Research Grant, #IRG-82-003-30, the National Center For Advancing Translational Sciences of the National Institutes of Health under Award Number UL1TR001086, and by the start-up funding package provided to CC by the Geisel School of Medicine at Dartmouth College.
Footnotes
Author contributions: CC, CL (Liu), CL (Lin) conceived of the project, MU, CS, CW, CL (Liu) and CC performed the computational drug prediction analysis, CH and CL (Lin) performed the mouse experiments, all authors contributed to the writing of the manuscript and interpretation of results.
Competing interests: The authors declare no conflict of interest.
References
- 1.Hurle MR, Yang L, Xie Q, Rajpal DK, Sanseau P, Agarwal P. Computational drug repositioning: from data to therapeutics. Clinical pharmacology and therapeutics. 2013;93(4):335–341. doi: 10.1038/clpt.2013.1. [DOI] [PubMed] [Google Scholar]
- 2.Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313(5795):1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 3.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483(7391):603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang W, Soares J, Greninger P, Edelman EJ, Lightfoot H, Forbes S, et al. Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic acids research. 2013;41(Database issue):D955–961. doi: 10.1093/nar/gks1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Welch JS, Ley TJ, Link DC, Miller CA, Larson DE, Koboldt DC, et al. The origin and evolution of mutations in acute myeloid leukemia. Cell. 2012;150(2):264–278. doi: 10.1016/j.cell.2012.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burnett A, Wetzler M, Lowenberg B. Therapeutic advances in acute myeloid leukemia. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2011;29(5):487–494. doi: 10.1200/JCO.2010.30.1820. [DOI] [PubMed] [Google Scholar]
- 7.Iorio F, Bosotti R, Scacheri E, Belcastro V, Mithbaokar P, Ferriero R, et al. Discovery of drug mode of action and drug repositioning from transcriptional responses. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(33):14621–14626. doi: 10.1073/pnas.1000138107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hassane DC, Guzman ML, Corbett C, Li X, Abboud R, Young F, et al. Discovery of agents that eradicate leukemia stem cells using an in silico screen of public gene expression data. Blood. 2008;111(12):5654–5662. doi: 10.1182/blood-2007-11-126003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sirota M, Dudley JT, Kim J, Chiang AP, Morgan AA, Sweet-Cordero A, et al. Discovery and preclinical validation of drug indications using compendia of public gene expression data. Science translational medicine. 2011;3(96):96ra77. doi: 10.1126/scitranslmed.3001318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dudley JT, Sirota M, Shenoy M, Pai RK, Roedder S, Chiang AP, et al. Computational repositioning of the anticonvulsant topiramate for inflammatory bowel disease. Science translational medicine. 2011;3(96):96ra76. doi: 10.1126/scitranslmed.3002648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang K, Sun J, Zhou S, Wan C, Qin S, Li C, et al. Prediction of drug-target interactions for drug repositioning only based on genomic expression similarity. PLoS computational biology. 2013;9(11):e1003315. doi: 10.1371/journal.pcbi.1003315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pacini C, Iorio F, Goncalves E, Iskar M, Klabunde T, Bork P, et al. DvD: An R/Cytoscape pipeline for drug repurposing using public repositories of gene expression data. Bioinformatics. 2013;29(1):132–134. doi: 10.1093/bioinformatics/bts656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ung MH, Varn FS, Cheng C. IDEA: Integrated Drug Expression Analysis - Integration of Gene Expression and Clinical Data for the Identification of Therapeutic Candidates. CPT: Pharmacometrics Syst Pharmacol. 2015 doi: 10.1002/psp4.51. Accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 15.Herold T, Metzeler KH, Vosberg S, Hartmann L, Rollig C, Stolzel F, et al. Isolated trisomy 13 defines a homogeneous AML subgroup with high frequency of mutations in spliceosome genes and poor prognosis. Blood. 2014;124(8):1304–1311. doi: 10.1182/blood-2013-12-540716. [DOI] [PubMed] [Google Scholar]
- 16.Appelbaum FR, Gundacker H, Head DR, Slovak ML, Willman CL, Godwin JE, et al. Age and acute myeloid leukemia. Blood. 2006;107(9):3481–3485. doi: 10.1182/blood-2005-09-3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilson CS, Davidson GS, Martin SB, Andries E, Potter J, Harvey R, et al. Gene expression profiling of adult acute myeloid leukemia identifies novel biologic clusters for risk classification and outcome prediction. Blood. 2006;108(2):685–696. doi: 10.1182/blood-2004-12-4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Valk PJ, Verhaak RG, Beijen MA, Erpelinck CA, Barjesteh van Waalwijk van Doorn-Khosrovani S, Boer JM, et al. Prognostically useful gene-expression profiles in acute myeloid leukemia. The New England journal of medicine. 2004;350(16):1617–1628. doi: 10.1056/NEJMoa040465. [DOI] [PubMed] [Google Scholar]
- 19.Wouters BJ, Lowenberg B, Erpelinck-Verschueren CA, van Putten WL, Valk PJ, Delwel R. Double CEBPA mutations, but not single CEBPA mutations, define a subgroup of acute myeloid leukemia with a distinctive gene expression profile that is uniquely associated with a favorable outcome. Blood. 2009;113(13):3088–3091. doi: 10.1182/blood-2008-09-179895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peiffer L, Poll-Wolbeck SJ, Flamme H, Gehrke I, Hallek M, Kreuzer KA. Trichostatin A effectively induces apoptosis in chronic lymphocytic leukemia cells via inhibition of Wnt signaling and histone deacetylation. Journal of cancer research and clinical oncology. 2014;140(8):1283–1293. doi: 10.1007/s00432-014-1689-0. [DOI] [PubMed] [Google Scholar]
- 21.Kosugi H, Towatari M, Hatano S, Kitamura K, Kiyoi H, Kinoshita T, et al. Histone deacetylase inhibitors are the potent inducer/enhancer of differentiation in acute myeloid leukemia: a new approach to anti-leukemia therapy. Leukemia. 1999;13(9):1316–1324. doi: 10.1038/sj.leu.2401508. [DOI] [PubMed] [Google Scholar]
- 22.Bolden JE, Peart MJ, Johnstone RW. Anticancer activities of histone deacetylase inhibitors. Nature reviews Drug discovery. 2006;5(9):769–784. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- 23.Massari NA, Medina VA, Cricco GP, Martinel Lamas DJ, Sambuco L, Pagotto R, et al. Antitumor activity of histamine and clozapine in a mouse experimental model of human melanoma. Journal of dermatological science. 2013;72(3):252–262. doi: 10.1016/j.jdermsci.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 24.Platta CS, Greenblatt DY, Kunnimalaiyaan M, Chen H. The HDAC inhibitor trichostatin A inhibits growth of small cell lung cancer cells. The Journal of surgical research. 2007;142(2):219–226. doi: 10.1016/j.jss.2006.12.555. [DOI] [PubMed] [Google Scholar]
- 25.Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Statistics in medicine. 1990;9(7):811–818. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
- 26.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 27.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic acids research. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Law V, Knox C, Djoumbou Y, Jewison T, Guo AC, Liu Y, et al. DrugBank 4. 0: shedding new light on drug metabolism. Nucleic acids research. 2014;42(Database issue):D1091–1097. doi: 10.1093/nar/gkt1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vigushin DM, Ali S, Pace PE, Mirsaidi N, Ito K, Adcock I, et al. Trichostatin A is a histone deacetylase inhibitor with potent antitumor activity against breast cancer in vivo. Clinical cancer research : an official journal of the American Association for Cancer Research. 2001;7(4):971–976. [PubMed] [Google Scholar]
- 30.Johnstone RW. Histone-deacetylase inhibitors: novel drugs for the treatment of cancer. Nature reviews Drug discovery. 2002;1(4):287–299. doi: 10.1038/nrd772. [DOI] [PubMed] [Google Scholar]
- 31.Shaker S, Bernstein M, Momparler LF, Momparler RL. Preclinical evaluation of antineoplastic activity of inhibitors of DNA methylation (5-aza-2′-deoxycytidine) and histone deacetylation (trichostatin A, depsipeptide) in combination against myeloid leukemic cells. Leukemia research. 2003;27(5):437–444. doi: 10.1016/s0145-2126(02)00222-9. [DOI] [PubMed] [Google Scholar]
- 32.Mrozek K, Heinonen K, de la Chapelle A, Bloomfield CD. Clinical significance of cytogenetics in acute myeloid leukemia. Seminars in oncology. 1997;24(1):17–31. [PubMed] [Google Scholar]
- 33.Slovak ML, Kopecky KJ, Cassileth PA, Harrington DH, Theil KS, Mohamed A, et al. Karyotypic analysis predicts outcome of preremission and postremission therapy in adult acute myeloid leukemia: a Southwest Oncology Group/Eastern Cooperative Oncology Group Study. Blood. 2000;96(13):4075–4083. [PubMed] [Google Scholar]
- 34.Zhou Y, Zhang Q, Stephens O, Heuck CJ, Tian E, Sawyer JR, et al. Prediction of cytogenetic abnormalities with gene expression profiles. Blood. 2012;119(21):e148–150. doi: 10.1182/blood-2011-10-388702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Merino D, Khaw SL, Glaser SP, Anderson DJ, Belmont LD, Wong C, et al. Bcl-2, Bcl-x(L), and Bcl-w are not equivalent targets of ABT-737 and navitoclax (ABT-263) in lymphoid and leukemic cells. Blood. 2012;119(24):5807–5816. doi: 10.1182/blood-2011-12-400929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tahir SK, Wass J, Joseph MK, Devanarayan V, Hessler P, Zhang H, et al. Identification of expression signatures predictive of sensitivity to the Bcl-2 family member inhibitor ABT-263 in small cell lung carcinoma and leukemia/lymphoma cell lines. Molecular cancer therapeutics. 2010;9(3):545–557. doi: 10.1158/1535-7163.MCT-09-0651. [DOI] [PubMed] [Google Scholar]
- 37.Yu C, Dasmahapatra G, Dent P, Grant S. Synergistic interactions between MEK1/2 and histone deacetylase inhibitors in BCR/ABL+ human leukemia cells. Leukemia. 2005;19(9):1579–1589. doi: 10.1038/sj.leu.2403868. [DOI] [PubMed] [Google Scholar]
- 38.Ozaki K, Kosugi M, Baba N, Fujio K, Sakamoto T, Kimura S, et al. Blockade of the ERK or PI3K-Akt signaling pathway enhances the cytotoxicity of histone deacetylase inhibitors in tumor cells resistant to gefitinib or imatinib. Biochemical and biophysical research communications. 2010;391(4):1610–1615. doi: 10.1016/j.bbrc.2009.12.086. [DOI] [PubMed] [Google Scholar]
- 39.Chang-Yew Leow C, Gerondakis S, Spencer A. MEK inhibitors as a chemotherapeutic intervention in multiple myeloma. Blood cancer journal. 2013;3:e105. doi: 10.1038/bcj.2013.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nishioka C, Ikezoe T, Yang J, Koeffler HP, Yokoyama A. Inhibition of MEK/ERK signaling synergistically potentiates histone deacetylase inhibitor-induced growth arrest, apoptosis and acetylation of histone H3 on p21waf1 promoter in acute myelogenous leukemia cell. Leukemia. 2008;22(7):1449–1452. doi: 10.1038/sj.leu.2405079. [DOI] [PubMed] [Google Scholar]
- 41.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium Nature genetics. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gupta AK, Grober JS, Hamilton TA, Ellis CN, Siegel MT, Voorhees JJ, et al. Sulfasalazine therapy for psoriatic arthritis: a double blind, placebo controlled trial. The Journal of rheumatology. 1995;22(5):894–898. [PubMed] [Google Scholar]
- 43.Wong DT, Bymaster FP, Engleman EA. Prozac (fluoxetine, Lilly 110140), the first selective serotonin uptake inhibitor and an antidepressant drug: twenty years since its first publication. Life sciences. 1995;57(5):411–441. doi: 10.1016/0024-3205(95)00209-o. [DOI] [PubMed] [Google Scholar]
- 44.Kane J, Honigfeld G, Singer J, Meltzer H. Clozapine for the treatment-resistant schizophrenic. A double-blind comparison with chlorpromazine. Archives of general psychiatry. 1988;45(9):789–796. doi: 10.1001/archpsyc.1988.01800330013001. [DOI] [PubMed] [Google Scholar]
- 45.Chowdhury AR, Mandal S, Mittra B, Sharma S, Mukhopadhyay S, Majumder HK. Betulinic acid, a potent inhibitor of eukaryotic topoisomerase I: identification of the inhibitory step, the major functional group responsible and development of more potent derivatives. Medical science monitor : international medical journal of experimental and clinical research. 2002;8(7):BR254–265. [PubMed] [Google Scholar]
- 46.Campoli-Richards DM, Lackner TE, Monk JP. Ceforanide. A review of its antibacterial activity, pharmacokinetic properties and clinical efficacy. Drugs. 1987;34(4):411–437. doi: 10.2165/00003495-198734040-00001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Correlation of TSA DRS with Bcl2 mRNA expression.