Abstract
Sigma factors, the key regulatory components of the bacterial RNA polymerase (RNAP), direct promoter DNA binding and DNA melting. The σ54-RNAP forms promoter complexes in which DNA melting is only triggered by an activator and ATP hydrolysis-driven reorganisation of an initial σ54-RNAP–promoter complex. We report that an initial bacterial RNAP–DNA complex can be reorganised by an activator to form an intermediate transcription initiation complex where full DNA melting has not yet occurred. Using σ54 as a chemical nuclease we now show that the reorganisation of the initial σ54-RNAP–promoter complex occurs upon interaction with the activator at the transition point of ATP hydrolysis. We demonstrate that this reorganisation event is an early step in the transcription initiation pathway that occurs independently of RNAP parts normally associated with stable DNA melting and open complex formation. Using photoreactive DNA probes, we provide evidence that within this reorganised σ54-RNAP–promoter complex, DNA contacts across the ‘to be melted' sequences are made by the σ54 subunit. Strikingly, the activator protein, but not core RNAP subunits, is close to these DNA sequences.
Keywords: AAA activator, DNA cleavage, photo-crosslinking, RNA polymerase, sigma factors
Introduction
Transcription is the first step in gene expression and is a major point of regulation. The DNA-dependent RNA polymerase (RNAP) is the central enzyme of gene expression whose core machinery is conserved throughout evolution (Ebright, 2000). In bacteria, the five-subunit core RNAP (subunit composition α2ββ′ω; E) has to collaborate with a sixth subunit, the sigma (σ) factor, to form the RNAP holoenzyme (subunit composition α2ββ′ωσ; Eσ) for promoter-specific transcription initiation. The common form of bacterial RNAP contains the σ70-type σ factors (so-called after the prototypical housekeeping σ70 factor of Escherichia coli) and utilises promoters characterised by conserved sequences centred at positions −35 and −10 from the transcription start point at +1 (Paget and Helmann, 2003). The variant RNAP form contains the σ54 factor. Eσ54 binds to promoters characterised by conserved sequences at positions −24 (the GG-region) and −12 (the GC-region) from the transcription start point (Buck et al, 2000).
In contrast to Eσ70, when Eσ54 binds promoters it forms transcriptionally inactive closed promoter complexes. Within the Eσ54 closed promoter complex, the bases immediately downstream of the consensus −12 GC-region are transiently distorted, thereby creating a fork junction DNA structure (Buck et al, 2000). This fork junction structure is used by Eσ54 to set up a network of interactions that prevent DNA melting in the absence of activation. DNA melting and formation of a transcription-competent Eσ54 open complex requires a specialised activator of the AAA (ATPases Associated with various cellular Activities) family (Buck et al, 2000; Zhang et al, 2002). Activators of Eσ54 bind to enhancer-like sequences located ∼150 bp upstream of the transcription start point and use ATP hydrolysis to interact (via a DNA looping event) with a nucleoprotein interface at the promoter GC-region of the Eσ54 closed promoter complex, referred to as the Eσ54 regulatory centre. The Eσ54 regulatory centre constitutes a tripartite interface involving regulatory σ54 residues (notably the amino-terminal Region I and residues within Region III; Figure 1A), DNA sequences immediately downstream of the consensus GC-region and surfaces of the core RNAP upstream of the RNAP catalytic centre (Wigneshweraraj et al, 2001; 2002; 2003). Activators of Eσ54 bring about conformational changes in the Eσ54 regulatory centre, altering the interaction between regulatory σ54 residues and the fork junction DNA structure that results in DNA melting and open complex formation (Buck et al, 2000). In contrast, open complex formation by Eσ70 occurs at most promoters spontaneously without the requirement of activators or energy source (Paget and Helmann, 2003).
Figure 1.
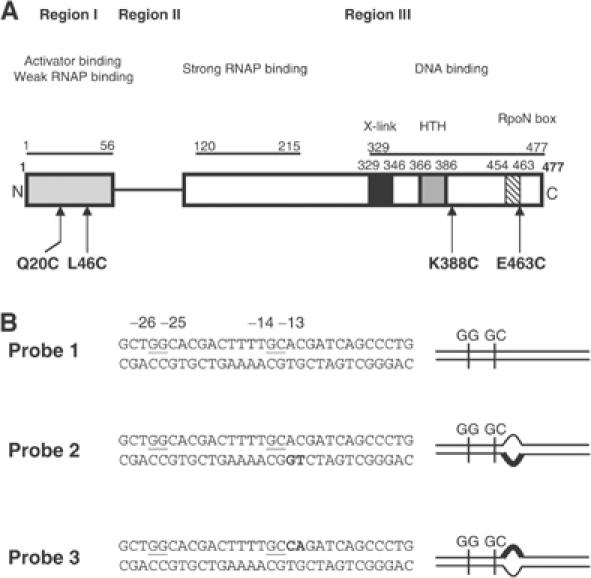
σ54 and S. meliloti nifH promoter probe variants used in this study. (A) Domain organisation of the Klebsiella pneumoniae σ54. The domains and residues involved in core RNAP, activator and DNA binding are indicated. In Region III, boxed are residues with potential roles in DNA interactions: the putative helix–turn–helix (HTH) motif (grey box), a region (X-link) shown to crosslink to DNA (black box) and the RpoN-box motif (hatched box) (reviewed in Buck et al, 2000). The residues targeted for FeBABE modification are indicated in bold typeface. (B) Nucleotide sequences of the three S. meliloti nifH promoter probes. Underlined are the two promoter consensus elements GG (positions −25 and −26; with respect to the transcription start site at +1) and GC (positions −13 and −14), which characterise the σ54 binding sites. Bold typeface indicates the changed residues on either the template strand (probe 2) or nontemplate strand (probe 3) to form heteroduplex probes. The symbols used to represent probes 1–3 are shown in the right panel.
Region I and regulatory residues within Region III of σ54 constitute a major target for the AAA activator within the Eσ54 regulatory centre. We have previously demonstrated that the relationship between σ54 Region I and Region III regulatory residues and the promoter GC-region is different within closed and open promoter complexes, whereas the relationship between other σ54 residues and the promoter GG-region remains unchanged (Casaz and Buck, 1999; Wigneshweraraj et al, 2001; Burrows et al, 2003). We reasoned that the change in the relationship between regulatory σ54 parts and promoter GC-region reflects a step in the ‘unlocking' of the silencing mechanism within the Eσ54 regulatory centre by the AAA activator (Casaz and Buck, 1999; Guo et al, 2000; Wigneshweraraj et al, 2001; Burrows et al, 2003). However, information about the AAA activator-driven reorganisation of the Eσ54 regulatory centre in the Eσ54 transcription initiation pathway that is associated with the change in the relationship between σ54 regulatory parts and the promoter GC-region is sparse. To address this issue, we have converted σ54 to a proximity-based DNA cleavage reagent through the conjugation with ((p-bromoacetamidobenzyl)-EDTA Fe) (FeBABE) (Ishihama, 2000) at four defined positions (Figure 1A). We now show that the reorganisation of the Eσ54 regulatory centre during open complex formation, measured as a change in the relationship between σ54 regulatory Region I, a regulatory residue in Region III and the promoter GC-region, is an early event that occurs upon interaction of the Eσ54 closed complex with the AAA activator at the transition point of ATP hydrolysis. The reorganisation of the Eσ54 regulatory centre is dependent on core RNAP subunits, but does not require β and β′ parts (β-subunit residues 186–433 and R454 and β′-subunit residues 215–220 and 1149–1190) normally associated with later stages of stable open promoter complex formation. Using photoreactive promoter DNA probes, we provide evidence for an interaction between the AAA activator and promoter DNA positions within and downstream of the Eσ54 regulatory centre when the AAA activator reorganises the Eσ54 regulatory centre at the transition point of ATP hydrolysis. Importantly, we describe for the first time how an initial bacterial RNAP–DNA complex is reorganised into a putative intermediate transcription initiation complex by an AAA ATPase where full DNA melting has not yet occurred.
Results
Interaction with AAA activator changes the relationship between regulatory σ54 Region I residue L46 and the promoter GC-region
We have recently shown using Eσ54 reconstituted with FeBABE-conjugated (via a sulphydryl group at position 46) σ54 (Cys46*Eσ54) that the relationship between regulatory σ54 Region I residue L46 and the promoter GC-region is different within closed and open promoter complexes (Burrows et al, 2003; Supplementary Figure 1). Since the regulatory Region I of σ54 is necessary for the Eσ54 silencing mechanism and constitutes a major interaction target for the AAA activator (Chaney et al, 2001), we wanted to determine at which point during the activation process the change in the relationship between σ54 Region I and the promoter GC-region takes place. As FeBABE is a proximity-based cleavage reagent (∼12–15 Å cleavage range from its attachment site; for review, see Ishihama, 2000), we rationalised that information about the point at which the change in the relationship between Region I and the promoter GC-region occurs can be obtained by monitoring changes in the DNA cleavage pattern within intermediate promoter complexes formed by Cys46*Eσ54. Previously, we showed that intermediate Eσ54 promoter complexes can be studied by using the poorly hydrolysable ATP analogue ATPγS or the ATP hydrolysis transition state analogue ADP·AlFx-bound AAA activator PspF (Phage shock protein F) (Cannon et al, 2003). For experimental simplicity, we used a truncated form of PspF, PspF1–275, that lacks the enhancer DNA-binding helix–turn–helix motif and can activate Eσ54 from solution (Cannon et al, 2003). The assay uses Eσ54 bound to a heteroduplex promoter probe, which mimics the conformation of the promoter DNA in the closed complex (probe 2 in Figure 1B; Cannon et al, 2003). Exposure of the Eσ54–probe 2 complex to either PspF1–275 and ATPγS (PspF1–275:ATPγS) or PspF1–275 and ADP·AlFx (PspF1–275:ADP·AlFx) induces conformational changes at the promoter GC-region, which can be monitored with ortho-copper phenanthroline, a probe for detecting minor groove specific conformational changes (Cannon et al, 2003). Using ATPγS and ADP·AlFx in combination with Cys46*Eσ54, we investigated whether PspF1–275:ATPγS, PspF1–275·ADP·AlFx or both can induce conformational changes in the Cys46*Eσ54–probe 2 complex that lead to changes in the DNA cleavage pattern by the FeBABE reagent at position L46 in regulatory σ54 Region I.
First, we confirmed by native-PAGE analysis and ortho-copper phenanthroline footprinting that Cys46*Eσ54 can (1) bind to probe 2 (Figure 2A) and (2) undergo PspF1–275:ATPγS- and PspF1–275:ADP·AlFx-dependent conformational changes in a manner similar to the wild-type Eσ54 (Cannon et al, 2003 and data not shown). Next, we tested whether Cys46*Eσ54 can cleave the DNA within Eσ54–probe 2 complexes upon the addition of ascorbate and hydrogen peroxide, which leads to the generation of hydroxyl radicals at the FeBABE moiety. To do so, promoter complexes were purified from a native-PAGE gel and the DNA was analysed by denaturing PAGE (see Materials and methods). As shown in Figure 2B, strong cleavage of the nontemplate DNA strand between positions −10 and −6 is seen within the Cys46*Eσ54–probe 2 complex (purified from Figure 2A, lane 4) (compare lanes 1 and 2). Similarly, template strand positions −9 to −5 were cleaved within the Cys46*Eσ54–probe 2 complex in the presence of ascorbate and hydrogen peroxide (see later). Control reactions containing Cys46Eσ54 (i.e. in the absence of FeBABE reagent) or the use of Eσ54 reconstituted with a mock-conjugated cysteine-free σ54 variant (Cys-Eσ54) (Burrows et al, 2003; Supplementary Figure 1) did not result in discernible DNA cleavage on either DNA strand (data not shown). This result extends previous studies and indicates that in the closed promoter complex, σ54 Region I position L46 is near the GC-region and the Eσ54 regulatory centre. To investigate the effect of PspF1–275:ATPγS and PspF1–275:ADP·AlFx on the proximity relationship between σ54 Region I and the promoter GC-region, we exposed the Cys46*Eσ54–probe 2 complex to PspF1–275 in the presence of ATPγS (Figure 2A, lane 6) or ADP·AlFx (Figure 2A, lane 5). As shown in Figure 2B, no detectable changes in the nontemplate strand (compare lanes 2 and 4) or template strand (data not shown) DNA cleavage pattern or intensity were observed in the presence of PspF1–275:ATPγS. In contrast, no DNA cleavage was detected on either the nontemplate (Figure 2B, compare lanes 2 and 6) or the template strand (see later) within the ternary Eσ54–probe 2–PspF1–275·ADP·AlFx complex. Analysis of the binary Eσ54–probe 2 complex from Figure 2A, lane 5, revealed the expected DNA cleavage pattern (Figure 2B, lanes 7 and 8), suggesting that the absence of DNA cleavage within the ternary Eσ54–probe 2–PspF1–275:ADP·AlFx is not due to the absence of hydroxyl radicals generated by the FeBABE under the conditions used. Since PspF1–275:ATPγS represents the state of the AAA activator when it is bound by ATP and PspF1–275:ADP·AlFx represents a state of the activator at the point of ATP hydrolysis when the AAA activator can form a stable ternary complex with the Eσ54–probe 2 complex (Figure 2A, compare lanes 2 and 5 with 3 and 6; Chaney et al, 2001; Cannon et al, 2003), the results strongly suggest that a change in relationship between regulatory σ54 Region I residue L46 and the promoter GC-region, and thus a step in the reorganisation of the Eσ54 regulatory centre en route to open complex formation, occurs at the transition point of ATP hydrolysis. In agreement with this conclusion, the DNA cleavage patterns on both strands were unchanged in control reactions containing PspF1–275 and ADP, AMP·PNP or AMP (data not shown).
Figure 2.
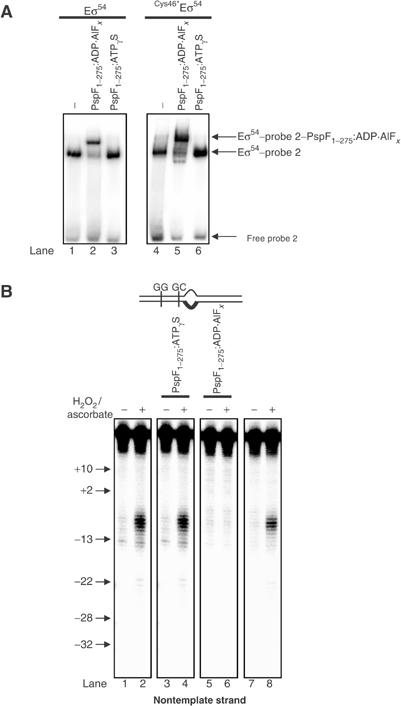
FeBABE cleavage of the S. meliloti nifH probe 2 by FeBABE-conjugated Cys46*Eσ54. (A) Activities of the wild-type Eσ54 and the FeBABE-conjugated Cys46*Eσ54 to bind the S. meliloti nifH probe 2 in the absence and presence of PspF1–275:ATPγS and PspF1–275:ADP·AlFx as judged by native-PAGE. The migration positions of the free probe 2, the Eσ54–probe 2 and the Eσ54:PspF1–275:ADP·AlFx–probe 2 complexes are indicated. (B) FeBABE cleavage profiles of S. meliloti nifH nontemplate strand by Cys46*Eσ54 (lanes 1 and 2), in the presence of PspF1–275:ATPγS (lanes 3 and 4) and PspF1–275:ADP·AlFx (lanes 5 and 6) and from DNA isolated from the Cys46*Eσ54–probe 2 (A, lane 5) (lanes 7 and 8). Reactions to which ascorbate and hydrogen peroxide were added to initiate DNA cleavage are marked with +; control reactions to which no ascorbate or hydrogen peroxide was added are marked with −. The molecular mass marker positions are indicated.
ADP·AlFx form of the AAA activator is required for reorganisation of the Eσ54 regulatory centre
In the next set of assays, we conducted a series of reactions to establish that reorganisation of the Eσ54 regulatory centre indeed occurs at the transition point of ATP hydrolysis, that is, in the presence of PspF1–275:ADP·AlFx. As expected, we did not detect any changes in the DNA cleavage pattern (between positions −9 and −5 on the template DNA strand and positions −10 and −6 on the nontemplate DNA strand) in reactions containing (1) ADP·AlFx in the absence of PspF1–275 (Figure 3A, compare lanes 3, 4, 13 and 14 with 7, 8, 17 and 18) and (2) PspF1–275 in the absence of ADP·AlFx (Figure 3A, compare lanes 3, 4, 13 and 14 with 9, 10, 19 and 20). A change in the DNA cleavage pattern was only evident in reactions containing both PspF1–275 and ADP·AlFx (Figure 3A, compare lanes 3, 4, 13 and 14 with 5, 6, 15 and 16). These data confirm that a conformational change involving σ54 Region I and the promoter GC-region requires the presence of PspF1–275 and ADP·AlFx. Similarly, in assays using homoduplex promoter probe (probe 1; Figure 1B), a change in the relationship between σ54 Region I residue L46 and the promoter GC-region only occurred in the presence of PspF1–275:ADP·AlFx (Figure 3B and data not shown). Overall, our new result extends previous observations (Burrows et al, 2003) and firmly establishes that activator interaction(s) induces an organisational change in the Eσ54 closed promoter complex that occurs at the transition point of ATP hydrolysis. By inference, we suggest that this organisational change reflects a step in the ‘unlocking' of the Eσ54 silencing mechanism.
Figure 3.
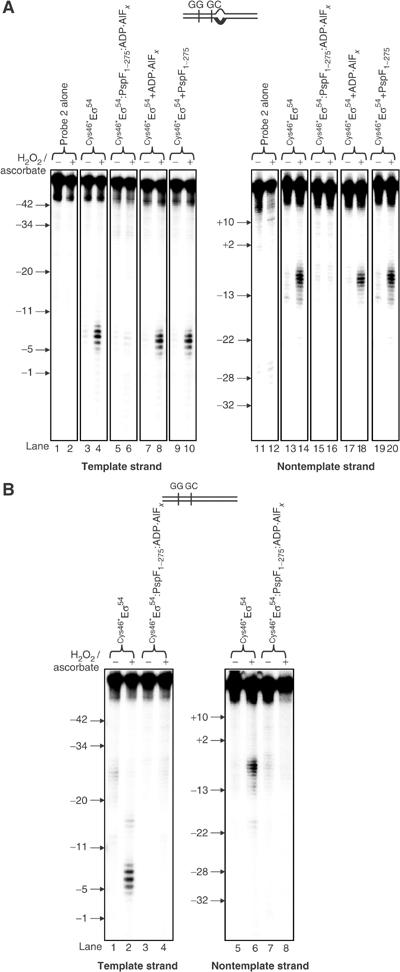
FeBABE cleavage of the S. meliloti nifH probes 1 and 2 by FeBABE-conjugated Cys46*Eσ54. (A) FeBABE cleavage profiles of the S. meliloti nifH probe 2 template and nontemplate DNA strands by Cys46*Eσ54 (lanes 3, 4, 13 and 14), in the presence of PspF1–275:ADP·AlFx (lanes 5, 6, 15 and 16), ADP·AlFx only (lanes 7, 8, 17 and 18) and PspF1–275 only (lanes 9, 10, 19 and 20). (B) FeBABE cleavage profiles of the S. meliloti nifH probe 1 template and nontemplate DNA strands by Cys46*Eσ54 (lanes 1, 2, 5 and 6) and PspF1–275:ADP·AlFx (lanes 3, 4, 7 and 8). In both (A) and (B), reactions to which ascorbate and hydrogen peroxide were added to initiate cleavage are marked with +; control reactions to which no ascorbate or hydrogen peroxide was added are marked with −. The molecular mass marker positions are indicated.
The organisational change induced by the ADP·AlFx form of the AAA activator is restricted to the Eσ54 regulatory centre
To further ascertain that the ADP·AlFx-bound form of the AAA activator induces an organisational change within the Eσ54 regulatory centre, we conducted assays with Eσ54 reconstituted with σ54 containing FeBABE at positions Q20 (Cys20*Eσ54) and K388 (Cys388*Eσ54). To investigate whether the organisational change is restricted to the Eσ54 regulatory centre, we conducted assays with Eσ54 reconstituted with σ54 containing FeBABE at position E463 (Cys463*Eσ54). σ54 Q20 is located within the regulatory Region I; K388 was previously reported to be involved in regulation of Eσ54 open complex formation (Wang and Gralla, 2001) and E463 is part of the highly conserved RpoN-box motif (Figure 1A) and has been shown to be proximal to the transcription start-site distal conserved promoter element, the GG-region (Figure 1B) (Burrows et al, 2003). Eσ54 reconstituted with Cys20*, Cys46*, Cys388* and Cys463* σ54 mutants show wild-type levels of transcription activity from the Sinorhizobium meliloti nifH promoter (Supplementary Figure 1). Within the Cys20*Eσ54–probe 2 complex, only the DNA nontemplate strand is cleaved between positions −15 and −11 (Figure 4A, lanes 2 and 5). Within the Cys388*Eσ54–probe 2 complex, cleavage of the nontemplate strand DNA between positions −18 and −15 and the template strand DNA between positions −11 and −4 was observed (Figure 4A, lanes 8 and 9, respectively). Within the Cys463*Eσ54–probe 2 complex, cleavage of both DNA strands at positions −24 and −25 was observed (Figure 4B, lane 2 and data not shown). All three enzymes, Cys20*Eσ54, Cys388*Eσ54 and Cys463*Eσ54, formed ternary complexes with PspF1–275:ADP·AlFx and probe 2 as judged by native-PAGE (data not shown). We tested whether the FeBABE-mediated DNA cleavage pattern by Cys20*Eσ54 and Cys388*Eσ54 at the promoter GC-region has changed within ternary complexes with PspF1–275:ADP·AlFx when compared to the binary Eσ54–probe 2 complexes. As shown in Figure 4A, DNA cleavage was not detected or was greatly reduced in ternary complexes formed by Cys20*Eσ54 or Cys388*Eσ54 (compare lanes 4, 5 and 7–10). In contrast, no change in the DNA cleavage pattern between the binary Cys463*Eσ54–probe 2 and ternary Cys463*Eσ54–probe2–PspF1–275:ADP·AlFx complexes was observed (Figure 4B, lanes 2 and 3). Similar assays using PspF1–275:ATPγS revealed no changes in the DNA cleavage pattern with either of the three enzymes (data not shown). Overall, the results obtained using Eσ54 containing FeBABE at three different regulatory positions (L46, Q20 and K388) firmly establish that an organisational change within the Eσ54 closed complex that occurs upon the interaction with the AAA activator at the transition point of ATP hydrolysis is restricted to the Eσ54 regulatory centre. Further, the results withCys463*Eσ54 also confirm that the presence of PspF1–275 and/or ADP·AlFx does not per se inhibit DNA cleavage.
Figure 4.
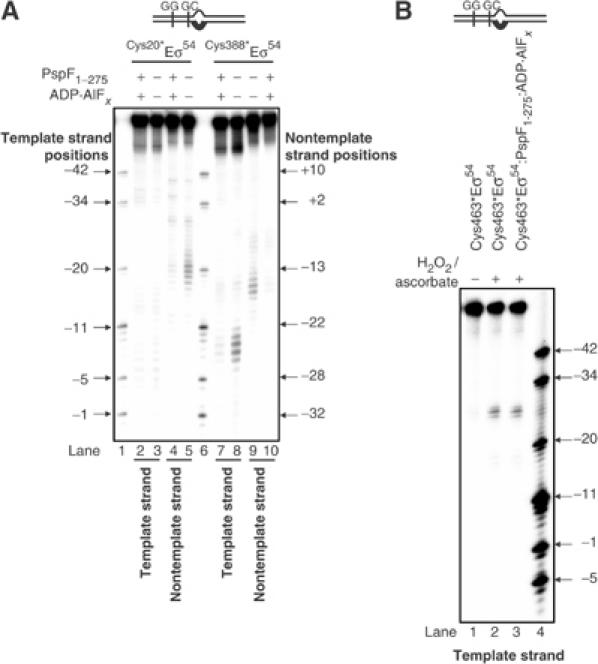
FeBABE cleavage of the S. meliloti nifH probe 2 by FeBABE-conjugated Cys20*Eσ54, Cys388*Eσ54 and Cys463*Eσ54. (A) FeBABE cleavage profiles of the S. meliloti nifH probe 2 template and nontemplate DNA strands by Cys20*Eσ54 (lanes 3 and 5), in the presence of PspF1–275:ADP·AlFx (lanes 2 and 4), Cys388*Eσ54 (lanes 8 and 9) and PspF1–275:ADP·AlFx (lanes 7 and 10). Lanes 1 and 6 contain a mixture of end-labelled S. meliloti nifH promoter DNA fragments as a molecular mass marker. All reactions contain ascorbate and hydrogen peroxide to initiate cleavage. (B) FeBABE cleavage profiles of the S. meliloti nifH probe 2 template DNA strand by Cys463*Eσ54 (lanes 1 and 2) and in the presence of PspF1–275:ADP·AlFx (lane 3). Reactions to which ascorbate and hydrogen peroxide were added to initiate cleavage are marked with +; control reactions to which no ascorbate or hydrogen peroxide was added are marked with −. Lane 4 contains a mixture of end-labelled S. meliloti nifH promoter DNA fragments as a molecular mass marker.
The organisational change induced by the ADP·AlFx form of the AAA activator at the promoter GC-region is dependent on core RNAP
In the next set of assays, we wanted to investigate whether the change in the relationship between regulatory σ54 parts and the promoter GC-region is dependent on the core RNAP. Since the wild-type σ54 alone does not bind either promoter probe 1 or 2 efficiently, we used promoter probe 3 for these assays (Cannon et al, 2003). Promoter probe 3 is identical in conformation to probe 2 and contains a heteroduplex segment at positions −12 and −11 that mimics the conformation of DNA within closed promoter complexes (Figure 1B) (Cannon et al, 2000). In contrast to probe 2, probe 3 contains wild-type template strand DNA sequence within the heteroduplex segment that allows tight binding of σ54 in the absence of core RNAP (see Figure 1B). Initially, we conducted experiments with Cys46*Eσ54 to confirm that FeBABE-mediated DNA cleavage near the promoter GC-region (between positions −10 and −6 on the nontemplate strand and positions −9 and −6 on the template strand) was seen within Cys46*Eσ54–probe 3 complex (Figure 5, lane 3 and data not shown). As expected from experiments using probe 1 (Figure 3B) and probe 2 (Figure 3A), no DNA cleavage was detected within the Cys46*Eσ54–probe 3–PspF1–275:ADP·AlFx complex (Figure 5, lane 5), revealing that the conformational change between σ54 Region I residue L46 and the promoter GC-region of promoter probe 3 occurs in the presence of PspF1–275:ADP·AlFx. Strikingly, this conformational change is not detected in the absence of core RNAP, since no changes in the DNA cleavage pattern are detected when Cys46*σ54–probe 3 and Cys46*σ54–probe 3–PspF1–275:ADP·AlFx complexes are compared (Figure 5, lanes 8 and 10). Similar results were obtained with Cys20*σ54 on probe 3 in the absence of core RNAP (data not shown). Thus, the AAA activator- and ADP·AlFx dependent reorganisation of the Eσ54 regulatory centre at the promoter GC-region requires core RNAP, implying a role of core RNAP surfaces in controlling the organisation of the Eσ54 regulatory centre.
Figure 5.
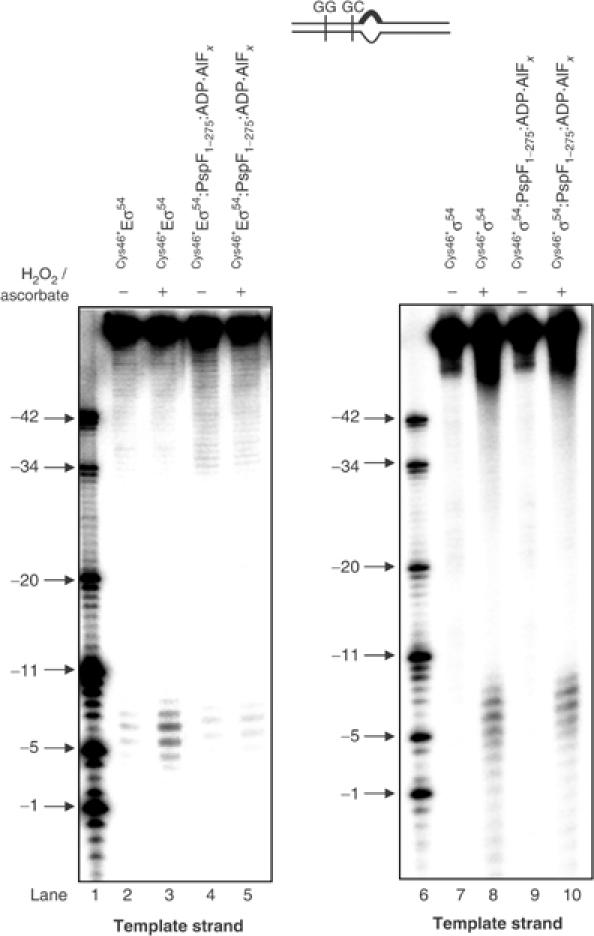
FeBABE cleavage of the S. meliloti nifH probe 3 by FeBABE-conjugated Cys46*σ54 in the presence and absence of core RNAP. FeBABE cleavage profiles of the S. meliloti nifH probe 3 template DNA strand by Cys46*Eσ54 (lanes 2 and 3), in the presence of PspF1–275:ADP·AlFx (lanes 4 and 5), Cys46*σ54 (lanes 7 and 8), in the presence of PspF1–275:ADP·AlFx (lanes 9 and 10). Reactions to which ascorbate and hydrogen peroxide were added to initiate cleavage are marked with +; control reactions to which no ascorbate or hydrogen peroxide was added are marked with −. Lanes 1 and 6 contain a mixture of end-labelled S. meliloti nifH promoter DNA fragments as a molecular mass marker.
Core RNAP surfaces associated with open complex formation do not contribute to the reorganisation of the Eσ54 regulatory centre by the AAA activator
In order to identify core RNAP surfaces involved in reorganisation of the Eσ54 regulatory centre upon interaction with AAA activator at the point of ATP hydrolysis, we reconstituted Cys46*Eσ54 using core RNAP mutants harbouring deletions/substitutions in parts involved in open promoter complex formation in the context of Eσ70 and/or Eσ54. These were (i) core RNAP lacking the β-subunit downstream lobe domain (βΔ186−433E; Nechaev et al, 2000; Wigneshweraraj et al, 2002), (ii) impaired for β-subunit downstream lobe functionality (βRH454E; Nechaev et al, 2000; Wigneshweraraj et al, 2002), (iii) lacking the β′-subunit jaw domain (β′Δ1149–1190E; Ederth et al, 2002; Wigneshweraraj et al, in preparation) and (iv) defective for intermediate promoter complex formation (β′Δ215–220E; Bartlett et al, 1998) (Figure 6A). First, we confirmed by native-PAGE that mutant Cys46*Eσ54s can form the binary Cys46*Eσ54–probe 2 and ternary Cys46*Eσ54–probe 2–PspF1–275:ADP·AlFx complexes. The Cys46*Eσ54 containing the βRH454 and β′Δ1149–1190 mutations bound probe 2 with reduced activity when compared to the wild-type and the other two mutant Cys46*Eσ54s (Figure 6B, compare lanes 1, 3 and 9 with 5 and 7). However, all mutant Cys46*Eσ54s formed relatively comparable amounts of Cys46*Eσ54–probe 2–PspF1–275:ADP·AlFx complex (Figure 6B, lanes 2, 4, 6, 8 and 10). DNA cleavage between positions −9 and −5 on the template strand and −10 and −6 on the nontemplate strand was detected within the mutant Eσ54–probe 2 complexes (Figure 6C, lanes 1, 3, 5 and 8 and data not shown). Within mutant ternary Eσ54–probe 2–PspF1–275:ADP·AlFx complexes, DNA cleavage was not detected as seen within the wild-type ternary Eσ54–probe 2–PspF1–275:ADP·AlFx complex (Figure 6C, lanes 2, 4, 6 and 7). Therefore, we suggest that the β-subunit downstream lobe, β′-subunit jaw domain and the β′-subunit residues 215–220 are not involved in the initial organisation or AAA activator-driven reorganisation of the Eσ54 regulatory centre. Overall, the results suggest that core RNAP surfaces that are normally associated with later stages of open complex formation do not contribute to the reorganisation of the Eσ54 regulatory centre, and by inference, do not contribute to the Eσ54 silencing mechanism. It therefore appears that AAA activator- and ADP·AlFx-dependent reorganisation of the Eσ54 regulatory centre is an early event in the Eσ54 activation pathway prior to the open complex formation event.
Figure 6.
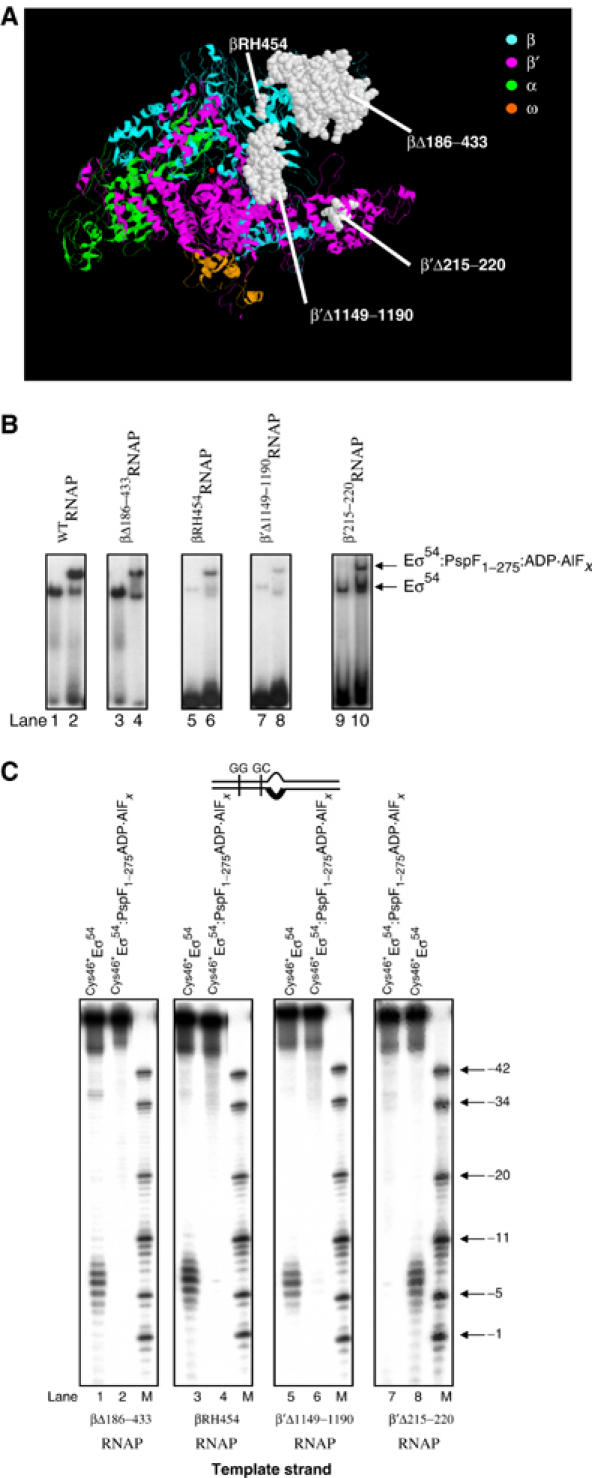
The contribution of core RNAP parts to the FeBABE cleavage pattern of the S. meliloti nifH probe 2 by FeBABE-conjugated Cys46*Eσ54. (A) Mutations used in this work are presented on the ribbon diagram of the three-dimensional structure of Thermus aquaticus core RNAP (Zhang et al, 1999). The active centre Mg2+ is shown as a red dot and the subunits of the RNAP are colour-coded as shown. The view is roughly parallel with the main axis of the RNAP channel. The amino acids of T. aquaticus β and β′ subunits corresponding to E. coli amino acids β186–433, βR454, β′215–220 and β′1149–1190 that were deleted/mutated are shown in white space-fill. (B) Activities of the Cys46*Eσ54 reconstituted with WTRNAP (lanes 1 and 2), βΔ186–433RNAP (lanes 3 and 4), βRH454RNAP (lanes 5 and 6), β′Δ1149–1190RNAP (lanes 7 and 8) and β′Δ215–220RNAP (lanes 9 and 10) to bind the S. meliloti nifH probe 2 in the absence and presence of PspF1–275:ADP·AlFx (lanes 2, 4, 6, 8 and 10) as judged by native-PAGE. The migration positions of the free probe 2, the Eσ54–probe 2 and the Eσ54:PspF1–275:ADP·AlFx–probe 2 complexes are as indicated. (C) FeBABE cleavage profiles of S. meliloti nifH probe 2 template DNA strand by Cys46*Eσ54 reconstituted with mutant core RNAP forms (lanes 1, 3, 5 and 8) and in the presence of PspF1–275:ADP·AlFx (lanes 2, 4, 6 and 7). All reactions contained ascorbate and hydrogen peroxide to initiate cleavage. Lanes M contain a mixture of end-labelled S. meliloti nifH promoter DNA fragments as a molecular mass marker.
Promoter GC-region is protected from cleavage by hydroxyl radicals within the Eσ54–probe 2–PspF1–275:ADP·AlFx complex
We studied the binary Eσ54–probe 2 complex and ternary Eσ54–probe 2–PspF1–275:ADP·AlFx complex by solution hydroxyl radical footprinting in order to distinguish whether the basis for the change in the relationship, implied from the loss of DNA cleavage by tethered FeBABE forms of σ54, between regulatory σ54 parts and the promoter GC-region is due to (1) a change in proximity between σ54 and the promoter DNA and/or (2) protection of promoter GC-region by other protein part(s). As shown in Figure 7A, binding of Eσ54 to probe 2 detectably protects promoter DNA between positions −32 and −11 from hydroxyl radical cleavage (lanes 4 and 10). Two hyper-reactive sites centred at positions −20 (on the template strand, lane 4) and −20 and −6 (on the nontemplate strand, lane 10) are also evident. Within the ternary Eσ54–probe 2–PspF1–275:ADP·AlFx complex, a strong protection of the DNA is seen between positions −11 and −6 (box II) on the template strand and between positions −13 and −4 (box III) on the nontemplate strand (Figure 7A, box II, lanes 3 and 4 and box III, lanes 10 and 11, respectively). Interestingly, the protected positions (boxes II and III) overlap with positions that are subject to localised FeBABE-mediated cleavage within the binary Eσ54–probe 2 complex and that are absent in the ternary Eσ54–probe 2–PspF1–275:ADP·AlFx complex (see Figure 3). Therefore, we suggest that the failure to see FeBABE-mediated DNA cleavage within the ternary Eσ54–probe 2–PspF1–275:ADP·AlFx complex is due to the promoter GC-region being protected by protein sequences—possibly other σ54, AAA activator and/or core RNAP parts. However, we cannot exclude the possibility that the proximity relationship between regulatory σ54 parts and the promoter GC-region has also changed within the reorganised Eσ54 regulatory centre to cause loss of DNA cleavage. Further, we note increased cleavage of both DNA strands downstream of position −20 within the ternary Eσ54–probe 2–PspF1–275:ADP·AlFx complex (Figure 7A, box I, lanes 3 and 4 and box IV, lanes 10 and 11). This observation further supports the view that the binary Eσ54–probe 2 and the ternary Eσ54–probe 2–PspF1–275:ADP·AlFx complexes are conformationally different.
Figure 7.
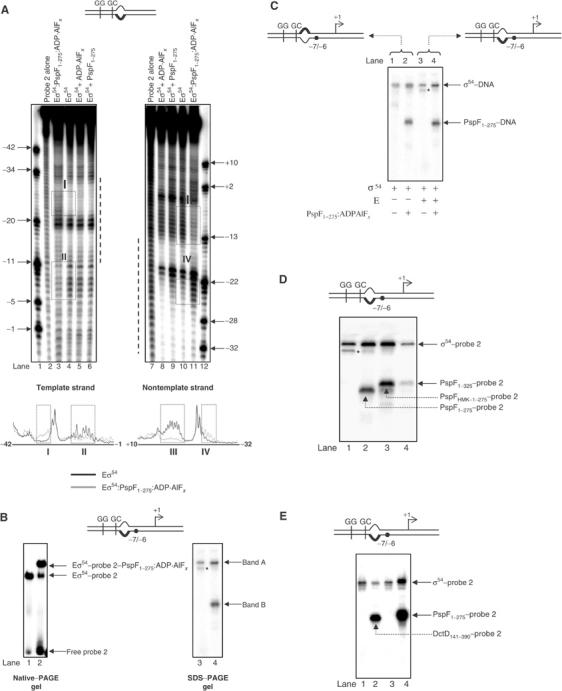
Promoter GC-region is protected from cleavage by hydroxyl radicals within the Eσ54–probe 2–PspF1–275:ADP·AlFx complex. (A) (Top panel) Hydroxyl radical footprints of the S. meliloti nifH probe 2 template and nontemplate DNA strands by Eσ54 (lanes 4 and 10), in the presence of PspF1–275:ADP·AlFx (lanes 3 and 11), ADP·AlFx (lanes 5 and 8) and PspF1–275 (lanes 6 and 9). The dotted line represents the region of DNA (approximately positions −32 to −11) protected from hydroxyl radical cleavage within the Eσ54–probe 2 complex. Boxes I–IV highlight regions in the footprinting profiles where marked differences are observed. In the lower panel, PhosphorImager traces of Eσ54:PspF1–275:ADP·AlFx (light grey lines) and Eσ54 (black lines) of the reactions shown in lanes 3 and 4 (template DNA strand) and 10 and 11 (nontemplate DNA strand) are shown. (B) Native-PAGE gel showing the binding of Eσ54 in the absence (lane 1) and presence (lane 2) of PspF1–275:ADP·AlFx to APB-modified probe 2 (left panel). SDS–PAGE analysis of the Eσ54–probe 2 (lane 3) and Eσ54–probe 2–PspF1–275:ADP·AlFx (lane 4) complexes following UV irradiation (right panel). The migration positions of the two crosslinked protein–DNA complexes (band A and B) are indicated. (C) SDS–PAGE analysis of the σ54–probe 3 (lane 1), σ54–probe 3–PspF1–275:ADP·AlFx (lane 2), Eσ54–probe 2 (lane 3) and Eσ54–probe 2–PspF1–275:ADP·AlFx (lane 4) complexes following UV irradiation. Reactions to which core RNAP was added are indicated. (D) SDS–PAGE analysis of Eσ54–probe 2–activator:ADP·AlFx complexes (lanes 2–4) formed using three different molecular weight forms of PspF following UV irradiation. Lane 1 contains Eσ54–probe 2. (E) SDS–PAGE analysis of Eσ54–probe 2–DctD141–390:ADP·AlFx complex following UV irradiation. In (B–D), the asterisk indicates a proteolytic degradation product of the σ54 subunit introduced adventitiously during sample preparation (our unpublished observations). In (B–E), on the schematic representation of the DNA probes, the black dot indicates the position that was modified with APB.
AAA activator can be crosslinked to the promoter GC-region within the reorganised Eσ54 regulatory centre
We used a proximity-based protein–DNA crosslinking approach to further study the relationship between regulatory σ54 residues and the promoter GC-region upon ADP·AlFx-dependent interaction with activator. Our aim was to investigate which protein component of the ternary Eσ54–probe 2–PspF1–275:ADP·AlFx complex (i.e. α, β, β′, ω, σ54 and/or PspF1–275) contributes to the protection of the promoter GC-region from FeBABE and solution hydroxyl radical-mediated cleavage within ternary promoter complexes. Synthetic photoactive 32P-tagged probe 2 was made by derivatising a single, specifically placed phosphorothioate with p-azidophenacyl bromide (APB) (see Materials and methods). Upon UV irradiation, the aryl azide group of APB reacts with protein residues within ∼12 Å range from its attachment site on the DNA, which results in the formation of covalently crosslinked protein–DNA complexes (Mayer and Barany, 1995).
Since FeBABE-mediated cleavage by Cys46*Eσ54 occurs between positions −10 and −6 on the template DNA strand and between positions −9 and −5 on the nontemplate DNA strand; Figure 3A), we initially APB-derivatised phosphorothioates incorporated between nucleotide positions −6 and −7 on both DNA strands. First, we used the wild-type Eσ54 to establish that APB derivatisation did not prevent formation of the binary Eσ54–probe 2 and the ternary Eσ54–probe 2–PspF1–275:ADP·AlFx complexes (data not shown). Next, we used Cys46*Eσ54 to confirm that the change in relationship between σ54 Region I residue L46 and the promoter GC-region in the presence of PspF1–275:ADP·AlFx occurs in the APB-derivatised probe 2 as seen with the unmodified probe 2 (data not shown). To identify which protein component(s) of the binary Eσ54–probe 2 and ternary Eσ54–probe 2–PspF1–275:ADP·ALFx complexes is proximal to DNA position −7/−6, we formed promoter complexes in the presence and absence of PspF1–275:ADP·AlFx on 32P-tagged APB-derivatised probe 2 (in each case, the APB-containing DNA strand was tagged with 32P). Following UV irradiation, the promoter complexes were purified from a native-PAGE gel (Figure 7B, left panel) and analysed by SDS–PAGE (see Material and methods). As shown in Figure 7B (right panel), SDS–PAGE analysis of the crosslinked binary Eσ54–probe 2 complex (from Figure 7B, lane 1) revealed a single radioactive band (band A, lane 3). SDS–PAGE analysis of the crosslinked ternary Eσ54–probe 2–PspF:ADP·AlFx complex (from Figure 7B, lane 2) revealed two bands: one that migrated at the identical position as band A (Figure 7B, compare lanes 3 and 4) and a second faster migrating band (band B, Figure 7B, lane 4). Bands A and B were not detected within control reactions, which were not subjected to UV irradiation or unmodified probe 2 was used (data not shown). Analysis of the SDS–PAGE gel shown in Figure 7B using antibodies against β, β′, σ54 and anti-His-tag antibodies (to detect PspF) identified band A as σ54 and band B as PspF1–275 (data not shown).
Since σ54 alone, in the absence of core RNAP, does not bind probe 2 well but can bind tightly to probe 3, we repeated the reactions on 32P-tagged APB-derivatised probe 3 in the absence of core RNAP subunits in order to further confirm that band A is indeed a crosslinked σ54–DNA complex. As shown in Figure 7C, bands A and B were evident in the presence or in the absence of core RNAP on 32P-tagged APB-derivatised probes 2 and 3, respectively (compare lanes 1 and 2 with 3 and 4, respectively). Complementary results were obtained in reactions where APB was placed on the nontemplate strand DNA of probes 2 and 3 (data not shown).
To further ascertain that band B is a crosslinked PspF1–275–DNA complex, we used three different molecular weight forms of PspF (PspF1–275, PspFHMK-1–275 and PspF1–325) to form the ternary Eσ54–probe 2–PspF:ADP·AlFx complex. SDS–PAGE analysis of ternary Eσ54–probe 2–PspF:ADP·AlFx complexes formed using the three different forms of PspF confirmed that band B is indeed a crosslinked PspF–DNA complex (Figure 7D, lanes 2–4). Assays with the Eσ54-dependent AAA activator DctD141–390 (C4-dicarboxylic acid transport protein D) from S. meliloti (Xu et al, 2004) further confirmed that band B is a bona fide crosslinked AAA activator–DNA complex (Figure 7E, lane 2). Additional assays using probe 2 with APB at positions −15/−14, −14/−13, −13/−12, −12/−11, −11/−10 and −1/+1 gave similar results in that σ54 and PspF1–275 became crosslinked in an ADP·AlFx-dependent manner (data not shown). Further, additional control reactions in which only PspF1–275 and ADP·AlFx were present did not result in the formation of a crosslinked PspF1–275–DNA complex (data not shown).
Overall, the crosslinking data complement the solution hydroxyl radical footprinting results and suggest that both σ54 and the AAA activator contribute to the protection of the promoter GC-region from FeBABE and solution hydroxyl radical-mediated cleavage within ternary promoter complexes with the AAA activator. Interestingly, we failed to detect crosslinks between core RNAP subunits (notably β and β′ subunits) and the DNA within the binary Eσ54–DNA or ternary Eσ54–DNA–activator:ADP·AlFx complexes (see Discussion). Importantly, however, the crosslinking data report that the AAA activator is proximal to the promoter GC-region at an intermediate step in the Eσ54 transcription initiation pathway.
Discussion
DNA melting and transcription-competent open complex formation by σ54 RNAP only occur upon an ATP hydrolysis-dependent interaction with an AAA activator. Tight regulation of transcription initiation by Eσ54 requires a complex network of protein–DNA interactions between regulatory residues of σ54 and a fork junction DNA structure at the promoter GC-region within initial (closed) Eσ54 promoter complexes referred to as the Eσ54 regulatory centre. The regulatory centre is reorganised by the AAA activator in an ATP hydrolysis-dependent manner during open complex formation. This study was prompted by our previous observation that the relationship between regulatory σ54 residues and the promoter GC region is changed upon the transition from closed promoter complex to open promoter complex (Wigneshweraraj et al, 2001; Burrows et al, 2003). We have used an ATP analogue, ADP·AlFx, to capture an intermediate Eσ54 promoter complex and establish that at least one step in reorganisation of the Eσ54 regulatory centre occurs upon interaction with the activator at the transition point of ATP hydrolysis. Guo et al (2000) suggested that during open complex formation by Eσ54, the AAA activator causes tight interactions between regulatory σ54 residues and the template strand of the fork junction DNA structure at the promoter GC-region to be broken and new interactions with the nontemplate DNA strand to be established. This so-called ‘strand switching' event is an integral part of the DNA melting process by the bacterial RNAP and is expected to be an early event in the bacterial transcription initiation pathway (Guo et al, 2000; Fenton and Gralla, 2003). We suggest that reorganisation of the Eσ54 regulatory centre by the ADP·AlFx form of the AAA activator reflects at least one aspect of the ‘strand switching' event and by inference is an early event in the Eσ54 transcription initiation pathway.
We have demonstrated a role for core RNAP surfaces in reorganisation of the Eσ54 regulatory centre. Since β and β′ parts known to be associated with stable DNA melting and open complex formation do not appear to contribute to the reorganisation event, this observation further emphasises that the step at which the Eσ54 regulatory centre is reorganised by the AAA activator is an early event in the Eσ54 transcription initiation pathway. In agreement with this view, DNA melting is not detected within Eσ54 promoter complexes formed on probes 1–3 or a supercoiled plasmid harbouring the S. meliloti nifH promoter in the presence of the AAA activator and ADP·AlFx (Cannon et al, 2003; our unpublished observations). An interaction between the AAA domain of Eσ54 activators and the ‘to be melted' DNA sequences downstream of the promoter GC-region could be significant for causing changes in the DNA structure important for making open promoter complexes. In support of this view, two motifs within the AAA domain of the E. coli helicase RuvB interact with, and modulate, DNA in an ATP hydrolysis-dependent manner (Iwasaki et al, 2000).
The most striking observation is that within the intermediate Eσ54 promoter complex, the AAA activator is proximal to DNA sequences that are melted within normal open promoter complexes. Therefore, at least one intermediate Eσ54 promoter complex is organised in such a way that the AAA activator is within 12 Å (recall that the maximum distance between the phosphorothioate and the photoreactive site on APB is ∼12 Å) of promoter DNA positions −15 to −1. In support of a putative interaction between the AAA activator and the promoter GC-region DNA, the ATP hydrolysis activity of the AAA activator is stimulated in the presence of promoter DNA and σ54 promoter complexes (Cannon et al, 2004).
The proximity-based photo-crosslinking also indicates that DNA across the core promoter GC-region (from positions −15 to −10) and promoter DNA sequences that are melted within open complexes (from positions −12 to +1) are contacted exclusively by the σ54 subunit within the closed and at least one intermediate Eσ54 promoter complex. Consistent with this observation, the crystal structure of the EσA promoter complex revealed that major promoter DNA contacts were exclusively made by σA (Murakami et al, 2002). An analogous situation exists in eukaryotic RNAP II transcription, where no or very little contact of promoter DNA and RNAP II exists prior to DNA melting (Bushnell et al, 2004). Since closed promoter complex formation by RNAP occurs in a conformation referred to as the ‘closed clamp state' where access to the catalytic cleft of the RNAP is restricted by the β′ clamp and β′ jaw domain and thus is not wide enough for interaction with double-stranded DNA, we envisage that DNA contacts with β and β′ subunits are only established once DNA melting occurs or has occurred. In support of this view, we can photo-crosslink β and β′ subunits to several positions (from −15 to +1) on the S. meliloti nifH promoter within bona fide open promoter complexes, that is, when the ‘open clamped state' conformation is established (Wigneshweraraj et al, in preparation). Thus, it appears that within bacterial transcription complexes, the path of the promoter DNA is defined by interactions with the σ subunit and AAA activator (in the case of Eσ54) prior to open complex formation. Similarly, within the RNAP II transcription initiation complex, the path of the promoter DNA is defined by accessory transcription factors Tfg2 subunit of TFIIF (the homologue of the bacterial σ factor), TFIIE and TFIIH (Bushnell et al, 2004). Interestingly, TFIIH is an ATPase that is required to trigger DNA melting and thus is conceptually analogous to the AAA activator within intermediate Eσ54 transcription complexes.
The use of σ54-containing RNAP in combination with the ADP·AlFx-bound AAA activator has enabled us to capture and study one intermediate bacterial RNAP–DNA complex in which full DNA melting has not yet occurred. This intermediate complex is clearly different from the initial Eσ54 closed promoter complex and likely reflects the ‘unlocked' conformation where full ‘strand-switching' and ‘clamp opening' events have not yet occurred and where Eσ54 has not acquired full heparin stability—the latter being a hallmark feature of bona fide open promoter complexes. Since the ADP·AlFx-bound AAA activator is locked in the ATP hydrolysis transition state conformation, we envisage that release of ADP and/or Pi by the AAA activator and DNA melting are linked events. Thus, we postulate that product(s) release (ADP and/or Pi) by the AAA activator following ATP hydrolysis is accompanied by full ‘strand-switching' and ‘clamp opening' events. In agreement with the latter, we recently demonstrated that ATP hydrolysis-dependent interaction of the AAA activator with the Eσ54 regulatory centre (notably σ54 Region I) induces conformational changes in the β′ jaw domain (see above), which precedes open complex formation (Wigneshweraraj et al, in preparation). It appears that modulation of accessory factors, like the σ subunit, which bind to the so-called ‘upstream face' of the RNAP (in this case, modulation of the σ54 subunit by the AAA activator) enables RNAP (and σ factor) to adopt several different conformations needed for transcription initiation.
Materials and methods
Promoter DNA probes and proteins
S. meliloti nifH promoter probes 1–3 were constructed as described by Cannon et al (2003) and Wigneshweraraj et al (2003. K. pneumoniae single-cysteine variants of σ54, Q20C, L46C, K388C and E463C (Figure 1A) were constructed, purified and modified with FeBABE as described by Burrows et al (2003). The AAA domain (residues 1–275) of PspF (PspF1–275) and the heart-muscle-kinase tagged version thereof (PspFHMK-1–292) were constructed and purified as detailed by Chaney et al (2001) and Wigneshweraraj et al (2003). Wild-type E. coli core RNAP was purchased from Epicentre Technologies. Full-length PspF (PspF1–325) was purified as described by Wigneshweraraj et al (2003). The AAA domain of the S. meliloti DctD protein (residues 141–390) was purified as described by Xu et al (2004). The E. coli core RNAPs harbouring β-subunit mutations Δ186–433 and RH454 were purified as described by Wigneshweraraj et al (2003). The E. coli core RNAPs containing the β′-subunit deletions Δ1149–1190 and Δ215–220 were purified as described by Ederth et al (2002) and Bartlett et al (1998), respectively.
DNA cleavage assays
DNA cleavage assays were conducted at 37°C in STA buffer (25 mM Tris acetate, pH 8.0, 8 mM Mg-acetate, 100 mM KCl and 3.5% (w/v) PEG-6000). Reactions contained 100 nM promoter DNA probe, 200 nM Eσ54 (reconstituted using a 1:3 ratio of core RNAP to σ54) or 1 μM conjugated σ54. Where indicated, freshly prepared ascorbate (pH 7.0) and hydrogen peroxide were used at final concentrations of 2 and 1 mM, respectively. The promoter DNA probes were prepared as described by Wigneshweraraj et al (2003). Following initial incubation (for 5 min) of the labelled promoter DNA probe with Eσ54, ternary complexes (Eσ54–DNA–activator:ADP·AlFx) were formed by adding 1 μl of 0.2 mM ADP, 1 μl of 0.2 mM AlCl3, 1 μl of 5 mM NaF and 10 μM activator for 10 min at 37°C. DNA cleavage was initiated by the sequential addition of ascorbate and hydrogen peroxide, allowed to proceed for 10 min and quenched with 2 μl of native loading dye (from a 5 × stock: 50% (v/v) glycerol and 0.05% (w/v) bromophenol blue). Promoter complexes were separated from the free DNA probe by native-PAGE. Promoter complexes were gel purified as detailed by Cannon et al (2003) and recoveries of DNA were determined by dry Cerenkov counting. Equal numbers of counts were loaded onto a 10% denaturing gel. Dried gels were visualised and analysed using a PhosphorImager. The cleavage sites were determined by using γ-32P end-labelled fragments of the S. meliloti nifH promoter DNA.
Solution hydroxyl radical footprinting assays
Hydroxyl radical foortprinting assays were conducted at 30°C in STA buffer without PEG-6000. The 10 μl reaction mixture contained 50 nM DNA template and 100 nM Eσ54 in the presence of either PspF1–275 alone, ADP·AlFx or PspF1–275:ADP·AlFx (formed in situ as outlined above). Since glycerol is a radical scavenger, it was removed from the protein preparations by dialysis. The hydroxyl radical footprinting reaction was initiated by the sequential addition of 1 μl of 0.4 mM (NH4)2Fe(SO4)2·6H2O, 0.8 mM EDTA, 1 μl of freshly prepared 10 mM ascorbate (pH 7.0) and 1 μl of 0.3% (v/v) hydrogen peroxide. The cutting reactions were quenched after 1 min by addition of 2 μl of native loading dye. The promoter complexes were separated by native-PAGE and analysed as described above.
Photo-crosslinking assays
Phosphorothioated oligodeoxyribonucleotides (Qiagen) were derivatised with p-azidophenacyl bromide (Sigma) as described by Mayer and Barany (1995), purified through a Sephadex G-50 column in 10 mM Tris–HCl (pH 7.5), 32P-labelled and annealed to the complementary strand as described by Wigneshweraraj et al (2003). Promoter complexes were formed on modified promoter probes as detailed above. Reactions were then UV irradiated at 365 nm for 30 s using a UV-Stratalinker 1800 (Stratagene) and quenched by addition of 2 μl 5 × native loading dye. Promoter complexes were then separated from free DNA probe by native-PAGE using a 4.5% (w/v) native-PAGE gel (see above). Promoter complexes were then excised from the gel and eluted into 100 μl of 1 × SDS loading buffer (Sigma) for 1 h at 37°C. The samples were then heated at 95°C for 3 min and 20 μl was loaded onto a 12.5% SDS–PAGE gel and run at 200 V for 100 min. The gels were then dried and crosslinked protein–DNA complexes were visualised by PhosphorImager analysis.
Supplementary Material
Supplementary Figure 1
Acknowledgments
We thank A Ishihama for providing advice on using FeBABE and J Yuzenkova for purifying the β′Δ1149–1190RNAP. We are grateful to W Cannon for purifying the DctD141–390 protein, S Pharon for laboratory assistance and F Werner and P Bordes for comments on the manuscript. This work was supported by a BBSRC project grant to MB.
References
- Bartlett MS, Gaal T, Ross W, Gourse RL (1998) RNA polymerase mutants that destabilize RNA polymerase–promoter complexes alter NTP-sensing by rrn P1 promoters. J Mol Biol 279: 331–345 [DOI] [PubMed] [Google Scholar]
- Buck M, Gallegos MT, Studholme DJ, Guo Y, Gralla JD (2000) The bacterial enhancer-dependent sigma(54) (sigma(N)) transcription factor. J Bacteriol 182: 4129–4136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrows PC, Severinov K, Ishihama A, Buck M, Wigneshweraraj SR (2003) Mapping sigma 54–RNA polymerase interactions at the −24 consensus promoter element. J Biol Chem 278: 29728–29743 [DOI] [PubMed] [Google Scholar]
- Bushnell DA, Westover KD, Davis RE, Kornberg RD (2004) Structural basis of transcription: an RNA polymerase II–TFIIB cocrystal at 4.5 Angstroms. Science 303: 983–988 [DOI] [PubMed] [Google Scholar]
- Cannon W, Bordes P, Wigneshweraraj SR, Buck M (2003) Nucleotide-dependent triggering of RNA polymerase–DNA interactions by an AAA regulator of transcription. J Biol Chem 278: 19815–19825 [DOI] [PubMed] [Google Scholar]
- Cannon WV, Gallegos MT, Buck M (2000) Isomerization of a binary sigma–promoter DNA complex by transcription activators. Nat Struct Biol 7: 594–601 [DOI] [PubMed] [Google Scholar]
- Cannon WV, Schumacher J, Buck M (2004) Nucleotide-dependent interactions between a fork junction-RNA polymerase complex and an AAA+ transcriptional activator protein. Nucleic Acids Res 32: 4596–4608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casaz P, Buck M (1999) Region I modifies DNA-binding domain conformation of sigma 54 within the holoenzyme. J Mol Biol 285: 507–514 [DOI] [PubMed] [Google Scholar]
- Chaney M, Grande R, Wigneshweraraj SR, Cannon W, Casaz P, Gallegos MT, Schumacher J, Jones S, Elderkin S, Dago AE, Morett E, Buck M (2001) Binding of transcriptional activators to sigma 54 in the presence of the transition state analog ADP-aluminum fluoride: insights into activator mechanochemical action. Genes Dev 15: 2282–2294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebright RH (2000) RNA polymerase: structural similarities between bacterial RNA polymerase and eukaryotic RNA polymerase II. J Mol Biol 304: 687–698 [DOI] [PubMed] [Google Scholar]
- Ederth J, Artsimovitch I, Isaksson LA, Landick R (2002) The downstream DNA jaw of bacterial RNA polymerase facilitates both transcriptional initiation and pausing. J Biol Chem 277: 37456–37463 [DOI] [PubMed] [Google Scholar]
- Fenton MS, Gralla JD (2003) Roles for inhibitory interactions in the use of the −10 promoter element by sigma 70 holoenzyme. J Biol Chem 278: 39669–39674 [DOI] [PubMed] [Google Scholar]
- Guo Y, Lew CM, Gralla JD (2000) Promoter opening by sigma(54) and sigma(70) RNA polymerases: sigma factor-directed alterations in the mechanism and tightness of control. Genes Dev 14: 2242–2255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihama A (2000) Molecular anatomy of RNA polymerase using protein-conjugated metal probes with nuclease and protease activities. Chem Commun 13: 1091–1094 [Google Scholar]
- Iwasaki H, Han YW, Okamoto T, Ohnishi T, Yoshikawa M, Yamada K, Toh H, Daiyasu H, Ogura T, Shinagawa H (2000) Mutational analysis of the functional motifs of RuvB, an AAA+ class helicase and motor protein for holliday junction branch migration. Mol Microbiol 36: 528–538 [DOI] [PubMed] [Google Scholar]
- Mayer AN, Barany F (1995) Photoaffinity cross-linking of TaqI restriction endonuclease using an aryl azide linked to the phosphate backbone. Gene 153: 1–8 [DOI] [PubMed] [Google Scholar]
- Murakami KS, Masuda S, Campbell EA, Muzzin O, Darst SA (2002) Structural basis of transcription initiation: an RNA polymerase holoenzyme–DNA complex. Science 296: 1285–1290 [DOI] [PubMed] [Google Scholar]
- Nechaev S, Chlenov M, Severinov K (2000) Dissection of two hallmarks of the open promoter complex by mutation in an RNA polymerase core subunit. J Biol Chem 275: 25516–25522 [DOI] [PubMed] [Google Scholar]
- Paget MS, Helmann JD (2003) The sigma70 family of sigma factors. Genome Biol 4: 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Gralla JD (2001) Roles for the C-terminal region of sigma 54 in transcriptional silencing and DNA binding. J Biol Chem 276: 8979–8986 [DOI] [PubMed] [Google Scholar]
- Wigneshweraraj SR, Burrows PC, Severinov K, Buck M (2004) Contribution of the β′ jaw domain to σ54-RNA polymerase dependent transcription (in preparation)
- Wigneshweraraj SR, Chaney MK, Ishihama A, Buck M (2001) Regulatory sequences in sigma 54 localise near the start of DNA melting. J Mol Biol 306: 681–701 [DOI] [PubMed] [Google Scholar]
- Wigneshweraraj SR, Kuznedelov K, Severinov K, Buck M (2003) Multiple roles of the RNA polymerase beta subunit flap domain in sigma 54-dependent transcription. J Biol Chem 278: 3455–3465 [DOI] [PubMed] [Google Scholar]
- Wigneshweraraj SR, Nechaev S, Severinov K, Buck M (2002) Beta subunit residues 186–433 and 436–445 are commonly used by Esigma54 and Esigma70 RNA polymerase for open promoter complex formation. J Mol Biol 319: 1067–1083 [DOI] [PubMed] [Google Scholar]
- Xu H, Gu B, Nixon BT, Hoover TR (2004) Purification and characterization of the AAA+ domain of Sinorhizobium meliloti DctD, a sigma54-dependent transcriptional activator. J Bacteriol 186: 3499–3507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Campbell EA, Minakhin L, Richter C, Severinov K, Darst SA (1999) Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 Å resolution. Cell 98: 811–824 [DOI] [PubMed] [Google Scholar]
- Zhang X, Chaney M, Wigneshweraraj SR, Schumacher J, Bordes P, Cannon W, Buck M (2002) Mechanochemical ATPases and transcriptional activation. Mol Microbiol 45: 895–903 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1


