Abstract
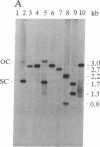
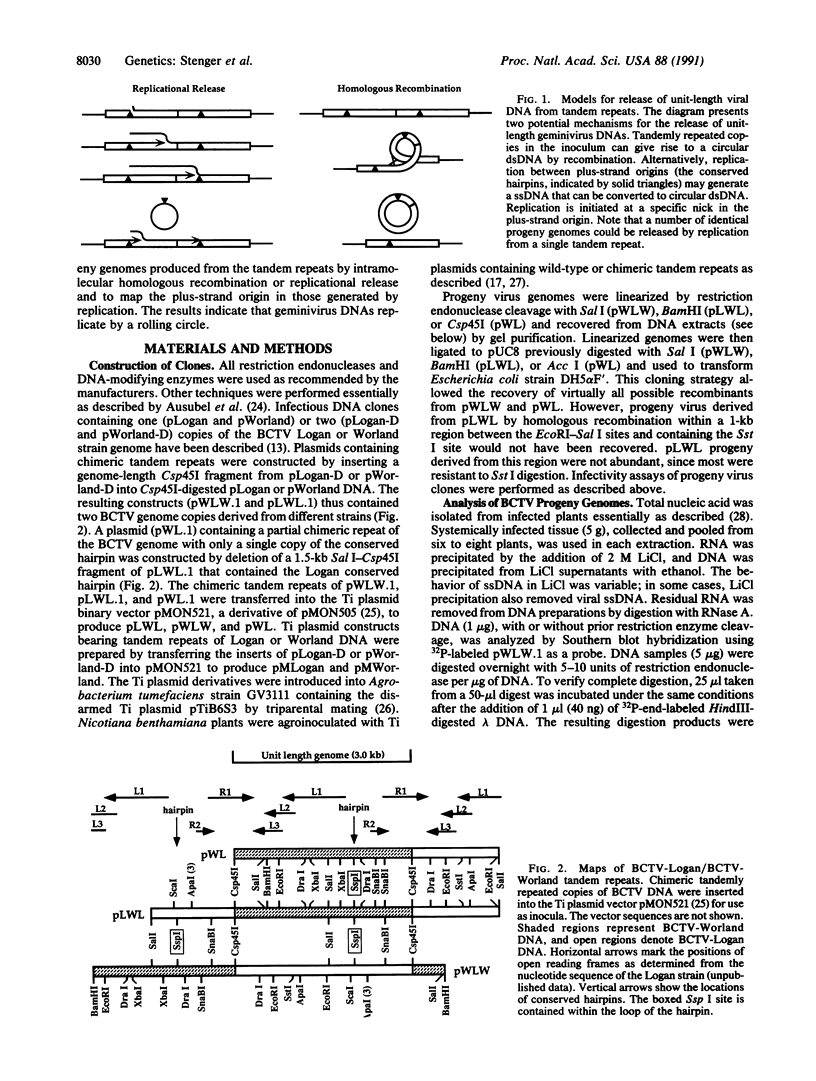
Agrobacterium-mediated inoculation of Nicotiana benthamiana plants with Ti plasmids containing tandem genome repeats derived from different strains of the gemini-virus beet curly top virus (BCTV) resulted in the production of unit-length recombinant progeny genomes in systemically infected plants. When two putative plus-strand origins of replication were present in constructs used as inocula, a replicational escape mechanism was favored that resulted in progeny genomes of a single predominant genotype. The genotype was dependent upon the arrangement of repeated parental genomes in the inocula. Sequencing across the junction between parental BCTV strains in the recombinant progeny allowed mapping of the plus-strand origin of replication to a 20-base-pair sequence within the conserved hairpin found in all geminivirus genomes. In contrast, when inocula contained tandemly repeated BCTV genome sequences but only a single conserved hairpin, a number of different progeny genotypes were simultaneously replicated in infected plants, a result expected if unit-length viral genomes were generated by random intramolecular recombination events. These results and other considerations indicate that geminivirus DNA replication occurs by a rolling-circle mechanism.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Briddon R. W., Watts J., Markham P. G., Stanley J. The coat protein of beet curly top virus is essential for infectivity. Virology. 1989 Oct;172(2):628–633. doi: 10.1016/0042-6822(89)90205-5. [DOI] [PubMed] [Google Scholar]
- Carrington J. C., Morris T. J. Complementary DNA cloning and analysis of carnation mottle virus RNA. Virology. 1984 Nov;139(1):22–31. doi: 10.1016/0042-6822(84)90326-x. [DOI] [PubMed] [Google Scholar]
- Davies J. W., Stanley J. Geminivirus genes and vectors. Trends Genet. 1989 Mar;5(3):77–81. doi: 10.1016/0168-9525(89)90030-9. [DOI] [PubMed] [Google Scholar]
- Donson J., Accotto G. P., Boulton M. I., Mullineaux P. M., Davies J. W. The nucleotide sequence of a geminivirus from Digitaria sanguinalis. Virology. 1987 Nov;161(1):160–169. doi: 10.1016/0042-6822(87)90182-6. [DOI] [PubMed] [Google Scholar]
- Donson J., Morris-Krsinich B. A., Mullineaux P. M., Boulton M. I., Davies J. W. A putative primer for second-strand DNA synthesis of maize streak virus is virion-associated. EMBO J. 1984 Dec 20;3(13):3069–3073. doi: 10.1002/j.1460-2075.1984.tb02260.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotto G. P., Horiuchi K. Replication of a plasmid containing two origins of bacteriophage. J Mol Biol. 1981 Nov 25;153(1):169–176. doi: 10.1016/0022-2836(81)90532-5. [DOI] [PubMed] [Google Scholar]
- Grimsley N., Hohn T., Hohn B. Recombination in a plant virus: template-switching in cauliflower mosaic virus. EMBO J. 1986 Apr;5(4):641–646. doi: 10.1002/j.1460-2075.1986.tb04261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gros M. F., te Riele H., Ehrlich S. D. Rolling circle replication of single-stranded DNA plasmid pC194. EMBO J. 1987 Dec 1;6(12):3863–3869. doi: 10.1002/j.1460-2075.1987.tb02724.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton W. D., Bisaro D. M., Buck K. W. Identification of novel DNA forms in tomato golden mosaic virus infected tissue. Evidence for a two component viral genome. Nucleic Acids Res. 1982 Aug 25;10(16):4901–4912. doi: 10.1093/nar/10.16.4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidekamp F., Baas P. D., Jansz H. S. Nucleotide sequences at the phi X gene A protein cleavage site in replicative form I DNAs of bacteriophages U3, G14, and alpha 3. J Virol. 1982 Apr;42(1):91–99. doi: 10.1128/jvi.42.1.91-99.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarowitz S. G., Pinder A. J., Damsteegt V. D., Rogers S. G. Maize streak virus genes essential for systemic spread and symptom development. EMBO J. 1989 Apr;8(4):1023–1032. doi: 10.1002/j.1460-2075.1989.tb03469.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Michel B., Ehrlich S. D. Illegitimate recombination occurs between the replication origin of the plasmid pC194 and a progressing replication fork. EMBO J. 1986 Dec 20;5(13):3691–3696. doi: 10.1002/j.1460-2075.1986.tb04701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revington G. N., Sunter G., Bisaro D. M. DNA sequences essential for replication of the B genome component of tomato golden mosaic virus. Plant Cell. 1989 Oct;1(10):985–992. doi: 10.1105/tpc.1.10.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers S. G., Bisaro D. M., Horsch R. B., Fraley R. T., Hoffmann N. L., Brand L., Elmer J. S., Lloyd A. M. Tomato golden mosaic virus A component DNA replicates autonomously in transgenic plants. Cell. 1986 May 23;45(4):593–600. doi: 10.1016/0092-8674(86)90291-6. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slomka M. J., Buck K. W., Coutts R. H. Characterisation of multimeric DNA forms associated with tomato golden mosaic virus infection. Arch Virol. 1988;100(1-2):99–108. doi: 10.1007/BF01310911. [DOI] [PubMed] [Google Scholar]
- Sozhamannan S., Dabert P., Moretto V., Ehrlich S. D., Gruss A. Plus-origin mapping of single-stranded DNA plasmid pE194 and nick site homologies with other plasmids. J Bacteriol. 1990 Aug;172(8):4543–4548. doi: 10.1128/jb.172.8.4543-4548.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley J., Markham P. G., Callis R. J., Pinner M. S. The nucleotide sequence of an infectious clone of the geminivirus beet curly top virus. EMBO J. 1986 Aug;5(8):1761–1767. doi: 10.1002/j.1460-2075.1986.tb04424.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley J., Townsend R. Characterisation of DNA forms associated with cassava latent virus infection. Nucleic Acids Res. 1985 Apr 11;13(7):2189–2206. doi: 10.1093/nar/13.7.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenger D. C., Carbonaro D., Duffus J. E. Genomic characterization of phenotypic variants of beet curly top virus. J Gen Virol. 1990 Oct;71(Pt 10):2211–2215. doi: 10.1099/0022-1317-71-10-2211. [DOI] [PubMed] [Google Scholar]
- van der Ende A., Teertstra R., Weisbeek P. J. Initiation and termination of the bacteriophage phi X174 rolling circle DNA replication in vivo: packaging of plasmid single-stranded DNA into bacteriophage phi X174 coats. Nucleic Acids Res. 1982 Nov 11;10(21):6849–6863. doi: 10.1093/nar/10.21.6849. [DOI] [PMC free article] [PubMed] [Google Scholar]