Abstract
Objective
Secretin-stimulated pancreatic juice contains DNA shed from cells lining the pancreatic ducts. Genetic analysis of this fluid may form a test to detect pancreatic ductal neoplasia.
Design
We employed digital next-generation sequencing (‘digital NGS’) to detect low-abundance mutations in secretin-stimulated juice samples collected from the duodenum of subjects enrolled in Cancer of the Pancreas Screening studies at Johns Hopkins Hospital. For each juice sample, digital NGS necessitated 96 NGS reactions sequencing nine genes. The study population included 115 subjects (53 discovery, 62 validation) (1) with pancreatic ductal adenocarcinoma (PDAC), (2) intraductal papillary mucinous neoplasm (IPMN), (3) controls with non-suspicious pancreata.
Results
Cases with PDAC and IPMN were more likely to have mutant DNA detected in pancreatic juice than controls (both p<0.0001); mutant DNA concentrations were higher in patients with PDAC than IPMN (p=0.003) or controls (p<0.001). TP53 and/or SMAD4 mutations were commonly detected in juice samples from patients with PDAC and were not detected in controls (p<0.0001); mutant TP53/SMAD4 concentrations could distinguish PDAC from IPMN cases with 32.4% sensitivity, 100% specificity (area under the curve, AUC 0.73, p=0.0002) and controls (AUC 0.82, p<0.0001). Two of four patients who developed pancreatic cancer despite close surveillance had SMAD4/TP53 mutations from their cancer detected in juice samples collected over 1 year prior to their pancreatic cancer diagnosis when no suspicious pancreatic lesions were detected by imaging.
Conclusions
The detection in pancreatic juice of mutations important for the progression of low-grade dysplasia to high-grade dysplasia and invasive pancreatic cancer may improve the management of patients undergoing pancreatic screening and surveillance.
Keywords: PANCREATIC CANCER, PANCREATIC PATHOLOGY, MUTATIONS, ENDOSCOPIC ULTRASONOGRAPHY
Significance of this study.
What is already known on this subject?
▸ Mutations arising from the pancreatic ductal system can be detected in secretin-stimulated pancreatic juice samples.
▸ The molecular progression of pancreatic ductal neoplasia involves the acquisition of mutations in multiple genes, found in low-grade pancreatic intraepithelial neoplasias and intraductal papillary mucinous neoplasms (IPMNs) (such as KRAS and GNAS) and others that emerge as these lesions progress to higher-grade precursors and to invasive pancreatic ductal adenocarcinoma (such as TP53 and SMAD4).
▸ A test that could detect these mutations in pancreatic juice could have diagnostic utility.
What are the new findings?
▸ We have developed a novel next-generation sequencing (NGS) method, termed digital NGS to detect low concentrations (0.1% to 1%) of mutations in nine genes in pancreatic juice.
▸ We find overall juice mutation concentrations, particularly mutations affecting TP53 and SMAD4 could distinguish juice samples from patients with pancreatic cancer from those with IPMNs and disease controls without pancreatic neoplasia.
▸ Two of four patients who developed pancreatic cancer while under surveillance had SMAD4 or TP53 mutations from their cancer detected in pancreatic juice samples collected within 18 months of their cancer detection when no suspicious pancreatic lesions were detected by imaging.
How might it impact on clinical practice in the foreseeable future?
▸ These results highlight the potential of using pancreatic juice analysis to detect worrisome mutations that could help in the surveillance and risk stratification of patients undergoing pancreatic screening and surveillance.
Introduction
Pancreatic cancer is expected to be the second leading cause of cancer-related deaths in the USA by 2030.1 Most patients with pancreatic ductal adenocarcinoma (PDAC) present with advanced-stage cancers and have rapidly progressive disease.
Endoscopic ultrasonography (EUS) and/or MRI/MR cholangiopancreatography (MRCP) can accurately identify subcentimetre pancreatic cysts2 and are used to screen individuals with a strong family history of pancreatic cancer to try to detect asymptomatic Stage I pancreatic cancers and significant precancerous lesions.2–10 Most pancreatic cysts detected in patients undergoing screening are thought to be intraductal papillary mucinous neoplasms (IPMNs).2 However, the most common precancerous lesions, pancreatic intraepithelial neoplasias (PanINs), are generally too small (<5 mm diameter by definition) to be identified by imaging modalities.11 New tests are needed to identify clinically significant precursor lesions and early curable invasive cancers, and one potential approach is to analyse pancreatic juice. Analysis of secretin-stimulated pancreatic juice collected endoscopically from the duodenum of patients enrolled in the Cancer of the Pancreas Screening (CAPS) trials revealed that the detection of GNAS mutations closely correlated with having IPMNs, and the detection of TP53 mutations in 67% of cases with PDAC and 50% of cases with advanced precursor lesions.2 4 12–14 Similarly, KRAS mutations are commonly detected in juice samples from patients with pancreatic cancer and patients undergoing screening. KRAS mutations detected in the pancreatic juice of high-risk individuals without pancreatic imaging abnormalities are thought to arise from small PanIN lesions.15 These studies used digital melt-curve analysis and pyrosequencing to detect mutations but next-generation sequencing (NGS) is being widely used in clinical laboratories to detect somatic mutations in cancer tissues.16 17
NGS is being evaluated as a test to detect low-abundance somatic mutations in secondary fluids such as plasma,18 but the rate of sequencing errors generated by NGS assays poses challenges to the detection of low-abundance mutations. Sequence variants identified using NGS-based tests need to be present at sufficient concentrations (>1%) to be considered as true mutations rather than background sequencing errors.19 Somatic mutation concentrations in duodenal collections of pancreatic juice are generally quite low (usually 0.1–1%) even among patients with PDAC15 and thus their detection by NGS requires modifications to standard NGS protocols. Molecular strategies such as ‘SafeSeq’ have been employed in research settings to help distinguish true low-abundance somatic mutations from low-level errors related to NGS.20 In principle, the ability of NGS to accurately detect low-abundance mutations could be improved by using ‘digital’ strategies, analogous to digital PCR. We developed a digital NGS method for this purpose by performing discrete NGS analyses on many (typically 96) individual aliquots of DNA from a single biological sample where each aliquot contains only a few genome equivalents of DNA. Each aliquot can then be expected to have either zero or one mutation-containing DNA template at each nucleotide of interest in addition to small numbers of wild type templates. True somatic mutations should be detectable in more than one aliquot.21 In this study, we evaluated digital NGS as a strategy to detect low-abundance mutations and then applied the method to detect mutations in duodenal collections of pancreatic juice obtained from patients with and without pancreatic ductal neoplasia to evaluate the diagnostic accuracy of this test.
Materials and methods
Patients and specimens
Pancreatic juice samples for this study were obtained from participants enrolled in the CAPS studies (http://clinicaltrials.gov, NCT00438906, NCT00714701 and NCT02000089).2 4 One hundred and fifteen prospectively enrolled subjects (53 discovery, 62 validation), were included to represent a variety of diagnostic possibilities (see online supplementary table S1). Patient groups included those diagnosed with (1) PDAC (n=34), including selected patients who developed pancreatic cancer while under surveillance (n=4), (2) IPMN (n=57), diagnosed by surgical pathology or imaging findings, including cases undergoing pancreatic screening, or (3) controls (n=24) with normal pancreata undergoing EUS for other indications, or chronic pancreatitis. Archived primary pancreatic cancer or IPMN tissue was sequenced from some patients to compare mutations detected in juice samples with those present in tumours.
gutjnl-2015-311166supp002.pdf (153.6KB, pdf)
Pancreatic juice secretion was stimulated by infusing human synthetic secretin (ChiRhoClin)(0.2 µg/kg intravenously over a minute). Juice was collected from the duodenal lumen for ∼5 min (typically, 5–10 mL).13 In addition, several samples of pancreatic cyst fluid aspirated during EUS were sequenced.22
All elements of this study were approved by the Johns Hopkins institutional review board, and written informed consent was obtained from all patients.
Digital NGS
All digital NGS assays were performed blinded to patient information. An Ion AmpliSeq Custom Panel was employed to multiplex PCR and sequence nine genes (122 amplicons in two primer pools, see online supplementary table S2) mutated in pancreatic ductal neoplasms (KRAS, GNAS, TP53, SMAD4, CDKN2A, RNF43, TGFBR2, BRAF, PIK3CA).23–26 Ninety-six aliquots of DNA from each patient's juice were made and each aliquot was subjected to NGS. A mutation score of one was given for each mutation-containing aliquot.
Estimating digital NGS accuracy
Digital NGS was performed on wild type fibroblast DNA samples and three reference pools containing low mutation concentrations (20 pancreatic cancer cell lines mixed with fibroblast DNA) (see online supplementary tables S3 and S4).
Digital-droplet PCR
Digital-droplet PCR (ddPCR) was performed blinded to digital NGS results to help evaluate the accuracy of digital NGS.
(See online supplementary materials for additional methods).
gutjnl-2015-311166supp001.pdf (93.7KB, pdf)
Statistics
Median mutation scores between PDAC, IPMN and control groups were compared by the Mann-Whitney U and χ2 tests. Receiver operating characteristic (ROC) curves were generated to evaluate candidate gene's mutations and the area under the curve (AUC) was computed by the trapezoidal method. SPSS software and GraphPad Prism6 were used. A two-tailed p<0.05 was considered statistically significant.
Results
Detection of low-abundance mutations in DNA reference pools by digital NGS
We found digital NGS could detect all 28 missense and nonsense mutations in pancreatic cancer DNA reference pools present at concentrations ranging from >0.1% to 1% relative to wild type DNA and 90% of mutations present at the 0.1% level relative to wild type DNA (see online supplementary table S4). To avoid calling NGS-related sequencing errors mutations, we required the detection of the same sequence variant in three independent digital NGS reactions, as a criterion for calling non-hot spot sequence variants identified by digital NGS as true mutations (in addition to the usual metrics for calling mutations). We also compared digital NGS to ddPCR for their ability to detect KRAS mutations in pancreatic juice and found almost complete concordance between the two methods (see also online supplementary materials).
Somatic mutations detected in pancreatic juice samples
Using digital NGS, we compared the mutation profiles of patients diagnosed with PDAC versus IPMN versus controls first in a discovery set (Cases #1–53) and then in a validation set (Cases #54–115). The results of the combined set are summarised in figures 1–3 and see online supplementary table 5 table 1. Discovery and validation set results are presented in online supplementary figures S2–S4 and supplementary materials. Mutational analysis of primary pancreatic cancer or IPMN was also performed for cases with sufficient neoplastic tissue (see online supplementary tables S6 and S5). Most juice samples with mutations had digital NGS scores of <10 (range 1–87) (table 1).
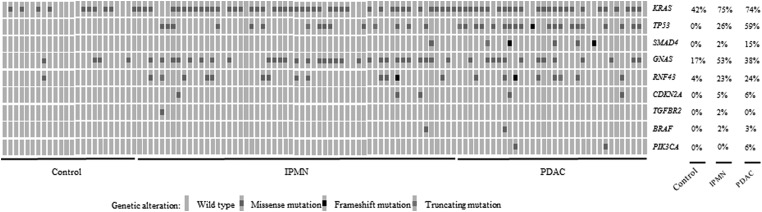
Figure 1.
Prevalence of genes identified as mutated in pancreatic juice by digital next-generation sequencing. Some cases with pancreatic ductal adenocarcinoma (PDAC) also had intraductal papillary mucinous neoplasm (IPMN).
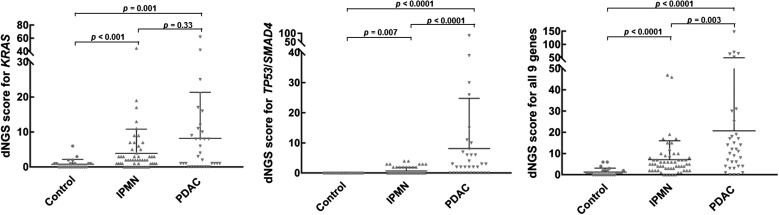
Figure 2.
Pancreatic juice mutation concentrations (digital next-generation sequencing (dNGS) scores) by disease group. IPMN, intraductal papillary mucinous neoplasm; PDAC, pancreatic ductal adenocarcinoma.
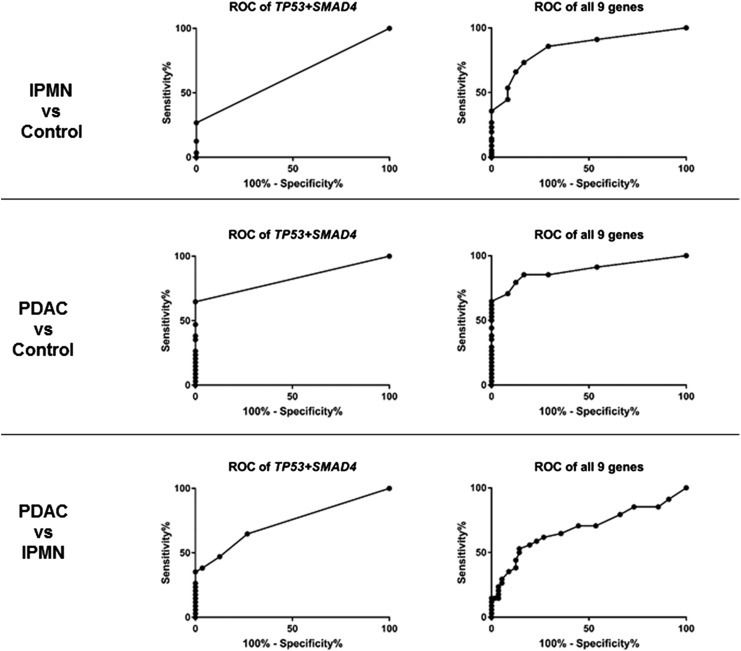
Figure 3.
Receiver operating characteristic (ROC) curve analysis evaluating how pancreatic juice gene mutation scores to distinguished disease groups. IPMN, intraductal papillary mucinous neoplasm; PDAC, pancreatic ductal adenocarcinoma.
Table 1.
Somatic mutations identified in pancreatic juice by digital NGS*
|
KRAS |
GNAS |
RNF43 |
TP53 |
SMAD4 |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case# | M/F | Age | Disease group | Risk** | dNGS# | SNV | dNGS# | SNV | dNGS# | SNV | dNGS# | SNV | dNGS# | SNV |
| #01 | F | 27 | Control | na | 0 | 0 | 0 | 0 | 0 | |||||
| #02 | F | 79 | Control | na | 1 | p.G12D | 0 | 0 | 0 | 0 | ||||
| #03 | F | 77 | Control | na | 0 | 0 | 0 | 0 | 0 | |||||
| #04 | M | 50 | Control | na | 1 | p.G12D | 0 | 0 | 0 | 0 | ||||
| #05 | F | 78 | Control | na | 0 | 0 | 0 | 0 | 0 | |||||
| #06 | M | 76 | Control | na | 0 | 0 | 0 | 0 | 0 | |||||
| #07 | F | 49 | Control | na | 1,2,2 | p.G12D/V,G13D | 0 | 0 | 0 | 0 | ||||
| #08 | F | 65 | Control | na | 0 | 3 | p.R201C | 3 | p.Q402R | 0 | 0 | |||
| #09 | M | 45 | Control | na | 1,2 | p.G12D/V | 0 | 0 | 0 | 0 | ||||
| #10 | F | 68 | Control | na | 0 | 0 | 0 | 0 | 0 | |||||
| #11 | F | 48 | Control | na | 0 | 0 | 0 | 0 | 0 | |||||
| #12 | F | 43 | Control | na | 0 | 0 | 0 | 0 | 0 | |||||
| #13 | F | 44 | Control | na | 0 | 0 | 0 | 0 | 0 | |||||
| #54 | M | 46 | Control | na | 0 | 0 | 0 | 0 | 0 | |||||
| #55 | F | 72 | Control | na | 2 | p.Q61L | 0 | 0 | 0 | 0 | ||||
| #56 | M | 34 | Control | na | 1 | p.Q61L | 0 | 0 | 0 | 0 | ||||
| #57 | F | 52 | Control | na | 1 | p.G12S | 1 | p.R201H | 0 | 0 | 0 | |||
| #58 | M | 66 | Control | na | 0 | 2,2 | p.R201C/H | 0 | 0 | 0 | ||||
| #59 | F | 59 | Control | na | 1 | p.G12S | 0 | 0 | 0 | 0 | ||||
| #60 | M | 59 | Control | na | 1,1 | p.G12D/S | 0 | 0 | 0 | 0 | ||||
| #61 | F | 73 | Control | na | 0 | 0 | 0 | 0 | 0 | |||||
| #62 | M | 48 | Control | na | 0 | 0 | 0 | 0 | 0 | |||||
| #63 | M | 40 | Control | na | 0 | 1 | p.R201C | 0 | 0 | 0 | ||||
| #64 | F | 32 | Control | na | 1 | p.G12D | 0 | 0 | 0 | 0 | ||||
| #15 | M | 64 | IPMNs | f | 1 | p.G12V | 0 | 0 | 0 | 0 | ||||
| #23 | F | 64 | IPMNs | s | 3,4 | p.G12D/V | 0 | 0 | 0 | 0 | ||||
| #24 | M | 21 | IPMNs | s | 3 | p.G13D | 29 | p.R201C | 10,3 | p.R343H, Y550C | 0 | 0 | ||
| #25 | M | 67 | IPMNs | s | 0 | 0 | 0 | 0 | 0 | |||||
| #26 | M | 51 | IPMNs | f | 0 | 4,1 | p.R201C/H | 4,3 | p.P634S, Q402X | 3 | p.R196L | 0 | ||
| #27 | M | 62 | IPMNs | f | 0 | 0 | 0 | 2 | p.R248Q | 0 | ||||
| #28 | M | 70 | IPMNs | s | 1 | p.G12D | 4,1 | p.R201C/H | 0 | 4 | p.K291R | 0 | ||
| #29 | F | 75 | IPMNs | s | 1 | p.G12V | 2 | p.Q227R | 0 | 0 | 0 | |||
| #30 | F | 63 | IPMNs | s | 1,2,4 | p.G12D/V/R | 0 | 3 | p.S446F | 0 | 0 | |||
| #65 | F | 58 | IPMN | s | 1,1,1 | p.G12D/V,Q61L | 1 | p.Q227R | 0 | 0 | 0 | |||
| #66 | M | 67 | IPMN | s | 9 | p.G12C | 0 | 0 | 0 | 0 | ||||
| #67 | M | 70 | IPMN | s | 1,1 | p.G12D/V | 2 | p.R201C | 0 | 0 | 0 | |||
| #68 | M | 59 | IPMN | s | 1 | p.G12V | 0 | 3 | p.S464G | 0 | 0 | |||
| #69 | F | 65 | IPMN | s | 1,1 | p.G12S, Q61R | 1 | p.R201C | 3 | p.S478P | 0 | 0 | ||
| #70 | M | 62 | IPMN | s | 1 | p.Q61R | 0 | 0 | 3 | p.M246V | 0 | |||
| #71 | F | 84 | IPMN | s | 0 | 0 | 0 | 0 | 0 | |||||
| #72 | M | 66 | IPMN | s | 1,2 | p.G12V, Q61H | 1 | p.R201H | 0 | 0 | 0 | |||
| #73 | F | 60 | IPMN | s | 1 | p.Q61L | 0 | 0 | 0 | 0 | ||||
| #74 | F | 66 | IPMN | s | 1,1,1 | p.G13D, Q61H/R | 0 | 0 | 0 | 0 | ||||
| #75 | M | 70 | IPMN | s | 0 | 0 | 0 | 0 | 0 | |||||
| #76 | M | 41 | IPMN | s | 1,1 | p.G12V, Q61R | 0 | 0 | 2 | p.R175H | 0 | |||
| #77 | F | 55 | IPMN | s | 1 | p.Q61R | 0 | 0 | 0 | 0 | ||||
| #78 | F | 74 | IPMN | s | 1,1,1 | p.G12C, G13D, Q61R | 0 | 3 | p.A148V | 3 | p.R175H | 0 | ||
| #79 | M | 76 | IPMN | s | 0 | 1 | p.Q227R | 5 | p.A629T | 0 | 0 | |||
| #80 | F | 64 | IPMN | s | 1,8 | p.G12V, Q61H | 8 | p.R201C | 0 | 0 | 0 | |||
| #81 | F | 44 | IPMN | s | 2,1,1,1,1 | p.G12D/V/R, p.Q61H/R | 1 | p.R201H | 0 | 0 | 0 | |||
| #82 | F | 77 | IPMN | s | 1 | p.Q61H | 1 | p.R201H | 0 | 0 | 0 | |||
| #83 | F | 67 | IPMN | s | 2,1,1,2,1 | p.G12D/V/R/S, G13D | 0 | 0 | 0 | 0 | ||||
| #84 | M | 84 | IPMN | s | 1,1 | p.G12C/S | 1 | p.R201H | 3 | p.G263E | 0 | 0 | ||
| #85 | M | 49 | IPMN | s | 1,2 | p.G12D/V | 0 | 0 | 0 | 0 | ||||
| #86 | F | 74 | IPMN | s | 1,1,1 | p.G12D, Q61H/R | 1 | p.R201C | 3 | p.L122P | 3 | p.Q165X | 0 | |
| #87 | F | 77 | IPMN | s | 1,2,1,1 | p.G12D/V/S, Q61L | 0 | 0 | 0 | 0 | ||||
| #88 | F | 83 | IPMN | s | 2 | p.G12V | 1 | p.R201H | 0 | 2 | p.R282W | 0 | ||
| #89 | F | 83 | IPMN | s | 0 | 1 | p.R201H | 0 | 4 | p.R248W | 0 | |||
| #90 | M | 70 | IPMN | s | 1,1 | p.G12D, Q61H | 1 | p.Q227P | 0 | 2 | p.G245S | 0 | ||
| #91 | F | 62 | IPMN | s | 1,1,2 | p.G12V/R/S | 3 | p.R201C | 0 | 0 | 0 | |||
| #92 | F | 70 | IPMN | s | 5,4,2 | p.G12D/V/C | 3,1 | p.R201C, Q227R | 0 | 0 | 0 | |||
| #93 | M | 62 | IPMN | s | 1,1,1 | p.G12C/S, Q61H | 0 | 0 | 0 | 0 | ||||
| #94 | F | 66 | IPMN | s | 0 | 0 | 0 | 0 | 0 | |||||
| #95 | M | 64 | IPMN | s | 0 | 2 | p.R201C | 0 | 0 | 0 | ||||
| #96 | M | 76 | IPMN | s | 0 | 0 | 0 | 2 | p.R248W | 0 | ||||
| #97 | F | 83 | IPMN | s | 13 | p.G12V | 1,1 | p.R201C/H | 0 | 0 | 0 | |||
| #98 | F | 68 | IPMN | s | 0 | 0 | 0 | 2 | p.Q136X | 0 | ||||
| #99 | F | 60 | IPMN | s | 1,1 | p.Q61L/R | 1 | p.R201H | 3 | p.S111G | 0 | 0 | ||
| #14 | M | 48 | LG-IPMN | s | 0 | 1 | p.R201H | 4 | p.A618V | 0 | 0 | |||
| #16 | F | 63 | LG-IPMN | f | 2 | p.G12D | 0 | 0 | 0 | 0 | ||||
| #17 | F | 60 | LG-IPMN | f | 5,8,1,3,2 | p.G13D, G12C/R/V, Q61R | 8 | p.R201H | 15 | p.L12Rfs | 2 | p.R248Q | 0 | |
| #18 | M | 75 | LG-IPMN | f | 1 | p.G12V | 2,1 | p.Q227R, R201C | 0 | 0 | 0 | |||
| #100 | M | 78 | LG-IPMN | f | 1,2 | p.G13D, Q61R | 1 | p.Q227R | 0 | 2 | p.R175H | 0 | ||
| #101 | M | 72 | LG-IPMN | f | 0 | 0 | 0 | 0 | 0 | |||||
| #19 | F | 67 | IM-IPMN | s | 2 | p.G13D | 2 | p.R201C | 0 | 0 | 0 | |||
| #20 | F | 73 | IM-IPMN | s | 0 | 2 | p.R201C | 0 | 2 | p.R282W | 0 | |||
| #21 | F | 67 | IM-IPMN | s | 1 | p.G12V | 1 | p.R201C | 0 | 0 | 3 | p.W524R^ | ||
| #22 | M | 65 | IM-IPMN | f | 2 | p.G12V | 0 | 0 | 0 | 0 | ||||
| #102 | M | 72 | IM-IPMN | s | 5 | p.Q61H | 0 | 3 | p.R127Q | 0 | 0 | |||
| #103 | F | 75 | IM-IPMN | f | 7,1,18,1,18 | p.G12A/V/R, G13D, Q61H | 1 | p.R201C | 0 | 0 | 0 | |||
| #104 | M | 71 | HG-IPMN | f | 1,1,2,1,7,4,1 | p.G12D/R/S, G13D, p.Q61L/H/P | 0 | 0 | 0 | 0 | ||||
| #31 | M | 79 | PDAC/IPMNs | s | 1 | p.G12V | 0 | 0 | 2 | p.R196X | 0 | |||
| #32 | M | 69 | PDAC/IPMNs | s | 1 | p.Q61R | 0 | 0 | 3 | p.R175H | 0 | |||
| #33 | F | 74 | PDAC/IPMNs | s | 4,2 | p.G13D, G12R | 1 | p.R201H | 0 | 0 | 0 | |||
| #34 | F | 55 | PDAC/IPMNs | s | 23,6,3,2,2,2,2,2 | p.G12D/S/R/C/V, G13C/D | 0 | 3 | p.R454H | 26 | p.R248Q | 0 | ||
| #35 | M | 65 | PDAC/IPMNs | f | 1 | p.G12V | 0 | 0 | 3 | p.R248Q | 0 | |||
| #36 | M | 74 | PDAC/IPMNs | f | 21 | p.G12D | 2 | p.R201C | 0 | 0 | 26,4 | p.Q311X, Q256X | ||
| #37 | M | 69 | PDAC/IPMN | s | 0 | 61 | p.R201C | 0 | 87 | p.L194R | 0 | |||
| #38 | M | 73 | PDAC/IPMNs | s | 12 | p.G12D | 1,1 | p.R201C/H | 0 | 0 | 0 | |||
| #105 | M | 68 | PDAC/IPMNs | s | 1 | p.G12D | 6 | p.R201C | 3,3 | p.F322S, H295R | 8 | p.C141G | 0 | |
| #106 | M | 44 | PDAC/IPMNs | s | 10 | p.Q61R | 0 | 0 | 12 | p.R175H | 3 | p.T273fs | ||
| #107 | M | 59 | PDAC/IPMN | s | 8,8 | p.G12D/S | 0 | 10 | p.P470fs | 2 | p.R175H | 0 | ||
| #115 | M | 68 | PDAC/IPMN | s | 6,1,1 | p.G12C/S, Q61L | 1 | p.R201H | 0 | 3 | p.R181C | 0 | ||
| #39 | M | 70 | PDAC | f | 0 | 0 | 0 | 0 | 0 | |||||
| #40 | M | 53 | PDAC | s | 0 | 0 | 0 | 6 | p. L130Sfs | 0 | ||||
| #41 | F | 46 | PDAC | s | 3,6 | p.G12D/R | 2 | p.R201H | 4 | p.C471R | 0 | 0 | ||
| #42 | M | 78 | PDAC | s | 6,2 | p.G13D, G12V | 1 | p.R201C | 0 | 0 | 0 | |||
| #43 | F | 59 | PDAC | s | 3,5 | p.G12S/V | 0 | 0 | 2 | p.R248W | 0 | |||
| #44 | F | 77 | PDAC | s | 1 | p.G12D | 2 | p.R201C | 5 | p.H556R | 3 | p.R175H | 7 | p.A457V^ |
| #45 | M | 56 | PDAC | s | 6,2 | p.G12D/V | 0 | 4 | p.R519X | 7 | p.Y220C | 0 | ||
| #46 | M | 58 | PDAC | s | 17 | p.G12D | 0 | 0 | 0 | 0 | ||||
| #47 | F | 75 | PDAC | s | 32,22,4,4 | p.G13D, Q61H, G12D/R | 0 | 0 | 2 | p.R273C | 0 | |||
| #48 | M | 59 | PDAC | f | 0 | 0 | 3,3 | p.L109P, V480A | 3 | p.C277R | 3 | p.M543T^ | ||
| #49 | F | 69 | PDAC | s | 23,2 | p.G12V/S | 2 | p.R201C | 0 | 39 | p.R196X | 0 | ||
| #50 | F | 68 | PDAC | s | 0 | 0 | 0 | 0 | 0 | |||||
| #51 | F | 66 | PDAC | s | 0 | 0 | 0 | 0 | 11 | p.K50Kfs | ||||
| #52 | F | 62 | PDAC | f | 4 | p.G12D | 0 | 0 | 0 | 0 | ||||
| #53 | F | 79 | PDAC | f | 1,1 | p.G12D/R | 0 | 0 | 4,1,1 | p.Y220C, R248W/Q | 0 | |||
| #108 | M | 63 | PDAC | s | 0 | 1 | p.Q227R | 0 | 0 | 0 | ||||
| #109 | M | 71 | PDAC | s | 1 | p.Q61H | 0 | 0 | 0 | 0 | ||||
| #110 | M | 70 | PDAC | f | 0 | 0 | 3 | p.H472R | 2 | p.R248W | 0 | |||
| #111 | M | 62 | PDAC | s | 2,1 | p.G12D, G13D | 0 | 0 | 0 | 0 | ||||
| #112 | F | 71 | PDAC | s | 1 | p.Q61H | 1 | p.R201C | 0 | 2,2 | p.R248W, Y163C | 0 | ||
| #113 | F | 77 | PDAC | s | 11 | p.G12V | 1 | p.R201H | 0 | 2 | p.Y220C | 0 | ||
| #114 | M | 57 | PDAC | s | 0 | 0 | 0 | 0 | 0 | |||||
Case #20 had a BRAF K601K mutation (score 4), Case #26 had a TGFBR2 mutation D317Y (score 4), Case #53 had PIK3CA mutations H1047L/R (score 3, 6), Case#105 had a BRAF F595L mutation (score 4), Case # had a Case #107 had a PIK3CA H1047R mutation (score 3).
*, Other four genes shown in supplementary table;**, s: sporadic; f: familial. dNGS#:digital NGS score (mutation concentration); SNV:single nucleotide variant (i.e. somatic mutation). ^suspected pathogenic.
gutjnl-2015-311166supp003.pdf (257.5KB, pdf)
In the combined set of 115 patients, 31 of 34 (91.2%) patients with PDAC and 51 of 56 (91.1%) diagnosed with an IPMN (without PDAC) had at least one mutation detected in their pancreatic juice sample, compared with 13 of 24 (54.2%) controls without evidence of pancreatic neoplasia (p=0.001 and p<0.001). KRAS mutations (with both digital NGS and ddPCR for the discovery set) were detected in juice samples of 10 of 24 controls (41.7%), 42 of 56 (75.0%) patients with IPMNs and 25 of 34 (73.5%) patients with PDAC. Several patients, particularly those with PDAC, had multiple KRAS mutations detected in their juice samples.15
Thirty-five cases had deleterious TP53 mutations, including 20 with PDAC and 15 with IPMN. Deleterious SMAD4 mutations were detected in seven patients; three had missense mutations. SMAD4 missense mutations are often deleterious.27 28 Six of these patients had pancreatic cancer. The one non-PDAC case (Case#21) with a deleterious SMAD4 mutation underwent resection for high-risk findings;29 a dilated (∼1 cm) main pancreatic duct associated with a 6 cm IPMN with intermediate-grade dysplasia. None of the other 80 cases had a SMAD4 mutation (p<0.001, vs PDAC). Mutations in TP53 and/or SMAD4, the two most specific markers, were not detected in the juice samples of controls but were found in 22 of 34 (64.7%) cases with PDAC and 16 of 56 (30.4%) cases with IPMN (p<0.0001, p=0.003, respectively, PDAC vs IPMN, p=0.0007, χ2).
Twelve other patients with IPMN underwent pancreatic resection including three cases with TP53 mutations: these cases had intermediate-grade dysplasia in their IPMN and/or PanIN-2 in their resection specimen. Thirteen IPMN cases with low TP53 mutations (digital NGS scores ≤4) are still under surveillance without evidence of progression ≥1 year after their juice sample was obtained. Case#20 underwent distal pancreatectomy for an IPMN. She had GNAS and BRAF mutations in addition to a TP53 detected in her preoperative juice sample. She was diagnosed with metastatic PDAC 6 years later, despite having an unremarkable pancreas by surveillance CT including 6 months prior to presenting with metastatic disease. Overall, (including progressing cases described below) and in addition to the 3 TP53-mutation-positive cases that underwent resection with intermediate-grade dysplasia/PanIN-2, 4 of the 17 patients with IPMN with low TP53 mutation concentrations in their pancreatic juice (digital NGS scores≤4) that continued surveillance progressed to pancreatic cancer during the study period.
Thirty-six (53%) of the 68 cases diagnosed as having IPMN (including 12 cases with PDAC that also had IPMN) had GNAS mutations detected in their pancreatic juice samples. Of the 21 cases that had RNF43 mutations in their pancreatic juice, 13 also had a GNAS mutation, and 16 arose in patients diagnosed with IPMN, consistent with evidence that RNF43 and especially GNAS are mutated more often in IPMNs than in usual PDACs.23 Cases diagnosed with IPMN were more likely to have mutations detected in their pancreatic juice than controls (51 of 56, 91.1%) (p<0.001), and more likely than controls to have mutations other than KRAS and GNAS detected in their pancreatic juice samples (26 of 56 vs 1 of 24) (p=0.0005).
CDKN2A, PIK3CA, TGFBR2 and BRAF mutations were detected in a minority of juice samples from patients with PDAC or IPMN consistent with the low prevalence of these mutations in primary pancreatic cancers.
Overall, pancreatic juice mutation concentrations were significantly higher in juice samples from patients with PDAC compared with controls (p<0.0001), as were concentrations of mutant KRAS alone (p=0.001) and concentrations of mutant TP53 and/or SMAD4 (p<0.001) (figure 2, table 2). By ROC curve analysis, overall digital NGS mutation scores could distinguish PDAC cases from controls and IPMN cases with AUCs of 0.89 (p<0.0001) and 0.69, p=0.003), respectively (figure 3, table 3). Pancreatic juice concentrations of mutated TP53 and/or SMAD4 were higher among cases diagnosed with PDAC than IPMN (Mann-Whitney, p<0.0001). By ROC curve analysis, digital NGS scores for mutant TP53 and/or SMAD4 could distinguish PDAC cases from IPMN cases without PDAC with 32.4% sensitivity and 100% specificity (AUC 0.73, p=0.0002), and from controls with an AUC of 0.82 (p<0.001, 100% specificity, 64.7% sensitivity). Among PDAC cases with TP53 and/or SMAD4 mutations, 12 of 22 had digital NGS scores of ≥5 compared with 0 of 16 with IPMN (p=0.001). By ROC analysis, overall digital NGS scores could also distinguish IPMN cases from controls with an AUC of 0.85 (p<0.0001).
Table 2.
Statistical analysis of pancreatic juice mutation concentrations by digital NGS scores
|
KRAS only |
GNAS only |
TP53 only |
TP53+SMAD4 |
All 9 genes |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| dNGS # | n | Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD |
| Controls | 24 | 0.8 | 1.4 | 0.4 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.3 | 1.8 |
| IPMN | 56 | 3.9 | 6.9 | 1.1 | 1.8 | 0.7 | 1.2 | 0.8 | 1.3 | 7.1 | 8.9 |
| PDAC | 34 | 8.2 | 13 | 2.4 | 10.4 | 6.5 | 16.3 | 8.2 | 16.6 | 20.7 | 29.6 |
| KRAS only | GNAS only | TP53 only | TP53+SMAD4 | All 9 genes | |||||||
| Mann-Whitney test | p Value | p Value | p Value | p Value | p Value | ||||||
| PDAC vs controls | 0.001 | 0.098 | <0.0001 | <0.0001 | <0.0001 | ||||||
| IPMN v controls | <0.001 | 0.006 | 0.007 | 0.007 | <0.0001 | ||||||
| PDAC vs IPMN | 0.335 | 0.281 | 0.0003 | <0.0001 | 0.003 | ||||||
dNGS#, digital NGS score (mutation concentration); IPMN, intraductal papillary mucinous neoplasm; NGS, next-generation sequencing; PDAC, pancreatic ductal adenocarcinoma.
Table 3.
Receiver operating characteristic (ROC) curve analysis of pancreatic juice mutation concentrations (overall 115#)
| KRAS only | GNAS only | TP53 only | TP53+SMAD4 | All 9 genes | ||
|---|---|---|---|---|---|---|
| PDAC vs controls | AUC | 0.74 | 0.61 | 0.79 | 0.82 | 0.89 |
| 95% CI of AUC | 0.62 to 0.87 | 0.46 to 0.76 | 0.68 to 0.91 | 0.72 to 0.93 | 0.80 to 0.97 | |
| Specificity | 91.7% | 83.4% | 100.0% | 100.0% | 83.4% | |
| Sensitivity | 50.0% | 38.2% | 58.8% | 64.7% | 85.3% | |
| p Value | 0.002 | 0.174 | 0.0002 | <0.0001 | <0.0001 | |
| IPMN vs controls | AUC | 0.74 | 0.67 | 0.63 | 0.63 | 0.85 |
| 95% CI of AUC | 0.62 to 0.85 | 0.55 to 0.80 | 0.51 to 0.76 | 0.51 to 0.76 | 0.76 to 0.93 | |
| Specificity | 83.4% | 83.4% | 100.0% | 100.0% | 83.4% | |
| Sensitivity | 57.1% | 51.8% | 26.8% | 26.8% | 73.2% | |
| p Value | 0.001 | 0.015 | 0.058 | 0.058 | <0.0001 | |
| PDAC vs IPMN | AUC | 0.56 | 0.56 | 0.69 | 0.73 | 0.69 |
| 95% CI of AUC | 0.43 to 0.70 | 0.43 to 0.68 | 0.58 to 0.81 | 0.62 to 0.85 | 0.56 to 0.81 | |
| Specificity | 42.9% | 51.8% | 73.2% | 73.2% | 85.7% | |
| Sensitivity | 52.9% | 61.8% | 58.8% | 64.7% | 52.9% | |
| p Value | 0.290 | 0.324 | 0.002 | 0.0002 | 0.003 |
AUC, area under the curve; IPMN, intraductal papillary mucinous neoplasm; PDAC, pancreatic ductal adenocarcinoma.
One case of interest (Case#24, table 1) with McCune-Albright syndrome (which is caused by postzygotic GNAS mutations) underwent EUS to evaluate further innumerable pancreatic cysts detected by CT scan. His pancreatic juice sample had a high mutant GNAS concentration (digital NGS score=29) and RNF43.
Detection of somatic mutations in pancreatic juice before a pancreatic cancer diagnosis
We also analysed serial pancreatic juice samples from four patients who had undergone pancreatic cancer screening and surveillance and later developed pancreatic cancer. Case #36 (tables 1 and 4) had undergone surveillance over 5 years and had pancreatic juice collected 61 months, 16 months and 4 months before being diagnosed with pancreatic cancer. For 2+ years prior to his PDAC diagnosis, four small stable-appearing pancreatic body and tail cysts (diameters 6–10 mm) and focal main pancreatic duct (3.7 mm) dilation were detected by EUS and MRI/MRCP. At diagnosis MRI/MRCP identified a new 3 cm pancreatic head mass. Pancreatic-protocol CT (3D) also identified this mass with duodenal wall distortion. EUS confirmed duodenal distortion and fine-needle aspiration (FNA) confirmed cancer. His resection specimen harboured Stage IIB moderately differentiated PDAC, multifocal PanIN-2 and PanIN-3. Digital NGS analysis of the pancreatic juice sample collected 4 months before diagnosis found high mutant KRAS (G12D) and mutant SMAD4 (Q311X) concentrations: both mutations were detected in his resected pancreatic cancer. This juice sample also had a GNAS (R201C) mutation likely from his IPMN. His juice sample collected 16 months before diagnosis had low SMAD4 Q311X mutation concentrations, as well as mutations not found in his cancer. Low levels of KRAS G12D and GNAS R201C were detected in the 61-month prediagnostic juice sample, but not the SMAD4 mutation.
Table 4.
Serial pancreatic juice collections from a patient who developed pancreatic cancer during surveillance
| Case# | Time (months before PDAC diagnosis) |
GNAS |
KRAS |
TP53 |
SMAD4 |
||||
|---|---|---|---|---|---|---|---|---|---|
| dNGS# | SNV | dNGS# | SNV | dNGS# | SNV | dNGS# | SNV | ||
| Case#36_1 | 61 | 3 | p.R201C | 2 | p.G12D | 1 | p.R175H | 1 | p.Q256X |
| Case#36_2 | 16 | 0 | 3 | p.G12R | 4 | p.R175H | 1 | p.Q311X | |
| Case#36_3 | 4 | 2 | p.R201C | 21 | p.G12D | 1 | p.R175H | 26,4 | p.Q311X/p.Q256X |
| Case#36_ PDAC_tumour tissue | 0 | 1 | p.R201C | 75 | p.G12D | 0 | 90 | p.Q311X | |
dNGS#, digital NGS score (mutation concentration); NGS, next-generation sequencing; PDAC, pancreatic ductal adenocarcinoma; SNV, single nucleotide variant (ie, somatic mutation).
Case#35 (see online supplementary table S7) developed pancreatic cancer in his tail while under surveillance for pancreatic body cysts. This patient had a TP53 R248Q mutation identified in his resected primary cancer that was detectable by digital NGS in the juice samples collected 13 months before diagnosis and at diagnosis. We previously reported using digital melt-curve analysis with pyrosequencing to detect this mutation in the prediagnostic juice sample of this case.13
Case#48 (see online supplementary table S7) had mutations detected in his juice sample collected at the time of his diagnosis, but not in a juice sample collected 15 months before diagnosis. At this 15-month visit, a fatty atrophic pancreas and a small pancreatic-body cyst (4.3 mm) were detected by EUS and MRI. He underwent total pancreatectomy after neoadjuvant therapy.
Case#53 underwent five EUS surveillance visits over 4 years (see online supplementary table S7). For the first 3 years, EUS, MRI and CT all revealed multiple subcentimetre pancreatic body and tail cysts, and a 2.4 cm pancreatic neck cyst with minimal adjacent focal duct dilation but no concerning features. This cyst, sampled on three EUS visits, had unremarkable cytology. The merits of surgical resection were discussed at our multidisciplinary CAPS conference and the consensus was MRI within 6 months, EUS in 12 months. Instead, the patient had an unremarkable abdominal CT scan after 12 months, and 4 months later, presented with recent weight loss and abdominal pain. Pancreatic-protocol CT and 3D reconstruction identified new duodenal compression. EUS revealed a pancreatic head mass causing proximal duodenal compression that limited duodenal intubation and precluded pancreatic juice collection with no change in the pancreatic cysts. FNA confirmed cancer. The patient underwent pancreaticoduodenectomy for a 2.5 cm diameter poorly differentiated, T3N1, margin-negative, PDAC and multifocal PanIN-2. Surgical pathology concluded the cancer arose from an IPMN, consistent with imaging findings. Laser capture microdissection was performed to isolate tumour DNA from four regions of the resected pancreatic cancer (∼0.5 cm to 1 cm apart). Ninety-six digital NGS reactions performed on these four cancer samples and an EUS/FNA sample of the cancer revealed mutations in KRAS (G12D), TP53 (Q100Rframeshift) and SMAD4 (Q388X) in all four cancer specimens, as well as subclonal mutations (SMAD4 G386R, H290R) in some of the samples. Digital NGS of the EUS/FNA sample identified the KRAS G12D mutation and the SMAD4 H290R mutation, but other mutations detected in the resected cancer were not found despite having an adequate FNA sample (with 13% KRAS G12D concentration). In addition, the FNA sample had a mutation not found in the resected cancer samples (TP53 L344P), consistent with genetic heterogeneity in geographical regions of pancreatic and other carcinomas.30 31 We also analysed three pancreatic cyst fluid samples collected 16–36 months before pancreatic cancer diagnosis. Each cyst fluid sample had only GNAS R201C, KRAS G12V and KRAS G12D mutations. Notably, the pancreatic cancer did not have a GNAS mutation, additional evidence that the cancer did not arise from IPMN. Digital NGS analysis was also performed on four pancreatic juice samples. The baseline juice sample collected 4 years prediagnosis contained only low levels of mutant KRAS. The juice sample collected closest to her pancreatic cancer diagnosis (19 months prediagnosis) had KRAS codon 12 mutations and a TP53 mutation (Y220C) but not the SMAD4 or TP53 mutations detected in the cancer (see online supplementary table S6).
Discussion
Pancreatic imaging is used to identify evidence of pancreatic neoplasia, but currently used tests have limitations: they cannot identify PanIN, and they often cannot adequately evaluate the neoplastic nature of pancreatic cysts. EUS is considered an excellent test to detect small solid pancreatic masses,32 but EUS can miss isoechoic lesions, and the accuracy of EUS for detecting very small (subcentimetre) PDACs has not been extensively studied.
Molecular analysis of pancreatic juice could, in theory, provide evidence for the presence of pancreatic neoplasia that may not be evident using pancreatic imaging tests. Overall, the prevalence of mutations detected by digital NGS in our study population is consistent with the expected prevalence of mutations in precursor neoplasms and pancreatic cancer.11 24 33 We have previously demonstrated that KRAS and GNAS mutations can be detected in duodenal collections of pancreatic juice14 15 34 and that these mutations are commonly found in patients with pancreatic cancer, and in patients undergoing screening for their family history of pancreatic cancer. We also found that when mutations (usually mutant KRAS or GNAS) were detected in controls without evidence of pancreatic neoplasia, their concentrations were usually low, consistent with prior reports.14 15 Since most patients with low-grade PanINs and IPMNs will not develop an invasive pancreatic cancer,35 additional diagnostic markers are needed that are more specific for the presence of high-grade dysplasia and early invasive pancreatic cancer. We find that mutations in TP53 which are thought to emerge as PanIN and IPMN progress from low-grade to high-grade dysplasia and invasive pancreatic cancer,11 are commonly found in the pancreatic juice samples of patients with invasive pancreatic cancer but were not detected in controls without neoplasia. Low concentrations of TP53 mutations were detected in a minority of IPMN cases, but higher concentrations (mutation score ≥5) were only detected in cases with pancreatic cancer. Long-term follow-up will be required to determine if low levels of TP53 mutations in pancreatic juice is associated with neoplastic progression. We found that total mutant DNA concentrations could distinguish PDAC cases from controls with high accuracy (AUC 0.89, p<0.001). SMAD4 mutations were the most specific for pancreatic cancer; only 1 of 80 cases without pancreatic cancer had a SMAD4 mutation; an IPMN case with high-risk features.29
The pancreatic juice digital NGS results of the high-risk individuals under surveillance who subsequently developed an invasive pancreatic cancer highlight the potential clinical utility of this test. Two of these cases had SMAD4 or TP53 mutations detectable in their pancreatic juice samples over 1 year prior to their pancreatic cancer diagnosis, at a time when no suspicious lesion was evident by imaging. These mutations matched those found in their cancers. These cases also provide some insight into how quickly a pancreatic cancer can emerge (and reach Stage IIB disease) after a non-concerning EUS exam. While mathematical modelling of gene alterations of pancreatic cancers has been used to estimate that it takes many years for an initiating pancreatic cancer cell to spread beyond the pancreas,31 other reports, that have investigated pancreatic cancer growth and progression in patients, emphasise the rapid progression of pancreatic cancer.36 37 Thus, analyses using patient age and tumour stage at diagnosis,36 observations of the extent of tumour progression in some patients awaiting resection of their pancreatic cancer,37 as well as the experience of screening high-risk patients all support the notion that the time it takes for pancreatic cancers to grow from an undetectable to a detectable stage (representing the growth through the T1 stage to higher T stages) is short (perhaps 1 year or less). These reports are also consistent with newer mathematical models of primary malignant tumour growth that account for tumour cell migration within the tumour.38 Since the ultimate goal of pancreatic screening is to prevent death by identifying either Stage I pancreatic cancer or if possible, PanIN-3 lesions, current pancreatic screening protocols recommend annual surveillance even when there are lesions detected by imaging.39
The limitations of our pancreatic juice sampling and analysis in its current form is suggested by the lack of any detectable mutations in several PDAC cases, as well as the lack of any detectable mutations in the juice sample from Case#48 collected 15 months before their PDAC diagnosis. The results of Case#53 also gave insight into the limitations of pancreatic juice analysis—the SMAD4 and TP53 mutations identified in the cancer were not detected in the pancreatic juice sample collected 19 months prediagnosis, nor was the GNAS R201C mutation that was detected in IPMN cyst fluid samples. Although SMAD4 mutations and high TP53 mutation concentrations could distinguish pancreatic cancer cases from those with IPMN with high specificity, improvements in the diagnostic sensitivity of our pancreatic juice test are needed. Such improvements might require obtaining a better pancreatic juice sample. Pancreatic juice samples collected from the duodenum has much higher mutation concentrations than pancreatic juice collected from the pancreatic duct.12 An ideal pancreatic juice test would distinguish cases with either PDAC, IPMN with high-grade dysplasia or PanIN-3 from those with low-grade dysplasia/PanIN-1, particularly in the high-risk population where the prevalence of IPMN is common.2 Results in one case also highlighted the limitations of using FNAs for deep sequencing to detect mutations in the cancer; several of the mutations present in the resected tumour sample were not detected in the FNA sample. This case highlights the challenges tumour heterogeneity poses for performing comprehensive mutational analysis of pancreatic tumour samples. One strength of this study was that we were able to investigate the pancreatic juice mutation profiles of several patients whose samples were collected months to years before pancreatic cancer diagnosis. More of these patients need to be studied to better evaluate pancreatic cancer early detection tests, but this requires large numbers of patients to undergo regular screening and biospecimen collections.
Pancreatic cancers that occur in patients with pre-existing IPMN often do not arise from their IPMN. This was true of the four cases reported that developed invasive pancreatic cancer while under surveillance. Since pancreatic juice samples contain markers shed from throughout the pancreatic ductal system, analysis of juice samples from patients with pancreatic cysts has the potential to identify evidence of PanIN or invasive cancer apart from their cyst(s). In this regard, pancreatic juice may have a complementary role to pancreatic cyst fluid analysis, which is a better sample to analyse to identify the neoplastic nature of a pancreatic cyst.22 The need to evaluate the whole pancreas is likely to be particularly important for patients with a family history of pancreatic cancer who often have multifocal PanINs and IPMNs in their pancreas.40 Indeed, despite the fact that IPMNs are commonly detected in patients undergoing pancreatic screening for their family history of pancreatic cancer, most pancreatic cancers that develop in these patients are thought to arise through the PanIN pathway.11 Consistent with this hypothesis, a histological review of over 1000 pancreatic cancers found no significant differences (such as more IPMN-associated cancers) between familial and sporadic cases.41 Recognising the importance of PanIN in pancreatic cancer development, even among patients who have pancreatic cysts, has implications for how we screen patients with pancreatic cysts, particularly those with an extensive family history of pancreatic cancer.42 Subcentimetre pancreatic cysts may have low risk of progression to cancer, but patients with these cysts may still benefit from regular surveillance to detect PanIN-related progression.
The digital NGS assay could be used to identify low-abundance mutations in other biological samples where mutation concentrations are expected to be lower than the limit of detection of conventional NGS assays, and since it can be performed with standard NGS reagents, could be readily incorporated into molecular diagnostic laboratory protocols after inhouse evaluation.
Overall, these results point to the potential clinical utility of a pancreatic juice NGS test for selected patients undergoing pancreatic evaluation. Before NGS-based tests of pancreatic juice can become a clinical test, further evaluation of the utility of NGS analysis of pancreatic juice in patients undergoing pancreatic evaluation is needed, particularly in clinical scenarios where the test would be most useful, such as patients undergoing pancreatic screening and surveillance for their familial risk of pancreatic cancer with incidentally identified pancreatic cysts.
Footnotes
Contributors: Conceived and designed the experiments (MG and JY); acquisition of data (JY, YS, KS, JANA, MB, MIC, AML, TB and MF); analysis and interpretation of data (JY, MG, MIC, AML, MS, KS, AB, JANA, MB and SF); drafted the manuscript (JY and MG); statistical analysis (JY); revised the manuscript and agreed with the manuscript's results and conclusions (all the authors); material support (ChiRhoClin, Burtonsville, MD); obtained funding (MG), study supervision (MG).
Funding: This work was supported by Susan Wojcicki and Dennis Troper, NIH grants (R01CA176828 and CA62924), the Pancreatic Cancer Action Network, the Lustgarten Foundation for Pancreatic Cancer Research, the Jimmy V Foundation, the Rolfe Pancreatic Cancer Foundation, and Hugh and Rachel Victor.
Competing interests: None declared.
Patient consent: Obtained.
Ethics approval: Johns Hopkins University IRB.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Rahib L, Smith BD, Aizenberg R, et al. . Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res 2014;74:2913–21. 10.1158/0008-5472.CAN-14-0155 [DOI] [PubMed] [Google Scholar]
- 2.Canto MI, Hruban RH, Fishman EK, et al. . Frequent detection of pancreatic lesions in asymptomatic high-risk individuals. Gastroenterology 2012;142:796–804; quiz e14–5 10.1053/j.gastro.2012.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brentnall TA, Bronner MP, Byrd DR, et al. . Early diagnosis and treatment of pancreatic dysplasia in patients with a family history of pancreatic cancer. Ann Intern Med 1999;131:247–55. 10.7326/0003-4819-131-4-199908170-00003 [DOI] [PubMed] [Google Scholar]
- 4.Canto MI, Goggins M, Hruban RH, et al. . Screening for early pancreatic neoplasia in high-risk individuals: a prospective controlled study. Clin Gastroenterol Hepatol 2006;4:766–81; quiz 665 10.1016/j.cgh.2006.02.005 [DOI] [PubMed] [Google Scholar]
- 5.Langer P, Kann PH, Fendrich V, et al. . Five years of prospective screening of high-risk individuals from families with familial pancreatic cancer. Gut 2009;58:1410–18. 10.1136/gut.2008.171611 [DOI] [PubMed] [Google Scholar]
- 6.Ludwig E, Olson SH, Bayuga S, et al. . Feasibility and yield of screening in relatives from familial pancreatic cancer families. Am J Gastroenterol 2011;106:946–54. 10.1038/ajg.2011.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poley JW, Kluijt I, Gouma DJ, et al. . The yield of first-time endoscopic ultrasonography in screening individuals at a high risk of developing pancreatic cancer. Am J Gastroenterol 2009;104:2175–81. 10.1038/ajg.2009.276 [DOI] [PubMed] [Google Scholar]
- 8.Vasen HF, Wasser M, van Mil A, et al. . Magnetic resonance imaging surveillance detects early-stage pancreatic cancer in carriers of a p16-leiden mutation. Gastroenterology 2011;140:850–6. 10.1053/j.gastro.2010.11.048 [DOI] [PubMed] [Google Scholar]
- 9.Verna EC, Hwang C, Stevens PD, et al. . Pancreatic cancer screening in a prospective cohort of high-risk patients: a comprehensive strategy of imaging and genetics. Clin Cancer Res 2010;16:5028–37. 10.1158/1078-0432.CCR-09-3209 [DOI] [PubMed] [Google Scholar]
- 10.Al-Sukhni W, Borgida A, Rothenmund H, et al. . Screening for pancreatic cancer in a high-risk cohort: an eight-year experience. J Gastrointest Surg 2012;16: 771–83. 10.1007/s11605-011-1781-6 [DOI] [PubMed] [Google Scholar]
- 11.Hruban RH, Maitra A, Goggins M. Update on pancreatic intraepithelial neoplasia. Int J Clin Exp Pathol 2008;1:306–16. [PMC free article] [PubMed] [Google Scholar]
- 12.Sadakari Y, Kanda M, Maitani K, et al. . Mutant KRAS and GNAS DNA concentrations in secretin-stimulated pancreatic fluid collected from the pancreatic duct and the duodenal lumen. Clin Transl Gastroenterol 2014;5:e62 10.1038/ctg.2014.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanda M, Sadakari Y, Borges M, et al. . Mutant TP53 in duodenal samples of pancreatic juice from patients with pancreatic cancer or high-grade dysplasia. Clin Gastroenterol Hepatol 2013;11:719–30. 10.1016/j.cgh.2012.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kanda M, Knight S, Topazian M, et al. . Mutant GNAS detected in duodenal collections of secretin-stimulated pancreatic juice indicates the presence or emergence of pancreatic cysts. Gut 2013;62:1024–33. 10.1136/gutjnl-2012-302823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eshleman JR, Norris AL, Sadakari Y, et al. . KRAS and guanine nucleotide-binding protein mutations in pancreatic juice collected from the duodenum of patients at high risk for neoplasia undergoing endoscopic ultrasound. Clin Gastroenterol Hepatol 2015;13:963–9.e4. 10.1016/j.cgh.2014.11.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Debeljak M, Freed DN, Welch JA, et al. . Haplotype counting by next-generation sequencing for ultrasensitive human DNA detection. J Mol Diagn 2014;16:495–503. 10.1016/j.jmoldx.2014.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diekstra A, Bosgoed E, Rikken A, et al. . Translating sanger-based routine DNA diagnostics into generic massive parallel ion semiconductor sequencing. Clin Chem 2015;61:154–62. 10.1373/clinchem.2014.225250 [DOI] [PubMed] [Google Scholar]
- 18.Bettegowda C, Sausen M, Leary RJ, et al. . Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 2014;6:224ra24 10.1126/scitranslmed.3007094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quail MA, Smith M, Coupland P, et al. . A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genomics 2012;13:341 10.1186/1471-2164-13-341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kinde I, Wu J, Papadopoulos N, et al. . Detection and quantification of rare mutations with massively parallel sequencing. Proc Natl Acad Sci USA 2011;108:9530–5. 10.1073/pnas.1105422108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vogelstein B, Kinzler KW. Digital PCR. Proc Natl Acad Sci USA 1999;96: 9236–41. 10.1073/pnas.96.16.9236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Springer S, Wang Y, Molin MD, et al. . A combination of molecular markers and clinical features improve the classification of pancreatic cysts. Gastroenterology 2015;4:01067–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu J, Jiao Y, Dal Molin M, et al. . Whole-exome sequencing of neoplastic cysts of the pancreas reveals recurrent mutations in components of ubiquitin-dependent pathways. Proc Natl Acad Sci USA 2011;108:21188–93. 10.1073/pnas.1118046108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones S, Zhang X, Parsons DW, et al. . Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 2008;321:1801–6. 10.1126/science.1164368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Biankin AV, Waddell N, Kassahn KS, et al. . Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature 2012;491:399–405. 10.1038/nature11547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu J, Matthaei H, Maitra A, et al. . Recurrent GNAS mutations define an unexpected pathway for pancreatic cyst development. Sci Transl Med 2011;3:92ra66 10.1126/scitranslmed.3002543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carr JC, Dahdaleh FS, Wang D, et al. . Germline mutations in SMAD4 disrupt bone morphogenetic protein signaling. J Surg Res 2012;174:211–14. 10.1016/j.jss.2011.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. http://www.arup.utah.edu/database/smad4/SMAD4_display.php.
- 29.Tanaka M, Fernandez-Del Castillo C, Adsay V, et al. . International consensus guidelines 2012 for the management of IPMN and MCN of the pancreas. Pancreatology 2012;12:183–97. 10.1016/j.pan.2012.04.004 [DOI] [PubMed] [Google Scholar]
- 30.Vogelstein B, Papadopoulos N, Velculescu VE, et al. . Cancer genome landscapes. Science 2013;339:1546–58. 10.1126/science.1235122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yachida S, Jones S, Bozic I, et al. . Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature 2010;467:1114–17. 10.1038/nature09515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khashab MA, Yong E, Lennon AM, et al. . EUS is still superior to multidetector computerized tomography for detection of pancreatic neuroendocrine tumors. Gastrointest Endosc 2011;73:691–6. 10.1016/j.gie.2010.08.030 [DOI] [PubMed] [Google Scholar]
- 33.Bailey P, Chang DK, Nones K, et al. . Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 2016;531:47–52. 10.1038/nature16965 [DOI] [PubMed] [Google Scholar]
- 34.Kanda M, Matthaei H, Wu J, et al. . Presence of somatic mutations in most early-stage pancreatic intraepithelial neoplasia. Gastroenterology 2012;142: 730–3.e9. 10.1053/j.gastro.2011.12.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bassi C, Sarr MG, Lillemoe KD, et al. . Natural History of Intraductal Papillary Mucinous Neoplasms (IPMN): Current Evidence and Implications for Management. J Gastrointest Surg 2008;12:645–50. 10.1007/s11605-007-0447-x [DOI] [PubMed] [Google Scholar]
- 36.Yu J, Blackford A, Dal Molin M, et al. . Time to progression of pancreatic ductal adenocarcinoma from low-to-high tumour stages. Gut 2015;64: 1783–9. 10.1136/gutjnl-2014-308653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raman SP, Reddy S, Weiss MJ, et al. . Impact of the time interval between MDCT imaging and surgery on the accuracy of identifying metastatic disease in patients with pancreatic cancer. AJR Am J Roentgenol 2015;204:W37–42. 10.2214/AJR.13.12439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Waclaw B, Bozic I, Pittman ME, et al. . A spatial model predicts that dispersal and cell turnover limit intratumour heterogeneity. Nature 2015;525:261–4. 10.1038/nature14971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Canto MI, Harinck F, Hruban RH, et al. . International Consensus Recommendations on the Management of Patients with Increased Risk for Familial Pancreatic Cancer (The Cancer of the Pancreas Screening (CAPS) Consortium Summit). Gut 2013;62:339–47. 10.1136/gutjnl-2012-303108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brune K, Abe T, Canto M, et al. . Multifocal neoplastic precursor lesions associated with lobular atrophy of the pancreas in patients having a strong family history of pancreatic cancer. Am J Surg Pathol 2006;30:1067–76. doi:pas.0000213265.84725.0b [PMC free article] [PubMed] [Google Scholar]
- 41.Singhi AD, Ishida H, Ali SZ, et al. . A histomorphologic comparison of familial and sporadic pancreatic cancers. Pancreatology 2015;15:387–91. 10.1016/j.pan.2015.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vege SS, Ziring B, Jain R, et al. . American gastroenterological association institute guideline on the diagnosis and management of asymptomatic neoplastic pancreatic cysts. Gastroenterology 2015;148:819–22. 10.1053/j.gastro.2015.01.015 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
gutjnl-2015-311166supp002.pdf (153.6KB, pdf)
gutjnl-2015-311166supp001.pdf (93.7KB, pdf)
gutjnl-2015-311166supp003.pdf (257.5KB, pdf)