Abstract
The heat shock transcription factor (HSF) family consists of three members in mammals and regulates expression of heat shock genes via a heat shock element. HSF1 and HSF2 are required for some developmental processes, but it is unclear how they regulate these processes. To elucidate the mechanisms of developmental regulation by HSFs, we generated mice in which the HSF4 gene is mutated. HSF4-null mice had cataract with abnormal lens fiber cells containing inclusion-like structures, probably due to decreased expression of γ-crystallin, which maintains protein stability. Furthermore, we found increased proliferation and premature differentiation of the mutant lens epithelial cells, which is associated with increased expression of growth factors, FGF-1, FGF-4, and FGF-7. Unexpectedly, HSF1 competed with HSF4 for the expression of FGFs not only in the lens but also in other tissues. These findings reveal the lens-specific role of HSF4, which activates γ-crystallin genes, and also indicate that HSF1 and HSF4 are involved in regulating expression of growth factor genes, which are essential for cell growth and differentiation.
Keywords: crystallin, FGF, heat shock, lens, transcription
Introduction
All organisms respond to elevated temperatures by inducing a set of heat shock proteins (Hsps). This response is regulated mainly at the level of transcription by heat shock transcription factor (HSF), which binds to a heat shock element (HSE), which is composed of at least three inverted repeats of consensus sequence nGAAn (Wu, 1995). The HSF family consists of four members (HSF1–4) in vertebrates, whereas a single HSF is present in yeast, nematode, and the fruit fly (Morimoto, 1998). HSF1 and HSF2 are ubiquitous among vertebrate species, whereas HSF3 and HSF4 have been characterized only in avians and in mammals, respectively (Nakai, 1999). All of the HSFs have a DNA-binding domain and a leucine zipper-like trimerization domain (HR-A/B), and HSF trimers bind to HSE with high affinity. There is another leucine zipper-like domain (HR-C) that suppresses trimer formation except in HSF4. Therefore, HSFs stay as monomers or dimers under normal growth conditions and are converted to trimers when they are activated (Morimoto, 1998). In mammals, HSF1 is necessary for induction of heat shock genes under heat shock conditions and for acquisition of thermotolerance (McMillan et al, 1998; Zhang et al, 2002).
In addition to the role in the activation of gene expression in response to various stresses, HSFs have been shown to be involved in cell growth and differentiation (Morano and Thiele, 1999; Pirkkala et al, 2001). In Drosophila, a single HSF is necessary for oogenesis and early larval development (Jedlicka et al, 1997). Likewise, mouse HSF1 is required for oogenesis, placental development, and normal growth (Xiao et al, 1999; Christians et al, 2000; Zhang et al, 2002). Furthermore, HSF1 plays a role in eliminating injured male germ cells when these cells are exposed to stress (Nakai et al, 2000; Izu et al, 2004). Remarkably, these developmental functions of HSF1 are not mediated through the induction of Hsps. HSF2 is also involved in oogenesis, spermatogenesis, and brain formation (Kallio et al, 2002; Wang et al, 2003). Furthermore, HSF1-null or HSF2-null male mice are fertile (Xiao et al, 1999; Kallio et al, 2002; McMillan et al, 2002; Zhang et al, 2002; Wang et al, 2003; Izu et al, 2004), whereas mice deficient in both HSF1 and HSF2 are sterile with severe defects in spermatogenesis (Wang et al, 2004). This observation indicates the complementary roles of HSF1 and HSF2 in spermatogenesis. Although comprehensive chromatin immunoprecipitation (ChIP) analysis revealed that HSF1 binds to many genes in vivo in human cells under normal growth conditions (Trinklein et al, 2004), it is yet unclear how HSF1 and HSF2 regulate the developmental processes and which target genes are responsible for the development.
In contrast to HSF1 and HSF2 proteins, which are expressed in most tissues, the expression level of HSF4 protein is too low to be detected in many tissues except in the brain and lung (Tanabe et al, 1999). There are at least two isoforms, HSF4a and HSF4b, which are derived by alternative RNA splicing events. HSF4b has the potential to activate transcription, whereas HSF4a does not have this potential (Nakai et al, 1997; Tanabe et al, 1999; Frejtag et al, 2001; Zhang et al, 2001). Both isoforms lack an HR-C domain that inhibits the trimer formation. Therefore, HSF4 is constitutively a trimer that binds to HSE, suggesting that HSF4 may have physiological roles during development (Nakai, 1999). It was shown recently that mutations of HSF4 are associated with dominant inherited cataracts in human (Bu et al, 2002). In this study, we found that HSF4 has a major HSE-binding activity specifically in the lens extract. As the lens consists of only two cell populations, it provides an excellent model of cell growth and differentiation (McAvoy et al, 1999). To elucidate the roles of HSFs in these processes, we generated mice in which the HSF4 gene was mutated. We found anomalies of the lens, and revealed novel HSF4 target genes that are essential for cell growth and differentiation.
Result
HSF4 consists of a major HSE-binding activity in the lens
In the absence of stress, there are weak, but distinct, HSE-binding activities in many tissues. Antibody supershift experiments showed that most of the HSE-binding activities consist of HSF1 and HSF2 (Fiorenza et al, 1995). HSF4 mRNA is expressed ubiquitously at low levels (Nakai et al, 1997; Tanabe et al, 1999), and HSF4 protein has been detected in the brain and lung, and cell lines such as C2C12 (Tanabe et al, 1999). To detect HSE-binding activity of HSF4, we performed gel shift assay using extracts isolated from various tissues including the skin, inner ear, and nasal cavity. HSE-binding activity was composed mostly of HSF1 in the tissue extracts as in the brain and lung extracts (Figure 1A, data not shown). Exceptionally, the HSE-binding activity in the lens extract was exclusively composed of HSF4 (Figure 1A). Furthermore, the expression level of HSF4 protein was much higher in the lens compared to those in the brain and lung, whereas the expression level of HSF1 protein was lower in the lens (Figure 1B). These results suggest unique roles of HSF4 in the lens.
Figure 1.
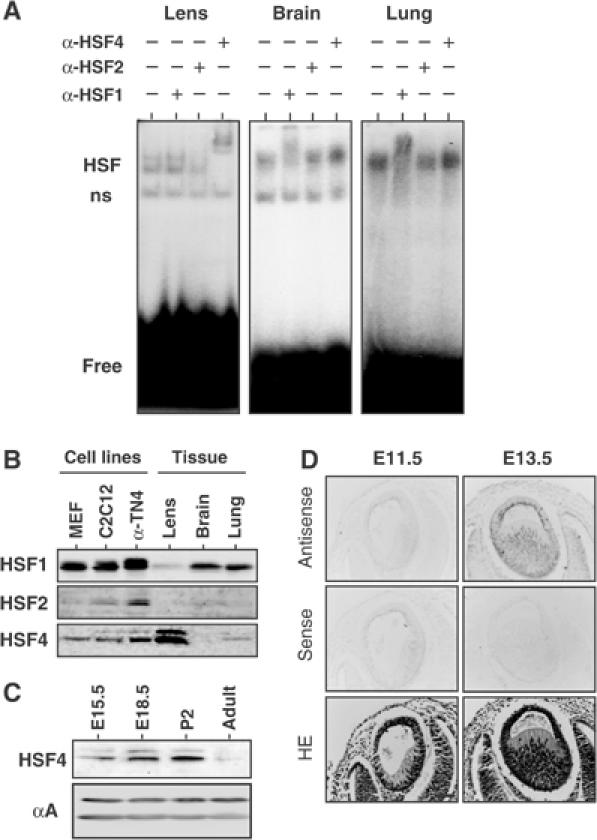
HSF4 consists of a major HSE-binding activity in the mouse lens. (A) Gel shift assay in the presence or absence of antibody for HSF1, HSF2, or HSF4. Whole tissue extracts were prepared from the lens, brain, and lung in 6-week-old mice, and were mixed with the binding reaction containing a 32P-labeled HSE oligonucleotide and antibody. HSF indicates the complexes of HSFs and an HSE probe, ns indicates nonspecific binding activity, and free indicates an unbound HSE probe. (B) Protein levels of HSF1, HSF2, and HSF4 in cells and tissues. Cell extracts (40 μg) prepared from MEF, C2C12, and α-TN4, and tissue extracts (80 μg) prepared from the lens, brain, and lung in 2-day-old mice were subjected to Western blot analysis using each specific antibody. (C) Protein levels of HSF4 in the lens at E15.5, E18.5, p2, and 6-week-old (adult) were analyzed by Western blot. Level of αA-crystallin was constant during development. (D) In situ hybridization was performed on the eye sections of E11.5 and E13.5 embryos using sense and antisense probes specific for HSF4. The sections were also stained with hematoxylin and eosin (HE). Bar, 50 μm.
We next examined the profile of HSF4 expression during lens development. We detected only a b-isoform of HSF4 (HSF4b) at any developmental stages by Western blot analysis (Figures 1C and 2D) and RT–PCR analysis (data not shown). Consistent with a previous work showing HSE-binding activity in the lens (Somasundaram and Bhat, 2000), the level of HSF4 protein reached a peak early after birth, and then decreased (Figure 1C). At embryonic day 13.5 (E13.5), cells in the posterior half of the lens vesicle elongated and differentiated to form the primary fibers. HSF4 mRNA is not expressed at E11.5, but is expressed in both epithelial cells and fiber cells at E13.5 (Figure 1D). HSF4 mRNA continued to be expressed in both cell types of the lens at least until 6 weeks after birth (data not shown).
Figure 2.
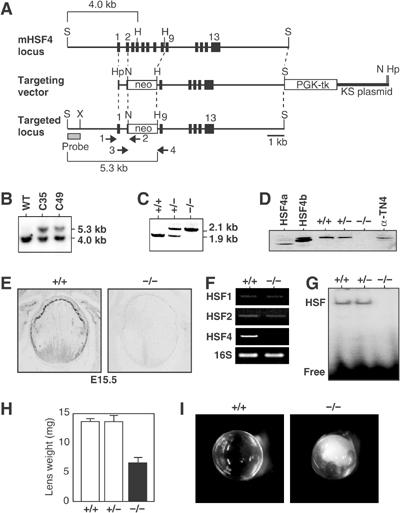
Cataract formation in HSF4-null mice. (A) Schematic representation of wild-type and mutant HSF4 loci together with targeting vector. The targeting vector was constructed to replace a part of exon 2 and exons 3–8 with a neomycin-resistant gene cassette. Locations of an external probe used to confirm correct targeting, and locations of PCR primers used to screen targeted ES clones (primers 1 and 2) and to identify genotype (primers 3 and 4) are shown. S, SalI; H, HindIII; Hp, HpaI; N, NotI; X, XbaI. (B) Southern blot of SalI/HindIII-digested genomic DNAs isolated from wild-type and targeted two ES clones (C39 and C49) using 32P-labeled probes described in (A). (C) PCR genotypic analysis for targeted locus. Mouse tail genomic DNA was isolated and was used to amplify DNA fragments by PCR using primers 3 and 4. (D) Western blot analysis of extracts of the lens of 6-week-old mice (+/+, +/−, −/−), 293 cells ectopically expressing HSF4a and HSF4b, and mouse lens epithelial α-TN4 cells. (E) In situ hybridization on the eye sections of E15.5 wild-type (+/+) and HSF4-null (−/−) mice using an antisense probe specific for HSF4. (F) mRNA levels of HSFs in wild-type(+/+) and HSF4-null (−/−) lens were examined by RT–PCR. (G) Whole-cell extracts were prepared from the lenses, and gel shift assay was performed using a 32P-labeled HSE oligonucleotide. Positions of free probe and probe bound by HSF are shown. (H) Lens weights in 6-week-old wild-type and mutant mice. Means and standard deviations were estimated by analyzing each of the three mice. (I) Lens opacity in 6-week-old wild-type and HSF4-null mice.
Inactivation of the HSF4 gene causes abnormal lens fiber cells containing inclusion-like structures
To examine the in vivo functions of HSF4, we generated targeted disruptions of the mouse HSF4 gene by homologous recombination in TT2 embryonic stem (ES) cells (Inouye et al, 2003). In the targeting vector, a region containing a part of exon 2 and exons 3–8 was substituted with a neomycin-resistant gene cassette (Figure 2A). This substitution removes amino acids at position 68 to position 248 in mouse HSF4, which contains part of the DNA-binding domain and an oligomerization domain (Tanabe et al, 1999). The ES cells were electroporated with the targeting vector, and placed under positive–negative selection (Inouye et al, 2003). The surviving 176 ES clones were screened for homologous recombination by PCR and Southern blotting (Figure 2B), and correct gene targeting occurred in 22 clones. Two lines of knockout mice were generated by injecting C35 and C49 ES clones into eight-cell embryos of ICR mice, and were maintained by mating with ICR mice. Progeny of matings between heterozygotes were analyzed by PCR to identify wild-type (+/+), heterozygous (+/−), and homozygous (−/−) mutant offspring (Figure 2C), and were found to be present at the expected Mendelian ratio (49:82:43, respectively; n=174). Body weights of HSF4-null mice were as same as those of wild-type mice at all developmental stages (data not shown). Male and female HSF4-null mice were fertile, and the histology of the testis and ovary was normal (data not shown). We detected no abnormality in the brain or lung in HSF4-null mice (data not shown).
We next examined the lens, and confirmed that HSF4 expression was absent in HSF4-null mice (Figure 2D and E). Expression levels of HSF1 and HSF2 mRNAs were normal in HSF4-null lens (Figure 2F). We detected no HSE-binding activity in an extract isolated from HSF4-null lens (Figure 2G). Weight of the HSF4-null lens was lower than that of the wild-type lens (Figure 2H), and HSF4-null mice had cataract (Figure 2I). We then examined histology of the lens. In 6-week-old mice, wild-type lens fiber cells were flattened by dehydration, whereas HSF4-null fiber cells swelled (Figure 3A). Furthermore, HSF4-null fiber cells in the center of the lens contained inclusion-like structures, which were stained heavily with eosin and were possibly protein aggregates (Figure 3A, arrows in d). These abnormalities can be observed even at 2 days after birth (data not shown). However, HSF4-null lens cells normally elongated and differentiated into fiber cells until E15.5 (data not shown). The inclusion-like structures were stained with antibodies for abundant lens proteins, αB-crystallin (Figure 3Af) and αA-crystallin (data not shown). Hsp70 was not accumulated in the inclusion (data not shown). These results suggest that loss of HSF4 function may cause protein aggregates in the center of the lens.
Figure 3.
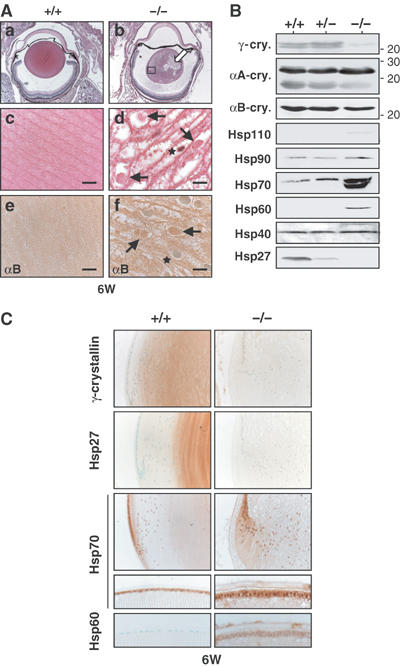
The lens fiber cells contain inclusion-like structures in HSF4-null mice. (A) Histological examination of the lens sections of the 6-week-old wild-type (a, c) and HSF4-null (b, d) mice stained with HE. Immunostaining of αB-crystallin was performed using the lens sections of wild-type (e) and HSF4-null (f) mice. The open arrow in (b) indicates the lesion where the structure of the fiber cells was not recognized. The legion indicated by a square in (b) was enlarged into (d). Arrows in (d, f) indicate cytoplasmic inclusion-like structures and stars indicate the nucleus. Magnification: (a, b) × 25; (c–f) × 1000. Bars, 10 μm. (B) Expression of Hsps and αA-, αB-, and γ-crystallins. Proteins isolated from the lens in 6-week-old wild-type (+/+) and mutant (+/− and −/−) mice were analyzed by Western blot analysis using each specific antibody. In the Hsp70 column, an upper band represents Hsc70 and a lower band represents an inducible Hsp70. (C) Immunohistochemistry of 6-week-old mice using antiserum for each specific antiserum. The bow regions (upper columns) and the epithelial layers (lower columns) are shown.
Expression of γ-crystallins decreases in HSF4-null mice
As heat shock genes are candidate targets of HSF4, we examined the expression of major Hsps in the lens. We found that Hsp27 expression in the inner layer of the lens reduced in HSF4-null mice (Figure 3B and C). However, it is unclear whether reduced expression of Hsp27 leads to protein aggregates. Unexpectedly, we found that expression of Hsp60, Hsp70, Hsp90, and Hsp110 increased (Figure 3B). Immunohistochemical analysis also showed increased expression of Hsp70 and Hsp60 in the epithelial cells of HSF4-null lens (Figure 3C). These results indicate that HSF4 suppresses expression of major Hsps in epithelial cells.
As crystallins are major lens structural proteins in the lens, we examined expression of crystallins in the HSF4-null lens. The expressions of αA-crystallins, αB-crystallins, and β-crystallins were constant in the HSF4-null lens (Figure 3B, data not shown). Unexpectedly, we found that γ-crystallin expression was markedly reduced (Figure 3B and C). All six γ-crystallin gene expressions were markedly reduced in the lens of adult 6-week-old HSF4-null mice when these were estimated by semiquantitative RT–PCR analysis (Figure 4A). The expression levels were reduced even in the lens of 2-day-old HSF4-null mice, but were normal in the lens of E15.5 HSF4-null embryos. The stage at which abnormal morphology of fiber cells becomes apparent correlates with the initiation of decreased expression of the γ-crystallin genes. These results demonstrate that HSF4 is required to maintain γ-crystallin gene expression at later developmental stages. As the γ-crystallin is essential for protein stability in the lens fiber cells as described below, the abnormalities of the fiber cells are partly due to the reduced expression of γ-crystallin.
Figure 4.
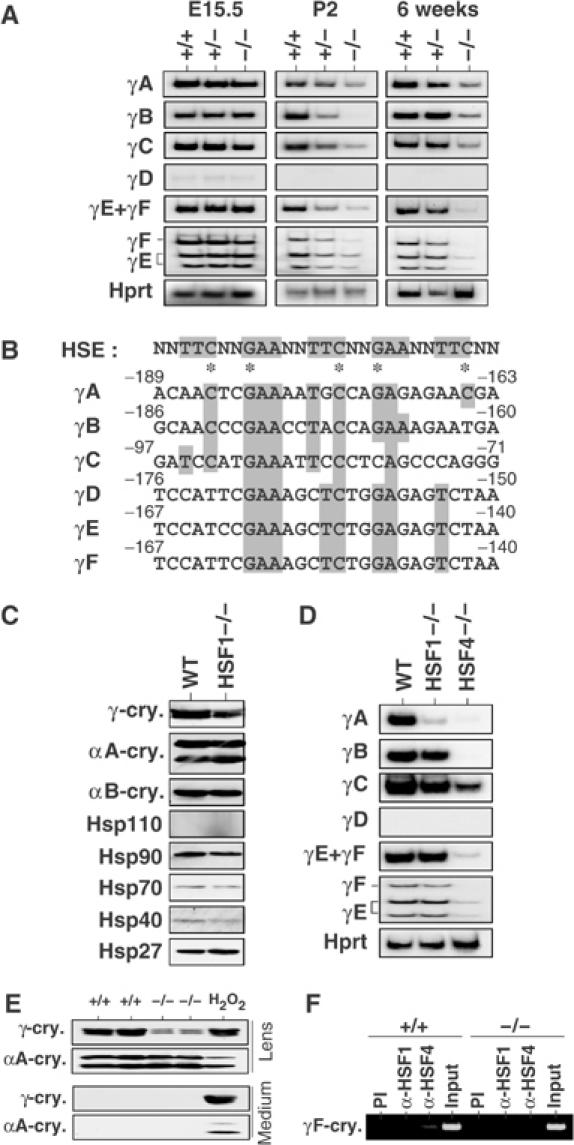
Expression of the γ-crystallin genes is markedly reduced in HSF4-null lens. (A) RT–PCR analysis of mRNAs of the γ-crystallin genes using specific primers. Total RNAs were isolated from lenses in 6-week-old mice, 2-day-old mice (P2), and 15.5 dpc embryos (E15.5). Images of autoradiography are shown. To identify γE and γF, the PCR products amplified with the same set of primers were digested with BglII. (B) Promoter sequence alignment of the six mouse γ-crystallin genes. Sequences identical to the consensus HSE sequences are shown in gray boxes. The asterisks indicate key nucleotides essential for HSF1 binding. (C) Expression of crystallins and Hsps in 6-week-old wild-type (WT) and HSF1-null (HSF1−/−) mice examined by Western blot analysis. (D) RT–PCR analysis of mRNAs of the γ-crystallin genes using total RNAs isolated from lenses in 6-week-old mice. Images of autoradiography are shown. (E) Lenses were removed from 2-week-old wild-type (+/+) and HSF4-null (−/−) mice, and were incubated in medium at 37°C for 24 h. To damage lens cells, the lenses from wild-type mice were incubated in the presence of 1 mM H2O2. Proteins in the lens and medium were analyzed by Western blot analysis using antiserum specific for αA- or γ-crystallin. (F) ChIP-enriched DNAs from 2-week-old wild-type (+/+) and HSF4-null (−/−) lenses using preimmune serum (PI), anti-HSF1 serum (α-HSF1), and anti-HSF4 serum (α-HSF4) as well as an input DNA were amplified using primers specific for the γF-crystallin gene by PCR analysis. The DNA fragment (−349 to +6) was amplified.
As we found HSE-like sequences in all six γ-crystallin genes near transcription start sites (Figure 4B), we also analyzed γ-crystallin expression in the lens of HSF1-null mice (Inouye et al, 2003). Cataract did not develop in adult HSF1-null mice (see Figure 7A), but the γ-crystallin level reduced a little (∼70% of control level) (Figure 4C). Especially, expression of the γA-crystallin gene reduced significantly (Figure 4D). The reduction of γ-crystallins was not due to the shift of γ-crystallin proteins into an insoluble fraction (data not shown) or the leakage of proteins from damaged fiber cells (Figure 4E) (Piatigorsky et al, 1978). We further analyzed the binding of HSF4 to the HSE-like sequence of the γF-crystallin gene. Chromatin immunoprecipitation analysis revealed that HSF4 binds to the upstream region (−349 to +6) of the γF-crystallin gene in vivo (Figure 4F). These results suggest that HSF4 directly binds and regulates expression of the γ-crystallin gene.
Figure 7.
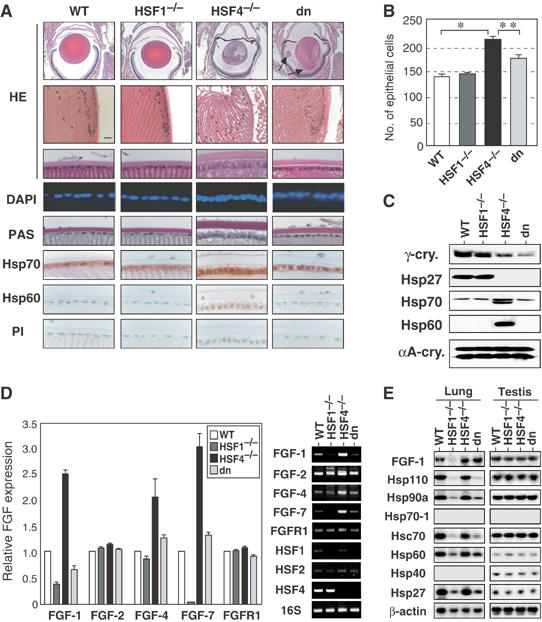
HSF1 competes with HSF4 for the expression of FGFs. (A) Histological examination of the lens sections of 6-week-old wild-type, HSF1-null, HSF4-null, and double-null (dn) mice. Sections were stained with HE and DAPI, PAS, or immunostained using a preimmune serum (PI) or an antiserum specific for Hsp70 or Hsp60. Lens extrusion was heavily accumulated in double-null mice (arrows). (B) Numbers of total epithelial cells per section in six lenses were shown on the right. The stars indicate P<0.01. (B) In situ hybridization was performed on 6-week-old wild-type (+/+), and HSF4-null (−/−) lenses using sense and antisense probes specific for FGF-1. (C) Proteins isolated from the lenses in 6-week-old mice were analyzed by Western blot analysis using each specific antibody. (D) RT–PCR analysis of mRNAs of the FGF-related genes in wild-type, HSF1-null, HSF4-null, and double-null (dn) mice. RT–PCR analysis was performed using total RNAs isolated from lenses in 6-week-old mice. Relative FGF expression levels are estimated from three experiments. Representative data are shown on the right. (E) Northern blot analysis of FGF-1 and Hsp mRNAs using total RNAs isolated from the lung and testis.
The γ-crystallin gene expression is directly regulated by many transcription factors, including Pax-6 (Kondoh, 2002), Sox-1 (Nishiguchi et al, 1998), c-Maf (Kim et al, 1999), and Prox-1 (Lengler et al, 2001), and their regulations are essential for differentiation into fiber cells. In the HSF4-null lens, however, expression levels of these transcription factors were normal (data not shown). It is unclear why HSF4 is not required for expression of γ-crystallin genes during embryonic development, although Sox-1, c-Maf, and Prox-1 transcription factors are essential for the expression.
HSF4 regulates proliferation and differentiation of lens epithelial cells by suppressing FGF expression
In addition to the abnormalities of the fiber cells, we observed abnormal epithelial cell morphology. Epithelial cells were found in the anterior of the lens and had cuboidal morphology in the wild-type lens. However, the epithelial cells in adult HSF4-null mice were columnar (Figure 5A). Furthermore, the epithelial cells in the bow region, where epithelial cells differentiated into fiber cells, were highly elongated in 2- and 6-week-old mice (Figure 5A). Electron micrographs showed the columnar morphology of the epithelial cells, and also showed that fiber cells near the epithelial layer were rich in organelles such as mitochondria (Figure 5B). Denucleation in fiber cells was also inhibited (Figure 5B). Furthermore, epithelial cells detached from the capsule that was stained with PAS (see Figure 7A). Numbers of epithelial cells per section in HSF4-null mice were the same as those in wild-type mice during embryonic development, but increased even at 2 days postpartum (Figure 5C). 5-bromo-2′-deoxyuridine (BrdU)-incorporated epithelial cells were observed throughout the epithelial layer in E18.5 mice and the number of the BrdU-positive cells increased in HSF4-null mice (Figure 5D). These results indicate increased proliferation of epithelial cells and premature differentiation of fiber cells in HSF4-null mice.
Figure 5.
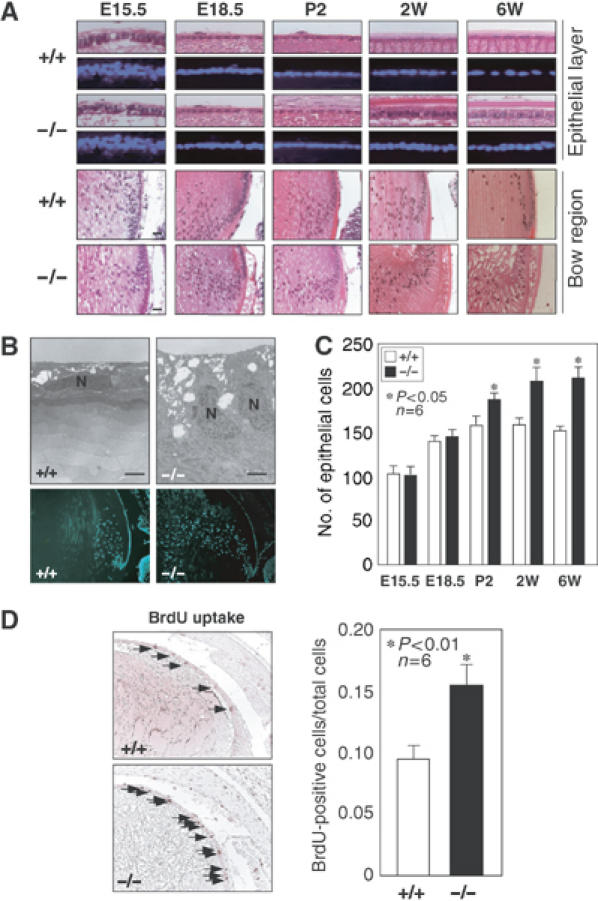
Increased proliferation and premature differentiation of the lens epithelial cells in HSF4-null mice. (A) Histological examination of the lens sections of 6-week-old, 2-week-old, 2-day-old, E18.5, and E15.5 wild-type (+/+) and HSF4-null (−/−) mice. Sections were stained with HE and DAPI. The epithelial layer (upper columns) is only found in the anterior of the lens. The bow regions, where epithelial cells differentiate into fiber cells, are also shown in the lower columns. (B) Transmission electron microscopic analysis (upper columns) and DAPI staining (lower columns) of 2-week-old wild-type and HSF4-null lens. The nuclei of the epithelial cells are indicated as N. Bar, 2 μm. (C) Numbers of total epithelial cells per section in six lenses were counted. The stars indicate P<0.05. (D) BrdU incorporation in the lens epithelial cells of E18.5 mice. The arrows indicate cells incorporated with BrdU. Percentages of BrdU-positive cells are shown. The stars indicate P<0.01.
Fibroblast growth factors (FGFs) regulate growth and differentiation of lens epithelial cells (McAvoy et al, 1999). Transgenic mice expressing a low dose of FGF-1 exhibited epithelial cell hyperplasia, which is associated with premature elongation of epithelial cells (Robinson et al, 1995). These phenotypes are also observed in transgenic mice expressing FGF-4, FGF-7, FGF-8, and FGF-9 (Lovicu and Overbeek, 1998), and are similar to those observed in HSF4-null mice. Therefore, we examined expression of the FGFs by RT–PCR analysis. It clearly revealed that FGF-1, FGF-4, FGF-7, but not FGF-2, were expressed at higher levels in HSF4-null lens than those in wild-type lens (Figure 6A). In situ hybridization analysis showed high expression of FGF1 mRNA in HSF4-null epithelial cells and fiber cells (Figure 6B). To examine the roles of HSF4 in the FGF expression in other cell types, we overexpressed HSF4 into mouse embryo fibroblast (MEF) cells using an adenovirus vector, and found that HSF4 suppressed FGF-7 expression (Figure 6C). Furthermore, ChIP analysis revealed that HSF4 bound to the upstream region (−615 to +10) of the FGF-7 gene in vivo in the lens (Figure 6D). This region contains five HSE consensus sequences (Finch et al, 1995; Fasciana et al, 1996). These data strongly suggest that HSF4 represses FGF expression by directly binding to FGF genes.
Figure 6.
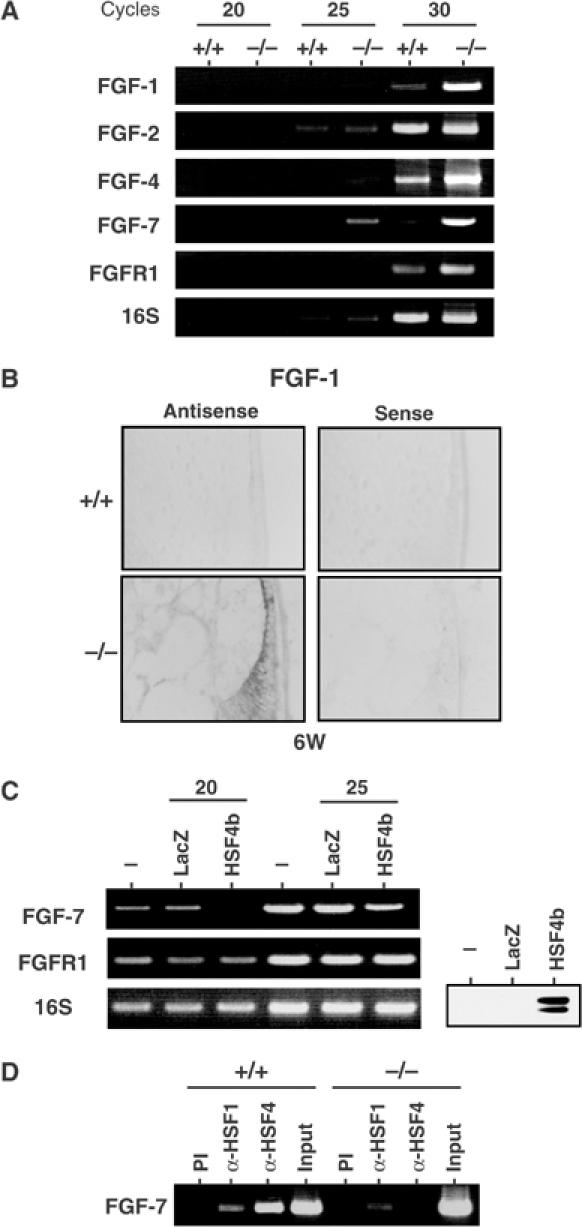
Expression of FGFs is high in HSF4-null lenses. (A) RT–PCR analysis of mRNAs of the FGF-related genes using specific primers. RT–PCR was performed using total RNAs isolated from lenses of 6-week-old wild-type (+/+) and HSF4-null (−/−) mice, and DNA bands were stained with ethidium bromide. Representative data are shown. (B) In situ hybridization was performed on 6-week-old wild-type (+/+) and HSF4-null (−/−) lenses using sense and antisense probes specific for FGF-1. (C) RT–PCR analysis of mRNAs of FGF-related genes in MEF cells overexpressing HSF4b or LacZ as a control. MEF cells were transfected with adenovirus expressing HSF4b or LacZ. At 48 h after transfection, mRNA levels of FGF-7 and FGFR1 were estimated by RT–PCR. Western blot analysis of HSF4 protein is shown. (D) ChIP-enriched DNAs from 2-week-old wild-type (+/+) and HSF4-null (−/−) lenses using preimmune serum (PI), anti-HSF1 serum (α-HSF1), and anti-HSF4 serum (α-HSF4) as well as an input DNA were amplified using primers specific for the FGF7 gene by PCR analysis. The DNA fragment (−615 to +10) was amplified.
HSF1 and HSF4 have opposing effects on FGF expression
We next examined involvement of HSF1 in the regulation of FGF expression. We found that the epithelial cell morphology was normal in the HSF1-null lens (Figure 7A), but the expression of FGF-1 and FGF-7 was decreased (Figure 7D). As HSF1 also bound directly FGF-7 gene (Figure 6D), HSF1 and HSF4 may regulate expression of FGF genes via HSE. To elucidate the interplay between HSF1 and HSF4 in FGF expression, we analyzed mice deficient for both HSF1 and HSF4 (double-null). We found that the high levels of FGF-1, FGF-4, and FGF-7 expression in the HSF4-null lens returned to normal levels in the double-null lens (Figure 7D). Consistent with the expression levels of FGFs, the increased number of epithelial cells in the HSF4-null lens reverted to almost the normal number and elongated epithelial cells returned to a fairly normal morphology in the double-null lens (Figure 7A and B). Furthermore, high levels of Hsp70 and Hsp60 expressions in HSF4-null epithelial cells returned to normal levels in the double-null cells (Figure 7A and C). These results suggest that HSF1 may compete with HSF4 for the expression of FGFs and Hsps.
To reveal whether HSFs regulate FGF expression in other tissues, we examined FGF expression in the lung, where HSF4 and HSF1 are expressed (Tanabe et al, 1999), as well as in the testis. We did not detect any distinct abnormality in the two tissues (data not shown). Although expression of both FGFs and Hsps was not altered in HSF4-null testis, FGF-1 expression was markedly reduced in HSF1-null lung (Figure 7E). Furthermore, the FGF-1 expression was reverted in double-null lung. Expression of major Hsps was also reduced in HSF1-null lung, and expression of some Hsps was partially reverted in double-null lung (Figure 7E). These results suggest that HSF1 competes with HSF4 for the expression of FGFs and Hsps in general. As only an HSF4b isoform is expressed in the lung (Tanabe et al, 1999), HSF4b may act as a repressor of constitutive gene expression in many cell types.
It is noteworthy that lens morphology was more affected in double-null mice than in HSF4-null mice. Double-null mice developed severe cataract (data not shown). Weight of the double-null lens (4.57±0.17 mg) in 6-week-old mice was similar to that of the HSF4-null lens (4.24±0.69 mg). As the posterior lens capsule is thin, capsular rupture was observed in about half of the HSF4-null lens (data not shown). In contrast, the capsule ruptured in all double-null lens, and lens extrusion was heavily accumulated in double-null mice (Figure 7A, arrows). This observation may be explained by the fact that both HSF1 and HSF4 act as activators at least for γ-crystallin genes.
Discussion
In this study, we showed that expression of HSF4 is uniquely high in the mouse lens among major tissues. Analysis of HSF4-null lens revealed a specific role of HSF4, that is, it is required to maintain lens-specific γ-crystallin gene expression in fiber cells. Furthermore, analysis of abnormal growth and differentiation of epithelial cells revealed the general role of HSF1 and HSF4, which is to competitively regulate expression of FGFs.
Specific role of HSF4 in lens development
The lens is characterized by its high degree of transparency, which is essential for refracting light and focusing it onto the retina. Development of the lens occurs by differentiation of peripheral epithelial cells into elongated fiber cells, which accumulate in concentric layers and lose their nuclei and other organelles (McAvoy et al, 1999; Kondoh, 2002). The concentration of proteins in the lens fiber cells is extremely high, as much as 450 mg/ml of proteins in the center of the lens (Fagerholm et al, 1981). As proteins in the center of the lens cannot turnover and must remain stable and soluble throughout the life of the organism (Zigler, 1994; Bhat, 2003), it is important to stabilize proteins.
To stabilize proteins in that extreme condition, crystallins constitute nearly 90% of the soluble protein in the lens fiber cells (Zigler, 1994). The ubiquitous vertebrate crystallins consist of two unrelated families of proteins, the α-crystallins and the β, γ-crystallins (Wistow and Piatigorsky, 1988). The former belongs to a small Hsp/α-crystallin superfamily. The latter belongs to a β, γ-crystallin superfamily that has a repeat of a characteristic Greek key βγ motif (Wistow and Piatigorsky, 1988). Members of this family in soil bacteria and in slime mold are both induced in response to stresses such as dehydrated environments, and have roles in protein stabilization under dehydrated conditions (Wistow, 1990). We showed that HSF4 expression is extremely high in the lens among major tissues and is required for maintaining γ-crystallin expression in postnatal fiber cells. HSF might have originally played a role in the regulation of protein folding by upregulating Hsps. In the lens, however, the HSF4 gene may have evolved to adapt to the unique dehydrated environment of the lens.
Six genes, γAγBγCγDγEγF-crystallin genes, have been identified in the same chromosome in the mouse and human, which might arise by multiple gene duplication, and their expressions are independently regulated during development (Zigler, 1994; Heon et al, 1999; Kondoh, 2002). Surprisingly, all six γ-crystallin genes have conserved HSE sequences near their transcription start sites. These HSE sequences consist of at least three nGAAn units, and are recognized by HSF4 and HSF1. In this regard, the HSE sequences in the developmentally regulated γ-crystallin genes are similar to those in heat shock genes. As HSF4 stays as a trimer that binds to HSE with high affinity (Nakai et al, 1997; Tanabe et al, 1999), it is easy to conclude that HSF4 plays a role in the constitutive expression of γ-crystallin genes by binding to conventional HSE. In contrast, vertebrate HSF1 is mostly a monomer in the absence of stress (Morimoto, 1998). Nevertheless, HSF1 regulates constitutive Hsp90α expression in chicken B lymphocytes (Tanabe et al, 1998; Nakai and Ishikawa, 2001), and regulates Hsp27 expression in the mouse heart (Yan et al, 2002). As the HSE sequences in the γ-crystallin genes consist of more than three nGAAn units, only a low quantity of HSF1 may bind to and regulate the γ-crystallin genes during development.
HSF1 and HSF4 competitively regulate FGF expression, which is essential for cell growth and differentiation
A low dose of FGFs stimulates proliferation of lens epithelial cells, whereas higher doses induce cell differentiation (McAvoy and Chamberlain, 1989; Robinson et al, 1995; Lovicu and Overbeek, 1998). Analysis of FGF expression revealed that expression of FGF-1, FGF-4, and FGF-7 increases in HSF4-null lens. Consistently, loss of HSF4 function resulted in increased proliferation and premature differentiation of the lens epithelial cells. Furthermore, HSF1 activates the similar set of FGFs in the lens that is repressed by HSF4. Therefore, abnormal levels of FGFs in HSF4-null or HSF1-null lens reversed to normal levels in double-null lens, and proliferation and differentiation of the epithelial cells were almost normal in double-null lens. We found that many HSE consensus sequences are located in the upstream region of the FGF-7 gene, and that the FGF-7 gene is a direct target of HSF4 and HSF1. These results demonstrate that HSF1 and HSF4 competitively regulate expression of FGFs that are essential for cell growth and differentiation. As FGFs regulate growth and differentiation of various cell types (Burgess and Maciag, 1989), regulation of FGFs by HSFs might be an important regulatory pathway for development in general.
We previously predicted the existence of two HSF4 isoforms, HSF4a and HSF4b, which are derived from alternatively spliced mRNA species (Tanabe et al, 1999). However, we could detect only HSF4b in the lens, as in other tissues. HSF4b has a weak but distinct potential to activate transcription in some cells (Tanabe et al, 1999). In fact, HSF4b can activate γ-crystallin and Hsp27 genes in the fiber cells. In marked contrast, it represses FGF and Hsp genes in epithelial cells. These results demonstrate that HSF4b can be an activator or a repressor depending on the cell type, and suggest that there may be a cell type-specific coregulator of HSF4. Although HSF1 inhibits induction of TNF-α or IL-1β in response to endotoxin shock (Xiao et al, 1999; Xie et al, 2002), this is the first demonstration that constitutive expression of target genes is repressed by a member of HSF family. In general, HSF4 may be a repressor that competes with HSF1 as an activator.
In this study, we showed abnormality of both the lens epithelial cells and fiber cells in HSF4-null mice. These abnormalities may be partly due to reduced expression of γ-crystallins and increased expression of FGFs. In addition, we observed that Hsp27 expression reduced significantly in the fiber cells and that Hsp70 and Hsp60 increased in the epithelial cells. It is possible that the changes in expression levels of Hsps cause abnormal lens development. Although HSF1-null and HSF2-null mice exhibit only restricted phenotypes, our results suggest that HSFs may be involved in fine regulation of Hsp genes and growth factor genes, the regulation of which is not strictly necessary for development. Rather, HSFs might delicately regulate expression of these genes, which maintain homeostasis of the body under normal physiological condition.
Materials and methods
Generation of HSF4-null mice
A DNA fragment containing the mouse HSF4 gene was isolated from a mouse 129 SvJ lambda genomic library (Stratagene) using an HSF4 cDNA probe and the gene structure was characterized by restriction mapping and sequencing. A 7.5 kb HindIII/SalI fragment and a 0.8 kb PCR-amplified fragment were inserted into a plasmid pKO (Inouye et al, 2003) to generate a targeting vector. The linearized targeting vector was electroporated into TT2 ES cells (C57Bl/6 × CBA F1 origin). We selected targeted ES cells with G418 and gancyclovir and identified homologous recombination with PCR and Southern blot. We injected HSF4+/− two ES clones (C35 and C49) into eight-cell embryos of ICR mice and crossed the chimeras into ICR females. We crossed first-generation heterozygote mice (F1) into ICR or C57Bl/6 mice and screened the progeny by PCR and Southern blot. The progeny that carried the targeted allele were maintained by crossing into ICR or C57Bl/6 mice. We analyzed the eye phenotypes of mice crossed for more than six generations.
HSF1-null mice maintained by crossing into ICR mice were used (Inouye et al, 2003). To generate mice deficient for both HSF1 and HSF4, male and female mice with a genotype of HSF1+/−; HSF4+/− were crossed. Wild-type (+/+), heterozygous (HSF4+/−), and homozygous (HSF4−/−) mutant offspring were present at the expected Mendelian ratio even in the presence of the HSF1 mutation.
Gel shift assay
Lenses and other tissues isolated from mice were frozen at −80°C until use. The lens extracts were prepared in buffer C as previously described (Nakai et al, 1995). Aliquots containing 80 μg of proteins were subjected to gel shift assay using an ideal HSE oligonucleotide as a probe in the presence or absence of antiserum for each HSF, α-HSF1γ, α-HSF2δ, or α-HSF4b (2.0 μl of 1:10 diluted antiserum in phosphate-buffered saline (PBS)) (Nakai et al, 1997).
Western blot analysis
To isolate soluble and insoluble lens extracts, lenses were homogenized in a buffer containing 0.1 M sodium phosphate (pH 7.4) and 0.1 M NaCl. After centrifugation at 13 000 g for 20 min at 4°C, the supernatants were removed and the pellets were dissolved in a sodium dodecyl sulfate (SDS) loading buffer. Aliquots containing equal amounts of proteins were subjected to Western bolt analysis using antibodies for Hsp90 (a kind gift from Dr Y Miyata), Hsp40 (StressGen, SPA-400), Hsp27, αA-crystallins, and αB-crystallins (Kato et al, 1998), and for γ-crystallin (a kind gift from Dr H Kondoh). To detect Hsp110, Hsp70, and Hsp60, we generated antiserum specific for each Hsp. Recombinant mouse Hsp110 (amino acids 577–877), human Hsp70 (amino acids 1–641), or mouse Hsp60 (amino acids 358–554) fused to glutathione-S-transferase was immunized into rabbits in a TiterMax (CrtRx Co., Georgia) water-in-oil emulsion.
To examine leakage of crystallins from lens, lenses were removed from 2-week-old mice and incubated in Dulbecco's modified Eagle's medium (DMEM) medium containing 10% fetal bovine serum at 37°C for 24 h in the presence or absence of 1 mM H2O2 as a control (Tumminia et al, 1994). Proteins leaked from the damaged lens cells into the medium were precipitated with 10% TCA, and subjected to Western blot analysis using antiserum specific for αA- or γ-crystallin.
MEFs, mouse myoblasts C2C12, and mouse lens epithelial cells α-TN4 were cultured in DMEM containing 10% fetal bovine serum. To examine protein levels of HSFs, the cells and tissues were homogenized in NP-40 lysis buffer (150 mM NaCl, 1.0% Nonidet P-40, 50 mM Tris (pH 8.0), 1 mM phenylmethylsulfonyl fluoride, 1 μg/ml leupeptin, 1 μg/ml pepstatin) by sonication, and centrifuged at 12 000 g for 10 min. Equal mounts of protein were loaded on SDS–PAGE, and transferred onto nitrocellulose membranes. The membranes were blotted with α-HSF2α (Nakai et al, 1995) and α-mHSF4t (Tanabe et al, 1999). To detect HSF1, we generated antiserum raised against recombinant mouse HSF1 (amino acids 227–503) as described above.
Histopathology and immunohistochemistry
The eyes were dissected, fixed in NEO-Fix (Merck) at room temperature for 16–24 h, embedded in paraffin and cut into sections 5 μm thick. Sections were stained with hematoxylin and eosin, or with 4′,6′-diamidino-2-phenyindole (DAPI). Immunostaining of the paraffin sections was performed as described previously (Nakai et al, 2000) using antibodies as described above. Signals were detected using a DAB substrate kit (Vector Laboratories, CA).
To examine the patterns of DNA replication, incorporation of BrdU was analyzed using immunohistochemistry. Pregnant mice at 18.5 days postcopulation (dpc) were injected intraperitoneally with 50 μg/g BrdU (Sigma) diluted with PBS. At 1 h after the injection, fetal heads were dissected, fixed, and embedded in paraffin as described above. Immunohistochemical staining was performed essentially as described (Nishiyama et al, 2002). Numbers of BrdU-positive cells in the lens epithelial cells were counted in three wild-type and three HSF4-null embryos.
In situ hybridization
In situ hybridization was performed as described previously (Izu et al, 2004). The plasmid pmHSF4b-N containing 5′-terminal 436 base pairs of mHSF4 cDNA was generated by removing a HindIII fragment from pmHSF4b-1 (Tanabe et al, 1999), and was used to generate a digoxigenin-labeled probe (Roche, Mannheim, Germany). An FGF7 cDNA fragment corresponding to the full protein coding region was amplified by RT–PCR, subcloned into a pGEM7 vector (Promega), and used to generate a digoxigenin-labeled probe.
RT–PCR analysis
Total RNA was isolated from lenses in 6-week-old mice, 2-day-old mice, 15.5 dpc embryos, and from MEF cells. cDNAs were synthesized from 1 μg of total RNA by using avian myeloblastosis virus-reverse transcriptase (Invitrogen) and random hexamer primers. The PCR reaction was performed as described previously using primers for amplifying HSF1 and HSF2 genes (Fiorenza et al, 1995), HSF4 and S16 ribosomal protein genes (Tanabe et al, 1999), and γ-crystallin genes (Nishiguchi et al, 1998) in the presence of [α-32P]dCTP. The reactions were subjected to electrophoresis on a 4% polyacrylamide gel, and the autoradiogram is shown. To examine expression of FGFs, PCR reactions were performed using specific primer sets for FGF genes (Hajihosseini and Heath, 2002). The amplified DNA was stained with ethidium bromide and photographed using Epi-Light UV FA1100 (Aisin Cosmos R&D Co., Japan).
Chromatin immunoprecipitation
Four lenses isolated from 6-week-old mice were treated at 37°C for 15 min with 10 ml of 1% formaldehyde/DMEM containing 10% FCS, and then incubated in 0.125 M glycine at room temperature for 5 min. After washing with PBS two times, the lenses were suspended in 1 ml of PBS, homogenized using a Dounce homogenizer, and centrifuged at 2000 r.p.m. for 2 min. The pellet fractions were suspended in 200 μl of SDS lysis buffer (1% SDS, 10 mM ethylenediamine tetraacetic acid, 50 mM Tris–HCl (pH 8.1)), incubated at 4°C for 10 min, and then sonicated. The lysates were added with 8 μl of 5 M NaCl and incubated at 65°C for 4 h. DNA was recovered by phenol/chloroform extraction. ChIP was performed using a ChIP assay kit (Upstate, New York) essentially according to the manufacturer's instructions. A total of 30 and 40 cycles of PCR reactions were performed to amplify γF-crystallin and FGF-7 DNA fragments, respectively. An anti-mHSF4t serum or an anti-cHSF1c serum (Tanabe et al, 1999) was used for immunoprecipitation. Primers used to amplify ChIP-enriched DNA were as follows: gamma F1, 5′-ATC TCC TTG GGT CAG CAG ATC C-3′; gamma F2, 5′-ATG GGA TGG TGC TGT TGA G-3′; FGF7 F1, 5′-GAT TAG ACG TGG CCA TTG ATT-3′; FGF7 F2, 5′-TGT GAG TGC GTG GAG CCT TT-3′.
Transmission electron microscopy
The lenses from 2-week-old mice were fixed with a fixative (2% paraformaldehyde, 2.5% glutaraldehyde in 0.1 M cacodylate buffer at pH 7.4) for 2 h at room temperature and stored at 4°C for several weeks. After washing with 0.1 M cacodylate buffer (pH 7.4) three times (5 min each), cells were postfixed with ice-cold 1% OsO4 in the same buffer for 2 h. The samples were rinsed with distilled water, stained with 0.5% aqueous uranyl acetate for 2 h at room temperature, dehydrated with ethanol, and embedded in Poly/Bed 812. Ultrathin sections were cut, doubly stained with uranyl acetate and lead citrate, and viewed with a JEM 1010 transmission electron microscope (JEOL) (Fujikura et al, 2002).
Statistical analysis
Significant values were determined by analyzing data with the Mann–Whitney's U test using StatView version 4.5J for Macintosh (Abacus Concepts, Berkley, CA). A level of P<0.05 was considered significant.
Acknowledgments
We thank Drs RI Morimoto, S Ishii, and S Katoh for discussion, H Kondoh, Y Miyata, N Fujii, and P Russell for reagents, Dr Y Shinkai for teaching us gene targeting, Drs H Hamada and H Ishikawa for analysis of BrdU incorporation, and Drs T Murata and K Yamaguchi for mouse maintenance. This work was supported in part by Grants-in-Aid for Scientific Research (B) (C), and on Priority Area-Cell Cycle, Life of Proteins, and Cancer, from the Ministry of Education, Culture, Sports, Science and Technology, Japan, the Naitoh Foundation, and the Mochida Foundation.
References
- Bhat SP (2003) Crystallins, genes and cataract. Prog Drug Res 60: 205–262 [DOI] [PubMed] [Google Scholar]
- Bu L, Jin Y, Shi Y, Chu R, Ban A, Eiberg H, Andres L, Jiang H, Zheng G, Qian M, Cui B, Xia Y, Liu J, Hu L, Zhao G, Hayden MR, Kong X (2002) Mutant DNA-binding domain of HSF4 is associated with autosomal dominant lamellar and Marner cataract. Nat Genet 31: 276–278 [DOI] [PubMed] [Google Scholar]
- Burgess WH, Maciag T (1989) The heparin-binding (fibroblast) growth factor family of proteins. Annu Rev Biochem 58: 575–606 [DOI] [PubMed] [Google Scholar]
- Christians E, Davis AA, Thomas SD, Benjamin IJ (2000) Maternal effect of Hsf1 on reproductive success. Nature 407: 693–694 [DOI] [PubMed] [Google Scholar]
- Fagerholm PP, Philipson BT, Lindstrom B (1981) Normal human lens—the distribution of protein. Exp Eye Res 33: 615–620 [DOI] [PubMed] [Google Scholar]
- Fasciana C, van der Made AC, Faber PW, Trapman J (1996) Androgen regulation of the rat keratinocyte growth factor (KGF/FGF7) promoter. Biochem Biophys Res Commun 220: 858–863 [DOI] [PubMed] [Google Scholar]
- Finch PW, Lengel C, Chedid M (1995) Cloning and characterization of the promoter region of the human keratinocyte growth factor gene. J Biol Chem 270: 11230–11237 [DOI] [PubMed] [Google Scholar]
- Fiorenza MT, Farkas T, Dissing M, Kolding D, Zimarino V (1995) Complex expression of murine heat shock transcription factors. Nucleic Acids Res 23: 467–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frejtag W, Zhang Y, Dai R, Anderson MG, Mivechi NF (2001) Heat shock factor-4 (HSF-4a) represses basal transcription through interaction with TFIIF. J Biol Chem 276: 14685–14694 [DOI] [PubMed] [Google Scholar]
- Fujikura J, Yamato E, Yonemura S, Hosoda K, Masui S, Nakao K, Miyazaki J, Niwa H (2002) Differentiation of embryonic stem cells is induced by GATA factors. Genes Dev 16: 784–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajihosseini MK, Heath JK (2002) Expression patterns of fibroblast growth factors-18 and -20 in mouse embryos is suggestive of novel roles in calvarial and limb development. Mech Dev 113: 79–83 [DOI] [PubMed] [Google Scholar]
- Heon E, Priston M, Schorderet DF, Billingsley GD, Girard PO, Lubsen N, Munier FL (1999) The γ-crystallins and human cataracts: a puzzle made clearer. Am J Hum Genet 65: 1261–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye S, Katsuki K, Izu H, Fujimoto M, Sugahara K, Yamada S, Shinkai Y, Oka Y, Katoh Y, Nakai A (2003) Activation of heat shock genes is not necessary for heat shock transcription factor 1 to protect cell death against a single exposure to high temperatures. Mol Cell Biol 23: 5882–5895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izu H, Inouye S, Fujimoto M, Shiraishi K, Naito K, Nakai A (2004) Heat-shock transcription factor 1 is involved in quality control mechanisms in male germ cells. Biol Reprod 70: 18–24 [DOI] [PubMed] [Google Scholar]
- Jedlicka P, Mortin MA, Wu C (1997) Multiple functions of Drosophila heat shock transcription factor in vivo. EMBO J 16: 2452–2462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallio M, Chang Y, Manuel M, Alastalo TP, Rallu M, Gitton Y, Pirkkala L, Loones MT, Paslaru L, Larney S, Hiard S, Morange M, Sistonen L, Mezger V (2002) Brain abnormalities, defective meiotic chromosome synapsis and female subfertility in HSF2 null mice. EMBO J 21: 2591–2601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato K, Ito H, Kamei K, Inaguma Y, Iwamoto I, Saga S (1998) Phosphorylation of αB-crystallin in mitotic cells and identification of enzymatic activities responsible for phosphorylation. J Biol Chem 273: 28346–28354 [DOI] [PubMed] [Google Scholar]
- Kim JI, Li T, Ho IC, Grusby MJ, Glimcher LH (1999) Requirement for the c-Maf transcription factor in crystallin gene regulation and lens development. Proc Natl Acad Sci USA 96: 3781–3785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondoh H (2002) Development of the eye. In Mouse Development, Rossant J, Tam PP (eds) pp 519–538. New York: Academic Press [Google Scholar]
- Lengler J, Krausz E, Tomarev S, Prescott A, Quinlan RA, Graw J (2001) Antagonistic action of Six3 and Prox1 at the γ-crystallin promoter. Nucleic Acids Res 29: 515–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovicu FJ, Overbeek PA (1998) Overlapping effects of different members of the FGF family on lens fiber differentiation in transgenic mice. Development 125: 3365–3377 [DOI] [PubMed] [Google Scholar]
- McAvoy JW, Chamberlain CG (1989) Fibroblast growth factor (FGF) induces different responses in lens epithelial cells depending on its concentration. Development 107: 221–228 [DOI] [PubMed] [Google Scholar]
- McAvoy JW, Chamberlain CG, de Iongh RU, Hales AM, Lovicu FJ (1999) Lens development. Eye 13: 425–437 [DOI] [PubMed] [Google Scholar]
- McMillan DR, Christians E, Forster M, Xiao X, Connell P, Plumier JC, Zuo X, Richardson J, Morgan S, Benjamin IJ (2002) Heat shock transcription factor 2 is not essential for embryonic development, fertility, or adult cognitive and psychomotor function in mice. Mol Cell Biol 22: 8005–8014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan DR, Xiao X, Shao L, Graves K, Benjamin IJ (1998) Targeted disruption of heat shock transcription factor 1 abolishes thermotolerance and protection against heat-inducible apoptosis. J Biol Chem 273: 7523–7528 [DOI] [PubMed] [Google Scholar]
- Morano KA, Thiele DJ (1999) Heat shock factor function and regulation in response to cellular stress, growth, and differentiation signals. Gene Expr 7: 271–282 [PMC free article] [PubMed] [Google Scholar]
- Morimoto RI (1998) Regulation of the heat shock transcriptional response: cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev 12: 3788–3796 [DOI] [PubMed] [Google Scholar]
- Nakai A (1999) New aspects in the vertebrate heat shock factor system: Hsf3 and Hsf4. Cell Stress Chaperones 4: 86–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai A, Ishikawa T (2001) Cell cycle transition under stress conditions controlled by vertebrate heat shock factors. EMBO J 20: 2885–2895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai A, Kawazoe Y, Tanabe M, Nagata K, Morimoto RI (1995) The DNA-binding properties of two heat shock factors, HSF1 and HSF3 are induced in the avian erythroblast cell line HD6. Mol Cell Biol 15: 5168–5178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai A, Tanabe M, Kawazoe Y, Inazawa J, Morimoto RI, Nagata K (1997) HSF4, a new member of the human heat shock factor family which lacks properties of a transcriptional activator. Mol Cell Biol 17: 469–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai A, Suzuki M, Tanabe M (2000) Arrest of spermatogenesis in mice expressing an active heat shock transcription factor 1. EMBO J 19: 1545–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiguchi S, Wood H, Kondoh H, Lovell-Badge R, Episkopou V (1998) Sox1 directly regulates the γ-crystallin genes and is essential for lens development in mice. Genes Dev 12: 776–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama Y, Hamada H, Nonaka S, Yamamoto H, Nanno M, Katayama Y, Takahashi H, Ishikawa H (2002) Homeostatic regulation of intestinal villous epithelia by B lymphocytes. J Immunol 168: 2626–2633 [DOI] [PubMed] [Google Scholar]
- Piatigorsky J, Fukui HN, Kinoshita JH (1978) Differential metabolism and leakage of protein in an inherited cataract and a normal lens cultured with ouabain. Nature 274: 558–562 [DOI] [PubMed] [Google Scholar]
- Pirkkala L, Nykanen P, Sistonen L (2001) Roles of the heat shock transcription factors in regulation of the heat shock response and beyond. FASEB J 15: 1118–1131 [DOI] [PubMed] [Google Scholar]
- Robinson ML, Overbeek PA, Verran DJ, Grizzle WE, Stockard CR, Friesel R, Maciag T, Thompson JA (1995) Extracellular FGF-1 acts as a lens differentiation factor in transgenic mice. Development 121: 505–514 [DOI] [PubMed] [Google Scholar]
- Somasundaram T, Bhat SP (2000) Canonical heat shock element in the αB-crystallin gene shows tissue-specific and developmentally controlled interactions with heat shock factor. J Biol Chem 275: 17154–17159 [DOI] [PubMed] [Google Scholar]
- Tanabe M, Kawazoe Y, Takeda S, Morimoto RI, Nagata K, Nakai A (1998) Disruption of the HSF3 gene results in the severe reduction of heat shock gene expression and loss of thermotolerance. EMBO J 17: 1750–1758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe M, Sasai N, Nagata K, Liu XD, Liu PC, Thiele DJ, Nakai A (1999) The mammalian HSF4 gene generates both an activator and a repressor of heat shock genes by alternative splicing. J Biol Chem 274: 27845–27856 [DOI] [PubMed] [Google Scholar]
- Trinklein ND, Murray JI, Hartman SJ, Botstein D, Myers RM (2004) The role of heat shock transcription factor 1 in the genome-wide regulation of the mammalian heat shock response. Mol Biol Cell 15: 1254–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumminia SJ, Qin C, Zigler JS, Russell P (1994) The integrity of mammalian lenses in organ culture. Exp Eye Res 58: 367–374 [DOI] [PubMed] [Google Scholar]
- Wang G, Ying Z, Jin X, Tu N, Zhang Y, Phillips M, Moskophidis D, Mivechi NF (2004) Essential requirement for both hsf1 and hsf2 transcriptional activity in spermatogenesis and male fertility. Genesis 38: 66–80 [DOI] [PubMed] [Google Scholar]
- Wang G, Zhang J, Moskophidis D, Mivechi NF (2003) Targeted disruption of the heat shock transcription factor (hsf)-2 gene results in increased embryonic lethality, neuronal defects, and reduced spermatogenesis. Genesis 36: 48–61 [DOI] [PubMed] [Google Scholar]
- Wistow G (1990) Evolution of a protein superfamily: relationships between vertebrate lens crystallins and microorganism dormancy proteins. J Mol Evol 30: 140–145 [DOI] [PubMed] [Google Scholar]
- Wistow GJ, Piatigorsky J (1988) Lens crystallins: the evolution and expression of proteins for a highly specialized tissue. Annu Rev Biochem 57: 479–504 [DOI] [PubMed] [Google Scholar]
- Wu C (1995) Heat shock transcription factors: structure and regulation. Annu Rev Cell Biol 11: 441–469 [DOI] [PubMed] [Google Scholar]
- Xiao X, Zuo X, Davis AA, McMillan DR, Curry BB, Richardson JA, Benjamin IJ (1999) HSF1 is required for extra-embryonic development, postnatal growth and protection during inflammatory responses in mice. EMBO J 18: 5943–5952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Chen C, Stevenson MA, Auron PE, Calderwood SK (2002) Heat shock factor 1 represses transcription of the IL-1β gene through physical interaction with the nuclear factor of interleukin 6. J Biol Chem 277: 11802–11810 [DOI] [PubMed] [Google Scholar]
- Yan LJ, Christians ES, Liu L, Xiao X, Sohal RS, Benjamin IJ (2002) Mouse heat shock transcription factor 1 deficiency alters cardiac redox homeostasis and increases mitochondrial oxidative damage. EMBO J 21: 5164–5172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Frejtag W, Dai R, Mivechi NF (2001) Heat shock factor-4 (HSF-4a) is a repressor of HSF-1 mediated transcription. J Cell Biochem 82: 692–703 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Huang L, Zhang J, Moskophidis D, Mivechi NF (2002) Targeted disruption of hsf1 leads to lack of thermotolerance and defines tissue-specific regulation for stress-inducible Hsp molecular chaperones. J Cell Biochem 86: 376–393 [DOI] [PubMed] [Google Scholar]
- Zigler JS Jr (1994) Lens proteins. In Principles and Practice of Ophthalmology, Albert DM, Jakobiec FA (eds) pp 97–113. Pennsylvania: WB Saunders Company [Google Scholar]


