Abstract
To construct an East Asia mitochondrial DNA (mtDNA) phylogeny, we sequenced the complete mitochondrial genomes of 672 Japanese individuals (http://www.giib.or.jp/mtsnp/index_e.html). This allowed us to perform a phylogenetic analysis with a pool of 942 Asiatic sequences. New clades and subclades emerged from the Japanese data. On the basis of this unequivocal phylogeny, we classified 4713 Asian partial mitochondrial sequences, with <10% ambiguity. Applying population and phylogeographic methods, we used these sequences to shed light on the controversial issue of the peopling of Japan. Population-based comparisons confirmed that present-day Japanese have their closest genetic affinity to northern Asian populations, especially to Koreans, which finding is congruent with the proposed Continental gene flow to Japan after the Yayoi period. This phylogeographic approach unraveled a high degree of differentiation in Paleolithic Japanese. Ancient southern and northern migrations were detected based on the existence of basic M and N lineages in Ryukyuans and Ainu. Direct connections with Tibet, parallel to those found for the Y-chromosome, were also apparent. Furthermore, the highest diversity found in Japan for some derived clades suggests that Japan could be included in an area of migratory expansion to Continental Asia. All the theories that have been proposed up to now to explain the peopling of Japan seem insufficient to accommodate fully this complex picture.
Recent analysis of global mitochondrial DNA diversity in humans based on complete mtDNA sequences has provided compelling evidence of a human mtDNA origin in Africa (Ingman et al. 2000). Less than 100,000 years ago, at least two mtDNA human lineages began to rapidly spread from Africa to the Old World (Maca-Meyer et al. 2001). The archaeological records attest that humans reached Japan, at the eastern edge of Asia, around 30,000 years ago (Glover 1980). At that time, Japan was connected to the Continent by both northern and southern land bridges, enabling two migratory routes. As early as 13,000 years ago, pottery appeared in Japan and Siberia for the first time in the world (Shiraishi 2002). Subsequent technical improvements gave rise to the Japanese Neolithic period known as the Jomon period, in which the population growth was considerable. Later, Continental people arrived in Japan from the Korean peninsula, initiating the Yayoi period, with this migration reaching its maximum at the beginning of the first millennium.
With this archaeological framework in mind, it was of anthropological interest to us to know whether the modern Japanese are the result of an admixture between the Paleolithic-Neolithic aborigines and more recent immigrant populations, whether the indigenous population gradually evolved to give rise to the modern Japanese, with subsequent colonizations having strong cultural influences but only minor demographic impact, or even whether the late Neolithic waves entirely replaced the indigenous residents. Morphometric data obtained from the remains of Japanese Paleolithic people are more in accordance with a southern origin for these first immigrants. Subsequent morphological studies on modern indigenous (northern Ainu and southern Ryukyuans) and mainland Japanese favored an admixture model in which the former would be descendants of the Paleolithic Japanese and the latter derived from the Continental immigrants who gave rise to the Yayoi period (Hanihara 1991). Genetic analysis using classical markers assigned a definitive northern origin to the Upper Paleolithic inhabitants of Japan; but whereas some authors favored a homogeneous background for all modern Japanese (Nei 1995), others claimed that although Upper Paleolithic and Yayoi period immigrants had probably a northern Asian origin, they were genetically differentiated (Omoto and Saitou 1997). The application of molecular markers to define maternal and paternal lineages to the peopling of Japan confirmed the dual admixture model but added some interesting novelties. For example, the study of Y-chromosome markers led to the discovery of remarkable Korean and Tibetan influences on the Japanese population (Hammer and Horai 1995); and mtDNA HVS-I sequences also confirmed the Korean input (Horai et al. 1996) and closer affinities of the Japanese to Tibetans than to southern Asians (Qian et al. 2001). In quantitative estimations of maternal admixture, it was found that ∼65% of the mainland Japanese gene pool was derived from Continental gene flow after the Yayoi period. However, the indigenous Ainu from the northern island of Hokkaido and the Ryukyuans from southern Okinawa showed <20% Continental specificity, pointing to them as the most probable descendants of the Jomon people. The fact that these indigenous groups were, in turn, genetically well differentiated indicated a notable degree of heterogeneity and/or isolation among the early Japanese immigrants (Horai et al. 1996). However, two handicaps of these studies are the incomplete representation of Asian populations and the relatively small sample size of those analyzed, which weakens the reliance on the relative affinities found by genetic distance methods (Helgason et al. 2001). For mtDNA there are currently enough HVI/HVII data from eastern Asia, including Japan, to test the validity of the above-mentioned results. However, these sequences have been assorted into different clades following different insufficient criteria or even have not been classified at all. Furthermore, the phylogenetic confidence of results based only on sequences from the noncoding region (HVI, HVII) has been recently questioned (Bandelt et al. 2000). This is mainly due to the frequent occurrence of parallel mutations in independent lineages that confuse the correct classification, a source of error that is increased because the basal motif in the noncoding region for the two macrolineages that expanded throughout Asia is the same (16223). In addition, as the noncoding region has not evolved at a constant rate across all human lineages, it is considered inappropriate to use this region for dating evolutionary events (Ingman et al. 2000; Finnilä et al. 2001).
To make reliable use of this important source of available data on the mtDNA noncoding region to contrast the maternal structure and to determine the most probable origin of the modern Japanese, we have undertaken the following approach: First, we used a set of complete mtDNA sequences of 672 Japanese individuals to create a phylogenetic network (Bandelt et al. 1999) that related them to other complete sequences, already published, belonging to the major haplogroups proposed by others (Torroni et al. 1992, 1996; Macaulay et al. 1999; Yao et al. 2002a). Discriminative positions in the noncoding region, defining additional Asian subhaplogroups, were then used to further classify 766 previously published Japanese partial sequences. For this purpose we also included other unambiguously assorted sequence data reported by other research groups (Derbeneva et al. 2002b; Yao et al. 2002a). These HVI sequences thus pooled were then compared with other published Asian sequences. Finally, using all of these classified sequences, we tested the relative affinities of modern Japanese and Continental Asians using global distance methods and phylogeographic approaches framed at different age levels.
RESULTS
Eastern Asia Phylogeny Based on Complete mtDNA Sequences
The phylogenetic network constructed with the complete mtDNA sequences fully coincides with those previously published at worldwide (Maca-Meyer et al. 2001; Herrnstadt et al. 2002) or regional scale (Kong et al. 2003). Moreover, their main branches are well supported by high bootstrap values on a neighbor-joining tree (Supplemental material, condensed by more than 40% bootstrap values).
From the L3 African trunk, two early branches came out of Africa and radiated extensively, originating superhaplogroups M and N, which were defined by the basic mutations depicted in Figures 1A and 2, respectively. Representatives of both superhaplogroups reached Japan. The construction of these phylogenetic trees by using our Japanese complete sequences and other published Asian sequences (Table 1) resulted in a better definition of the known haplogroups and in the identification of new clades at different phylogenetic levels. Characteristic HVI motifs and diagnostic RFLPs in the coding region, and coalescence ages for these haplogroups and subhaplogroups are given in Supplemental Tables A and B. To contribute to the unification of the mitochondrial nomenclature, we revised the previously proposed haplogroups by adding the following new information.
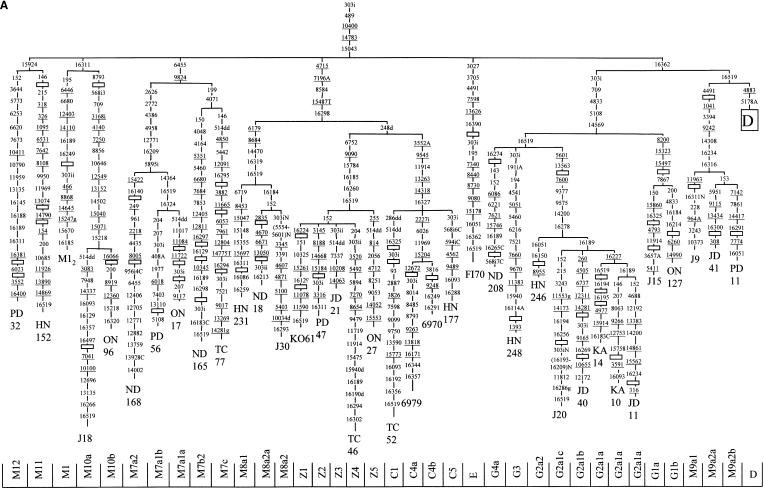
Figure 1.
Phylogenetic tree, based on complete mtDNA sequences, for macrohaplogroup M in general (A) and for subhaplogroup D (B) in particular. Subject origins are given in Table 1. The numbers along the links refer to nucleotide positions, arbitrarily written in ascending order. Open boxes are nodes from which other (not shown) sequences branch. A, C, G, and T indicate transversions; whereas “d” indicates deletions and “i” insertions. Nonrecurrent mutations are underlined.
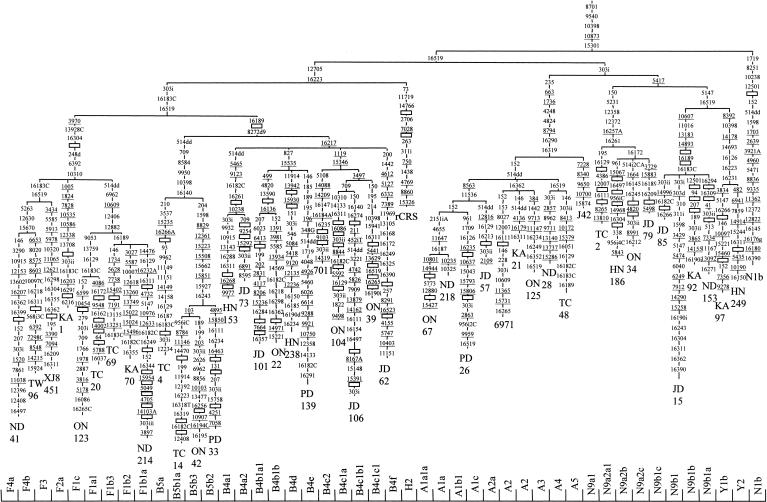
Figure 2.
Phylogenetic tree, based on complete mtDNA sequences, for macrohaplogroup N. Origins of subjects are explained in Table 1. The numbers along the links refer to nucleotide positions, arbitrarily written in ascending order. Open boxes are nodes from which other (not shown) sequences branch. A, C, G, and T indicate transversions; whereas “d” indicates deletions and “i” insertions. Nonrecurrent mutations are underlined.
Table 1.
| Sample | Haplogroup | Origin | References |
|---|---|---|---|
| PD32 | M12 | Japanese | This work |
| HN152 | M11 | Japanese | This work |
| M12 | M1 | Jordanian | Maca-Meyer et al. 2001 |
| J18 | M10a | Japanese | This work |
| ON96 | M10b | Japanese | This work |
| ND168 | M7a2 | Japanese | This work |
| PD56 | M7a1b | Japanese | This work |
| ON17 | M7a1a | Japanese | This work |
| ND165 | M7b2 | Japanese | This work |
| TC77 | M7c | Japanese | This work |
| HN231 | M8a1 | Japanese | This work |
| ND18 | M8a2a | Japanese | This work |
| J30 | M8a2 | Japanese | This work |
| KO61 | Z1 | Koryac | Ingman and Gyllensten 2003 |
| PD47 | Z2 | Japanese | This work |
| JD21 | Z3 | Japanese | This work |
| TC46 | Z4 | Japanese | This work |
| ON27 | Z5 | Japanese | This work |
| TC52 | C1 | Japanese | This work |
| 6979 | C4a | Evenki | Ingman et al. 2000 |
| 6970 | C4B | Buryat | Ingman et al. 2000 |
| HN177 | C5 | Japanese | This work |
| F170 | E | Philippine | Ingman and Gyllensten 2003 |
| ND208 | G4a | Japanese | This work |
| HN248 | G3 | Japanese | This work |
| HN246 | G2a2 | Japanese | This work |
| J20 | G2a1c | Japanese | This work |
| JD40 | G2a1b | Japanese | This work |
| KA14 | G2a1a | Japanese | This work |
| KA10 | G2a1a | Japanese | This work |
| JD11 | G2a1a | Japanese | This work |
| J15 | G1a1 | Japanese | This work |
| ON127 | G1a2 | Japanese | This work |
| J9 | M9a1 | Japanese | This work |
| JD41 | M9a2a | Japanese | This work |
| PD11 | M9a2b | Japanese | This work |
| JD47 | D6 | Japanese | This work |
| TC12 | D5a2 | Japanese | This work |
| PD97 | D5a1a | Japanese | This work |
| GC22 | D5a1b | Japanese | This work |
| ON46 | D5b1a | Japanese | This work |
| TC24 | D5b1b | Japanese | This work |
| TC30 | D5b2 | Japanese | This work |
| J21 | D5c1 | Japanese | This work |
| JD6 | D5c2 | Japanese | This work |
| ND140 | D4a | Japanese | This work |
| TC73 | D4b1a | Japanese | This work |
| PD48 | D4b1b | Japanese | This work |
| GC20 | D4b2a | Japanese | This work |
| KA83 | D4b2b | Japanese | This work |
| ON143 | D4c | Japanese | This work |
| JD36 | D4f | Japanese | This work |
| TC70 | D4d1a | Japanese | This work |
| KA96 | D4d1b | Japanese | This work |
| ND205 | D4d2 | Japanese | This work |
| 0N138 | D4e1 | Japanese | This work |
| J35 | D4e2 | Japanese | This work |
| J43 | D4g | Japanese | This work |
| KA90 | D4h | Japanese | This work |
| ND188 | D4i | Japanese | This work |
| KA58 | D4j | Japanese | This work |
| PD60 | D4k1 | Japanese | This work |
| ND212 | D4k2 | Japanese | This work |
| TC8 | D4k3 | Japanese | This work |
| JD100 | D4l | Japanese | This work |
| PD134 | D4m | Japanese | This work |
| HN194 | D4n | Japanese | This work |
| ND41 | F4a | Japanese | This work |
| TW96 | F4b | Indigenous Taiwanese | Ingman and Gyllensten 2003 |
| XJ8451 | F3 | Chinese | Kong et al. 2003 |
| KA1 | F2a | Japanese | This work |
| ON123 | F1c | Japanese | This work |
| TC20 | F1a1 | Japanese | This work |
| TC69 | F1b3 | Japanese | This work |
| KA70 | F1b2 | Japanese | This work |
| ND214 | F1b1a | Japanese | This work |
| TC4 | B5a | Japanese | This work |
| TC14 | B5b1a | Japanese | This work |
| ON42 | B5b3 | Japanese | This work |
| PD33 | B5b2 | Japanese | This work |
| HN153 | B4a1 | Japanese | This work |
| JD73 | B4a2 | Japanese | This work |
| JD101 | B4b1a1 | Japanese | This work |
| ON22 | B4b1b | Japanese | This work |
| HN238 | B4d | Japanese | This work |
| PD139 | B4e | Japanese | This work |
| 7011 | B4c2 | Uzbek | Ingman et al. 2000 |
| ON104 | B4c1a | Japanese | This work |
| JD106 | B4c1b1 | Japanese | This work |
| ON39 | B4c1c1 | Japanese | This work |
| JD62 | B4f | Japanese | This work |
| rCRS | H2 | English | Andrews et al. 1999 |
| N1b | N1b | Jordanian | Maca-Meyer et al. 2001 |
| ON67 | A1a1a | Japanese | This work |
| ND218 | A1a | Japanese | This work |
| PD26 | A1b1 | Japanese | This work |
| JD57 | A1c | Japanese | This work |
| 6971 | A2a | Chukchi | Ingman et al. 2000 |
| KA21 | A2 | Japanese | This work |
| ON125 | A2 | Japanese | This work |
| ND28 | A3 | Japanese | This work |
| TC48 | A4 | Japanese | This work |
| J42 | A5 | Japanese | This work |
| TC2 | N9a1 | Japanese | This work |
| HN186 | N9a2a1 | Japanese | This work |
| ON34 | N9a2b | Japanese | This work |
| JD79 | N9a2c | Japanese | This work |
| JD85 | N9b1c | Japanese | This work |
| JD15 | N9b1 | Japanese | This work |
| KA92 | N9b1b | Japanese | This work |
| ND153 | N9b1a | Japanese | This work |
| KA97 | Y1b | Japanese | This work |
| HN249 | Y2 | Japanese | This work |
Subdivisions Within Macrohaplogroup M
Haplogroup D
Haplogroup D has been defined by the specific RFLP -5176 AluI (Torroni et al. 1992). Studies on Native American HVI sequences permitted further subdivision of D into subgroups D1 by mutation 16325 and D2 by mutation 16271 (Forster et al. 1996). Additional subdivisions into subhaplogroups D4 and D5 have been proposed for Asian lineages (Yao et al. 2002a). These investigators characterized D4 by position 3010. Two additional mutations, 8414 and 14668, have been proposed to define D4 (Fig. 1B; Kivisild et al. 2002). Whereas these two latter mutations seem to be rare events, 3010 has also been independently detected in haplogroups H and J. A new branch at the same phylogenetic level as D4 and D5 has been detected in Japan (Fig. 1B). It is characterized by mutations 709, 1719, 3714, and 12654 and was named D6. The subdivision of D4 into subgroups D4a and D4b was proposed on the basis of the distinctive mutational motif 152, 3206, 14979, and 16129 for the first and 10181 and 16319 for the second (Kivisild et al. 2002). Both subclades have been detected in our Japanese sample. From our data it can be deduced that mutation 8473 is also basal for D4a. In relation to D4b it seems that its ancestral branch is defined by the 8020 substitution (Fig. 1B). Consequently, the D4b subgroup proposed by Yao et al. (2002a) should be renamed D4b1 harboring 15440 and 15951 as additional basic mutations. A new subgroup characterized by 1382C, 8964, and 9824A mutations and named D4b2, is represented by lineages GC20 and KA83 in Figure 1B. Furthermore, 12 new branches at the same phylogenetic level as subhaplogroups D4a and D4b can be identified in the network. Accordingly, they have been successively named from D4c to D4n. On the other hand, D5 was defined by mutations 150, 10397, and 16189 (Yao et al. 2002a); however, 16189 is not present in all D5 lineages. We have named D5a and D5b those lineages that share this mutation and 9180 and D5c those lacking them. Consequently, we propose to rename D5a of Yao et al. (2002a) as D5a1. Additional mutations (1107 and 5301) define D5 (Fig. 1B), as has been recently confirmed (Kong et al. 2003). Of the four mutations at the basal branch of this group, 10397 seems to be a unique event; and the group can be diagnosed by the RFLP polymorphism +10396 BsrI. Recently, the phylogeny of haplogroup D has been revised in the light of complete sequences from Aleuts (Derbeneva et al. 2002b). By comparing their nomenclature to ours, it is possible to equate their D2 lineage to our D4e1 and their D3 lineage to our D4b1. As a total, D is the most abundant haplogroup in people of central and eastern Asia including mainland Japanese but not in the Ainu and Ryukyuans. However, the geographic distributions of some subhaplogroups are peculiar. For example, D5 is prevalent in southern areas. D4a is abundant in Chukchi of northeast Siberia, but D4a1 has its highest frequency in the Ryukyuans and clade D4n in the Ainu (Table 2).
Table 2.
Frequency (in Percentage) of Each Haplogroup in Each Group of Populations
| Group | JPN | RYU | AIN | Ch1 | Ch2 | Ch3 | Ch4 | Ch5 | Ch6 | CA1 | CA2 | TWA | MAN | ITE | FIU | ALU | KAM | CHU | TUV | BUR | KOR | TIB | SAK | FIL | IND | SAB |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample | 1312 | 50 | 51 | 213 | 435 | 32 | 72 | 757 | 67 | 204 | 93 | 208 | 98 | 46 | 38 | 56 | 91 | 60 | 36 | 40 | 537 | 65 | 20 | 32 | 40 | 34 |
| L2 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 5 | - | - | - |
| M/N | 1.3 | 18 | 1.96 | 3.29 | 1.61 | - | 4.17 | 5.55 | - | 1.96 | 1.08 | 2.4 | 1.02 | - | 2.63 | - | - | - | - | 25 | 5.96 | 9.23 | - | 6.25 | 10 | 17.6 |
| I/W/N | - | - | - | - | - | - | - | - | - | 2.45 | - | - | - | - | 2.63 | - | - | - | - | - | 0.37 | - | - | - | - | - |
| A1 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.67 | - | - | 0.19 | - | - | - | - | - |
| A1a | 2.13 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 3.54 | - | - | - | - | - |
| A1b | 4.57 | 2 | 3.92 | 5.16 | 7.36 | - | 5.56 | 2.77 | 7.46 | 6.86 | 2.15 | 0.48 | 3.06 | 6.52 | - | 1.79 | 3.3 | - | 5.56 | 5 | 2.98 | 6.15 | - | - | - | - |
| A1b1 | 0.15 | - | - | 0.47 | 0.46 | - | 1.39 | 0.92 | - | - | - | - | - | - | - | - | - | - | - | - | 0.74 | - | - | - | - | - |
| A2a | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 10 | - | - | - | - | - | - | - | - |
| A2b | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 3.57 | 2.2 | 50 | - | - | 0.37 | 3.08 | - | - | - | - |
| N9a | 4.57 | - | - | 3.76 | 1.38 | - | 6.94 | 2.91 | - | 1.96 | - | - | - | - | - | - | - | - | - | - | 3.91 | - | - | - | 2.5 | - |
| N9b | 2.13 | 2 | 2 | - | - | - | - | 0.13 | - | - | - | - | - | - | - | - | - | - | - | - | 0.19 | - | - | - | - | - |
| Y | 0.38 | - | 21.6 | 3.29 | 1.38 | - | - | 0.53 | - | 0.49 | - | 0.96 | - | 4.35 | - | 7.14 | 9.89 | - | - | - | 2.23 | - | - | 3.13 | 2.5 | 2.94 |
| HV | 0.91 | 4 | - | 0.47 | 1.84 | - | - | 1.85 | - | 13.7 | 20.5 | - | 18.4 | - | 36.79 | - | 1.1 | - | 2.78 | 5 | 1.12 | - | - | 3.13 | 5 | - |
| JT | - | - | - | 2.82 | 1.15 | - | - | - | - | 5.4 | 8.61 | - | 19.4 | - | 23.65 | - | - | - | - | - | - | - | - | - | - | - |
| UK | - | - | - | - | 1.38 | - | - | 0.39 | - | 10.3 | 5.39 | - | 25.5 | - | 10.52 | - | - | - | 2.78 | - | - | - | - | - | 5 | - |
| R9a | 0.08 | - | - | 1.88 | 0.69 | - | - | 1.85 | - | - | - | 7.69 | - | - | - | - | - | - | - | - | - | - | - | 3.13 | - | 2.94 |
| R11 | - | - | - | 0.94 | 0.46 | 12.5 | - | 1.85 | - | 0.49 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| B | 1.3 | - | - | 0.47 | 1.84 | - | 4.17 | 1.19 | - | 0.98 | 1.08 | - | - | - | - | - | - | - | - | - | - | - | - | 3.13 | - | 2.94 |
| B4 | 0.76 | 2 | - | 1.88 | 2.53 | 9.38 | 1.39 | 1.98 | 5.97 | 1.96 | 1.08 | - | - | - | - | - | - | - | - | 2.5 | 0.74 | 1.54 | - | - | - | - |
| B4a | 0.84 | - | - | 2.35 | 1.61 | - | 1.39 | 4.36 | - | 0.49 | 1.08 | 14.9 | - | - | - | - | - | - | 2.78 | - | 0.56 | - | - | - | - | - |
| B4a1 | 0.84 | 2 | - | - | - | - | - | - | - | - | - | 0.48 | - | - | - | - | - | - | - | - | 0.56 | - | - | - | - | - |
| B4a2 | - | - | - | 0.47 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| B4a3 | - | - | - | 0.47 | - | 15.6 | 2.78 | 1.85 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| B4b | 0.53 | - | - | 1.88 | 1.61 | - | 6.94 | 1.72 | - | - | - | 4.33 | - | - | - | - | - | - | 11.1 | - | 0.74 | - | - | - | - | - |
| B4b1 | 2.13 | 2 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.56 | - | - | - | - | - |
| B4c1 | 1.6 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.74 | - | - | - | - | - |
| B4c1b | 0.61 | - | - | 1.88 | - | - | - | 1.06 | - | - | - | 4.81 | - | - | - | - | - | - | - | - | 0.56 | - | - | - | - | - |
| B4c2 | 0.08 | - | - | - | - | - | - | 1.19 | - | 0.49 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| B4f | 0.3 | 4 | 2 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.38 | - | - | - | - | - |
| B5 | - | - | - | - | - | - | - | 0.13 | - | - | - | 0.48 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| B5a | - | - | - | 1.88 | 0.69 | - | 1.39 | 6.74 | 5.97 | 0.98 | 1.08 | 6.25 | - | - | - | - | - | - | - | - | - | - | - | 3.13 | - | 11.8 |
| B5a1 | 0.61 | - | - | 0.47 | 1.38 | - | - | - | - | 0.49 | 1.08 | - | - | - | - | - | - | - | - | - | 3.54 | - | - | - | - | - |
| B5b | 0.3 | 2 | - | - | 1.38 | - | - | 0.4 | - | 0.49 | - | - | - | - | - | - | - | - | - | - | 0.19 | - | - | 12.5 | - | - |
| B5b1 | 0.99 | - | - | - | - | - | - | 0.13 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| B5b2 | 2.29 | - | - | 0.47 | 1.15 | - | 1.39 | 0.26 | - | 0.49 | - | - | - | - | - | - | - | - | - | - | 0.93 | - | - | - | - | - |
| B5b3 | 0.08 | - | - | - | - | - | - | 0.13 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| F | 0.23 | - | - | 0.94 | 2.3 | - | - | 2.25 | 1.49 | 1.96 | 2.15 | 1.92 | - | - | - | - | - | - | - | - | 0.37 | 1.54 | - | - | - | 2.94 |
| F1a | 0.15 | - | - | 0.47 | 1.61 | 15.6 | 4.17 | 4.62 | 1.49 | - | 1.08 | - | - | - | - | - | - | - | - | - | 0.74 | - | - | 12.5 | 35 | 2.94 |
| F1a1 | 1.52 | - | - | 2.82 | 1.84 | 15.6 | 2.78 | 7.93 | - | 1.47 | - | 7.21 | - | - | - | - | - | - | - | - | 1.12 | - | - | - | 5 | - |
| F1b | 3.13 | 2 | 2 | 2.82 | 2.99 | - | 2.78 | 1.19 | 2.99 | 3.92 | - | - | 1.02 | - | - | - | - | - | 8.33 | 5 | 2.05 | - | - | - | - | - |
| F1c | - | - | - | 1.41 | 1.15 | - | 1.39 | 0.4 | - | 0.49 | - | - | - | - | - | - | - | - | - | - | 0.19 | 1.54 | - | - | - | - |
| F2 | - | - | - | 0.47 | 0.46 | 3.13 | 6.94 | 0.4 | - | - | - | - | - | - | 5.26 | - | - | - | - | - | - | - | - | - | - | - |
| F2a | 0.15 | - | - | 0.47 | 0.92 | - | 1.39 | 0.79 | 7.46 | - | - | - | - | - | - | - | - | - | - | - | 0.19 | - | - | - | - | - |
| F2a1 | 0.08 | - | - | - | - | - | - | 0.66 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.5 | - |
| F3 | 0.08 | - | - | - | 0.46 | - | - | 0.53 | - | - | - | - | - | - | - | - | - | - | - | - | 0.19 | - | - | - | - | - |
| F4 | - | - | - | 0.47 | - | - | - | 0.92 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 3.13 | 2.5 | 14.7 |
| F4b | - | - | - | - | - | - | - | 0.13 | - | - | - | 10.1 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| M | 0.38 | 2 | 3.92 | - | - | - | - | 0.26 | - | 0.49 | - | - | - | - | - | - | - | - | - | - | 0.19 | - | - | - | - | - |
| M (PNG) | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 10 | - |
| M5/D4a/G1 | 0.46 | - | 13.7 | 0.94 | 0.23 | 18.8 | - | 0.26 | - | 0.49 | 2.15 | - | - | - | - | - | - | 3.33 | - | 2.5 | 1.68 | - | 5 | - | 2.5 | 2.94 |
| M7a | 7.39 | 12 | 15.7 | - | 0.23 | - | - | 0.53 | 4.48 | - | - | - | - | - | - | - | - | - | - | 2.5 | 3.35 | - | - | 6.25 | - | - |
| M7a1 | 0.08 | 14 | - | 1.41 | 1.38 | - | 1.39 | 2.25 | - | 0.98 | - | 0.48 | - | - | - | - | - | - | - | - | - | - | - | 3.13 | - | - |
| M7b | - | - | - | 0.47 | 0.46 | - | 1.39 | 2.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| M7b1 | 0.08 | - | - | 2.35 | 2.99 | - | - | 5.02 | - | 0.49 | - | 12 | - | - | - | - | - | - | - | - | 0.56 | - | - | - | 2.5 | - |
| M7b2 | 4.73 | 8 | 3.92 | 0.47 | 0.69 | - | 1.39 | 0.13 | - | - | - | - | 1.02 | - | - | - | - | - | - | - | 2.79 | - | - | - | - | - |
| M7c | 0.76 | 2 | - | 1.88 | 2.07 | - | 2.78 | 2.51 | - | 1.96 | - | 4.33 | - | - | - | - | - | - | - | 2.5 | 3.72 | 3.08 | - | 18.8 | 2.5 | 20.6 |
| M8 | 0.15 | - | - | 0.47 | - | - | - | 0.13 | - | - | - | - | - | - | - | - | - | 1.67 | - | - | 0.37 | - | - | - | - | - |
| M8a | 1.22 | - | - | 4.23 | 0.92 | - | 4.17 | 1.59 | - | 1.47 | - | - | - | - | - | - | - | - | - | 2.5 | 1.68 | - | - | - | - | - |
| M8a2 | - | - | - | - | 0.23 | - | - | - | - | - | 1.08 | - | - | 13 | - | - | 20.9 | 1.67 | 2.78 | - | 0.37 | - | - | - | - | - |
| C | - | - | - | 1.41 | 6.67 | - | 2.78 | 4.1 | 7.46 | 3.92 | 9.68 | - | 12.2 | - | - | 16.1 | 26.4 | 5 | 16.7 | 10 | 0.37 | 1.54 | - | - | - | - |
| C1 | 0.3 | - | - | - | 0.46 | - | - | - | - | - | 1.08 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| C4a | 0.08 | - | - | 0.47 | 0.92 | - | - | - | - | 0.98 | 1.08 | - | 4.08 | - | - | - | - | - | 13.9 | - | 0.37 | - | - | - | - | - |
| C5 | 0.08 | - | - | - | 0.23 | - | - | - | - | 1.96 | 2.15 | - | 1.02 | - | - | - | - | - | 2.78 | - | 0.74 | - | - | - | - | - |
| Z | 1.3 | - | - | 2.35 | 2.76 | - | 4.17 | 0.53 | 10.4 | 0.49 | 9.68 | - | - | 6.52 | - | - | 8.79 | - | - | - | 1.49 | 9.23 | - | - | - | - |
| G | 0.53 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.37 | - | - | - | - | - |
| G1a1/D | 2.13 | - | 5.88 | - | 0.23 | - | - | 0.4 | - | - | - | - | - | - | - | - | - | - | - | - | 2.05 | - | 10 | - | - | - |
| G2a | 1.68 | 4 | - | 1.88 | - | - | - | 0.26 | - | 0.49 | - | - | - | - | - | - | - | - | 2.78 | 5 | 1.86 | - | - | - | - | - |
| G2a1 | 2.52 | - | 3.92 | 5.16 | 0.92 | - | 1.39 | 0.66 | - | 7.35 | 1.08 | - | 6.12 | - | - | - | - | - | 5.56 | - | 1.49 | - | - | - | - | - |
| G5 | - | - | - | - | - | - | - | - | - | - | - | - | - | 69.6 | - | 67.9 | 27.5 | 6.67 | - | - | - | - | - | - | - | - |
| M9 | - | - | - | - | - | - | 2.78 | 1.59 | 2.99 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| M9a | 2.44 | 4 | 1.96 | 5.63 | 1.61 | - | 1.39 | 1.98 | - | 2.45 | 1.08 | - | - | - | - | - | - | - | - | 5 | 3.17 | 10.8 | - | - | - | - |
| M10 | 1.3 | - | - | 2.35 | 1.84 | - | - | 1.72 | - | 0.49 | 1.08 | - | - | - | - | - | - | - | - | 2.5 | 4.66 | 7.69 | - | - | - | 2.94 |
| M12 | 0.08 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.19 | 6.15 | - | - | - | - |
| D | 0.08 | - | - | 0.47 | 0.46 | - | 1.39 | 0.26 | - | - | - | - | 2.04 | - | - | - | - | - | - | - | 0.19 | - | - | - | - | - |
| D4 | 18.9 | 2 | 7.84 | 12.7 | 20.9 | 6.25 | 1.39 | 6.74 | 16.4 | 11.3 | 18.3 | 13.5 | 3.06 | - | 18.4 | - | - | 1.67 | 13.9 | 17.5 | 18.4 | 12.3 | - | 18.8 | 7.5 | 11.8 |
| D4a | 7.39 | - | 1.96 | 4.23 | - | - | 1.39 | 0.92 | - | 2.45 | 1.08 | 0.48 | - | - | - | - | - | 11.7 | - | - | 5.77 | - | 80 | - | - | - |
| D4a1 | 0.53 | 4 | - | 0.94 | 1.38 | - | 2.78 | 1.06 | - | - | - | - | - | - | - | - | - | - | - | - | 2.42 | 1.54 | - | - | - | - |
| D4b | 2.36 | 6 | - | 1.41 | 1.61 | - | 1.39 | 0.92 | - | 0.98 | 3.23 | - | 1.02 | - | - | 3.57 | - | 6.67 | 2.78 | 2.5 | 0.93 | 18.5 | - | - | - | - |
| D4d | 2.67 | - | - | 0.94 | 0.69 | 3.13 | - | - | - | 1.96 | 1.08 | - | - | - | - | - | - | - | 2.78 | 2.5 | 0.93 | - | - | - | - | - |
| D4k | 0.15 | - | - | - | 1.15 | - | 1.39 | 0.13 | 22.4 | - | - | - | - | - | - | - | - | - | - | - | 0.19 | 3.08 | - | - | - | - |
| D4n | 0.61 | - | 3.92 | - | 0.23 | - | 1.39 | - | 2.99 | - | - | - | - | - | - | - | - | - | - | - | 0.19 | - | - | - | 2.5 | - |
| D5 | 3.73 | 2 | - | 1.88 | 3.22 | - | 4.17 | 2.64 | - | 0.98 | - | 6.25 | 1.02 | - | - | - | - | - | - | 2.5 | 2.98 | 1.54 | - | 3.13 | 2.5 | 2.94 |
| D5a | 1.07 | - | 3.92 | 1.41 | 3.91 | - | 4.17 | 1.59 | - | 0.49 | - | - | - | - | - | - | - | - | 2.78 | - | 0.56 | 1.54 | - | - | - | - |
Haplogroup M9
It is confirmed that haplogroup M9 is characterized by mutation 4491 (Fig. 1A), as recently proposed (Kong et al. 2003). Subhaplogroup M9a, as redefined by Kong et al. (2003), was identified by positions 153, 3394, 14308, 16234, and 16316 (Yao et al. 2002a). Nevertheless, not all lineages have 153. Although M9 could be RFLP-diagnosed by +1038 NlaIII and +3391 HaeIII polymorphisms, the latter one should be avoided; as 3391 is also present in some D4d1 lineages (Fig. 1B) and thus could produce misclassification. We have grouped lineages with 11963 as M9a1 and those with 153 as M9a2. M9 has a central and eastern Asian geographic distribution, and it reaches its greatest frequency (11%) and diversity (87%) in Tibet. In Japan, in addition to mainland Japanese it has been detected in the indigenous Ainu and Ryukyuans (Horai et al. 1996).
Haplogroup G
This haplogroup was first detected by Ballinger et al. (1992) and later named G by Torroni et al. (1994). It was defined by the presence of the combined RFLP polymorphism +4830 HaeII/+4831 HhaI. In addition, the basal branch has mutations 709, 5108, and 14569 (Fig. 1; Kivisild et al. 2002). Subhaplogroup G1 was defined by transition 16017 (Schurr et al. 1999) and G2 by mutations 7600 and 16278 (Yao et al. 2002a). Recently, mutations 8200, 15323, and 15497 have been used for G1 status (Kong et al. 2003). This is confirmed with our Japanese sequences; consequently, we have defined G1a by 7867 (Fig. 1A). To avoid repetitions, the G1 group of Schurr et al. (1999) has been provisionally renamed as G5 (Table 2). At least two mutations (5601 and 13563) characterize G2; and five more, G2a (Fig. 1A; Kong et al. 2003). We have defined subclade G2a1 by the presence of 16189 and the derivative G2a1a by the addition of 16227, whereas 16051 and 16150 identify G2a2 lineages. Furthermore, two new subclades, G3 and G4, are also apparent in Japanese (Fig. 1A). Subgroup G5 is dominant in northeastern Siberia, but we have not detected it in our set of Japanese complete sequences. However, G1a1 has its highest frequencies in a cluster embracing Japanese, Ainu, Ryukyuan, and Koreans. On the contrary, G2 is relatively abundant in northern China and central Asia, reaching notable frequencies in the Mansi and in Tuvinians at the respective west and east ends of South Siberia (Table 2).
Haplogroup E
Haplogroup E was first RFLP-defined as having +16389 HinfI and -7598 HhaI by Ballinger et al. (1992), who named it G, and then later it was renamed E by Torroni et al. (1994). As a loss of restriction sites can be produced by different nucleotide mutations within the recognition sequence, since the beginning, some G2 sequences characterized by the 7600 transition were erroneously classified as belonging to haplogroup E. Recently, based on the complete sequences of coding regions, Herrnstadt et al. (2002) defined three Asiatic lineages as E, although only one (sequence 214) seems to be a genuine representative. It possesses transition 7598, which, similar to 7600, is also detectable with HhaI as a site loss; and it also harbors mutations 10834 and 869, which were found by Ballinger et al. (1992) as -10830 HinfI and +868 DdeI in all and some individuals respectively classified as E. However, the inclusion of a Philippine complete sequence (Ingman and Gyllensten 2003) in our global tree clearly demonstrates that the last two mutations might only define a branch of E, as the Philippine sequence lacks both of them. On the contrary, in addition to 7598 and 16390, some of the four E mutations represented in Figure 1A before the branching point might be basic mutations. In Herrnstadt et al. (2002), sequence 169 belongs to Haplogroup M9 because it has all coding-region positions defining this haplogroup; and sequence 287 to M1 because it has 6446 and 6680, the coding-region mutations that define the basic branch of M1 (Fig. 1). It must be mentioned that the ambiguous Korean lineage classified as E/G by Schurr et al. (1999), because it had both the -7598 HhaI characteristic E site and the +4830 HhaI characteristic G site, has been recently found again in a Korean sample (Snäll et al. 2002). All of them are, in fact, members of subhaplogroup G2. It seems that haplogroup E has a southern Asia distribution. Until now it has been detected in the Malay peninsula populations and in the Sabah of Borneo (Ballinger et al. 1992); and it is also present in coastal Papua New Guinea (Stoneking et al. 1990) as well as in some Pacific islands such as Guam (Herrnstadt et al. 2002) and the Philippines (Ingman and Gyllensten 2003). However, until now, it has not been detected in more northern Continental populations or islands such as the Japanese archipelago.
Haplogroup M8
A monophyletic clade (Fig. 1A) groups M8a, C, and Z lineages. Mutations 4715, 15487T, and 16298 have been proposed as diagnostic for this clade (Yao et al. 2002a). The transversion 7196A and the transition 8584 should also be included in its definition (Fig. 1A; Kivisild et al. 2002). However, as the 248d is also shared by all Z and C lineages (Fig. 1A), a basal node defined by this deletion and named CZ has been recently proposed (Kong et al. 2003). Subhaplogroup C was RFLP-defined by Torroni et al. (1992) by +13262 AluI. Yao et al. (2002a) added 248d, 14318, and 16327 as characteristic of C. In addition, positions 3552A, 9545, and 11914 are also diagnostic of this clade (Fig. 1A; Kivisild et al. 2002). The Japanese TC52 has the C1 status and the Buryat 6970 and the Evenky 6979 have the C4 status proposed by Kong et al. (2003). Subhaplogroup Z was defined by Schurr et al. (1999) by the presence of the following noncoding motifs: 16185, 16223, 16224, 16260, and 16298. Recently, it was considered that only 16185 and 16260 mutations should be counted as basic for the group (Yao et al. 2002a). However, in full agreement with the characterization proposed on the basis of complete Chinese Z sequences (Kong et al. 2003), three additional mutations (6752, 9090, and 15784) have been placed on the basal branch of Z (Fig. 1A). We detected four Japanese Z clades that, in addition, shared mutation 152 and another without it. Tentatively, they have been named from Z1 to Z5 (Fig. 1A). Yao et al. (2002a) defined M8a by 14470, 16184, and 16319 transitions. Two more mutations (6179 and 8684) are also characteristic of this subhaplogroup (Kong et al. 2003). In Japanese we have found that 16184 is not harbored by all M8a members. Consequently, lineages with this mutation have M8a2 status and those lacking it M8a1 status (Fig. 1A). The largest diversities for C are in Korea (100%), central Asia (86%), and northern China (78%-74%). Therefore, C can be considered a clade with a Northeast Asian radiation. Representatives of subhaplogroup Z extend from the Saami (Finnilä et al. 2001) and Russians (Malyarchuk and Derenko 2001) of west Eurasia to the people of the eastern peninsula of Kamchatka (Schurr et al. 1999). Its largest diversities are found in Koreans (88%), northern China (73%), and central Asia (67%), compatible with a central-East Asian origin of radiation for this group. Finally, M8a has its highest diversity in Koreans (100%), and southern (100%) and eastern Chinese, including Taiwanese (73%). Thus, southeastern China was a potential focus of radiation of this group. All these subhaplogroups are present in mainland Japanese but neither in Ryukyuans nor in Ainu.
Haplogroup M7
This haplogroup was defined by Bamshad et al. (2001) as having two branches, M7a characterized by 16209 and M7b by 16297 transitions. Yao et al. (2002a) assigned mutations 199 and 9824 as basic for M7. However, our phylogenetic tree points to 6455 and 9824 as the basal mutations for this group, whereas 199 is only common to the M7b and M7c subgroups (Fig. 1A), which coincides with the phylogeny proposed by Kivisild et al. (2002). M7 can be RFLP-diagnosed by the lack of the 6451 MboII restriction site. The M7a subgroup can be defined by several codingregion positions (Fig. 1A; Kivisild et al. 2002). The M7b classification remains as proposed in Kivisild et al. (2002); but M7c has, in addition to 146 and 16295, three more coding-region substitutions (4850, 5442, and 12091) in its basal branch (Fig. 1A). At this point, it is worthwhile pointing out that the ambiguously assigned sequence 536 in Herrnstadt et al. (2002) belongs to M7c, as it has the five identifying coding-region mutations distinctive of this subhaplogroup. As for the geographic distribution, M7a1 has its highest frequencies (14%) and diversities (86%) in the Ryukyuans, and it is also very common in the whole of China, with a mean diversity of ∼76%. But, curiously, it has not been detected in Koreans or in Ainu, and is rare in mainland Japanese. In a similar way, M7a has its highest diversity in Ryukyuans (83%). Both groups are rather common in the Philippines. Although M7b has its greatest diversity in northern China (75%-62%), its derivative M7b2, has it again in Ryukyuans (100%), Koreans (53%), and mainland Japanese (45%). On the contrary, M7c is absent in Ainu and rare in mainland Japanese but very common in Sabah and the Philippines, although its highest diversity is in the whole of China (76% ± 11%).
Haplogroup M10
This haplogroup has been defined by substitutions 10646 and 16311 (Yao et al. 2002a). In addition, Kong et al. (2003) have found several new mutations in its basal branch that we confirm here (Fig. 1A). Minor modifications are that a new Japanese lineage shares with M10 only the 8793 mutation, and that a new mutation, 13152, seems to be basal for our M10 Japanese lineages. Although its highest frequency is in Tibetans (8%), the largest diversities are found in China. It is present in Koreans and mainland Japanese but has not been detected in either Ainu or Ryukyuans (Table 2).
Haplogroup M11
This haplogroup has been defined by Kong et al. (2003) by seven coding-region mutations (1095, 6531, 7642, 8108, 9950, 11969, and 13074) and four mutations in HVS-II (146, 215, 318, and 326). We confirm the same characterization for our M11 Japanese lineages. A subclade defined by mutation 14340 was found in Chinese (Kong et al. 2003), but it has not been detected in Japanese. In turn, Japanese have a new subclade characterized by mutation 14790. Finally, our data suggest that mutation 15924 is at the root of M11 and the new clade M12.
Haplogroup M12
This haplogroup has been defined in the present study. It harbors a characteristic motif (16145-16188-16189-16223-16381) in its noncoding region and several unique mutations in its coding region (Fig. 1A). Overall, it is a rare haplogroup, being detected only in mainland Japanese, Koreans, and Tibetans, the lastmentioned sample showing its highest frequency (8%) and diversity (50%).
Haplogroup M1
Although not present in eastern Asia, this haplogroup has been included in the phylogenetic tree of macrohaplogroup M to ascertain its hierarchical level with respect to other M clades. It was first detected in Ethiopia (Quintana-Murci et al. 1999) and defined by four transitions in the HVSI region (16129, 16189, 16249, and 16311). After this, M1 was also detected in the Mediterranean basin including Jordan (Maca-Meyer et al. 2001). Several mutations in the coding region are distinctive of this haplogroup (Fig. 1A). Its RFLP diagnosis is possible by an MnlI site loss at position 12401.
Subdivisions Within Macrohaplogroup N
Representatives of two major superhaplogroup N migratory branches are present in Japan. Two main clades, that directly sprout from the basal N trunk (A and N9), have a prevailing northern Asia dispersion, whereas the other two (B and F), having a southern radiation focus, belong to the derivative R clade, characterized by the loss of 16223 and 12705 mutations. Although not detected in Japan, to compare their hierarchical levels with those of the Asian branches, we have included the rCRS sequence and a N1b sequence (Kivisild et al. 1999) as representatives of the western Eurasian R and N clades, respectively.
Haplogroup A
This haplogroup was defined by an HaeIII site gain at 663 (Torroni et al. 1992). It was subdivided on the basis of HVSI motifs in A1 (16223-16290-16319) and A2 (16111-16223-16290-16319) by Forster et al. (1996). In our Japanese sample, we have detected several A1 representatives characterized by two substitutions (8563, 11536). Two of these lineages (ON67 and ND218) have been ascribed to the A1a subgroup that is defined by 4655, 11647, and 16187 substitutions. Two additional A1 Japanese clusters (A1b and A1c) have also been phylogenetically defined (Fig. 2). The A2 subgroup is represented in the tree by a Chukchi (6971) and two (KA21 and ON125) Japanese lineages, all sharing the 16362 mutation. As the Chukchi harbors the 16111 and 16265 mutations, it has been labeled as an A2a representative, as tentatively proposed by Saillard et al. (2000), having four additional mutations (152, 153, 8027, and 12007) in its basal branch. Owing to their phylogenetic position, three more Japanese lineages (ND28, TC48, and J42) should be classified as representatives of three new A subhaplogroups, respectively named A3, A4, and A5 (Fig. 2). Geographically, whereas A1 has a wide northern and central Asian distribution, subclade A1a is confined to Korea and mainland Japan. The greatest diversity for A1 is in central Asia (79%). In Japan it is present in both mainland and indigenous populations. Subhaplogroup A2 is mainly present in northeast Siberia including the Kamchatka peninsula, although a lineage has also been detected in Tibet. The main diversity (30%) and frequency (60%) for this subhaplogroup are in the Chukchi.
Subhaplogroups Y, N9a, and N9b
Haplogroup N9 characterized by the 5417 substitution (Yao et al. 2002a) phylogenetically comprises three subhaplogroups. Subhaplogroup N9a was mentioned as another N subcluster with a distinctive HVSI motif (16223, 16257A, 16261) by Richards et al. (2000). It appears named as N9a in Yao et al. (2002a), who added as basal substitutions 150 and 5231. Recently, Kong et al. (2003) added mutations 12358 and 12372 at the basal branch of N9a, which is according to our Japanese phylogeny (Fig. 2). A Japanese N9a1 lineage (TC2) shares mutations 4386, 12007, 16111, and 16129 with the Chinese lineage GD7834 of Kong et al. (2003). Three more N9a Japanese clusters sharing 16172 as their basal mutation have been considered distinct N9a2 branches (Fig. 2). Subhaplogroup Y was first identified by a set of HVSI polymorphisms (16126, 16189, 16231, 16266, 16519), an HaeIII site loss at 8391 and MboI and DdeI site gains at 7933 and 10394, respectively (Schurr et al. 1999). However, according to the classification of Kong et al. (2003), all these mutations define the Y1a1 branch specifically. Our Japanese (Fig. 2) and the Chinese (Kong et al. 2003) phylogenies characterize Y by seven mutations (8392, 10398, 14178, 14693, 16126, and 16231 gains and a 16223 loss). The branch Y1 would be identified by mutations 3834 and 16266, and the Y1a subcluster by 7933 (Fig. 2; Kong et al. 2003). In Japan we have found a new subclade (Y1b) characterized by four mutations (146, 10097, 15221, 15460). Furthermore, a new branch (Y2) with the same phylogenetic consideration as Y1, and distinguished by six basal mutations must be aggregated to the Y phylogeny (Fig. 2). Finally, we have detected a sister branch of Y in Japan. This new lineage, named N9b, shares two basal mutations (5147 and 16519) with Y and is further characterized by four (10607, 11016, 13183, 14893) additional mutations in its basal branch. All N9b1 representatives seem to have the 16189 mutation, and three branches of this trunk (a, b, and c) have been provisionally defined (Fig. 2). The geographic distribution of subhaplogroup Y is predominantly in Northeast Asia. The highest frequency (22%) is in the Ainu, although only one lineage accounts for this frequency. The greatest diversities are in northern China (80%), and this group is also very diverse in the Nivkhs from northeast Siberia (Torroni et al. 1993a). As for N9a, it has a great diversity in the whole of China (83%) and Korea (79%). In Japan, only mainland Japanese have N9a representatives. Finally, N9b is very scarce, being detected in southern China and Korea. Surprisingly, it is most abundant in the Japanese including the indigenous Ryukyans and Ainu.
Haplogroup F
This haplogroup was first defined as group A by Ballinger et al. (1992), and later renamed as F by Torroni et al. (1994). This group was characterized by the lack of HincII and HpaI sites at 12406. According to the newly proposed nomenclature (Kivisild et al. 2002; Kong et al. 2003), 12406 is now one of the six mutations that specifically define subhaplogroup F1. Recently, haplogroup F has been phylogenetically included as a subcluster of haplogroup R9 (Yao et al. 2002a). Besides F1, two new subgroups (F2 and F3) have been defined by Kong et al. (2003). We have found a new subcluster, named F4 (Fig. 2), that is characterized by three coding-region mutations (5263, 12630, 15670). This group has a particularly high incidence in Southeast Asia (Ballinger et al. 1992), but only subhaplogroup F1b is well represented in the Japanese, including the indigenous Ainu and Ryukyuan. The highest diversities for this subgroup are in eastern China including Taiwan (100%).
Haplogroup B
Renamed as B after Torroni et al. (1992), this haplogroup was identified by the presence of a 9-bp deletion in the COII/tRNALys intergenic region of mtDNA. This polymorphism was first detected in Asia by RFLP analysis (Cann and Wilson 1983). It was used to classify Japanese on the basis of the presence/absence of this deletion (Horai and Matsunaga 1986). Even in Asia, the monophyletic status of this cluster has been repeatedly questioned (Ballinger et al. 1992; Yao et al. 2000b); but although the 9-bp deletion has a high recurrence, it seems that together with transition 16189 it defines fairly well a monophyletic cluster, at least in eastern Asia. Recently, a sister clade of B, keeping the 16189 mutation but lacking the 9-bp deletion, has been detected in China, being designated as R11 (Kong et al. 2003). Asian subhaplogroups of B have been named as B4, identified by the 16217 mutation and B5, characterized by 10398 and 16140 mutations (Yao et al. 2002a). It has been deduced from analysis of complete sequences that transitions 709, 8584, and 9950 are also in the basal branch defining B5 (Fig. 2; Kong et al. 2003). Lower-level subdivisions have also been proposed. Three subclades (B4a, B4b, and B4c) were defined within B4 (Kong et al. 2003). At the same phylogenetic level are our Japanese branches named B4d, B4e, and B4f; and several new secondary clusters have also been detected in Japan within B4a, B4b, and B4c (Fig. 2). It is worthwhile to mention that those lineages harboring 16189, 16217, 16247, and 16261, also known as the Polynesian motif (Soodyall et al. 1995), belong to a branch of B4a, having in addition to 16247, 146, 6719, 12239, 14022, and 15746 as basic mutations. The B5 cluster was also subdivided in B5a and B5b on the basis of the HVSI mutations 16266A and 16243, respectively (Yao et al. 2002a), and reinforced with several additional positions after the analysis of complete Chinese (Kong et al. 2003) and Japanese (Fig. 2) sequences. Within B5b, new subdivisions are necessary to accurately classify the Japanese sequences (Fig. 2). Finally, on the basis of characteristic HVSI motifs, we had tentatively defined as B4a3 those lineages with 16189, 16217, 16261, and 16292 transitions. However, the phylogenetic position of a Chinese complete sequence (GD7812) belonging to this HVSI group (Kong et al. 2003) shows that a future redefinition of B4a might be necessary. The geographic distribution of haplogroup B is very complex. As expected from its age, the ancestral motif is widely distributed in Asia excluding Koryacks and other Siberians. The likewise old subhaplogroup B4 has mainly a central-eastern Asian distribution with diversities near 100% from central Asia to Japan. B4a shows a similar distribution as B4, having branches prevalent in Ryukyuans, Lahu of Yunnan, and aborigine Taiwanese (Table 2). In a similar vein, some branches of B4c are more abundant in southern areas (B4c2), whereas others (B4c1) are mainly detected in Korea and Japan, with derivatives in Taiwan (B4c1b). On the other hand, subhaplogroup B5a has its greatest diversity in southern-eastern China (89%), including Taiwan aborigines (67%), but its B5a1 derivative shows the greatest diversity in northern China (71%), being present in mainland Japanese. In turn, subhaplogroup B5b has its major diversity in Korea (83%) and also reached the Philippines (50%). Curiously, the B5b1 derivative shows its highest diversity (67%) and frequency (1%) in mainland Japanese.
Lineage Sorting and Population Pooling
A total of 110 clades with different phylogenetic range have been proposed on the basis of the pool of the eastern Asian complete sequences (Figs. 1A,B and 2). Of these subdivisions (Table 2), 83 have been used to classify all Asian partial sequences analyzed in this study. As a test of accuracy in the sorting of partial sequences into haplogroups, we classified our 672 Japanese complete sequences by using only their HVSI motifs and found that 34 of them (5%) had an ambiguous status or were misclassified. The main sources of errors were those sequences that differed from CRS in only one or two mutations. For instance, the 16223 mutation was found in M and N backgrounds. The 16189, 16223 motif can be D6 or N9b. Within M, sorting into D or G was one of the main sources of ambiguity. Some 16223, 16325, 16362 lineages were D4 and some G1. The motif 16114A, 16223, 16362, classified as D4, was in reality G3. Sometimes further subdivision within a haplogroup is rather difficult; for example, there are 16189, 16223, 16362 representatives in D4 and in D5. Because of recurrency and isolation, it can be expected that this uncertainty level increases with geographic distance. For instance, we have found that several 16129, 16223 Japanese lineages belong to D4, but to infer from this that southern Asian sequences with the same HVSI motif are also D4 would be inappropriate. From a total of 4713 sequences analyzed, 9.2% had an ambiguous status. In spite of this percentage there are enough sequences left to carry out population analysis with statistical confidence.
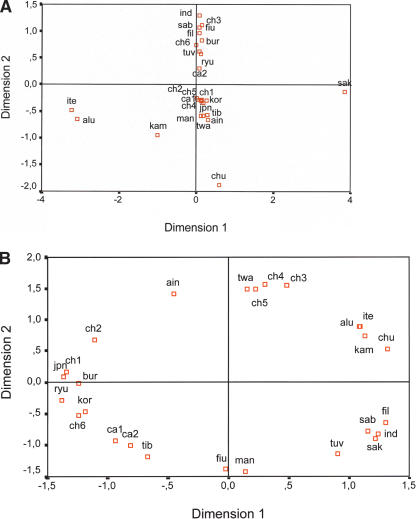
In a first approach, Japanese, Ainu, and Ryukyuan samples were compared with the rest of Asian samples shown in Table 3 by means of FST. The closest affinities of mainland Japanese were to three population groups. The first include Korean and Han from Shandong (mean P-value = 0.29 ± 0.06), the second Han from Liaoning and Xinjiang, and the Tu ethnic minority (0.20 ± 0.06), and the third Han from Xi'an and the Sali, a branch of the Yi ethnic group (0.15 ± 0.06). Ryukyuans and Ainu behave as outliers with significant differences with all the samples. Population groups resulting from the FST and CLUSTER analysis are defined in Table 3. Although mainland Japanese from Aichi were significantly different from other mainland Japanese because of their high frequency of haplogroup B, they were merged with them as JPN for comparisons with other areas. Control of the conglomerate number expected in CLUSTER analysis allows for a hierarchical grouping of populations. With two conglomerates, the first distinguished isolate was the aboriginal Sakai from Thailand (Fucharoen et al. 2001). This group was unique among other Thai people owing to its lack of lineages with the 9-bp deletion that characterizes haplogroup B, and to the high frequency of the authors' C6 cluster (included in our D4a). The lack of any representative of macrohaplogroup N in a population anthropologically considered one of the oldest groups in Thailand, if not caused by genetic drift, is compatible with the hypothesis that derivatives of macrohaplogroup N had, in southern Asia, a different route from macrohaplogroup M (Maca-Meyer et al. 2001). Also striking is the presence in Sakai of an unequivocal representative (16223-16274-16278-1629416309) of the sub-Saharan African L2a haplogroup (Torroni et al. 2001), which again is compatible with the physical characteristics of this Negrito group. Although the suggestion that the first spreading out of Africa of modern humans could have carried some L2 lineages in addition to the L3 ancestors (Watson et al. 1997) is a tempting explanation, a recent admixture is more in consonance with the phylogenetic proximity of this lineage to the present African ones. The next outsiders were the majority of the Siberian isolates, which could not be pooled because of big differences in the frequency of distinctive haplogroups (Table 2). This considerable differentiation was already emphasized (Schurr et al. 1999), with strong genetic drift being its most probable cause. Subsequent isolates belong to some Chinese minorities such as those of Lisu and Nu, Lahu, and Taiwanese aborigines. Unexpectedly, other Chinese minorities (Bai, Sali, and Tu) were left in Han Chinese northern clusters. The Bai belong to the Sino-Tibetan Tibeto-Burman ethnic linguistic group and have been strongly influenced by Han. The Sali are a minority within the Yi ethnic group whose most probable ancestors were the Qiang from northwest China. Finally, the Tu, although belonging to the Mongolian branch of the Altaic Family, show their main genetic affinities to the Han from Xi'an (P = 0.95), Xinjiang (P = 0.89), and Shanghai (P = 0.79), all of them clustered in the Ch2 group. On the other hand, Thais, Vietnamese, and Cambodians joined with southern Chinese. As already observed (Chunjie et al. 2000; Yao et al. 2002a), the Han Chinese do not comprise a homogeneous group. With the exception of cluster Ch4, that includes samples from Hubei and Guandong (Table 3), they appear geographically differentiated. The two central Asian groups detected mainly differ in their frequencies for A1b, Z, and G2a. With less than 14 conglomerates, the Japanese, including Ainu and Ryukyuans, were part of a big group formed by Korean, Buryat, Tibetans, and northern Chinese. Ainu was the first differentiated Japanese sample. Ryukyuans separated later, when mainland Japanese and Koreans still comprised a single group. The lack of homogeneity between Ainu and Ryukyuans was pointed out by Horai et al. (1996), who questioned that they shared a recent common ancestor. The main differences between them were attributed to two dominant clusters (C1 and C16, corresponding to our Y and M5/D4a/G1, respectively) present in Ainu but absent in Ryukuyans, and two Ryukyuan dominant clusters (C3 and C13, belonging to our R and M, respectively) absent in Ainu. In addition, applying the present haplogroup nomenclature to the same data, the high frequency of M7a1 and D4a1/D4b in Ryukyuans, but their absence in Ainu, stands out. The MDS plot (Fig. 3A), based on FST haplogroup frequency distances between final groups (data not shown), only partially reflects the sequential process described above, as only Sakai and Siberians are well differentiated from the rest. On the contrary, relationships obtained from haplotype matches (Fig. 3B) show populations highly structured by geography with the only exceptions being the Ainu and Tuvinian isolates.
Table 3.
Asian Populations Used in This Study
| Population | Locality | Ethnic group | Group | Sample | HVRI | HVRII | Othera | References |
|---|---|---|---|---|---|---|---|---|
| Japan | Tokyo | Japanese | JPN | 373 | 16024-16569 | 1-648 | 649-16023 | This work |
| Japan | Nagoya | Japanese | JPN | 299 | 16024-16569 | 1-648 | 649-16023 | This work |
| Japan | Japanese | JPN | 20 | 1600-16413 | — | Bamshad et al. 2001 | ||
| 19 | — | 71-270 | Jorde et al. 1995 | |||||
| Japan | Tokyo | Japanese | JPN | 162 | 16051-16365 | 73-340 | Imaizumi et al. 2002 | |
| Japan | Tokyo | Japanese | JPN | 150 | 16030-16481 | Nishimake et al. 1999 | ||
| Japan | Tokyo | Japanese | JPN | 13 | 16024-16569 | 1-648 | RFLPs | Abe et al. 1998 |
| Japan | Miyazaki | Japanese | JPN | 100 | 15998-16400 | 30-407 | Seo et al. 1998 | |
| Japan | Tottori | Japanese | JPN | 89 | 16026-16396 | Oota et al. 2002 | ||
| Japan | Shizuoka | Japanese | JPN | 62 | 16129-16569 | 1-41 | Horai et al. 1996 | |
| Japan | Aichi | Japanese | JPN | 50 | 16040-16375 | 20-430 | Koyama et al. 2002 | |
| Japan | Okinawa | Ryukyuan | RYU | 50 | 16129-16569 | 1-41 | Horai et al. 1996 | |
| Japan | Hokkaido | Ainu | AIN | 51 | 16129-16569 | 1-41 | Horai et al. 1996 | |
| Korea | Korean | KOR | 306 | 16020-16400 | 1-70 | Lee et al. 1997 | ||
| Korea | Korean | KOR | 4 | 16024-16370 | Torroni et al. 1993a,b | |||
| Korea | Korean | KOR | 60 | 16024-16365 | 73-340 | Pfeiffer et al. 1998 | ||
| Korea | Korean | KOR | 2 | 16000-16413 | — | Bamshad et al. 2001 | ||
| Korea | — | 71-270 | Jorde et al. 1995 | |||||
| Korea | Korean | KOR | 64 | 16129-16569 | 1-41 | Horai et al. 1996 | ||
| Korea | Korean | KOR | 3 | 16128-16408 | Horai and Hayasaka 1990 | |||
| Korea | Korean | KOR | 98 | 16075-16362 | 73-315 | 14747-15887 | Lee et al. 2002 | |
| China | Liaoning | Han | Ch1 | 51 | 16001-16497 | 30-47 | 10171-10659 and RFLPs | Yao et al. 2002a |
| China | Shandong | Han | Ch1 | 50 | 16001-16497 | 30-47 | 10171-10659 and RFLPs | Yao et al. 2002a |
| China | Yunnan | Bai | Ch1 | 31 | 16001-16495 | Yao et al. 2002b | ||
| China | Changsha | Han | Ch1 | 82 | 16026-16396 | Oota et al. 2002 | ||
| China | Xinjiang | Han | Ch2 | 47 | 16001-16497 | 30-47 | 10171-10659 and RFLPs | Yao et al. 2002a |
| China | Yunnan | Sali | Ch2 | 31 | 16001-16495 | Yao et al. 2002b | ||
| China | Qinghai | Tu | Ch2 | 35 | 16001-16495 | Yao et al. 2002b | ||
| China | Xi'an | Han | Ch2 | 84 | 16026-16396 | Oota et al. 2002 | ||
| China | Shanghai | Han | Ch2 | 120 | 13030-16481 | Nishimake et al. 1999 | ||
| Mongolia | Mongolian | Ch2 | 103 | 16020-16400 | RFLPs | Kolman et al. 1996 | ||
| Mongolia | Mongolian | Ch2 | 15 | 16001-16495 | Yao et al. 2002b | |||
| China | Yunnan | Lahu | Ch3 | 32 | 16048-16569 | 1-49 | Qian et al. 2001 | |
| China | Hubei | Han | Ch4 | 42 | 16001-16497 | 30-47 | 10171-10659 and RFLPs | Yao et al. 2002a |
| China | Guangdong | Han | Ch4 | 30 | 16001-16497 | 30-47 | 10171-10659 and RFLPs | Yao et al. 2002a |
| China | Yunnan | Han | Ch5 | 43 | 16001-16497 | 30-47 | 10171-10659 and RFLPs | Yao et al. 2002a |
| China | Taiwan | Ch5 | 6 | 16024-16370 | Torroni et al. 1993a,b | |||
| China | Taiwan | Ch5 | 3 | 15999-16413 | Bamshad et al. 2001 | |||
| China | Taiwan | Ch5 | 9 | 16065-16375 | Sykes et al. 1995 | |||
| China | Taiwan | Ch5 | 66 | 16129-16569 | 1-41 | Horai et al. 1996 | ||
| China | Taiwan | Han | Ch5 | 155 | 15997-16569 | 1-407 | Tsai et al. 2001 | |
| China | Yunnan | Dai | Ch5 | 21 | 16048-16569 | 1-49 | Qian et al. 2001 | |
| China | Yunnan | Wa | Ch5 | 22 | 16048-16569 | 1-49 | Qian et al. 2001 | |
| China | Yunnan | Dai | Ch5 | 38 | 16001-16495 | Yao et al. 2002b | ||
| China | Guangxi | Zhuang | Ch5 | 83 | 16001-16495 | Yao et al. 2002b | ||
| China | South China | Han | Ch5 | 28 | 16024-16399 | Betty et al. 1996 | ||
| Thailand | Ch5 | 32 | 16001-16495 | Yao et al. 2002b | ||||
| Thailand | See ref. | Ch5 | 121 | 16048-16569 | 1-41 | Fucharoen et al. 2001 | ||
| Thailand | See ref. | Native | Ch5 | 74 | 16048-16569 | 1-41 | Fucharoen et al. 2001 | |
| Vietnam | Ch5 | 35 | 16026-16396 | Oota et al. 2002 | ||||
| Vietnam | Ch5 | 9 | 15999-16413 | — | Bamshad et al. 2001 | |||
| — | 71-270 | Jorde et al. 1995 | ||||||
| Cambodia | Ch5 | 12 | 15999-16413 | — | Bamshad et al. 2001 | |||
| — | 71-270 | Jorde et al. 1995 | ||||||
| China | Yunnan | Lisu | Ch6 | 37 | 16001-16495 | Yao et al. 2002b | ||
| China | Yunnan | Nu | Ch6 | 30 | 16001-16495 | Yao et al. 2002b | ||
| China | Taiwan | Native | TWA | 28 | 15997-16400 | 30-407 | Melton et al. 1998 | |
| China | Taiwan | Native | TWA | 180 | 16048-16569 | 1-41 | Tajima et al. 2003 | |
| Central Asia | Uygur | CA1 | 46 | 16001-16495 | Yao et al. 2000a | |||
| Kazagstan | Kazakh | CA1 | 55 | 15997-16400 | Comas et al. 1998 | |||
| Kirgizistan | Talas | Kirghiz | CA1 | 48 | 15997-16400 | Comas et al. 1998 | ||
| Kazagstan | Uygur | CA1 | 55 | 15997-16400 | Comas et al. 1998 | |||
| Central Asia | Kazak | CA2 | 30 | 16001-16495 | Yao et al. 2000a | |||
| Kirgizistan | Sary-Tash | Kirghiz | CA2 | 46 | 15997-16400 | Comas et al. 1998 | ||
| Siberia | See ref. | Altai | CA2 | 17 | 16024-16383 | Shields et al. 1993 | ||
| Tibet | Tibetan | TIB | 1 | 16024-16370 | Torroni et al. 1993a,b | |||
| Tibet | Tibetan | TIB | 40 | 16001-16495 | Yao et al. 2000b | |||
| Tibet | Tibetan | TIB | 24 | 16048-16569 | 1-41 | Qian et al. 2001 | ||
| Russia | East Ural | Mansi | MAN | 98 | 16039-16519 | 64-295 | RFLPs | Derbeneva et al. 2002a |
| Siberia | Finno-Ugrian | FIU | 38 | 13021-16505 | Voevoda Accession nos. AF214068-AF214105 | |||
| South Siberia | Tuvinian | TUV | 36 | 16000-16400 | RFLPs | Derenko et al. 2000 | ||
| South Siberia | Buryat | BUR | 40 | 16000-16400 | RFLPs | Derenko et al. 2000 | ||
| Siberia | Chukchi | CHU | 60 | 16001-16405 | Voevoda et al. 1994 | |||
| Siberia | Aluitor | Koryak | ALU | 56 | 16000-16525 | Schurr et al. 1999 | ||
| Siberia | Karagin | Koryak | KAM | 37 | 16000-16525 | Schurr et al. 1999 | ||
| Siberia | Palan | Koryak | KAM | 54 | 16000-16525 | Schurr et al. 1999 | ||
| Siberia | Kovran | Itel men | ITE | 46 | 16000-16525 | Schurr et al. 1999 | ||
| Philippine | FIL | 32 | 16065-16375 | Sykes et al. 1995 | ||||
| Thailand | Trang | Sakai | SAK | 20 | 16048-16569 | 1-41 | Fucharoen et al. 2001 | |
| Malaysia | IND | 6 | 15999-16413 | — | Bamshad et al. 2001 | |||
| — | 71-270 | Jorde et al. 1995 | ||||||
| Indonesia | IND | 34 | 16024-16400 | 31-407 | Redd and Stoneking 1999 | |||
| Borneo | Sabah | SAB | 34 | 16065-16375 | Sykes et al. 1995 |
RFLPs and additional sequences.
Figure 3.
MDS plots based on (A) FST and (B) D match distances. Population groups are as detailed in Table 3.
The Peopling of Japan
To further know the relative affinities of the Japanese between themselves and with the different Asian groups formed, the data obtained from the global approaches based on haplogroup frequency distances and on sequence match identities are presented in Table 4. Both values are moderately correlated in the comparisons involving the mainland Japanese (r = -0.479; two-tail probability 0.012) but not at all in those involving aborigine Ryukyuans (r = -0.310; two-tail probability 0.115) and Ainu (r = 0.087; two-tail probability 0.667). This result can be explained by assuming that these aboriginal people have suffered important genetic drift effects with substantial changes in haplogroup frequencies and lineage losses or, less probably, that these populations have been isolated long enough to have accumulated new variation. Results based on haplogroup frequencies by far relate mainland Japanese to Koreans followed by northern Chinese. Ryukyuans present the smallest distances to Buryats from South Siberia, followed in short by southern Chinese. In turn, the Ainu have their closest affinities with mainland Japanese, Koreans, and northern Chinese. As regards sequence matches, mainland Japanese also joins first to Koreans and second to Buryats. Aborigine Ryukyuans are closest to Buryats and then to Koreans. Finally, Ainu show comparatively less shared sequences, their greater affinities being toward Chukchi and Koryaks of Kamchatka. This global picture is congruent with an important influence on mainland Japanese from northern Asian populations through Korea, that the Ryukyuans had a dual northern and southern Asian background previous to the new northern influences acquired by admixture with mainland Japanese, and that the Ainu represent the most isolated group in Japan in spite of the genetic input received from Kamchatka. Also noticeable is the great distance and low identity values obtained for the Ainu-Ryukyuan pair compared with those obtained in their respective comparison to mainland Japanese, which is another hint of its notable maternal isolation.
Table 4.
Frequency-Based FST and Sequence Match Identities (In Percentage) Between Japanese Samples and With Other Asian Populations
| JPN
|
RYU
|
AIN
|
||||
|---|---|---|---|---|---|---|
| FST | Matches | FST | Matches | FST | Matches | |
| RYU | 0.04 | 0.41 | ||||
| AIN | 0.04 | 0.33 | 0.05 | 0.04 | ||
| KOR | 0.00 | 1.10 | 0.04 | 0.57 | 0.04 | 0.25 |
| CH1 | 0.01 | 0.59 | 0.04 | 0.11 | 0.04 | 0.18 |
| CH2 | 0.01 | 0.51 | 0.05 | 0.19 | 0.05 | 0.21 |
| CH3 | 0.07 | 0.01 | 0.10 | 0.00 | 0.08 | 0.00 |
| CH4 | 0.03 | 0.06 | 0.03 | 0.00 | 0.05 | 0.03 |
| CH5 | 0.03 | 0.16 | 0.03 | 0.09 | 0.05 | 0.08 |
| CH6 | 0.04 | 0.01 | 0.08 | 0.00 | 0.08 | 0.09 |
| TWA | 0.04 | 0.23 | 0.07 | 0.08 | 0.08 | 0.04 |
| TIB | 0.04 | 0.36 | 0.04 | 0.18 | 0.08 | 0.06 |
| CA1 | 0.02 | 0.58 | 0.04 | 0.25 | 0.05 | 0.16 |
| CA2 | 0.04 | 0.73 | 0.07 | 0.20 | 0.08 | 0.19 |
| ITE | 0.29 | 0.00 | 0.39 | 0.00 | 0.40 | 0.26 |
| FIU | 0.06 | 0.50 | 0.08 | 0.32 | 0.10 | 0.10 |
| MAN | 0.06 | 0.24 | 0.06 | 0.24 | 0.08 | 0.04 |
| ALU | 0.29 | 0.01 | 0.39 | 0.00 | 0.39 | 0.46 |
| KAM | 0.14 | 0.01 | 0.16 | 0.00 | 0.15 | 0.45 |
| CHU | 0.17 | 0.01 | 0.21 | 0.00 | 0.22 | 0.00 |
| TUV | 0.03 | 0.09 | 0.07 | 0.17 | 0.07 | 0.05 |
| BUR | 0.03 | 0.97 | 0.02 | 2.75 | 0.07 | 0.15 |
| FIL | 0.03 | 0.11 | 0.05 | 0.13 | 0.06 | 0.00 |
| IND | 0.09 | 0.04 | 0.09 | 0.00 | 0.11 | 0.00 |
| SAK | 0.29 | 0.00 | 0.44 | 0.00 | 0.43 | 0.00 |
| SAB | 0.06 | 0.09 | 0.05 | 0.29 | 0.08 | 0.12 |
The distance and identity statistics used above are based on frequencies of haplogroups and haplotypes, respectively; however, frequencies are more affected by genetic drift than the number of different haplotypes present in a population. To measure the relative affinities of Japanese populations between them and to Continental Asia in a frequency-independent way, we chose a haplotype-sharing approach calculating the relative contribution of lineages shared with other areas to the number of different haplotypes present in each Japanese population. In these comparisons all other Asians were merged. Table 5 shows the results of this analysis. Note that despite the difference in sample size the haplotype frequency in mainland Japanese and Ainu is ∼50%, whereas in Ryukyuans it is 84%; which means that, if there was not a bias in the sampling process, in spite of its small size, the Ainu sample seems to be representative of that population. However, it would be desirable to enlarge that of the Ryukyuans (Helgason et al. 2000). Haplotypes present only in a given population account for 13% in Ainu but ∼50% in mainland Japanese (60%) and Ryukyuans (45%). This finding once more points to the existence of important drift effects in Ainu. Mainland Japanese exclusively share with Ryukyuans and Ainu only 3% and 2%, respectively, of its lineages, which could reach 6% and 3% if those also shared with Continental Asian populations are added. In comparison they shared 21% of its lineages with other Asians. On the contrary, Ryukyuans and Ainu share about 50% of their lineages with mainland Japanese and only 10% and 21%, respectively, with Continental populations, which may reflect other independent Asian influences on Japan. With respect to those lineages exclusively shared by Japanese and Continental Asian populations, it is worth mentioning that, again, Korea is the main contributor, participating in ∼50% of the haplotype sharing with mainland Japanese (55%), as much as with Ryukyuans (50%) and Ainu (50%). However, differences exist in the provenance of the rest of the shared lineages. Whereas in Ainu (northern China and Siberia) and in Ryukyuans (northern China and central Asia) they are from northern areas, the second region contributing to mainland Japanese is southern China (17.5%), followed, at the same level (12.5%), by northern China and central Asia. In addition, there exists a minor percentage of exclusive sharing with Indonesia (2.5%). On the other hand, all the matches with Siberia and Tibet are also shared with other populations. From these results, it can be deduced that the ancient Japanese inhabitants came from northern Asia and that southern areas affected the Japanese by later immigration. Nevertheless, it must be borne in mind that older influences could be undetectable by lineage sharing. With respect to the haplogroup affiliation of those lineages that Ainu and Ryukyuans exclusively shared with no Japanese samples, new differences appear between them. Ainu share derived lineages of haplogroups A, G, M9, and D5, all of them compatible with a rather recent Siberian influence. In contrast, those shared by Ryukyuans are basical M lineages, more congruent with an older radiation from southern China. These dual influences are also detected when the haplogroup affiliation of the Ainu and Ryukyuan unique lineages is studied. First, the percentage of lineages belonging to macrohaplogroup N is larger in Ainu (50%) than in Ryukyuans (15%) and from a different provenance, as those in Ainu are from haplogroups N, N9b, and Y, whereas those of Ryukyuans belong to the southern haplogroups F and B. The remaining 50% of the Ainu lineages equitably belong to different M haplogroups (M, M7c, G1, and D5a), but in Ryukyuans the remainder are mainly concentrated in M7a (41%) and M7b2 (18%), two groups that have their greatest Asian diversities precisely in Ryukyuans. Although an indigenous focus of radiation cannot be discarded, it is more conservative to suppose that the most probable origin of these lineages is again southern China. Thus, Ainu and Ryukyuans are not only largely isolated populations, but they most probably had different maternal origins.
Table 5.
Distribution of Unique and Shared Haplotypes in Japanese Populations
| Japanese populations
|
|||
|---|---|---|---|
| JPN | RYU | AIN | |
| Sample | 1318 | 50 | 51 |
| Haplotypes | 626 | 42 | 24 |
| Haplotype frequency | 0.48 | 0.84 | 0.47 |
| Singleton + Unique | 377 (0.60) | 19 (0.45) | 3 (0.13) |
| Shared | 249 (0.40) | 23 (0.55) | 21 (0.87) |
| JPN | 137 (0.22) | 20 (0.48) | 13 (0.54) |
| RYU | 20 (0.03) | 1 (0.02) | 1 (0.04) |
| AIN | 13 (0.02) | 1 (0.02) | 5 (0.21) |
| Othera | 130 (0.21) | 4 (0.10) | 5 (0.21) |
Other Asians.
Although no matches are involved, the geographic distribution of haplogroup frequency and diversities for some groups present in Japan and in other distinct Asian areas are also relevant to trace these older connections. For instance, haplogroups M9, M10, M12, D4b, and F1c have correlated geographic frequencies with a peak in an area that comprises Tibet (Table 2). Curiously, one of these haplogroups (M12) is today absent in China but present in Korea and Japan.
DISCUSSION
Although the recent out-of-Africa origin for all modern humans (Cann et al. 1987) is being widely supported (Takahata et al. 2001), the most probable time and routes chosen by these earliest migrants to reach eastern Asia is an open issue. In the following discussion we weigh the different alternatives proposed in light of the phylogenetic tree obtained from complete mtDNA sequences. One of the first questions raised was whether there was more than one out-of-Africa dispersion. All the mtDNA lineages detected in Old World populations belong to one of two M and N macrohaplogroups with only secondary representatives in Africa. The proposed radiation ages for both, 30,000 to 58,000 years ago and 43,000 to 53,000 years ago, respectively (Maca-Meyer et al. 2001), give a temporal frame compatible with only one main dispersion or two successive dispersions, in which case the M precursor is the most probable candidate for the older exit. Even if the one dispersion option is chosen, more than one geographical route to eastern Asia is possible. In fact, a northern Continental route through the Near East and western-central Asia and a southern coastal route through the Arabian and Indian peninsulas have been proposed (Cavalli-Sforza et al. 1994; Kivisild et al. 1999). The geographical distribution of these two macrohaplogroups, with lack of ancient M representatives and the presence of deep N lineages in western Asia, and the abundance of basal M lineages in India and southwestern Asia and concomitant lack of equivalent-age N clades, gave rise to the hypothesis that N represents the main footprint of the northern Continental expansion, whereas M is the equivalent footprint for the southern coastal expansion. The presence of N and M lineages in alternative areas has been explained to have been the result of secondary migrations (Maca-Meyer et al. 2001). However, another plausible explanation is that both M and N reached southern Asia at the same time, quickly expanding to Papua New Guinea (PNG) during maximal glacial ages when the permafrost boundary precluded a northern human occupation. During postglacial ages, subsequent migrations northward carried derivatives of both macrohaplogroups to northern Asia (Forster et al. 2001). Nevertheless, under this second hypothesis, the presence of basal N clusters should be expected in India, southern Asia, and PNG; but this is not the case. All N representatives in India belong to R, a clade derived from N by the loss of 16223 and 12705 mutations (Fig. 2). In addition, the bulk of these Indian lineages belong to western Caucasian haplogroups that, most probably, reached India as the result of secondary immigrations, as has already been proposed (Kivisild et al. 1999; Bamshad et al. 2001). Similarly, the N representatives in southern Asia belong to haplogroups F and B, two sister clades also derived from R (Fig. 2). Furthermore, when totally sequenced PNG N lineages (Ingman et al. 2000; Ingman and Gyllensten 2003) are added to the N phylogenetic tree (data not shown), they form three monophyletic clades that have their roots in the derived R trunk. On the contrary, the geographically northern Asian clades A, N9a, N9b, and Y (Fig. 2) and the western Eurasian clades W, N1b, I, and X all split from the basal N root (Maca-Meyer et al. 2001), although A, N9a, N9b, and Y radiations were delayed congruent with subsequent northern Asian expansions. Therefore, at present, mtDNA data are compatible with the supposition that the northern route, harboring mainly N precursors, met climatic difficulties and when they finally reached Southeast Asia, the M representatives, brought by the southern route, had already colonized the area. This southern expansion of N derivatives has, as a lower temporal boundary, the coalescence ages of F, B, and PNG R haplogroups being ∼46,000 ± 10,000 years ago. However, when recently published (Ingman et al. 2000; Ingman and Gyllensten 2003) Australian N lineages are taken into account, it seems evident that the real situation could be far more complex than the one migration-one lineage hypothesis. Australian N lineages directly sprout from the basal trunk (data not shown). They most probably differentiated in that continent, supporting the idea that ancestral N lineages reached Australia but not PNG, although the undemonstrable possibility of lineage extinctions and subsequent recolonization events in PNG can be an argument. Both hypotheses have difficulties to explain the presence of ancient N lineages in Australia. If the two, M and N lineages, were brought with the southern coastal dispersion, the lack of primitive N in India, southern Asia, and PNG has to be explained by the subsequent loss of all N lineages carried to Australia; if the northern Continental route of N is favored, the loss of N representatives in all populations formed in route to Australia has also to be explained. Recently, an N lineage has been detected in Chenchus, a southern Indian tribal group (Kivisild et al. 2003). From the information published, it can be deduced that this lineage only shares mutation 1719 with the western Eurasian Nb1/I and X clades. More extensive studies of populations in southern India and southern and central Asia would add empirical support to any of these theories.
Concerning macrohaplogroup M, it has already been commented that the star radiation of all the main Indian and southeast Asian M clades strongly suggests that this wide geographic colonization could have happened in a relatively short time (Maca-Meyer et al. 2001). This star radiation includes the Australian and PNG M complete sequences recently published (Ingman et al. 2000; Ingman and Gyllensten 2003). However, for those clades and subclades with later northward expansions, long radiation delays are observed. For instance, whereas M7 and M8 have coalescence ages ∼35,000 to 45,000 years ago, other groups such as G, D4, M7a, or M7c have coalescence ages ∼15,000 to 30,000 years ago, more in frame with those calculated for A, Y, and N9 derivates, which, although belonging to macrohaplogroup N, share with them a central-northern Asian geographic distribution (see Supplemental material). It seems that the simultaneous lineage bursts ∼60,000 to 70,000 years ago from Africa (Maca-Meyer et al. 2001), ∼30,000 to 55,000 years ago for macrohaplogroups M and N, and ∼15,000 to 30,000 years ago for clusters with prominent central-northern Asian radiations were related to main climatic changes. The role of selection in these expansions is an open question (Elson et al. 2004; Ruiz-Pesini et al. 2004).
The application of global pairwise-distance and detailed phylogeographic methods to the peopling of Japan shows that both approaches have different grasps but together demonstrate that the actual Japanese population is the result of a complex demographic history, from which the different theories proposed to explain it only emphasize partial aspects. Global distances and detailed haplotype comparisons confirm that Ainu and Ryukyuans are heterogeneous populations (Horai et al. 1996) and that both are well differentiated from the mainland Japanese. In spite of this, they have common peculiarities such as having the highest frequencies in Asia for M7a, M7b2, and N9b, shared with mainland Japanese. Furthermore, for both, their closest relatives are northern populations. At first sight, these results are against a supposed southern origin for the Paleolithic Japanese, favoring the replacement theory or even that the Paleolithic inhabitants of Japan came from northeastern Asia (Nei 1995). Although based on a single locus, our results are strikingly coincident with the previously proposed northern origin and influences received by the Japanese. In an early study using serum gammaglobulin polymorphisms, it was concluded that the homeland of all Japanese could have been in the Lake Baikal area in Siberia (Matsumoto 1988), which agrees with the close proximity found here between Buryats and Ryukyuans or mainland Japanese. More recently, classical markers (Omoto and Saitou 1997) and mtDNA (Horai et al. 1996) studies demonstrated that the Japanese are most closely related to the Koreans, which is also true in our global analysis. It can be added that a substantial part of this common maternal pool has recent roots, as Korea specifically shares with Ainu, mainland Japanese, and Ryukyuans 10%, 7%, and 5%, respectively, of their haplotypes. This particular affinity is increased with the existence of derived lineages only detected (A1a, B4c1, B4f) or mainly detected (N9b, B4a1, B4b1, G1a, M7b2, M12) in Japanese and Koreans. This Korean influence has been attributed to the archeologically well-documented Continental immigration to Japan during the Yayoi period (Horai et al. 1996). However, specific haplotype matches with other areas increases the geographic range of these recent influences. Thus, mainland Japanese share part of their haplotypes exclusively with South China (2.5%), North China (1.5%), Central Asia (1.5%), and Indonesia (0.3%); and, also, Ryukyuans have specific affinities with North China (2.4%) and Central Asia (2.4%). The recent Siberian input on the Ainu has also been stressed (Schurr et al. 1999). At least, another independent migratory wave from central Asia also affected mainland Japanese. It was first detected by the peculiar distribution of the Y-chromosome marker YAP+, and seems to have originated in an area including Tibet (Su et al. 2000). Haplogroup M12 is its mitochondrial counterpart. As with the Y-chromosome marker, its punctual presence in Tibet and eastern Asia might be explained as the result of subsequent migrations in the Continent that erased the route followed by the people harboring these markers. In addition, there are clues, at least in Ryukyuans, that a substantial part of their maternal pool had an ancient southern Asian provenance. This fraction is represented by the M, M7a, and M7a1 basic lineages (31%), which the Ryukyuans do not share with northern populations. This southern signal is, in part, congruent with the southern Asian origin for the Paleolithic Japanese proposed by the dual structure model (Hanihara 1991). Furthermore, the fact that the highest diversities for M7a, M7a1, and M7b2 have been found in Ryukyuans and for N9b and B5b2 in Japan raises the possibility that this area was within a focus of migratory radiations to northern and southern isles and even to the mainland from Paleolithic to recent times. The significant latitudinal clines detected in Japan for some genetic markers (Orito et al. 2001; Takeshita et al. 2001) could also be explained as the result of southern and northern influences on Japanese. Finally, some mtDNA results obtained from ancient Jomon remains (Horai et al. 1991; Shinoda and Kanai 1999; K.-I. Shinoda, unpubl.) are congruent with a genetically diverse background for the Paleolithic Japanese population (Horai et al. 1996). A tentative comparison of Jomon with present-day Japanese populations based on shared lineages (data not shown) significantly relates Jomon first to the indigenous Ainu and then to Ryukyuans and last to mainland Japanese. In summary, Japan could have received several northern and southern Asian maternal inputs since Paleolithic times, with notable northern Asian immigrations through Korea in the late Neolithic and more specific gene flows from western Asia, Siberia, and southern islands.
METHODS
Samples
Complete mtDNA sequences were obtained from a total of 672 unrelated Japanese including 373 from Tokyo and 299 from the Nagoya area. All subjects gave their written consent to participate in this study, which was approved by the Ethical Committees of the Gifu International Institute of Biotechnology and collaborative institutions. The sources of 11 additional complete sequences used to build the final phylogenetic trees are in Table 1. For the analysis of the peopling of Japan, we used a total of 1438 Japanese and 3275 central and eastern Asian HVI sequences, as detailed in Table 3.
Isolation and Amplification of DNA
Total DNA was extracted from the blood with either Dr. Gen TLE (Takara) or MagExtractor System MFX-2000 (Toyobo). The entire mitochondrial genome was amplified as six fragments (∼3000-3400 bp) by the first PCR and 60 overlapping segments (∼600-1000 bp) by the second PCR. The primer pairs and their nucleotide sequences were described previously (Tanaka et al. 1996). The conditions for the first and second PCR were the same: an initial denaturation step for 5 min at 94°C, followed by 40 cycles of denaturation for 15 sec at 94°C, annealing for 15 sec at 60°C, and extension for 3 min at 72°C, with a final extension for 10 min at 72°C. The amplified fragments were analyzed by electrophoresis on a 1% agarose gel and visualized by staining with ethidium bromide. These second PCR products were purified by use of the MultiScreen-PCR Plates (Millipore). The quality of DNA templates was examined by electrophoresis on a 1.2% agarose gel after staining with ethidium bromide by use of a Ready-To-Run Separation Unit (Amersham Pharmacia Biotech).
Sequence Analysis of Mitochondrial DNA
Sequence reactions were carried out with a BigDye terminator cycle sequencing FS ready reaction kit (Applied Biosystems). After excess dye terminators had been removed with MultiScreen-HV plates (Millipore) packed with Sephadex G50 superfine (Pharmacia), the purified DNA samples were precipitated with ethanol, dried, and suspended in the template suppression reagent (TSR) or formamide from Applied Biosystems. The dissolved DNA samples were heated for 2 min at 95°C for denaturation, then immediately cooled on ice. Sequences were analyzed with automated DNA sequencers 377 and 310 by use of Sequencing Analysis Program version 4.1 (Applied Biosystems). A computer program, Sequencher version 4.1 (Gene Codes Co.), was used to indicate possible single nucleotide polymorphism (SNP) loci. For verification, visual inspection of each candidate SNP was carried out. At least two overlapping DNA templates amplified with different primer pairs were used for identification of each SNP. Mitochondrial SNPs (mtSNPs) were identified by comparison with the revised Cambridge sequence (rCRS) reported by Andrews et al. (1999).
Phylogenetic Analysis of Complete Coding-Region mtDNA Sequences
In this present study, nucleotide positions were numbered as in the Cambridge Reference Sequence (CRS; Anderson et al. 1981), nucleotide substitutions were expressed as differences from the revised CRS (Andrews et al. 1999), transitions were denoted only by their nucleotide positions, and transversions were designated by their nucleotide positions followed by the changed base. A total of 942 complete coding-region mtDNA sequences, including our 672 Japanese; one additional Japanese (GenBank accession no. AB055387); 53 worldwide sequences (Ingman et al. 2000); 42 worldwide sequences (Maca-Meyer et al. 2001); two Finnish sequences having Asian relatives (Finnilä et al. 2001); 17 Asian sequences without concrete geographic assignation (Herrnstadt et al. 2002); 37 sequences from the Bering area (Derbeneva et al. 2002b); 70 Asian, New Guinean, and Australian sequences (Ingman and Gyllensten 2003); and 48 Chinese sequences (Kong et al. 2003) were aligned with the rCRS by CLUSTAL V software, and the coding region was used to construct a phylogenetic network (Bandelt et al. 1999) rooted with a chimpanzee sequence (GenBank accession no. D38113) as implemented in the Network 3.1 program (Fluxus Engineering; http://www.fluxus-engineering.com). The noncoding positions were added by hand using molecular weighted parsimony criteria (Bandelt et al. 2000). The phylogenetic relationships obtained were also confirmed by means of a neighbor-joining tree (1000× bootstrapped; Saitou and Nei 1987), built using MEGA2 (Kumar et al. 2001). From this network (see Supplemental material) we chose 102 Japanese and nine Asiatic sequences that represented the main clusters and subclusters within the two macrohaplogroups M and N that colonized Asia. To define these groups we followed the most generalized cladistic nomenclature actually used to classify mtDNA lineages (Richards et al. 1998). For the haplogroups previously detected, we maintained the same notation as their authors proposed (Richards et al. 2000; Bamshad et al. 2001; Kivisild et al. 2002; Yao et al. 2002a; Kong et al. 2003). Those haplogroups introduced here for the first time were named according to their phylogenetic range deduced from the tree of complete sequences.
Haplogroup Assorting of Published Partial mtDNA Sequences
The unambiguously classified complete mtDNA sequences were used as an initial pool that was hierarchically enlarged by the successive addition of those published partial mtDNA sequences with the largest coding information, ending with those for which information on only control-region sequences for both mtDNA hypervariable segments or just one (HVS-I and/or HVS-II) was available, always following sequence matches or, as default, sequence-relatedness criteria. Some of those partial sequences that could be assigned to more than one haplogroup were tentatively assorted in the most probable one deduced from their geographic origin and the relative haplogroup distribution.
Pooling Small Size Samples and Rare Clades
To avoid small sample sizes and rare alleles in population comparisons, samples with <20 individuals were pooled with others from the same geographic and ethnic group. Within populations, individuals belonging to rare clades were pooled with those classified in the nearest branch. Pairwise sample distances were calculated as linearized FST distances as implemented in the ARLEQUIN program (Schneider et al. 2000), taking mtDNA as one locus with as many alleles as the different subhaplogroups considered.
Quantitative Affinities of Japanese Samples
Relative affinities of Japanese samples to the other Asiatic populations were assessed by linearized FST distances, using subhaplogroup frequencies, and haplotype matches' distances (D) estimated simply as D = 1 - ∑(xiyi), xi and yi being the frequency of haplotype i in the two compared populations. To be statistically robust, these analyses require large sample sizes, thus further pooling was necessary. Previous studies in the area prevented us from pooling populations by geographic proximity (Schurr et al. 1999) and/or ethno-linguistic relationship (Comas et al. 1998; Chunjie et al. 2000; Yao et al. 2002a). For this reason, a genetic affinity criterion was chosen. Two approaches were used. In the first, all samples with no significant FST distances between them and with a similar behavior to the rest of the samples studied, were grouped. In the second, pooling was carried out by means of the CLUSTER algorithm implemented in the SPSS ver 9 package. We followed an iterative method specifying the number of conglomerates from 2 to 30. Different groupings were tested by AMOVA, and that with the least assigned variance within areas was chosen. The data were graphically represented by multidimensional scaling (MDS) plots (Kruskal and Wish 1978) using SPSS.
Qualitative Affinities of Japanese Samples
Particular sharing of subhaplogroups and particular haplotype matches of Japanese samples with concrete Continental areas were phylogeographically analyzed by taking into account the relative genetic diversities of the clades involved in the different areas, measured as relative haplotypic frequencies, and their minimum estimates of coalescence ages based on mean divergence among lineages for the coding region (Saillard et al. 2000). A constant evolutionary rate of 1.7 × 10-8 per site per year (Ingman et al. 2000) was used.
Acknowledgments
This work was supported in part by the Support Project for Database Development from the Japan Science and Technology Corporation (to M.T.), Grants-in-Aid for Scientific Research (C2-10832009, A2-15200051) and for Priority Areas from the Ministry of Education, Science, Sports and Culture of Japan (to M.T.), and by grants BMC2001-3511 and COF2002-015 (to V.M.C.).
Footnotes
[Supplemental material is available online at www.genome.org.]
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2286304.
References
- Abe, S., Usami, S., Shinkawa, H., Weston, M.D., Overbeck, L.D., Hoover, D.M., Kenyon, J.B., Horai, S., and Kimberling, W.J. 1998. Phylogenetic analysis of mitochondrial DNA in Japanese pedigrees of sensorineural hearing loss associated with the A1555G mutation. Eur. J. Hum. Genet. 6: 563-569. [DOI] [PubMed] [Google Scholar]
- Anderson, S., Bankier, A.T., Barrell, B.G., de Bruijn, M.H., Coulson, A.R., Drouin, J., Eperon, I.C., Nierlich, D.P., Roe, B.A., Sanger, F., et al. 1981. Sequence and organization of the human mitochondrial genome. Nature 290: 457-465. [DOI] [PubMed] [Google Scholar]
- Andrews, R.M., Kubacka, I., Chinnery, P.F., Lightowlers, R.N., Turnbull, D.M., and Howell, N. 1999. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat. Genet. 23: 147. [DOI] [PubMed] [Google Scholar]
- Ballinger, S.W., Schurr, T.G., Torroni, A., Gan, Y.Y., Hodge, J.A., Hassan, K., Chen, K.H., and Wallace, D.C. 1992. Southeast Asian mitochondrial DNA analysis reveals genetic continuity of ancient mongoloid migrations. Genetics 130: 139-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamshad, M., Kivisild, T., Watkins, W.S., Dixon, M.E., Ricker, C.E., Rao, B.B., Naidu, J.M., Prasad, B.V., Reddy, P.G., Rasanayagam, A., et al. 2001. Genetic evidence on the origins of Indian caste populations. Genome Res. 11: 994-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandelt, H.-J., Forster, P., and Röhl, A. 1999. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16: 37-48. [DOI] [PubMed] [Google Scholar]
- Bandelt, H.-J., Macaulay, V., and Richards, M. 2000. Median networks: Speedy construction and greedy reduction, one simulation, and two case studies from human mtDNA. Mol. Phylogenet. Evol. 16: 8-28. [DOI] [PubMed] [Google Scholar]
- Betty, D.J., Chin-Atkins, A.N., Croft, L., Sraml, M., and Easteal, S. 1996. Multiple independent origins of the COII/tRNA(Lys) intergenic 9-bp mtDNA deletion in aboriginal Australians. Am. J. Hum. Genet. 58: 428-433. [PMC free article] [PubMed] [Google Scholar]
- Cann, R.L. and Wilson, A.C. 1983. Length mutations in human mitochondrial DNA. Genetics 104: 669-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cann, R.L., Stoneking, M., and Wilson, A.C. 1987. Mitochondrial DNA and human evolution. Nature 325: 31-36. [DOI] [PubMed] [Google Scholar]
- Cavalli-Sforza, L.L., Menozzi, P., and Piazza, A. 1994. The history and geography of human genes. Princeton University Press, Princeton, NJ.
- Chunjie, X., Cavalli-Sforza, L.L., Minch, E., and Ruofu, D.U. 2000. Principal component analysis of gene frequencies of Chinese populations. Science in China Ser. C 43: 472-481. [Google Scholar]
- Comas, D., Calafell, F., Mateu, E., Perez-Lezaun, A., Bosch, E., Martinez-Arias, R., Clarimon, J., Facchini, F., Fiori, G., Luiselli, D., et al. 1998. Trading genes along the silk road: mtDNA sequences and the origin of central Asian populations. Am. J. Hum. Genet. 63: 1824-1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbeneva, O.A., Starikovskaya, E.B., Wallace, D.C., and Sukernik, R.I. 2002a. Traces of early Eurasians in the Mansi of northwest Siberia revealed by mitochondrial DNA analysis. Am. J. Hum. Genet. 70: 1009-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbeneva, O.A., Sukernik, R.I., Volodko, N.V., Hosseini, S.H., Lott, M.T., and Wallace, D.C. 2002b. Analysis of mitochondrial DNA diversity in the Aleuts of the commander islands and its implications for the genetic history of Beringia. Am. J. Hum. Genet. 71: 415-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derenko, M.V., Malyarchuk, B.A., Dambueva, I.K., Shaikhaev, G.O., Dorzhu, C.M., Nimaev, D.D., and Zakharov, I.A. 2000. Mitochondrial DNA variation in two South Siberian Aboriginal populations: Implications for the genetic history of North Asia. Hum. Biol. 72: 945-973. [PubMed] [Google Scholar]
- Elson, J.L., Turnbull, D.M., and Howell, N. 2004. Comparative genomics and the evolution of human mitochondrial DNA: Assessing the effects of selection. Am. J. Hum. Genet. 74: 229-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnilä, S., Lehtonen, M.S., and Majamaa, K. 2001. Phylogenetic network for European mtDNA. Am. J. Hum. Genet. 68: 1475-1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster, P., Harding, R., Torroni, A., and Bandelt, H.J. 1996. Origin and evolution of Native American mtDNA variation: A reappraisal. Am. J. Hum. Genet. 59: 935-945. [PMC free article] [PubMed] [Google Scholar]
- Forster, P., Torroni, A., Renfrew, C., and Röhl, A. 2001. Phylogenetic star contraction applied to Asian and Papuan mtDNA evolution. Mol. Biol. Evol. 18: 1864-1881. [DOI] [PubMed] [Google Scholar]
- Fucharoen, G., Fucharoen, S., and Horai, S. 2001. Mitochondrial DNA polymorphisms in Thailand. J. Hum. Genet. 46: 115-125. [DOI] [PubMed] [Google Scholar]
- Glover, I.C. 1980. Agricultural origins in East Asia. In The Cambridge encyclopedia of archaeology (ed. A. Sherratt), pp. 152-161. Crown, New York.
- Hammer, M.F. and Horai, S. 1995. Y chromosomal DNA variation and the peopling of Japan. Am. J. Hum. Genet. 56: 951-962. [PMC free article] [PubMed] [Google Scholar]
- Hanihara, K. 1991. Dual structure model for the population history of the Japanese. Japan Review 2: 1-33. [Google Scholar]
- Helgason, A., Sigureth Ardottir, S., Gulcher, J.R., Ward, R., and Stefansson, K. 2000. mtDNA and the origin of the Icelanders: Deciphering signals of recent population history. Am. J. Hum. Genet. 66: 999-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helgason, A., Hickey, E., Goodacre, S., Bosnes, V., Stefánsson, K., Ward, R., and Sykes, B. 2001. mtDNA and the islands of the North Atlantic: Estimating the proportions of Norse and Gaelic ancestry. Am. J. Hum. Genet. 68: 723-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrnstadt, C., Elson, J.L., Fahy, E., Preston, G., Turnbull, D.M., Anderson, C., Ghosh, S.S., Olefsky, J.M., Beal, M.F., Davis, R.E., et al. 2002. Reduced-median-network analysis of complete mitochondrial DNA coding-region sequences for the major African, Asian, and European haplogroups. Am. J. Hum. Genet. 70: 1152-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horai, S. and Hayasaka, K. 1990. Intraspecific nucleotide sequence differences in the major noncoding region of human mitochondrial DNA. Am. J. Hum. Genet. 46: 828-842. [PMC free article] [PubMed] [Google Scholar]
- Horai, S. and Matsunaga, E. 1986. Mitochondrial DNA polymorphism in Japanese. II. Analysis with restriction enzymes of four or five base pair recognition. Hum. Genet. 72: 105-117. [DOI] [PubMed] [Google Scholar]
- Horai, S., Kondo, R., Murayama, K., Hayashi, S., Koike, H., and Nakai, N. 1991. Phylogenetic affiliation of ancient and contemporary humans inferred from mitochondrial DNA. Phil. Trans. R Soc. Lond. B 333: 409-417. [DOI] [PubMed] [Google Scholar]
- Horai, S., Murayama, K., Hayasaka, K., Matsubayashi, S., Hattori, Y., Fucharoen, G., Harihara, S., Park, K.S., Omoto, K., and Pan, I.H. 1996. mtDNA polymorphism in East Asian Populations, with special reference to the peopling of Japan. Am. J. Hum. Genet. 59: 579-590. [PMC free article] [PubMed] [Google Scholar]
- Imaizumi, K., Parsons, T.J., Yoshino, M., and Holland, M.M. 2002. A new database of mitochondrial DNA hypervariable regions I and II sequences from 162 Japanese individuals. Int. J. Legal. Med. 116: 68-73. [DOI] [PubMed] [Google Scholar]
- Ingman, M. and Gyllensten, U. 2003. Mitochondrial genome variation and evolutionary history of Australian and New Guinean Aborigines. Genome Res. 13: 1600-1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingman, M., Kaessmann, H., Pääbo, S., and Gyllensten, U. 2000. Mitochondrial genome variation and the origin of modern humans. Nature 408: 708-713. [DOI] [PubMed] [Google Scholar]
- Jorde, L.B., Bamshad, M.J., Watkins, W.S., Zenger, R., Fraley, A.E., Krakowiak, P.A., Carpenter, K.D., Soodyall, H., Jenkins, T., and Rogers, A.R. 1995. Origins and affinities of modern humans: A comparison of mitochondrial and nuclear genetic data. Am. J. Hum. Genet. 57: 523-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kivisild, T., Kaldma, K., Metspalu, M., Parik, J., Papiha, S., and Villems, R. 1999. The place of the Indian mitochondrial DNA variants in the global network of maternal lineages and the peopling of the Old World. In Genomic diversity: Applications in human population genetics (eds. S. Papiha et al.), pp. 135-152. Plenum Press, New York.
- Kivisild, T., Tolk, H.-V., Parik, J., Wang, Y., Papiha, S.S., Bandelt, H.-J., and Villems, R. 2002. The emerging limbs and twigs of the East Asian mtDNA tree. Mol. Biol. Evol. 19: 1737-1751. [DOI] [PubMed] [Google Scholar]
- Kivisild, T., Rootsi, S., Metspalu, M., Mastana, S., Kaldma, K., Parik, J., Metspalu, E., Adojaan, M., Tolk, H.V., Stepanov, V., et al. 2003. The genetic heritage of the earliest settlers persists both in Indian tribal and caste populations. Am. J. Hum. Genet. 72: 313-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolman, C.J., Sambuughin, N., and Bermingham, E. 1996. Mitochondrial DNA analysis of Mongolian populations and implications for the origin of New World founders. Genetics 142: 1321-1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, Q.-P., Yao, Y.-G., Sun, C., Bandelt, H.-J., Zhu, C.-L., and Zhang, Y.-P. 2003. Phylogeny of East Asian mitochondrial DNA lineages inferred from complete sequences. Am. J. Hum. Genet. 73: 671-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama, H., Iwasa, M., Maeno, Y., Tsuchimochi, T., Isobe, I., Seko-Nakamura, Y., Monma-Ohtaki, J., Matsumoto, T., Ogawa, S., Sato, B., et al. 2002. Mitochondrial sequence haplotype in the Japanese population. Forensic Sci. Int. 125: 93-96. [DOI] [PubMed] [Google Scholar]
- Kruskal, J.B. and Wish, M. 1978. Multidimensional scaling. Sage Publications, Beverly Hills, CA.
- Kumar, S., Tamura, K., Jakobsen, I.B., and Nei, M. 2001. MEGA2: Molecular Evolutionary Genetics Analysis software. Bioinformatics 17: 1244-1245. [DOI] [PubMed] [Google Scholar]
- Lee, S.D., Shin, C.H., Kim, K.B., Lee, Y.S., and Lee, J.B. 1997. Sequence variation of mitochondrial DNA control region in Koreans. Forensic Sci. Int. 87: 99-116. [DOI] [PubMed] [Google Scholar]
- Lee, S.D., Lee, Y.S., and Lee, J.B. 2002. Polymorphism in the mitochondrial cytochrome B gene in Koreans. An additional marker for individual identification. Int. J. Legal Med. 116: 74-78. [DOI] [PubMed] [Google Scholar]
- Maca-Meyer, N., González, A.M., Larruga, J.M., Flores, C., and Cabrera, V.M. 2001. Major genomic mitochondrial lineages delineate early human expansions. BMC Genet. 2: 13-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaulay, V., Richards, M., Hickey, E., Vega, E., Cruciani, F., Guida, V., Scozzari, R., Bonne-Tamir, B., Sykes, B., and Torroni, A. 1999. The emerging tree of West Eurasian mtDNAs: A synthesis of control-region sequences and RFLP. Am. J. Hum. Genet. 64: 232-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malyarchuk, B.A. and Derenko, M.V. 2001. Mitochondrial DNA variability in Russians and Ukrainians: Implication to the origin of the Eastern Slavs. Ann. Hum. Genet. 65: 63-78. [DOI] [PubMed] [Google Scholar]
- Matsumoto, H. 1988. Characteristics of Mongoloid and neighboring populations based on the genetic markers of human immunoglobulins. Hum. Genet. 80: 207-218. [DOI] [PubMed] [Google Scholar]
- Melton, T., Clifford, S., Martinson, J., Batzer, M., and Stoneking, M. 1998. Genetic evidence for the proto-Austronesian homeland in Asia: mtDNA and nuclear DNA variation in Taiwanese aboriginal tribes. Am. J. Hum. Genet. 63: 1807-1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei, M. 1995. The origins of human populations: Genetic, linguistic, and archeological data. In The origin and past of modern humans as viewed from DNA (eds. S. Brenner and K. Hanihara), pp. 71-91. World Scientific, Singapore.
- Nishimaki, Y., Sato, K., Fang, L., Ma, M., Hasekura, H., and Boettcher, B. 1999. Sequence polymorphism in the mtDNA HV1 region in Japanese and Chinese. Legal Med. 1: 238-249. [DOI] [PubMed] [Google Scholar]
- Omoto, K. and Saitou, N. 1997. Genetic origins of the Japanese: A partial support for the dual structure hypothesis. Am. J. Phys. Anthropol. 102: 437-446. [DOI] [PubMed] [Google Scholar]
- Oota, H., Kitano, T., Jin, F., Yuasa, I., Wang, L., Ueda, S., Saitou, N., and Stoneking, M. 2002. Extreme mtDNA homogeneity in continental Asian populations. Am. J. Phys. Anthropol. 118: 146-153. [DOI] [PubMed] [Google Scholar]
- Orito, E., Ichida, T., Sakugawa, H., Sata, M., Horiike, N., Hino, K., Okita, K., Okanoue, T., Iino, S., Tanaka, E., et al. 2001. Geographic distribution of hepatitis B virus (HBV) genotype in patients with chronic HBV infection in Japan. Hepatology 34: 590-594. [DOI] [PubMed] [Google Scholar]
- Pfeiffer, H., Steighner, R., Fisher, R., Mornstad, H., Yoon, C.L., and Holland, M.M. 1998. Mitochondrial DNA extraction and typing from isolated dentin-experimental evaluation in a Korean population. Int. J. Legal Med. 111: 309-313. [DOI] [PubMed] [Google Scholar]
- Qian, Y.P., Chu, Z.T., Dai, Q., Wei, C.D., Chu, J.Y., Tajima, A., and Horai, S. 2001. Mitochondrial DNA polymorphisms in Yunnan nationalities in China. J. Hum. Genet. 46: 211-220. [DOI] [PubMed] [Google Scholar]
- Quintana-Murci, L., Semino, O., Bandelt, H.-J., Passarino, G., McElreavey, K., and Santachiara-Benereccetti, A.S. 1999. Genetic evidence of an early exit of Homo sapiens sapiens from Africa through eastern Africa. Nat. Genet. 23: 437-441. [DOI] [PubMed] [Google Scholar]
- Redd, A.J. and Stoneking, M. 1999. Peopling of Sahul: mtDNA variation in aboriginal Australian and Papua New Guinean populations. Am. J. Hum. Genet. 65: 808-828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards, M., Macaulay, V., Bandelt, H.-J., and Sykes, B. 1998. Phylogeography of mitochondrial DNA in western Europe. Ann. Hum. Genet. 62: 241-260. [DOI] [PubMed] [Google Scholar]
- Richards, M., Macaulay, V., Hickey, E., Vega, E., Sykes, B., Guida, V., Rengo, C., Sellitto, D., Cruciani, F., Kivisild, T., et al. 2000. Tracing European founder lineages in the Near Eastern mtDNA pool. Am. J. Hum. Genet. 67: 1251-1276. [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Pesini, E., Mishmar, D., Brandon, M., Procaccio, V., and Wallace, D.C. 2004. Effects of purifying and adaptive selection on regional variation in human mtDNA. Science 303: 223-226. [DOI] [PubMed] [Google Scholar]
- Saillard, J., Forster, P., Lynnerup, N., Bandelt, H.J., and Norby, S. 2000. mtDNA variation among Greenland Eskimos: The edge of the Beringian expansion. Am. J. Hum. Genet. 67: 718-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou, N. and Nei, M. 1987. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406-425. [DOI] [PubMed] [Google Scholar]
- Schneider, S., Roessli, D., and Excoffier, L. 2000. Arlequin ver. 2000: A software for population genetics data analysis. Genetic and Biometry Laboratory, University of Geneva, Switzerland.
- Schurr, T.G., Sukernik, R.I., Starikovskaya, Y.B., and Wallace, D.C. 1999. Mitochondrial DNA variation in Koryaks and Itel'men: Population replacement in Okhotsk Sea-Bering Sea region during the Neolithic. Am. J. Phys. Anthropol. 108: 1-39. [DOI] [PubMed] [Google Scholar]
- Seo, Y., Stradmann-Bellinghausen, B., Rittner, C., Takahama, K., and Schneider, P.M. 1998. Sequence polymorphism of mitochondrial DNA control region in Japanese. Forensic Sci. Int. 97: 155-164. [DOI] [PubMed] [Google Scholar]
- Shields, G.F., Schmiechen, A.M., Frazier, B.L., Redd, A., Voevoda, M.I., Reed, J.K., and Ward, R.H. 1993. mtDNA sequences suggest a recent evolutionary divergence for Beringian and northern North American populations. Am. J. Hum. Genet. 53: 549-562. [PMC free article] [PubMed] [Google Scholar]
- Shinoda, K.-I. and Kanai, S. 1999. Intracemetry genetic analysis at the Nakazuma Jomon site in Japan by mitochondrial DNA sequencing. Anthropol. Sci. 107: 129-140. [Google Scholar]
- Shiraishi, T. 2002. Wakoku tanjou (The formation of ancient Japanese society). In History of Japan 1 (ed. T. Shiraishi et al.), pp. 8-94. Yoshikawakobunkan, Tokyo, Japan (in Japanese).
- Snäll, N., Savontaus, M.-L., Kares, S., Lee, M.S., Cho, E.K., Rinne, J.O., and Huoponen, K. 2002. A rare mitochondrial DNA haplotype observed in Koreans. Hum. Biol. 74: 253-262. [DOI] [PubMed] [Google Scholar]
- Soodyall, H., Jenkins, T., and Stoneking, M. 1995. `Polynesian' mtDNA in the Malagasy. Nat. Genet. 10: 377-378. [DOI] [PubMed] [Google Scholar]
- Stoneking, M., Jorde, L.B., Bhatia, K., and Wilson, A.C. 1990. Geographic variation in human mitochondrial DNA from Papua New Guinea. Genetics 124: 717-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, B., Xiao, C., Deka, R., Seielstad, M.T., Kangwanpong, D., Xiao, J., Lu, D., Underhill, P., Cavalli-Sforza, L., Chakraborty, R., et al. 2000. Y chromosome haplotypes reveal prehistorical migrations to the Himalayas. Hum. Genet. 107: 582-590. [DOI] [PubMed] [Google Scholar]
- Sykes, B., Leiboff, A., Low-Beer, J., Tetzner, S., and Richards, M. 1995. The origins of the Polynesians: An interpretation from mitochondrial lineage analysis. Am. J. Hum. Genet. 57: 1463-1475. [PMC free article] [PubMed] [Google Scholar]
- Tajima, A., Sun, C.-S., Pan, I.-H., Ishida, T., Saitou, N., and Horai, S. 2003. Mitochondrial DNA polymorphisms in nine aboriginal groups of Taiwan: Implications for the population history of aboriginal Taiwanese. Hum. Genet. 113: 24-33. [DOI] [PubMed] [Google Scholar]
- Takahata, N., Lee, S.-H., and Satta, Y. 2001. Testing multiregionality of modern human origins. Mol. Biol. Evol. 18: 172-183. [DOI] [PubMed] [Google Scholar]
- Takeshita, T., Yasuda, Y., Nakashima, K., Mogi, K., Kishi, H., Shiono, K., Sagisaka, I., Yuasa, H., Nishimukai, H., and Kimura, H. 2001. Geographical north-south decline in DNASE*2 in Japanese populations. Hum. Biol. 73: 129-134. [DOI] [PubMed] [Google Scholar]
- Tanaka, M., Hayakawa, M., and Ozawa, T. 1996. Automated sequencing of mitochondrial DNA. Methods Enzymol. 264: 407-421. [DOI] [PubMed] [Google Scholar]
- Torroni, A., Schurr, T.G., Yang, C.C., Szathmary, E.J., Williams, R.C., Schanfield, M.S., Troup, G.A., Knowler, W.C., Lawrence, D.N., Weiss, K.M., et al. 1992. Native American mitochondrial DNA analysis indicates that the Amerind and the Nadene populations were founded by two independent migrations. Genetics 130: 153-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torroni, A., Sukernik, R.I., Schurr, T.G., Starikorskaya, Y.B., Cabell, M.F., Crawford, M.H., Comuzzie, A.G., and Wallace, D.C. 1993a. mtDNA variation of aboriginal Siberians reveals distinct genetic affinities with Native Americans. Am. J. Hum. Genet. 53: 591-608. [PMC free article] [PubMed] [Google Scholar]
- Torroni, A., Schurr, T.G., Cabell, M.F., Brown, M.D., Neel, J.V., Larsen, M., Smith, D.G., Vullo, C.M., and Wallace, D.C. 1993b. Asian affinities and continental radiation of the four founding Native American mtDNAs. Am. J. Hum. Genet. 53: 563-590. [PMC free article] [PubMed] [Google Scholar]
- Torroni, A., Miller, J.A., Moore, L.G., Zamudio, S., Zhuang, J., Droma, T., and Wallace, D.C. 1994. Mitochondrial DNA analysis in Tibet: Implications for the origin of the Tibetan population and its adaptation to high altitude. Am. J. Phys. Anthropol. 93: 189-199. [DOI] [PubMed] [Google Scholar]
- Torroni, A., Huoponen, K., Francalacci, P., Petrozzi, M., Morelli, L., Scozzari, R., Obinu, D., Savontaus, M.-L., and Wallace, D.C. 1996. Classification of European mtDNAs from an analysis of three European populations. Genetics 144: 1835-1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torroni, A., Rengo, C., Guida, V., Cruciani, F., Sellitto, D., Coppa, A., Calderon, F.L., Simionati, B., Valle, G., Richards, M., et al. 2001. Do the four clades of the mtDNA haplogroup L2 evolve at different rates? Am. J. Hum. Genet. 69: 1348-1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, L.C., Lin, C.Y., Lee, J.C., Chang, J.G., Linacre, A., and Goodwin, W. 2001. Sequence polymorphism of mitochondrial D-loop DNA in the Taiwanese Han population. Forensic Sci. Int. 119: 239-247. [DOI] [PubMed] [Google Scholar]
- Voevoda, M.I., Avksentyuk, A.V., Ivanova, A.V., Astakhova, T.I., Babenko, V.N., Kurilovich, S.A., Duffy, L.K., Segal, B., and Shields, G.F. 1994. Molecular genetic studies in the population of native inhabitants of Chukchee Peninsula. Analysis of polymorphism of mitochondrial DNA and of genes controlling alcohol metabolizing enzymes. Sibirskii Ekolog. Z. 1: 139-151. [Google Scholar]
- Watson, E., Forster, P., Richards, M., and Bandelt, H.J. 1997. Mitochondrial footprints of human expansions in Africa. Am. J. Hum. Genet. 61: 691-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, Y.G., Lu, X.M., Luo, H.R., Li, W.H., and Zhang, Y.P. 2000a. Gene admixture in the silk road region of China: Evidence from mtDNA and melanocortin 1 receptor polymorphism. Genes Genet. Syst. 75: 173-178. [DOI] [PubMed] [Google Scholar]
- Yao, Y.G., Watkins, W.S., and Zhang, Y.P. 2000b. Evolutionary history of the mtDNA 9-bp deletion in Chinese populations and its relevance to the peopling of east and southeast Asia. Hum. Genet. 107: 504-512. [DOI] [PubMed] [Google Scholar]
- Yao, Y.G., Kong, Q.P., Bandelt, H.J., Kivisild, T., and Zhang, Y.P. 2002a. Phylogeographic differentiation of mitochondrial DNA in Han Chinese. Am. J. Hum. Genet. 70: 635-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, Y.-G., Nie, L., Harpending, H., Fu, Y.-X., Yuan, Z.-G., and Zhang, Y.-P. 2002b. Genetic relationship of Chinese ethnic populations revealed by mtDNA sequence diversity. Am. J. Phys. Anthropol. 118: 63-76. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://www.fluxus-engineering.com; Network 3.1 program, Fluxus Engineering.
- http://www.giib.or.jp/mtsnp/index_e.html; authors' data.