Abstract
Using paired-end sequences from bacterial artificial chromosomes, we have constructed high-resolution synteny and rearrangement breakpoint maps among human, mouse, and rat genomes. Among the >300 syntenic blocks identified are segments of over 40 Mb without any detected interspecies rearrangements, as well as regions with frequently broken synteny and extensive rearrangements. As closely related species, mouse and rat share the majority of the breakpoints and often have the same types of rearrangements when compared with the human genome. However, the breakpoints not shared between them indicate that mouse rearrangements are more often interchromosomal, whereas intrachromosomal rearrangements are more prominent in rat. Centromeres may have played a significant role in reorganizing a number of chromosomes in all three species. The comparison of the three species indicates that genome rearrangements follow a path that accommodates a delicate balance between maintaining a basic structure underlying all mammalian species and permitting variations that are necessary for speciation.
The availability of the human (International Human Genome Sequencing Consortium 2001; Venter et al. 2001), mouse (Waterston et al. 2002), and rat (Rat Sequencing Project Consortium 2004) genomic sequences greatly facilitates mammalian genome evolution studies. Comparison of the three genomes reveals both conservation and variation, allowing the identification of both ancestral genome fragments and changes in chromosomal structure through evolution. Here, we have constructed a synteny and rearrangement breakpoint map for the three species using bacterial artificial chromosome (BAC) clones (Kim et al. 1996), which has allowed us to identify large-scale conservation and variation at a 100-300 kb resolution. The results indicate that whereas each genome tends to diverge, much of the basic structure is maintained. The study provides data to assess the balance of genome conservation and rearrangement in maintaining the fundamental functions and facilitating the evolutionary process including speciation.
RESULTS
Synteny and Rearrangement Breakpoint Map Construction
Various methods (e.g., BLASTZ, Multiz, PatternHunter, Pash) have been used to compare mammalian genome sequences (Ma et al. 2002; Schwartz et al. 2003; Kalafus et al. 2004) and several synteny maps are currently available for mouse, rat, and human (e.g. www.ensembl.org and www.genome.ucsc.edu). We have taken a complementary approach by using the BAC end sequence (BES) resource that was generated for these three species, as well as a number of primates (Mahairas et al. 1999; Zhao et al. 2000, 2001; Fujiyama et al. 2002). We have mapped BACs by placing their paired BESs on the genome assemblies through sequence matches and examined the chromosomal locations, orientation, and distance of each paired mates on the genome (Zhao 2001). Clones that were successfully and uniquely assigned served as anchors for synteny/breakpoint map construction on the basis of clone order, BAC orientation, and distance between the clones. Owing to the large size (usually 80-300 kb) and orientation of BACs, as well as the additional mapping information provided by paired mates (e.g., location, orientation, and spanned distance on the genome) along with the BES sequence-match strength, our approach offers advantages in discriminating false matches originating from genome repeats and local misassemblies (see Discussion). For instance, we have resolved the synteny of at least additional 2.5 Mb sequences flanking the centromeric region of the human chromosome 9 (H9) that is currently missing at both www.ensembl.org and www.genome.ucsc.edu sites (see Supplemental material).
With a total of 246,747 BACs aligned for human, 195,086 for mouse, and 160,683 for rat, the genome coverage by the clones is 12-, 14-, and 11-fold for the three species, respectively (see Supplemental material). With these BACs as markers, we performed two-way and three-way comparisons of the three species to identify syntenic blocks (a syntenic block is defined as a maximal chromosomal fragment in which a series of BACs are in the same order and with the same orientation in all species compared. Synteny here refers to the homology between chromosomal fragments from different species compared that form syntenic blocks). To reduce false results caused by local misassemblies and repeats matches, we excluded blocks with sizes <100 kb and a total clone number <6 for mouse versus rat, <3 for human versus mouse or rat, and <2 for the three species. In all cases, the syntenic blocks range from 100 kb to over 40 Mb (see Supplemental material), and are consistent with the fragile breakage model, but not with the random breakage model (Nadeau and Taylor 1984; Sankoff et al. 1997, 2000; Sankoff 2003; Pevzner and Tesler 2003a; Fig. 1), indicating regions with great conservation and regions with extensive variation. The block numbers found between the species (Table 1) are more or less comparable to the reported figures (Waterston et al. 2002; Kent et al. 2003; Pevzner and Tesler 2003a,b; Bourque et al. 2004; Rat Sequencing Project Consortium 2004). Although we do not have as many markers as in these other studies, our anchors have the highest sequence coverage for all three genomes. For instance, the ∼558,000 sequence anchors used for the human/mouse comparison by Waterson et al. (2002) comprise about 7.5% of the mouse genome, whereas the BAC anchors used in this study cover 55%-90% of the genomes (Table 1). The synteny data can be downloaded from the TIGR Web site at ftp://ftp.tigr.org/pub/data/Bac_Resource/HumanMouseRatSynteny.
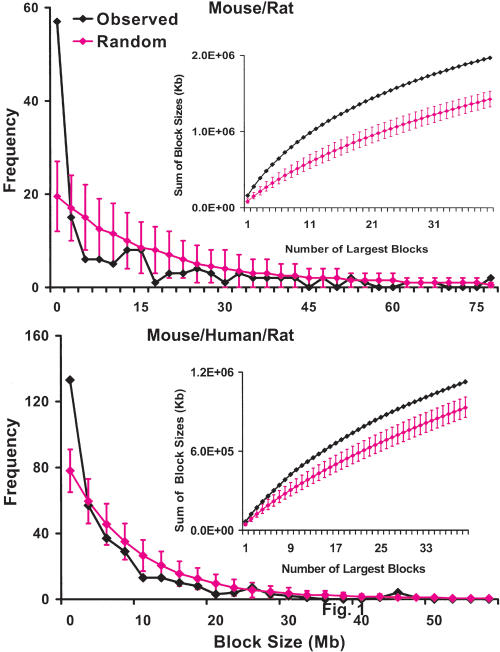
Figure 1.
Syntenic block length distribution. The numbers of blocks with length above 100 kb in 2.5-Mb bins are plotted in blue. Expected values at 95% confidence for each bin on the basis of the Monte Carlo simulation of the random breakage model are plotted in pink. The data indicate that there are significantly more small blocks than what are predicated by the random breakage model, further confirmed by examining the distributions of the sum of sizes of the largest 40 blocks (as shown in the insets, the predicated values are smaller). (Top) The synteny between mouse and rat. (Bottom) The synteny among mouse, rat, and human. (Insets) Distributions of the sum of the lengths (kb) of the largest 40 blocks for the observed data (blue) and the predicated values at 95% confidence based on the random model (pink).
Table 1.
Human, Mouse, and Rat Synteny
|
Syntenyb
|
||||
|---|---|---|---|---|
| Species | BACs placed | Genome covered by BACsa | Totalc | Per blockd |
| Mouse vs. Rat | 108,680 | 2.32 Gb mouse, 2.44 Gb rat, 90% | 146 blocks, 2.47 Gb mouse and 2.64 Gb rat. | 743 BACs, 16.9 Mb mouse and 18.5 Mb rat. |
| Human vs. Mouse | 29,463 | 1.65 Gb mouse, 1.83 Gb human, 64% | 299 blocks, 2.31 Gb mouse and 2.55 Gb human. | 98 BACs, 7.9 Mb mouse and 8.5 Mb human. |
| Human vs. Rat | 26,729 | 1.62 Gb rat, 1.71 Gb human, 60% | 295 blocks, 2.42 Gb rat and 2.48 Gb human. | 90 BACs, 8.2 Mb rat and 8.4 Mb human. |
| Human, mouse and rat | 21,363 | 1.41 Gb mouse, 1.47 Gb rat, 1.56 Gb human, 55% | 328 blocks, 2.2 Gb mouse, 2.3 Gb rat and 2.4 Gb human. | 64 BACs, 6.8 Mb mouse, 7.2 Mb rat, and 7.3 Mb human. |
The total number of bases (in Gb) and the percentage of the genome covered by the BACs for each genome involved.
All clones within a block are in the same order and orientation on each genome involved. Blocks with the same orientation were merged without considering gaps, after discarding blocks with size <100 kb or total clone # below 6 for mouse vs. rat, 3 for human vs. mouse or rat, 2 for the three species.
Total number of syntenic blocks identified, total amount of genome covered by the blocks on each genome.
Average number of BACs per block, average block size on each genome involved.
Mouse/Rat Synteny and Rearrangements
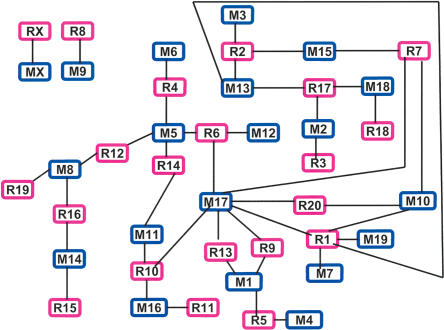
The synteny reveals interesting relationships among the chromosomes (Fig. 2). Except for mouse chromosome 9 (M9) and rat chromosome 8 (R8), as well as the X chromosomes that are exclusively syntenic to each other, all other chromosomes form a network, indicating that the exchange of genetic materials has occurred among the mouse or rat chromosomes within the network but not with outside members (e.g., M8/R9, MX/RX) since the mouse/rat divergence. Within the network, chromosomes also display varying degrees of complexity in synteny, with M17 having the most complex synteny (having blocks of synteny conserved with more rat chromosomes than do other mouse chromosomes; Fig. 2). However, these features are not apparent in other synteny plots such as those used by Rat Sequencing Project Consortium (2004), demonstrating the value of the presentation style shown in Figure 2. Our analyses indicate that some chromosomes are very conserved between mouse and rat. For instance, no other rearrangements were detected between M2 and R3, except for three blocks of 150-230 kb with 1-2 BACs inverted (possibly reflecting incorrect assemblies). Other chromosomes had more extensive rearrangements, for example, at least three major inversions involving regions as large as the entire R11 chromosome were found between M16 and R11. These inversions were not reported in nonsequence-based studies (Nilsson et al. 2001).
Figure 2.
The mouse/rat synteny. Each block represents a mouse (blue) or rat (pink) chromosome (e.g., M1, mouse chromosome 1; R1, rat chromosome 1). A line was drawn between a mouse block and a rat block if synteny was found between the two chromosomes. This plot better reveals the interactions between the chromosomes within a genome compared with other synteny plots used (Waterston et al. 2002; Kirkness et al. 2003; Rat Sequencing Project Consortium 2004) as demonstrated by the following. (1) Except for the M9/R8 and X chromosomes, the rest of the chromosomes have exchanged genetic materials with other chromosomes forming a complex synteny network. (2) Within the network, chromosomes also display varying degrees of complexity in synteny. For instance, mouse chromosomes M3, M4, M6, M7, M12, and M19 are syntenic only to a single rat chromosome. The same is true for R3, R11, R15, R18, and R19. The remaining chromosomes are syntenic to 2 to 3 chromosomes from the other species, except for R1, M5, and M17 with synteny to 5, 4, and 7 chromosomes, respectively. (3) M17 has the most complex synteny. Compared with its rat homologs, M17 has additionally rearranged with M10 and M1 multiple times, as well as with M5, M11, and M16 at least once. Nearly every mouse/rat syntenic block in M10 and M1 has rearranged with M17, indicating subsequent fusions of these blocks forming the two chromosomes. (4) Chromosomes within the network seem to group together on the basis of the conservation and rearrangement between the two species. For instance, chromosomes located at the left (M8/M14 vs. R19/R16/R15), where rat is more rearranged, differ from those at the bottom, right corner (M17/M1/M4 vs. R9/R13/R5), where mouse is more rearranged. Similarly, whereas a large variation was found between M17/M10/M7/M19 and R7/R20/R1 at the right, a great conservation was identified between M3/M13/M15/M2/M18 and R2/R17/R3/R18/R7 at the top.
As expected, the mouse and rat genomes are very conserved compared with each other. Beside this great conservation, we want to know what changes have occurred in each lineage since the two species diverged. To do this, we need a reference genome. This is because comparing the two species will only tell us the differences between them, but cannot discriminate which species is more ancestral and which species has rearranged in regions that involve the differences. The ideal reference would be the most recent mouse/rat common ancestor, which, however, is un-realistic, because the genomic sequences of any mammalian ancestor are unknown. Fortunately, human provides an excellent alternative. This is because, as a more distant species (human and rodents diverged ∼75 million years ago, whereas mouse and rat diverged ∼12-24 million years ago [Waterston et al. 2002; Rat Sequencing Project Consortium 2004]), human has an equal genetic distance from both rodents. More importantly, compared with mouse and rat, human has been evolving from the human/rodent common ancestor at a slower rearrangement rate (O'Brien et al. 1999); thus, overall, its genome is more ancestral.
To identify the differences in rearrangements between the two rodents, we categorized the mouse/rat breakpoints on the human genome as shown in Table 2. As expected, mouse and rat share the majority of the breakpoints (266 of 348 total, 76%). In addition, among the shared breakpoints, 96% (255 of 266 total) involve the same type of rearrangements (either intrachromosomal or interchromosomal). For the remaining 4%, neither rodent seems to favor one rearrangement type over the other (five for mouse inter and rat intra, six for mouse intra and rat inter). However, a different picture was observed for species-specific breakpoints. Among a total of 35 mouse-specific breakpoints, 14 (40%) are interchromosomal. In addition, eight of the 25 total intrachromosomal breakpoints are closely associated with these interchromosomal rearrangements (e.g., subsequent inversion breakpoints. See Table 3), and therefore, only 17 (49%) are due to pure intrachromosomal events. For rat, however, nearly all breakpoints (46 of 47) are intrachromosomal. It is unlikely that the observed differences in the rearrangement pattern between mouse and rat are caused by genome misassemblies or repeats matches, as all mouse-specific interchromosomal breakpoints and many of the rat-specific intrachromosomal breakpoints involve syntenic blocks of >1 Mb and with >10 BACs (Tables 3,4). In addition, further examination has indicated that intrachromosomal rearrangements in rat are concentrated on several nearmeta- or metacentric chromosomes, which further argues against genome misassemblies as the reason. These observations are somewhat consistent with a reciprocal chromosomal painting study reporting at least three times more interchromosomal rearrangements in the mouse genome compared with the rat genome (Stanyon et al. 1999). To investigate the differences between mouse and rat, we have focused on these species specific breakpoints.
Table 2.
Mouse/Rat Rearrangement Breakpoints on the Human Genomea
| M-Inter R-Inter | M-Intra R-Intra | M-Inter R-Intra | M-Intra R-Inter | M-Inter | R-Inter | M-Intra | R-Intra |
|---|---|---|---|---|---|---|---|
| 130b | 125b | 6 | 5 | 14c | 1d | 21e | 46f |
All breakpoints identified are incorporated. M-Inter R-Inter: shared interchromosomal breakpoints. M-Intra R-Intra: shared intrachromosomal breakpoints. M-Inter R-Intra: shared breakpoints, interchromosomal for mouse and intrachromosomal for rat. M-Inter: mouse-specific interchromosomal breakpoints.
About 58 are due to human specific intrachromosomal rearrangements.
All 14 breakpoints are described in details in Table 3.
R15/R16 fission, the only exception for the mouse fissions described in Table 3.
Eight are associated with M-Inter (mouse fission related inversions) as described in Table 3, involving syntenic blocks of >100 kb and with >3 BACs.
A total of 14 are due to inversions in R1p, R11, R19, R20, R10/M11/H17 as described in Table 4, involving synteny blocks of >100 kb and with >3 BACs; 30 involve simple inversions in the middle of the human/mouse syntenic blocks.
Table 3.
Mouse Interchromosomal Fissionsa
| Human fragment (Mb)b | Breakpoints for mouse (Mb)c | Mouse syntenic blocks (Mb) | Breakpoints for rat (Mb)d | Rat syntenic blocks (Mb) |
|---|---|---|---|---|
| H2 (10.7-52) | 26.3, 29 | M12 (17.3-3.1, tipe), M5 (28.4-30.7), M17 (70-90.5) | 35 | R6 (41.2-18.8) and (0.3-16.7, tipf) |
| H5 (8.9-96.2) | 43 | M15 (32.6-3.1, tip), M13 (116.2-70.8, endg) | 87.18, 87.47 | R2 (85.2-1.4) with a 400 kb inversion |
| H5 (154.3-173.5) | 171.5, 172.6 | M11 (55.5-33), M17h (25.2-25.5), M11 (31.7-32.2) | R10 (34-15.8) | |
| H5 (98.5-109.9) | 101.5, 102.8 | M1 (94.3-97) and (98.6-97.7), M17 (57.3-63.8) | R9 (93.6-104.3) | |
| H6 (150-171) | 155, 159, 159.4, 165, 170 | M10 (3.2-7.1, tip), M17 (6-3, tip), 7.24-7.05), (12.2-8.4), (6.7-7) and (13.8-14) | R1 (34.2-54) | |
| H6 (39.3-52.4) | 49 | M17 (48.2-39.6), M1 (18.5-21.1) | R9 (6.3-19.8) | |
| H7 (76.3-97) | 85, 92.2 | M5 (19.6-10.8) and (3.2-10.5, tip), M6 (3.1-7.4, tip) | R4 (9.3-32) with 148 kb inversion in the middle | |
| H8 (51.8-62.3) | 56.3 | M1 (7.7-3.1, tip), M4 (3.0-9.1, tip) | R5 (10.8-23.55) with 17 kb inversion in the middle | |
| H10 (0.5-27) | 6, 15.5 | M13 (9.4-3.3, tip), M2 (11.7-3.4, tip), and (12.1-23.1) | R17 (71.6-96.5, end) | |
| H11 (58.5-71.4) | 63, 64.4, 69 | M19 (12-7.8), (3.8-3.0, tip) and (3.99-6.8), M7 (133.7-132.8, end)i | R1 (214-204.8) | |
| H16 (0.3-15.9) | 3.2 | M17 (24.8-22.2), M16 (3.1-13.9, tip) | R10 (15.4-0) | |
| H21 (42.5-47) | 44 | M17 (29.7-30.7), M10 (78-76) | R20 (9.4-12.8) | |
| H22 (27.5-30.8) | 30.6 | M11 (5.4-3.0, tip), M5 (31.2-31.4) | R14 (86.2-83.5) |
All blocks shown are above 100 kb and with >3 BACs.
Each human fragment (indicated in parentheses) corresponds to a region in a single rat chromosome, but is syntenic to fragments from multiple mouse chromosomes. For instance, the first 27 Mb of H10 (0.5-27 Mb) broke twice, forming one syntenic block to M13's initial 6 Mb and two blocks in opposite direction to M2's initial 20 Mb, whereas it corresponded to R17's last 25 Mb as a single piece. In some cases, rat has an inversion, but still remains less rearranged (e.g., the H2/R6/M12/M5/M17 synteny). Only one exception was found where H10 (74.8-81.2 Mb) broke at 79.6 Mb to R15 (4.2-0 Mb) and R16 (0-1.4 Mb), whereas it was continuous for M14 (15.3-21 Mb).
Breakpoints on the human genome.
Breakpoints on the human genome.
Tip: the beginning of euchromatic sequences of the chromosomes, which starts at 3 Mb in the mouse assembly and 1 bp in the rat assembly. The tip is near the centromeric region in each mouse chromosome, and has been identified at the fission-related breakpoints for 13 mouse chromosomes.
Tip: the beginning of euchromatic sequences of the chromosomes, which starts at 3 Mb in the mouse assembly and 1 bp in the rat assembly. The tip is near the centromeric region in each mouse chromosome, and has been identified at the fission-related breakpoints for 13 mouse chromosomes.
End: the end of euchromatic sequences of the chromosomes in the assembly.
M17 was involved in seven of the 13 total fissions identified. The M11/M17/M11 breakages are likely due to a translocation.
M19 and M7 seem to have evolved from an ancestral chromosome closer to an R1q fragment through fission. Except for several inversions mostly associated with this fission event, no reorganizations were found in them.
Table 4.
Rat Intrachromosomal Rearrangementsa
| Human fragment (Mb)b | Breakpoints for rat (Mb) | Rat syntenic blocks (Mb)c | Breakpoints for mouse (Mb) | Mouse syntenic blocks (Mb) |
|---|---|---|---|---|
| H1 (1-58.3) | 26.2, 28 | R5 (173-152.7), (151.3-152.6) and (151-124.3) | M4 (152.7-101.3) | |
| H1 (231.1-236.4) | 232 | R17 (59.37-59.52) and (66.6-71.2) | M13 (13.5-9.8) | |
| H3 (169-179.2) | 170.5, 173 | R2 (113.6-115), (117.5-115.1) and (113.5-106.3) | 171.9 | M3 (28.8-31.3) and (28.6-20.7) |
| H3 (75.7-126) | 90-94.9 (centromeric region) | R11 (14.31-0.1, tip), (38.54-38.4) and (40-69.2) | M16 (76-33.2) | |
| H6 (123.2-150) | 128, 135 | R1 (24.3-29.2), (17.2-24) and (16.7-2.5, tip) | M10 (33.3-7.3) | |
| H10 (89.4-121.1) | 99, 102 | R1 (236.6-247), (249-247.2) and (249.6-268) | M19 (31-60.8) | |
| H10 (55.4-74.5) | 68, 70.3 | R20 (14.5-25), (26.8-25.2) and (30-27.2) | M10 (74.4-59) | |
| H12 (55-104.3) | 57 | R7 (2.2-1.1, tip) and (68-23.3) | M10 (130.5-86.1) | |
| H16 q-arm (46.5-end) | 55.6, 66.7 | R19 (23-15.1), (11.90-0.2, tip) and (34-53.8) addition to the 4 Mb inversion | M8 (84.6-123.2) with 4 Mb inversion | |
| H17p tip (0-1.2) and H17q tip (24.5-30)d | Three extra breakages | R10 (68.4-63.7) | ||
| H21 (14.6-42) | 31.2, 32.2 | R11 (14.4-29.4), (30.4-29.6) and (30.5-37.2) | M16 (76.1-98.5) |
All blocks shown are above 100 kb and with >3 BACs. Simple inversions below 1 Mb are not included.
Each human fragment corresponds to a single mouse fragment, but syntenic to several distinctive regions in the same rat chromosome. For example, a 27 Mb fragment of H6 (123.2-150 Mb) split twice to form blocks syntenic to three fragments of R1p, whereas it is contiguous to its M10 homolog. Sometimes mouse has an inversion but still remains less rearranged (e.g., H3/R2/M3 synteny).
R1p, R11, and R19 are more extensively rearranged with multiple extra major inversions. Other rat chromosomes (e.g., R5, R2) are simpler where an inversion of large segment (>1 Mb) was observed in rat but not in mouse (e.g., the H1/R5/M4 synteny).
H17p can be transformed to M11 (77-60 Mb) with seven breakages and ∼5 inversions, and H17q to M11 (77.5-122.2 Mb) with ∼10 breakages and nine inversions. However, extra breakages and inversions are required to transform H17q tip (24.5-30 Mb) and H17p tip (0-1.12 Mb) to R10 (68.4-63.7 Mb) beside the same amount of rearrangements for the transformation of H17p to R10 (63.1-46 Mb) and H17q to R10 (68.4-110.6 Mb).
Mouse Has More Interchromosomal Fission Rearrangements, Which Often Involve the Centromeric Regions
The examination of the mouse-specific breakpoints has revealed 13 human fragments with ∼273 Mb total and from 10 chromosomes, each of which corresponds to a region in a single rat chromosome but is syntenic to fragments from multiple mouse chromosomes (Table 3). Thus, mouse is more rearranged than rat in these cases due to the extra fission (and subsequent fusion) events. Because these mouse-specific fissions were identified in 14 mouse chromosomes with homology to nine rat chromosomes with only one exception, (see Table 3 and Supplemental material), they represent a major rearrangement mechanism specific to mouse. Interestingly, 70% (nine of 13 total) of these rearrangements involve the centromeric regions, as the beginning of the euchromatic sequence of a total of 13 mouse chromosomes (chromosomal tip) was identified at the breakpoints (four tip-tip fissions, two tip-end fissions, and three tip-middle fragment fissions. See Table 3). This indicates that the centromeric regions of mouse chromosomes are very active for interchromosomal rearrangements, consistent with a study reporting that numerous Robertsonian fusions have resulted in six distinct chromosomal races (e.g., 2N = 20, 2N = 22, etc., instead of the normal 2N = 40) in the wild populations of M. musculus found in Maderia (Britton-Davidian et al. 2000).
The M5/M6 fission was previously identified by cytogenetic studies; recent duplications associated with this event were found in the pericentromeric regions of multiple mouse chromosomes, and two novel mouse satellite repeats were identified (Walentinsson et al. 2001; Thomas et al. 2003a). To determine whether other fissions in Table 3 also involve such duplications and repeats, we performed a preliminary study of the sequences flanking these breakpoints in each lineage. The analyses indicate that often the human sequences are rich in SINEs (usually >20%), and the mouse/rat sequences are rich in L1 (usually >30%) or LTR repeats (usually >20%). Except for the gene phosphatidylserine de-carboxylase (encoded in 30 Mb of H22) that is partially duplicated in M11 (tip), M17 (6Mb), and M5 (31Mb) (the gene is partial in M11 and M17, but complete in M5), as well as the gene Notch3 (encoded in 15 Mb of H19) that is partially duplicated in M10 (78 Mb, partial) and M17 (31.8 Mb, complete), no other significant duplications were found. The 27-bp novel mouse satellites described by Thomas et al. (2003a) have been identified in a tandem, head-to-tail fashion in the chromosomal tip of M17 (52 copies at 3.06 Mb and 103 copies at 3.07 Mb), M2 (15 copies at 3 Mb), and M4 (57 copies at 3.35 Mb). The 36-bp satellites are less frequent; beside what have been described by Thomas et al. (2003a), no significant amounts were found in other places, except 13 copies at M4 3.27 Mb and five copies at M7 3.06 Mb.
Rat Has More Intrachromosomal Rearrangements, With Several Nearmeta- or Metacentric Chromosomes More Extensively Rearranged
We have found a number of rat chromosomal fragments that appeared to be more rearranged than mouse, mainly resulting from intrachromosomal events, by examining the rat-specific breakpoints. Often these breakpoints reside in human chromosomal regions corresponding to a single mouse fragment, but syntenic to several distinctive regions in the same rat chromosome. About 11 such human fragments with 330 Mb total and from eight chromosomes were identified, with synteny to 10 rat chromosomes and nine mouse chromosomes (Table 4). Thus, these intrachromosomal changes represent a major rearrangement mechanism specific to rat. Interestingly, rat chromosomes that are more extensively rearranged than mouse with multiple inversions are usually acro- (R11) or metacentric (R1p, R19, R20), whereas those with a simple inversion are usually telocentric (see Table 4 and Supplemental material). For a few exceptions in which mouse has more intrachromosomal changes, the corresponding rat chromosomes are either telocentric or only the q-arm is involved (see Supplemental material). As in the case in mouse, these observations once again suggest the involvement of centromeres in these species-specific rearrangements, which is more clearly seen in R11 and R19 (see Centromeres and Rearrangements section). This is further supported by the enrichment of satellite repeats near the breakpoints (e.g., 3% at R1 34 Mb, 13% at R20 25 Mb, 7.3% at R19 34.8 Mb, 37% at R17 49-50 Mb, and >2% at R7 9-12 Mb). Similar to mouse, the rat-specific breakpoints are rich in repeats (often >25% SINEs in human, >15% LTR, but <10% SINEs in mouse, and >40% L1s or >20% LTRs in rat).
Human Rearrangements
Intrachromosomal Rearrangements are Common in Human
Both intra- and interchromosomal rearrangements are identified in human by examining its mouse/rat synteny (Table 5; see Supplemental material), however, intrachromosomal events are more frequent by using other species as an outgroup. For instance, by placing the same set of the BAC-end mate pairs that have been mapped to the human/mouse/rat genomes to the recently released dog genome, we have found that among a total of 20 mouse/rat breakpoints identified in H7, eight are mouse/rat specific, seven are human specific due to rearrangements within H7 chromosomal arms or between the arms, and five are common to mouse/rat/dog (Fig. 3). The observed intrachromosomal changes in H7 are consistent with studies indicating that the present day H7 is a fusion product of an acrocentric chromosome and one arm of a submetacentric chromosome in an ancestral placental mammal, followed by paracentric and pericentric inversions (Richard et al. 2000). Frequent cytogenetic changes associated with cancer and a high percentage of segmental duplication (8.3% vs. 5% of the entire genome) have been reported in H7 (Hillier et al. 2003), and it would be useful to find out whether these features are related to the observed rearrangements.
Table 5.
The Human Genome Heterogeneity Revealed by the Human/Mouse/Rat Synteny
| Properties | Human chr | Mouse/rat synteny |
|---|---|---|
| Chromosomal associationsa | H3/H21, H4q/H8p, H12/H22, H21/H22, H13/H14b H1/H10, H7/H22, and H19p/H16q | Fragments from the two human chromosomes are adjacent to each other syntenic to the same mouse/rat chromosomal region |
| Intra-chromosomal Rearrangementsc | H1q, H2, H3, H4, H7, H8p, H9, H10, H11, H12, H13, H15, H16, H17, H18, and H20 | Obtained with dog as the outgroup, where dog is continuously syntenic to mouse/rat without interruption, but human breaks into two syntenic blocks |
| Two arms with unrelated synteny | H16, H8, H10, and H19d | P-arm and q-arm are syntenic to fragments from completely different mouse/rat chromosomes |
| Synteny uninterrupted by the centromere | H4, H5, H6, H11, and H12 | A single mouse/rat syntenic block continues from p-arm to q-arm without any rearrangements detected around the centromeric region |
| Synteny broken at the centromere | H7, H2, H8, H9, H10, H16, H18, H19, HX, and H20 | Different syntenic blocks were found on the two sides of the centromere |
| Intermediate rearrangements at the centromere | H1, H3 | Except two inversions at the first 780 kb of H1q, H1's centromere is within a single syntenic block. H3 splits at its centromere for rat, but not for mouse (see Table 4). |
Some of the associations are not exactly the same in the two rodents. For instance, further reorganization has occurred to H3/H21 in rat (see Table 4 and Supplemental material). In addition, we found several associations different from reported (Richard et al. 2000; Murphy et al. 2001; Stanyon et al. 2003). For instance, instead of H7partial/H16p and H16q/H19q identified in a number of placental mammals (Richard et al. 2000), we observed H19p(partial)/H16q and did not find any associations between H7 and H16.
The tip of H14 is adjacent to the tip of H13 corresponding to the same rodent chromosomes, whereas H15 is syntenic to rodent chromosomes that have no homology to either H13 or H14 (see Supplementary material).
These rearrangements account for about 58 of the 255 mouse/rat shared breakpoints shown in Table 2.
Evidence has indicated that the two arms of these chromosomes were unlinked in a primate ancestor (Haig 1999; Richard et al. 2000, 2003; Murphy et al. 2001; Carbone et al. 2002; Misceo et al. 2003).
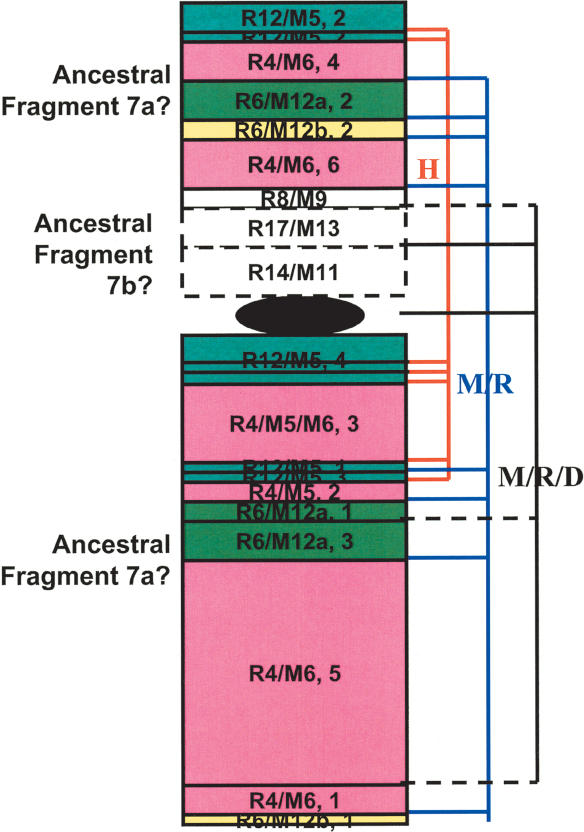
Figure 3.
Intrachromosomal rearrangements in H7. H7p's first 32 Mb and the entire H7q are syntenic to R4(1-86 Mb)/M5(27-3)/M6(3-57), R12(9.7-27)/M5(143-128), R6(49-64)/M12(25-39)a, and R6(143-148)/M12(110-115)b (numbers in parentheses are in megabases). However, each of these continuous mouse/rat fragments broke and formed multiple syntenic blocks that scatter on both arms of H7. Blocks belonging to the same mouse/rat fragment are in the same color and numerically numbered on the basis of their orders in the fragment. For example, fragment R12/M5 split into four blocks with the second (labeled as “R12/M5, 2” in the plot) syntenic to the beginning of H7p and the other three to H7q. These portions of H7 are possibly derived from the large ancestral fragment H7a (blocks with solid lines). The rest of H7p, syntenic to R8(22-25Mb)/M9(22-25), R17(51.6-58)/M13(20-15), and R14(87-99)/M11(6.3-17), are possibly from the small ancestral fragment H7b (blocks with dashed lines). Block sizes are somewhat arbitrary and do not reflect the actual fragment length. Using dog as outgroup, among a total of 20 breakpoints shown, seven are due to human-specific intrachromosomal rearrangements (red lines on the right), eight are mouse/rat specific (blue lines), and five are shared by mouse/rat/dog (black lines, solid lines indicating where mouse/rat and dog break at the same site, dashed lines indicating where mouse/rat and dog break at slightly different [within 1-2 Mb] sites).
Similar to H7, intrachromosomal rearrangements have been identified in many other human chromosomes with the exception of 5, 6, 14, 19, 21, 22, and X. Using dog as the outgroup, in total, we have identified about 58 intrachromosomal breakpoints and four interchromosomal breakpoints that are specific to human. In addition, interarm rearrangements have been found in a number of chromosomes including 3, 7, 9, 11, 17, 18, and 20. The use of chicken as the outgoup also indicates significantly more intrachromosomal events. Thus, intrachromosomal rearrangements are more common in human.
Synteny Across the Human Centromeres
Human chromosomes exhibit interesting rearrangement features around the centromeric regions, some with the mouse/rat synteny broken, whereas others with the synteny continuing from the p-arm to the q-arm without any interruptions (see Table 5 and Supplemental material). Possibly, this is related to the origins of the centromere. Chromosomes with the broken synteny (e.g., H16, H10) may have their centromere developed at a later time, and those with no or fewer rearrangements (e.g., H6, H1) may have kept the ancestral centromere. This is somewhat supported by studies indicating that H16 is a fusion product of two chromosomes in a primate ancestor (Misceo et al. 2003) and H1 represents an intact ancestral chromosome that has been fissioned in most placental species (Murphy et al. 2003). This is also consistent with studies that detected extensive duplications and rearrangements in sequences flanking the H10 centromere among primates (Jackson et al. 1999), and studies that found the H6 centromere in three distinctive regions among primates without rearrangements detected to account for its movement (termed centromere repositioning; Eder et al. 2003).
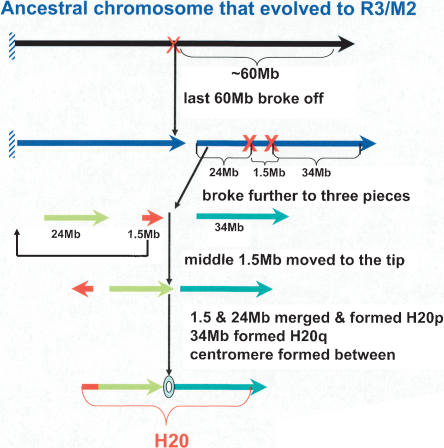
An interesting association of rearrangements with the centromere formation has been found in H20. If we replace the centromeric region of H20 with its beginning 1.5 Mb inverted, the entire chromosome forms a single block syntenic to the last 50/54 Mb of M2/R3. On this basis, we propose a possible path for the evolution of H20 (Fig. 4), which suggests that the centromere in H20 appeared after primates diverged from rodents, and that the process was apparently related to the shuffling of the 1.5-Mb fragment, encoding at least 30 genes, including SOX12 and TCF15 (Deloukas et al. 2001).
Figure 4.
A possible evolution path of H20, based on its synteny to M2/R3. The last 50-60-Mb fragment was broken off an ancestral chromosome that evolved to M2/R3. The fragment further split into three pieces with an approximate size of 24, 1.5, and 34 Mb, respectively. Then, the middle 1.5-Mb piece was moved to the tip, inverted, and fused with the 24-Mb piece, forming the p-arm, whereas the 34-Mb piece formed the q-arm. A centromere formed between the two arms where the original middle 1.5 Mb was located.
DISCUSSION
Comparative genomic studies have greatly advanced our understanding of biology (Mural et al. 2002; Waterston et al. 2002; Eichler and Sankoff 2003; Kirkness et al. 2003; Sankoff 2003; Thomas et al. 2003b; Rat Sequencing Project Consortium 2004) and provided opportunities to address questions that were unanswerable just a few years ago. Mouse and rat are important model systems for biomedical research, and a thorough comparison of their genomes in relation to the human genome is an important advance toward understanding differences and similarities of these species. Here, we report for the first time an analysis of chromosome-based whole-genome sequence comparison of the three species, focusing on large-scale rearrangements. Our study, together with other sequence analyses (e.g., micro-insertions and micro-deletions, nucleotide substitutions, duplications, etc.) provides the basis for a more complete picture of the evolutionary relationships of these species.
BES Mate Pairs for Mammalian Genome Comparison
This approach complements those based on direct comparison of the assembled genome sequences (Ma et al. 2002; Schwartz et al. 2003; Kalafus et al. 2004) for several reasons. First, repetitive sequences comprise a large portion of these genomes, which complicates both the assembly and interspecies comparison. Considering the complexity of the repeats and the number of matches that need to be analyzed, it is certainly not trivial to discriminate false positives, especially when comparing human with rodent, because of the lower sequence identity. Even though programs such as RepeatMasker (http://ftp.genome.washington.edu/RM/RepeatMasker.html) are used to mask many repeats, the current database does not include all repeats on the genomes, especially for mouse and rat. Second, the current mouse and rat assemblies are draft sequences, thus local misassemblies are possible, which further complicates the rearrangement analyses.
This approach greatly lessens these problems. BACs are large (usually 80-300 kb) and we only used their two ends (∼400-600 bp each) for the placement, thus avoiding possible repeat-rich regions and local misassemblies between the end sequences. In addition, we selected BACs on the basis of location, orientation, and distance of their paired ends, along with sequence matches, which not only effectively reduces false matches, but also provides additional information for the map construction. For example, the determination of synteny block orientation, required for studying inversions, was based not only on the order of clones within a block, but also on the orientation of each individual clone. This indeed helped in resolving local misassemblies. The size of BACs not only provides a sequence coverage significantly higher than most other anchors (Waterston et al. 2002; Rat Sequencing Project Consortium 2004), but also adds additional information. Under usual situations, a BAC should span a slightly different distance on the three genomes in the order of human > rat > mouse, reflecting the differences in genome size. Thus, the accuracy of the synteny can be evaluated to a certain extent on the basis of block size. As a result of these features, our data are interpretable even in the centromeric and other repeats-rich regions (see Supplemental material). Moreover, many breakpoints can be further confirmed by identifying bridging clones whose two ends span the same breakpoint on one genome with correct orientation and distance, but are in two separate syntenic blocks on the other genome. Importantly, BAC clones are substrates for biological studies and clones spanning interesting regions are immediately available for the research community (e.g., through http://bacpac.chori.org; Osogawa et al. 2000, 2001 and 2004).
Stability vs. Speciation
Human, mouse, and rat genomes have evolved from a common ancestral genome by balancing conservation that is required for stability and variations that are necessary for speciation. The comparison of the three genomes at the chromosomal level provides a more refined view of how these needs have been accommodated in the three species. Independent mathematical modeling of the syntenic block length distribution by us (Fig.1) and others (Pevzner and Tesler 2003a) supports the fragile breakage model, but not the random breakage model (Nadeau and Taylor 1984) for mammalian genome evolution, indicating that the genomes consist of regions that are conserved and regions that are prone to variations. Regions as large as >40 Mb with no detected rearrangements, as well as regions with frequently broken synteny and extensive rearrangements are both identified in the analyses. Large blocks are preserved most likely because they play a role in maintaining fundamental genome stability for all three species, whereas regions that tolerate structural rearrangements provide opportunities for evolution.
Our findings suggest that there is a basic structure underlying the three mammalian genomes that retains the fundamental function, and individual species tend to diverge from others provided this structure is maintained. As closely related species, mouse and rat share many of the breakpoints in the human genome and often have the same types of rearrangements. However, the breakpoints not shared between them indicate that mouse rearrangements tend to be more interchromosomal, whereas rat rearrangements are more often intrachromosomal. As a result (at least partially), compared with rat, mouse chromosomes display a large variation in the extent of rearrangement. X chromosomes provide another example (Fig. 5). Whereas the start and the end of MX correspond to HXp, a majority of its middle portion is syntenic to HXq. With rat, the opposite is observed. Considering that sex chromosomes rarely rearrange with autosomes, apparently mouse and rat differ from each other to the maximum allowed extent by a differential use of the two arms of HX.
Figure 5.
An apparent differential use of the two arms of HX in mouse and rat. Through several breakages and inversions, HX can be transformed to five fragments of MX as well as five fragments of RX (numerically numbered in blocks as MX1, MX2, or RX1, RX2...). Specifically, HXp corresponds to three fragments of MX (the last 20 Mb, a middle 17 Mb, and the start 16 Mb), and to two middle fragments of RX. HXq corresponds to three fragments of RX (the last 78 Mb, a middle 9 Mb, and the start 12 Mb), and to two middle fragments of MX. Blocks with the same fragment number in both mouse and rat are indicated with the same color (e.g., MX1 and RX1 are both represented as white blocks). Block sizes are somewhat arbitrary and do not reflect the actual fragment length. MX1: 3-19 Mb, MX2: 19.3-61, MX3: 80-63, MX4: 80-130, MX5a: 133-130, and MX5b: 150-135. RX1: 12-0.8, RX2: 29-12, RX3: 38-29, RX4a: 42-40, RX4b: 44-82, RX5a: 82-129, and RX5b: 132-160 (fragment ranges are all in Mb).
The comparison between human and mouse/rat also supports this “stability versus speciation” theory. As described, H20 is beautifully preserved among the three species. A few other chromosomes, on the other hand, display highly rearranged synteny (e.g., H19 [Dehal et al. 2001] and H22, see Supplemental material). Large chromosomes usually mix both stable and fragile regions within themselves. For instance, whereas the p-arm and the last 103 Mb of H2 are conserved with segments as large as 55 Mb without rearrangements found, frequently broken synteny was detected at its 95-139 Mb region, where a head-to-head fusion of two ancestral primate chromosomes (Yunis and Prakash 1982; Ijdo et al. 1991) as well as intra- and interchromosomal duplications (Fan et al. 2002) were found. Thus, the broken synteny is likely also due to rearrangements within the human lineage.
Centromeres and Rearrangements
Centromeres vary by size and composition as well as nearby sequence features (repeats, duplications, etc.; Jackson 2003), and play an essential role in mitosis and meiosis (Henikoff et al. 2001; Henikoff and Malik 2002; Malik and Henikoff 2002). Due to its sequence complexity, except for a few cytogenetic and other physical mapping studies (Jackson et al. 1999; Ventura et al. 2001, 2003; Wong and Choo 2001; Amor and Choo 2002; Eder et al. 2003; Murphy et al 2003), rearrangement analyses (Nadeau and Taylor 1984; Sankoff et al. 1997, 2000; Nadeau and Sankoff 1998; Waterston et al. 2002; Kent et al. 2003; Pevzner and Tesler 2003a,b; Sankoff 2003; Bourque et al. 2004; Rat Sequencing Project Consortium 2004) have been largely focused on noncentromeric regions. Although none of the chromosomes has its centromere sequence determined, the comparison of the three species reveals useful information regarding the centromere evolution nonetheless, which suggests that centromeres play a substantial role in reorganizing the genomes of the three species.
As described in previous sections, mouse- or rat-specific rearrangements are closely associated with centromeres. Rat acro- and metacentric chromosomes usually have a more complex synteny profile to mouse compared with the telocentrics, and several of them are more intrachromosomally rearranged (e.g., R1p, R19, R11, and R20). Mouse chromosomes are all acrocentric, and a majority of them are the products of extra interchromosomal fissions involving the centromeric regions (e.g., M1-M2, M4-M6, M10-M13, M15-M17, and M19. See Table 3). These observations raise many questions regarding the chromosomal morphology of the mouse/rat common ancestor and the centromere evolution in each rodent. For instance, R11 and R19 are presumably derived from ancestral chromosomes that were closer to M16 and M8, respectively. As the corresponding fragments of M16/M8 are noncentromeric, R11/R19 must have their centromere formed after the mouse/rat divergence. Did this cause the reorganization of these two chromosomes? If so, R18 (also metacentric) must follow a different mechanism, as it is also syntenic to noncentromeric regions in M18 (Fig. 2) but without rearrangements found, except for a few regions below 400 kb inverted. Did a new centromere form in R18, or did the old centromere disappear in M18? Rat metacentric chromosomes are usually more rearranged than the corresponding mouse chromosomes, suggesting that the mouse/rat ancestor consists of mostly acro- or telocentric chromosomes. If so, then why has the selection favored metacentrics in rat, and will more such chromosomes be formed in the future? As female meiosis drive can quickly change a species' karyotype (Pardo-Manuel de Villena and Sapienza 2001), have metacentric chromosomes emerged in rat through this mechanism?
Human chromosomes also display interesting features across their centromeric regions, some with the mouse/rat synteny broken, whereas others with the synteny uninterrupted (Table 5). These observations could be partially explained by the ancient centromere silencing and neocentromere emergence theory proposed for the centromere repositioning phenomenon observed in H6, H15, and HX in primates (Ventura et al. 2001, 2003; Eder et al. 2003), especially when ∼60 examples of constitutional human neocentromere distributed in 16 chromosomes have been reported, and neocentromere have been detected in at least two types of human cancers (Amor and Choo 2002). Our analyses indicate that this theory might also apply to a few mouse/rat chromosome pairs (e.g., M18/R18, R1/M7/M19). However, this theory alone cannot adequately explain the apparent association between the centromere evolution and rearrangements found in a number of chromosomes (e.g., H20, R11, and R19). Thus, additional mechanisms are involved.
Rat Closer to Human?
Compared with interchromosomal rearrangements, intrachromosomal events ought to be less harmful to an organism under usual situations, as only one chromosome is changed, which probably explains why intrachromosomal rearrangements are more frequently observed in these species. As for the mouse-specific interchromosomal rearrangements shown in Table 3, the majority of them involve the centromeric regions, most likely because changes in these regions are less harmful and can survive the selection, compared with those in the euchromatic regions, where more genes are encoded.
The rearrangement differences between the two rodents suggest that, except for the small inversions, overall, the rat genome might have a structure on a large scale closer to the human genome than the mouse genome. This is because, lacking the extra interchromosomal changes of mouse (Table 3), many rat fragments are closer to human. In addition, for those that are more rearranged (R1p, R11, and R19), the changes are mostly intra-chromosomal (Table 4), a mechanism that is also frequently seen in human (Table 5). In terms of chromosome morphology (and possibly the genome size as well), rat is also between mouse and human. However, a definite answer requires more complete sequencing (e.g., R12p and R13p may not be complete in the assembly), accurate assemblies, and thorough analyses.
Because the mouse and rat genomes are highly rearranged, it has been argued that these species may not mirror the genome organization of other mammals or other rodents (O'Brien et al. 2001). Although this is a valid point, we believe that mouse and rat can provide useful information that may not be obtained by focusing on more conserved species. This is because any ancestral fragments identified in their genomes are likely to be highly significant in maintaining the basic functional stability for all mammalian genomes. It is of concern that the current available mouse and rat genome sequences are from inbred strains (Waterston et al. 2002; Rat Sequencing Project Consortium 2004). It would be useful to analyze the genome of a wild strain and find out whether inbreeding has caused significant changes in the genome under the artificial selection pressure.
METHODS
BAC Clone Placement
The analyses were performed using genome assemblies of the human NCBI33 build, the dog genome canFam1 version, and the chicken genome version 2 downloaded from www.genome.ucsc.edu, the mouse NCBI30 build downloaded from ensembl (www.ensembl.org/Mus_musculus/), and the rat 3.1 version downloaded from www.ratgenome.hgsc.bcm.tmc.edu/. We compared ∼280,000 human BAC end sequence (BES) pairs, 192,000 mouse pairs, 138,000 rat pairs, as well as ∼100,000 pairs from other primate species (∼79,000 chimp pairs were downloaded from GenBank generated by RIKEN [Fujiyama et al. 2002]) to the three genomes as follows. BESs were first repeat-masked using Repeats-Masker (http://ftp.genome.washington.edu/RM/RepeatMasker.html) and then compared with the genome sequences using WU BLAST (http://sapiens.wustl.edu/blast/). Matches were selected on the basis of the alignment strength. When comparing BESs to the genome of the same species (e.g., human BESs to the human genome), matches with ≥90% sequence identities and at least half of the BES aligning onto the genome were selected. In cases involving different species (e.g., human BESs to the mouse genome), the best hit was selected for each BES from matches that had error rate <0.01 and length >100bp. For each clone that had both ends successfully placed on the genome, we examined three pieces of information as follows: (1) chromosome locations (its two ends on the same chromosome or not), (2) orientation (the two ends pointing toward each other or not, if on the same chromosome), and (3) distance between the two ends (if on the same chromosome, with the right orientation). We only selected clones that had two ends on the same chromosome, pointing toward each other, and with a distance of 1-400 kb between them.
Synteny Map Construction
From successfully mapped clones, we excluded those that had multiple placements and only selected those that were uniquely mapped to every genome involved for the map construction. We identified the synteny between the genomes using each BAC as a anchor, and blocks were built on the basis of the clone order, BAC orientation, and distance between the clones. For example, within a block, all BACs must be in the same order, with the same BAC orientation (forward or reverse), and gaps between the clones below the certain selected value on every genome involved. A block proceeded only if all these criteria were satisfied.
Acknowledgments
We thank the US Department of Energy and the National Institutes of Health for funding the large-scale BAC end-sequencing project for human, mouse, and rat. We are grateful to the human, mouse, dog, rat, and chicken sequencing consortiums for sequencing these genomes.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2663304. Article published online before print in September 2004.
References
- Amor, D.J. and Choo, K.H. 2002. Neocentromeres: Role in human disease, evolution, and centromere study. Am. J. Hum. Genet. 71: 695-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourque, G., Pevzner, P.A., and Tesler, G. 2004. Reconstructing the genomic architecture of ancestral mammals: Lessons from human, mouse, and rat genomes. Genome Res. 14: 507-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton-Davidian, J., Catalan, J., da Graca Ramalhinho, M., Ganem, G., Auffray, J.C., Capela, R., Biscoito, M., Searle, J.B., and da Luz Mathias, M. 2000. Rapid chromosomal evolution in island mice. Nature 403: 158. [DOI] [PubMed] [Google Scholar]
- Carbone, L., Ventura, M., Tempesta, S., Rocchi, M., and Archidiacono, N. 2002. Evolutionary history of chromosome 10 in primates. Chromosoma 111: 267-272. [DOI] [PubMed] [Google Scholar]
- Dehal, P., Predki, P., Olsen, A.S., Kobayashi, A., Folta, P., Lucas, S., Land, M., Terry, A., Ecale Zhou, C.L., Rash, S., et al. 2001. Human chromosome 19 and related regions in mouse: Conservative and lineage-specific evolution. Science 293: 104-111. [DOI] [PubMed] [Google Scholar]
- Deloukas, P., Matthews, L.H., Ashurst, J., Burton, J., Gilbert, J.G., Jones, M., Stavrides, G., Almeida, J.P., Babbage, A.K., Bagguley, C.L., et al. 2001. The DNA sequence and comparative analysis of human chromosome 20. Nature 414: 865-871. [DOI] [PubMed] [Google Scholar]
- Eder, V., Ventura, M., Ianigro, M., Teti, M., Rocchi, M., and Archidiacono, N. 2003. Chromosome 6 phylogeny in primates and centromere repositioning. Mol. Biol. Evol. 20: 1506-1512. [DOI] [PubMed] [Google Scholar]
- Eichler, E.E. and Sankoff, D. 2003. Structural dynamics of eukaryotic chromosome evolution. Science 301: 793-797. [DOI] [PubMed] [Google Scholar]
- Fan, Y., Newman, T., Linardopoulou, E., and Trask, B.J. 2002. Gene content and function of the ancestral chromosome fusion site in human chromosome 2q13-2q14.1 and paralogous regions. Genome Res. 12: 1663-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiyama, A., Watanabe, H., Toyoda, A., Taylor, T.D., Itoh, T., Tsai, S.F., Park, H.S., Yaspo, M.L., Lehrach, H., Chen, Z., et al. 2002. Construction and analysis of a human-chimpanzee comparative clone map. Science 295: 131-134. [DOI] [PubMed] [Google Scholar]
- Haig, D. 1999. A brief history of human autosomes. Philos. Trans. R Soc. Lond. B Biol. Sci. 354: 1447-1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff, S. and Malik, H.S. 2002. Centromeres: Selfish drivers. Nature 417: 227. [DOI] [PubMed] [Google Scholar]
- Henikoff, S., Ahmad, K., and Malik, H.S. 2001. The centromere paradox: Stable inheritance with rapidly evolving DNA. Science 293: 1098-1102. [DOI] [PubMed] [Google Scholar]
- Hillier, L.W., Fulton, R.S., Fulton, L.A., Graves, T.A., Pepin, K.H., Wagner-McPherson, C., Layman, D., Maas, J., Jaeger, S., Walker, R., et al. 2003. The DNA sequence of human chromosome 7. Nature 424: 157-164. [DOI] [PubMed] [Google Scholar]
- Ijdo, J., Baldini, A., Ward, D.C., Reeders, S.T., and Wells, R.A. 1991. Origin of human chromosome 2: An ancestral telomere-telomere fusion. Proc. Natl. Acad. Sci. 88: 9051-9055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860-921. [DOI] [PubMed] [Google Scholar]
- Jackson, M. 2003. Duplicate, decouple, disperse: The evolutionary transience of human centromeric regions. Curr. Opin. Genet. Dev. 13: 629-635. [DOI] [PubMed] [Google Scholar]
- Jackson, M.S., Rocchi, M., Thompson, G., Hearn, T., Crosier, M., Guy, J., Kirk, D., Mulligan, L., Ricco, A., Piccininni, S., et al. 1999. Sequences flanking the centromere of human chromosome 10 are a complex patchwork of arm-specific sequences, stable duplications and unstable sequences with homologies to telomeric and other centromeric locations. Hum. Mol. Genet. 8: 205-215. [DOI] [PubMed] [Google Scholar]
- Kalafus, K.J., Jackson, A.R., and Milosavljevic, A. 2004. Pash: Efficient genome-scale sequence anchoring by positional hashing. Genome Res. 14: 672-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent, W.J., Baertsch, R., Hinrichs, A., Miller, W., and Haussler, D. 2003. Evolution's cauldron: Duplication, deletion, and rearrangement in the mouse and human genomes. Proc. Natl. Acad. Sci. 100: 11484-11489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, U.J., Birren, B., Sheng, Y.L., Slepak, T., Mancino, V., Boysen, C., Kang, H.L., Simon, M.I., and Shizuya, H. 1996. Construction and characterization of a human Bacterial Artificial Chromosome library. Genomics 34: 213-218. [DOI] [PubMed] [Google Scholar]
- Kirkness, E.F., Bafna, V., Halpern, A.L., Levy, S., Remington, K., Rusch, D.B., Delcher, A.L., Pop, M., Wang, W., Fraser, C.M., et al. 2003. The dog genome: Survey sequencing and comparative analysis. Science 301: 1898-1903. [DOI] [PubMed] [Google Scholar]
- Ma, B., Tromp, J., and Li, M. 2002. PatternHunter: Faster and more sensitive homology search. Bioinformatics 18: 440-445. [DOI] [PubMed] [Google Scholar]
- Mahairas, G.G., Wallace, J.C., Smith, K., Swartzell, S., Holtzman, T., Keller, A., Shaker, R., Furlong, J., Young, J., Zhao, S., et al. 1999. Sequence tagged connectors: A sequence approach to mapping and scanning the human genome. Proc. Natl. Acad. Sci. 96: 9739-9744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik, H.S. and Henikoff, S. 2002. Conflict begets complexity: The evolution of centromeres. Curr. Opin. Genet. Dev. 12: 711-718. [DOI] [PubMed] [Google Scholar]
- Misceo, D., Ventura, M., Eder, V., Rocchi, M., and Archidiacono, N. 2003. Human chromosome 16 conservation in primates. Chromosome Res. 11: 323-326. [DOI] [PubMed] [Google Scholar]
- Mural, R.J., Adams, M.D., Myers, E.W., Smith, H.O., Miklos, G.L., Wides, R., Halpern, A., Li, P.W., Sutton, G.G., Nadeau, J., et al. 2002. A comparison of whole-genome shotgun-derived mouse chromosome 16 and the human genome. Science 296: 1661-1671. [DOI] [PubMed] [Google Scholar]
- Murphy, W.J., Stanyon, R., and O'Brien, S.J. 2001. Evolution of mammalian genome organization inferred from comparative gene mapping. Genome Biol. 2: REVIEWS0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, W.J., Fronicke, L., O'Brien, S.J., and Stanyon, R. 2003. The origin of human chromosome 1 and its homologs in placental mammals. Genome Res. 13: 1880-1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau, J.H. and Sankoff, D. 1998. Counting on comparative maps. Trends Genet. 14: 495-501. [DOI] [PubMed] [Google Scholar]
- Nadeau, J.H. and Taylor, B.A. 1984. Lengths of chromosomal segments conserved since divergence of man and mouse. Proc. Natl. Acad. Sci. 81: 814-818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson, S., Helou, K., Walentinsson, A., Szpirer, C., Nerman, O., and Stahl, F. 2001. Rat-mouse and rat-human comparative maps based on gene homology and high-resolution zoo-FISH. Genomics 74: 287-298. [DOI] [PubMed] [Google Scholar]
- O'Brien, S.J., Menotti-Raymond, M., Murphy, W.J., Nash, W.G., Weinberg, J., Stanyon, R., Copeland, N.G., Jenkins, N.A., Womack, J.E., and Marshall Graves, J.A. 1999. The promise of comparative genomics in mammals. Science 286: 458-462, 479-481. [DOI] [PubMed] [Google Scholar]
- O'Brien, S.J., Eizirik, E., and Murphy, W.J. 2001. Genomics. On choosing mammalian genomes for sequencing. Science 292: 2264-2266. [DOI] [PubMed] [Google Scholar]
- Osoegawa, K., Tateno, M., Woon, P.-Y., Frengen, E., Mammoser, A.G., Catanese, J.J., Hayashizaki, Y., and de Jong, P.J. 2000. Bacterial artificial chromosome libraries for mouse sequencing and functional analysis. Genome Res. 10: 116-128. [PMC free article] [PubMed] [Google Scholar]
- Osoegawa, K., Mammoser, A.G., Wu, C., Frengen, E., Zeng, C., Catanese, J.J., and de Jong, P.J. 2001. A bacterial artificial chromosome library for sequencing the complete human genome. Genome Res. 11: 483-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osoegawa, K., Zhu, B., Li Shu, C., Ren, T., Cao, Q., Vessere, G.M., Lutz, M.M., Jensen-Seaman, M.I., Zhao, S., and de Jong, P. 2004. BAC resources for the rat genome project. Genome Res. 14: 780-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo-Manuel de Villena, F. and Sapienza, C. 2001. Female meiosis drives karyotypic evolution in mammals. Genetics 159: 1179-1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pevzner, P. and Tesler, G. 2003a. Human and mouse genomic sequences reveal extensive breakpoint reuse in mammalian evolution. Proc. Natl. Acad. Sci. 100: 7672-7677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 2003b. Genome rearrangements in mammalian evolution: Lessons from human and mouse genomes. Genome Res. 13: 37-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rat Sequencing Project Consortium. 2004. Genome sequence of the brown Norway rat yields insights into mammalian evolution. Nature 428: 493-521. [DOI] [PubMed] [Google Scholar]
- Richard, F., Lombard, M., and Dutrillaux, B. 2000. Phylogenetic origin of human chromosomes 7, 16, and 19 and their homologs in placental mammals. Genome Res. 10: 644-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 2003. Reconstruction of the ancestral karyotype of eutherian mammals. Chromosome Res. 11: 605-618. [DOI] [PubMed] [Google Scholar]
- Sankoff, D. 2003. Rearrangements and chromosomal evolution. Curr. Opin. Genet. Dev. 13: 583-587. [DOI] [PubMed] [Google Scholar]
- Sankoff, D., Parent, M.N., Marchand, I., and Ferretti, V. 1997. In Eighth annual symposium on combinatorial pattern matching, lecture notes in computer science, Vol. 1264, pp. 262-274. Springer, New York. [Google Scholar]
- Sankoff, D., Parent, M.N., and Bryant, D. 2000. In Comparative genomics: Empirical and analytical approaches to gene order dynamics, map alignment, and the evolution of gene families, (eds. D. Sankoff and J. Nadeau), pp. 299-305. Kluwer, Dordrecht, The Netherlands.
- Schwartz, S., Kent, W.J., Smit, A., Zhang, Z., Baertsch, R., Hardison, R.C., Haussler, D., and Miller, W. 2003. Human-mouse alignments with BLASTZ. Genome Res. 13: 103-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanyon, R., Yang, F., Cavagna, P., O'Brien, P.C., Bagga, M., Ferguson-Smith, M.A., and Wienberg, J. 1999. Reciprocal chromosome painting shows that genomic rearrangement between rat and mouse proceeds ten times faster than between humans and cats. Cytogenet. Cell Genet. 84: 150-155. [DOI] [PubMed] [Google Scholar]
- Stanyon, R., Stone, G., Garcia, M., and Froenicke, L. 2003. Reciprocal chromosome painting shows that squirrels, unlike murid rodents, have a highly conserved genome organization. Genomics 82: 245-249. [DOI] [PubMed] [Google Scholar]
- Thomas, J.W., Schueler, M.G., Summers, T.J., Blakesley, R.W., McDowell, J.C., Thomas, P.J., Idol, J.R., Maduro, V.V., Lee-Lin, S.Q., Touchman, J.W., et al. 2003a. Pericentromeric duplications in the laboratory mouse. Genome Res. 13: 55-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, J.W., Touchman, J.W., Blakesley, R.W., Bouffard, G.G., Beckstrom-Sternberg, S.M., Margulies, E.H., Blanchette, M., Siepel, A.C., Thomas, P.J., McDowell, J.C., et al. 2003b. Comparative analyses of multi-species sequences from targeted genomic regions. Nature 424: 788-793. [DOI] [PubMed] [Google Scholar]
- Venter, J.C., Adams, M.D., Myers, E.W., Li, P.W., Mural, R.J., Sutton, G.G., Smith, H.O., Yandell, M., Evans, C.A., et al. 2001. The sequence of the human genome. Science 291: 1304-1351. [DOI] [PubMed] [Google Scholar]
- Ventura, M., Archidiacono, N., and Rocchi, M. 2001. Centromere emergence in evolution. Genome Res. 11: 595-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura, M., Mudge, J.M., Palumbo, V., Burn, S., Blennow, E., Pierluigi, M., Giorda, R., Zuffardi, O., Archidiacono, N., Jackson, M.S., et al. 2003. Neocentromeres in 15q24-26 map to duplicons which flanked an ancestral centromere in 15q25. Genome Res. 13: 2059-2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walentinsson, A., Helou, K., and Levan, G. 2001. A dual-color FISH gene map of the proximal region of rat Chromosome 4 and comparative analysis in human and mouse. Mamm. Gen. 12: 900-908. [DOI] [PubMed] [Google Scholar]
- Waterston, R.H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J.F., Agarwal, P., Agarwala, R., Ainscough, R., Alexandersson, M., An, P., et. al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520-562. [DOI] [PubMed] [Google Scholar]
- Wong, L. and Choo, K. 2001. Centromere on the Move. Genome Res. 11: 513-516. [DOI] [PubMed] [Google Scholar]
- Yunis, J.J. and Prakash, O. 1982. The origin of man: A chromosomal pictorial legacy. Science 215: 1525-1530. [DOI] [PubMed] [Google Scholar]
- Zhao, S. 2001. A comprehensive BAC resource. Nucleic Acids Res. 29: 141-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, S., Malek, J., Mahairas, G., Fu, L., Nierman, W., Venter, J.C., and Adams, M.D. 2000. Human BAC ends quality assessment and sequence analyses. Genomics 63: 321-332. [DOI] [PubMed] [Google Scholar]
- Zhao, S., Shatsman, S., Ayodeji, B., Geer, K., Tsegaye, G., Krol, M., Gebregeorgis, E., Shvartsbeyn, A., Russell, D., Overton, L., et al. 2001. Mouse BAC ends quality assessment and sequence analyses. Genome Res. 11: 1736-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- ftp://ftp.tigr.org/pub/data/Bac_Resource/HumanMouseRatSynteny; mouse/rat/human synteny data used in this study.
- http://bacpac.chori.org; providing BAC clones.
- http://ftp.genome.washington.edu/RM/RepeatMasker.html; providing the RepeatMasker program.
- http://sapiens.wustl.edu/blast/; providing the WUBLAST program.
- www.ensembl.org; providing genomic sequence resource for various species.
- www.ensembl.org/Mus_musculus/; providing genomic sequence resource for mouse.
- www.genome.ucsc.edu; providing genomic sequence resource for various species.
- www.ratgenome.hgsc.bcm.tmc.edu/; providing the rat genomic sequence resource.