Abstract
We have performed the first genome-wide analysis of the Inverted Repeat (IR) structure in the human genome, using a novel and efficient software package called Inverted Repeats Finder (IRF). After masking of known repetitive elements, IRF detected 22,624 human IRs characterized by arm size from 25 bp to >100 kb with at least 75% identity, and spacer length up to 100 kb. This analysis required 6 h on a desktop PC. In all, 166 IRs had arm lengths >8 kb. From this set, IRs were excluded if they were in unfinished/unassembled regions of the genome, or clustered with other closely related IRs, yielding a set of 96 large IRs. Of these, 24 (25%) occurred on the X-chromosome, although it represents only ∼5% of the genome. Of the X-chromosome IRs, 83.3% were ≥99% identical, compared with 28.8% of autosomal IRs. Eleven IRs from Chromosome X, one from Chromosome 11, and seven already described from Chromosome Y contain genes predominantly expressed in testis. PCR analysis of eight of these IRs correctly amplified the corresponding region in the human genome, and six were also confirmed in gorilla or chimpanzee genomes. Similarity dot-plots revealed that 22 IRs contained further secondary homologous structures partially categorized into three distinct patterns. The prevalence of large highly homologous IRs containing testes genes on the X- and Y-chromosomes suggests a possible role in male germ-line gene expression and/or maintaining sequence integrity by gene conversion.
The recent completion of the human DNA sequence (Lander et al. 2001) provides a unique historical opportunity to fully describe the complete catalog of DNA structural genomic elements. It is now clear that the human genome contains a remarkably complex pattern of both ancient and recent duplications, with as much as 5% of our genome consisting of recent segmental duplications (Bailey et al. 2002; Samonte and Eichler 2002). These generally range in size from 1 to >200 kb, have 90%-100% sequence identity, and have been identified on every human chromosome, mainly in pericentromeric and subtelomeric regions. These segmental duplications are observed both within chromosomes and between nonhomologous chromosomes, and represent important regions for genome evolution and plasticity.
Inverted Repeats (IRs) make up one class of human duplications, which consist of two arms of similar DNA—with one inverted and complemented relative to the other—around a central, usually nonhomologous spacer. Large IRs have been observed in physical maps of the X-chromosome, and have been associated with chromosomal rearrangements and gene deletions (Lafreniere et al. 1993; Small et al. 1997; McDonell et al. 2000; Aradhya et al. 2001). Recently, the finished sequence of the human Y-chromosome has revealed the presence of several remarkably large and highly homologous IRs, up to 1.4 Mb in size and 99.97% identity, which contain Y-specific genes expressed in testes and thought to be required for spermatogenesis (Skaletsky et al. 2003). Gene conversion is evidently maintaining the homology between the arms of these palindromes, and thus the sequence integrity and function of the genes in the absence of meiotic recombination between homologs (Rozen et al. 2003).
IRs are widespread in both prokaryotic and eukaryotic genomes, and have been associated with a myriad of possible functions, reviewed in Pearson et al. (1996). Some IRs are capable of extruding into DNA cruciforms, structures in which the normal double-stranded DNA denatures, and complementary arms in the same strand form intrastrand double helices, or stems. The spacer regions become unpaired loops at the top of each stem, and the four-way junction where the bases of the stems meet is indistinguishable from a Holliday structure. The ability of particular IRs to extrude into cruciforms depends on the size and sequence composition of both the arms and spacer region. The energy barrier required to denature the DNA and extrude into cruciforms is reduced by the unwinding torsional stress induced by local negative supercoiling (Shlyakhtenko et al. 2000; Benham et al. 2002).
To facilitate the study of genomic IRs, a novel and efficient computer program called Inverted Repeats Finder (IRF) was developed, and the first genome-wide analysis of IRs in the human genome was performed. The largest and most homologous IRs found in the human genome, described in detail in this report, showed a disproportionately high representation on the X-chromosome. The program IRF, as well as updated descriptions of the IR structure of future assemblies of the human genome, will be made publicly available at the Inverted Repeat Data Base (IRDB) (http://tandem.bu.edu).
RESULTS
Inverted Repeats Finder (IRF) Reveals a Preponderance of Large, Highly Homologous IRs on the X-Chromosome
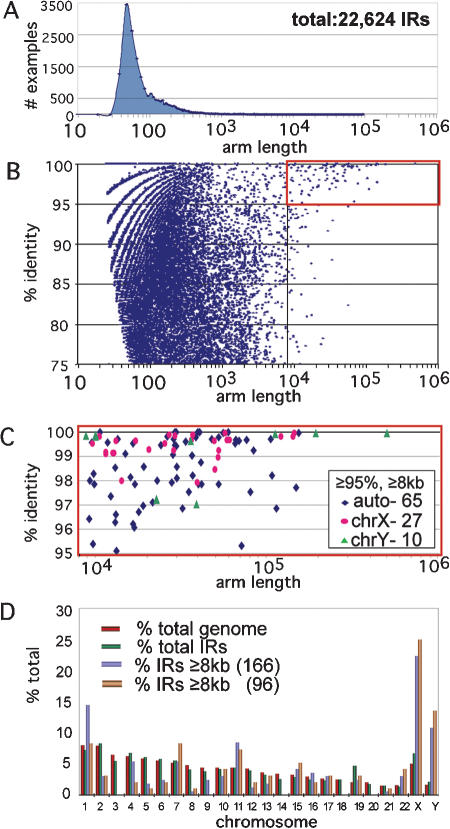
IRF was run against each human chromosome from the latest available version of the human genome sequence (hg16). Repeat-Masker (Jurka 2000; A.F.A. Smit and P. Green, unpubl.; http://ftp.genome.washington.edu/RM/RepeatMasker.html) was used to exclude repetitive elements during identification of candidate IRs, thereby blocking detection of hundreds of thousands of biologically uninteresting IRs that consist of two nearby homologous, oppositely oriented interspersed repeats. However, during alignment and extension of IRs, repetitive elements were included. Short and low-identity IRs initially detected by IRF were filtered out (see Methods), leaving a set of 22,624 IRs with spacer lengths up to 100 kb that were ≥75% identical between arms (Fig. 1A,B). These IRs had arm lengths from 25 bp up to 500 kb, albeit skewed toward lengths under 100 bp (Fig. 1A). The spacer lengths were skewed toward lengths <500 bp (mean of 9358 bp, median of 327 bp; data not shown).
Figure 1.
Results from Inverted Repeat Finder. (A) Distribution of arm lengths of human IRs detected by IRF, mean of 551 bp, median of 85 bp. Note the log scale on the x-axis. (B) Distribution of arm length (x-axis) by percent identity between arms (y-axis). The x-axes in A and B correspond for direct comparison. The “birdcrest” pattern observed resulted from limited ranges of percent identity values for the shortest most similar repeats, for example, 1/25 bp, 1/26 bp,..., 2/25 bp, 2/26 bp,.... As IR length increases, the pattern becomes less constrained. In all, 166 IRs had arm lengths ≥8000 bp (vertical line), of which 37 were on the X-chromosome and 18 on the Y-chromosome. (C) IRs detected by IRF that are ≥8000 bp, ≥95% identical, of which 27 are from the X-chromosome and 10 from the Y-chromosome. (D) Comparison on each human chromosome of percent total IRs (22,624), percent IRs ≥8 kb (166) detected by IRF, and percent IRs ≥8 kb after exclusion (96; see Supplemental S1). Values are compared to the percent of total assembled genome (3.07 × 109 bp) for each chromosome.
Figure 1B suggests that in general there is no correlation between arm length and percent similarity. However, there did appear to be a distinct outlying subset of large IRs (arm lengths ≥8 kb) and high arm-to-arm identity (≥95%; Fig. 1C). Therefore, in this report we concentrated on the set of 166 large IRs detected by IRF (>8 kb; Fig. 1B,C). The effectiveness of IRF is demonstrated by the detection of nine of the 10 large IRs recently described on the Y-chromosome (Fig. 1C, green triangles). The two largest and most similar IRs detected by IRF corresponded precisely to the 496-kb IR (P5) and 190-kb IR (P4) on the Y-chromosome (Table 1; Skaletsky et al. 2003). IRF detected but failed to extend fully two of the Chromosome Y IRs (P1 and P2), owing to large insertions/deletions in the arms. Three X-chromosome IRs detected by IRF with arm identities >99% and arm lengths of 119.3 kb, 35.8 kb, and 11.4 kb were previously observed in physical maps (IRX-70.95, IRX152.06, and IRX-152.30; Table 1; Lafreniere et al. 1993; Small et al. 1997; Aradhya et al. 2001).
Table 1.
Inverted Repeats in the Human Genome, >8000 bp, >99% Homology
| IR name (chromosome-pos)a | Chromo band | Arm (kb) | Spacer (kb) | % identity | Gene | Tissue | RT PCR/Northernb | Secondary structurec |
|---|---|---|---|---|---|---|---|---|
| X chromosome | ||||||||
| IRX-47.303 | Xp11.23 | 28.3 | 2.796 | 99.55 | SSX4 | Testes, CTA gene | + (Gure et al. 2002) | + (4) |
| IRX-50.36 | Xp11.22 | 25.1 | 7.392 | 99.53 | None | — | ||
| IRX-50.79 | Xp11.22 | 36.4 | 99.854 | 99.89 | MAGE-D4 | Testes, ubiquitous | + (Chomez et al. 2001) | — |
| IRX-51.17g | Xp11.22 | 142.2 | 8.34 | 99.97 | GAGE-D2,3 | Testes, CTA gene | + (Zendman et al. 2003a) | — |
| IRX-51.4958 | Xp11.22 | 28.7 | 21.642 | 99.87 | GAGE-D2 | Testes, CTA gene | + (Zendman et al. 2003a) | + (2) |
| IRX-51.73 | Xp11.22 | 59.8 | 0.239 | 99.67 | SSX2 | Testes, CTA gene | + (Gure et al. 2002) | + (1) |
| IRX-54.4798 | Xp11.21 | 26.7 | 12.909 | 99.89 | BC035907 | Breast mRNA | — | |
| IRX-61.28 | Xq11.2 | 56.6 | 8.145 | 99.95 | AK056835 | Prostate mRNA | — | |
| IRX-69.83 | Xq13.1 | 57.7 | 0.002 | 99.70 | AK093553 | Thymus, mRNAs | + (1) | |
| IRX-70.95 | Xq13.1 | 119.3 | 0.417 | 99.80 | DMRTC1 | Testes, kidney, pancreas | + (Ottolenghi et al. 2002) | — |
| IRX-71.13 | Xq13.2 | 9.5 | 71.919 | 99.55 | BC041956 | Brain mRNA | + (2) | |
| IRX-100.37 | Xq22.1 | 140.6 | 10.764 | 99.84 | NXF2 | Testes | + (Wang et al. 2001) | — |
| IRX-102.05 | Xq22.3 | 20.3 | 59.982 | 99.27 | AK091321 | Brain mRNA | + (2) | |
| IRX-104.30 | Xq22.3 | 12.6 | 8.677 | 99.13 | None | - | ||
| IRX-118.01 | Xq24 | 48.9 | 62.046 | 99.65 | PEPP-2(OTEX) | Testes | + (Wayne et al. 2002) | — |
| IRX-133.62 | Xq26.1 | 52.0 | 13.852 | 99.25 | MGC27005 | Testes mRNA | +(4) | |
| IRX-139.37 | Xq27 | 12.7 | 3.938 | 99.13 | SPANXA1,2 | Testes, CTA gene | + (Zendman et al. 2003b) | + (4) |
| IRX-144.56 | Xq27.3 | 10.5 | 0.004 | 99.84 | AW235137 | ESTs only | — | |
| IRX-147.49 | Xq28 | 28.3 | 41.009 | 99.23 | MAGE-A9f | Testes, CTA gene | + (Chomez et al. 2001) | — |
| IRX-150.52 | Xq28 | 51.2 | 8.994 | 98.96e | MAGA-A2,3,6 | Testes, CTA gene | + (Chomez et al. 2001) | + (2) |
| IRX-152.06 | Xq28 | 11.4 | 37.625 | 99.11 | None | — | ||
| IRX-152.30 | Xq28 | 35.8 | 21.759 | 99.69 | LAGE2A | Testes, ovary, CTA gene | + (Chen et al. 1997) | — |
| Y chromosome | ||||||||
| IRY-15.142 | P8 | 35.8 | 3.408 | 99.64 | VCY | Testes | — | |
| IRY-16.95 | P7 | 8.7 | 12.638 | 99.86 | None | |||
| IRY-17.35 | P6 | 110.0 | 46.229 | 99.92 | None | |||
| IRY-19.01 | P5 | 495.5 | 3.457 | 99.97 | CDY | Testes | ||
| IRY-19.72 | P4 | 190.2 | 39.642 | 99.97 | HSFY | Testes | ||
| IRY-23.2 | P3d | 283 | 169 | 99.94 | PRY | Testes | + (2) | |
| IRY-24.201 | P2d | 122.0 | 2.1 | 99.97 | DAZ | Testes | ||
| IRY-25.12 | P1.1 | 9.9 | 3.945 | 99.84 | None | |||
| IRY-25.81 | P1d | 1450 | 2.1 | 99.97 | DAZ | Testes | + (1) | |
| IRY-26.55 | P1.2 | 9.9 | 3.945 | 99.89 | None | |||
| Autosomes | ||||||||
| IR1-16.41 | 1p36.13 | 28.8 | 58.601 | 99.45 | None | — | ||
| IR1-103.57 | 1p21.1 | 29.4 | 16.216 | 99.96 | AMY1,2 | Salivary gland | + (6) | |
| IR1-147.04 | 1q21.12 | 26.1 | 6.739 | 99.71 | Histones | Ubiquitous | — | |
| IR2-95.69 | 2q11.1 | 82.7 | 82.801 | 99.14 | TRIM-43 | Placenta mRNA | + (2) | |
| IR6-26.8590 | 6p22.1 | 29.2 | 23.853 | 99.37 | No genes | + (2) | ||
| IR7-62.25 | 7q11.21 | 75.5 | 16.941 | 99.65 | BC036215 | Testes mRNA | — | |
| IR7-64.48 | 7q11.21 | 51.7 | 57.283 | 99.72 | None | + (6) | ||
| IR7-142.79 | 7q34 | 65.4 | 75.992 | 99.67 | BC043153 | Testes mRNA | + (3) | |
| IR7-142.86 | 7q34 | 28.3 | 99.969 | 99.70 | None | — | ||
| IR7-143.36 | 7q35 | 67.9 | 41.373 | 99.56 | AF327904 | Kidney, liver mRNAs | + (3) | |
| IR7-143.41 | 7q35 | 23.8 | 57.199 | 99.46 | None | + (3) | ||
| IR11-89.34 | 11q14.3 | 103.9 | 3.225 | 99.57 | RNF18 | Testes, kidney, spleen | + (Yoshikawa et al. 2000) | + (1) |
| IR13-62.12 | 13q21.31 | 10.6 | 31.553 | 99.88 | No genes | + (3) | ||
| IR15-28.49 | 15q13.2 | 58.6 | 93.355 | 99.57 | AK09040 | Spleen mRNA | + (5) | |
| IR17-19.19 | 17p11.2 | 38.0 | 48.103 | 99.74 | GRAP | Various | + (3) | |
| IR17-58.63 | 17q23.2 | 16.5 | 75.248 | 99.35 | No genes | — | ||
| IR22-20.15 | 22q11.21 | 64.5 | 48.672 | 99.16 | HIC2, AL133030 | Brain, Testes mRNA | — |
IR name-IR chromosome-genome position (in megabase pairs); for example, IRX-47.303 is the IR from the X-chromosome, center of spacer, at genomic position 47.303 Mb.
RT-PCR or Northern blot confirming testes expression. Reference in parentheses. Y-chromosome genes not included.
Secondary structure. (+) Secondary structure detected by dot-plot.—No secondary structure detected. (1) IR in IR (e.g., Fig. 3A). (2) Spacer homologous to arm (e.g., Fig. 3B). (3) IR intertwined with another IR (e.g., Fig. 3C). (4) Multigene family structure. (5) IR15-28.49 and IR15-30.53 (Supplemental data S2), separated by ∼2 Mb, are homologous to each other. (6) Secondary structure not one of the patterns (1) to (5).
As reported in Skaletsky et al. (2003). IRY-23.2 included in Table 1 replaces an internal IR (IRY-23.3047) (Supplemental data S1 and S2).
IRX-150.52 included for complete description of confirmed testes genes, although slightly less than 99% sequence identity (98.96%).
The MAGE-A9 genes were actually found in the arms of a large IR (IRX-147.36) which contains IRX-147.49 in its spacer. IRX-147.36 (arm length: 29.1 kb, spacer 164.9 kb, % identity 99.74) was detected by IRF when a spacer ≤500 kb was used (Supplemental data S2).
The percentage of the total set of 22,624 IRs (% total IRs) detected by IRF on each chromosome was approximately proportional to the chromosome size (% total genome), suggesting no chromosome-specific difference in the density of IRs in general (Fig. 1D). However, for IRs ≥8 kb, the percentage was significantly increased on both the X- and Y-chromosomes, shown for IRs with ≥95% arm identity in Figure 1C. Of the 166 IRs detected by IRF that are ≥8 kb, ≥75% arm identity, 37 (22.3%) were detected on the X-chromosome and 18 (10.8%) on the Y-chromosome, although these chromosomes represent only ∼5% and ∼1.6% of the genome, respectively (Fig. 1D).
To produce the most robust list of large IRs possible, the 166 largest IRs detected by IRF were evaluated by visualizing them on the UCSC Genome Browser using the custom track file provided by IRF (see Methods). Note that when viewing these custom tracks, a striking mirror symmetrical pattern for the arms of each IR is seen in the RepeatMasker Tracks of the UCSC Genome Browser. We took a conservative approach and removed all IRs that were possible false positives due to assembly errors, such as IRs that span gaps (12) or abut gaps (9) (Supplemental data S1). However, several of the IRs excluded as potential false positives may be confirmed as true IRs upon further refinement of the sequence assembly. Conversely, it is possible that the arms of highly homologous IRs may have been inadvertently combined into a single sequence, resulting in false negatives undetected by IRF. It will be important to compare the results of IRF from different builds of the human genome available at IRDB (http://tandem.bu.edu) to re-evaluate IRs that were excluded as potential false positives in the current set.
Overall, 70 of the 166 large IRs initially detected by IRF in hg16 build 34 were excluded based on several criteria (see Methods; Supplemental data S1), yielding a final set of 96 IRs (Supplemental data S2). This exclusion process did not significantly alter the distribution of IRs across the chromosomes and specifically did not affect the significance of the high proportion of IRs found on the X-chromosome (24 IRs, 25%) and Y-chromosome (13 IRs, 13.6%; Fig. 1D). The high proportion of X-chromosome IRs did not appear to be caused by any bias in the quality of the DNA sequence, as the X-chromosome has a relatively high proportion of gaps (Kent et al. 2002). Table 1 lists the IRs that have ≥99% arm identity. Remarkably, 20 of the 24 (83.3%) X-chromosome IRs were ≥99% identical, whereas only 17 of the 59 (28.8%) autosomal IRs were ≥99% identical (Table 2).
Table 2.
Total Human IRs >8000 bp on X- and Y-chromosomes and Autosomes
| ≥99%
|
≥95% to 99%
|
≥75% to 95%
|
Total (≥75%)
|
|||||
|---|---|---|---|---|---|---|---|---|
| Arm identity | Total | Known testes gene | Total | Known testes gene | Total | Known testes gene | Total | Known testes gene |
| X-chromosome | 20 | 10 | 3 | 1 | 1 | 0 | 24 | 11 |
| Y-chromosomea | 10 | 6 | 0 | 0 | 3 | 1 | 13 | 7 |
| Autosomes | 17 | 1 | 17 | 0 | 25 | 0 | 59 | 1 |
| Totals | 43 | 17 | 22 | 1 | 30 | 1 | 96 | 19 |
Human IRs with longer spacer length (up to 500 kb) were also examined. IRF detected a total of 31,163 IRs with spacer length ≤500 kb, ≥75% arm identity representing an additional 8539 IRs (27.4%). Of these IRs, 486 had a ≥8-kb arm length. Not surprisingly, a much higher proportion of these (151/486, 31.1%) span gaps in the sequence than do IRs with spacers ≤100 kb (12/166, 7.2%). A lower percentage of large IRs with spacer lengths ≤500 kb were found on the X-chromosome (56/486, 11.6%) as compared with IRs with spacer size ≤100 kb (37/166, 22.3%; see Fig. 1D). For all IRs with a spacer ≤500 kb, the average spacer size for X-chromosome IRs (mean 100 kb, median 61 kb) was significantly smaller than for autosomes (mean 210 kb, median 187 kb). However, IRs with spacers ≤500 kb and arm identity ≥99% were overrepresented on the X-chromosome (29/103, 28.2%). The high percentage of IRs with spacers ≤500 kb that span gaps suggests that these may represent a less reliable data set than IRs with spacers ≤100 kb. IRs with spacer sizes ≤500 kb will be included in the IRF analyses of the current and future assemblies of the human genome available in IRDB (http://tandem.bu.edu).
Large IRs on the X-Chromosome Predominantly Contain Testes Genes
Analysis of the genes contained within the set of 96 large IRs revealed a striking preference for genes that are predominantly or exclusively expressed in testes (Table 1), organized in opposite orientation on either arm of the IR, similar to the Y-chromosome (Skaletsky et al. 2003). Eleven large IRs from Chromosome X and one from Chromosome 11 contain a gene for which either an RT-PCR or a Northern blot directly demonstrating testes expression has been published (Table 1). Eight of these genes are expressed exclusively in the testes (from normal tissues), and four were expressed in at least one other tissue (Table 1). Eight of the Chromosome X genes were identified as Cancer-testes antigen (CTA) genes (Table 1), which are expressed predominantly or exclusively from testes, and in certain cancers (Zendman et al. 2003a; Scanlan et al. 2004). These results demonstrate that the human X-chromosome contains a preponderance of large, highly homologous IRs that contain testes genes (Table 2). Approximately 20% of the ∼52 genes on the human X-chromosome that are expressed predominantly in testes (from the GNF atlas 2 database; see Methods) are represented in these IRs. Many of the remaining IRs listed in Table 1 contained mRNAs cloned from various cDNA libraries. A representative mRNA and tissue was listed for each, with testes mRNAs preferentially identified when present, although these were not counted as known testes genes in Tables 1 and 2.
Analysis of IRs in the Mouse Genome
IRF will be useful for the analysis of any sequenced genome for which RepeatMasking is possible. For comparison to a non-primate mammal, we ran IRF on the current version of the mouse genome (NCBI build 32; Waterston et al. 2002). The large proportion of unfinished sequence and many small and large gaps in the current mouse assembly reduce confidence in the reliability of the set of IRs detected by IRF. For example, IRF detected 303 large IRs (spacer ≤100 kb, arm ≥8 kb, ≥75% arm identity) of which 196 (64.6%) span gaps, as compared with 12/166 (7.2%) of large IRs that span gaps in the human analysis. Furthermore, many of the internal IR arm/spacer boundaries were found exactly at the end of an assembled BAC sequence, which further suggests the possibility of an assembly error. Nevertheless, of the 107 large IRs detected in the mouse that do not span gaps, 46 (42.9%) are found on the X-chromosome. Of the nine large IRs on the mouse X-chromosome that contain genes, six contain testes genes, including mouse SSX genes (SSX4 and 5 and SSXB7), testes-expressed homeobox genes (TgiFx1 and TEX2), a testes-specific ferritin-like gene (FH17), and BC061169 (Xmr), which contains a Cor1 domain, a component of the chromosome core in the meiotic prophase chromosomes. Thus, this preliminary analysis of the IR structure of the mouse genome strongly suggests that the mouse X-chromosome also contains a preponderance of large IRs that contain testes genes. However, the analysis must be revisited as the mouse genome sequence and assembly are improved. IRF analyses of the current and future assemblies of mouse genome will be included in the IRDB.
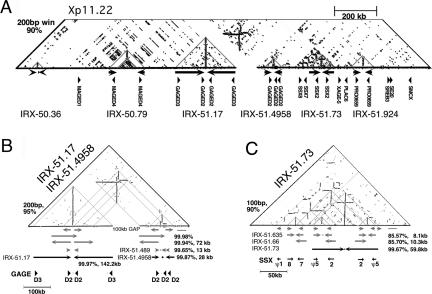
Analysis of IR Structure in Human Chromosome Xp11.22
Similarity dot-plot analysis was performed on each large IR and surrounding genomic DNA, to reveal details about repetitive DNA organization and potential for secondary structure formation (Kuroda-Kawaguchi et al. 2001). Figure 2A shows this analysis for the 2-Mb region that contained the highest density of large IRs in the genome, located on Chromosome Xp11.22. IRX51.17 directly abuts the distal edge of a 100-kb gap that remains in Xp11.22 in hg16, and this IR was not included in the final set of 96 large IRs (Supplemental data S2). However, it is the longest and most homologous IR on the X-chromosome, clearly seen as a long vertical line (Fig. 2B). It is also highly homologous to IRX-51.4958, both of which contain GAGE D2 genes (Fig. 2B), and one of these IRs may be eliminated when the intervening gap is closed. Notably, the Y-chromosome palindromes P1 and P2 containing the DAZ genes are also highly homologous, and appear to share a similar structure (although there is no intervening gap; Kuroda-Kawaguchi et al. 2001; Skaletsky et al. 2003).
Figure 2.
IRs in Xp11.2. (A) Similarity dot-plot of 2 Mb from Xp11.2. Homologous regions indicated by horizontal lines (direct repeats) or vertical lines (inverted repeats). Six large IRs were detected in this region as vertical lines (gray triangles). No other significant sequence similarities were seen in the top portion of the dot-plot not shown. The window size and percent identity for the dot-plot are indicated. (B) Higher-resolution view of the region containing IRX-51.17 and IRX-51.4958, which both contain the GAGE-D genes. Arrows on the same line represent homologous regions as indicated by the dot-plot; percent identity indicated when calculated. Internal IRs detected by IRF are indicated, for example, IRX-51.489 (Supplemental data S1), and the midpoint of the spacer indicated by a dot. IRX-51.17 was not included in the final set of 96 large IRs. (C) Higher-resolution view of IRX-51.73 and surrounding region, which contains an SSX gene cluster. Both arms of the IRX-51.73 contain inverted copies of SSX2 and SSX pseudo5. As in B, internal IRs detected by IRF are indicated, for example, IRX-51.635, and the midpoint of the spacer of the IR for which details are provided is indicated by a dot (Supplemental data S1).
Other features of the similarity dot-plot of Xp11.22 (Fig. 2A) include some relatively dense regions indicative of repetitive low-complexity DNA around IRX-50.79 and IRX-51.73. In the case of IRX-50.79, this region simply represents a high density of full-length LINE elements in tandem and inverted orientation within the spacer region. In the case of IRX-51.73, this region represents a cluster of SSX genes and pseudogenes (Fig. 2C), arranged in at least three inverted pairs that are ∼85% identical, consistent with previous genomic mapping (Gure et al. 2002). Notably, two of these pairs (SSX2 and SSXpseudo5) are found within either arm of the 59-kb IRX-51.73, and therefore are inverted relative to themselves (Fig. 2C).
Classification of Large IRs Into Three Distinct Patterns of Genomic Organization
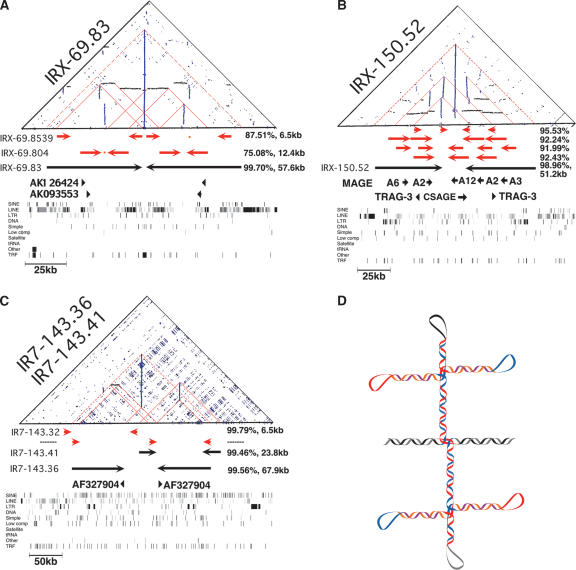
Similarity dot-plot analysis of the IRs in Table 1 showed that 22 displayed a complex pattern of inverted and/or tandem regions of similarity, whereas the remainder simply showed a single vertical line. Of these IRs, 16 could be classified into three distinct patterns based on the genomic organization of inverted and tandem repeats within the large IRs, which each suggest different potential secondary structures (Fig. 3). The first pattern of genomic organization consists of IRs that contain mirror image IRs within both arms of the larger IR (Fig. 3A), seen in three large IRs (Table 1). This type of pattern was also present in the largest Y-chromosome palindrome P1, which contains within each arm the 9.9-kb P1.1 and P1.2 (Kuroda-Kawaguchi et al. 2001; Skaletsky et al. 2003). Notably, these types of IRs would have the potential to extrude into unusual double cruciform structures (Fig. 3D).
Figure 3.
Secondary structure patterns of IRs. (A) Dot-plot similarity analysis of IRX-69.83, which contains smaller, less homologous IRs in each arm. Internal IRs detected by IRF and percent identity are indicated (Supplemental data S1 and S2). Below each dot-plot are the RepeatMasker and TRF (Benson 1999) tracks from the UCSC Genome Browser, where the IR and internal secondary structures can be observed visually as mirror symmetries. (B) Dot-plot similarity analysis of IRX-150.52. The spacer of this IR is homologous to a region in the arms. (C) Analysis of IR7-143.36 and IR7-143.41. One arm of both of these IRs is found in the spacer of the other. IRs with similar secondary structure to A, B, and C are indicated in Table 1. Internal IRs detected in these regions were eliminated from the final data set (Supplemental data S1 and S2). The dot-plot similarity analysis was performed with a window size of 50 bp and mimimum identity of 85%. (D) Possible double cruciform structure suggested by IRs that contain internal IRs, for example, in A and Figure 2C. (Black and gray DNA strands) outside of IR region; (blue and red DNA strands) large, high homology IR; (purple and orange DNA strands) internal, lower homology IRs.
The second pattern of genomic organization consists of the spacer region of the IR containing regions of similarity to the arm regions (Fig. 3B), seen in seven large IRs (Table 1). The internal IRs characteristic of patterns 1 and 2 (Fig. 3, A and B, respectively) such as IRX-69.804 (Fig. 3A) were originally detected by IRF but excluded from the final set of 96 IRs because they contributed to secondary structure (Table 1; Supplemental data S1). The third pattern of genomic organization was seen for six IRs that formed three intertwined pairs, where an arm of each IR is in the spacer of the other IR, shown for IRs IR7-143.36 and IR7-143.41 in Figure 3C. Four other IRs (Table 1) contained secondary structure patterns caused by the presence of a multigene cluster, which, however, could not be classified into one of the three other common patterns.
Conservation of IR Arm Boundaries in Great Apes
To confirm these IRs in the human genome, PCR primers were designed from the human genome sequence (hg16) to define arm-specific STSs for several examples. One primer (primer A, Fig. 4) was designed to hybridize to sequences present in both arms, on either side of the spacer. Individual primers were designed within the spacer to be specific for either the left or right arm/spacer boundary (primers L and R, respectively). In each case (Fig. 4), PCR amplification of human genomic DNA using primer pairs A-L and A-R resulted in PCR amplification of the predicted size fragments (∼300-900 bp), and thus confirmed the presence of both arms of the IR in the human genome. Sequence analysis of these PCR products confirmed their correspondence to the appropriate arm-spacer boundaries in the human genome sequence (Supplemental data S3).
Figure 4.
PCR analysis of internal arm/spacer boundary. For each IR indicated, a single primer (A) was designed to hybridize to both arms. Individual primers (L and R) were designed to hybridize to the spacer region. PCR amplification was performed using primer pairs A-L and A-R with human, gorilla, and chimpanzee genomic DNA. For each IR, a + indicates that PCR products were amplified using both primer pairs. All amplified IRs are >99% homologous (Table 1), except IRX-51.924, which is 97% homologous (Fig. 2A; Supplemental data S1 and S2). The PCR primers and DNA sequence of PCR products are shown in Supplemental data S3. The high homology between the arm and spacer of IRX-51.17 and IRX-51.924 (see Fig. 2B) does not allow for specific STSs, although two distinct sets of primers pairs were used (Supplemental data S3).
A high degree of similarity between the arms of these IRs (Table 1) suggests that they are either relatively recent duplications, or are undergoing arm-to-arm homogenization. Therefore, we attempted PCR amplification using our human arm-specific STSs on gorilla and chimpanzee genomic DNA to assess whether these IRs were present in a common ancestor. In six of the eight examples positive in human, both arm/spacer boundaries were amplified in gorilla, and four of these were also amplified in chimp (Fig. 4). These PCR products were the same size as predicted from the human genome, and sequence analysis showed that they corresponded to the appropriate regions in human (Supplemental data S3). Great apes and humans diverged from a common ancestor ∼5 million years ago, and in general show ∼1%-2% sequence divergence between species (Rozen et al. 2003). Thus, the presence of IRs that have >99.5% arm-to-arm identity in humans (Table 1), yet are also present in great apes, suggests that they are considerably older than their percent identity indicates. Notably, in such a cross-species PCR-based assay in which primers designed to human DNA sequence are tested in other species, a negative result does not necessarily demonstrate that the IR is absent from great apes, as there may simply be a change in the primer sequence.
DISCUSSION
We have performed the first genome-wide analysis of the IR structure of the current version (build 34) of the human genome DNA sequence, using a novel efficient software package called IRF, which is available for ongoing future analyses at IRDB (http://tandem.bu.edu). We found that the human X-chromosome contained a disproportionately high number of large, highly homologous IRs that contained testes genes. This is highly analogous to the IRs found on the human Y-chromosome, which are evidently undergoing conversion to preserve gene integrity and the function of male fertility genes in the absence of meiotic pairing and crossing over (Rozen et al. 2003; Skaletsky et al. 2003). Arm-to-arm gene conversion would not be as critical for maintaining gene integrity on autosomes and the X-chromosome, because they do undergo meiotic crossing over. However, the X-chromosome does so only in females, and thus at a 50% reduced frequency relative to autosomes. Although our arm-specific STSs did not provide enough DNA sequence to assess gene conversion between arms in this report, gene conversion has been previously described between the arms of at least two X-chromosome IRs in Xq28, IRX-152.06 and IRX-152.30 (Small et al. 1997; Aradhya et al. 2001).
The highly homologous IRs on the X-chromosome predominantly contain genes expressed in testes, suggesting a possible role in male germ-line gene expression. The accumulation of sex-linked genes on the X-chromosome appears to be dependent on their timing of expression in meiosis. The mammalian X- and Y-chromosomes undergo male germ-line sex chromosome inactivation (MSCI), which prevents expression of X- and Y-linked genes during meiotic pachytene. The X-chromosome appears to accumulate spermatogenesis genes that are expressed prior to MSCI (Wang et al. 2001; Wu and Xu 2003; Khil et al. 2004). These genes are consistent with a model of sexually antagonistic alleles in which recessive genes beneficial to XY males but detrimental to XX females would accumulate on the X chromosomes because the detrimental effects would initially be masked in females due to heterozygosity (Wang et al. 2001; Lercher et al. 2003). However, as these genes accumulate on the X-chromosomes in the population, modifiers would be expected to arise that limit the genes' expression to the male. The formation of IR-based structures could provide one mechanism to limit expression to the male germ line, preventing deleterious expression of these X-linked alleles in the female (Wu and Xu 2003). The IRs on the Y-chromosome may play a similar role in male germ-line expression.
Most genes expressed during later stages of spermatogenesis have been found on autosomes (Eddy and O'Brien 1998; Emerson et al. 2004; Khil et al. 2004). Furthermore, MSCI of many essential X-linked genes appears to be compensated by the testes-specific expression of autosomal retrotransposed copies (Wu and Xu 2003; Emerson et al. 2004; Wang 2004). However, none of the X-linked testes genes found in IRs (Table 1) have retrotransposed autosomal counterparts (Emerson et al. 2004; Wang 2004), although expression of at least some of them, for example, MAGE-A, GAGE-D, and SPANX, may be required during or after MSCI (Zendman et al. 2003a). Thus, the IRs on which these genes are found may, through formation of cruciforms or other unusual chromatin structures, permit escape from meiotic X inactivation and permit expression of critical spermatogenesis genes that remain exclusively on the X-chromosome (Skaletsky et al. 2003).
The largest autosomal IR observed (IR11-89.34), which was present in great apes by our PCR assay (Fig. 4), also contains the testes gene RNF18 (Table 1). IR1-147.04, also present in great apes (Fig. 4), contains the only human histone gene cluster organized in an IR, which, however, does not contain histone H2B, for which a testes-specific homolog has been identified (Zalensky et al. 2002). Thus, if autosomal IRs facilitate male germ-cell expression, this IR may represent a subset of replication-dependent histone genes used during germ cell development or meiosis (Marzluff et al. 2002).
The highly homologous IRs described here suggest the formation of large DNA cruciform structures, the arms of which would be indistinguishable from normal double-stranded DNA. Such large cruciforms could both replicate and be transcribed essentially normally. They would be exquisite structures for regulating the topological state of chromosomal regions, especially during chromatin remodeling and/or nucleosome replacement. Removal of nucleosomes from DNA creates negative superhelical twist, which could be relaxed by extrusion into a cruciform. Some IRs with complex secondary structures (Fig. 3; Table 1) could form distinct cruciform structures, such as the double cruciform shown in Figure 3D. In IRs in which the spacer is homologous to the arm (Fig. 3B; Table 1), multiple cruciforms are possible, each pairing different sets of homologous genes and permitting conversion between them. And when one IR is surrounded by another IR (Fig. 3C; Table 1), two mutually exclusive cruciforms are possible.
The large and highly homologous IRs described here could potentially lead to aberrant sister-chromatid exchange and chromosome rearrangements. Notably, several human isodicentric Xq chromosome breakpoints have been mapped to Xp11.22 (Wolff et al. 1996), the region of densest large IR occurrence (Fig. 2), and at least three isodicentric Xp chromosome breakpoints have been precisely mapped to the 119-kb IRX-70.95 (Table 1; McDonell et al. 2000). Furthermore, rearrangements between the arms of at least two IRs in Xq28, IRX152.06 and IRX-152.30, have been implicated in deletions of the emerin gene and the NEMO gene, respectively (Small et al. 1997; Aradhya et al. 2001). Thus, the IRs described in this report may represent regions that are particularly prone to genomic rearrangements.
To summarize, we have examined the IR structure of the human genome, and revealed a remarkable preponderance of large, highly homologous IRs on the human X-chromosome, in regions containing testes genes. These IRs may play an important evolutionary or regulatory role in controlling sex-specific gene expression critical during germ-cell development or meiosis.
METHODS
IRF (http://tandem.bu.edu/cgi-bin/irdb/irdb.exe) is a prototype tool for identifying approximate inverted repeats in nucleotide sequences that is similar in concept to the Tandem Repeats Finder (Benson 1999). Candidate IRs are detected by finding short, exact, reverse-complement matches of 4-7 nt (k-tuples) between nonoverlapping fragments of a sequence. A “center” position is defined for each k-tuple match. Short k-tuples are used to detect short IRs with short spacers, and longer k-tuples are used to detect longer IRs with potentially larger spacers, typically 10-100 kb. The program detects “clusters” of k-tuple matches having the same or nearly the same center and falling within a small interval of sequence. Several interval sizes are prespecified, typically between 30 and 2000 nt long.
Candidate IRs are confirmed (aligned and extended) or rejected by computing Smith-Waterman style similarity alignment. An efficient “narrowband” technique is used (Benson 1999), which computes alignment scores in a band of specified width around the presumed correct alignment, shifting the band as the location of the alignment shifts. When an alignment exceeds a prespecified minimum alignment score, the IR pair is reported. The alignment will terminate at the border of an insertion/deletion in one of the repeat copies when it is longer than the bandwidth. The remaining matching parts may be detected as an independent IR pair with a significantly shifted center. In the IRF version used here, the maximum alignment was 500,000 bp, with a bandwidth of 200 bp.
IRF was run against human genome sequences of each chromosome (hg16, obtained from NCBI), using parameters 2,3,5,40::match, mismatch, indel, minimum score. Chromosomes were run simultaneously on a computing cluster consisting of four nodes, each with a single 2.4 GHz Pentium 4 processor, 2 GB RAM, and 80 GB hard disk storage. Analysis took 1.5 h, the equivalent of 6 h on a single computer. The data were then inserted into a Microsoft SQL Server database.
At these settings of IRF, the shortest IR detected has 20 nt in each arm with a identity of 100% between the arms. For all runs, repetitive elements masked by RepeatMasker (Jurka 2000; A.F.A. Smit and P. Green, unpubl.; http://ftp.genome.washington.edu/RM/RepeatMasker.html) were excluded during initial candidate detection but were included in subsequent alignment and extension of IRs. The initial set of IRs detected by IRF was filtered to remove redundant IRs, defined as those that shared a positional identity ≥60%, removing the IRs with the lower “homology times length” score. These IRs were then filtered to retain those with arm identity ≥75% and a homology-times-length score of 25 bp, yielding the final data set of 22,624 IRs (spacer ≤100 kb; Fig. 1A,B) or 31,163 IRs (spacer ≤500 kb). Analysis of these IRs was performed using the data download feature of IRF and Microsoft Excel.
The complete set of IRs detected by IRF or any subsequently filtered subset can be displayed on the assembled human genome sequence using the UCSC Genome Browser (Kent et al. 2002) by downloading a custom track gff file generated by IRF onto your computer and uploading this file into the UCSC custom track option. IRs were confirmed by visual analysis of the mirror symmetrical pattern of the RepeatMasker track, and by BLAT searches with DNA sequences from one arm, which always identified the other arm. Each IR is named by the chromosome on which it is found and the approximate genomic coordinates (in megabase pairs) of the center of the spacer, for example, IRX-47.303 (Table 1). Exclusion of IRs from the data set was performed as described in Supplemental data S1. Those that were located in unfinished or unassembled regions of the current genome assembly were excluded, for example, spanning or abutting gaps. IRs that share a common center were collapsed into single IRs. In cases in which multiple IRs occurred in complex clusters containing multigene families, for example, the UGT genes on Chromosome 4, or in regions with patterns of secondary structure (Figs. 2 and 3), only the largest most similar IR in the cluster was included. Approximately the same proportion of IRs were excluded from the X- (13) and Y-chromosomes (five) relative to the autosomes (52) as were initially detected by IRF, and thus did not contribute any bias to the overall conclusions of this study (Fig. 1D).
To estimate the number of X-chromosome genes expressed predominantly from testes, we queried the human transcriptome represented in the GNF atlas 2 data base (Su et al. 2004) using the “gene sorter” feature of the UCSC Genome Browser for genes on the X-chromosome with a minimum testes expression ratio of 1.0 and a maximum expression ratio of 1.5 for all other tissues.
Two specific IRs with spacer >100 kb are listed in Table 1. The 283-kb IRY-23.2 has a 169-kb spacer (P3; Skaletsky et al. 2003) that contained the 9.8-kb IRY-23.30274. IRY-23.2 has replaced IRY-23.30274 in Table 1 (Supplemental data S1 and S2). The 29.1-kb IRX-147.36 has a 164.9-kb spacer that contained IRX-147.49. The MAGE-A9 genes were actually found in the arms of IRX-147.36 (Table 1; Supplemental data S2). Both of these IRs were correctly detected when the spacer length was set at ≤500 kb. Similarity dot-plots were performed using MacVector 7.0 and analyzed with the help of Canvas 5.0. Additional alignments were performed using CLUSTAL 1.81 (Thompson et al. 1994).
PCR was performed using standard protocols. Gorilla and chimpanzee genomic DNA were obtained from Coriell Cell Repositories. DNA sequencing was performed using standard protocols.
Acknowledgments
We thank Laura Carrel (Penn State, Hershey, PA) for comments on the manuscript and Alfredo Rodriquez (Boston University) for helpful advice. This work was supported in part by grants from the NSF DBI-0413462 (to G.B.) and the NIH R21 HG002919 (to P.E.W.).
Footnotes
[Supplemental material is available online at www.genome.org.]
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2542904.
References
- Aradhya, S., Bardaro, T., Galgoczy, P., Yamagata, T., Esposito, T., Patlan, H., Ciccodicola, A., Munnich, A., Kenwrick, S., Platzer, M., et al. 2001. Multiple pathogenic and benign genomic rearrangements occur at a 35 kb duplication involving the NEMO and LAGE2 genes. Hum. Mol. Genet. 10: 2557-2567. [DOI] [PubMed] [Google Scholar]
- Bailey, J.A., Gu, Z., Clark, R.A., Reinert, K., Samonte, R.V., Schwartz, S., Adams, M.D., Myers, E.W., Li, P.W., and Eichler, E.E. 2002. Recent segmental duplications in the human genome. Science 297: 1003-1007. [DOI] [PubMed] [Google Scholar]
- Benham, C.J., Savitt, A.G., and Bauer, W.R. 2002. Extrusion of an imperfect palindrome to a cruciform in superhelical DNA: Complete determination of energetics using a statistical mechanical model. J. Mol. Biol. 316: 563-581. [DOI] [PubMed] [Google Scholar]
- Benson, G. 1999. Tandem Repeats Finder: A program to analyze DNA sequences. Nucleic Acids Res. 27: 573-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y.T., Scanlan, M.J., Sahin, U., Tureci, O., Gure, A.O., Tsang, S., Williamson, B., Stockert, E., Pfreundschuh, M., and Old, L.J. 1997. A testicular antigen aberrantly expressed in human cancers detected by autologous antibody screening. Proc. Natl. Acad. Sci. 94: 1914-1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomez, P., De Backer, O., Bertrand, M., De Plaen, E., Boon, T., and Lucas, S. 2001. An overview of the MAGE gene family with the identification of all human members of the family. Cancer Res. 61: 5544-5551. [PubMed] [Google Scholar]
- Eddy, E.M. and O'Brien, D.A. 1998. Gene expression during mammalian meiosis. Curr. Top. Dev. Biol. 37: 141-200. [PubMed] [Google Scholar]
- Emerson, J.J., Kaessmann, H., Betran, E., and Long, M. 2004. Extensive gene traffic on the mammalian X chromosome. Science 303: 537-540. [DOI] [PubMed] [Google Scholar]
- Gure, A.O., Wei, I.J., Old, L.J., and Chen, Y.T. 2002. The SSX gene family: Characterization of 9 complete genes. Int. J. Cancer 101: 448-453. [DOI] [PubMed] [Google Scholar]
- Jurka, J. 2000. Repbase update: A database and an electronic journal of repetitive elements. Trends Genet. 16: 418-420. [DOI] [PubMed] [Google Scholar]
- Kent, W.J., Sugnet, C.W., Furey, T.S., Roskin, K.M., Pringle, T.H., Zahler, A.M., and Haussler, D. 2002. The Human Genome Browser at UCSC. Genome Res. 12: 996-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khil, P.P., Smirnova, N.A., Romanienko, P.J., and Camerini-Otero, R.D. 2004. The mouse X chromosome is enriched for sex-biased genes not subject to selection by meiotic sex chromosome inactivation. Nat. Genet. 36: 642-646. [DOI] [PubMed] [Google Scholar]
- Kuroda-Kawaguchi, T., Skaletsky, H., Brown, L.G., Minx, P.J., Cordum, H.S., Waterston, R.H., Wilson, R.K., Silber, S., Oates, R., Rozen, S., et al. 2001. The AZFc region of the Y chromosome features massive palindromes and uniform recurrent deletions in infertile men. Nat. Genet. 29: 279-286. [DOI] [PubMed] [Google Scholar]
- Lafreniere, R.G., Brown, C.J., Rider, S., Chelly, J., Taillon-Miller, P., Chinault, A.C., Monaco, A.P., and Willard, H.F. 1993. 2.6 Mb YAC contig of the human X inactivation center region in Xq13: Physical linkage of the RPS4X, PHKA1, XIST and DXS128E genes. Hum. Mol. Genet. 2: 1105-1115. [DOI] [PubMed] [Google Scholar]
- Lander, E.S., Linton, L.M., Birren, B., Nusbaum, C., Zody, M.C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860-921. [DOI] [PubMed] [Google Scholar]
- Lercher, M.J., Urrutia, A.O., and Hurst, L.D. 2003. Evidence that the human X chromosome is enriched for male-specific but not female-specific genes. Mol. Biol. Evol. 20: 1113-1116. [DOI] [PubMed] [Google Scholar]
- Marzluff, W.F., Gongidi, P., Woods, K.R., Jin, J., and Maltais, L.J. 2002. The human and mouse replication-dependent histone genes. Genomics 80: 487-498. [PubMed] [Google Scholar]
- McDonell, N., Ramser, J., Francis, F., Vinet, M.C., Rider, S., Sudbrak, R., Riesselman, L., Yaspo, M.L., Reinhardt, R., Monaco, A.P., et al. 2000. Characterization of a highly complex region in Xq13 and mapping of three isodicentric breakpoints associated with preleukemia. Genomics 64: 221-229. [DOI] [PubMed] [Google Scholar]
- Ottolenghi, C., Fellous, M., Barbieri, M., and McElreavey, K. 2002. Novel paralogy relations among human chromosomes support a link between the phylogeny of doublesex-related genes and the evolution of sex determination. Genomics 79: 333-343. [DOI] [PubMed] [Google Scholar]
- Pearson, C.E., Zorbas, H., Price, G.B., and Zannis-Hadjopoulos, M. 1996. Inverted repeats, stem-loops, and cruciforms: Significance for initiation of DNA replication. J. Cell Biochem. 63: 1-22. [DOI] [PubMed] [Google Scholar]
- Rozen, S., Skaletsky, H., Marszalek, J.D., Minx, P.J., Cordum, H.S., Waterston, R.H., Wilson, R.K., and Page, D.C. 2003. Abundant gene conversion between arms of palindromes in human and ape Y chromosomes. Nature 423: 873-876. [DOI] [PubMed] [Google Scholar]
- Samonte, R.V. and Eichler, E.E. 2002. Segmental duplications and the evolution of the primate genome. Nat. Rev. Genet. 3: 65-72. [DOI] [PubMed] [Google Scholar]
- Scanlan, M.J., Simpson, A.J.G., and Old, L.J. 2004. The cancer/testis genes: Review, standardization, and commentary. Cancer Imm. 4: 1-15. [PubMed] [Google Scholar]
- Shlyakhtenko, L.S., Hsieh, P., Grigoriev, M., Potaman, V.N., Sinden, R.R., and Lyubchenko, Y.L. 2000. A cruciform structural transition provides a molecular switch for chromosome structure and dynamics. J. Mol. Biol. 296: 1169-1173. [DOI] [PubMed] [Google Scholar]
- Skaletsky, H., Kuroda-Kawaguchi, T., Minx, P.J., Cordum, H.S., Hillier, L., Brown, L.G., Repping, S., Pyntikova, T., Ali, J., Bieri, T., et al. 2003. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 423: 825-837. [DOI] [PubMed] [Google Scholar]
- Small, K., Iber, J., and Warren, S.T. 1997. Emerin deletion reveals a common X-chromosome inversion mediated by inverted repeats. Nat. Genet. 16: 96-99. [DOI] [PubMed] [Google Scholar]
- Su, A.I., Wiltshire, T., Batalov, S., Lapp, H., Ching, K.A., Block, D., Zhang, J., Soden, R., Hayakawa, M., Kreiman, G., et al. 2004. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. 101: 6062-6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. 1994. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, P.J. 2004. X chromosomes, retrogenes and their role in male reproduction. Trends Endocrin. Metab. 15: 79-83. [DOI] [PubMed] [Google Scholar]
- Wang, P.J., McCarrey, J.R., Yang, F., and Page, D.C. 2001. An abundance of X-linked genes expressed in spermatogonia. Nat. Genet. 27: 422-426. [DOI] [PubMed] [Google Scholar]
- Waterston, R.H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J.F., Agarwal, P., Agarwala, R., Ainscough, R., Alexandersson, M., An, P., et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520-562. [DOI] [PubMed] [Google Scholar]
- Wayne, C.M., MacLean, J.A., Cornwall, G., and Wilkinson, M.F. 2002. Two novel human X-linked homeobox genes, hPEPP1 and hPEPP2, selectively expressed in the testis. Gene 301: 1-11. [DOI] [PubMed] [Google Scholar]
- Wolff, D.J., Miller, A.P., Van Dyke, D.L., Schwartz, S., and Willard, H.F. 1996. Molecular definition of breakpoints associated with human Xq isochromosomes: Implications for mechanisms of formation. Am. J. Hum. Genet. 58: 154-160. [PMC free article] [PubMed] [Google Scholar]
- Wu, C.I. and Xu, E.Y. 2003. Sexual antagonism and X inactivation—The SAXI hypothesis. Trends Genet. 19: 243-247. [DOI] [PubMed] [Google Scholar]
- Yoshikawa, T., Seki, N., Azuma, T., Masuho, Y., Muramatsu, M., Miyajima, N., and Saito, T. 2000. Isolation of a cDNA for a novel human RING finger protein gene, RNF18, by the virtual transcribed sequence (VTS) approach(1). Biochim. Biophys. Acta 1493: 349-355. [DOI] [PubMed] [Google Scholar]
- Zalensky, A.O., Siino, J.S., Gineitis, A.A., Zalenskaya, I.A., Tomilin, N.V., Yau, P., and Bradbury, E.M. 2002. Human testis/sperm-specific histone H2B (hTSH2B). Molecular cloning and characterization. J. Biol. Chem. 277: 43474-43480. [DOI] [PubMed] [Google Scholar]
- Zendman, A.J., Ruiter, D.J., and Van Muijen, G.N. 2003a. Cancer/testis-associated genes: Identification, expression profile, and putative function. J. Cell Physiol. 194: 272-288. [DOI] [PubMed] [Google Scholar]
- Zendman, A.J., Zschocke, J., van Kraats, A.A., de Wit, N.J., Kurpisz, M., Weidle, U.H., Ruiter, D.J., Weiss, E.H., and van Muijen, G.N. 2003b. The human SPANX multigene family: Genomic organization, alignment and expression in male germ cells and tumor cell lines. Gene 309: 125-133. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://ftp.genome.washington.edu/RM/RepeatMasker.html; RepeatMasker.
- http://tandem.bu.edu/cgi-bin/irdb/irdb.exe; Inverted Repeat Finder.
- http://tandem.bu.edu; Inverted Repeat Data Base (IRDB).