Abstract
In addition to their central function in protein biosynthesis, tRNAs also play a pervasive role in genome evolution and architecture because of their extensive ability to serve as templates for retroposition. Close to half of the human genome consists of discernible transposable elements, a vast majority of which are derived from RNA via reverse transcription and genomic integration. Apart from the presence of direct repeats (DRs) that flank the integrated sequence of retroposons, genomic integrations are usually marked by an oligo(A) tail. Here, we describe a novel class of retroposons that lack A-tails and are therefore termed tailless retropseudogenes. Analysis of ∼2500 tRNA-related young tailless retropseudogene sequences revealed that they comprise processed and unprocessed (pre-)tRNAs, 3′-truncated in their loop regions, or truncated tRNA-derived SINE RNAs. Surprisingly, their mostly nonrandom integration is dependent on the priming of reverse transcription at sites determined by their 3′-terminal 2-18 nucleotides and completely independent from oligoadenylation of the template RNA. Thus, tailless retropseudogenes point to a novel, variant mechanism for the biogenesis of retrosequences.
tRNAs play an ancient, but still central biological role in protein biosynthesis (Brosius 2001). Initial sequencing and analysis of the human genome revealed 497 tRNA genes and 324 pseudogenes (Lander et al. 2001). Because SINEs (Short INterspersed Elements) are chiefly tRNA-derived retroposons (Daniels and Deininger 1985; Sakomoto and Okada 1985; Okada 1991), the true number of tRNA pseudogenes exceeds tens, even hundreds of thousands in vertebrate genomes. Their role in genome architecture and evolution is pervasive (Brosius 1999a).
Mature tRNAs can serve as templates for retroposition, presumably after atypical polyadenylation (Schmid and Maraia 1992). The same is true for tRNAs at various stages of posttranscriptional processing. Here, we show that mammalian tRNAs can also serve as templates in various stages of decay, even when processing has not been completed. Retroposons are generated by reverse transcription of RNA molecules with concomitant genomic integration of the cDNA and can be subdivided into non-autonomous SINEs and autonomous LINEs (Long INterspersed Elements; Deininger et al. 2003). LINE-encoded RT can be used in trans for SINE retroposition (Kazazian 2000; Weiner 2000), where it is thought to prime on the oligo(A) tract of, for example, SINE-RNAs (Eickbush 1992). Apart from the presence of an oligo(A) tail, genomic integrations of younger retroposons are marked by discernible short direct repeats (DRs) flanking the integrated sequence. Once reverse transcribed and integrated into a new chromosomal location, the fate of retroposons essentially depends on the chromosomal environment. On rare occasions, internal and cis-acting external promoter elements are combined, resulting in the birth of one or a few transcribable master SINEs. These constitute the sole sources for template RNAs for further retroposition events (Deininger et al. 1992; Schmid and Maraia 1992; Brosius 1999b; Kuryshev et al. 2001). Without proper external and internal control elements, the integrated sequence is condemned to life as an inactive retropseudogene.
We have discovered a novel class of multiple-source tRNA-related tailless retropseudogenes that lack the classic oligo(A) tail, but usually feature the 5′ portion of the original tRNA or SINE founder RNA. They preferentially terminate in the region corresponding to the anticodon, variable, or TψC loop. The availability of large numbers of tailless retropseudogene sequences and their genomic environments, including analyses of orthologous loci devoid of such retroposons, enabled us to document the likely events from processing to RT priming and genomic integration, and allowed us to propose a potential mechanism of tailless retropseudogene generation.
RESULTS AND DISCUSSION
Search Results and Species Distribution
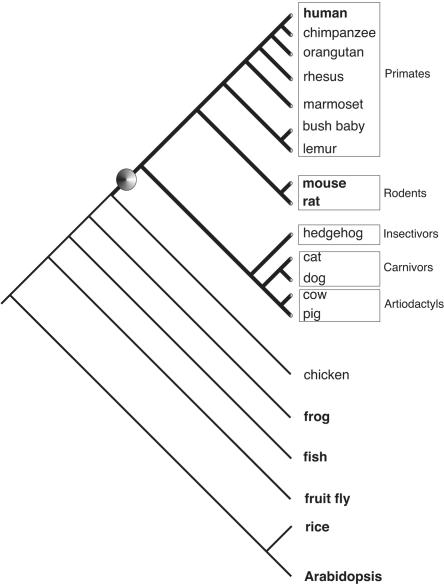
Using a computational, genome-wide search strategy for extracting preferentially young, 30-100 nucleotide-long integrations marked by perfect DRs, we found close to 2.7 million candidates in the genomes of human, mouse, and rat. To specifically focus on truncated tRNA-related sequences, we performed local BLAST searches (NCBI) of the extracted short sequences against a compilation of available tRNAs from any known species and identified ∼2500 matches (Supplemental Fig. 1). In contrast, we were able to detect less than a handful of young retroposed full-length tRNAs with perfect DRs. Moreover, we also detected a lineage-specific distribution of tailless retropseudogenes (Fig. 1). Searches in complete genomes (see Methods) of fish, fruit fly, nematode, and plants, or in large sequenced genomic portions of birds and amphibians, failed to detect tailless retropseudogenes. However, we readily discovered tailless retropseudogenes in similarly large sequenced portions of additional mammals such as pig, cow, dog, cat, and hedgehog, but not marsupials and monotremes, possibly due to a lack of available sequences. Thus, a variant mechanism appears to exist for generating retroposons, which seems to be restricted to mammals.
Figure 1.
Taxonomic distribution of tailless retropseudogenes based on GenBank searches. Tailless retropseudogenes were retrieved exclusively from mammalian representatives (bold lines) indicating a mammalian-specific distribution with the presumed origin marked by a dot. Species represented in GenBank with their entire genome sequences are shown in bold letters. Branches of the phylogenetic tree are not drawn to scale.
The novel class of tailless retropseudogenes described here are a highly abundant class of tRNA-related retroposons derived from diverse 3′ truncated RNA templates. Hence, all tRNA-derived tailless retropseudogenes represent the internal A-box promotor region, but lack a B-box sequence. With the exception of tRNAPro, thus far, all tRNA classes, as well as SINE RNA-derived templates were found to yield tailless retropseudogenes. Our findings reveal that tailless retropseudogenes are generated via a common mechanism of truncation and/or retroposition that arose during an early stage of mammalian evolution.
tRNA-Derived Tailless Retropseudogenes
We found 225, 58, and 116 tailless retropseudogenes directly derived from tRNAs or pre-tRNAs, 3′ truncated at recurrent positions, primarily in single-stranded domains in the human, mouse, and rat genomes, respectively. Relaxing the conditions to more moderately conserved flanking DRs (see Methods) revealed an additional 583, 514, and 456 tailless retropseudogenes, respectively. Among the 225 tailless retropseudogenes with perfect DRs extracted from the human genome, we distinguished two subgroups; 33 tailless retropseudogenes feature the mature 5′ ends of the corresponding tRNA templates, and the other contain a few additional 5′ terminal nucleotides. Although only 49 of those 5′ extensions matched 2-8 nucleotides of the 5′ unique leader sequences of the corresponding pre-tRNAs, the remaining extensions could be explained by sequence variation or nontemplated additions.
SINE-RNA Derived Tailless Retropseudogenes
A large number of rodent tailless retropseudogenes were detected that are not derived directly from either tRNAs or pre-tRNAs. We found 446 tailless retropseudogenes in mouse and 1597 in rat that originated, instead, from similarly 3′-truncated tRNA-derived SINE transcripts. All 446 mouse and 1453 of the rat sequences are derived from 4.5SI RNA (Gogolevskaya and Kramerov 2002), whereas the other 144 rat tailless retropseudogenes are derived from BC1 RNA (DeChiara and Brosius 1987) or transcribed ID elements (Kim et al. 1995).
Breakpoints of Tailless Retropseudogenes
Of the tRNA-derived tailless retropseudogenes with discernible priming sites (Supplemental Table), 77.4% of the truncations occur in the anticodon loop, 13.8% in the variable loop, 4.2% in the anticodon stem, 1.7% in the TψC stem region, and 2.9% in the TψC loop. Interestingly, 85.8% of all tailless retropseudogenes terminal nucleotides are purines (Figs. 2, 3; Supplemental data; see Methods). Surprisingly, this demonstrates that 3′-truncated, tRNA-derived RNAs, at various stages of processing, can serve quite efficiently as templates for retroposition. Although we cannot completely rule out internal priming on single-stranded regions of RNA templates (Cost et al. 2002), we currently favor priming at free processed 3′ ends, as the nicking is expected to release single-stranded priming sites from previously inaccessible secondary/tertiary structures.
Figure 2.
Genomic integration mechanism of tRNA-derived tailless retropseudogenes. (A) RNA break point (arrow) and priming site (boxed nucleotides) in an example in which a fragment of pre-tRNACys serves as a tailless retropseudogene template. Tailless retropseudogene-specific substitutions differing from the presumed tRNA precursor template are encircled. Nucleotides representing the remainder of the tRNA precursor, but not part of the tailless retropseudogenes, are depicted as “thin” letters. (B) The genomic integration site with the consensus sequence TT/WX1-17 (in this particular case, TT/WX6) is shown above the bracket. The RNA priming site is boxed, the direction of reverse transcription is indicated by an arrow. The presumed single-stranded, staggered end is shown by a double-headed arrow, part of which (seven nucleotides in this case) acts as a DNA primer for reverse transcription. (C) Sequence of chromosomal locus after integration of the tailless retropseudogenes. Direct repeats (DR) are indicated by arrows. Note that the priming site (within the boxed tailless retropseudogenes) is part of the DR.
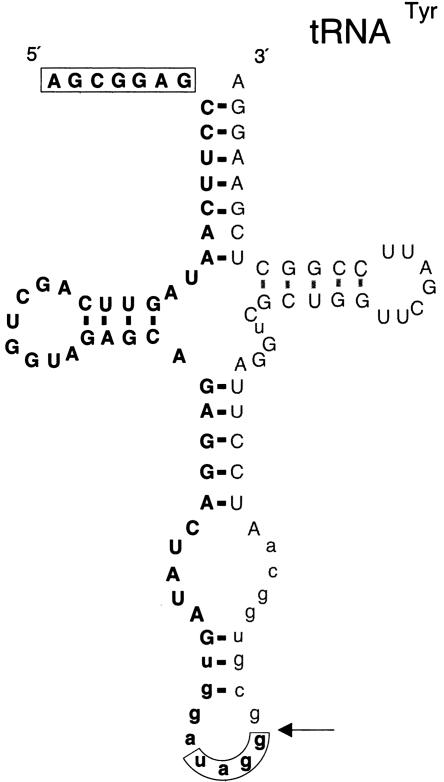
Figure 3.
Precursor and cleavage point of a tailless retropseudogene from an intron-containing tRNA. In an example where human pre-tRNATyr serves as a precursor for a tailless retropseudogene, the RNA breakpoint 3′ to a purine residue (arrow) is located in the intron (lower case letters). The priming site, also located in the intron, is boxed. The boxed nucleotides at the 5′-end represent unprocessed nucleotides that are present in the integrated tailless retropseudogene. Nucleotides representing the remainder of the tRNA precursor, but not part of the tailless retropseudogene, are depicted by “thin” letters.
Genomic Priming Sites
Reverse transcription of (pre)-tRNAs should be mediated by LINE-1 retroelements, as they encode the only known endonuclease/RT that is active in trans (Dewannieux et al. 2003). A characteristic feature of LINE-mediated enzymatic activity is nicking of 5′ TT/AAAA 3′ recognition motifs in genomic sequences. The stretch of several T-residues on the opposite strand is the presumed primer for SINE founder RNAs that contain adenosine-rich sequences (Jurka 1997; Cost and Boeke 1998). However, in 1839 of the 2442 tailless retropseudogenes, we identified discernible priming sites all complementary to 2-18 nucleotides of the 3′ termini of the truncated template RNAs (Fig. 2A; Supplemental Table). The priming sites are distinguishable in 183 different sequence motifs. Thus, on the basis of Jurka's kinkable DNA sites (Jurka 1997), we formulated a more heterogeneous integration motif characteristic for tailless retropseudogenes, TT/WX(1-17) (Fig. 2B,C; Supplemental data; see Methods). The two target-specific T residues in this motif are individually conserved in 53% and 66% of all analyzed integration sites, the subsequent W-residue is 85% conserved, about equally as either a T- or an A-residue (Fig. 2). X(1-17) denotes the remainder of the DNA target, part of which acts as a DNA primer for reverse transcription, and the first five nucleotides of which are predominantly adenosines (Supplemental Fig. 3).
Unspliced tRNA-Templates
In eight examples, the priming site is complementary to intronic sequences of the pre-tRNA (Fig. 3; Supplemental data; see Methods). Three additional tailless retropseudogenes feature complete internal tRNA-specific introns. These and examples with additional 5′-precursor sequences demonstrate that truncation can take place prior to completion of splicing and/or RNase P-catalyzed 5′ tRNA processing (Fig. 3).
The use of pre-existing DNA nicks in LINE1 endonuclease-independent retroposition has been described in vitro and in cell lines (Cost et al. 2002; Morrish et al. 2002). Unlike the mechanism corroborated by Morrish et al. (2002), which does not yield direct repeats, tailless retropseudogene integrations at the genomic level are always accompanied by DRs typically longer than the staggered-end DNA-priming site. As a precautionary note, it is worth mentioning that the events in vitro and in cell lines may not reflect the dynamics and germ-line events that actually occur in the genomes of organisms.
Presence/Absence Analyses
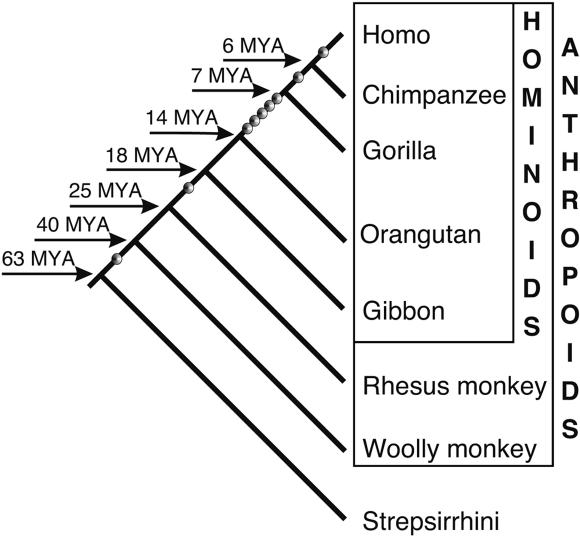
To obtain further information on genomic target sites for the integration of tailless retropseudogenes, we PCR amplified tailless retropseudogene loci for representatives of all primate infraorders (Fig. 4). Presence/absence analyses in primates (Shedlock and Okada 2000; Schmitz et al. 2001), spanning about 63 million years (Myr), revealed one human-specific tailless retropseudogene that is probably younger than 6 Myr, one tailless retropseudogene at orthologous loci in only human and chimpanzee, five tailless retropseudogenes restricted to the human, chimpanzee, and gorilla branch, one tailless retropseudogene common to all hominoids (apes), and one tailless retropseudogene common to all anthropoids that integrated between 40 and 63 Myr ago (Goodman et al. 1998). Thus, our data are consistent with continuous tailless retropseudogenes' retroposition in primates over a period of more than 40 Myr (Fig. 5).
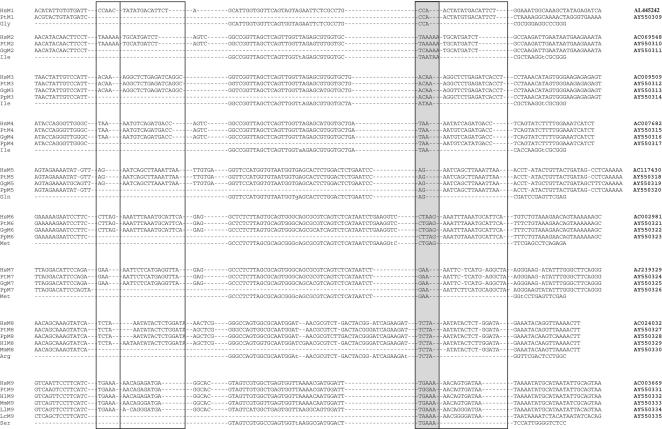
Figure 4.
Tailless retropseudogene presence/absence alignments for primates. In the alignments of orthologous loci for M1-M9 markers (one or several with tailless retropseudogenes and one or more orthologous loci without tailless retropseudogenes) all direct repeats are boxed and priming sites are shaded. The nonorthologous, most closely related tRNA species is given in the last line of each of the nine alignments. Hyphens represent gaps in the sequences. Homo sapiens (Hs); Pan troglodytes (Pt); Gorilla gorilla (Gg); Pongo pygmaeus (Pp); Hylobates lar (Hl) for Hominoidea; Macaca mulatta (Mm) for Cercopithecoidea; Lagothrix lagotricha (Ll) for Platyrrhini; Lemur catta (Lc) for Strepsirrhini. Accession nos. or internal reference numbers are shown in bold.
Figure 5.
Timeframes of integration of primate tailless retropseudogenes defined by presence/absence analyses. Dots on the lineage leading to human mark the nine established single-integration events. Species splitting times are adapted from Goodman and Page (Goodman et al. 1998).
Lack of Oligo(A) Tails
The most striking feature of tailless retropseudogenes is their complete lack of oligo(A) tails. Previously, Saba et al. (1983) described one 4.5SI-derived, oligo(A)-less, retroposed sequence displaying features of our tailless retropseudogenes. Roy-Engel et al. (2002) recently reported that not only is the presence of an oligo(A) tail important for efficient retroposition of Alu SINE RNA, but that its length largely determines whether an Alu element is retropositionally active as a master gene or not (but see Hagan et al. 2003). Among the tailless retropseudogenes, there are tRNA-related templates, including 4.5SI RNA or BC1 RNA, that do contain short internal stretches of A-residues and that prime at such sites. Thus, in a fraction of cases, reverse transcription primes exclusively on A-residues. However, in these cases, the retroposition process does not extend the length of the A-tail.
In conclusion, we have discovered a novel class of retroposons in mammals, termed tailless retropseudogenes, derived from truncated tRNAs and tRNA-related SINE RNAs and generated by a hitherto unknown variant retroposition mechanism, probably guided by the LINE reverse transcriptase, in which neither the presence of an oligo(A) tail on the RNA template or its length are important for retroposition. Variable tailless retropseudogenes priming sites also imply that the proposed competitive template switching involving the A-rich regions of LINE and SINE RNAs (Cost et al. 2002) is not a general requirement for SINE retroposition. Finally, nicking sites acting as primers search perfectly matched complementary RNA sequences for integration sites. Recently, we detected thousands of additional tailless retropseudogenes, in which other RNAs such as ribosomal 5S RNA or small nuclear RNAs (snRNAs) served as efficient templates (data not shown). This underscores the numerically significant proliferation of tailless retropseudogenes in mammalian genomes.
METHODS
Computer-Based Extraction of Young Tailless Retropseudogenes
We developed a computer algorithm to identify recent, tRNA-derived short integrations in entire genomes. We restricted our initial screening of the entire human, mouse, and rat genomes to 30-100 nucleotide-long sequences flanked by at least 10 nucleotide-long perfect direct repeats. The sequences retrieved from this search were blasted against all published tRNA sequences, and the resulting matches cross-checked by the RepeatMasker program. We subsequently retrieved all available sequence information from the NCBI GenBank to establish the taxonomic distribution of tailless retropseudogene sequences in the entire genomes of fish (Takifugu rubripes, Danio rerio), fruit fly (Drosophila melanogaster), nematode (Caenorhabditis elegans), and plants (Oryza sativa, Arabidopsis thaliana), or in large sequenced genomic portions of birds (Gallus gallus) and amphibians (Xenopus tropicalis). A flowchart of the computational strategy is shown as Supplemental Figure 1. We carried out BLAT searches for a rough estimation of the human genome-wide tailless retropseudogene frequency using the table browser option with the chrN_rmsk table and the tRNA repClass. We then separated tailless retropseudogenes from full-length tRNA genes and pseudogenes using the RepeatMasker.
Presence/Absence Analyses in Primates
To perform presence/absence analyses in primates, we subjected entire tRNA sequences to a genome BLAST of the human NCBI nucleotide databases (NCBI BLASTN 2.2.6). From the graphic output of the BLAST results, we randomly selected 50 fragmented tRNA sequences displaying tailless retropseudogene characteristics. Flanking PCR primers were designed for representatives of all major primate clades including Hominoidea (Homo sapiens, Pan troglodytes, Gorilla gorilla, Pongo pygmaeus, Hylobates lar); Cercopithecoidea (Macaca mulatta); Platyrrhini (Lagothrix lagotricha); Strepsirrhini (Lemur catta). In ZOO-PCRs, small products indicate the absence of respective tailless retropseudogenes. Thus, we established nine clear presence/absence patterns. From the species distribution of tailless retropseudogenes, we deduced the time of integration and verified the presence/absence data by sequence analyses (Figs. 4,5).
Sixteen additional presence/absence patterns were retrieved computationally from GenBank by searching mouse and rat genomes (Supplemental Fig. 2A,B). Sequence comparisons enabled us to reconstruct nicking sites, staggered ends—parts of which are priming sites on template RNAs—and the formation of DR.
URLs
Our data are available at: http://zmbe2.uni-muenster.de/expath/addmat/tailless_retropseudogenes1.htm. Genomic sequences were downloaded from ftp://ftp.ncbi.nih.gov/genomes. We used the RepeatMasker (A.F.A. Smit and P. Green; RepeatMasker at http://www.repeatmasker.org/cgi-bin/WEBRepeatMasker) to screen out repetitive sequences. Structural analysis of the human tRNA integrations were compared with the predicted tRNA secondary structures compiled at http://rna.wustl.edu/GtRDB/Hs/Hs-align.html. UCSC Genome Bioinformatics searches (BLAT) were performed using (http://genome.ucsc.edu/cgi-bin/hgGateway; http://genome.ucsc.edu/cgi-bin/hgBlat)
Acknowledgments
We thank Christian Roos for providing samples, Martina Ohme for assistance in amplification of the primate presence/absence loci, and Carsten Raabe and Timofey Rozhdestvensky for discussions. Furthermore, we thank Marsha Bundman and Kira Gee for editorial help. This work was supported by the German Human Genome Project through the BMBF (01KW9966) to J.B.
Footnotes
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2720104. Article published online before print in September 2004.
[Supplemental material is available online at www.genome.org and http://zmbe2.uni-muenster.de/expath/addmat/tailless_retropseudogenes1.htm. The sequence data from this study have been submitted to GenBank under accession nos. AY550309-AY550335. The following individuals kindly provided reagents, samples, or unpublished information as indicated in the paper: C. Roos.]
References
- Brosius, J. 1999a. Transmutation of tRNA over time. Nat. Genet. 22: 8-9. [DOI] [PubMed] [Google Scholar]
- ____. 1999b. RNAs from all categories generate retrosequences that may be exapted as novel genes or regulatory elements. Gene 238: 115-134. [DOI] [PubMed] [Google Scholar]
- ____. 2001. tRNAs in the spotlight during protein biosynthesis. Trends Biochem. Sci. 26: 653-656. [DOI] [PubMed] [Google Scholar]
- Cost, G.J. and Boeke, J.D. 1998. Targeting of human retrotransposon integration is directed by the specificity of the L1 endonuclease for regions of unusual DNA structure. Biochemistry 37: 18081-18093. [DOI] [PubMed] [Google Scholar]
- Cost, G.J., Feng, Q., Jacquier, A., and Boeke, J.D. 2002. Human L1 element target-primed reverse transcriptase in vitro. EMBO 21: 5899-5910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels, G.R. and Deininger, P.L. 1985. Repeat sequence families derived from mammalian tRNA genes. Nature 317: 819-822. [DOI] [PubMed] [Google Scholar]
- DeChiara, T.M. and Brosius, J. 1987. Neural BC1 RNA: cDNA clones reveal nonrepetitive sequence content. Proc. Natl. Acad. Sci. 84: 2624-2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deininger, P.L., Batzer, M.A., Hutchison III, C.A., and Edgell, M.H. 1992. Master genes in mammalian repetitive DNA amplification. Trends Genet. 8: 307-311. [DOI] [PubMed] [Google Scholar]
- Deininger, P.L., Moran, J.V., Batzer, M.A., and Kazazian, H.H. 2003. Mobile elements and mammalian genome evolution. Curr. Opin. Genet. Dev. 13: 651-658. [DOI] [PubMed] [Google Scholar]
- Dewannieux, M., Esnault, C., and Heidmann, T. 2003. LINE-mediated retrotransposition of marked Alu sequences. Nat. Genet. 35: 41-48. [DOI] [PubMed] [Google Scholar]
- Eickbush, T.H. 1992. Transposing without ends: The non/LTR retrotransposable elements. New Biol. 4: 430-440. [PubMed] [Google Scholar]
- Gogolevskaya, I.K. and Kramerov, D.A. 2002. Evolutionary history of 4.5SI RNA and indication that it is functional. J. Mol. Evol. 54: 354-364. [DOI] [PubMed] [Google Scholar]
- Goodman, M., Porter, C.A., Czelusniak, J., Page, S.L., Schneider, H., Shoshani, J., Gunnell, G., and Groves, C.P. 1998. Toward a phylogenetic classification of primates based on DNA evidence complemented by fossil evidence. Mol. Phylogen. and Evol. 9: 585-598. [DOI] [PubMed] [Google Scholar]
- Hagan, C.R., Sheffield, R.F., and Rudin, C.M. 2003. Human Alu element retrotransposition induced by genotoxic stress. Nat. Genet. 35: 219-220. [DOI] [PubMed] [Google Scholar]
- Jurka, J. 1997. Sequence patterns indicate an enzymatic involvement in integration of mammalian retroposons. Proc. Natl. Acad. Sci. 94: 1872-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian, H.H. 2000. L1 Retrotransposons shape the mammalian genome. Science 18: 1152-1153. [DOI] [PubMed] [Google Scholar]
- Kim, J., Kass, D.H., and Deininger, P.L. 1995. Transcription and processing of the rodent ID repeat family in germline and somatic cells. Nucleic Acids Res. 23: 2245-2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuryshev, V.Y., Skryabin, B.V., Kremerskothen, J., Jurka, J., and Brosius, J. 2001. Birth of a gene: Locus of neuronal BC200 snmRNA in three prosimians and human BC200 pseudogenes as archives of change in the Anthropoidea lineage. J. Mol. Biol. 309: 1049-1066. [DOI] [PubMed] [Google Scholar]
- Lander, E., Lander, E.S., Linton, L.M., Birren, B., Nusbaum, C., Zody, M.C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860-921. [DOI] [PubMed] [Google Scholar]
- Morrish, T.A., Gilbert, N., Myers, J.S., Vincent, B.J., Stamato, T.D., Taccioli, G.E., Batzer, M.A., and Moran, J.V. 2002. DNA repair mediated by endonuclease-independent LINE-1 retrotransposition. Nat. Genet. 31: 159-165. [DOI] [PubMed] [Google Scholar]
- Okada, N. 1991. SINEs: Short interspersed repeated elements of the eukaryotic genome. Trends Ecol. Evol. 6: 358-361. [DOI] [PubMed] [Google Scholar]
- Roy-Engel, A.M., Salem, A.H., Oyeniran, O.O., Deininger, L., Hedges, D.J., Kilroy, G.E., Batzer, M.A., and Deininger, P.L. 2002. Active Alu element “A-Tails”: Size does matter. Genome Res. 12: 1333-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saba, J.A., Busch, H., and Reddy, R. 1983. A new moderately repetitive rat DNA sequence detected by a cloned 4.5 SI DNA. J. Biol. Chem. 260: 1354-1357. [PubMed] [Google Scholar]
- Sakamoto, K. and Okada, N. 1985. Rodent type 2 Alu family, rat identifier sequence, rabbit C family, and bovine or goat 73-bp repeat may have evolved from tRNA genes. J. Mol. Evol. 22: 134-140. [DOI] [PubMed] [Google Scholar]
- Schmid, C. and Maraia, R. 1992. Transcriptional regulation and transpositional selection of active SINE sequences. Curr. Opin. Genet. Dev. 2: 874-882. [DOI] [PubMed] [Google Scholar]
- Schmitz, J., Ohme, M., and Zischler, H. 2001. SINE insertions in cladistic analyses and the phylogenetic affiliations of Tarsius bancanus to other primates. Genetics 157: 777-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shedlock, A.M. and Okada, N. 2000. SINE insertions: Powerful tools for molecular systematics. Bioessays 22: 148-160. [DOI] [PubMed] [Google Scholar]
- Weiner, A.M. 2000. Do all SINEs lead to LINEs? Nat. Genet. 24: 332-333. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://zmbe2.unimuenster.de/expath/addmat/tailless_retropseudogenes1.htm; supplementary data, including sequences and additional priming sites.
- http://www.repeatmasker.org/cgi-bin/WEBRepeatMasker; A.F.A. Smit and P. Green, RepeatMasker.
- http://rna.wustl.edu/GtRDB/Hs/Hs-align.html; tRNAscan-SE online tRNA Analysis.
- http://genome.ucsc.edu/cgi-bin/hgGateway; UCSC Genome Browser Gateway.
- http://genome.ucsc.edu/cgi-bin/hgBlat; UCSC BLAT Search Genome.
- ftp://ftp.ncbi.nih.gov/genomes; NCBI Genome Assembly.