Abstract
Maize (Zea mays L. ssp. mays), one of the most important agricultural crops in the world, originated by hybridization of two closely related progenitors. To investigate the fate of its genes after tetraploidization, we analyzed the sequence of five duplicated regions from different chromosomal locations. We also compared corresponding regions from sorghum and rice, two important crops that have largely collinear maps with maize. The split of sorghum and maize progenitors was recently estimated to be 11.9 Mya, whereas rice diverged from the common ancestor of maize and sorghum ∼50 Mya. A data set of roughly 4 Mb yielded 206 predicted genes from the three species, excluding any transposon-related genes, but including eight gene remnants. On average, 14% of the genes within the aligned regions are noncollinear between any two species. However, scoring each maize region separately, the set of noncollinear genes between all four regions jumps to 68%. This is largely because at least 50% of the duplicated genes from the two progenitors of maize have been lost over a very short period of time, possibly as short as 5 million years. Using the nearly completed rice sequence, we found noncollinear genes in other chromosomal positions, frequently in more than one. This demonstrates that many genes in these species have moved to new chromosomal locations in the last 50 million years or less, most as single gene events that did not dramatically alter gene structure.
The grass family (Gramineae) contains many agronomically important plants such as rice (Oryza sativa), maize (Zea mays), barley (Hordeum vulgare), oats (Avena sativa), sorghum (Sorghum bicolor), wheat (Triticum spp.), and rye (Secale cereale). Despite their recent origin ∼50-65 million years ago (Mya), the grasses have diversified into nearly 10,000 different taxa, thus providing examples of many closely related species (Kellogg 2001). This close relationship has also been evident from comparative mapping studies (Hulbert et al. 1990; Ahn and Tanksley 1993; Moore et al. 1995; Gale and Devos 1998; Keller and Feuillet 2000; Paterson et al. 2000), indicating that gene content and order are highly conserved (collinear) and that most of their wide genome size variation is caused by polyploidy and transposable element amplification (SanMiguel et al. 1998; reviewed in Bennetzen 2000).
These initial comparative mapping studies were based on a small number of genetic markers and relatively few progeny, and thus they could only resolve large chromosomal rearrangements like translocations, intrachromosomal inversions, segmental genome duplications, and chromosomal fusions. Although a few such rearrangements were observed, they did not prevent the presentation of the genomes of different species in the form of circles with conserved genetic markers located in radial locations. The circle diagram also depicted the relatively small invariance in genetic map sizes, despite huge differences in physical sizes (Gale and Devos 1998). Another important feature of the comparative map was its clear presentation of a whole-genome duplication (WGD) that occurred in an ancestor of maize. Although the rice genome is presented as a single circle, the maize genome is presented as two circles. Wheat, in contrast, is a more recent polyploid species that provides three almost perfectly collinear orthologous (“homoeologous”) genomes (Akhunov et al. 2003). Wheat has maintained entire homoeologous chromosomes while the two progenitor genomes that formed maize have undergone major rearrangements to form chromosomes with composite structures of homoeologous regions (McClintock 1930; Ting 1966; Helentjaris et al. 1988; Gaut 2001).
With the generation of large-insert genomic libraries using artificial bacterial chromosomes (BACs), studies were undertaken to compare orthologous regions among different grass species to study the conservation of gene content and order (Chen et al. 1997; Tikhonov et al. 1999; Bennetzen 2000; Keller and Feuillet 2000; Ramakrishna et al. 2002a; Song et al. 2002; Ilic et al. 2003). In some cases, collinearity could be found between several species, and in others many exceptions were found. Still, a major limitation in these studies was that the compared intervals might have been sufficient for a small genome like rice with an average gene density of 1 gene per 8 kb (Song et al. 2002), but not for larger genomes like maize, barley, and wheat, unless gene-rich regions could be identified. Furthermore, chromosome-walking techniques were frequently hampered by the presence of repetitive sequences. However, DNA fingerprinting of maize BAC clones has overcome the problems with repeat sequences and linked clones via their common restriction fragments (Coe et al. 2002; Engler et al. 2003). For instance, a 435-kb region containing a cluster of storage protein genes provided a large interval from maize that could be compared with intervals of sorghum and rice. These studies proposed a mosaic organization of collinear and noncollinear genes (Song et al. 2002).
Recent comparisons of genomic intervals in maize have shown dramatic intraspecific variation in sequence content within different inbred lines (Fu and Dooner 2002; Song and Messing 2003). These differences arose both from massive insertions of transposable elements and from insertion and deletion of genes. In one case, a putative disease-defense-related gene (ORF3) expressed during maize endosperm development was present in one inbred line but missing in another one. Furthermore, expression analysis from hybrids indicated that noncollinearity could also result in nondosage types of interactions between such different genomes, which might explain overdominance, a hallmark of heterosis (Song and Messing 2003). All these comparisons therefore indicate that rapid changes in the location of genes within the grass family could occur within just a few million years or less. Understanding the nature and rates of these rearrangements is important for understanding the significance of chromosomal organization relative to genome function and for the implementation of future crop improvement strategies.
To gain a more representative data set for comparative genomic analysis, we selected 12 different regions of the maize genome on different chromosomes with known genetic markers representing known duplicate loci (Rhoades 1951; Goodman et al. 1980; McMillin and Scandalios 1980; Wendel et al. 1986). The same data set has been used to determine that the two progenitors of maize and the progenitor of sorghum split at about the same time, 11.9 Mya (Swigonova et al. 2004). Furthermore, we could determine a time interval where the two progenitors of maize hybridized, between 11.9 and 4.8 Mya. In this study, we find that the homoeologous regions provide an interleaving pattern of collinear genes on both homoeologous regions in the maize genome, indicating a massive loss of duplicated genes. Furthermore, we also demonstrate substantial positional instability of cereal genes.
RESULTS
Selection and Annotation of BAC Clones
A total of 24 BAC clones have been sequenced, six from Sorghum bicolor cv. Btx623 and 18 from maize inbred B73 (Swigonova et al. 2004). These sequences represent five chromosomal regions in sorghum and 10 different regions in the maize genome. The 10 regions in maize can be aligned as five duplicated regions that are marked by known genetic markers that are conserved in sorghum as well: orp1 and orp2 (abbreviated orp1/2), r1/b1, c1/pl1, tb1/2, and tbp1/2. The orp1/2 regions are the largest sequences because they represent overlapping regions with two additional mapped markers, zmfie1/2. The maize genetic markers were also used to identify orthologous clones from the Oryza sativa ssp. japonica cv. Nipponbare genome, using a BLAST search (Altschul et al. 1997), which has been sequenced by the International Rice Genome Sequencing Project, IRGSP (http://rgp.dna.affrc.go.jp). All sequences that were aligned are listed in Table 1. Each region was then analyzed for gene content using a combination of gene-finding programs and homology searches (Methods). A putative gene is only counted if the predicted gene has a match in GenBank with a BLASTP E-value smaller than e-10. Furthermore, BLAST searches were performed to exclude genes that have homology with transposable elements (TE). The complete annotation of each sequenced BAC clone has been submitted to GenBank.
Table 1.
Gene Content Statistics
| Loci | Marker genes | Chromosome location | Length (kb) | Gene count | Average length (kb/gene) | Genes in window | Noncollinear genes | Noncollinear genes % |
|---|---|---|---|---|---|---|---|---|
| orp1/orp2 | orp1 | 4S | 358 | 5 | 71.6 | 5 | 1 | 20% |
| orp2 | 10S | 286 | 13 | 22.0 | 8 | 1 | 13% | |
| Sorghum ortholog | Unknown | 202 | 25 | 8.1 | 13 | 2 | 15% | |
| Rice ortholog | 8 | 133 | 20 | 6.7 | 10 | 2 | 20% | |
| r1/b1 | r1 | 10L | 290 | 10 | 29.0 | 6 | 2 | 33% |
| b1 | 2S | 206 | 5 | 41.2 | 5 | 0 | 0% | |
| Sorghum ortholog | Unknown | 157 | 17 | 9.2 | 11 | 2 | 18% | |
| Rice ortholog | 4 | 250 | 21 | 11.9 | 9 | 0 | 0% | |
| c1/pl1 | c1 | 9S | 331 | 8 | 41.4 | 6 | 1 | 17% |
| pl1 | 6L | 316 | 8 | 39.5 | 7 | 1 | 14% | |
| Sorghum ortholog | Unknown | 144 | 8 | 18.0 | 8 | 0 | 0% | |
| Rice ortholog | 6 | 123 | 7 | 17.6 | 7 | 0 | 0% | |
| tb1/tb2 | tb1 | 1L | 220 | 3 | 73.3 | 2 | 0 | 0% |
| tb2 | 5S | 141 | 7 | 20.1 | 5 | 0 | 0% | |
| Sorghum ortholog | Unknown | 78 | 2 | 39.0 | 2 | 0 | 0% | |
| Rice ortholog | 3 | 139 | 9 | 15.4 | 8 | 3 | 38% | |
| tbp1/tbp2 | tbp1 | 1L | 212 | 8 | 26.5 | 2 | 0 | 0% |
| tbp2 | 5S | 194 | 8 | 24.3 | 8 | 3 | 38% | |
| Sorghum ortholog | Unknown | 100 | 10 | 10.0 | 9 | 1 | 11% | |
| Rice oetholog | 3 | 153 | 13 | 11.8 | 9 | 1 | 11% | |
| Maize | 2554 | 75 | 34.1 | 54 | 9 | 17% | ||
| Sorghum | 681 | 62 | 11.0 | 43 | 5 | 12% | ||
| Rice | 798 | 70 | 11.4 | 43 | 6 | 14% | ||
| Total | 4033 | 207 | n/a | 140 | 20 | 14% |
The totals for each species are in bold.
Alignment of Chromosomal Regions Using Genetic Markers
To illustrate the relative positions of all genes, all six genetic markers have been used to align the five chromosomal regions from the two duplicated regions in maize with the regions in sorghum and rice (Fig. 1A-E). Because the location and the density of genes could not be predicted from the selection of BAC clones, in many cases not all genes within each interval can be aligned. However, in one case in sorghum and in several cases in maize, additional overlapping BAC clones have been selected and sequenced to extend the alignments by identifying additional conserved genes. There are several salient features emerging from these alignments. A summary of the sequence analysis is shown in Table 1. A total of ∼4 Mb of sequences from all four aligned regions has been analyzed, predicting 206 genes including eight gene remnants that are either heavily truncated or interrupted by a transposon. Although one might expect as many genes in both regions of maize as rice and sorghum combined, the actual number of genes found in the maize regions is 47% lower, whereas the physical length of both regions of maize is 73% longer than sorghum and rice combined. These differences between maize and the other two species are further exemplified by the differences in average gene density. The gene density in rice and sorghum is nearly the same. Previously, the gene density in rice has been estimated to be 8 kb per gene, 30% lower than seen here (Song et al. 2002). We would therefore assume that these intervals in rice are rather gene-poor or/and that our gene predictions have been more conservative. Gene density in maize is three times lower than in either rice or sorghum. However, if 35 kb per gene is an accurate average gene density in maize inbred B73, then the ∼2.365-Gb maize genome (http://pgir.rutgers; http://www.genome.arizona.edu/fpc/maize) would contain ∼68,000 genes. However, this number is certainly too high because these BACs were all chosen because they contained at least one gene. Maize, like other complex plant genomes, might have large areas that are relatively gene-poor, and we did not attempt to sample any of them in this study. Even in the gene-containing regions that we chose, average gene density differed dramatically. An example is the maize duplicated regions containing the orp1/2 and zmfie1/2 markers; the region on Chromo some 4 is fairly gene-poor at 71.6 kb/gene, but the orthologous Chromosome 10 region is relatively generich with 22 kb/gene.
Figure 1.
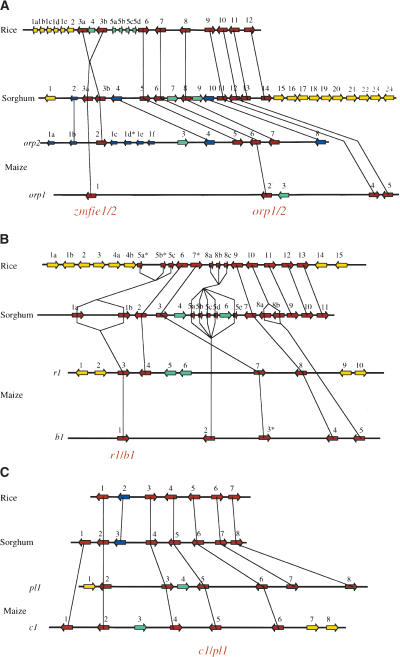
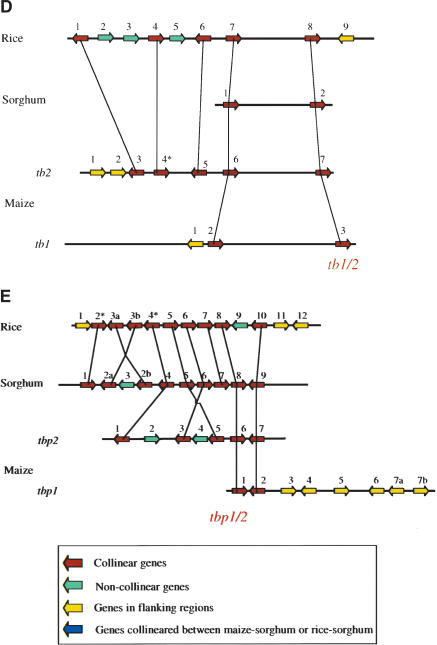
Graphic representation of the alignment, position, and polarity of all predicted genes within the selected chromosomal intervals of maize, sorghum, and rice. The relative positions of all predicted and known genes of the orthologous regions, defined by the six genetic markers highlighted under the bottom interval in red, are graphically presented. Each interval is drawn as a horizontal bar with the gene polarity indicated by an arrow. The origin of each region is labeled to the left of the bar. The physical length of each interval and other properties are reported in Table 1. Predicted and known genes are numbered from the left to the right for each interval. Duplicated genes are given the same number but ordered by a, b, c,.... Gene numbers with an * indicate gene remnants. Conservation of genes between different intervals is indicated by vertical connecting lines. A key is provided as an inset to show the conserved genes (collinear genes), and also the noncollinear genes, genes in flanking regions, and gene remnants. (A) An alignment of regions containing the zmfie1/2 and orp1/2 genes and their orthologs in sorghum and rice. (B) The r1/b1 genes and their orthologs. (C) The c1/pl1 genes and their orthologs. (D) The tb1/2 genes and their orthologs. (E) The tbp1/2 genes and their orthologs.
Synteny by Combining Both Duplicated Regions of Maize
To estimate the conservation of gene order in maize, sorghum, and rice, a gene count was performed for those genes that fall within an interval of both duplicated regions of maize and the regions in sorghum and rice. This process prevented us from using genes in the flanking regions, but permitted us to clearly define the orthologous chromosomal intervals (Fig. 1A-E). Discounting gene amplification but counting conservation of genes between at least two species, the number of genes in the orthologous intervals drops from 206 to 139. From those 139 genes, 19 (14%) are noncollinear under these conditions (Table 1). Interestingly, the percentages of the noncollinear genes in rice, sorghum, and maize are very constant, with 15% in maize by combining the two duplicated regions, 12% in sorghum, and 14% in rice. However, if we apply the most stringent conditions and count only genes collinear across all four intervals, the two in maize and the ones in sorghum, and rice, then the number of noncollinear genes would increase to 68%. This dramatic difference illustrates the enormous changes that must have taken place after the two progenitors of maize split from the progenitor of sorghum (Swigonova et al. 2004).
In some instances, where genes are conserved in their position between rice and sorghum, they are present in one of the duplicated regions of maize, but not the other one. For example, gene 5 (ocl5-like gene) and gene 7 (unknown function) on maize Chromosome 10 are collinear with rice and sorghum; however, both of them are missing on maize Chromosome 4 in the zmfie-orp region (Fig. 1A). The same is true in the r1/b1 region; for gene 4 on Chromosome 10, the genetic modifier adjacent to the r1 gene, is present in rice and sorghum, but absent in the b1 region (Fig. 1B); whereas gene 2 in the b1 region, a cis-zeatin O-glucosyltransferase gene, is missing in the r1 region. Although there are some additional cases in the r1/b1 and c1/pl1 regions, they require confirmation by having more flanking sequence information in maize. Nevertheless, such a pattern indicates that, after the hybridization of the two progenitors of maize, many duplicated genes have been lost.
Species-Specific Gene Additions or Subtractions
Every interval has noncollinear genes, and in some cases genes are also tandemly amplified. Moreover, there are examples where orthologous genes are missing between the two duplicated regions of maize or between different species. In the zmfie-orp region, there are three genes that are only conserved between sorghum and maize (Fig. 1A). These could be genes that were inserted into a common ancestor of these two species or were deleted from the rice region. There is one cf2-like gene in sorghum (gene 2) and six copies of the orthologous cf2-like gene in maize (gene 1a-1f); gene 4 of sorghum is collinear with gene 4 of maize and gene 10 of sorghum with gene 8 of maize. In the c1/pl1 region there is one gene whose position is conserved between sorghum and rice (gene 2 of rice and gene 3 of sorghum; see Fig. 1C), but missing in both duplicated regions of maize. Even BLAST searches against EST and GSS databases have been unsuccessful in finding a homolog elsewhere in the maize genome, whereas a similar search in sorghum provided two EST matches. One complication is that typical gene-finding programs would not recognize pseudogenes that are heavily truncated. There are a total of eight gene remnants. The FGENESH program predicted four, of which three (gene 1d in the maize orp1 region; gene 5a, 5b in the rice r1 region) would produce truncated proteins; gene 4 in the tb2 region has a retroelement insertion. Another four were only detected by a BLAST search: gene 7 in the rice r1 region and gene 3 in the maize b1 region. The tbp2 gene 1 in maize Chromosome 5 and the sorghum ortholog gene 4 can still be aligned to a short sequence in rice gene remnant 4* (Fig. 1E). The same is true for the sorghum gene 1 and the rice gene remnant 2*, which is severely truncated. Although many non-collinearities could be explained by gene deletions, some are most simply explained by novel gene insertions. For instance, gene 3 in the sorghum tbp region is likely to be an insertion because it has no homologs in the orthologous maize or rice regions, whereas gene 4 in the pl1 region and gene 3 in the c1 region are most easily explained as insertions in maize.
If one uses shared genes between sorghum and rice to define the “ancestral” grass genome, then there are 11 orthologous gene sets that can be scored across all four cereal genomes (rice, sorghum, and two from maize). This number ignores the six genes (fie, orp, r1/b1, c1/pl, tb, and tbp) that were used to define the regions, as we chose them because they were all present in two orthologous regions of maize. Of the 24 maize genes predicted at the time of the tetraploid origin of maize, only 18 now remain intact (Table 2). Hence, ∼50% of the duplicated copies of genes have been removed or severely damaged over the last ∼12 million years. In most cases, only one homoeologous copy has been removed, but in the case of rice gene 2 (orthologous to sorghum gene 3) in the c1/pl1 region, both maize copies appear to be missing.
Table 2.
Gene Losses in Maize Homoeologous Regions
| Regions | orp 1/2 | orp 1/2 | r1/b1 | r1/b1 | r1/b1 | r1/b1 | c1/pl1 | c1/pl1 | c1/pl1 | c1/pl1 | tb1/2 | tbp1/2 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Rice | gene6 | gene8 | gene6 | gene7 | gene8 | gene9 | gene1 | gene2 | gene3 | gene5 | gene7 | gene10 |
| Sorghum | gene5 | gene8 | gene2 | gene3 | gene5 | gene7 | gene2 | gene3 | gene4 | gene6 | gene1 | gene9 |
| Maize (orp1/b1/pl1/tb2/tbp2) | gene5 | gene7 | No | No | gene2 | gene4 | gene2 | No | gene3 | gene6 | gene6 | gene8 |
| Maize (orp1/r1/c1/tb1/tbp1) | No | No | gene4 | gene7 | No | gene8 | gene2 | No | gene4 | gene6 | gene2 | gene2 |
Distribution of Noncollinear Genes in the Rice Genome
It is not unexpected that the WGD in maize has led to preservation and/or loss of individual duplicates. This phenomenon has long been hypothesized, and increasing evidence using genome data supports this idea (Nadeau and Sankoff 1997; Prince and Pickett 2002). However, what is the significance of the insertion of new genes into an otherwise collinear region? This question might be answered if the genomes of all the three species compared with each other had already been completely sequenced. Nevertheless, we can use the rice genome, where a map-based sequence has now been generated, as a reference for asking the question whether genes present in sorghum and maize are absent only in the orthologous regions of rice. Therefore, BLAST searches were performed for those noncollinear genes in sorghum and maize that were missing in the rice orthologous regions to identify homologous genes and their positions in the rice genome. A large percentage (85%) of orthologous genes were not simply lost, but are now found somewhere else in the rice genome (Table 3). Distribution of these homologous sequences is not clustered but appears to be positioned on different rice chromosomes. Moreover, for a majority of them, multiple copies of the noncollinear gene can be found on different chromosomes. With just this two-point comparison, the rice lineage versus the maize/sorghum lineage, we cannot determine whether these gene movements occurred in an ancestor of rice or an ancestor of the Andropogoneae that gave rise to maize and sorghum.
Table 3.
Paralogous Sequences in the Rice Genome
| Gene annotation | Expected location | Actual location | |||
|---|---|---|---|---|---|
| Fie_orp region | Maize (orp2 region) | gene1 | cf2-like protein | Ch8 | Ch1, Ch12, Ch4 |
| gene3 | Unknown protein | Ch8 | Ch3, Ch7, Ch10 | ||
| gene4 | Unknown protein | Ch8 | no hit | ||
| gene8 | Endonuclease/exonuclease/phosphatase family | Ch8 | no hit | ||
| Maize (orp1 region) | gene3 | Putative tubby-like protein | Ch8 | Ch2 | |
| Sorghum | gene1 | Receptor-like kinase | Ch8 | no hit | |
| gene2 | cf2-like protein | Ch8 | Ch1, Ch12, Ch4 | ||
| gene4 | Unknown protein | Ch8 | no hit | ||
| gene7 | Unknown protein | Ch8 | Ch6, Ch12, Ch7, Ch4, Ch1, Ch11 | ||
| gene9 | Unknown protein | Ch8 | Ch7, Ch2, Ch9 | ||
| gene10 | Endonuclease/exonuclease/phosphatase family | Ch8 | Ch12 | ||
| r1/b1 region | Maize (r1 region) | gene1 | NADP-dependent malic enzyme | Ch4 | Ch1 |
| gene2 | Putative pinhead protein | Ch4 | Ch4, Ch2, Ch5, Ch6 | ||
| gene5 | Putative S-receptor kinase | Ch4 | Ch9 | ||
| gene6 | Putative aldose reductase-related protein | Ch4 | Ch6 | ||
| gene8 | Response regulator Cipl like | Ch4 | no hit | ||
| gene9 | 4-coumarate-CoA ligase-like protein | Ch4 | Ch4 | ||
| Sorghum | gene4 | No apical meristem (NAM) family protein | Ch4 | Ch5, Ch6 | |
| gene6 | WD-40 repeat family protein | Ch4 | Ch2, Ch7, Ch12, Ch3, Ch9 | ||
| c1/pl1 region | Maize (c1 region) | gene3 | Epsilon-COP | Ch6 | Ch4 |
| gene7 | 40S ribosomal protein S8 | Ch6 | Ch2, Ch4 | ||
| gene8 | Putative casein kinase I | Ch6 | Ch2, Ch4, Ch10 | ||
| Maize (pl1 region) | gene1 | Putative RNA-binding protein | Ch6 | Ch2, Ch3, Ch8, Ch4 | |
| gene4 | Heme oxygenase I | Ch6 | Ch2, Ch1, Ch5, Ch9, Ch10 | ||
| tb1/tb2 region | Maize (tb1 region) | gene1 | Putative metal-Transporting ATPase | Ch3 | Ch3 |
| Maize (tb2 region) | gene1 | Glycyl-tRNA synthetase | Ch3 | Ch8, Ch9, Ch3, Ch6 | |
| gene2 | Unknown protein | Ch3 | Ch10 | ||
| tbp1/tbp2 region | Maize (tbp2 region) | gene2 | Putative casein kinase I | Ch3 | Ch4 |
| gene5 | Gamma-tubulin I | Ch3 | Ch5 | ||
| Sorghum | gene3 | Unknown protein | Ch3 | Ch12 |
DISCUSSION
Gene Deletions Versus Insertions
The studies described here have an impact on our understanding of two important questions in plant genomics. One is related to polyploidization, and the other is related to the dynamic features of plant chromosomal sequences. Although it has been known for some time that sequences in chromosomes are not as static as previous genetic analysis of model species has suggested, this knowledge was mainly based on studies of transposable elements. It came as a surprise that DNA sequences might move around the genome, but it still was believed that the genes themselves were usually fixed in their positions, providing the foundation of reproducible genetic maps. Over short time periods, less than a few million years, conserved gene map location still holds true after all the new insights from genomic studies. However, we can detect that the positions of some genes within chromosomes can be quite different in some closely related species. This insight became possible largely because the rice genome has now been sequenced in its entirety. For the first time, we can ask if noncollinear genes are simply lost in the other species or have a copy somewhere else. This suggests that most genes present in the ancestral chromosome can be lost if there is a backup somewhere else in the genome. The mechanisms behind this phenomenon are not clear.
One possible step toward gene movement would involve amplification prior to insertion at a new location. This could guarantee that a copy remains, at least transiently, in any heterozygote containing the newly inserted gene. WGD is one mechanism whereby all genes are doubled in number, providing a possible opportunity for additional gene movement. Recently, a comparison was made of a storage protein gene family in maize, consisting of 41 members in seven nonduplicated genomic locations (Song and Messing 2002, 2003). Comparison to sorghum suggested that upon the split of the two progenitors of maize and the progenitor of sorghum only one or two copies existed. Even this copy has diverged so much from any genes in rice that hybridization fails to detect any such sequence in the rice genome. However, amplification of these genes to now constitute 41 members occurred within the last 4.5 million years (Song et al. 2001). At the same time, the gene family expanded by placing copies in six additional genomic locations including different chromosomes. Because such an expansion apparently involved random insertion of genes, it may bear some similarity to the action of transposable elements. However, there may be different mechanisms of gene movement and amplification, and the frequency can vary enormously. For instance, some complex disease-resistance genes can average changes in specificity, copy number, and other organizational features as often as 1% of meiosis (Richter et al. 1995; Ramakrishna et al. 2002b,c). Furthermore, recent studies (Fu and Dooner 2002; Song and Messing 2003) have shown that gene collinearity between maize lines can also deviate. In our data set, sequences of all five regions are from one inbred line, B73, a line that was chosen for the public maize BAC library resources. Although the extent of intraspecific violation of gene collinearity in maize inbreds, races, and wild relatives remains to be determined, their diversity will add another level of complexity to understanding the dynamic nature of grass genomes.
Differentiation of Paralogous Sequences
One interesting feature of our results is the high percentage of genes that have undergone changes in position. Nearly a seventh of the genes have moved for the three species investigated here. This number would increase even further if we consider genes in the flanking regions as well. When those in the sorghum and maize flanking regions were also compared with the rice genome, they were also found in unlinked locations (Table 3). Such an extent of nonorthologous genes is consistent with earlier experimental data, in which 20%-40% of highly homologous markers failed to map to orthologous locations in different grass species (Bennetzen and Freeling 1993). Nevertheless, the remaining 86% collinear genes (Table 1) explain why the comparison of the genetic maps provides syntenic alignment over large chromosomal segments. The question that arises is not only by what mechanism genes might have moved but whether any evolutionary or environmental pressures might have triggered this movement. In the case of disease-resistance genes, it is clear that rearrangements can yield new variants of gene products that may better defend the host against new pathogen variants (Richter et al. 1995; Mondragon-Palomino et al. 2002). However, many transposed genes, like adh1 of maize (Ilic et al. 2003) and the diverse set of proteins that we see in our study, do not fit these criteria (Table 3).
In the zein region of maize, a recent amplification of storage proteins led to a change in their transcriptional regulation (Song et al. 2001), wherein the new gene copies are regulated by a different transcription factor. Therefore, one could envision that the positional changes of genes could also be associated with the acquisition of a change in their transcriptional regulation. This could be caused by the insertion of the rearranged gene into a region with novel genetic or epigenetic features that might alter gene expression, as has often been observed for the complimentary phenomenon in which a transposable element inserts near a gene and thereby alters its regulation (Barkan and Martienssen 1991). In many cases, we find that reinserted genes are present on entirely different chromosomes, guaranteeing that they will be in a new chromosomal environment. Such a feature could be far more ubiquitous than previously thought because the recent analysis of rice Chromosome 10 has shown that tandemly arranged gene families represent a larger percentage of the total genes (25%) in rice than they do in Arabidopsis (17%; The Rice Chromosome 10 Sequencing Consortium 2003).
Gene Instability Because of WGD
Comparison between closely related diploid genomes and the duplicated regions of the maize genome that have arisen by WGD provides novel insights into the intraspecies collinearity of ancient tetraploid plant species. It has already been suggested that genes are lost in other species because of polyploidization (Song et al. 1995; Feldman et al. 1997; Wolfe and Shields 1997; Ozkan et al. 2001; Wolfe 2001). The striking feature is that, although homoeologous regions can easily be detected, interruption of collinearity is far greater than in interspecies comparison. However, such a structural difference seen between the two homoeologous regions cannot be explained by rapid gene subfunctionalization of duplicated genes (Lynch and Conery 2000). For example, lack of any coding sequence in the corresponding region of the ocl5 gene in the orp2 region (Fig. 1A) and the genetic modifier gene, which is next to the r1 gene, demonstrate that gene losses occurred after tetraploidization. However, if one takes into consideration both homoeologous regions in maize, one can reconstruct the gene order from the ancestral chromosomes. An earlier analysis in maize (Ilic et al. 2003) indicated ∼20% retention of functional duplicated gene copies between the two homoeologous chromosome sets of maize, but this was based on only one analyzed region from two different inbred lines. Our more comprehensive analysis suggests ∼50% retention of duplicated copies, but we do not know how many of these are functional. A mosaic structure of collinearity between two homoeologous regions has recently also been described for the WGD of yeast. In that case, the genome of Kluyveromyces waltii contains the composite gene order of the duplicated regions of yeast (Kellis et al. 2004), similar to the way rice and sorghum provide a guide for understanding ancestral gene order and composition in maize. However, the resulting gene loss in yeast of 90% of duplicated copies is far beyond what has occurred to this date in the maize genome. Nevertheless, considering that the WGD of the ancestral progenitors of yeast may have occurred 150 Mya compared with 4.8-11.9 Mya for the progenitors of maize, the gene loss in maize appeared to have occurred within a relatively short time.
METHODS
BAC Clone Isolation and Sequencing
High-density filters for the maize (Zea mays L. cv. B73) BAC libraries, described previously by Yim et al. (2002), were screened with probes made from PCR products of maize r1, c1, orp1, tb1, and tbp1 genes based on their sequences in GenBank. Similarly, filters for sorghum (Sorghum bicolor cv. BTx623) BAC libraries were screened using the same probes. All positive clones resulting from the screen were further digested by NotI and HindIII for fingerprint analysis. The clones were further analyzed by restriction mapping using the same probes used for screening. BAC DNA was isolated using a BAC DNA isolation kit (QIAGEN). The purified BAC DNA was physically sheared and then ligated into a pUC vector for shotgun libraries as previously reported by Song et al. (2001). Sequencing was done on an ABI 3700 DNA sequencer using the ABI PRISM BigDye Terminator Cycle Sequencing Ready Reaction kit (Applied BioSystems). Base-calling and assembly were based on phred/phrap programs (Ewing et al. 1998). About 10× coverage was generated for all of the BACs, and sequence gaps were finished by specific primers identified with the help of consed (Gordon et al. 1998) or by transposon minilibraries for subclones that cover the gaps. The transposon minilibraries were made according to the manufacturer's instructions (Finnzyme).
Sequence Analysis
After assembly of the sequences of individual BAC clones, overlapping clones were merged to form individual contigs. Single BAC clones and contigs were subjected to the gene prediction program, FGENESH, set at the monocot option (http://www.softberry.com). Predicted genes were further verified with homology searches, using BLASTP (Altschul et al. 1997) against the GenBank protein and DNA databases. Only matches with a BLASTP E-value smaller than e-10 were accepted. Both nr and HTGS databases were used in searches for homologs in the rice genome.
Maize Accessions
Some accessions may contain more than one clone: orp1/zmfie1: AY555142, AY560576; orp2/zmfie2: AY555143, AY560578 (two clones); r1: AF466202 (two clones); b1: AF466203, AY542310; c1: AY530950, AY530951; pl1: AY530952, AY560577; tb1: AF464738, AY325816; tb2: AF466646; tbp1: AY542798; tbp2: AY542797.
Sorghum Accessions
Some accessions may contain more than one clone: orp/zmfie: AF466200; r1/b1: AY542311; c1/pl1: AF466199; tb1/2: AF466204; tbp1/2: AF466201.
Rice Accessions
orp/zmfie: AP003896, AP005620; r1/b1: AL606682, AL606647; c1/pl1: AP005652; tb1/2: AC091775; tbp1/2: AC133859.
Figure 2.
Acknowledgments
We thank J. Emberton, G. Fuks, Arvind Bharti, A. Bronzino-Nelson, S. Kavchok, G. Keizer, and S. Young for technical assistance. We are also grateful to Andrew Fernandez pointing out an artifact of the assembly of AY542797. This work was supported by NSF grant 9975618.
Footnotes
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2701104.
References
- Ahn, S. and Tanksley, S.D. 1993. Comparative linkage maps of the rice and maize genomes. Proc. Natl. Acad. Sci. 90: 7980-7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhunov, E.D., Akhunova, A.R., Linkiewicz, A.M., Dubcovsky, J., Hummel, D., Lazo, G., Chao, S., Anderson, O.D., David, J., Qi, L., et al. 2003. Synteny perturbations between wheat homoeologous chromosomes caused by locus duplications and deletions correlate with recombination rates. Proc. Natl. Acad. Sci. 100: 10836-10841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schäffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan, A. and Martienssen, R.A. 1991. Inactivation of maize transposon Mu suppresses a mutant phenotype by activating an outward-reading promoter near the end of Mu1. Proc. Natl. Acad. Sci. 88: 3502-3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen, J.L. 2000. Comparative sequence analysis of plant nuclear genomes: Microcolinearity and its many exceptions. Plant Cell 12: 1021-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen, J.L. and Freeling, M. 1993. Grasses as a single genetic system: Genome composition, collinearity and compatibility. Trends Genet. 9: 259-261. [DOI] [PubMed] [Google Scholar]
- Chen, M., SanMiguel, P., Oliveira, A.C.D., Woo, S.S., Zhang, H., Wing, R., and Bennetzen, J.L. 1997. Microcollinearity in sh2-homologous regions of the maize, rice, and sorghum genomes. Proc. Natl. Acad. Sci. 94: 3431-3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coe, E., Cone, K., McMullen, M., Chen, S.S., Davis, G., Gardiner, J., Liscum, E., Polacco, M., Paterson, A., Sanchez-Villeda, H., et al. 2002. Access to the maize genome: An integrated physical and genetic map. Plant Physiol. 128: 9-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler, F.W., Hatfield, J., Nelson, W., and Soderlund, C.A. 2003. Locating sequence on FPC maps and selecting a minimal tiling path. Genome Res. 13: 2152-2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing, B., Hillier, L., Wendl, M.C., and Green, P. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8: 175-185. [DOI] [PubMed] [Google Scholar]
- Feldman, M., Liu, B., Segal, G., Abbo, S., Levy, A.A., and Vega, J.M. 1997. Rapid elimination of low-copy DNA sequences in polyploid wheat: A possible mechanism for differentiation of homoeologous chromosomes. Genetics 147: 1381-1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, H. and Dooner, H.K. 2002. Intraspecific violation of genetic colinearity and its implications in maize. Proc. Natl. Acad. Sci. 99: 9573-9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale, M.D. and Devos, K. 1998. Plant comparative genetics after 10 years. Science 282: 656-659. [DOI] [PubMed] [Google Scholar]
- Gaut, B.S. 2001. Patterns of chromosomal duplication in maize and their implications for comparative maps of the grasses. Genome Res. 11: 55-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman, M.M., Stuber, C.W., Newton, K., and Weissinger, H.H. 1980. Linkage relationships of 19 enzyme loci in maize. Genetics 96: 697-710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon, D., Abajian, C., and Green, P. 1998. Consed: A graphical tool for sequence finishing. Genome Res. 8: 195-202. [DOI] [PubMed] [Google Scholar]
- Helentjaris, T., Weber, D., and Wright, S. 1988. Identification of the genomic locations of duplicate nucleotide sequences in maize by analysis of restriction fragment length polymorphism. Genetics 118: 353-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulbert, S.H., Richter, T.E., Axtell, J.D., and Bennetzen, J.L. 1990. Genetic mapping and characterization of sorghum and related crops by means of maize DNA probes. Proc. Natl. Acad. Sci. 87: 4251-4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilic, K., SanMiguel, P.J., and Bennetzen, J.L. 2003. A complex history of rearrangement in an orthologous region of the maize, sorghum and rice genomes. Proc. Natl. Acad. Sci. 100: 12265-12270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller, B. and Feuillet, C. 2000. Colinearity and gene density in grass genomes. Trends Plant Sci. 5: 246-251. [DOI] [PubMed] [Google Scholar]
- Kellis, M., Birren, B.W., and Lander, E.S. 2004. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature 428: 617-624. [DOI] [PubMed] [Google Scholar]
- Kellogg, E.A. 2001. Evolutionary history of the grasses. Plant Physiol. 125: 1198-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, M. and Conery, J.S. 2000. The evolutionary fate and consequences of duplicate genes. Science 290: 1151-1155. [DOI] [PubMed] [Google Scholar]
- McClintock, B. 1930. A cytological demonstration of the location of an interchange between two non-homologous chromosomes of Zea mays. Proc. Natl. Acad. Sci. 16: 791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillin, D.E. and Scandalios, J.G. 1980. Duplicated cytosolic malate dehydrogenase genes in Zea mays. Proc. Natl. Acad. Sci. 77: 4866-4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondragon-Palomino, M., Meyers, B.C., Michelmore, R.W., and Gaut, B.S. 2002. Patterns of positive selection in the complete NBS-LRR gene family of Arabidopsis thaliana. Genome Res. 12: 1305-1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, G., Devos, K., Wang, Z., and Gale, M.D. 1995. Grasses, line up and form a circle. Curr. Biol. 5: 737-739. [DOI] [PubMed] [Google Scholar]
- Nadeau, J.H. and Sankoff, D. 1997. Comparable rates of gene loss and functional divergence after genome duplications early in vertebrate evolution. Genetics 147: 1259-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozkan, H., Levy, A.A., and Feldman, M. 2001. Allopolyploidy-induced rapid genome evolution in the wheat (Aegilops-Triticum) group. Plant Cell 13: 1735-1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson, A.H., Bowers, J.E., Burow, M.D., Draye, X., Elsik, C.G., Jiang, C.X., Katsar, C.S., Lan, T.H., Lin, Y.R., Ming, R., et al. 2000. Comparative genomics of plant chromosomes. Plant Cell 12: 1523-1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince, V.E. and Pickett, F.B. 2002. Splitting pairs: The diverging fates of duplicated genes. Nat. Rev. Genet. 3: 827-837. [DOI] [PubMed] [Google Scholar]
- Ramakrishna, W., Dubcovsky, J., Park, Y.J., Busso, C., Emberton, J., SanMiguel, P., and Bennetzen, J.L. 2002a. Different types and rates of genome evolution detected by comparative sequence analysis of orthologous segments from four cereal genomes. Genetics 162: 1389-1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishna, W., Emberton, J., Ogden, M., SanMiguel, P., and Bennetzen, J.L. 2002b. Structural analysis of the maize rp1 complex reveals numerous sites and unexpected mechanisms of local rearrangement. Plant Cell 14: 3213-3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishna, W., Emberton, J., SanMiguel, P., Ogden, M., Llaca, V., Messing, J., and Bennetzen, J.L. 2002c. Comparative sequence analysis of the sorghum Rph region and the maize Rp1 resistance gene complex. Plant Physiol. 130: 1728-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades, M.M. 1951. Duplicated genes in maize. Am. Nat. 85: 105-110. [Google Scholar]
- The Rice Chromosome 10 Sequencing Consortium. 2003. In-depth view of structure, activity, and evolution of rice chromosome 10. Science 300: 1566-1569. [DOI] [PubMed] [Google Scholar]
- Richter, T.E., Pryor, T.J., Bennetzen, J.L., and Hulbert, S.H. 1995. New rust resistance specificities associated with recombination in the Rp1 complex in maize. Genetics 141: 373-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SanMiguel, P., Gaut, B.S., Tikhonov, A., Nakajima, Y., and Bennetzen, J.L. 1998. The paleontology of intergene retrotransposons of maize. Nat. Genet. 20: 43-45. [DOI] [PubMed] [Google Scholar]
- Song, R. and Messing, J. 2002. Contiguous genomic DNA sequence comprising the 19-kD zein gene family from maize. Plant Physiol. 130: 1626-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 2003. Gene expression of a gene family in maize based on noncollinear haplotypes. Proc. Natl. Acad. Sci. 100: 9055-9060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, K., Lu, P., Tang, K., and Osborn, T.C. 1995. Rapid genome change in synthetic polyploids of Brassica and its implications for polyploid evolution. Proc. Natl. Acad. Sci. 92: 7719-7723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, R., Llaca, V., Linton, E., and Messing, J. 2001. Sequence, regulation, and evolution of the maize 22-kD zein gene family. Genome Res. 11: 1817-1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, R., Llaca V., and Messing, J. 2002. Mosaic organization of orthologous sequences in grass genomes. Genome Res. 12: 1549-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swigonova, Z., Lai, J., Ma, J., Ramakrishna, W., Llaca, V., Bennetzen, J.L., and Messing, J. 2004. Close split of maize and sorghum genome progenitors. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- Tikhonov, A.P., SanMiguel, P.J., Nakajima, Y., Gorenstein, N.D., Bennetzen, J.L., and Avramova, Z. 1999. Colinearity and its exceptions in orthologous adh regions of maize and sorghum. Proc. Natl. Acad. Sci. 96: 7409-7414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting, Y.C. 1966. Duplications and meiotic behavior of the chromosomes in haploid maize (Zea mays L.). Cytologia 31: 324-329. [Google Scholar]
- Wendel, J.F., Stuber, C.W., Edwards, M.D., and Goodman, M.M. 1986. Duplicated chromosomal segments in Zea mays L.: Further evidence from hexokinase isozymes. Theor. Appl. Genet. 72: 178-185. [DOI] [PubMed] [Google Scholar]
- Wolfe, K.H. 2001. Yesterday's polyploids and the mystery of diploidization. Nat. Rev. Genet. 2: 333-341. [DOI] [PubMed] [Google Scholar]
- Wolfe, K.H. and Shields, D.C. 1997. Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387: 708-713. [DOI] [PubMed] [Google Scholar]
- Yim, Y.S., Davis, G.L., Duru, N.A., Musket, T.A., Linton, E.W., Messing, J.W., McMullen, M.D., Soderlund, C.A., Polacco, M.L., Gardiner, J.M., et al. 2002. Characterization of three maize bacterial artificial chromosome libraries toward anchoring of the physical map to the genetic map using high-density bacterial artificial chromosome filter hybridization. Plant Physiol. 130: 1686-1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://rgp.dna.affrc.go.jp; International Rice Genome Sequencing Project, IRGSP.
- http://www.genome.arizona.edu/fpc/maize; maize genome.
- http://www.softberry.com; FGENESH.
- http://pgir.rutgers.edu; maize genome.