Abstract
Quantitative gene expression analysis aims to define the gene expression patterns determining cell behavior. So far, these assessments can only be performed at the population level. Therefore, they determine the average gene expression within a population, overlooking possible cell-to-cell heterogeneity that could lead to different cell behaviors/cell fates. Understanding individual cell behavior requires multiple gene expression analyses of single cells, and may be fundamental for the understanding of all types of biological events and/or differentiation processes. We here describe a new reverse transcription-polymerase chain reaction (RT-PCR) approach allowing the simultaneous quantification of the expression of 20 genes in the same single cell. This method has broad application, in different species and any type of gene combination. RT efficiency is evaluated. Uniform and maximized amplification conditions for all genes are provided. Abundance relationships are maintained, allowing the precise quantification of the absolute number of mRNA molecules per cell, ranging from 2 to 1.28×109 for each individual gene. We evaluated the impact of this approach on functional genetic read-outs by studying an apparently homogeneous population (monoclonal T cells recovered 4 d after antigen stimulation), using either this method or conventional real-time RT-PCR. Single-cell studies revealed considerable cell-to-cell variation: All T cells did not express all individual genes. Gene coexpression patterns were very heterogeneous. mRNA copy numbers varied between different transcripts and in different cells. As a consequence, this single-cell assay introduces new and fundamental information regarding functional genomic read-outs. By comparison, we also show that conventional quantitative assays determining population averages supply insufficient information, and may even be highly misleading.
Functional genomic analysis is fundamental for understanding how genomic expression profiles influence cell fate. Such studies are usually performed by using either micro-arrays or a real-time quantitative reverse transcription polymerase chain reaction (RT-PCR). These methodologies can determine multiple gene expression, but have a major limitation. They only allow studies at the population level and thus only determine average gene expression. They cannot evaluate variations of gene expression between individual cells. However, in many types of biological events, individual cells within apparently homogeneous populations have different fates. It is likely that these different fates are conditioned by different patterns of gene expression. Because the events occurring in each individual cell are unknown, current methods may fail to identify the gene expression balance that ultimately determines cell behavior. This latter information requires multiple gene expression analysis of single cells, which may be a fundamental step for the understanding of all types of biological events and/or differentiation processes.
Multiple analysis of gene expression at the single-cell level requires major technological advances. Most techniques are qualitative and only allow studies of the expression of a few genes (Phillips and Lipski 2000; Veiga-Fernandes et al. 2000; Walter et al. 2000; Lambolez et al. 2002). When more genes were to be tested, these methods were reported to have inherent biases (Phillips and Lipski 2000; Walter et al. 2000). Indeed, in more extensive gene expression studies, the efficiency of detection was simply not controlled (Ruano et al. 1995; Plant et al. 1997; Zawar et al. 1999; Gallopin et al. 2000).
In principle, there are no sensitivity limitations for single-cell gene expression analysis. Single-cell methods can detect genomic DNA, that is, two gene copies, when two successive PCR amplifications of the same gene are performed (Loffert et al. 1996). However, the modification of such methodology to allow multiple mRNA expression studies involves serious difficulties. First, the amount of mRNA extracted from a single cell is so minute that samples cannot be split. The expression of multiple genes must be investigated in the same sample and in the same RT-PCR round. This implies the presence of multiple primers and the generation of multiple amplicons in a single PCR round, which may induce serious competition between different amplifications. It was claimed that analysis of the coexpression of more than five genes in one cell simultaneously would necessarily lead to nonspecific inhibitions of amplification (Walter et al. 2000). These potential competition events may induce false-negative results that are particularly difficult to control in single-cell assays. Indeed, as each individual cell is potentially different, it is not possible to determine whether a negative result is due to the absence of gene expression or to the absence of amplification due to competition.
Further difficulties are involved in attempting to quantify gene expression in single cells. Such quantification would require the demonstration of the maintenance of abundance relationships between multiple genes and throughout multiple reactions: from mRNA to cDNA, and throughout two successive PCR amplifications. The template switching required by two-step amplifications may introduce potential bias (Phillips and Lipski 2000). Moreover, it was postulated that abundance relationships could not be maintained throughout exponential amplification, as theoretical mathematic analysis showed that hybridization kinetics during thermal cycling could induce both sequence- and copy number-dependent bias (Peccoud and Jacob 1996). However, certain techniques of enhanced reverse transcription faithfully maintained relative abundance relationships using exponential amplification (Iscove et al. 2002; Makrigiorgos et al. 2002). These new findings open the possibility that RT-PCR methods could be modified in such a way that abundance relationships could still be maintained, allowing quantification of gene expression in individual cells.
Here we describe a new method in which all previous limitations have been overcome since the expression of 20 different genes can be quantified simultaneously in each cell. We further demonstrate that this powerful technique imparts fundamental new information on cell behavior. In contrast, we also show that gene expression studies performed at the population level do not impart sufficient information and may even be highly misleading.
RESULTS
General Aspects of Quantitative Single-Cell Multiplex RT-PCR
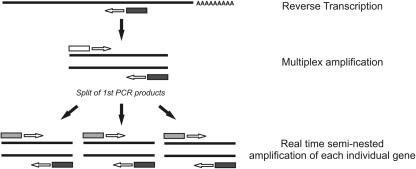
Sorted cells are lysed and the mRNA is retrotranscribed using specific 3′ primers. A first PCR follows, where both 3′ and 5′ primers for all 20 different genes are present in the same reaction (Fig. 1). The products of this first amplification are next split into individual wells where a second seminested real-time PCR amplifies each individual gene separately (Fig. 1). To quantify the number of mRNA copies of different genes, the cycle threshold (CT) value obtained for each different gene product is then compared with a known quantified RNA standard that followed the same rules of retrotranscription and amplification of the tested samples. This comparison allows a precise determination of mRNA copy numbers of different genes from a single individual cell.
Figure 1.
Outline of the quantitative multiplex single-cell RT-PCR. Single-cell mRNA is retrotranscribed using a 3′-specific primer for each individual gene of interest (dark gray box). Next, single-cell cDNA is amplified on a first multiplex PCR where both 3′ (dark gray box) and 5′ (white box) primers of all different genes are present (15 cycles). Products of the first amplification are next split for a second seminested real-time PCR where a nested 5′ primer (light gray box) and the 3′ primer (dark gray box) are used to amplify each gene separately. This second round of amplification allows a precise quantification because test samples are compared with an RNA standard submitted to the same RT and amplification protocol.
The feasibility of this method is strictly dependent on multiple parameters: a precise experimental strategy, which includes the use of specific reverse transcription (see Discussion), the use of precise rules for primer design, and particular amplification conditions (see Methods).
Validation of Primer Design Strategy
Efficiency
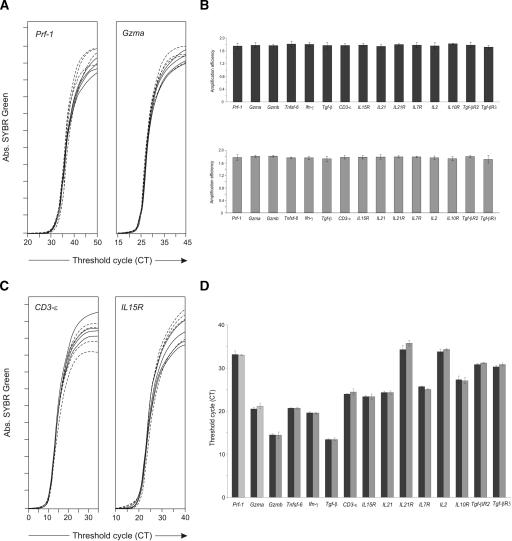
To allow comparison of the expression of different genes, PCR reactions amplifying different cDNA fragments must have the same efficiency. We tested the efficiency of our PCR amplifications on the cDNA from gut intraepithelial T lymphocytes (IELs), because this template contains all of the cDNAs coding for the 20 different genes we investigated. Aliquots of this template were amplified separately for each gene product, using primer combinations from either the first or the second PCR. The PCR accumulation slope on the linear phase of all of these PCRs (Fig. 2A) allowed us to evaluate PCR efficiency (Ramakers et al. 2003). We show that all 40 types of PCR had equal efficiency (Fig. 2B).
Figure 2.
Efficiency and competition of PCR amplifications. (A,B) Aliquots of cDNA from mouse IEL were amplified separately for each gene and each type of PCR reaction to determine PCR efficiency. (A) Quadruplicate amplification slopes for Prf-1, Gzma. Set of primers from the first PCR (solid lines) the second PCR (dashed lines).The same tests were performed for all genes, giving the same results (B) Slope values from the quadruplicates were assessed on the exponential phase of the real-time amplification reaction, and PCR efficiencies were determined using LinRegPCR 7.0 software. Means ± SD of PCR efficiencies are shown for the first (upper panel) and second (bottom panel) PCRs. The significance of these differences was evaluated by ANOVA. Within each PCR, all primer combinations had the same efficiency. We also found no significant difference in the variance between the first and second PCR amplifications (ANOVA, P > 0.1). Data are from one of three independent experiments. (C,D) Competition: Aliquots of cDNA from IELs were amplified separately for each gene, or in multiplex in the first PCR round. Quadruplicates of these reactions were further amplified in a second real-time PCR. (C) Quadruplicates of amplification for Cd3-ε; IL15r, genes amplified in multiplex (solid line) or alone (dashed line). (D) Comparison of threshold cycle mean values (CT) obtained for each different gene amplified in multiplex (black bars) or separately (gray bars). No significant differences were observed between the two amplification conditions for each different gene (t-test, P > 0.1).
Competition
A key feature of our method is the first PCR reaction, where all 40 primers amplifying different cDNAs are present and the 20 cDNA types are amplified simultaneously. This is required for assessment of multiple gene expressions in the same cell. However, this multiple amplification may result in PCR inhibition and/or reduced PCR efficiency that may invalidate the data (Phillips and Lipski 2000; Walter et al. 2000). To evaluate competition, the same amount of IEL cDNA was amplified on the first PCR round: either separately for each individual gene, or in combination with all other genes. Next, the PCR products generated in these two conditions were amplified on a second quantitative PCR. As shown in Figure 2C,D, the relative levels of expression of each gene were the same when the gene was amplified separately or in combination.
We conclude that our method provides a variety of PCR amplifications with similar efficiencies. Moreover, the 20 primer combinations of the first PCR can be associated in the same reaction, showing that our methodology prevents competition between different amplifications, a major handicap of previous methods for multiple simultaneous amplifications (Phillips and Lipski 2000; Walter et al. 2000). Our strategy thus allows simultaneous multiple gene analysis in the same sample. Because all PCRs have the same efficiency, this method also allows the comparison of relative levels of expression of different genes between themselves.
Broad Spectra Application
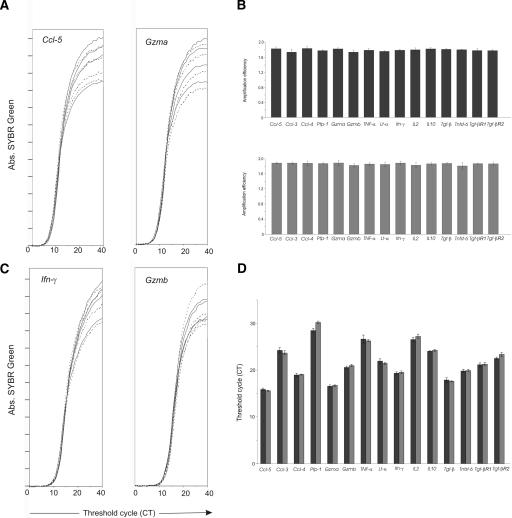
We next investigated whether the strategy we used for this particular multiplex single-cell analysis could be applied for any other type of gene combination, and in different species. For that purpose we used the same rules described in the Methods section to select primers and amplicons that study multiple T-cell functions (including cytokine and chemokine expression) in human cells. We tested the efficiency of our PCR amplifications on cDNA from human small intestine, because this template also expresses all of the cDNAs we investigated. Aliquots of this template were amplified separately for each gene product, using primer combinations from either the first or the second PCR (Fig. 3A). Evaluation of PCR efficiency (Ramakers et al. 2003) revealed that all 40 of these individual PCR reactions had the same efficiency (Fig. 3B). To evaluate competition, the same amount of human small intestine cDNA was amplified on the first PCR round, either separately for each individual gene or in combination with all other genes. Next, the PCR products generated in these two conditions were amplified on a second quantitative PCR. As with the mouse multiple gene analysis (Fig. 2B,C), we found no inhibition due to primer/amplicon competition, as relative levels of expression were identical whether each gene was amplified separately or in combination (Fig. 3C,D).
Figure 3.
Broader applications for the quantitative single-cell multiplex RT-PCR. Aliquots of cDNA from human small intestine were tested as described in Figure 2. (A,B) Efficiency of PCR amplification for different gene products. Aliquots were amplified separately for each gene and each type of PCR. (A) Quadruplicate amplification slopes for Ccl-5, Gzma, first PCR (solid lines) and second PCR (dash lines). Amplification of all genes gave the same results (B) Slope values from the quadruplicates were assessed on the exponential phase; efficiency and significance of variation were performed as described in Figure 2. Results show mean ± SD of PCR efficiencies of the first (upper panel) and second (bottom panel) PCRs. All primer combinations had the same efficiency. We also found no significant difference in the variance between the first and second PCR amplifications (ANOVA, P > 0.05). Data are from one of three independent experiments. (C,D) Competition: Aliquots of cDNA from human small intestine were amplified separately for each gene, or in multiplex in the first PCR round. Quadruplicates of these reactions were further amplified in a second real-time PCR. (C) Quadruplicates of amplification for Ifn-γ, Gzmb genes amplified in multiplex (solid line) or alone (dashed line). Comparison of CT values obtained for each different gene amplified in multiplex (black bars) or separately (gray bars). No significant differences were observed between the two amplification conditions for each different gene (t-test, P > 0.1).
We conclude that by using the primer/amplicon selection strategy and the amplification conditions described in the Methods section, this methodology can be applied to the simultaneous quantification of any 20 mRNA combinations. This validates the basic principles of the method for multiparameter analysis and provides proof of its broad spectrum of applications.
From Population Studies to Single-Cell Studies
Template Switching
The quantification of the limited material recovered from single cells as well as the study of multiple parameters in a single cell requires the use of a two-step PCR amplification, and the consequent template switching from the first to the second PCR round. Such template switching may introduce potential bias (Phillips and Lipski 2000). To prevent such bias, the first amplification round should amplify rare templates in such a way that aliquot switching should exclude tube-to-tube variability. The initial amplification should also guarantee that highly expressed genes do not reach a saturation plateau that would exclude accurate quantitative assessments on the second PCR. In other words, to prevent biased assessments, the first round of amplification must preserve the initial representation of rare genes, simultaneously excluding excessive amplification of highly abundant gene copies.
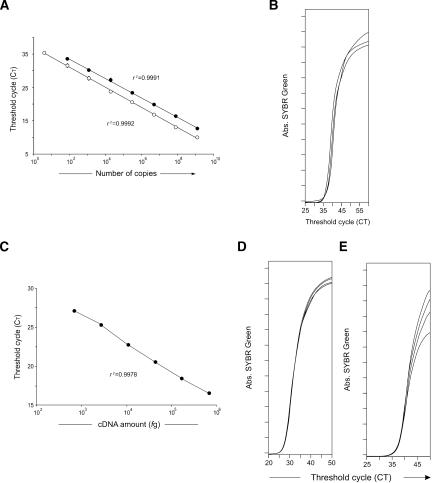
We tested for bias introduced by the template switching between the first and second PCRs using several approaches. First, we studied the amplification of a synthesized double-stranded template, corresponding to the mouse Gzma sequence we amplified in our PCRs (Fig. 4A). Because the molecular weight of this template was known, we could calculate the absolute number of DNA molecules that was present in each reaction. In this way, we could study possible artifacts of two-step reactions at both high and low copy numbers of starting material. Decreasing concentrations of this template, by a factor of 16, from 1.28×109 to four molecules, were amplified in single or double-step amplification (Fig. 4A). Upon a single amplification, the linear regression curve of this standard had a high correlation coefficient (r2 = 0.999). These results show that we can assess a vast range of template copy numbers while maintaining linearity. Next, we studied the amplification of the same double-stranded template using a two-step amplification. The number of amplification cycles in the first PCR ranged from five to 30. When the first PCR was 15 cycles, high correlation coefficients (r2 = 0.999) were maintained in the second PCR. Furthermore, using this 15-cycle preamplification, linearity was maintained at both high and low template concentrations (Fig. 4A). We tested these same parameters for other synthesized cDNA sequences, namely mouse Gzmb and Prf-1 and human CD3-ε, and similar results were obtained (data not shown). We further investigated tube-to-tube variability at very low copy numbers directly. Triplicates of synthesized mouse Gzma sequence (four molecules) were amplified in a first 15-cycle PCR, and then aliquots of these preamplified products were amplified, using a quantitative PCR. We show that all samples had equal CT values (Fig. 4B).
Figure 4.
Linearity of double-round PCR amplification. (A) A double-stranded synthesized DNA sequence from Gzma was amplified in quadruplicate by a single PCR (•) or by a two-step PCR of 15 preamplification cycles (○). Decreasing template concentrations by a factor of 16 were used. Means ± SD of quadruplicates are included, although SDs frequently overlap with the symbols as we found little or no variation. Linear regression curves of these amplified standards are indicated and have similar high correlation coefficients (r2 = 0.999). Similar results were obtained with three different types of synthesized cDNA. (B) Four molecules of a double-stranded synthesized Gzma sequence were amplified in triplicate using 15 cycles on the first PCR amplification followed by a second real-time PCR. PCR accumulation curves are shown. (C) A bulk cDNA population (7 × 105 to 680 fg) was used to amplify 28S (Mrp-S21) gene in quadruplicate by a two-step PCR of 15 preamplification cycles. Decreasing template concentrations by a factor of 4 were used. Mean values and standard deviations of quadruplicates are included, but SDs overlap with the symbols, as we found little or no variation. The linear regression curve of this amplified standard is indicated (r2 = 0.9978). (D) Single cells were retrotranscribed and preamplified (15 cycles) for Hprt-1. A second quantitative PCR was used for the evaluation of the relative expression level (CT) of Hprt-1 on each individual cell. Quadruplicates of amplification are shown. (E) Single individual cells were sorted, and treated for genomic DNA amplification (see Methods). The Gzma gene was next amplified in a seminested PCR (15 cycles of preamplification). PCR accumulation curves are shown for four individual cells. Data are from one of three independent experiments.
These results show that when a 15-cycle amplification is used in the first PCR, we can exclude the existence of bias or randomness introduced by template switching at template copy numbers from 1.28 × 109 to 4 molecules. It must be noted that at higher or lower amplification cycles on the first PCR, the second PCR does not follow the same rules of linearity (data not shown). When the first PCR has less than 15 cycles, low template concentrations were not amplified efficiently in the second PCR, as tube-to-tube variability was observed. Conversely, when the first PCR had more than 15 cycles we observed saturation when high template concentrations were used.
We next investigated whether preamplification was biasing quantitative estimates in cDNA extracted from normal cells. We first analyzed bias on highly expressed genes by studying mouse 28S (Mrp-S21) mRNA. We used serial dilutions of cDNA that was amplified in a double-round PCR (the first PCR was 15 cycles). We show that the correlation coefficient was high even when this highly expressed gene was tested, and high amounts of total cDNA were used (Fig. 4C). These results demonstrate that our double-round PCR conditions maintained linearity even when high template amounts were amplified.
We next analyzed possible bias in the amplification of rare messages. We studied tube-to-tube variability in mouse Hprt-1 expression, considered a low-expressed gene (Pannetier et al. 1993). We retrotranscribed and amplified (15 cycles) Hprt-1 of individual cells. Quadruplicates of the first PCR round were amplified in a second PCR. As shown in Figure 4D, quadruplicate aliquots of this message gave the same results, showing no tube-to-tube variability. We studied 15 individual cells using these conditions (data not shown), and all had the same quadruplicate CT values, which also excluded the possibility of template switching inducing randomness and consequent tube-to-tube variability.
We next investigated whether we could reproducibly amplify two gene copies, by amplifying genomic DNA from individual cells. For that purpose, single cells were sorted, lysed, and processed for DNA extraction. In these conditions, mRNA is degraded and only DNA can be amplified. We used primer combinations spanning a small intron. In these conditions, amplicons generated after DNA amplification were only slightly longer than those generated when cDNA was amplified, ensuring that DNA and cDNA amplification had similar efficiency. We show that even with a two-copy template in the first PCR round, all samples amplified in the second PCR had the same CT (Fig. 4E), excluding quantitative randomness effects and demonstrating the high sensitivity of our amplification procedure.
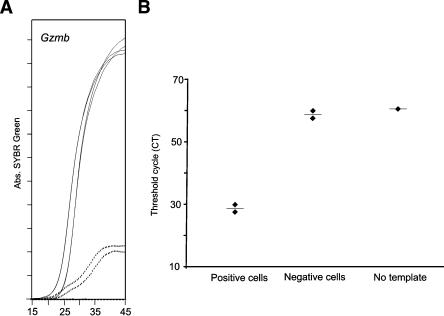
Finally, we tested the relative contribution of nonspecific signaling to our PCR read-outs. Indeed, SYBR Green incorporation in primers dimers could induce some type of background amplification. To evaluate this possibility, we studied several expresser and nonexpresser single cells for a particular gene product. As expected, some SYBR Green accumulation was sometimes detected in negative cells, but exponential accumulation (as found in positive cells) was never observed (Fig. 5A). Because CT values are evaluated in the linear phase of SYBR Green accumulation, CT determination in negative samples was not significantly different from samples where template was not included (Fig. 5B). These results demonstrate that negative cells did not originate significant background in our analysis.
Figure 5.
Impact of nonspecific signal in PCR quantification read-outs. Individual cells expressing or not Gzmb mRNA were amplified simultaneously. (A) SYBR Green signal in positive (solid lines) and negative (dotted lines) cells. (B) CT evaluation, using the Sequence Detector 1.7 software. The CT value of negative cells and wells not containing template was not significantly different (t-test: P = 0.18). This program's upper limit of detection is 60 cycles; that is, samples without template score with a CT of 60.
In summary, these results demonstrate that our method of amplification preserves the initial representation of rare genes, simultaneously excluding excessive amplification of highly abundant gene copies and therefore allowing precise quantification assessments of copy numbers ranging from 1.28×109 to 2 molecules.
Maintenance of Abundance Relationships
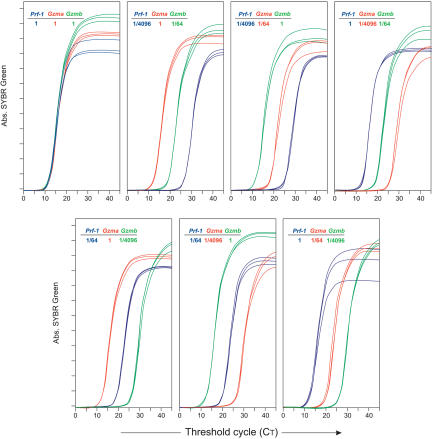
Exponential amplification has generally been considered to bias abundance relationships, as cDNAs of differing lengths and composition would be amplified with differing efficiencies (Freeman et al. 1999; Dixon et al. 2000; Phillips and Lipski 2000; Baugh et al. 2001). Our primers were designed to amplify cDNAs of similar length and composition, which should favor the maintenance of abundance relationships. To test directly how abundance relationships were maintained, different synthesized cDNA sequences (from mouse Gzma, Gzmb, and Prf-1) were quantified and mixed in known proportions (true ratios). These mixtures were then amplified in our two-step PCR. The CT values obtained in the second PCR were used to determine the corresponding ratios after amplification (test ratios). To compare the maintenance of abundance relationships at very different template concentrations, these three templates were mixed at 1/1, 1/64, and 1/4096 ratios.
When the three sequences were all mixed at the 1/1 ratio, all PCRs had the same CT (Fig. 6, upper left). When the different sequences were mixed at 1/1, 1/64, or 1/4096 ratios, differences of CT values between different dilutions of different genes reflected initial dilutions, that is, six cycles for a 64-fold difference and 12 cycles for a 4096-fold difference. This occurred for all genes, tested in all types of ratio combinations (Fig. 6). Therefore, despite very different initial template proportions, our PCR procedure provided a measure (test ratios) that was faithful to the original template proportions. These data demonstrate that the maintenance of abundance relationships is guaranteed even on a large dilution range of the target template in a sample.
Figure 6.
Maintenance of abundance relationships in double-step amplification. Different synthesized DNA sequences (Prf-1, blue; Gzma, red; Gzmb, green) were mixed in known proportions at different ratios indicated in each panel (true ratios). These mixtures were next amplified in quadruplicate by a two-step PCR of 15 preamplification cycles. Triplicates of amplification curves for each dilution are shown. CT values between different dilutions of different genes reflected initial dilution conditions; that is, six cycles for a 64-fold difference and 12 cycles for a 4096-fold difference (data not shown).
Reverse Transcription
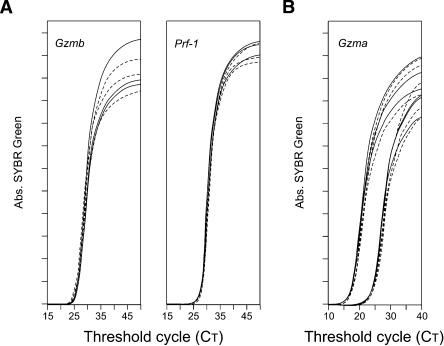
To ensure maximal efficiency in capturing mRNA molecules present in individual cells, we used specific reverse transcription (RT), and the 5′ extreme of the amplified gene fragments of the first PCR was designed to be located between 300 and 400 bp from the 3′ RT origin (see Discussion). To validate our approach, we assessed the efficiency of the RT in these conditions. RNA fragments from different genes and respective complement cDNA sequences were produced and purified. We compared the direct amplification of a precise number of cDNA molecules with the amplification of the same number of synthesized RNA copies after their reverse transcription. We found that both cDNA and reverse-transcribed RNA template were amplified with similar efficiency (Fig. 7). This occurred when RNA templates coding for different genes were tested (Fig. 7A), and such efficient RT was detected at all RNA concentrations studied (Fig. 7B). These results demonstrate that we maintain copy numbers in the RNA to cDNA transition, and that this RT approach is highly efficient.
Figure 7.
Efficiency of the reverse transcription. Gzma, Gzmb, and Prf-1 RNA and DNA molecules were synthesized and purified. RNA was first retrotranscribed and subsequently amplified; DNA sequences were amplified directly following the same conditions as for cDNA. For each gene the same number of DNA and RNA molecules was compared. Results show triplicate amplifications of RNA (solid lines) and DNA (dashed lines). (A) Comparison of RT efficiency of the same number of RNA molecules coding for different genes. Gzmb (left) or Prf-1 (right). (B) RT efficiency at different RNA concentrations. Different concentrations of Gzma RNA (ranging from 7.5 × 107 to 3 × 105 molecules) and corresponding DNA concentrations were compared. Results show RNA/DNA amplifications at two of the concentrations tested: 1.9 × 107 and 4.7 × 106 molecules. The same results were obtained for all other concentrations.
Population Versus Single-Cell Studies: The Impact of Single-Cell Analysis in the Evaluation of Functional Genetic Profiles
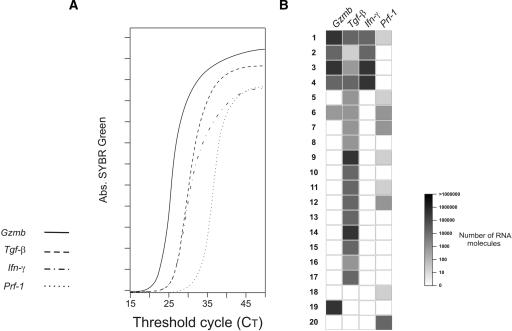
To compare the gene expression profiles obtained by single-cell analysis to those evaluated in bulk populations, we studied the same T-cell population using both methods. Mouse monoclonal CD8 T cells, 4 d after in vivo antigen stimulation (Tanchot et al. 1998; Veiga-Fernandes et al. 2000) were sorted either as 20 cells/well for population studies or as 20 single cells. All samples were retrotranscribed and amplified as described above. For simplicity, only four genes are shown.
Real-time PCR at a population level (Fig. 8A) showed a hierarchy of gene expression: Gzmb > Tgf-β = Ifn-γ > Prf-1. This data would suggest that Gmzb is the most expressed gene, and that this CD8 population differentiates similarly into Tgf-β- and Ifn-γ-expressing cells. In addition, these data suggest that these cells should be highly cytotoxic, due to the expression of high levels of Gzmb (an enzyme important for cytotoxicity) and express Prf-1. Indeed, CD8 killer activity requires the coexpression of both Gzmb and Prf-1 (Russell and Ley 2002).
Figure 8.
Impact of quantitative single-cell analysis on gene expression profiles. Monoclonal CD8 T cells, 4 d after in vivo antigen stimulation were sorted (A) at 20 cells/well for population studies, and (B) as 20 single cells at one cell/well. RT and PCR conditions were the same in both A,B. Results show: (A) Real-time PCR amplification of Gzmb (solid line), Tgf-β (dashed line), Inf-γ (dash-dotted line), Prf-1 (dotted line). (B) Quantification of gene expression in individual cells, using quantitative single-cell multiplex PCR. Expression levels of each gene in each cell are shown as shades of gray, compared to the log scale in the left. The absolute number of mRNA molecules was obtained by comparing amplifications with a standard of a known number of RNA molecules that followed the same rules of RT and amplification.
Results of single-cell studies revealed a very different scenario (Fig. 8B). First, most CD8 T cells differentiate into Tgf-β-expressing cells (17/20), whereas Ifn-γ and Gzmb expression was quite rare (4-6/20 cells). Moreover, Gzmb and Prf-1 were usually expressed by different cells. These findings indicate that this CD8 population should be virtually devoid of killer activity, because individual cells do not coexpress the two genes required to kill target cells.
We conclude that single-cell multiparameter studies of gene expression reveal fundamental new insights into cell behavior. Conversely, the studies performed in bulk populations may be highly misleading.
DISCUSSION
The analysis of heterogeneity within cellular populations has a major impact on cell biology. The final aim of this approach is to reveal the gene expression patterns that ultimately characterize and define the fate of individual cells. Different cell fates likely rely on both qualitative and quantitative differences of gene expression that affect multiple genes simultaneously, but tests allowing the assessment of these features in individual cells are lacking. Here we describe a single-cell multiplex RT-PCR that allows simultaneous quantitative analysis and comparison of the expression of 20 genes in each individual cell. We demonstrate that this method substantially improves functional genomic read-outs. Conversely, quantitative studies performed at the population level may be very misleading.
It is not surprising that single-cell and population studies do not overlap. Quantitative studies at the population level only determine average rates of gene expression. They do not evaluate the frequency of expressing cells. The same mRNA amount can correspond to rare cells expressing high mRNA levels or to a much higher cell number expressing lower mRNA levels. These two situations may have very different biological meanings. The impact of this potential bias has probably been underestimated, as the range of identical mRNA molecules each cell could express was not known. Here, for the first time, we were able to quantify messages at the single-cell level and found that expression of a single gene in individual cells could vary by 10,000 fold (data not shown). This extensive variation seriously undermines the interpretation of any quantitative studies that are not accompanied by frequency determinations. Indeed, in studies performed at the population level, very rare events (even at 10-4 frequencies) may score similarly to frequent events. Therefore, in population readouts, events that are not representative of a global population behavior may appear as very significant events. This bias is evident in the study we include, where Gzmb expression appears to be a dominant function in the studied population, whereas studies at the single-cell level reveal that only a few cells expressed this mRNA.
Another major potential impact of single-cell studies is the possibility to determine gene coexpression. We were surprised to verify that different genes (Prf-1 and Gzmb), which need to be coexpressed for CD8 cytotoxicity, could segregate into different individual cells. This finding emphasizes that studies at the population level are not sufficient to identify cell properties. Rather, coexpression studies at the single-cell level are fundamental for the interpretation of functional read-outs.
Concerning the present methodology, quantification of multiple gene expression in the same cell is only possible if several rules are followed simultaneously: the same efficiency of PCRs; the absence of primer and amplicon competition during the first PCR round, and an efficient RT to PCR transition requiring the use of specific RT. Moreover, all of these requirements are strictly interdependent.
Comparison of the expression of different genes between themselves requires that all individual PCRs have the same efficiency. This aim alone is not difficult to achieve. Primer combinations claiming to amplify multiple genes with similar efficiency are beginning to be available commercially. However, in these commercial kits (where controls for many important parameters are lacking), each individual gene must be studied separately, using one independent sample for each PCR. The ability to compare the expression of different genes between themselves and attribute coexpression of 20 genes to the same cell requires that all 20 different PCRs reactions be performed in the same tube and in the same PCR round. This imposes the requirement that, besides similar efficiency, the 40 primers and 20 amplicons of the first PCR also do not compete with one another.
It was claimed that analyses of more than five genes in one cell would necessarily lead to nonspecific inhibitions of amplification, which affect amplifications randomly (Walter et al. 2000). When using previous methods of single-cell amplification, we confirmed this claim, and all of our previous studies were restricted to four to five gene amplifications (Veiga-Fernandes et al. 2000; Lambolez et al. 2002). We found later that the modification of the PCR amplification conditions we describe herein, associated with a careful study of primer/amplicon competition, could prevent inhibition.
The constraints on primer selection impose another strategy: the use of specific reverse transcription which targets the mRNA sequence that is retrotranscribed and subsequently amplified. This is achieved by designing the 5′ extreme of the amplified gene fragments to be located between 300 and 400 bp from the 3′ RT origin. This strategy is both necessary and optimal for the maintenance of abundance relationships in the mRNA to cDNA transition.
The use of specific RT rather than poly-AAA reverse transcription is necessary to prevent 3′ bias that would modify abundance relationships in the transition from mRNA to cDNA. It is well known that poly-AAA reverse transcription preferentially transcribes mRNA fragments localized in the 3′ termini. This bias should be a major problem in our type of approach. Indeed, to ensure similar efficiency of amplification and prevent competition (essential aspects of our methodology), primers must be selected throughout the gene and not at the 3′ end only. Our strategy thus improves on previous methods used to achieve readings of gene expression in small samples such as modified poly-AAA reverse transcription methods (Dixon et al. 1998; Brail et al. 1999) or poly-AAA reverse transcription, followed by cDNA polyadenylation at the 3′ and subsequent single-primer amplification of the tailed cDNAs (Brail et al. 1999). In contrast to our method, all of these other methods induce 3′-biased abundance relationships in the sample, which are difficult to control (Brail et al. 1999). Another limitation of poly-AAA RT methods is that the 5′ sequences of the retrotranscribed genes might be incomplete, compromising any further precise assessment. We prevented this limitation by designing the 5′ extreme of the amplified gene fragments to be located between 300 and 400 bp from the 3′ RT origin. We demonstrate that this strategy provides a maximized and uniform amplification of retrotranscribed genes, because RNA fragments are fully retrotranscribed and abundance relationships are maintained in the mRNA to cDNA transition. By comparing to a standard of RNA that is simultaneously reverse-transcribed and amplified, we can calculate the absolute number of mRNA molecules expressed per cell. These additional controls are lacking in all previous RT strategies that do not attempt to determine RT efficiency. A methodology was recently described that allows exponential amplification of cDNA yields, preserving the relative gene expression patterns of the initial sample (Iscove et al. 2002). However, this methodology also requires that gene-specific primers and probes are restricted to 3′ transcript termini (Iscove et al. 2002). Therefore, this strategy is also totally incompatible with multigene comparative quantification in individual cells that require primer and amplicon selection throughout the gene and not only on the 3′ end.
Because all efforts to achieve readings of gene expression in small samples were directed to increase cDNA yields, and thus are incompatible with gene coexpression studies, we used an alternative approach to measure the minute mRNAs recovered from one cell. Instead of generating very high amounts of cDNA, we exponentially amplified the low cDNA yields we obtained from each individual cell, and used this exponential amplification to quantify transcripts. It is usually assumed that this approach can bias the information content of the sample, as theoretical mathematic analysis showed that hybridization kinetics during thermal cycling could cause both sequence- and copy number-dependent bias (Peccoud and Jacob 1996). We show here that if adequate primer combinations are used, and optimized PCR conditions are applied, double-strand exponential amplification yields reproducible results from 1.28×109 to 4 copies of mRNA, which should cover all ranges of gene expression at the single-cell level.
This technique brings new perspectives to the understanding of biological processes. Most differentiation events have been studied on the basis of a population phenotype which does not necessarily reflect heterogeneity among the population. Conversely, single-cell analysis will allow further dissecting of cell decisions that ultimately influence a population phenotype. This technical approach also has a broader interest for diagnosis of minute samples. Indeed, in several pathologies and infections, only very small tissue samples can be obtained for diagnosis or continuous follow-up of disease progression. This method overcomes all restrictions in sample size by allowing the quantitative assessment of multiple different parameters from just a few cells. Indeed, we are presently using this method to characterize HIV-specific CD8 T cells that were divided into eight subtypes by cell surface markers, each subtype representing less than 0.1% of Peripheral Blood Lymphocyte (PBL). This approach allows the determination of 20 cell functions simultaneously, even in such small sample sizes. Preliminary evidence suggests that we will be able to quantify the expression of up to 40 genes/cell.
In conclusion, we here describe a method of quantitative multiplex PCR that can be applied to an extended number of genes expressed in a single cell. We also show that the ability to quantify multiple gene usage by individual cells provides fundamental insights into cell physiology and functional genomics.
METHODS
FACS Sorting
Cells were sorted using a FACS Vantage equipped with an automatic cell deposition unit (Becton Dickinson). Cells were collected in individual PCR tubes containing 5 μL of PBS-DEPC 0.1%, and stored at -80°C.
Reverse Transcription
Cells were lysed by cooling at -80°C followed by heating to 65°C for 2 min. After cooling to 4°C, RNA was specifically retrotranscribed for 1 h at 37°C by adding 10 μL of a mix containing 0.13 μM specific 3′ primers (see Supplemental Material I and II), 50 mM KCl, and 10 mM Tris-HCl at pH 8.3 (Applied Biosystems), 3.3 mM MgCL2 (Applied Biosystems), 1 mM dNTPs (Pharmacia Biotech), 39 units of RNAse block (Stratagene), and 11.5 units of MuLV Reverse Transcriptase (Applied Biosystems), in a 15-μL reaction. The reaction was stopped by 3-min incubation at 95°C.
Primer Design
Gene sequence data and exon/intron boundaries were obtained from the Ensembl Project database (http://www.ensembl.org). The primers we selected for these PCR reactions are listed in Supplemental materials I and II.
Our primers were manually designed in order to avoid genomic amplification, by choosing 3′ and 5′ primers that hybridize with different exons. To achieve similar amplification efficiencies, we designed primers of 20 bp size targeting nonrepetitive sequences, with similar melting temperatures (Tm) calculated according to the formula (Tm = 64.9°C + 41°C × (number of G's and C's in the primer - 16.4)/number of bp of the primer) and amplifying fragments of a similar size. The composition of amplified fragments (50.61% ± 5.01% of GC content) was similar, which is required to obtain uniform amplification efficiency for all different mRNAs. To prevent nonspecific amplification, all individual primer sequences were used in a BLAST search (http://www.ncbi.nlm.nih.gov/genome/seq/MmBlast.html) of the mouse genome in order to check potential nonspecific hybridization of primers against other genes besides the targeted gene of interest. No significant hybridization was found with other genes.
To prevent primer competition, we selected primers and potential amplicons that did not cross-hybridize. Primer compatibility and size of the amplified fragments were assessed using the freely available software Amplify 1.2 (Engels 1993; http://engels.genetics.wisc.edu/amplify). The formation of primer dimers was also excluded, because the energies of primer associations that could lead to primer dimerization were considerably weaker than the 3′ binding energies of primer to template associations. Furthermore, the use of high annealing temperatures in our PCR protocol also contributed to exclude nonspecific amplification or inhibition. These aspects are of major importance, because during the first PCR round all genes are amplified simultaneously, and the disregard of such rules results in PCR inhibition.
First PCR Amplification
The cDNAs resulting from the reverse transcription reaction were next amplified. The first round of PCR consisted of one step of denaturation at 95°C for 10 min and 15 cycles of amplification (45 sec at 95°C, 1 min at 60°C, and 1 min 30 sec at 72°C) with 50 mM KCl, and 10 mM Tris-HCl at pH 8.3 (Applied Biosystems), 2 mM MgCl2 (Applied Biosystems), 0.2 mM GeneAmp dNTPs (Applied Biosystems), 3 units of AmpliTaq Gold DNA Polymerase (Applied Biosystems), and 0.015 μM of specific primers (see Supplemental materials I and II) in an 85-μL reaction volume. When PCR was performed from single-cell genomic DNA, cells were previously treated for 45 min at 55°C and 10 min at 95°C with 7.5 μg of proteinase K (Merck) and 50 mM KCl, and 10 mM Tris-HCl at pH 8.3 (Applied Biosystems) in a final volume of 15 μL.
Real-Time Quantitative PCR
Real-time quantitative PCR was performed by adding 10 μL of 2× SYBR Green PCR Master Mix (Applied Biosystems) to each well containing 4 μL of template and 6 μL of a primer mix with 0.25 μM of each specific primer (see Supplemental materials I and II) in a 20-μL reaction volume using the ABI PRISM 7700 Sequence Detection System (Applied Biosystems). After a denaturation step at 95°C for 10 min, the cycle profile used was 30 sec at 95°C, 30 sec at 60°C, and 45 sec at 72°C for 60 cycles of amplification. An aliquot of 4 μL from the first PCR of positive cells was used to quantify the expression level of each different gene. Threshold cycle (CT) was determined on the linear phase of PCRs using the software Sequence Detector version 1.7 (Applied Biosystems). PCR products were resolved on a 1.5% agarose ethidium bromide gel, and were all sequenced to confirm specificity (ABI PRISM 3100, Applied Biosystems). The PCR efficiency of each individual sample was assessed in the linear phase of a real-time PCR reaction using LinRegPCR version 7.0 software. This program uses the raw real-time PCR data of each individual sample and performs an assumption-free analysis (Ramakers et al. 2003).
Synthesis of Double-Strand DNA Sequences
cDNAs from Prf-1, Gzma, and Gzmb mouse genes were amplified using 5′-TCACACTGCCAGCGTAATGT-3′ and 5′-CTGTGGTAAGCATGCTCTGT-3′, 5′-TCAAATACCATCTGTGC TGG-3′ and 5′-AGAGGGAGCTGACTTATTGC-3′, and 5′-GTCAATGTGAAGCCAGGAGA-3′ and 5′-AGGATCCGATGTT GCTTCTG-3′, respectively. Amplified fragments were resolved on a 1.5% agarose ethidium bromide gel and purified using Wizard SV Gel and PCR clean-up System (Promega). DNA was quantified by incorporation of Picogreen (Molecular Probes) according to the manufacturer's instructions.
Molecular Cloning and In Vitro Transcription
cDNA was obtained from RNA extracted from gut intraepithelial T lymphocytes (IELs) of C57Bl/6 mice and human small intestine using the RNeasy mini-kit (QIAGEN). We used these cells because they express all of the genes we studied. The cDNA was synthesized by incubating for 1 h at 37° C using 2.2 mM poly-(T) (Applied Biosytems) in a 45-μL volume reaction containing 50 mM KCl, 10 mM Tris-HCl at pH 8.3 (Applied Biosystems), 3.3 mM MgCl2 (Applied Biosystems), 2.5 mM dNTPs (Pharmacia Biotech), 39 units RNAse block (Stratagene), and 3 units MuLV Reverse Transcriptase (Applied Biosystems). The reaction was stopped by 10-min incubation at 95°C. cDNAs from Prf-1, Gzma, and Gzmb mouse genes were first amplified using 5′-TCACACTGCCAGCG TAATGT-3′ and 5′-CTGTGGTAAGCATGCTCTGT-3′, 5′-TCAAATACCATCTGTGCTGG-3′ and 5′-AGAGGGAGCTGACT TATTGC-3′, and 5′-GTCAATGTGAAGCCAGGAGA-3′ and 5′-AGGATCCGATGTTGCTTCTG-3′, respectively. Amplified fragments were resolved on a 1.5% agarose ethidium bromide gel and purified using the Wizard SV Gel and PCR clean-up system. Sequences were next cloned as described (Poirel et al. 1997). Cloned fragments of Prf-1, Gzma, and Gzmb were used as templates for the in vitro transcription reaction. This reaction was performed using the MEGAscript T7 transcription kit (Ambion) using 1.5 μg of previously Hind III-linearized plasmid in a reaction volume of 80 μL. After transcription, the samples were treated with 4 units of DNase I for 15 min at 37°C and purified using the MEGAclear purification kit (Ambion). The purified RNA was eluted in 50 μL of Tris-EDTA, and an aliquot was run on a native gel (TBE1×, 2% agarose) and controlled for the presence of contaminating plasmid DNA and unfinished products of the in vitro transcription reaction using an Agilent 2100 bioanalyzer (Agilent Technologies) according to the manufacturer's instructions. In vitro transcriptions were >70% pure, and no DNA contaminations were detected. RNA and DNA were quantified by incorporation of Ribogreen and Picogreen (Molecular Probes) respectively, according to the manufacturer's instructions and using the ABI PRISM 7700 Sequence Detection System.
Acknowledgments
We thank O. Bernard for molecular cloning, C. Cordier and G. Megret for cell sorting, B. Schaeffer and A. Le Campion for statistics, and J. Lauber, A. Freitas, A. Eaton, F. Lambolez, E. Treiner, U. Walter, O. Azogui, and P. Vieira for helpful discussions. Supported by Association pour la Recherche sur le Cancer, Ligue pour la Recherche sur le Cancer, Fondation pour la Recherche Medical (H.V.-F.) and Science Technology Foundation (Portugal) (A.P., M.M., and H.V.-F.). This method is covered by a patent deposed by the Institut Necker (patent number 0208593).
Footnotes
[Supplemental material is available online at www.genome.org.]
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2890204.
References
- Baugh, L.R., Hill, A.A., Brown, E.L., and Hunter, C.P. 2001. Quantitative analysis of mRNA amplification by in vitro transcription. Nucleic Acids Res. 29: E29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brail, L.H., Jang, A., Billia, F., Iscove, N.N., Klamut, H.J., and Hill, R.P. 1999. Gene expression in individual cells: Analysis using global single cell reverse transcription polymerase chain reaction (GSC RT-PCR). Mutat. Res. 406: 45-54. [DOI] [PubMed] [Google Scholar]
- Dixon, A.K., Richardson, P.J., Lee, K., Carter, N.P., and Freeman, T.C. 1998. Expression profiling of single cells using 3′ end amplification (TPEA) PCR. Nucleic Acids Res. 26: 4426-4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon, A.K., Richardson, P.J., Pinnock, R.D., and Lee, K. 2000. Gene-expression analysis at the single-cell level. Trends Pharmacol. Sci. 21: 65-70. [DOI] [PubMed] [Google Scholar]
- Engels, W.R. 1993. Contributing software to the internet: The Amplify program. Trends Biochem. Sci. 18: 448-450. [DOI] [PubMed] [Google Scholar]
- Freeman, T.C., Lee, K., and Richardson, P.J. 1999. Analysis of gene expression in single cells. Curr. Opin. Biotechnol. 10: 579-582. [DOI] [PubMed] [Google Scholar]
- Gallopin, T., Fort, P., Eggermann, E., Cauli, B., Luppi, P.H., Rossier, J., Audinat, E., Muhlethaler, M., and Serafin, M. 2000. Identification of sleep-promoting neurons in vitro. Nature 404: 992-995. [DOI] [PubMed] [Google Scholar]
- Iscove, N.N., Barbara, M., Gu, M., Gibson, M., Modi, C., and Winegarden, N. 2002. Representation is faithfully preserved in global cDNA amplified exponentially from sub-picogram quantities of mRNA. Nat. Biotechnol. 20: 940-943. [DOI] [PubMed] [Google Scholar]
- Lambolez, F., Azogui, O., Joret, A.M., Garcia, C., von Boehmer, H., Di Santo, J., Ezine, S., and Rocha, B. 2002. Characterization of T cell differentiation in the murine gut. J. Exp. Med. 195: 437-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loffert, D., Ehlich, A., Muller, W., and Rajewsky, K. 1996. Surrogate light chain expression is required to establish immunoglobulin heavy chain allelic exclusion during early B cell development. Immunity 4: 133-144. [DOI] [PubMed] [Google Scholar]
- Makrigiorgos, G.M., Chakrabarti, S., Zhang, Y., Kaur, M., and Price, B.D. 2002. A PCR-based amplification method retaining the quantitative difference between two complex genomes. Nat. Biotechnol. 20: 936-939. [DOI] [PubMed] [Google Scholar]
- Pannetier, C., Delassus, S., Darche, S., Saucier, C., and Kourilsky, P. 1993. Quantitative titration of nucleic acids by enzymatic amplification reactions run to saturation. Nucleic Acids Res. 21: 577-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peccoud, J. and Jacob, C. 1996. Theoretical uncertainty of measurements using quantitative polymerase chain reaction. Biophys. J. 71: 101-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips, J.K. and Lipski, J. 2000. Single-cell RT-PCR as a tool to study gene expression in central and peripheral autonomic neurones. Auton. Neurosci. 86: 1-12. [DOI] [PubMed] [Google Scholar]
- Plant, T., Schirra, C., Garaschuk, O., Rossier, J., and Konnerth, A. 1997. Molecular determinants of NMDA receptor function in GABAergic neurones of rat forebrain. J. Physiol. 499:(Pt. 1) 47-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel, H., Oury, C., Carron, C., Duprez, E., Laabi, Y., Tsapis, A., Romana, S.P., Mauchauffe, M., Le Coniat, M., Berger, R., et al. 1997. The TEL gene products: Nuclear phosphoproteins with DNA binding properties. Oncogene 14: 349-357. [DOI] [PubMed] [Google Scholar]
- Ramakers, C., Ruijter, J.M., Deprez, R.H., and Moorman, A.F. 2003. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 339: 62-66. [DOI] [PubMed] [Google Scholar]
- Ruano, D., Lambolez, B., Rossier, J., Paternain, A.V., and Lerma, J. 1995. Kainate receptor subunits expressed in single cultured hippocampal neurons: Molecular and functional variants by RNA editing. Neuron 14: 1009-1017. [DOI] [PubMed] [Google Scholar]
- Russell, J.H. and Ley, T.J. 2002. Lymphocyte-mediated cytotoxicity. Annu. Rev. Immunol. 20: 323-370. [DOI] [PubMed] [Google Scholar]
- Tanchot, C., Guillaume, S., Delon, J., Bourgeois, C., Franzke, A., Sarukhan, A., Trautmann, A., and Rocha, B. 1998. Modifications of CD8+ T cell function during in vivo memory or tolerance induction. Immunity 8: 581-590. [DOI] [PubMed] [Google Scholar]
- Veiga-Fernandes, H., Walter, U., Bourgeois, C., McLean, A., and Rocha, B. 2000. Response of naive and memory CD8+ T cells to antigen stimulation in vivo. Nat. Immunol. 1: 47-53. [DOI] [PubMed] [Google Scholar]
- Walter, U., Franzke, A., Sarukhan, A., Zober, C., von Boehmer, H., Buer, J., Lechner, O., and Frantzke, A. 2000. Monitoring gene expression of TNFR family members by β-cells during development of autoimmune diabetes. Eur. J. Immunol. 30: 1224-1232. [DOI] [PubMed] [Google Scholar]
- Zawar, C., Plant, T.D., Schirra, C., Konnerth, A., and Neumcke, B. 1999. Cell-type specific expression of ATP-sensitive potassium channels in the rat hippocampus. J. Physiol. 514(Pt. 2): 327-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://www.ensembl.org; Ensembl Project homepage.
- http://engels.genetics.wisc.edu/amplify; Amplify 1.2 Software homepage.
- http://www.ncbi.nlm.nih.gov/genome/seq/MmBlast.html; Mouse Genome BLAST homepage.