Abstract
Here we describe the establishment of microscope-based functional screening assays in intact cells that allow us to systematically identify new proteins involved in secretory membrane traffic, and proteins that can influence the integrity of the Golgi complex. We were able to identify 20 new proteins that affected either secretory transport, Golgi morphology, or both, when overexpressed in cells. Control experiments with human orthologs to yeast proteins with a role in membrane traffic, or already well characterized mammalian regulators of the secretory pathway, confirmed the specificity and significance of our results. Proteins localized to the Golgi complex or endoplasmic reticulum (ER) showed preferential interference in our assays. Bioinformatic analysis of the new proteins interfering with membrane traffic and/or Golgi integrity revealed broad functional variety, but demonstrated a bias towards proteins with predicted coiled-coil domains and repeat structures. Extending our approach to a much larger set of novel proteins in the future will be an important step toward a more comprehensive understanding of the molecular basis of the secretory pathway. It will also serve as an example for similar microscope-based screens addressing different biological questions.
Automated DNA sequencing of the genomes from various species has now generated an enormous wealth of data that represents the basis for a comprehensive understanding of the molecular organization and function of living cells or organisms. The next essential steps toward this challenging goal will be to use these sequence data to deduce the respective gene coding regions, their regulatory elements, and ultimately the function of the encoded proteins in a cellular context. However, taking into account the 30,000 genes predicted in the human genome (Venter et al. 2001) and additionally the plethora of proteins resulting from alternative splicing and posttranslation modifications, it becomes evident that the development and availability of parallel analysis methods is critical to ensure an efficient functional dissection of the human proteome.
In recent years, genetic and biochemical approaches have been applied to identify and characterize single molecular components of the secretory pathway, a process that is central to cell organization. Secretory membrane traffic enables cells to distribute newly synthesized proteins, lipids, and carbohydrates, and thus ensures cell growth, homeostasis, and differentiation. More recently, systematic approaches such as organelle proteomics and yeast two-hybrid screening have attempted to identify new components of the pathway with the goal of reaching a more complete description of the molecular regulation of membrane traffic (Bell et al. 2001; Calero et al. 2002; Monier et al. 2002). However, despite their potential these techniques have limitations, and it is therefore essential to develop and apply alternative systematic approaches that further the goal of identifying all the molecules participating in membrane traffic regulation and biogenesis of associated organelles.
Here we describe the development and application of a screening strategy in intact cells that aims at the targeted identification of new proteins regulating membrane traffic. To this end we have exploited a collection of novel human full-length cDNAs (Wiemann et al. 2001), the open reading frames (ORFs) of which have been fused to green fluorescent protein (GFP)-variants at their N- and C-termini (Simpson et al. 2000). These ORF-GFP fusions have been initially characterized according to the subcellular localization of the respective proteins in mammalian cells (Simpson et al. 2001). Further functional information about these proteins is obtained by overexpression of the GFP-tagged ORFs in living cells, followed by subsequent monitoring of their interference with secretory membrane transport and the integrity of the Golgi complex. Although only a limited number of proteins were investigated in this pilot screen, we have been able to identify 20 new proteins with a putative role in membrane traffic and/or Golgi integrity. Extending our approach to large sets of proteins in the future will be an essential contribution to a more comprehensive understanding of the secretory pathway at the molecular level.
RESULTS
Application of the Transport Assay
We recently described the development and characterization of a fully automated high-content screening (HCS) platform (Liebel et al. 2003) which is capable of automatically acquiring and analyzing high-resolution multicolor images of immunostained cells. Using this system we developed a quantitative single-cell-based transport assay (Liebel et al. 2003; see also Methods), which we applied here to reveal interference of ectopically expressed GFP-tagged proteins with membrane traffic in the secretory pathway. This assay is based upon the monitoring of the transport of a well characterized cargo molecule, namely the ts-O45-G temperature-sensitive variant of the vesicular stomatitis viral G-protein VSV-G (Zilberstein et al. 1980). This transmembrane protein has the feature of accumulating in the endoplasmic reticulum (ER) at 39.5°C, but moves vectorially through the secretory pathway to the plasma membrane (PM) at the permissive temperature of 32°C, where an antibody recognizing an external epitope can detect it. This has the advantages that transport in individual cells is highly synchronized, and that background PM staining that may be caused by transport of the marker before and during expression of the GFP-tagged proteins under examination does not occur.
In control experiments, with cells expressing soluble cyano fluorescent protein (CFP) or yellow fluorescent protein (YFP), ts-O45-G transport to the PM occurred with an efficiency similar to that observed in neighboring nontransfected cells (Fig. 1A-C; data not shown). In contrast, when a GTP-restricted dominant negative mutant of the Sar1p GTPase (SAR1p [H79G]; Aridor et al. 1995) was expressed as a CFP- or YFP-tagged variant, the amount of ts-O45-G transported to the PM was drastically decreased (data not shown; Liebel et al. 2003). This is consistent with data from the literature showing that the Sar1p GTPase plays an essential role in ER exit (Aridor et al. 1995; Rowe et al. 1996; Pepperkok et al. 1998) and further demonstrates the capacity of the transport assay to reveal proteins interfering with secretory membrane traffic when overexpressed.
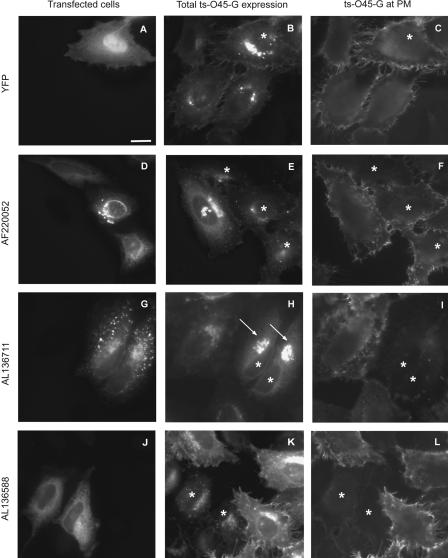
Figure 1.
Examples of the transport assay. Cells were treated, immunostained, and imaged as described in Methods. Cells transfected with (A) YFP alone, (D) CFP-AF220052 encoding the putative ER SNARE Use1p which has been proposed to be involved in retrograde Golgi to ER transport (Dilcher et al. 2003), CFP-AL136711 encoding the putative vacuole membrane protein (VMP1) which has been shown to be expressed in the conditions of the acute pancreatitis and inducing the formation of intracellular vacuoles (Dusetti et al. 2002), and (J) CFP-AL136588, an uncharacterized transmembrane protein. (B,E,H,K) Cells expressing ts-O45-G-YFP. (C,F,I,L) Immunostainings of ts-O45-G at the PM. Asterisks indicate transfected cells, arrows point to cells with ts-O45-G accumulated in the Golgi complex. Bar = 20 μm.
We next selected 100 GFP-tagged novel ORFs to be included in the transport assay. This selection was based upon their subcellular localization. Specifically, we chose proteins localizing to membranes of the secretory pathway, cytoskeletal structures, or the cytoplasm (Simpson et al. 2000; see also http://gfp-cdna.embl.de/). The assumption was that proteins showing these localizations would be the most likely to affect ts-O45-G transport. To compare the different experiments with different GFP-tagged proteins in a quantitative manner, the relative surface expression of ts-O45-G in transfected cells was normalized in each experiment to the average values obtained for nontransfected cells, giving an immediate indication of the effect of the expressed GFP-tagged proteins on ts-O45-G transport (see Table 1 and Methods for details). However, significant variations in different experiments due to varying average amounts of protein expressed were observed. To account for this problem and to make the data evaluation more robust, the population of transfected cells was divided into three expression classes (low, medium, and high), and the average normalized surface expression of ts-O45-G for each class was determined. Then a linear regression was fitted to the values obtained for these three classes and that of nontransfected cells to account for the changes in surface expression with growing amounts of expressed GFP-tagged protein (see Methods for details). In this way, variations in the average expression of the same protein in different experiments were less critical. Overexpression of soluble YFP and CFP resulted in average slope values of -0.0015 ± 0.0009 and -0.0002 ± 0.0002 (see Fig. 2A), respectively (averaging nine independent experiments each). Only those GFP-tagged proteins showing slopes of the fits that deviated from the values obtained for YFP or CFP more than the respective standard deviation of the population (± 0.0026 and ± 0.0055 for YFP and CFP, respectively) were considered as displaying an effect on ts-O45-G transport (Fig. 3).
Table 1.
The Clones With a Stong Effect in the Transport and Golgi Integrity Assays
| Relative surface expression of ts-O45-Ga
|
Effect on Golgi integrity
|
Featuresb
|
Orthologs
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Accession No. and clone IDs | N-terminusc | C-terminus | N-terminus | C-terminus | TM/soluble | domains, motifs | M. musculus | D. melanogaster | C. elegans | S. cerevisiae |
| Secretory pathway | ||||||||||
| AL080121 |
−0.0096e ± 0.0017f
|
−0.0116 ± 0.0073
|
no effect | no effect | 1 TM | AAH06694 | no | no | no | |
| hfbr2_2308d | 0.551 ± 0.271 | 0.371 ± 0.152 | 63%g | |||||||
| DKFZp564O0823 | ||||||||||
| AL136711 |
−0.0100 ± 0.0027
|
n.d.h | no effect | n.d. | 6-7 TM | AAH04013 | AAF46618 | CAA21543 | no | |
| hfkd2_3i13 | 0.398 ± 0.150 | 97% | 74% | 68% | ||||||
| DKFZp566l133 | ||||||||||
| Golgi complex | ||||||||||
| AL713668 | n.d. |
−0.0125 ± 0.0005
|
Int.: 0.55i | no effect | 5 TM | YIP domain | NM_145550 | NM_132613 | NM_062038 | YGL161cj |
| hamy2_10p70 | 0.378 ± 0.182 | 93% | 59% | 66% | 50% | |||||
| DKFZp761P7010 | ||||||||||
| AL136916 |
−0.0122 ± 0.0009
|
no effect | no effect | no effect | 4 TM | C terminal | BAB23556 | AAL48096 | no | no |
| hute1_19g19 | 0.377 ± 0.214 | “KKxx” motif | 97% | 52% | ||||||
| DKFZp586G1919 | ||||||||||
| AL136628 | n.d. |
−0.0100 ± 0.0028
|
n.d. | Frag.: 2.12i | soluble | coiled-coil | XP_131196 | no | no | no |
| hfbr2_2c18 | 0.421 ± 0.154 | 94% | ||||||||
| DKFZp564C182 | ||||||||||
| BC009485 |
−0.0091 ± 0.0018
|
−0.0097 ± 0.0023
|
Frag.: 1.97 | Frag.: 5.1 | soluble | XP_131196 | no | no | no | |
| IRALp962K1924 | 0.710 ± 0.341 | 0.719 ± 0.318 | 83% | |||||||
| IMAGE:3623656 | ||||||||||
| AL136588 |
−0.0095 ± 0.0019
|
n.d. | Int.: 0.63 | no effect | 2 TM | no | no | no | no | |
| hamy2_2d11 | 0.528 ± 0.0151 | |||||||||
| DKFZp761D112 | ||||||||||
| Cytoplasm | ||||||||||
| AL136775 |
−0.0106 ± 0.0031
|
n.d. | Frag.: 1.52 | no effect | soluble | XP_138135 | no | no | no | |
| htes3_17f10 | 0.537 ± 0.250 | 41% | ||||||||
| DKFZp434F1017 | ||||||||||
| AL080119 |
−0.0109 ± 0.0023
|
n.d. | Int.: 0.62 | no effect | soluble | HABP4 and RBP1 domains | AAH13665 | AAF44918 | AAB66119 | no |
| hfbr2_23m24 | 0.440 ± 0.186 | 98% | 44% | 41% | ||||||
| DKFZp564M2423 | ||||||||||
| AL136618 |
−0.0098 ± 0.0023
|
n.d. | no effect | n.d. | soluble | 6 ANK repeats | BAC26572 | no | no | no |
| hfbr2_23n16 | 0.579 ± 0.253 | 3 MORN repeats | 54% | |||||||
| DKFZp564N1623 | ||||||||||
| AL050156 |
−0.0097 ± 0.0024
|
−0.0056 ± 0.0015
|
no effect | Frag.: 1.88 | soluble | 3 TPR repeats | BAC30486 | AAF53617 | AAA83170 | YOR007C |
| hute1_20n10 | 0.603 ± 0.221 | 0.747 ± 0.156 | 94% | 49% | 57% | 45% | ||||
| DKFZp586N1020 | ||||||||||
| Cytoskeleton | ||||||||||
| AL13660418 |
−0.0089 ± 0.0022
|
n.d. | Frag.:2.4 | n.d. | soluble | coiled-coil | AAH46960 | no | no | no |
| hfbr2_22f21 | 0.642 ± 0.0340 | 72% | ||||||||
| DKFZp564F2122 | ||||||||||
Upper numbers are the slope b, those below are the ratio of relative surface expression in transfected to nontransfected cells. For more details see the Results and Methods sections.
All listed clones, except AL713668, which is predicted to be involved in secretory membrane transport, are novel and do not contain predictable signal peptides.
All clones exist in two variants: CFP fused to the N-terminus and YFP fused to the C-terminus.
Clone IDs used by http://gfp-cdna.embl.de/ (upper line) and http://www.dkfz-heidelberg.de/LIFEdb (lower line) databases, respectively.
Average value derived from independent experiments. For more details see Methods.
standard error value derived from independent experiments. For more details see Methods.
% of similarity on amino acid level derived by using “BLAST 2 sequences” program.
Not determined.
Change of average intensity or number of Golgi fragments. For more details see Methods.
Name of yeast ORF as assigned by SGD Yeast Database.
Figure 2.
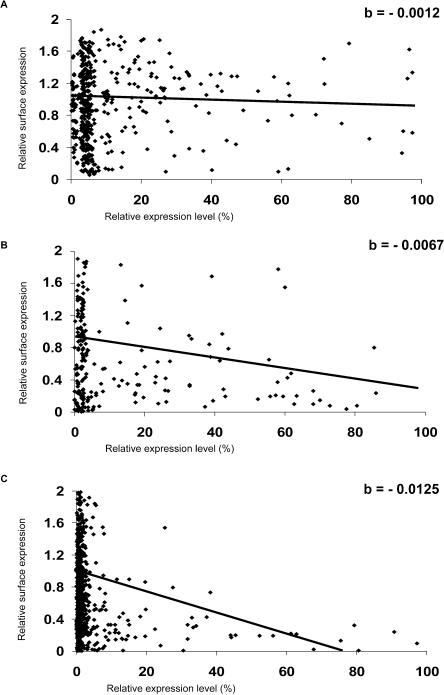
Quantification of the transport assay. Image acquisition of transfected cells, quantitation of fluorescence, data analysis, and classification of inhibitors were as described in Methods. The normalized amounts of ts-O45-G arriving at the PM (relative surface expression) are plotted as a function of the relative amounts of GFP protein expressed. (A) Example scatterplot of YFP-transfected cells. (B) Example scatterplot of cells transfected with the weak effector AF220052. (C) Example scatterplot of cells transfected with the strong effector AL136711. The linear curves were fitted to the data as described in Methods. The slopes (“b”) of these curves are given in the right upper corner of each graph. The steepness of the slope reflects the progression of the strength of inhibition with increasing amounts of transfected GFP-tagged protein.
Figure 3.
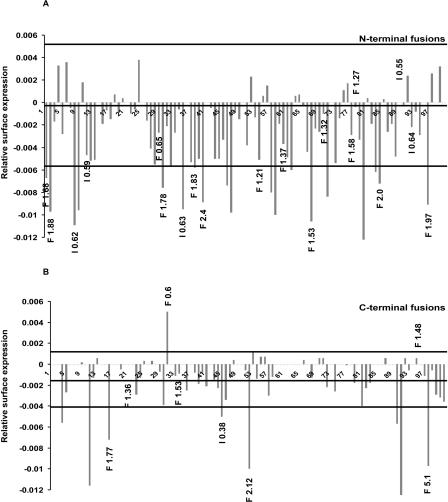
Summary of the results of the transport and Golgi integrity assays. Graphical representation of slope values of all for N-terminal CFP (A) and C-terminal YFP fusions (B) tested in the transport assay. The three horizontal lines in each graph depict the following: The middle line represents the average slope value of the negative controls transfected with CFP or YFP alone (-0.0002 and -0.0015 in A and B, respectively); the upper and lower lines indicate the interval, which is defined by the respective standard deviations of the population for the controls (from -0.0057 to 0.0053 and from -0.0041 to 0.0011 for A and B, respectively). Only those clones with slope values outside the respective intervals were assigned as effectors in the transport assay. The clones which induced alterations of the Golgi complex are depicted by the black numbers at the tips of the respective bars. “F” and “I” indicate changes in fragment number and intensity, respectively. The clones are numbered from 1 to 100 in ascending order of their accession numbers.
Of the 100 ORFs screened, 24 could be classified as inhibitors and one as an accelerator of ts-O45-G transport (Fig. 3, Table 1, Supplemental Table 1). Eleven of these showed a strong transport inhibition that was already apparent by visual inspection of the transfected cells (Figs. 1G-L, 2C; Table 1). Their slopes were typically greater than half of the difference between the slopes obtained for GFP variants and the corresponding GFP-tagged Sar1p [H79G] (slopes for YFP- and CFP-tagged SAR1p [H79G] were -0.0102 ± 0.0012 and -0.0141 ± 0.0034, respectively). The 13 remaining transport inhibitors caused a weaker effect on ts-O45-G transport, which was less obvious to appreciate by visual inspection (Fig. 1D-F), but could be clearly quantified by automated image analysis (Fig. 2B, Supplemental Table 1). Seventy-five of the proteins analyzed were classified as not inhibiting ts-O45-G transport (Supplemental Table 2).
Wherever possible, both N- and C-terminal GFP fusions were screened (51 clones). For 38 clones, only the N-terminal fusions and for 11 clones only the C-terminal fusions were tested. Proteins were considered to be effectors regardless of whether one or both tagged versions of the protein were identified as effectors in our assays.
Application of the Golgi Integrity Assay
In order to have independent confirmation of our results on transport inhibition, we next extended our screens to determine the effect of the GFP-tagged proteins on the morphology of the Golgi complex. Being a central organelle in the secretory pathway, the Golgi complex is directly dependent on bidirectional flow of membranes and proteins into and out of it. It is thus reasonable to postulate that the overexpression of those proteins which act in transport steps linked to the Golgi complex also affects the morphology of this organelle. To investigate this, we made use of the automated assay that we developed to quantify morphological changes of the Golgi (Liebel et al. 2003).
Similar to the experiments addressing ts-O45-G transport, each GFP-tagged ORF was expressed for 24 h. As a Golgi marker, the matrix protein GM130 (Nakamura et al. 1995) localizing pre-dominantly to the cis-side of the Golgi complex was used. This is a convenient marker protein for quantitative measurements, as it relocates from the juxtanuclear area into numerous distinct structures scattered throughout the cytoplasm when ER to Golgi transport is inhibited (Seemann et al. 2000; Miles et al. 2001). In total 60 GFP-tagged ORFs, including all of the ts-O45-G transport effectors, were tested for potential effects on Golgi morphology. The number of distinct GM130-positive structures and their average intensity were determined in transfected and nontransfected cells as described (Liebel et al. 2003). This revealed in total 25 proteins affecting Golgi morphology (Fig. 3, see examples in Fig. 4), with 15 of them being already identified as transport effectors. The remaining 10 proteins were affecting Golgi morphology only. For 35 of the proteins tested, as well as for soluble CFP or YFP, no effect on the Golgi complex, compared to nontransfected cells, could be observed.
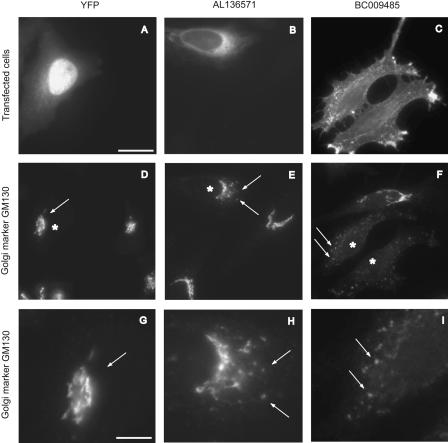
Figure 4.
Examples of the Golgi integrity assay. Cells were transfected with YFP alone (A), CFP-AL136571 (putative tethering factor Got1a; Conchon et al. 1999) (B), and the uncharacterized soluble protein BC009485-YFP (C), which cause no obvious alteration (D,G), weak alteration in Golgi complex morphology (E,H), or a strong scattering of the Golgi complex (F,I). Asterisks indicate transfected cells. Arrows in (D-F) point to Golgi structures and a higher magnification of these respective areas in (G-I). Arrows in G-I correspond to the arrows in D-F, respectively. Bar in A = 20 μm and is representative for images (A-F). Bar in (G) = 8 μm and is representative for images (G-I).
For 15 of the Golgi effectors, the Golgi morphology was only weakly affected, with an increased number of Golgi fragments varying by a factor between 1.2 and 2.0 compared to nontransfected cells. No significant loss of the total Golgi fluorescence occurred in cells transfected with these fusion proteins. In three of the Golgi effectors, the number of scattered GM130-positive Golgi structures was increased by more than a factor of 2.0 compared to nontransfected cells, and hence they were defined as having a strong effect on Golgi morphology (see examples in Fig. 4C,F,I and Table 1). Interestingly, for seven of the effectors the total Golgi-specific fluorescence or the number of Golgi fragments was reduced (Table 1, Supplemental Table 1). This might be due to a possible relocation of Golgi matrix proteins into the ER under conditions of an ER export block, as was recently demonstrated to occur (Puri and Linstedt 2003).
Relevance of Subcellular Localization and Targeting Signals
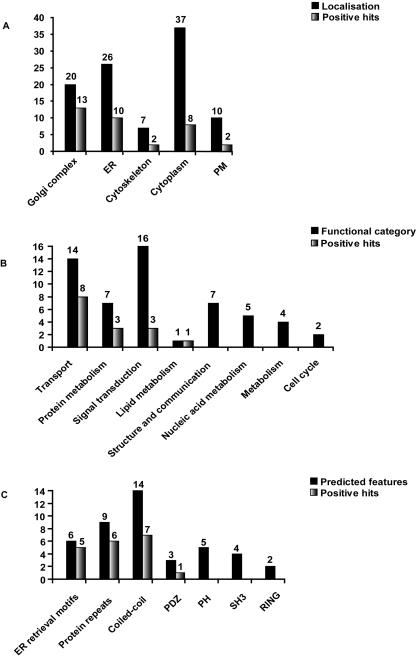
Among the 100 proteins tested here, those localizing to the ER or the Golgi complex showed the most frequent interference with ts-O45-G transport and/or Golgi morphology compared to those localizing to the PM, the cytoplasm, or to the cytoskeleton (Fig. 5A).
Figure 5.
Correlation of the data of functional analysis with the classes of localization and bioinformatic predictions. (A) Number of the positive hits according to classes of subcellular localization. Black bars show the number of proteins tested (100) and the gray bars the number of transport or Golgi morphology effectors identified. (B) Repartition of 56 proteins into the predicted functional categories (black) and hits revealed in these groups (gray). The remaining 44 proteins of the 100 screened were classified as unknown and are thus not shown. Among those 20 were effectors in our screens. (C) Repartition of (including the ones with predicted function) proteins into groups according to their sequence features (black) and hits revealed in these groups (gray). The remaining 57 proteins carried no predictable sequence features and are not shown; 16 of those were effectors in our screens. Proteins were considered to be effectors regardless of whether the N- or C-terminal or both tagged versions of the protein were identified as effectors in our assays.
It is well known that the position of the GFP tag has an effect on the subcellular localization of the fusion proteins (Simpson et al. 2001), and is therefore also likely to influence their cellular activities. Indeed, fusions with the GFP close to, for example, N-terminal myristoylation or C-terminal prenylation sites interfered with the proper localization of the proteins, and as a result these fusions showed no effect on transport or Golgi complex morphology. In contrast, when in the same proteins the GFP variants were fused to the opposite end, the localization was correct and a corresponding effect in our assays was observed (examples are the MARCKs-like protein AL713653 and the Rab GTPases AL136727 and AL136904).
Similarly, membrane proteins with predicted di-lysine and di-arginine ER-retrieval motifs in their cytoplasmic tails showed only an effect on ts-O45-G transport or Golgi morphology when these motifs were not masked by the GFP.
Analysis of Results According to Functional Predictions
For 56 of the proteins that were tested in these assays, it was possible to make functional predictions based on available literature data, the predicted domain structure, or similarities to already characterized yeast orthologs (Fig. 5B,C, Table 1, Supplemental Tables 1,2). Based on these criteria 14 of these proteins (AL136571, AL136727, AL136904, AL136617, AL713668, AL834226, AL713697, AL136594, AL136711, AF220052, AL136715, AL390215, AL834211, AL136559) are known or could be predicted to be involved in secretory membrane traffic, and eight of these (AL136571, AL136727, AL136904, AL136617, AL713668, AL834226, AL136711, AF220052) were indeed revealed as effectors in the transport or Golgi integrity assays. One of these (AL136571) is the human ortholog of the yeast Got1p protein, which has been described as being involved in ER to Golgi transport through concerted action with the syntaxin Sed5p (Conchon et al. 1999). A second example is an ortholog of the yeast unconventional ER SNARE Use1p (AF220052; Dilcher et al. 2003), apparently involved in retrograde Golgi to ER transport. The third example of this group (AL713668) is an ER-localized transmembrane protein that belongs to the YIP family (Calero et al. 2002), members of which have been shown to interact with various Rab GTPases, thus regulating various steps in membrane traffic (Tang et al. 2001; Heidtman et al. 2003; Sivars et al. 2003).
The remaining six proteins known to be involved in membrane traffic (AL713697, AL136594, AL136715, AL390215, AL834211, AL136559) have not been described as being directly relevant to ts-O45-G transport or Golgi morphology. Thus they should be expected to have no effect in our assays, as confirmed by our results. Examples of these proteins are amphiphysin II and synaptotagmin III. The first is involved in the clathrin-mediated endocytosis of synaptic vesicles (Zhang and Zelhof 2002), and the second participates in calcium-dependent regulated exocytosis (Ullrich et al. 1994).
Other effectors (transport or Golgi integrity assay) could be annotated as being involved in events such as signal transduction (e.g., the protein AL117466, which is a predicted LIM2 kinase) and protein metabolism (e.g., AL136807, homologous to RAMP4, which itself is involved in protein glycosylation during translocation). Specificity of our assays could be demonstrated by the finding that none of the proteins predicted to be involved in general cellular or nucleic acid metabolism (e.g., a predicted NADH ubiquinone oxidoreductase, AL080056, and an ortholog of the glucocorticoid-induced transcription activator AL110191) caused any interference with transport or Golgi morphology (Fig. 5B).
Analysis of Results According to Conserved Domains and Motifs With a Role in Membrane Traffic
For the remaining 44 proteins tested in our screens, no functional predictions were possible, although domains and motifs consistent with a role in membrane traffic could be identified in their sequences. Six of such proteins (AL136567, AL136916, AL136658, AL136927, AL136610, AL136617) carry the ER retrieval motifs “KKxx” or “xxRR,” which are known to interact with components of the vesicular coat complexes, COPI and COPII (Cosson and Letourneur 1994; Dominguez et al. 1998; Yuan et al. 2003). They are all localized to the ER and/or the Golgi complex and contain 1-8 potential transmembrane (TM) domains. In all of these examples the respective ER retrieval motifs are predicted to face the cytoplasm. Overexpression of five of these proteins caused various perturbations on ts-O45-G transport and/or Golgi morphology (Table 1, Supplemental Table 1). Interestingly, one of the proteins (AL136567) accelerated ts-O45-G transport.
A number of the effectors revealed here are predicted to contain repeated regions (armadillo, ankyrin, TPR, and WD40). Proteins with such domains are frequently reported to undergo protein-protein interactions with partners relevant to different cellular pathways, including membrane transport (Andrade et al. 2001). Six of nine of such proteins were identified as effectors in our assays. Frequently, proteins with predicted coiled-coil domains were also revealed as effectors in our assays. Seven of 14 of proteins with such domains were identified as effectors in our assays with a preferential perturbation of Golgi complex morphology. For 10 of the effectors in our assays, no particular sequence features could be predicted.
DISCUSSION
We have described the testing and application of two microscopy-based functional assays, which probe constitutive transport through the secretory pathway and Golgi morphology in intact cells. Automated image acquisition and data analyses, carried out on a high-content screening platform (Liebel et al. 2003), helped to identify subtle changes in ts-O45-G transport or Golgi morphology that were not apparent by visual inspection of single cells. Furthermore, automation allowed measurements on large populations of cells, ensuring data reliability and quality.
The proteins used in our screens were preselected with respect to their subcellular localization to membranes of the secretory pathway, cytoskeletal structures, or the cytoplasm. This preselection should result in an increased likelihood of identifying candidate proteins involved in the regulation of diverse steps in the secretory pathway. Indeed, of 100 proteins screened, 35 showed an effect on either Golgi morphology or ts-O45-G transport, or both (Fig. 3), with 20 being uncharacterized to date. Furthermore, analyzing the subcellular localizations of the effectors in our assays showed that those proteins associated with the ER or Golgi complex affected ts-O45-G transport or Golgi morphology more frequently than those proteins localizing to the PM or the cytoplasm (Fig. 5A). Eleven of the effectors could be classified as strong inhibitors, and we consider them most likely to be directly involved in membrane transport or Golgi morphology. For those classified as weak effectors, further independent experiments [such as involving gene knockdowns by RNA interference (RNAi)] might be necessary to obtain more definite evidence for a direct involvement in membrane transport or Golgi morphology. Several reasons exclude the possibility that transport inhibition and/or disturbance of Golgi morphology were unspecific under conditions of overexpression. First, some effectors in our assays are orthologs of well characterized yeast proteins and, consistent with the data available in yeast, these proteins interfered with transport or Golgi complex morphology. Second, in addition to the proteins predicted to act in membrane traffic, only those having been classified into the functional groups of “signal transduction” or “protein metabolism” perturbed transport or Golgi morphology. In contrast, proteins assigned to act on DNA or RNA, for example, had no effects in our experiments (Fig. 5B). Third, several proteins with a clear role in membrane traffic, but not involved in the transport steps analyzed by our assays (e.g., amphiphysin II and synaptotagmin III) also showed no effect. Finally, many of the proteins that scored as positives contained domains that are characteristic of proteins involved in the secretory pathway (e.g., “KKxx” or “xxRR” motifs in their cytoplasmic tails, see also Fig. 5C). Analysis of the sequence features of the screened proteins also demonstrates the specificity of our assays. For example, the likelihood of inhibiting transport or affecting Golgi morphology did not appear to depend on the presence or number of TM domains. Altogether, this demonstrates that the strategy presented here is able to identify novel proteins with specific roles in secretory membrane traffic and/or Golgi morphology. It will thus be an important tool in future large-scale screens to identify more novel proteins with a role in membrane traffic. The assays and microscopy techniques developed will also be powerful tools in functional screens using cellular knockdowns by RNAi, which are complementary to our overexpression strategy used here. In addition, one limitation of our overexpression approach could be overcome, namely that membrane proteins or those with a signal peptide could be affected when tagged at their N-terminal end, and thus in the absence of information on the C-terminal tagged protein, would either not be identified as effectors in our screens, or would show an effect on ts-O45-G transport due to the GFP tag interfering with their proper translocation into the ER.
About half of the effectors (15) revealed in our screens were positive in both assays. Ten affected only ts-O45-G transport, and 10 interfered with Golgi morphology only (Fig. 3). One possible explanation for these results is that those proteins affecting only ts-O45-G transport act in steps not directly related to Golgi morphology, such as post-TGN (trans-Golgi network) traffic. In contrast, those proteins affecting only Golgi morphology are most likely to be involved in the maintenance of the structure of this organelle. Interestingly, Golgi effectors of this kind frequently carried coiled-coil domains, consistent with the hypothesis that proteins with such domains play a role in Golgi structure (Gillingham and Munro 2003).
The Golgi matrix protein GM130 used in our studies as a Golgi marker is known to relocate into punctuate structures throughout the cytoplasm when transport between the ER and Golgi complex is impaired (Seemann et al. 2000; Miles et al. 2001). Consistent with this, the majority of the Golgi effectors (18 of 25) caused such scattering of GM130. However, overexpression of seven proteins induced reduction of the Golgi fluorescence and/or number of Golgi fragments. Recent work showed that under the conditions of direct ER exit block, GM130 is relocated to the ER (Puri and Linstedt 2003). Therefore, it is possible that these seven proteins could play a role in the regulation of ER exit. Consistent with this hypothesis, two of them (AL136567 and AL136610) contain ER retrieval signals and could be molecules cycling between ER and the Golgi. Another (AL713668) is a close homolog of Yip1p, a protein participating in the formation of COPII vesicles (Heidtman et al. 2003) at ER exit sites.
Recent data on the localization of almost the entire yeast genome (Kumar et al. 2002; Huh et al. 2003) predicts that about 10% (600) of all yeast proteins localize to the ER or Golgi complex. Based on data provided by the Gene Ontology Consortium (see Methods), about 20% of these ER- and Golgi-localized proteins have to date been assigned a function in the early secretory pathway. In contrast to this, 30% and 56% of the ER- or Golgi-localized human proteins screened here have an effect on ts-O45-G transport when overexpressed (Golgi morphology effectors were excluded from this calculation). One explanation for this divergence might be that the number of yeast proteins known to date to be involved in the secretory pathway may be an underestimate, because the functions of one-third of the yeast ORFs are still unknown. Also, it could be that a more complex regulatory network underlying the secretory pathway exists in humans compared to yeast. For example, tissue-specific regulatory molecules absent in yeast could be encoded in the human genome, increasing the number of potential proteins functioning in the early secretory pathway. Consequently, one would therefore assume a higher percentage of human proteins functionally associated with the early secretory pathway compared to yeast. In line with this hypothesis, systematic subcellular localization of novel human proteins (Simpson et al. 2000, see also http://gfp-cdna.embl.de/) show that 18% of 650 randomly selected human proteins associate with the ER or Golgi complex, which is clearly higher than in yeast (10%). An increased complexity of membrane traffic in humans compared to yeast cells may also be reflected by the fact that most of the ER- and Golgi-localized proteins inhibiting transport in our assays do not have homologs in yeast.
In summary, our approach described here is able to efficiently identify new proteins with a role in membrane transport and Golgi morphology. In combination with organelle proteomics (Bell et al. 2001) and large-scale bioinformatic predictions (Clark et al. 2003), this will ultimately lead to a more comprehensive understanding of the secretory pathway at the molecular level.
METHODS
Materials
GFP-tagged ORFS were generated and prepared as described (Simpson et al. 2000). Recombinant adenoviruses encoding the secretory marker protein ts-O45-G tagged with either CFP or YFP were as described (Keller et al. 2001). The mouse monoclonal anti-ts-O45-G antibody “VG” was a gift from Kai Simons (MPI-CBG).
Transport and Golgi Integrity Assays
HeLa cells ATCC (CCL-2) were plated at a density of 10,000 cells/well in LabTek eight-well chamber slides (Nalge Nunc) containing 300 μL medium/well. For each well, 0.6 μL plasmid DNA at a concentration of 500 ng/μL was added to the transfection mix consisting of 0.9 μL FuGENE6 (Roche) and 29 μL OPTIMEM 1 (Gibco/Invitrogen). After 6 h, transfected cells were overlaid with recombinant adenoviruses encoding the secretory marker protein ts-O45-G tagged with CFP or YFP to complement the color of the transfected CFP- or YFP-tagged protein. After a 45-min incubation, the cells were washed with culture medium and ts-O45-G was accumulated in the ER by incubating the cells at 39.5°C for 16 h. Thereafter, cells were shifted to 32°C in the presence of 100 μg/mL cycloheximide (Calbiochem) to prevent further protein synthesis and to release ts-O45-G from the ER. At 60 min thereafter, cells were fixed with 3% paraformaldehyde, and ts-O45-G on the cell surface was detected by immunostaining with a monoclonal antibody recognizing an extracellular ts-O45-G epitope at the PM (VG) followed by staining with Cy5-labeled secondary anti-mouse antibodies (Amersham Biosciences). Cell nuclei were labeled with Hoechst 33342 stain (Sigma) diluted in PBS to a final concentration of 0.1 μg/mL.
To measure the integrity of the Golgi complex, HeLa cells were transfected for 24 h with the appropriate plasmids. Thereafter they were fixed in 3% paraformaldehyde, permeabilized with 0.1% Triton X-100, and the Golgi complex was stained with mouse monoclonal anti-GM130 antibody (BD Bioscience) followed by Cy5 conjugated anti-mouse secondary antibodies. Cell nuclei were labeled with Hoechst stain as described above.
Multicolor images of stained cells were acquired using the microscope-based screening platform described (Liebel et al. 2003).
Quantification of the Transport Assay
For each experiment at least 500 cells were imaged, and fluorescence was quantified as described (Liebel et al. 2003). Briefly: The position and shape of cell nuclei were deduced from the Hoechst staining by thresholding. Thereafter, the area of the nuclei was dilated digitally. The fluorescence relating to the transfected GFP-tagged proteins or ts-O45-G at the plasma membrane were measured in this extended area. The fluorescence of the CFP- or YFP-ts-O45-G-specific fluorescence was measured in the ring-like structure of this extended area outside the nucleus (see Liebel et al. 2003 for details). Comparison of the results obtained with this automated data analysis and analyses involving the manual selection of the entire cell area as region of interest for the measurements showed that the results did not deviate more than the standard deviation of the mean obtained by automated data analysis.
To each individual cell were then assigned two parameters: the relative amount of the GFP-fusion protein expressed, and the relative surface expression of ts-O45-G defined as the ratio of ts-O45-G-specific plasma membrane fluorescence (“VG” antibody staining) and the CFP- or YFP-ts-O45-G-specific fluorescence. Measurements for all cells in a single experiment were summarized in a scatter plot with the x-axis relating to the amount of expressed GFP-tagged protein (normalized to the maximum fluorescence obtained in the particular experiment and expressed in percentage; see for example Fig. 2). Cells with a GFP fluorescence <10% were scored as nontransfected cells, and their average relative surface expression was determined. All values obtained for the relative surface expression in one experiment were then normalized to the value obtained for the relative surface expression in nontransfected cells (set as “1”; see Fig. 2).
Transfected cells (>10% of GFP fluorescence) were further divided into three expression classes representing low (10%-40%), medium (40%-70%), and high (70%-100%) expressing cells according to their fluorescence relating to the amount of transfected GFP-fusion protein. For each expression class, the average value of the relative surface expression was calculated. This allowed us to deduce from these values directly inhibitory (<1) or stimulatory (>1) effects of the expressed GFP-tagged protein on ts-O45-G transport compared to nontransfected cells. Also, this way of scoring the results allows discrimination of strong perturbing proteins, already showing an inhibition in the “low” expression class compared to weak inhibitors showing an inhibitory effect preferentially in the “high” expression class (lower values in Table 1).
The resulting values obtained for the average relative surface expression for the four expression classes were also fitted by the least square method to a linear function of the kind: y = a + bx. The progression of the effect of each GFP-tagged protein on ts-O45-G transport with increasing amounts of GFP-tagged protein expressed is thus characterized as the steepness of the slope (expressed by “b”). Positive “b”-values represent transport acceleration, and negative values represent transport inhibition with respect to nontransfected cells (upper values in Table 1).
At least two experiments were performed for each ORF tested, resulting in at least 200 transfected cells scored. For transport effectors, the experiments were repeated three or four times, and the respective standard errors of the means were calculated. If available, both N- and C-terminal GFP fusions of every ORF product were tested and processed separately.
Quantification of the Golgi Integrity Assay
On average, 100 cells were scored with ∼30 of them being transfected with the respective plasmid. Only cells with normal interphase morphology were taken for analysis. The population of imaged cells was divided into two groups; transfected (>10% of maximum GFP fluorescence obtained, see paragraph above) and nontransfected (<10% of maximum GFP fluorescence obtained) cells, and an average number of Golgi fragments and intensity of each Golgi fragment within each group were calculated as described (Liebel et al. 2003). The effect of GFP-tagged ORF overexpression was expressed as a normalized ratio of transfected and nontransfected cells for each of the two parameters measured. Identified changes of Golgi morphology were confirmed by visual inspection. When no changes in Golgi morphology were detected visually, the pooled standard error of the differences between the means for transfected and nontransfected cells was calculated (sp √1/n1+1/n2; see Hassard 1991; where sp is the pooled standard deviation, n1 the number of transfected cells and n2 the number of nontransfected cells). Only those ORFs for which the difference between the mean values for transfected and nontransfected cells was greater than two times the pooled standard error were considered significant Golgi effectors.
Databases and Analyses
Localization data were taken from http://gfp-cdna.embl.de/, and bioinformatic data of the clones were collected by Harvester (Liebel et al. 2004; http://harvester.embl.de/) and http://www.dkfz-heidelberg.de/LIFEdb. Assignment of the putative function of the proteins was based on the comparison of the output of SMART (http://smart.embl-heidelberg.de/), SCOPE (http://scop.berkeley.edu/), CDART (http://www.ncbi.nlm.nih.gov), and Pfam (http://www.sanger.ac.uk/Software/Pfam/) databases. Information about orthologs in other model species was taken from http://www.ncbi.nlm.nih.gov/ and http://www.yeastgenome.org/. Information about yeast genes involved in secretory membrane traffic was derived from the Gene Ontology Consortium (http://www.geneontology.org/) under the queries “secretory pathway” and “intracellular protein transport”.
Acknowledgments
We thank Brigitte Joggerst and Thi Bach Nga Ly for technical assistance, Kai Simons for antibodies, Steffi Bechtel for tagging ORFs to GFP, and Joanne Young for comments on the manuscript. We also thank Olympus, Olympus BioSystems, and Perkin Elmer for continuous support throughout the project. This work has been supported by a grant to R.P. from the Verbundforschung Baden-Wuerttemberg (No. 24-720.431-1-2/2) and grants from the BMBF 01KW0013 (R.P.) and NGFN 01GR1001 (R.P. and S.W.), DHGP 01KW0012 (S.W.).
Footnotes
[Supplemental material is available online at www.genome.org.]
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2658304.
References
- Andrade, M.A., Perez-Iratxeta, C., and Ponting, C.P. 2001. Protein repeats: Structures, functions, and evolution. J. Struct. Biol. 134: 117-131. [DOI] [PubMed] [Google Scholar]
- Aridor, M., Bannykh, S.I., Rowe, T., and Balch, W.E. 1995. Sequential coupling between COPII and COPI vesicle coats in endoplasmic reticulum to Golgi transport. J. Cell Biol. 131: 875-893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell, A.W., Ward, M.A., Blackstock, W.P., Freeman, H.N., Choudhary, J.S., Lewis, A.P., Chotai, D., Fazel, A., Gushue, J.N., Paiement, J., et al. 2001. Proteomics characterization of abundant Golgi membrane proteins. J. Biol. Chem. 276: 5152-5165. [DOI] [PubMed] [Google Scholar]
- Calero, M., Winand, N.J., and Collins, R.N. 2002. Identification of the novel proteins Yip4p and Yip5p as Rab GTPase interacting factors. FEBS Lett. 515: 89-98. [DOI] [PubMed] [Google Scholar]
- Clark, H.F., Gurney, A.L., Abaya, E., Baker, K., Baldwin, D., Brush, J., Chen, J., Chow, B., Chui, C., Crowley, C., et al. 2003. The secreted protein discovery initiative (SPDI), a large-scale effort to identify novel human secreted and transmembrane proteins: A bioinformatics assessment. Genome Res. 13: 2265-2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conchon, S., Cao, X., Barlowe, C., and Pelham, H.R. 1999. Got1p and Sft2p: Membrane proteins involved in traffic to the Golgi complex. EMBO J. 18: 3934-3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosson, P. and Letourneur, F. 1994. Coatomer interaction with di-lysine endoplasmic reticulum retention motifs. Science 263: 1629-1631. [DOI] [PubMed] [Google Scholar]
- Dilcher, M., Veith, B., Chidambaram, S., Hartmann, E., Schmitt, H.D., and Fischer von Mollard, G. 2003. Use1p is a yeast SNARE protein required for retrograde traffic to the ER. EMBO J. 22: 3664-3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez, M., Dejgaard, K., Fullekrug, J., Dahan, S., Fazel, A., Paccaud, J.P., Thomas, D.Y., Bergeron, J.J., and Nilsson, T. 1998. gp25L/emp24/p24 protein family members of the cis-Golgi network bind both COP I and II coatomer. J. Cell Biol. 140: 751-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dusetti, N.J., Jiang, Y., Vaccaro, M.I., Tomasini, R., Azizi Samir, A., Calvo, E.L., Ropolo, A., Fiedler, F., Mallo, G.V., Dagorn, J.C., et al. 2002. Cloning and expression of the rat vacuole membrane protein 1 (VMP1), a new gene activated in pancreas with acute pancreatitis, which promotes vacuole formation. Biochem. Biophys. Res. Commun. 290: 641-649. [DOI] [PubMed] [Google Scholar]
- Gillingham, A.K. and Munro, S. 2003. Long coiled-coil proteins and membrane traffic. Biochim. Biophys. Acta 1641: 71-85. [DOI] [PubMed] [Google Scholar]
- Hassard, T.H. 1991. Understanding biostatistics, pp. 156-157. Mosby-Year Book, St. Louis.
- Heidtman, M., Chen, C.Z., Collins, R.N., and Barlowe, C. 2003. A role for Yip1p in COPII vesicle biogenesis. J. Cell Biol. 163: 57-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh, W.K., Falvo, J.V., Gerke, L.C., Carroll, A.S., Howson, R.W., Weissman, J.S., and O'Shea, E.K. 2003. Global analysis of protein localization in budding yeast. Nature 425: 686-691. [DOI] [PubMed] [Google Scholar]
- Keller, P., Toomre, D., Diaz, E., White, J., and Simons, K. 2001. Multicolour imaging of post-Golgi sorting and trafficking in live cells. Nat. Cell Biol. 3: 140-149. [DOI] [PubMed] [Google Scholar]
- Kumar, A., Agarwal, S., Heyman, J.A., Matson, S., Heidtman, M., Piccirillo, S., Umansky, L., Drawid, A., Jansen, R., Liu, Y., et al. 2002. Subcellular localization of the yeast proteome. Genes & Dev. 16: 707-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebel, U., Starkuviene, V., Erfle, H., Simpson, J.C., Poustka, A., Wiemann, S., and Pepperkok, R. 2003. A microscope-based screening platform for large-scale functional protein analysis in intact cells. FEBS Lett. 554: 394-398. [DOI] [PubMed] [Google Scholar]
- Liebel, U., Kindler, B., and Pepperkok, R. 2004. “Harvester” a fast meta search engine of human protein resources. Bioinformatics 20: 1962-1963. [DOI] [PubMed] [Google Scholar]
- Miles, S., McManus, H., Forsten, K.E., and Storrie, B. 2001. Evidence that the entire Golgi apparatus cycles in interphase HeLa cells: Sensitivity of Golgi matrix proteins to an ER exit block. J. Cell Biol. 155: 543-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monier, S., Jollivet, F., Janoueix-Lerosey, I., Johannes, L., and Goud, B. 2002. Characterization of novel Rab6-interacting proteins involved in endosome-to-TGN transport. Traffic 3: 289-297. [DOI] [PubMed] [Google Scholar]
- Nakamura, N., Rabouille, C., Watson, R., Nilsson, T., Hui, N., Slusarewicz, P., Kreis, T.E., and Warren, G. 1995. Characterization of a cis-Golgi matrix protein, GM130. J. Cell Biol. 131: 1715-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepperkok, R., Lowe, M., Burke, B., and Kreis, T.E. 1998. Three distinct steps in transport of vesicular stomatitis virus glycoprotein from the ER to the cell surface in vivo with differential sensitivities to GTP γ S. J. Cell Sci. 111: 1877-1888. [DOI] [PubMed] [Google Scholar]
- Puri, S. and Linstedt, A.D. 2003. Capacity of the golgi apparatus for biogenesis from the endoplasmic reticulum. Mol. Biol. Cell. 14: 5011-5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe, T., Aridor, M., McCaffery, J.M., Plutner, H., Nuoffer, C., and Balch, W.E. 1996. COPII vesicles derived from mammalian endoplasmic reticulum microsomes recruit COPI. J. Cell Biol. 135: 895-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann, J., Jokitalo, E., Pypaert, M., and Warren, G. 2000. Matrix proteins can generate the higher order architecture of the Golgi apparatus. Nature 407: 1022-1026. [DOI] [PubMed] [Google Scholar]
- Simpson, J.C., Wellenreuther, R., Poustka, A., Pepperkok, R., and Wiemann, S. 2000. Systematic subcellular localization of novel proteins identified by large-scale cDNA sequencing. EMBO Rep. 1: 287-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson, J.C., Neubrand, V.E., Wiemann, S., and Pepperkok, R. 2001. Illuminating the human genome. Histochem Cell Biol. 115: 23-29. [DOI] [PubMed] [Google Scholar]
- Sivars, U., Aivazian, D., and Pfeffer, S.R. 2003. Yip3 catalyses the dissociation of endosomal Rab-GDI complexes. Nature 425: 856-859. [DOI] [PubMed] [Google Scholar]
- Tang, B.L., Ong, Y.S., Huang, B., Wei, S., Wong, E.T., Qi, R., Horstmann, H., and Hong, W. 2001. A membrane protein enriched in endoplasmic reticulum exit sites interacts with COPII. J. Biol. Chem. 276: 40008-40017. [DOI] [PubMed] [Google Scholar]
- Ullrich, B., Li, C., Zhang, J.Z., McMahon, H., Anderson, R.G., Geppert, M., and Sudhof, T.C. 1994. Functional properties of multiple synaptotagmins in brain. Neuron 13: 1281-1291. [DOI] [PubMed] [Google Scholar]
- Venter, J.C., Adams, M.D., Myers, E.W., Li, P.W., Mural, R.J., Sutton, G.G., Smith, H.O., Yandell, M., Evans, C.A., Holt, R.A., et al. 2001. The sequence of the human genome. Science 291: 1304-1351. [DOI] [PubMed] [Google Scholar]
- Wiemann, S., Weil, B., Wellenreuther, R., Gassenhuber, J., Glassl, S., Ansorge, W., Bocher, M., Blocker, H., Bauersachs, S., Blum, H., et al. 2001. Toward a catalog of human genes and proteins: Sequencing and analysis of 500 novel complete protein coding human cDNAs. Genome Res. 11: 422-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, H., Michelsen, K., and Schwappach, B. 2003. 14-3-3 dimers probe the assembly status of multimeric membrane proteins. Curr. Biol. 13: 638-646. [DOI] [PubMed] [Google Scholar]
- Zhang, B. and Zelhof, A.C. 2002. Amphiphysins: Raising the BAR for synaptic vesicle recycling and membrane dynamics. Bin-Amphiphysin-Rvsp. Traffic 3: 452-460. [DOI] [PubMed] [Google Scholar]
- Zilberstein, A., Snider, M.D., Porter, M., and Lodish, H.F. 1980. Mutants of vesicular stomatitis virus blocked at different stages in maturation of the viral glycoprotein. Cell 21: 417-427. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://gfp-cdna.embl.de/; localization database of novel GFP-tagged human cDNA products to subcellular compartments of the eukaryotic cell.
- http://harvester.embl.de/; collection of text-based information on human proteins from various public databases and prediction servers.
- http://www.dkfz.de/LIFEdb(qp1v2h55zigf4b551prescen)/LIFEdb.aspx; integrative database of human novel full-length cDNAs generated by German cDNA Consortium.
- http://smart.embl-heidelberg.de/; tool for the identification and annotation of functional protein domains and their architectures.
- http://scop.berkeley.edu/; database of protein classification according to their evolutionary and structural relationships.
- http://www.ncbi.nlm.nih.gov/Structure/lexington/lextington.cgi?cmd=rps;CDART—Conserved Domain Architecture Retrieval Tool for similarity search based on domain architecture.
- http://www.sanger.ac.uk/Software/Pfam/; database of the protein families and domains.
- http://www.yeastgenome.org/; database of the molecular biology and genetics of the yeast Saccharomyces cerevisia.
- http://www.geneontology.org/; collection of gene products in terms of their associated biological processes, cellular components and molecular functions.