Abstract
Western medicine often aims to specifically treat diseased tissues or organs. However, the majority of current therapeutics failed to do so owing to their limited selectivity and the consequent “off-target” side effects. Targeted therapy aims to enhance the selectivity of therapeutic effects and reduce adverse side effects. One approach toward this goal is to utilize disease-specific ligands to guide the delivery of less-specific therapeutics, such that the therapeutic effects can be guided specifically to diseased tissues or organs. Among these ligands, aptamers, also known as chemical antibodies, have emerged over the past decades as a novel class or targeting ligands that are capable or specific binding to disease biomarkers. Compared with other types or targeting ligands, aptamers have an array of unique advantageous features, which make them promising for developing aptamer–drug conjugates (ApDCs) for targeted therapy. In this Review, we will discuss ApDCs for targeted drug delivery in chemotherapy, gene therapy, immunotherapy, photodynamic therapy, and photothermal therapy, primarily of cancer.
Graphical abstract
INTRODUCTION TO APTAMERS AND APDCS
Cancer has been one of the major threats to human health for a long time. However, current cancer therapy often suffers from limited efficacy with adverse side effects that harm normal tissues.1 Targeted therapy aims to increase toxicity specifically in tumor tissues while reducing toxicity in healthy tissues. To this end, small molecule drugs were screened to specifically bind to cancer-associated biomarkers and inhibit their biological functions in cancer development. Alternatively, the advancement of biotechnologies allowed the development of drug– ligand conjugates for targeted therapy. In these conjugates, the ligands specifically recognize cancer-associated biomarkers and deliver conjugated drugs to target cancer cells but may not necessarily have direct therapeutic effects; drugs are conjugated to ligands via functional linkers that ensure the stability of conjugates and also allow conditional drug release in cancerous tissues or cells. Among ligands for targeted therapy, antibodies are well-established for specific recognition and/or biological regulation, and antibody–drug conjugates (ADCs)2 have attracted tremendous attention for targeted cancer therapy. Another type of ligands, aptamers, also known as chemical antibodies, are also attractive for developing aptamer–drug conjugates (ApDCs) for targeted therapy.
Nucleic acid aptamers are single-stranded oligonucleotides with specific recognition abilities to targets. Aptamers are screened through Systematic Evolution of Ligands by Exponential Enrichment (SELEX),3, 4 in combination with techniques such as capillary electrophoresis, microfluidics, and fluorescence-activated cell sorting. The targets of aptamers range from small molecules3, 8, 9 and proteins4, 10–16 to intact cells.17–27 A wide array of aptamers have been developed for biomarkers of high therapeutic interest (Table 1). Aptamers bind strongly to targets, with binding affinities (Kds of pM–nM) comparable to, or sometimes stronger than, those of other molecular ligands.28 Advancement of biochemistry and biophysics has facilitated the resolution of the crystal structures of aptamer–target complexes,29–32 which not only has elucidated that aptamer–target interactions are attributed to noncovalent interactions including hydrogen bonding, van der Waals force, and stacking interactions,29–32 but may also facilitate the development of aptamers and ApDCs through rational drug design and screening, as well as the discovery of new drugable sites on biomarkers.33–42 In addition to target binding, aptamers may also serve as therapeutics per se to modulate the biological functions of biomarkers. Pegaptanib (marketed as Macugen), an anti-VEGF aptamer that recognizes the majority of human VEGF-A isoforms, has been approved by the FDA for the treatment of age-related macular degeneration (AMD).43 Another example, AS1411, is a nucleolin-targeting aptamer44 currently under Phase II clinical investigation for the treatment of acute myeloid leukemia (AML).
Table 1.
Examples of Nucleic Acid Aptamers to Targets of Therapeutic Interest
| aptamers | biomarker targets | refs |
|---|---|---|
| Vascular endothelial growth factor | Age-related macular degeneration (AMD) | 43 |
| Nucleolin | Cancer | 45 |
| Protein tyrosine kinase 7 | Cancer | 46 |
| Immuno globulin µ heavy chains (IGHM) | Cancer | 47 |
| Prostate-specific membrane antigen | Cancer | 48 |
| Tenascin C | Cancer | 11 |
| Mucin 1 | Cancer | 49 |
| αvβ3 integrin | Cancer | 50 |
| NF-κB | Cancer | 51 |
| E2F transcription factor | Cancer | 52, 53 |
| HER2 | Cancer | 54–57 |
| HER3 | Cancer | 15 |
| EGFR | Cancer | 58–60 |
| Epithelial cell adhesion molecule (EpCAM) | Cancer | 61, 62 |
| CD30 | Cancer | 63 |
| Plasminogen activator inhibitor 1 | 64 | |
| α-thrombin | Thrombosis | 13 |
| HIV gpl20 | Viral infection | 16 |
| Immuno globulin E | Allergy | 65 |
| Osteoblasts | Metabolic skeletal disorders | 66 |
| Interleukin 6 receptor | Inflammatory diseases | 61 |
| Transferrin receptor | Cancer, lysosomal storage diseases, etc. | 68 |
Aptamers have some characteristic features or advantages over other recognition ligands.17 Current technolgies for aptamer identification are efficient and cost-effective. The current techniques for nucleic acid manufacture are capable of cost-effective synthesis and functionalization. Aptamers are also well-known for their ability for reversible conformational configuration after thermal or chemical denaturation and correspondingly long shelf life, rapid tissue penetration, low immunogeneity,69 and simplicity of antidote development.70, 71 Moreover, aptamers can be developed against toxins or low-immunogenicity agents that are beyond the capabilities of current antibody development technologies. Thus, aptamers may be generated for toxins or low-immunogenicity agents and used as carriers in ApDC-based targeted drug delivery. These advantages make aptamers attractive for developing ApDCs for targeted therapy.
As another major type of recognition ligands, antibodies have been extensively explored to develop ADCs in the past few decades.72, 73 Aptamers, owing to their characteristic features, are also promising candidates for applications in targeted therapy. Their high specificity and selectivity make them excellent candidates for targeted delivery of therapeutics. Their general chemical and thermal stability and conformational reversibility enable versatile designs of ApDCs. Similar to ADCs, an ApDC typically consists of three molecular parts: a ligand (aptamer), a linker, and a warhead (drug). Aptamers serve as recognition ligands to guide the delivery of therapeutics to target diseased sites and/or regulate the biological functions of target biomarkers. The chemical stability, simplicity of chemical modification, and ready molecular engineering of aptamers enable easy and programmable conjugation with many therapeutics, ranging from chemotherapeutics to photo-therapeutics, toxins, gene therapeutics, and vaccines (Table 2). ApDCs have been explored as both molecular ApDCs and nanoscale ApDCs.74–82 In this Review, we will mainly discuss molecular ApDCs.
Table 2.
Examples of Aptamers Used for ApDCs in Targeted Drug Delivery
| aptamers | therapeutic cargoes | refs |
|---|---|---|
| sgc8 | Chemotherapeutics, photosensitizers, photothermal agents, immunotherapeutics | 79, 80, 83–95 |
| TD05 | Photosensitizers | 96, 97 |
| AS1411 | Chemotherapeutics, photosensitizers | 81, 98 |
| A10 | Chemotherapeutics, siRNA/shRNA | 82, 99–106 |
| APT | Chemotherapeutics | 107–109 |
| CH6 | siRNA | 66 |
| Anti-gp120 aptamer | siRNA | 110 |
| Min.2 | Antigens | 111 |
| AIR-3A | Photosensitizers | 67 |
| GS24 | Enzymes | 68 |
APTAMER–CHEMOTHERAPEUTIC CONJUGATES
Chemotherapy is one of the main treatments for cancer therapy. Conventional chemotherapy is limited by adverse side effects in healthy tissues and low maximum tolerated dosage, due to the off-target effects of many chemotherapeutics. The ability to deliver chemotherapeutics selectively to target cancer tissues is promising to overcome these complications. Aptamers are amenable to coupling with a wide range of chemo-therapeutics, owing much to their chemical stability and the ability of current technologies to design and site-specifically modify aptamers with desired functional groups.
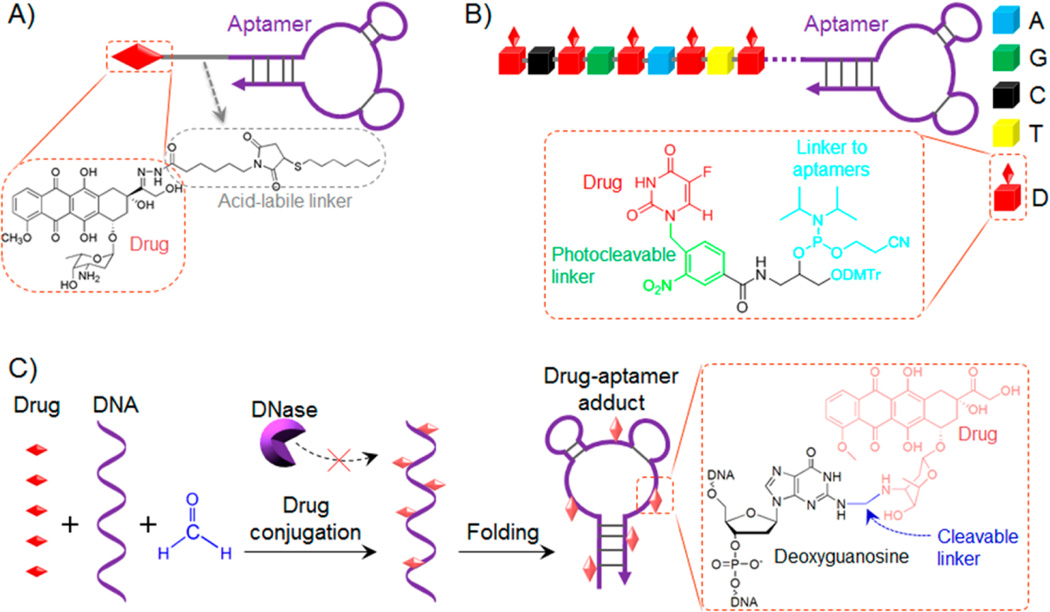
Aptamers can be covalently conjugated with chemotherapeutics. In an early trial, Huang et al. conjugated doxorubicin (Dox), one of the most commonly prescribed chemotherapeutics in cancer therapy, with a DNA aptamer sgc8, which selectively targets protein tyrosine kinase 7 (PTK7) overexpressed on many types of cancers (Figure 1A).85, 112 The conjugation was achieved via a hydrazone linker that allows conditional bond cleavage and subsequent drug release in acidic environment, such as endosomes or lysosomes. The resulting conjugate was studied in vitro in CEM, a human lymphoblastic leukemia cell line. This ApDC delivered Dox selectively to CEM cells, released Dox in endosome, and inhibited cancer cell proliferation. In contrast, the efficiency of drug delivery and cell proliferation in nontarget cells was dramatically reduced using this ApDC. This demonstrated the principle of ApDC-based targeted delivery of chemotherapeutics. Despite the successful demonstration of the concept, this strategy faces challenges such as low copy number of drugs conjugated onto each aptamer (1:1 ratio) and suboptimal synthesis yield. To overcome these challenges, Wang et al. developed a modular ApDC, in which multiple copies of drugs can be site-specifically conjugated onto each aptamer molecule (Figure 1B).94 Particularly, a phosphoramidite was designed and synthesized to carry an anticancer drug, 5-fluorouracil (5-FU), a widely prescribed drug for the treatment of cancers, such as colorectal cancer and pancreatic cancer. To enable spatiotemporal controllable drug release, a photocleavable (PC) linker was used to link the drug moiety with the backbone of the phosphoramidite. The phosphoramidite then served as a modular building block that can be conjugated with aptamers simply using automated solid-phase DNA synthesis. Therefore, ApDCs with desired copy number and positions of drugs can be logically designed and synthesized. In vitro study was conducted in a colon cancer cell line, HCT116, which also overexpresses PTK7 for specific sgc8 binding. The corresponding ApDC, sgc8-(5-FU)5, in which one sgc8 aptamer carries 5 copies of 5-FU, was proven to selectively deliver the chemotherapeutic into HCT116 cells overexpressing PTK, but not to cells that do not have PTK expression. Under light irradiation, the cleavage of the PC linker released the tethered 5-FU molecules from the aptamer backbone, and consequently inhibited cancer cell proliferation. In this study, one aptamer was able to carry multiple copies of drugs, thus increasing the drug loading capacity and promising to decrease the cost.
Figure 1.
Chemotherapeutic ApDCs developed by covalent conjugation of aptamers and drugs. (A) Aptamer sgc8 was covalently conjugated via hydrazone, an acid-labile linker, with Dox for targeted delivery and intracellular release of Dox (structures shown in inlet). (B) ApDCs were constructed by automated synthesis using a phosphoramidite that carried a 5-FU prodrug moiety via a photocleavable linker (structures shown in inlet). This phosphoramidite enabled site-specific conjugation of multiple copies of drugs on one aptamer, with additional photocontrollable drug release upon light irradiation after these ApDCs reached or were internalized into target cells. (C) ApDCs developed via a simple biocompatible reaction for covalent and heat-labile conjugation of one aptamer with multiple copies of drugs. (Shaded color of schematic drugs in ApDCs indicates the quenching of drug fluorescence upon conjugation.) The ApDCs, or drug–aptamer adducts in this case, were also resistant to nuclease degradation. (Reprinted with permission from refs 85 (copyright 2009 John Wiley & Sons, Inc.), 93 (copyright 2014 American Chemistry Society), and 94 (copyright 2015 Macmillan Publishers Limited).)
In a recent study, Zhu et al. utilized a simple biocompatible reaction to construct ApDCs with multiple copies of drug molecules on one aptamer. It is well-known that a variety of anticancer chemotherapeutics inhibit cell proliferation by forming adducts with genome DNA in cells to disrupt cell division and induce cell apoptosis (Figure 1C).93 Examples of these chemotherapeutics include anthracycline drugs (e.g., Dox, Epirubicin, and Daunorubicin) and cisplatin. Inspired by this, they optimized the reaction condition and synthesized adducts of anthracyclines with synthetic DNA, including aptamers. The conjugation was mediated using formaldehyde, which is commonly used in tissue fixation, as a cross-linker to form a methylene linker between the 3-NH2 group of Dox on one side and the 2-NH2 on deoxyguanosine (dG) on the other side. These ApDCs, or drug–aptamer adducts (DAAs) in this case, were featured as follows: (1) the synthesis of these ApDCs was performed simply in physiological buffer at relatively low temperature (10 °C) and the products were stable at this temperature or lower for storage; (2) one aptamer carried multiple drugs in the ApDCs, with the majority of drugs conjugated with the 2-NH2 on dG; and (3) the temperature-sensitive cleavage of the methylene linker allowed gradual drug release at physiological temperature. Using sgc8 and nucleolin-targeting aptamer AS1411, the corresponding ApDCs were proven to inhibit tumor growth and reduce side effects in a leukemia and a liver cancer xenograft model, respectively. Even though this approach is still confronted with the challenge of heterogeneity of drug copies on one aptamer and low spatial controllability of drug release, it presents a simple ApDC technology and provides insight into optimizing it for economical production of ApDCs with high drug loading capacity and drug release controllability.
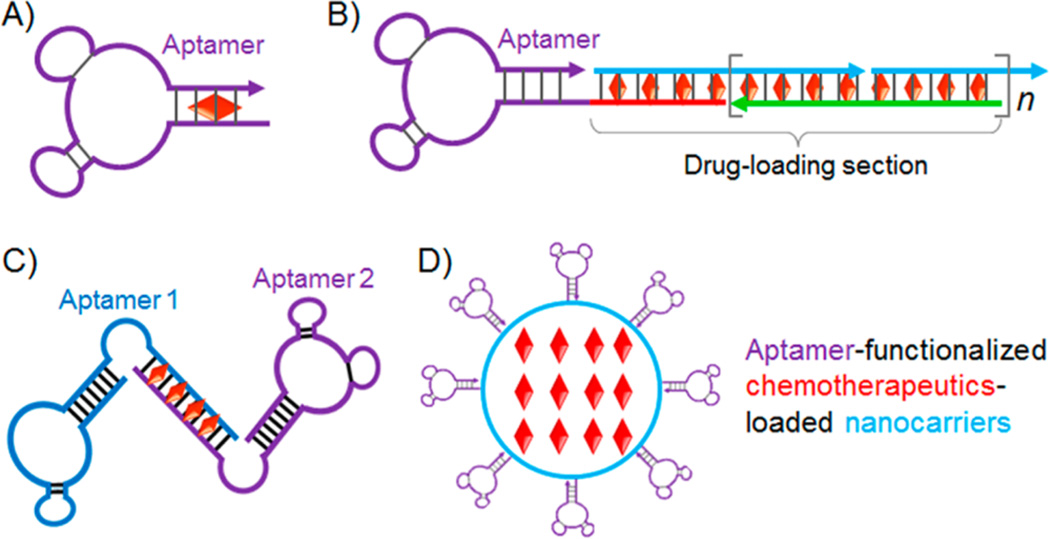
In addition to covalent drug conjugation, noncovalent drug conjugation has also been extensively explored to develop ApDCs for chemotherapeutics. One of the most common approaches exploits the ability of anthracycline drugs to be intercalated into tandem 5′-(GC)-3′ or 5′-(CG)-3′ sites in a dsDNA/dsRNA. Combined with the simplicity of programmable nucleic acid engineering, these drug-intercalating sites have been exploited intrinsically in aptamers or modified on aptamers to form ApDCs.113 In 2006, an ApDC was reported to explore a PSMA-targeting RNA aptamer, A10, which had an intrinsic drug-intercalating site for Dox, for targeted delivery of Dox (Figure 2A).114 Following this work, a series of studies have been reported to engineer aptamers with additional drug-intercalating sites, with the intention to increase drug loading capacity and maintain the recognition abilities of aptamers. In one example, aptamers were engineered to be tethered with a long repetitive drug-intercalating dsDNA, to form ApDCs, also named as aptamer-tethered DNA nanotrains, with high drug loading capacity for each aptamer (Figure 2B).80 The ApDCs demonstrated selectivity in anticancer drug delivery, effectively inhibited tumor growth, and reduced side effects in a leukemia xenograft model. By taking advantage of programmable DNA engineering, a bispecific aptamer was also developed, in which two aptamers, sgc8 and sgd5a, specifically recognizing different subtypes of cancer, were engineered into one entity, with a drug-intercalating dsDNA as both the linker and the drug loading sites (Figure 2C).115 The bivalent aptamer demonstrated the bispecificity in cancer cell detection and anticancer drug delivery. Despite extensive effort in the past decade, development of bivalent antibodies116, 117 has been challenged by the relatively low predictability and high cost of bivalent antibody engineering. In contrast, it is highly predictable and cost-effective to engineer bivalent aptamers.
Figure 2.
Chemotherapeutic ApDCs developed via noncovalent complexation of aptamers and drugs. (A) ApDC formed by physical intercalation of Dox with PSMA-targeting aptamer A10, which has an intrinsic drug intercalation site. (B) ApDC formed by physical intercalation of Dox with an aptamer-tethered DNA nanotrain, which was engineered to have up to 160 drug-intercalating sites (n = 1, 2, 3, …) on average. (C) Bispecific ApDC developed by linking two aptamers targeting different biomarkers via a drug-intercalating dsDNA linker. (Shaded color of schematic drugs in ApDCs in A–C indicates the quenching of drug fluorescence upon conjugation.) (D) PLGA-b-PEG nanoparticulate ApDC developed by modifying the nanoparticles with PSMA-targeting aptamers and encapsulating drugs inside nanoparticles. (Reprinted with permission from refs 80 (copyright 2013 National Academy of Sciences), 114 (copyright 2006 John Wiley & Sons, Inc.), (copyright 2012 John Wiley & Sons, Inc.), 118 (copyright 2008 National Academy of Sciences), and 119 (copyright 2011 National Academy of Sciences).)
Nanomaterials, owing to their typical high drug loading capacity and the characteristic feature of enhanced permeation and retention (EPR) effect in many types of tumors and inflammation, have emerged as promising drug carriers for passively targeted drug delivery in the treatment of related diseases. Combined with active-targeting aptamers, aptamer–nanocarrier conjugates have multifaceted values. Particularly for targeted delivery of chemotherapeutics, a wide variety of nanomaterials have been explored as the carriers, including liposomes, micelles, polymers, DNAs, gold, and magnetic nanoparticle. Among these nanocarriers, poly(lactic-co-glycolic-acid) (PLGA) nanoparticles have been explored for decades.75, 82, 118–120 Being biocompatible and biodegradable, PLGA has been well established and approved by the FDA for a range of clinical applications, including sustained drug release. To enhance the cell, tissue, and disease specificity in targeted drug delivery, aptamer functionalized PLGA nanoparticles have been extensively studied over the past decade.120 In addition, poly(ethylene glycol) (PEG) has also been integrated into these nanoparticles, by using amphiphilic PLGA–PEG diblock and PLGA–PEG–aptamer triblock polymers for nanoparticle self-assembly (Figure 2D).118, 119 The aptamer-functionalized PLGA–PEG nanoparticles are capable of sustained drug release, immune evasion, and specific targeting. Particularly, the tumor-specific aptamers modified on nanocarriers enhanced the drug delivery efficiency and subsequently improved tumor therapy efficacy.119 These nanoparticle ApDCs have demonstrated effective delivery of cisplatin into PSMA-positive prostate cancer.119 Liposomes, another well-established drug delivery system in the clinic, can also be conjugated with aptamers for improved targeting and hence improved drug delivery efficiency.86, 121, 122 In a study reported by Cao et al., nucleolin-targeting aptamer AS1411 was modified on the surfaces of cisplatin-loaded liposome nanoparticles for targeted drug delivery in a breast cancer cell line.121 The nanoconjugate delivery system selectively delivered the encapsulated drugs into LNCaP cells. Furthermore, by taking advantage of the as the antidote, the authors demonstrated the principle of inhibiting the drug efficacy by using cDNA of the aptamers.
APTAMER–NUCLEIC ACID THERAPEUTIC CONJUGATES
With the mapping of human genome and human disease genome over the past decades, gene therapy has been extensively studied to specifically regulate the expression of certain genes related to pathogenesis. Gene therapy has been shown to be promising for the treatment of a wide variety of diseases. There have also been many approaches to the delivery of genes or gene regulators, including virus-based gene delivery, RNA interference (RNAi), antisense oligonucleotides, and most recently genome editing using clustered regularly interspaced short palindromic repeats (CRISPR)-Cas systems. Despite their promising efficacy, these therapeutics also need guidance for selective delivery to target diseased tissues or organs. Toward this end, aptamers have the potential to play an important role in targeted gene therapy, by selective delivery of nucleic acid gene therapeutics.
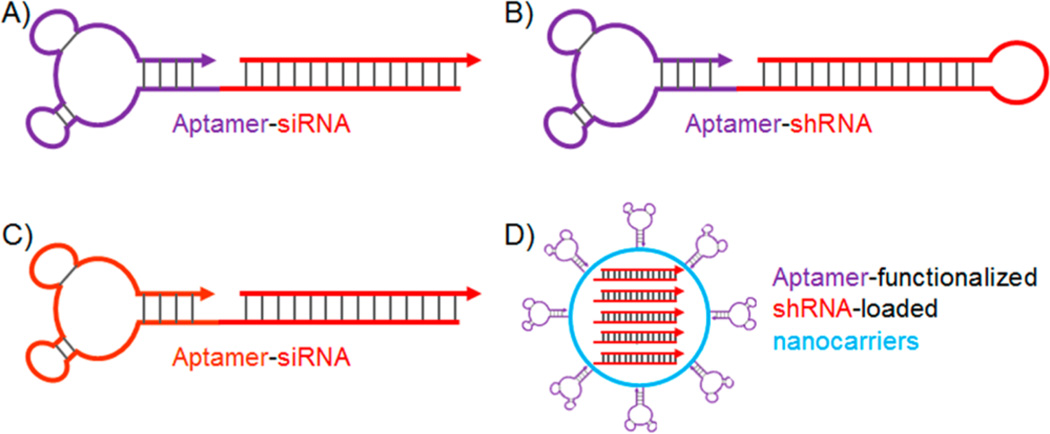
Again, by taking advantage of the simplicity of programmable nucleic acid engineering, aptamers have been widely explored to be conjugated with small interfering RNA (siRNA), small hairpin RNA (shRNA), and microRNA (miRNA) for targeted delivery of these therapeutics for RNAi-based gene therapy of diseases such as cancer and acquired immune deficiency syndrome (AIDS).55, 123–126 In one exceptional example, McNamara II et al. developed aptamer-siRNA chimeric RNAs for specific binding of target cancer cells and subsequent specific delivery of siRNA into target cells (Figure 3A).125 Particularly, PSMA specific A10 RNA aptamer was conjugated with siRNA against polo-like kinase 1 (Plk1) and B-cell lymphoma 2 (Bcl2). In the study using LNCaP cells that overexpress PSMA, the aptamer-siRNA conjugates (i.e., A10-Plk1 and A10-Bcl2) were internalized in the absence of transfection agents, followed by the processing of siRNA by Dicer and eventually led to the silencing of these survival genes and inducing apoptosis in LNCaP cancer cells. These conjugates did not induce apoptosis in PC-3 cells, which did not overexpress PSMA, suggesting aptamer-mediated potency. These aptamer–siRNA conjugates, when intratumorally injected to LNCaP tumor mice, specifically inhibited tumor growth. Further, Dassie et al. from the same group optimized A10-Plk1 for systemic injection, which showed improved regression of LNCaP tumors.126 In particular, these optimizations included (1) truncation of aptamer from 71 (A10) to 39 (A10–3.2) nucleotides in order to reduce the synthesis and treatment cost; (2) modification with 2′-fluoropyrimidines to increase the biostability; (3) systematic engineering of the A10–3.2-Plk1 conjugate to enhance the processing efficiency by RNA-induced silencing complex (RISC) machinery and further increase the silencing activity and specificity; and (4) modification of the conjugate with a 20 kDa PEG to increase the in vivo circulation half-life. As a benefit of these optimizations, the A10–3.2-Plk1 conjugate silenced the Plk1 gene at a reduced dosage over a prolonged period. In a similar manner, A10-Bcl2 aptamer-siRNA conjugate was selectively delivered into HER2-positive breast cancer cells, in which the expression of anti-apoptotic Bcl2 was suppressed, leading to the sensitization of these cells to the chemotherapeutic drug cisplatin.55
Figure 3.
ApDCs developed for targeted delivery of nucleic acid therapeutics in the treatment of cancer, AIDS, as well as metabolic skeletal disorders. (A) Chimeric aptamer–siRNA as ApDC for targeted delivery of siRNA and silencing of Plk1 and Bcl2 in PSMA-positive prostate cancer. (B) Chimeric aptamer–shRNA as ApDC for targeted delivery of shRNA and silencing of DNA-activated protein kinase (DNAPK) as a target for radiosensitization, which resulted in enhanced therapeutic response of PSMA-positive prostate tumor to ionization radiation. (C) ApDC developed for targeted delivery of (1) gp120-targeting aptamers and (2) siRNA for silencing HIV-1 tat/rev common exon sequence, for dual inhibition of HIV infection in T lymphocytes. (D) Lipid nanoparticulate ApDCs, CH6-siRNA-LNPs, for targeted silencing of osteogenic pleckstrin homology domain-containing family O member 1 (Plekho1) gene in osteoblast. (Reprinted with permission from refs 66 (copyright 2015 Macmillan Publishers Limited), 106 (copyright 2011 American Society for Clinical Investigation), 110 (copyright 2006 Macmillan Publishers Limited), and 115 (copyright 2008 American Society of Gene & Cell Therapy).)
Another form of RNAi agent, shRNA, has also attracted much intention and has been widely studied for gene therapy. In addition to shRNA generated from transfected plasmids or viral genome for a relatively prolonged gene regulation, synthetic shRNA is also a class of potent gene regulator for transient gene regulation.106, 127 In a similar manner to siRNA, shRNA has been engineered with aptamers to form aptamer–shRNA conjugates for targeted gene therapy. In one study reported by Ni et al., a truncated version of PSMA-targeting aptamer, A10–3, was conjugated with a shRNA that selectively sensitized PSMA-positive prostate cancer cells, but not surrounding normal tissues, to ionizing radiation (IR) during dose-escalated radiation therapy for localized prostate cancer (Figure 3B).106 Particularly, they identified a DNA-activated protein kinase (DNAPK) as a target for radiosensitization. Therefore, the DNAPK shRNA was selectively delivered by A10–3 into target prostate cancer cells and suppressed the expression of DNAPK in cultured prostate cancer cells, xenograft prostate tumors, and ex vivo human prostate tissues. More importantly, combined with IR, the aptamer–shRNA conjugate specifically enhanced the therapeutic response of PSMA-positive tumor to IR.
For intracellular delivery of nucleic acid therapeutics, endosome escape is a critical step for these therapeutics to reach cytoplasm or even nucleus and then execute therapeutic intervention. The mechanism for the endosome escape of the above aptamer–nucleic acid therapeutic conjugates has not been fully understood yet.55 The molecular pathway of the internalization and endosome escape of aptamers or ApDCs likely depends on the specific aptamer targets.
In addition to cancer therapy, aptamer–siRNA/shRNA conjugates have also been explored for the treatment of infectious diseases, such as AIDS, and for transfection of immune cells that are typically hard to transfect.110, 115, 127, 128 For example, Zhou et al. reported a cell-type specific delivery of aptamer-siRNA conjugate to dually inhibit HIV infection by both aptamers and siRNA in T lymphocytes (Figure 3C).110 The authors used an RNA aptamer that not only binds to, but also inhibits the biological functions of glycoprotein 120 (gp120), which is exposed on the surface of the HIV envelope and on the surfaces of HIV-1-infected cells. For more potent inhibition of HIV infection, a siRNA that inhibits HIV infection by targeting HIV-1 tat/rev common exon sequence was also fused with the anti-gp120 aptamer to construct an aptamer-siRNA conjugate.
Virus-based gene therapy has been extensively explored in the past decades. Despite the controversy over its safety, it is still promising for the treatment of many diseases. Similar to RNAi-based gene therapy and chemotherapy, the “off-target” effect is also often an issue in virus-based gene delivery. Tong et al. reported a method to covalently modify aptamers on the surfaces of viral capsids as multivalent drug delivery vehicles.83 This study provides insight into constructing aptamer-virus conjugates for targeted delivery of therapeutic viruses in gene therapy.
In addition, aptamer-functionalized nanoconjugates with siRNA/shRNA have offered a number of advantages, such as high payload capacity, protection of loaded drugs, as well as possible sustained release.66, 129, 130 In a recent study, Liang et al. reported the aptamer-mediated delivery of siRNA loaded in lipid nanoparticles (LNPs) selectively to osteoblast, as a potential bone anabolic strategy (Figure 3D).66 Particularly, an aptamer, CH6, was selected by cell-SELEX to specifically bind to both rat and human osteoblast. CH6 was functionalized onto LNPs that were loaded with osteogenic pleckstrin homology domain-containing family O member 1 (Plekho1) siRNA, and the CH6-siRNA-LNPs selectively delivered siRNA into osteoblast, mediated the uptake of the loaded siRNA into osteoblast, induced membrane ruffling and lysosome escape of siRNA, and subsequently induced specific silencing of Plekho1, which promoted bone formation, improved bone microarchitecture, increased bone mass, and eventually enhanced mechanical properties in rodents. It is noteworthy that the osteoblast-specific aptamer significantly decreased the uptake of nanotherapeutics in liver and kidney and enhanced the uptake in bone, which contributed to the enhanced therapeutic efficacy compared to the nontargeting counterparts.66
APTAMER–PROTEIN/PEPTIDE CONJUGATES
Proteins, as a main class of biologics, have tremendous potential as therapeutics or vaccines. Except for antibodies (e.g., HER2-targeting trastuzumab and EGFR-targeting cetuximab) or some peptides (e.g., GLP-1-targeting exendin-4), many other proteins or peptides lack the ability of specific targeting. It is thus desirable to guide them specifically to target diseased tissues or organs. In addition to the use of bivalent or chimeric antibodies, an alternative potential approach is to take advantage of aptamers’ ability of specific recognition for targeted delivery of therapeutic proteins or peptides.68, 100, 131
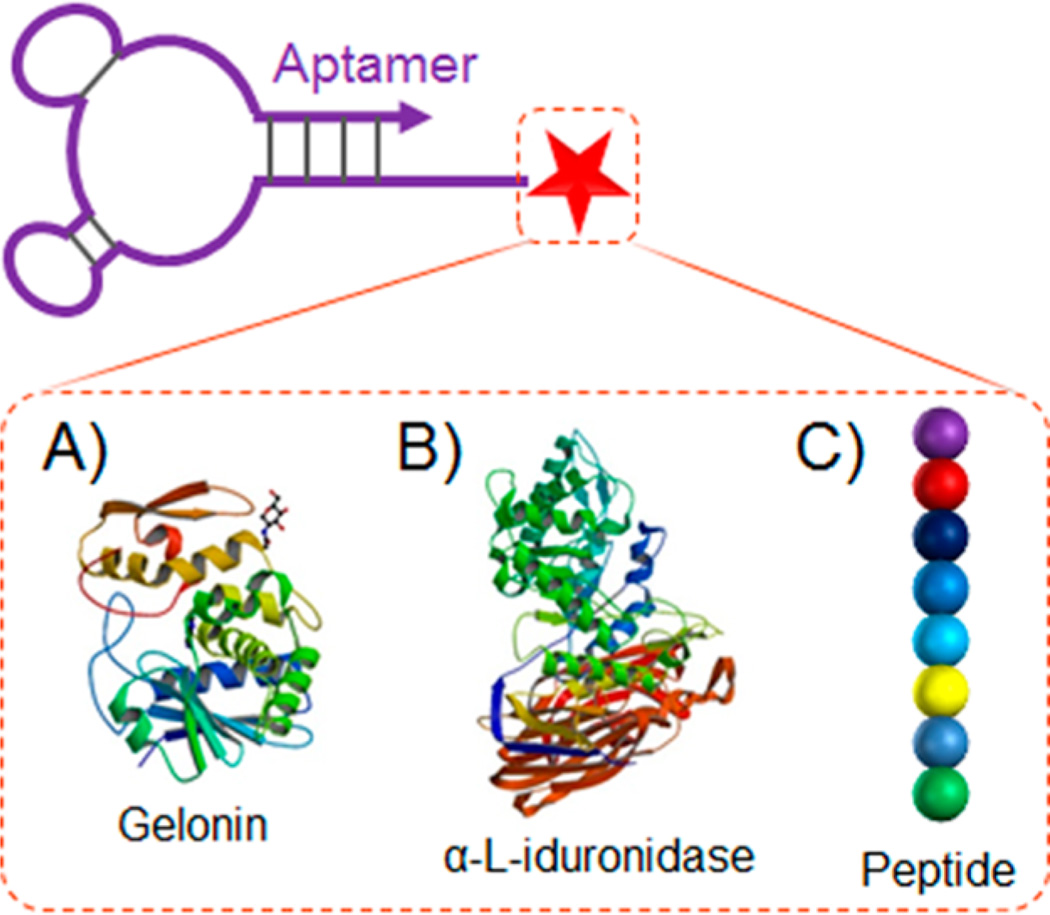
Toward this end, Chu et al. reported an aptamer-gelonin conjugate for targeted delivery of gelonin into PSMA-positive prostate cancer cells (Figure 4A).100 Gelonin is a small N-glycosidase and a ribosomal toxin that can cleave a specific glycosidic bond in ribosome RNA (rRNA) to disrupt protein synthesis and consequently induce cell death. Gelonin alone does not efficiently penetrate cells, thus demanding an external mechanism to facilitate the cell internalization. To do this, aptamer A10 was conjugated with recombinant gelonin (rGel). The A10-rGel conjugate selectively delivered rGel into PSMA-positive LNCaP cells (target), but not to PSMA-negative PC3 cells (nontarget). Compared with free gelonin, the toxicity of the conjugate was enhanced by 180-fold in target cells.
Figure 4.
ApDCs developed by conjugation of aptamers with therapeutic proteins, such as recombinant gelonin (A), lysosomal enzymes (B), and peptides, such as antigenic ovalbumin peptide SIINFEKL (C). (Reprinted with permission from refs 68 (copyright 2008 National Academy of Sciences), 100 (copyright 2006 American Association for Cancer Research), 111 (copyright 2014 American Society of Gene & Cell Therapy).)
Protein enzymes of therapeutic interest have also been developed into ApDCs and specifically delivered into cells for the treatment of lysosomal storage diseases.132 Lysosomal storage diseases are currently treated via enzyme replacement therapy, in which lysosomal enzymes are delivered into diseased cells through receptor-mediated endocytosis. However, a daunting challenge of the related therapeutics is the limited efficiency of enzyme uptake into cells. In an attempt to address this challenge, Chen et al. developed ApDCs by taking advantage of the natural transcytosis pathway (Figure 4B).68 Particularly, aptamers were selected to specifically bind to the extracellular domain of mouse transferrin receptor (TrR). One DNA aptamer, GS24, was verified to be able to hitchhike TrR and be taken up by fibroblasts. GS24 was then conjugated with α-l-iduronidase, a lysosomal enzyme. The resulting ApDC was taken up into lysosomes of fibroblasts by endocytosis, which eventually corrected defective glycosaminoglycan (GAG) degradation in these cells.
In another study, Wengerter et al. developed an aptamer-antigen conjugate and specifically delivered the attached ovalbumin peptide SIINFEKL, as a model antigen, to target CD8+ dendritic cells (Figure 4C).111 Aptamer-functionalized nanocarriers have also been explored for targeted delivery of proteins and peptides.
APTAMER–PHOTOSENSITIZER CONJUGATES
Photodynamic therapy (PDT) is an emerging treatment modality for a variety of diseases. It exploits the ability of photosensitizers to convert energy from photons to generate reactive oxygen species (ROS) from tissue oxygen.133 These ROS, including free radicals and singlet oxygen, are cytotoxic as they can attack and harm their surrounding biomolecules, leading to inhibition of cell proliferation, cell cycle arrest, apoptosis, or necrosis in cells. Even though the selectivity of the therapeutic activity can be controlled by light (using light to turn on photosensitization) to some extent, it is still demanding to specifically deliver photosensitizers into targeted diseased tissues or organs, especially those inside the human body that can be reached by light. Toward this end, disease biomarker-targeting aptamers are promising for targeted delivery of photosensitizers.67, 95, 134–140
In an early attempt, a photosensitizer, chlorin e6 (Ce6), was conjugated to aptamer TD05, which targets IgM µ heavy chain overexpressed in cancers such as B cell lymphoma (Figure 5A).97 TD05-Ce6 showed selective cytotoxicity in Ramos cancer cells, demonstrating the principle of ApDC-based PDT. In a more recent study, an aptamer-photosensitizer conjugate was developed using Ce6 and an RNA aptamer, AIR-3A, that specifically binds to interleukin 6 receptor (IL-6R).67 IL-6 and IL-6R are involved in various diseases ranging from inflammatory diseases such as Crohn’s disease, rheumatoid arthritis, psoriasis, systemic sclerosis, and some cancers. IL-6R-targeted drug delivery would thus be promising to selectively destroy the related targeted cells. Particularly in this study, the AIR-3A-ce6 conjugate was rapidly and specifically internalized by IL-6R-positive BaF3/gp130/IL6R/TNF cells. Compared to unconjugated Ce6, the aptamer-mediated internationalization of the conjugate also led to prolonged retention of the photosensitizers in target cells. Under light irradiation, targeted cells treated with AIR-3A-ce6 conjugates were selectively killed, in contrast to negligible cytotoxicity in target cells induced by free Ce6 or the negligible cytotoxicity in IL-6R-negative cells treated with AIR-3A-Ce6 conjugates. Therefore, by conjugation with aptamers, the photosensitizer showed cell-type selectivity in PDT, which is promising to reduce the “off-target” side effects.
Figure 5.
ApDCs for targeted delivery of photosensitizers in photodynamic therapy (PDT). (A) ApDC developed by direct conjugation of IgM-targeting aptamer, TD05, with photosensitizer Ce6, for targeted PDT. (B) Nanoparticulate ApDC developed by (1) modifying the exterior surfaces of engineered viral capsid (bacteriophage MS2) with PTK7-targeting aptamer sgc8 using an oxidative coupling reaction with an non-natural amino acid on capsids, and (2) modifying the interior capsid surfaces with photosensitizer porphyrins for PDT in leukemia cells. (Reprinted with permission from refs 92 (copyright 2010 American Chemical Society), and 97 (copyright John Wiley & Sons, Inc.).)
In addition to Ce6, other photosensitizers that have been studied for ApDC-based PDT include 5,10,15,20-tetrakis(1-methyl-4-pyridyl)-21H,23H–porphine (TMPyP4)89 and porphyrin.141 For example, TMPyP4 was encapsulated into a G-quadruplex, and the PTK7-targeting aptamer sgc8 was engineered to be fused with a TMPyP4-encapsulating G-quadruplex, resulting in a chimeric aptamer-TMPyP4 conjugate.89 The conjugate selectively delivered the carried photosensitizers into target cancer cells and, upon light irradiation, induced selective cytotoxicity in target cells, but not in nontarget cells.
A variety of aptamer–photosensitizer nanoconjugates have also been developed for targeted PDT.76, 81, 92, 137, 139, 142 These nanovehicles range from inorganic nanomaterials 76, 81, 142 to organic nanomaterials, including viral capsids.92 For instance, Stephanopoulos et al. dually modified bacteriophage MS2, a 27 nm spherical virus composed of 180 identical protein monomers, with aptamers and photosensitizers for targeted PDT (Figure 5B).92 Specifically, PTK7-targeting aptamer sgc8 was modified on the exterior surfaces of MS2, resulting in a targeted multivalent vehicle with about 20 copies of aptamers on each MS2 using an oxidative coupling reaction targeting an unnatural amino acid on MS2. The interior surfaces of MS2 were modified with porphyrins, with approximately 180 porphyrins loaded in one MS2 capsid, for the generation of cytotoxic singlet oxygen during PDT. The dual-modified nanoconjugate was applied for selective PDT in PTK7-overexpressing Jurkat leukemia cells.
APTAMER–PHOTOTHERMAL AGENT CONJUGATES
In addition to serving as drug carriers, nanomaterials can also serve as therapeutics and be conjugated with aptamers for ApDC nanoconjugates in targeted therapy. In this regard, photothermal nanomaterials have been extensively studied for targeted photothermal therapy (PTT). PTT exploits the ability of the related therapeutic materials to generate heat upon electromagnetic irradiation and induce toxicity by hyperthermia. Photothermal nanomaterials include gold nanomaterials143, 144 (e.g., nanorods, nanoshells, nanocages, nanoparticles, and larger-scale nanocomplexes), polydopamine-containing nanomaterials,145 carbon nanomaterials146 (e.g., nanographene, nanographene-oxide, and carbon nanotubes), and hybrid nanocomplexes147 (e.g., gold-graphene oxide complexes). These materials are generally amenable for bioconjugation with aptamers for active targeted delivery, in combination with EPR effect-mediated passive targeting in targeted PTT.
Huang et al. have developed conjugates of sgc8 and gold nanorods (AuNRs). AuNRs have a high hyperthermia effect due to high absorption efficiency of near-infrared (NIR) light.87 The sgc8-AuNR conjugate selectively bound to CEM cells and induced selective cytotoxicity by hyperthermia. Due to the strong surface plasmon absorption in the NIR region, AuNRs have also been utilized simultaneously as (1) efficient energy quenchers to control the selective action of photosensitizers, and (2) photothermal agents. As such, the AuNR–photo-sensitizer conjugates can be used for combinatorial PDT/PTT. In this manner, Wang et al. designed and synthesized aptamer-AuNR-Ce6 complexes.76 In these complexes, aptamers were engineered to aptamer switch probes (ASPs) comprising aptamers, linkers, and aptamer cDNA. One end of ASP was conjugated on the surfaces of AuNRs, and the other end was modified with Ce6, such that, in the absence of target cells, Ce6 was brought into close proximity and quenched by the gold surface, and the conjugate showed negligible phototoxicity due to an effective energy transfer quenching from the excited Ce6 to the AuNR. However, upon aptamer binding to target cells, the ASP changed conformation to switch Ce6 away from the gold surfaces, thereby producing singlet oxygen upon light irradiation for PDT. Furthermore, light irradiation also enabled AuNRs to induce hyperthermia effect. The combined hyperthermia and photodynamic effects led to an enhanced therapeutic potency in combined PTT/PDT, thus presenting a promising therapeutic regimen in targeted cancer therapy.
CHALLENGES OF APTAMERS AND APDCS
Alternatively known as chemical antibodies, nucleic acid aptamers have an array of characteristic advantages over their counterparts, especially antibodies.17, 113 Current technologies allow facile and simple aptamer screening within days to a few months; aptamers can be generated to some targets that are difficult or impossible to generate antibodies, for which some examples include toxins or less-immunogenic antigens; current solid-phase nucleic acid synthesis technologies allow easy and cost-effective aptamer manufacture with little batch-to-batch variation and, more impressively, a wide array of site-specific customized modifications either during or after synthesis for optimization of chemistry, pharmacological properties, and therapeutic abilities; aptamers are typically less susceptible to irreversible thermal and chemical (except for nucleases) denaturation than antibodies owing to their ability for easy conformational reconfiguration, enabling aptamer-based products with likely long shelf-life for storage and transportation at ambient temperature. In comparison to antibodies, the generally small sizes of aptamers endow them with faster tissue penetration, which is critical for therapeutic applications in targeted drug delivery to solid tissues; and finally, aptamers typically have negligible immunogenicity, ruling out the risk of side effects resulting from adverse immune responses. Driven by these unique features, the past few decades have witnessed the rapid advancement in development of aptamers and ApDCs for targeted therapy. In ApDCs, aptamers provide drugs with specific targeting ability, and in turn, the conjugated drugs enhance the therapeutic potency of aptamers.
Nonetheless, despite these attractive features, the development of aptamers or ApDCs for targeted therapy has been lagging. Until now, only one aptamer-based drug, pegaptanib (marketed as Macugen), has been approved by the US FDA. Pegaptanib is a PEGylated anti-VEGF aptamer for the treatment of age-related macular degeneration (AMD). Other than pegaptanib, the most advanced aptamer for cancer therapy, AS1411, is currently under Phase II investigation.44 The slow development of aptamers or ApDCs for therapeutic applications could be attributed to some limitations associated with conventional aptamers or ApDCs. First, aptamers generated using conventional SELEX technology may not have potent therapeutic effects, although they have high binding affinities. Conventional SELEX technology or the derived technologies screen aptamers solely on the binding abilities, without the assessment of their abilities to regulate biological functions. Consequently, the obtained aptamers are likely to bind tightly to cognate targets but may have suboptimal therapeutic effects. For targeted therapy, ApDCs are promising for addressing this limitation by conjugating potent therapeutics to aptamers. Second, nucleic acid aptamers and ApDCs are often susceptible to cleavage by ubiquitous nucleases in vivo.148 Fortunately, the simplicity of customized modifications on nucleic acids has enabled a number of approaches to prevent the fast degradation of nucleic acid aptamers and ApDCs. These approaches can be mainly classified into two categories: (1) chemical modification of nucleic acids with nuclease-resistant moieties,17, 148–150 such as 2′-O-methyl and 2′-fluoro on nucleotides, internuleotide phosphorothioated linkers, terminal PEGylations, and l-nucleotides (in the case of Spiegelmers); and (2) protection of nucleic acids using larger-scale shields, such as nanomaterials that encapsulate nucleic acids or immobilize nucleic acids on the surfaces.79, 151 Third, unmodified nucleic acids or ApDCs generally have suboptimal pharmacokinetics (PK) and biodistribution resulting from short serum half-lives associated with nuclease susceptibility, fast blood clearance, renal filtration, and uptake by reticuloendothelial system (RES) organs such as liver and spleen. Even though the short blood half-lives of systemically administered nucleic acid aptamers are beneficial to generate high signal-to-background contrast in whole body imaging,152 they also result in suboptimal efficacy of drug delivery into target tissues.62 To address this issue, modification of aptamers with chemical moieties or nanomaterials has again been leveraged. For example, aptamers modified with PEG (40 kDa) and 2′-omethyl resulted in prolonged half-lives.17, 149, 150, 153 This progress holds promise for overcoming the above challenges and developing aptamers and ApDCs for targeted therapy.
CONCLUDING REMARKS
Extensive genome sequencing and proteomic exploration in the past few decades have generated a wealth of information at the genomic and proteomic levels. Particularly for disease biology, a large part of the information is the mutations at the genetic level and correspondingly the changes (overexpression or downregulation) of certain biomolecules in a particular disease. The overexpressed biomolecules, often proteins, are used as biomarkers not only to “profile” a disease, but also for specific delivery of therapeutics into biomarker-overexpressing cells or tissues or organs. In addition to small molecule targeted therapeutics that act by specifically regulating disease-associated biomarkers, the conjugates of less selective therapeutics with biomarker-targeting ligands have emerged as an alternatively promising approach for targeted therapy. Aptamers and antibodies are two classes of these ligands. For targeted therapy, aptamers and ApDCs are attractive owing to such attributes as high binding affinity and selectivity, low manufacturing cost, high thermal and chemical stability or conformational reconfiguration, low immunogenicity, and low molecular weight and fast tissue penetration.
Meanwhile, many challenges for therapeutic aptamers and ApDCs have come to the forefront, particularly issues of manufacture cost, therapeutic formulation, biostability, bio-availability, and pharmacokinetics. Nonetheless, the attractive advantages of aptamers over their counterparts, including antibodies, still offer tremendous opportunities for ApDCs. For instance, the development of ADCs has been of tremendous interest in both academia and the pharmaceutical industry. The development of ADCs typically involves relatively complicated and costly genetic engineering, whereas the engineering of ApDCs is fairly straightforward owing to facile, cost-effective, and well-defined modifications and high programmability. With further understanding and optimization of aptamers and ApDCs, ApDCs are promising to play an important role in the next generation of targeted therapeutics.
Footnotes
Notes
The authors declare no competing financial interest.
REFERENCES
- 1.American Cancer Society. Cancer Facts & Figures 2013. 2013. [Google Scholar]
- 2.Ducry L, Stump B. Antibody-Drug Conjugates: Linking Cytotoxic Payloads to Monoclonal Antibodies. Bioconjugate Chem. 2009;21:5–13. doi: 10.1021/bc9002019. [DOI] [PubMed] [Google Scholar]
- 3.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 4.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 5.Mosing RK, Mendonsa SD, Bowser MT. Capillary Electrophoresis-SELEX Selection of Aptamers with Affinity for HIV-1 Reverse Transcriptase. Anal. Chem. 2005;77:6107–6112. doi: 10.1021/ac050836q. [DOI] [PubMed] [Google Scholar]
- 6.Mosing RK, Bowser MT. Microfluidic selection and applications of aptamers. J. Sep. Sci. 2007;30:1420–1426. doi: 10.1002/jssc.200600483. [DOI] [PubMed] [Google Scholar]
- 7.Mayer G, Ahmed M-SL, Dolf A, Endl E, Knolle PA, Famulok M. Fluorescence-activated cell sorting for aptamer SELEX with cell mixtures. Nat. Protoc. 2010;5:1993–2004. doi: 10.1038/nprot.2010.163. [DOI] [PubMed] [Google Scholar]
- 8.Huizenga DE, Szostak JW. A DNA Aptamer That Binds Adenosine and ATP. Biochemistry. 1995;34:656–665. doi: 10.1021/bi00002a033. [DOI] [PubMed] [Google Scholar]
- 9.Hermann T, Patel DJ. Adaptive Recognition by Nucleic Acid Aptamers. Science. 2000;287:820–825. doi: 10.1126/science.287.5454.820. [DOI] [PubMed] [Google Scholar]
- 10.Mallikaratchy P, Stahelin RV, Cao Z, Cho W, Tan W. Selection of DNA ligands for protein kinase C-delta. Chem. Commun. 2006:3229–3231. doi: 10.1039/b604778e. [DOI] [PubMed] [Google Scholar]
- 11.Daniels DA, Chen H, Hicke BJ, Swiderek KM, Gold L. A tenascin-C aptamer identified by tumor cell SELEX: Systematic evolution of ligands by exponential enrichment. Proc. Natl. Acad. Sci. U.S.A. 2003;100:15416–15421. doi: 10.1073/pnas.2136683100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parekh P, Tang Z, Turner PC, Moyer RW, Tan W. Aptamers Recognizing Glycosylated Hemagglutinin Expressed on the Surface of Vaccinia Virus-Infected Cells. Anal. Chem. 2010;82:8642–8649. doi: 10.1021/ac101801j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bock LC, Griffin LC, Latham JA, Vermaas EH, Toole JJ. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature. 1992;355:564–566. doi: 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- 14.Hu J, Wu J, Li C, Zhu L, Zhang WY, Kong G, Lu Z, Yang CJ. A G-Quadruplex Aptamer Inhibits the Phosphatase Activity of Oncogenic Protein Shp2 in vitro. ChemBioChem. 2011;12:424–430. doi: 10.1002/cbic.201000470. [DOI] [PubMed] [Google Scholar]
- 15.Chen C-h. B, Chernis GA, Hoang VQ, Landgraf R. Inhibition of heregulin signaling by an aptamer that preferentially binds to the oligomeric form of human epidermal growth factor receptor-3. Proc. Natl. Acad. Sci. U.S.A. 2003;100:9226–9231. doi: 10.1073/pnas.1332660100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khati M, Schüman M, Ibrahim J, Sattentau Q, Gordon S, James W. Neutralization of Infectivity of Diverse R5 Clinical Isolates of Human Immunodeficiency Virus Type 1 by gp120-Binding 2′F-RNA Aptamers. J. Virol. 2003;77:12692–12698. doi: 10.1128/JVI.77.23.12692-12698.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keefe AD, Pai S, Ellington A. Aptamers as therapeutics. Nat. Rev. Drug Discovery. 2010;9:537–550. doi: 10.1038/nrd3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo K-T, SchÄfer R, Paul A, Gerber A, Ziemer G, Wendel HP. A New Technique for the Isolation and Surface Immobilization of Mesenchymal Stem Cells from Whole Bone Marrow Using High-Specific DNA Aptamers. Stem Cells. 2006;24:2220–2231. doi: 10.1634/stemcells.2006-0015. [DOI] [PubMed] [Google Scholar]
- 19.Shangguan D, Li Y, Tang Z, Cao Z, Chen HW, Mallikaratchy P, Sefah K, Yang CJ, Tan W. Aptamers evolved from live cells as effective molecular probes for cancer study. Proc. Natl. Acad. Sci. U.S.A. 2006;103:11838–11843. doi: 10.1073/pnas.0602615103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang Z, Shangguan D, Wang K, Shi H, Sefah K, Mallikaratchy P, Chen HW, Li Y, Tan W. Selection of aptamers for molecular recognition and characterization of cancer cells. Anal. Chem. 2007;79:4900–4907. doi: 10.1021/ac070189y. [DOI] [PubMed] [Google Scholar]
- 21.Shangguan D, Meng L, Cao ZC, Xiao Z, Fang X, Li Y, Cardona D, Witek RP, Liu C, Tan W. Identification of liver cancer-specific aptamers using whole live cells. Anal. Chem. 2008;80:721–728. doi: 10.1021/ac701962v. [DOI] [PubMed] [Google Scholar]
- 22.Sefah K, Tang Z, Shangguan D, Chen H, Lopez-Colon D, Li Y, Parekh P, Martin J, Meng L, Phillips JA, et al. Molecular recognition of acute myeloid leukemia using aptamers. Leukemia. 2009;23:235–244. doi: 10.1038/leu.2008.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen HW, Medley CD, Sefah K, Shangguan D, Tang Z, Meng L, Smith JE, Tan W. Molecular recognition of small-cell lung cancer cells using aptamers. ChemMedChem. 2008;3:991–1001. doi: 10.1002/cmdc.200800030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bayrac AT, Sefah K, Parekh P, Bayrac C, Gulbakan B, Oktem HA, Tan W. In Vitro Selection of DNA Aptamers to Glioblastoma Multiforme. ACS Chem. Neurosci. 2011;2:175–181. doi: 10.1021/cn100114k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simaeys DV, López-Colón D, Sefah K, Sutphen R, Jimenez E, Tan W. Study of the Molecular Recognition of Aptamers Selected through Ovarian Cancer Cell-SELEX. PLoS One. 2010;5 doi: 10.1371/journal.pone.0013770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao Z, Xu L, Shi X, Tan W, Fang X, Shangguan D. Recognition of subtype non-small cell lung cancer by DNA aptamers selected from living cells. Analyst. 2009;134:1808–1814. doi: 10.1039/b904476k. [DOI] [PubMed] [Google Scholar]
- 27.Sefah K, Yang Z, Bradley KM, Hoshika S, Jimenez E, Zhang L, Zhu G, Shanker S, Yu F, Turek D, et al. In vitro selection with artificial expanded genetic information systems. Proc. Natl. Acad. Sci. USA. 2014;111:1449–1454. doi: 10.1073/pnas.1311778111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O’Donoghue M, Shi X, Fang X, Tan W. Single-molecule atomic force microscopy on live cells compares aptamer and antibody rupture forces. Anal. Bioanal. Chem. 2012;402:3205–3209. doi: 10.1007/s00216-011-5667-y. [DOI] [PubMed] [Google Scholar]
- 29.Padmanabhan K, Padmanabhan KP, Ferrara JD, Sadler JE, Tulinsky A. The structure of alpha-thrombin inhibited by a 15-mer single-stranded DNA aptamer. J. Biol. Chem. 1993;268:17651–17654. doi: 10.2210/pdb1hut/pdb. [DOI] [PubMed] [Google Scholar]
- 30.Long SB, Long MB, White RR, Sullenger BA. Crystal structure of an RNA aptamer bound to thrombin. RNA. 2008;14:2504–2512. doi: 10.1261/rna.1239308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lebars I, Legrand P, Aime A, Pinaud N, Fribourg S, Di Primo C. Exploring TAR-RNA aptamer loop-loop interaction by X-ray crystallography, UV spectroscopy and surface plasmon resonance. Nucleic Acids Res. 2008;36:7146–7156. doi: 10.1093/nar/gkn831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang DB, Vu D, Cassiday LA, Zimmerman JM, Maher LJ, Ghosh G. Crystal structure of NF-kappa B (p50)(2) complexed to a high-affinity RNA aptamer. Proc. Natl. Acad. Sci. U.SA. 2003;100:9268–9273. doi: 10.1073/pnas.1632011100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Green LS, Bell C, Janjic N. Aptamers as reagents for high-throughput screening. Biotechniques. 2001;30:1094. doi: 10.2144/01305dd02. [DOI] [PubMed] [Google Scholar]
- 34.Srivatsan SG, Famulok M. Functional nucleic acids in high throughput screening and drug discovery. Comb. Chem. High Throughput Screening. 2007;10:698–705. doi: 10.2174/138620707782507359. [DOI] [PubMed] [Google Scholar]
- 35.Yamazaki S, Tan L, Mayer G, Hartig JS, Song J-N, Reuter S, Restle T, Laufer SD, Grohmann D, Kraeusslich, et al. Alternative small-molecule target sites aptamer displacement identifies that escape viral resistance. Chem. Biol. 2007;14:804–812. doi: 10.1016/j.chembiol.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 36.Hafner M, Vianini E, Albertoni B, Marchetti L, Gruene I, Gloeckner C, Famulok M. Displacement of protein-bound aptamers with small molecules screened by fluorescence polarization. Nat. Protoc. 2008;3:579–587. doi: 10.1038/nprot.2008.15. [DOI] [PubMed] [Google Scholar]
- 37.Famulok M. Exploring Chemical Space with Aptamers. J. Med. Chem. 2009;52:6951–6957. doi: 10.1021/jm9014789. [DOI] [PubMed] [Google Scholar]
- 38.Mayer G, Faulhammer D, Graettinger M, Fessele S, Blind M. A RNA-Based Approach towards Small-Molecule Inhibitors. ChemBioChem. 2009;10:1993–1996. doi: 10.1002/cbic.200900325. [DOI] [PubMed] [Google Scholar]
- 39.Yamazaki S, Famulok M. Screening of Novel Inhibitors of HIV-1 Reverse Transcriptase with a Reporter Ribozyme Assay. In: Mayer G, editor. Methods in Molecular Biology. 2009. pp. 187–199. [DOI] [PubMed] [Google Scholar]
- 40.Niebel B, Lentz C, Pofahl M, Mayer G, Hoerauf A, Pfarr KM, Famulok M. ADLOC: An Aptamer-Displacement Assay Based on Luminescent Oxygen Channeling. Chem.—Eur. J. 2010;16:11100–11107. doi: 10.1002/chem.201001192. [DOI] [PubMed] [Google Scholar]
- 41.Auslaender D, Wieland M, Auslaender S, Tigges M, Fussenegger M. Rational design of a small molecule-responsive intramer controlling transgene expression in mammalian cells. Nucleic Acids Res. 2011;39 doi: 10.1093/nar/gkr829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mueller M, Ackermann D, Famulok M. Nucleic acid based tools for pharmacology and nano-engineering. C. R. Chim. 2011;14:819–825. [Google Scholar]
- 43.Gragoudas ES, Adamis AP, Cunningham ET, Feinsod M, Guyer DR. Pegaptanib for Neovascular Age-Related Macular Degeneration. N. Engl. J. Med. 2004;351:2805–2816. doi: 10.1056/NEJMoa042760. [DOI] [PubMed] [Google Scholar]
- 44.Ireson CR, Kelland LR. Discovery and development of anticancer aptamers. Mol. Cancer Ther. 2006;5:2957–2962. doi: 10.1158/1535-7163.MCT-06-0172. [DOI] [PubMed] [Google Scholar]
- 45.Bates PJ, Laber DA, Miller DM, Thomas SD, Trent JO. Discovery and development of the G-rich oligonucleotide AS1411 as a novel treatment for cancer. Exp. Mol. Pathol. 2009;86:151–164. doi: 10.1016/j.yexmp.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shangguan D, Cao Z, Meng L, Mallikaratchy P, Sefah K, Wang H, Li Y, Tan W. Cell-specific aptamer probes for membrane protein elucidation in cancer cells. J. Proteome Res. 2008;7:2133–2139. doi: 10.1021/pr700894d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mallikaratchy P, Tang Z, Kwame S, Meng L, Shangguan D, Tan W. Aptamer Directly Evolved from Live Cells Recognizes Membrane Bound Immunoglobin Heavy Mu Chain in Burkitt’s Lymphoma Cells. Mol. Cell. Proteomics. 2007;6:2230–2238. doi: 10.1074/mcp.M700026-MCP200. [DOI] [PubMed] [Google Scholar]
- 48.Lupold SE, Hicke BJ, Lin Y, Coffey DS. Identification and Characterization of Nuclease-stabilized RNA Molecules That Bind Human Prostate Cancer Cells via the Prostate-specific Membrane Antigen. Cancer Res. 2002;62:4029–4033. [PubMed] [Google Scholar]
- 49.Ferreira CSM, Matthews CS, Missailidis S. DNA aptamers that bind to MUC1 tumour marker: Design and characterization of MUC1-binding single-stranded DNA aptamers. Tumor Biol. 2006;27:289–301. doi: 10.1159/000096085. [DOI] [PubMed] [Google Scholar]
- 50.Mi J, Zhang X, Giangrande PH, McNamara Ii JO, Nimjee SM, Sarraf-Yazdi S, Sullenger BA, Clary BM. Targeted inhibition of αvβ3 integrin with an RNA aptamer impairs endothelial cell growth and survival. Biochem. Biophys. Res. Commun. 2005;338:956–963. doi: 10.1016/j.bbrc.2005.10.043. [DOI] [PubMed] [Google Scholar]
- 51.Lebruska LL, Maher LJ. Selection and Characterization of an RNA Decoy for Transcription Factor NF-kB. Biochemistry. 1999;38:3168–3174. doi: 10.1021/bi982515x. [DOI] [PubMed] [Google Scholar]
- 52.Martell RE, Nevins JR, Sullenger BA. Optimizing Aptamer Activity for Gene Therapy Applications Using Expression Cassette SELEX. Mol. Ther. 2002;6:30–34. doi: 10.1006/mthe.2002.0624. [DOI] [PubMed] [Google Scholar]
- 53.Ishizaki J, Nevins JR, Sullenger BA. Inhibition of cell proliferation by an RNA ligand that selectively blocks E2F function. Nat. Med. 1996;2:1386–1389. doi: 10.1038/nm1296-1386. [DOI] [PubMed] [Google Scholar]
- 54.Chi-hong BC, George AC, Van QH, Ralf L. Inhibition of heregulin signaling by an aptamer that preferentially binds to the oligomeric form of human epidermal growth factor receptor-3. Proc. Natl. Acad. Sci. U.SA. 2003;100:9226–9231. doi: 10.1073/pnas.1332660100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thiel KW, Hernandez LI, Dassie JP, Thiel WH, Liu X, Stockdale KR, Rothman AM, Hernandez FJ, McNamara JO, Giangrande PH. Delivery of chemo-sensitizing siRNAs to HER2+-breast cancer cells using RNA aptamers. Nucleic Acids Res. 2012;40:6319–6337. doi: 10.1093/nar/gks294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Varmira K, Hosseinimehr SJ, Noaparast Z, Abedi SM. A HER2-targeted RNA aptamer molecule labeled with 99mTc for single-photon imaging in malignant tumors. Nucl. Med. Biol. 2013;40:980–986. doi: 10.1016/j.nucmedbio.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 57.Varmira K, Hosseinimehr SJ, Noaparast Z, Abedi SM. An improved radiolabelled RNA aptamer molecule for HER2 imaging in cancers. J. Drug Target. 2014;22:116–122. doi: 10.3109/1061186X.2013.839688. [DOI] [PubMed] [Google Scholar]
- 58.Wang D-L, Song Y-L, Zhu Z, Li X-L, Zou Y, Yang H-T, Wang J-J, Yao P-S, Pan R-J, Yang CJ, Kang D-Z. Selection of DNA aptamers against epidermal growth factor receptor with high affinity and specificity. Biochem. Biophys. Res. Commun. 2014;453:681–685. doi: 10.1016/j.bbrc.2014.09.023. [DOI] [PubMed] [Google Scholar]
- 59.Esposito CL, Passaro D, Longobardo I, Condorelli G, Marotta P, Affuso A, de Franciscis V, Cerchia L. A neutralizing RNA aptamer against EGFR causes selective apoptotic cell death. PLoS One. 2010;6 doi: 10.1371/journal.pone.0024071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Carla Lucia E, Diana P, Immacolata L, Gerolama C, Pina M, Andrea A, Vittorio de F, Laura C. A Neutralizing RNA Aptamer against EGFR Causes Selective Apoptotic Cell Death. PLoS One. 2011;6 doi: 10.1371/journal.pone.0024071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Song Y, Zhu Z, An Y, Zhang W, Zhang H, Liu D, Yu C, Duan W, Yang CJ. Selection of DNA aptamers against epithelial cell adhesion molecule for cancer cell imaging and circulating tumor cell capture. Anal. Chem. 2013;85:4141–4149. doi: 10.1021/ac400366b. [DOI] [PubMed] [Google Scholar]
- 62.Xiang D, Shigdar S, Qiao G, Wang T, Kouzani AZ, Zhou S-FF, Kong L, Li Y, Pu C, Duan W. Nucleic acid aptamer-guided cancer therapeutics and diagnostics: the next generation of cancer medicine. Theranostics. 2014;5:23–42. doi: 10.7150/thno.10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang P, Zhao N, Zeng Z, Feng Y, Tung C-HH, Chang C-CC, Zu Y. Using an RNA aptamer probe for flow cytometry detection of CD30-expressing lymphoma cells. Lab. Invest. 2009;89:1423–1432. doi: 10.1038/labinvest.2009.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Blake CM, Sullenger BA, Lawrence DA, Fortenberry YM. Antimetastatic Potential of PAI-1-Specific RNA Aptamers. Oligonucleotides. 2009;19:117–128. doi: 10.1089/oli.2008.0177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wiegand T, Williams P, Dreskin S, Jouvin M, Kinet J, Tasset D. High-affinity oligonucleotide ligands to human IgE inhibit binding to Fc epsilon receptor I. J. Immunol. 1996;157:221–230. [PubMed] [Google Scholar]
- 66.Liang C, Guo B, Wu H, Shao N, Li D, Liu J, Dang L, Wang C, Li H, Li S, et al. Aptamer-functionalized lipid nanoparticles targeting osteoblasts as a novel RNA interference-based bone anabolic strategy. Nat. Med. 2015;21:288–294. doi: 10.1038/nm.3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kruspe S, Meyer C, Hahn U. Chlorin e6 conjugated interleukin-6 receptor aptamers selectively kill target cells upon irradiation. Mol. Ther. 2014;3:e143. doi: 10.1038/mtna.2013.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen C-h. B. H, Dellamaggiore KR, Ouellette CP, Sedano CD, Lizadjohry M, Chernis GA, Gonzales M, Baltasar FE, Fan AL, Myerowitz R, Neufeld EF. Aptamer-based endocytosis of a lysosomal enzyme. Proc. Natl. Acad. Sci. USA. 2008;105:15908–15913. doi: 10.1073/pnas.0808360105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Eyetech Study Group. Preclinical and phase 1A clinical evaluation of an anti-VEGF pegylated aptamer (EYE001) for the treatment of exudative age-related macular degeneration. Retina. 2002;2:143–152. doi: 10.1097/00006982-200204000-00002. [DOI] [PubMed] [Google Scholar]
- 70.Rusconi CP, Roberts JD, Pitoc GA, Nimjee SM, White RR, Quick G, Scardino E, Fay WP, Sullenger BA. Antidote-mediated control of an anticoagulant aptamer in vivo. Nat. Biotechnol. 2004;22:1423–1428. doi: 10.1038/nbt1023. [DOI] [PubMed] [Google Scholar]
- 71.Oney S, Lam RTS, Bompiani KM, Blake CM, Quick G, Heidel JD, Liu JY-C, Mack BC, Davis ME, Leong KW, Sullenger BA. Development of universal antidotes to control aptamer activity. Nat. Med. 2009;15:1224–1228. doi: 10.1038/nm.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hughes B. Antibody-drug conjugates for cancer: poised to deliver? Nat. Rev. Drug Discovery. 2010;9:665–667. doi: 10.1038/nrd3270. [DOI] [PubMed] [Google Scholar]
- 73.Webb S. Pharma interest surges in antibody drug conjugates. Nat. Biotechnol. 2011;29:297–298. doi: 10.1038/nbt0411-297. [DOI] [PubMed] [Google Scholar]
- 74.Hu R, Zhang X, Zhao Z, Zhu G, Chen T, Fu T, Tan W. DNA nanoflowers for multiplexed cellular imaging and traceable targeted drug delivery. Angew. Chem., Int. Ed. 2014;53:5821–5826. doi: 10.1002/anie.201400323. [DOI] [PubMed] [Google Scholar]
- 75.Huang F, You M, Chen T, Zhu G, Liang H, Tan W. Self-assembled hybrid nanoparticles for targeted co-delivery of two drugs into cancer cells. Chem. Commun. 2014;50:3103–3105. doi: 10.1039/c3cc49003c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang J, Zhu G, You M, Song E, Shukoor MI, Zhang K, Altman MB, Chen Y, Zhu Z, Huang CZ, et al. Assembly of Aptamer Switch Probes and Photosensitizer on Gold Nanorods for Targeted Photothermal and Photodynamic Cancer Therapy. ACS Nano. 2012;6:5070–5077. doi: 10.1021/nn300694v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zheng J, Zhu G, Li Y, Li C, You M, Chen T, Song E, Yang R, Tan W. A spherical nucleic acid platform based on self-assembled DNA biopolymer for high-performance cancer therapy. ACS Nano. 2013;7:6545–6554. doi: 10.1021/nn402344v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhou C, Chen T, Wu C, Zhu G, Qiu L, Cui C, Hou W, Tan W. Aptamer CaCO3 nanostructures: a facile, pH-responsive, specific platform for targeted anticancer theranostics. Chem.—Asian J. 2014;10:166–171. doi: 10.1002/asia.201403115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhu G, Hu R, Zhao Z, Chen Z, Zhang X, Tan W. Noncanonical self-assembly of multifunctional DNA nanoflowers for biomedical applications. J. Am. Chem. Soc. 2013;135:16438–16445. doi: 10.1021/ja406115e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhu G, Zheng J, Song E, Donovan M, Zhang K, Liu C, Tan W. Self-assembled, aptamer-tethered DNA nano-trains for targeted transport of molecular drugs in cancer theranostics. Proc. Natl. Acad. Sci. U.SA. 2013;110:7998–8003. doi: 10.1073/pnas.1220817110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shieh Y-A, Yang S-J, Wei M-F, Shieh M-J. Aptamer-based tumor-targeted drug delivery for photodynamic therapy. ACS Nano. 2010;4:1433–1442. doi: 10.1021/nn901374b. [DOI] [PubMed] [Google Scholar]
- 82.Gu F, Zhang L, Teply BA, Mann N, Wang A, Radovic-Moreno AF, Langer R, Farokhzad OC. Precise engineering of targeted nanoparticles by using self-assembled biointegrated block copolymers. Proc. Natl. Acad. Sci. U.SA. 2008;105:2586–2591. doi: 10.1073/pnas.0711714105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tong GJ, Hsiao SC, Carrico ZM, Francis MB. Viral capsid DNA aptamer conjugates as multivalent cell-targeting vehicles. J. Am. Chem. Soc. 2009;131:11174–11178. doi: 10.1021/ja903857f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Douglas SM, Bachelet I, Church GM. A Logic-Gated Nanorobot for Targeted Transport of Molecular Payloads. Science. 2012;335:831–834. doi: 10.1126/science.1214081. [DOI] [PubMed] [Google Scholar]
- 85.Huang Y-F, Shangguan D, Liu H, Phillips JA, Zhang X, Chen Y, Tan W. Molecular Assembly of an Aptamer-Drug Conjugate for Targeted Drug Delivery to Tumor Cells. ChemBioChem. 2009;10:862–868. doi: 10.1002/cbic.200800805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kang H, O’Donoghue MB, Liu H, Tan W. A liposome-based nanostructure for aptamer directed delivery. Chem. Commun. 2010;46:249–251. doi: 10.1039/b916911c. [DOI] [PubMed] [Google Scholar]
- 87.Huang Y-F, Sefah K, Bamrungsap S, Chang H-T, Tan W. Selective Photothermal Therapy for Mixed Cancer Cells Using Aptamer-Conjugated Nanorods. Langmuir. 2008;24:11860–11865. doi: 10.1021/la801969c. [DOI] [PubMed] [Google Scholar]
- 88.Luo Y-L, Shiao Y-S, Huang Y-F. Release of Photoactivatable Drugs from Plasmonic Nanoparticles for Targeted Cancer Therapy. ACS Nano. 2011;5:7796–7804. doi: 10.1021/nn201592s. [DOI] [PubMed] [Google Scholar]
- 89.Wang K, You M, Chen Y, Han D, Zhu Z, Huang J, Williams K, Yang CJ, Tan W. Self-Assembly of a Bifunctional DNA Carrier for Drug Delivery. Angew. Chem., Int. Ed. 2011;50:6098–6101. doi: 10.1002/anie.201008053. [DOI] [PubMed] [Google Scholar]
- 90.Kang H, Trondoli AC, Zhu G, Chen Y, Chang Y-J, Liu H, Huang Y-F, Zhang X, Tan W. Near-Infrared Light-Responsive Core-Shell Nanogels for Targeted Drug Delivery. ACS Nano. 2011;5:5094–5099. doi: 10.1021/nn201171r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhu G, Meng L, Ye M, Yang L, Sefah K, O’Donoghue MB, Chen Y, Xiong X, Huang J, Song E, Tan W. Self-Assembled Aptamer-Based Drug Carriers for Bispecific Cytotoxicity to Cancer Cells. Chem—Asian J. 2012;7:1630–1636. doi: 10.1002/asia.201101060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Stephanopoulos N, Tong GJ, Hsiao SC, Francis MB. Dual-surface modified virus capsids for targeted delivery of photodynamic agents to cancer cells. ACS Nano. 2010;4:6014–6020. doi: 10.1021/nn1014769. [DOI] [PubMed] [Google Scholar]
- 93.Zhu G, Cansiz S, You M, Qiu L, Han D, Zhang L, Mei L, Fu T, Chen Z, Tan W. Nuclease-resistant synthetic drug-DNA adducts: programmable drug-DNA conjugation for targeted anticancer drug delivery. NPG Asia Materials. 2015;7 [Google Scholar]
- 94.Wang R, Zhu G, Mei L, Xie Y, Ma H, Ye M, Qing F-L, Tan W. Automated modular synthesis of aptamer-drug conjugates for targeted drug delivery. J. Am. Chem. Soc. 2014;136:2731–2734. doi: 10.1021/ja4117395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Han D, Zhu G, Wu C, Zhu Z, Chen T, Zhang X, Tan W. Engineering a cell-surface aptamer circuit for targeted and amplified photodynamic cancer therapy. ACS Nano. 2013;7:2312–2319. doi: 10.1021/nn305484p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wu Y, Sefah K, Liu H, Wang R, Tan W. DNA aptamer-micelle as an efficient detection/delivery vehicle toward cancer cells. Proc. Natl. Acad. Sci. U.SA. 2010;107:5–10. doi: 10.1073/pnas.0909611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mallikaratchy P, Tang Z, Tan W. Cell specific aptamer-photosensitizer conjugates as a molecular tool in photodynamic therapy. ChemMedChem. 2008;3:425–428. doi: 10.1002/cmdc.200700260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cao Z, Tong R, Mishra A, Xu W, Wong GCL, Cheng J, Lu Y. Reversible Cell-Specific Drug Delivery with Aptamer-Functionalized Liposomes. Angew. Chem., Int. Ed. 2009;48:6494–6498. doi: 10.1002/anie.200901452. [DOI] [PubMed] [Google Scholar]
- 99.Bagalkot V, Farokhzad OC, Langer R, Jon S. An Aptamer-Doxorubicin Physical Conjugate as a Novel Targeted Drug-Delivery Platform. Angew. Chem., Int. Ed. 2006;45:8149–8152. doi: 10.1002/anie.200602251. [DOI] [PubMed] [Google Scholar]
- 100.Chu TC, Marks JW, Lavery LA, Faulkner S, Rosenblum MG, Ellington AD, Levy M. Aptamer:toxin conjugates that specifically target prostate tumor cells. Cancer Res. 2006;66:5989–5992. doi: 10.1158/0008-5472.CAN-05-4583. [DOI] [PubMed] [Google Scholar]
- 101.McNamara JO, Andrechek ER, Wang Y, Viles KD, Rempel RE, Gilboa E, Sullenger BA, Giangrande PH. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat. Biotechnol. 2006;24:1005–1015. doi: 10.1038/nbt1223. [DOI] [PubMed] [Google Scholar]
- 102.Kim D, Jeong YY, Jon S. A Drug-Loaded Aptamer-Gold Nanoparticle Bioconjugate for Combined CT Imaging and Therapy of Prostate Cancer. ACS Nano. 2010;4:3689–3696. doi: 10.1021/nn901877h. [DOI] [PubMed] [Google Scholar]
- 103.Bagalkot V, Zhang L, Levy-Nissenbaum E, Jon S, Kantoff PW, Langer R, Farokhzad OC. Quantum Dot-Aptamer Conjugates for Synchronous Cancer Imaging, Therapy, and Sensing of Drug Delivery Based on Bi-Fluorescence Resonance Energy Transfer. Nano Lett. 2007;7:3065–3070. doi: 10.1021/nl071546n. [DOI] [PubMed] [Google Scholar]
- 104.Dhar S, Kolishetti N, Lippard SJ, Farokhzad OC. Targeted delivery of a cisplatin prodrug for safer and more effective prostate cancer therapy in vivo. Proc. Natl. Acad. Sci. USA. 2011;108:1850–1855. doi: 10.1073/pnas.1011379108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Dassie JP, Liu X-y, Thomas GS, Whitaker RM, Thiel KW, Stockdale KR, Meyerholz DK, McCaffrey AP, McNamara JO, Giangrande PH. Systemic administration of optimized aptamer-siRNA chimeras promotes regression of PSMA-expressing tumors. Nat. Biotechnol. 2009;27:839–846. doi: 10.1038/nbt.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ni X, Zhang Y, Ribas J, Chowdhury WH, Castanares M, Zhang Z, Laiho M, DeWeese TL, Lupold SE. Prostate-targeted radiosensitization via aptamer-shRNA chimeras in human tumor xenografts. J. Clin. Invest. 2011;121:2383–2390. doi: 10.1172/JCI45109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pieve CD, Perkins AC, Missailidis S. Anti-MUC1 aptamers: radiolabelling with 99mTc and biodistribution in MCF-7 tumour-bearing mice. Nucl. Med. Biol. 2009;36:703–710. doi: 10.1016/j.nucmedbio.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 108.Savla R, Taratula O, Garbuzenko O, Mnko T. Tumor targeted quantum dot-mucin 1 aptamer-doxorubicin conjugate for imaging and treatment of cancer. J. Controlled Release. 2011;153:16–22. doi: 10.1016/j.jconrel.2011.02.015. [DOI] [PubMed] [Google Scholar]
- 109.Tan L, Neoh KG, Kang E-T, Choe W-S, Su X. Designer tridentate mucin 1 aptamer for targeted drug delivery. J. Pharma. Sci. 2012;101:1672–1677. doi: 10.1002/jps.23101. [DOI] [PubMed] [Google Scholar]
- 110.Zhou J, Li H, Li S, Zaia J, Rossi JJ. Novel dual inhibitory function aptamer-siRNA delivery system for HIV-1 therapy. Mol. Ther. 2008;16:1481–1489. doi: 10.1038/mt.2008.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wengerter BC, Katakowski JA, Rosenberg JM, Park CG, Almo SC, Palliser D, Levy M. Aptamer-targeted antigen delivery. Mol. Ther. 2014;22:1375–1387. doi: 10.1038/mt.2014.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Meng L, Sefah K, O’Donoghue MB, Zhu G, Shangguan D, Noorali A, Chen Y, Zhou L, Tan W. Silencing of PTK7 in colon cancer cells: caspase-10-dependent apoptosis via mitochondrial pathway. PLoS One. 2010;5 doi: 10.1371/journal.pone.0014018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhu G, Ye M, Donovan MJ, Song E, Zhao Z, Tan W. Nucleic acid aptamers: an emerging frontier in cancer therapy. Chem. Commun. 2012;48:10472–10480. doi: 10.1039/c2cc35042d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bagalkot V, Farokhzad OC, Langer R, Jon S. An aptamer-doxorubicin physical conjugate as a novel targeted drug-delivery platform. Angew. Chem., Int. Ed. 2006;45:8149–8152. doi: 10.1002/anie.200602251. [DOI] [PubMed] [Google Scholar]
- 115.Zhu G, Meng L, Ye M, Yang L, Sefah K, O’Donoghue MB, Chen Y, Xiong X, Huang J, Song E, Tan W. Self-assembled aptamer-based drug carriers for bispecific cytotoxicity to cancer cells. Chem—Asian J. 2012;7:1630–1636. doi: 10.1002/asia.201101060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wolf E, Hofmeister R, Kufer P, Schlereth B, Baeuerle PA. BiTEs: bispecific antibody constructs with unique antitumor activity. Drug Discovery Today. 2005;10:1237–1244. doi: 10.1016/S1359-6446(05)03554-3. [DOI] [PubMed] [Google Scholar]
- 117.Beck A, Wurch T, Bailly C, Corvaia N. Strategies and challenges for the next generation of therapeutic antibodies. Nat. Rev. Immunol. 2010;10:345–352. doi: 10.1038/nri2747. [DOI] [PubMed] [Google Scholar]
- 118.Dhar S, Gu FX, Langer R, Farokhzad OC, Lippard SJ. Targeted delivery of cisplatin to prostate cancer cells by aptamer functionalized Pt(IV) prodrug-PLGA-PEG nanoparticles. Proc. Natl. Acad. Sci. U.S.A. 2008;105:17356–17361. doi: 10.1073/pnas.0809154105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Dhar S, Kolishetti N, Lippard SJ, Farokhzad OC. Targeted delivery of a cisplatin prodrug for safer and more effective prostate cancer therapy in vivo. Proc. Natl. Acad. Sci. U.S.A. 2011;108:1850–1855. doi: 10.1073/pnas.1011379108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Farokhzad OC, Cheng J, Teply BA, Sherifi I, Jon S, Kantoff PW, Richie JP, Langer R. Targeted nanoparticle-aptamer bioconjugates for cancer chemotherapy in vivo. Proc. Natl. Acad. Sci. U.S.A. 2006;103:6315–6320. doi: 10.1073/pnas.0601755103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Cao Z, Tong R, Mishra A, Xu W, Wong GC, Cheng J, Lu Y. Reversible cell-specific drug delivery with aptamer-functionalized liposomes. Angew. Chem., Int. Ed. 2008;48:6494–6498. doi: 10.1002/anie.200901452. [DOI] [PubMed] [Google Scholar]
- 122.Alshaer W, Hillaireau H, Vergnaud J, Ismail S, Fattal E. Functionalizing Liposomes with anti-CD44 Aptamer for Selective Targeting of Cancer Cells. Bioconjugate Chem. 2014 doi: 10.1021/bc5004313. [DOI] [PubMed] [Google Scholar]
- 123.Li X, Zhao Q, Qiu L. Smart ligand: aptamer-mediated targeted delivery of chemotherapeutic drugs and siRNA for cancer therapy. J. Controlled Release. 2013;171:152–162. doi: 10.1016/j.jconrel.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 124.Hong Yan L, Xiaohu G. A Universal Protein Tag for Delivery of SiRNA-Aptamer Chimeras. Sci. Rep. 2013;3 doi: 10.1038/srep03129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.McNamara JO, Andrechek ER, Wang Y, Viles KD, Rempel RE, Gilboa E, Sullenger BA, Giangrande PH. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat. Biotechnol. 2006;24:1005–1015. doi: 10.1038/nbt1223. [DOI] [PubMed] [Google Scholar]
- 126.Dassie JP, Liu X-YY, Thomas GS, Whitaker RM, Thiel KW, Stockdale KR, Meyerholz DK, McCaffrey AP, McNamara JO, Giangrande PH. Systemic administration of optimized aptamer-siRNA chimeras promotes regression of PSMA-expressing tumors. Nat. Biotechnol. 2009;27:839–849. doi: 10.1038/nbt.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Song P, Chou YK, Zhang X, Meza-Romero R, Yomogida K, Benedek G, Chu C-QQ. CD4 aptamer-RORγt shRNA chimera inhibits IL-17 synthesis by human CD4(+) T cells. Biochem. Biophys. Res. Commun. 2014;452:1040–1045. doi: 10.1016/j.bbrc.2014.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zhou J, Rossi JJ. Aptamer-targeted RNAi for HIV-1 therapy. Methods Mol. Biol. 2010;721:355–371. doi: 10.1007/978-1-61779-037-9_22. [DOI] [PubMed] [Google Scholar]
- 129.Subramanian N, Kanwar JR, Athalya P, Janakiraman N, Khetan V, Kanwar RK, Eluchuri S, Krishnakumar S. EpCAM aptamer mediated cancer cell specific delivery of EpCAM siRNA using polymeric nanocomplex. J. Biomed. Sci. 2015;22:4. doi: 10.1186/s12929-014-0108-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Shu D, Shu Y, Haque F, Abdelmawla S, Guo P. Thermodynamically stable RNA three-way junction for constructing multifunctional nanoparticles for delivery of therapeutics. Nat. Nanotechnol. 2011;6:658–667. doi: 10.1038/nnano.2011.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Bruno JG, Carrillo MP, Crowell R. Preliminary development of DNA aptamer-Fc conjugate opsonins. J. Biomed. Sci. Res. A. 2009;90:1152–1161. doi: 10.1002/jbm.a.32182. [DOI] [PubMed] [Google Scholar]
- 132.Desnick RJ, Schuchman EH. Enzyme replacement and enhancement therapies: lessons from lysosomal disorders. Nat. Rev. Genet. 2002;3:954–966. doi: 10.1038/nrg963. [DOI] [PubMed] [Google Scholar]
- 133.Niagara Muhammad I, Muthu Kumara G, Jing Z, Paul CH, Ratha M, Yong Z. In vivo photodynamic therapy using upconversion nanoparticles as remote-controlled nanotrans-ducers. Nat. Med. 2012;18:1580–1585. doi: 10.1038/nm.2933. [DOI] [PubMed] [Google Scholar]
- 134.Cohen BA, Bergkvist M. Targeted in vitro photodynamic therapy via aptamer-labeled, porphyrin-loaded virus capsids. J. Photochem. Photobiol., B. 2013;121:67–74. doi: 10.1016/j.jphotobiol.2013.02.013. [DOI] [PubMed] [Google Scholar]
- 135.Ferreira CSM, Cheung MC, Missailidis S. Phototoxic aptamers selectively enter and kill epithelial cancer cells. Nucleic Acids Res. 2009;37:866–76. doi: 10.1093/nar/gkn967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Han D, Zhu G, Wu C, Zhu Z, Chen T, Zhang X, Tan W. Engineering a cell-surface aptamer circuit for targeted and amplified photodynamic cancer therapy. ACS Nano. 2013;7:2312–2319. doi: 10.1021/nn305484p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Stephanopoulos N, Tong GJ, Hsiao SC, Francis MB. Dual-surface modified virus capsids for targeted delivery of photodynamic agents to cancer cells. ACS Nano. 2010;4:6014–6020. doi: 10.1021/nn1014769. [DOI] [PubMed] [Google Scholar]
- 138.Wang K, You M, Chen Y, Han D, Zhu Z, Huang J, Williams K, Yang CJ, Tan W. Self-Assembly of a Bifunctional DNA Carrier for Drug Delivery. Angew. Chem., Int. Ed. 2011;50:6098–6101. doi: 10.1002/anie.201008053. [DOI] [PubMed] [Google Scholar]
- 139.Yuan Q, Wu Y, Wang J, Lu D, Zhao Z, Liu T. Targeted Bioimaging and Photodynamic Therapy Nanoplatform Using an Aptamer-Guided G-Quadruplex DNA Carrier and Near-Infrared Light. Angew. Chem., Int. Ed. 2013;52:13965–13969. doi: 10.1002/anie.201305707. [DOI] [PubMed] [Google Scholar]
- 140.You M, Zhu G, Chen T, Donovan MJ, Tan W. Programmable and multiparameter DNA-based logic platform for cancer recognition and targeted therapy. J. Am. Chem. Soc. 2015;137:667–674. doi: 10.1021/ja509263k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Sitaula S, Reed SM. Porphyrin conjugated to DNA by a 2′-amido-2′-deoxyuridine linkage. Bioorg. Med. Chem. Lett. 2008;18:850–855. doi: 10.1016/j.bmcl.2007.11.033. [DOI] [PubMed] [Google Scholar]
- 142.Wang J, You M, Zhu G, Shukoor MI, Chen Z, Zhao Z, Altman MB, Yuan Q, Zhu Z, Chen Y, Huang CZ, Tan W. Photosensitizer-Gold Nanorod Composite for Targeted Multimodal Therapy. Small. 2013;9:3678–3684. doi: 10.1002/smll.201202155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Huang P, Lin J, Li W, Rong P, Wang Z, Wang S, Wang X, Sun X, Aronova M, Niu G, et al. Biodegradable gold nanovesicles with an ultrastrong plasmonic coupling effect for photoacoustic imaging and photothermal therapy. Angew. Chem., Int. Ed. 2013;52:13958–13964. doi: 10.1002/anie.201308986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Huang X, El-Sayed IH, Qian W, El-Sayed MA. Cancer Cell Imaging and Photothermal Therapy in the Near-Infrared Region by Using Gold Nanorods. J. Am. Chem. Soc. 2006;128:2115. doi: 10.1021/ja057254a. [DOI] [PubMed] [Google Scholar]
- 145.Lin L-S, Cong Z-X, Cao J-B, Ke K-M, Peng Q-L, Gao J, Yang H-H, Liu G, Chen X. Multifunctional Fe3O4@ polydopamine core-shell nanocomposites for intracellular mRNA detection and imaging-g uided photothermal therapy. ACS Nano. 2014;8:3876–3883. doi: 10.1021/nn500722y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Rong P, Yang K, Srivastan A, Kiesewetter DO, Yue X, Wang F, Nie L, Bhirde A, Wang Z, Liu Z, et al. Photosensitizer loaded nano-graphene for multimodality imaging guided tumor photodynamic therapy. Theranostics. 2013;4:229–239. doi: 10.7150/thno.8070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Yang L, Tseng Y-T, Suo G, Chen L, Yu J, Chiu W-J, Huang C-C, Lin C-H. Photothermal Therapeutic Response of Cancer Cells to Aptamer-Gold Nanoparticle-Hybridized Graphene Oxide under NIR Illumination. ACS Appl. Mater. Interfaces. 2015;7:5097–5106. doi: 10.1021/am508117e. [DOI] [PubMed] [Google Scholar]
- 148.Wilson C, Keefe AD. Building oligonucleotide therapeutics using non-natural chemistries. Curr. Opin. Chem. Biol. 2006;10:607–614. doi: 10.1016/j.cbpa.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 149.Da Pieve C, Blackshaw E, Missailidis S, Perkins AC. PEGylation and biodistribution of an anti-MUC1 aptamer in MCF-7 tumor-bearing mice. Bioconjugate Chem. 2012;23:1377–1381. doi: 10.1021/bc300128r. [DOI] [PubMed] [Google Scholar]
- 150.McCauley TG, Kurz JC, Merlino PG, Lewis SD, Gilbert M, Epstein DM, Marsh HN. Pharmacologic and pharmacokinetic assessment of anti-TGFbeta2 aptamers in rabbit plasma and aqueous humor. Pharm. Res. 2006;23:303–311. doi: 10.1007/s11095-005-9305-2. [DOI] [PubMed] [Google Scholar]
- 151.Rosi NL, Giljohann DA, Thaxton CS, Lytton-Jean AKR, Han MS, Mirkin CA. Oligonucleotide-Modified Gold Nanoparticles for Intracellular Gene Regulation. Science. 2006;312:1027–1030. doi: 10.1126/science.1125559. [DOI] [PubMed] [Google Scholar]
- 152.Jacobson O, Yan X, Gang N, Weiss ID, Ma Y, Szajek LP, Shen B, Kiesewetter DO, Chen X. PET Imaging of Tenascin-C with a Radiolabeled Single-Strand DNA Aptamer. J. Nucl. Med. 2015;56:616–621. doi: 10.2967/jnumed.114.149484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Burmeister PE, Lewis SD, Silva RF, Preiss JR, Horwitz LR, Pendergrast PS, McCauley TG, Kurz JC, Epstein DM, Wilson C, et al. Direct in vitro selection of a 2′-O-methyl aptamer to VEGF. Chem. Biol. 2004;12:25–33. doi: 10.1016/j.chembiol.2004.10.017. [DOI] [PubMed] [Google Scholar]