FIG 6 .
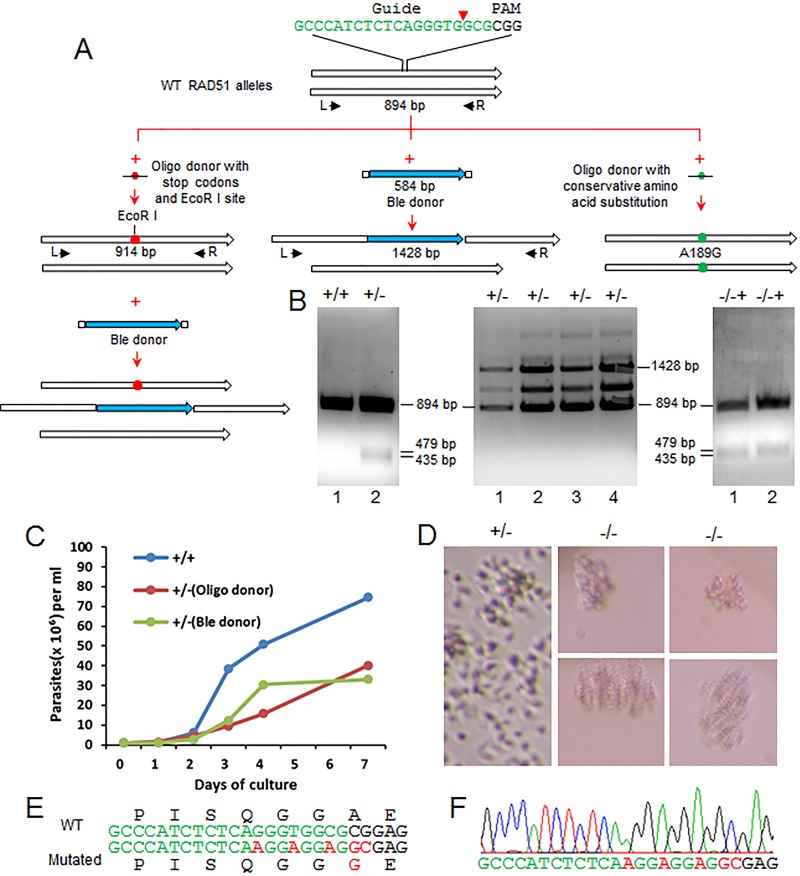
RAD51 is essential for L. donovani. (A) Strategies used to generate RAD51 disruption mutants and a mutant with a single conserved amino acid substitution. To generate various RAD51 mutants, L. donovani cells were transfected with Cas9- and RAD51-targeting gRNA expression vectors followed by transfection of oligonucleotide donors (with stop codons and an EcoRI site or with a conservative amino acid substitution) and/or the bleomycin selection marker donor. Genomic DNA from these L. donovani cells (clones) were subjected to PCR, restriction enzyme digestion, and sequencing analysis. (B) PCR and restriction enzyme analysis of RAD51 single- and double-allele disruption mutants. (Left) PCR amplification of the RAD51 sequence with primers L and R, followed by EcoRI digestion. Lane 1, wild-type L. donovani; lane 2, RAD51+/− mutant with a single RAD51 allele disrupted by the oligonucleotide donor containing stop codons and an EcoRI site. Note that although the EcoRI-digested bands (479 and 435 bp) were detected, the 894-bp wild-type RAD51 allele band remained in this single RAD51 disruption mutant. (Middle) PCR analysis of phleomycin resistance clones (RAD51+/− mutants) with primers L and R. Both the 1,428-bp bleomycin marker insertion band and the 894-bp wild-type RAD51 allele band were detected in all these phleomycin resistance clones. Note that sequencing indicates that the additional bands detected are rearrangements of the 1,428-bp ble insertion bands. (Right) PCR and EcoRI digestion analysis of Rad51−/−+ mutants with one allele disrupted with stop codons and an EcoRI site containing oligonucleotide donor and the other allele with a bleomycin selection marker donor. PCR bands (not shown) similar to those in the middle panel, including the 1,428-bp ble insertion bands and the approximately 900-bp bands, were obtained from these −/−+ mutants. The approximate 900-bp bands were then extracted from the gel and subjected to complete EcoRI digestion. Note that the 894-bp wild-type (WT) RAD51 allele band remained in these −/−+ mutants. These are representative data of more than 100 clones analyzed. (C) Growth curves of L. donovani cells targeted by RAD51 gRNA: RAD51+/− (Oligo donor), RAD51+/− mutant with the oligonucleotide donor (stop codons) insertion, RAD51+/− Ble donor, RAD51+/− mutant with bleomycin selection marker donor insertion, and RAD51+/+, wild-type L. donovani cells expressing a control gRNAa targeting the LdMT gene. The data are representative of three independent experiments. (D) Microscope images showing that disruption of all RAD51 alleles is lethal for L. donovani. The RAD51+/− mutant cells, which continue expressing RAD51-targeting gRNA were cloned in 96-well plates, and cell growth was monitored by microscopy. The image for RAD51+/− cells was taken 1 week after cloning; the images for Rad51−/− cells were taken 3 weeks after cloning. (E) Partial sequence of the oligonucleotide donor with mutations resulting in a single conservative amino acid substitution of the RAD51 protein (A189G) and inactivation of the RAD51 gRNA-targeting site. (F) Direct sequencing of the PCR product amplified from an L. donovani clone, showing both alleles of RAD51 have been mutated to the sequence of the oligonucleotide donor (see panels A and E).