Abstract
To study the mechanism of gene targeting, we examined heteroduplex DNA (hDNA) formation during targeting of two separate chromosomal locations in Saccharomyces cerevisiae. We examined both replacement of the entire gene with a heterologous selectable marker and correction of a single base pair insertion mutation by gene targeting, and in all cases our results were consistent with separate strand invasion/resolution at the two ends of the targeting fragment as the dominant mechanism in wild-type cells. A small subset of transformants was consistent with assimilation of a single strand of targeting DNA encompassing both flanking homology regions and the marker into hDNA. hDNA formation during correction of a point mutation by targeted integration was conspicuously altered in a mismatch repair-deficient background and was consistent with single-strand invasion/assimilation without mismatch correction, confirming that gene targeting by this pathway is actively impeded in wild-type yeast. Finally, inversion of one targeted locus and mutation of an active origin of DNA replication at the other locus affected hDNA formation significantly, suggesting that formation of productive interactions between the targeting DNA and the targeted site in the chromosome is sensitive to local DNA dynamics.
Targeted gene disruption was introduced in the late 1970s and since has become one of the most useful methods available for investigating gene function. In early studies in yeast, genes were disrupted by inserting a piece of the targeted gene into a plasmid to facilitate homologous recombination of the foreign plasmid DNA into the yeast chromosome (1, 2). Subsequently, it was found that cutting the plasmid in the region of homology to the chromosome greatly increased the frequency of recombination in both yeast (3) and mammalian (4, 5) cells. This finding led to the proposal that this type of gene disruption, known as “ends-in” gene targeting, occurs by a process analogous to double-strand break repair (DSBR), whereby integration of the broken plasmid into the chromosome occurs during repair of the break by gene conversion associated with crossing over (3). The ends-in method disrupts gene function by inserting foreign DNA at the target site without deleting the targeted gene.
To address the limitations of insertional mutagenesis, gene replacement strategies were developed in yeast (6) and later implemented in mammalian cells (7) by using recombinant linear DNA in which the two ends of the fragment are homologous to the regions flanking the targeted gene but the gene itself is replaced by a selectable marker. The ends of the linear targeting fragment are recombinogenic, facilitating replacement of the targeted gene with the selectable marker. This type of gene targeting is called “ends-out” because the edges of the targeting DNA correspond to two divergent, discontinuous stretches of chromosomal DNA. In effect, the two ends of the targeting fragment are separate, unrelated broken ends, so the mechanism of integration probably involves initially uncoordinated interactions between each end of the targeting fragment and its corresponding region of homology in the chromosome. Thus, integration of ends-out targeting DNA must occur by some mechanism other than simple gene conversion accompanied by crossing over.
This notion is reinforced by the observation that, whereas mitotic gene conversion is reduced 50- to 100-fold in mutants in which the Rad51 recombinase is absent (8, 9), targeted integration frequency is reduced only ≈8-fold (10). Despite limited dependence on RAD51, the mechanism of ends-out gene targeting in yeast clearly requires the broader recombination machinery because targeted integration is extremely deficient in rad52 mutants (10). This observation at first glance seems paradoxical, because the primary role of Rad52 in recombination is generally believed to be accessory to Rad51, but Rad52 also participates in a variety of RAD51-independent recombination events (reviewed in ref. 11). In vitro, Rad52 anneals complementary oligonucleotides (12), suggesting a possible role for this protein in base-pairing one strand of the targeting DNA with the targeted chromosomal locus in the absence of Rad51.
Despite these obvious differences from DSBR, it has been postulated that ends-out gene targeting occurs by separate crossovers at the two ends of the targeting fragment (13), but supporting data are limited and important mechanistic questions remain unanswered. Li et al. (14) examined heteroduplex DNA (hDNA) formation during targeted gene replacement in a mouse hybridoma cell line and found evidence of hDNA in both flanking homology regions in 9 of 26 transformants, eight of which were consistent with the model featuring invasion of the chromosome by different strands of targeting DNA at the two ends, as shown in Fig. 1a. Leung et al. (15) raised the idea that gene targeting can occur by assimilation of a single strand of the targeting fragment into heteroduplex with the corresponding chromosomal locus followed by mismatch correction (Fig. 1b). However, clear evidence for this mechanism was found only in mismatch repair-defective pms1 mutants, not in wild-type cells. Furthermore, targeting fragments used in that study (15) differed from the targeted chromosomal locus by only a few base pairs, not the large central heterologies typical of targeted gene replacement. It appears that even when single-strand assimilation intermediates were able to form, subsequent mismatch repair was overwhelmingly in favor of the chromosomal allele. Thus, gene targeting by this mechanism would be unlikely in wild-type cells. A separate study in yeast showed large increases in targeted gene replacement in mutants from which a different mismatch repair gene, MSH2, was deleted (16), suggesting that the mismatch repair machinery in wild-type cells acts specifically to prevent integration of a selectable marker, even when flanked by large regions of homology.
Fig. 1.
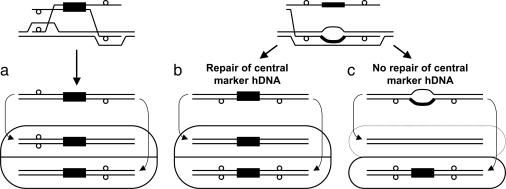
Models of gene targeting. (a) Two-end invasion/resolution. (b and c) Single-strand assimilation followed by mismatch correction (b) and without mismatch correction (c). Different mechanisms of gene targeting give rise to distinctive colonies of cells. (a and b) Flanking markers in trans (a) and in cis (b) are shown. (c) No sectoring of flanking markers with respect to the central marker is shown. Solid black boxes represent the selectable marker, small open circles indicate restriction site polymorphisms in the targeting DNA, rounded rectangles represent the colonies arising from gene targeting by the indicated mechanism, and curved arrows show how the two constituent cell genotypes arise from replication of the two different strands of DNA in the targeted cell and segregation during the next cell division. Because the marker on the targeting DNA is selected, the daughter cell that lacks the marker in c will not grow (as indicated by the dotted line), whereas the daughter cell with the marker will give rise to a colony that is not sectored for the flanking markers.
To investigate the mechanism of gene targeting in wild-type yeast, we developed targeting fragments that allow us to physically examine transformants for evidence of hDNA in the flanking homology regions during targeted integration. Our results suggest that gene targeting is typically initiated by separate strand invasions at the two ends of the targeting molecule (Fig. 1a) and infrequently occurs by assimilation of a single strand of targeting DNA encompassing both flanking homology regions into hDNA followed by mismatch correction (Fig. 1b). The mechanism and frequency of targeted integration in a mismatch repair-deficient pms1 background were radically altered and were consistent with single-strand assimilation without mismatch correction (Fig. 1c), confirming that this is a minor, secondary pathway in wild-type yeast.
Materials and Methods
Yeast Strains. All strains are derived from LSY678, a RAD5 derivative of W303-1A (17). Strains LSY697 and LSY698, containing the met17-sna and met17::ADE2 alleles, respectively, have been described previously (8). To create a trp1::URA3 derivative, LSY678 was first transformed to Trp+ with a SnaBI/ApaLI fragment from plasmid pRS414 (18), creating strain LSY1099. The TRP1 ORF of LSY1099 was replaced with the URA3 marker by PCR-mediated gene disruption (19) to create strain LSY1103. TRP1-ars1- strain LSY1508 was made by transforming LSY1103 with an SspI/BsaBI fragment from pLL111-ars1-. Trp+ Ura- clones were checked by PCR for the presence of a XhoI mutation at ars1. LSY1509, a trp1::URA3-ars1- derivative of LSY1508, was made by PCR-mediated gene disruption of LSY1508 using the same primers used to make LSY1103. Inverted met17::ADE2 strain LSY1539 was created by transforming LSY678 with BstEII-digested pLL152, and the inversion was confirmed by PCR. LSY946-1C was made by crossing LSY697 to pms1::TRP1 strain HKY594-1B (a gift from Hannah Klein, New York University School of Medicine, New York).
Plasmids. Plasmid pLL140 contains a 3.3-kb MET17 fragment cloned into plasmid pGEM-T (Promega). Plasmid pLL147 is a derivative of pLL140 with the following interrupted palindromic inserts cloned into unique restriction sites in the MET17 flanking regions: upstream insert at SwaI, 5′-ATTTctagctaggccttttttaggcctagctagAAAT-3′; downstream insert at BglII, 5′-AcgtcgatccggattttttccggatcgacgGATCT-3′.
Plasmid pLL111 contains a 2.2-kb TRP1 fragment cloned into plasmid pGEM-T. Plasmid pLL113 is a derivative of pLL111 with the following interrupted palindromic inserts cloned into unique restriction sites in the TRP1 flanking regions: upstream insert at SnaBI, 5′-TACcgagctaggccttttttaggcctagctcgGTA-3′; downstream insert at BglII, 5′-AcgtcgatccggattttttccggatcgacgGATCT-3′.
An ars1 mutant derivative of pLL111, pLL111-ars1-, was made by replacing the A site with a XhoI linker by site-directed mutagenesis using the Gene Editor kit (Promega) (20). An ars1 derivative of pLL113, pLL113-ars1-, was made by subcloning sequences from pLL113 into pLL111-ars1-. Plasmids pLL153 and pLL154 were created by using the same primers and technique used to create pLL111, except that the template was genomic DNA from strains LSY1103 (trp1::URA3 and ARS1) and LSY1509 (trp1::URA3 and ars1-), respectively. All plasmids containing ARS1 were able to confer high-frequency transformation of yeast whereas both ars1- derivative plasmids were not, indicating a disruption of origin activity.
To invert the met17::ADE2 disruption, a 7.1-kb fragment containing met17::ADE2 from LSY698 was PCR-amplified and cloned into the NotI and BamHI sites of pRS423 (18), creating pLL151. To flip met17::ADE2, plasmid pLL151 was digested with SpeI and religated. Clones with the 2.9-kb SpeI fragment in inverted orientation with respect to the surrounding sequences were identified by restriction analysis and designated pLL152.
Analysis of hDNA. For gene targeting, plasmids pLL113 and pLL113-ars1- were digested with SspI and BsaBI to release the TRP1 fragment, and plasmid pLL147 was digested with SpeI to release the MET17 fragment. Transformation of yeast cells was done by the lithium acetate method using ≈100–300 ng of linear targeting DNA with 50 μg of salmon sperm carrier DNA. Transformed cells were plated onto synthetic complete (SC) medium lacking Trp (SC -Trp) or Met (SC -Met), as appropriate. To exclude transformants arising by end-joining of the fragment or random integration, Trp+ colonies were replicaplated to medium containing 5-fluoroorotic acid (5-FOA) to identify clones that had lost the trp1::URA3 marker. Similarly, Met+ transformants that had replaced the met17::ADE2 marker were identified by colony color or by replica plating to SC -Met -Ade. The Met+ Ade+ transformants accounted for <10% of total events. To exclude random integration events in the met17-sna background, genomic DNA from Met+ clones was amplified by PCR with primers flanking the met17-sna mutation. PCR products were digested with SnaBI, which cuts MET17 sequences but not met17-sna. PCR products that failed to digest to completion were not analyzed further. Transformation efficiencies of the different target strains were similar for a given marker (data not shown) except for the pms1 strain, as described in Results. The transformation efficiency at TRP1 was ≈10-fold lower than at MET17 (data not shown).
After identifying correct clones by phenotype, whole individual colonies were suspended in 5 ml of synthetic complete (SC) medium and grown overnight. Ten microliters of each culture was spotted onto selective media to preserve the strains for further analysis, and ≈96 individual colonies were analyzed for each transformed strain. Genomic DNA from each strain was amplified by PCR with primers specific to the upstream or downstream flanking region and then digested with restriction enzymes diagnostic of the chromosomal allele or the targeting allele for each flanking region (see Fig. 2). PCR products that digested to completion with the restriction enzyme specific to the targeting allele were scored as “vector site only” and PCR products that digested to completion with the restriction enzyme specific to the chromosomal allele were scored as “chromosomal site only.” PCR products that showed partial digestion with both enzymes were scored as “hDNA.”
Fig. 2.
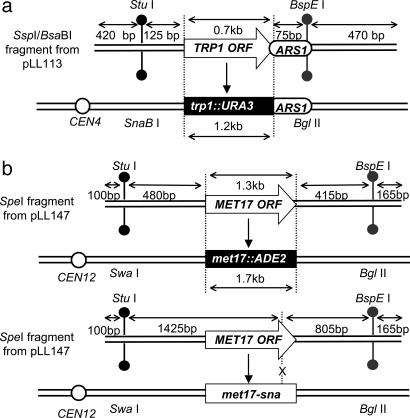
Schematics of targeting DNA and targeted yeast strains. Open arrows show the selectable markers, TRP1 or MET17, black boxes show the corresponding disrupted allele in the chromosome, and open circles indicate the position of the centromere with respect to the targeted locus (not to scale). (a) Targeting at the TRP1 locus. (b) Targeting at the MET17 locus of a complete disruption (Upper) or a point mutation (Lower). Restriction site polymorphisms in the flanking homology regions are indicated.
Clones that were scored as hDNA in both flanking regions were subcultured to segregate the two cell types and isolate pure clonal populations. Six to eight individual subclones were characterized by PCR and restriction digestion. Two patterns of subclones were expected. In one, both the upstream and downstream PCR products from all subclones were cleaved by enzymes of the same specificity, either chromosome-specific or targeting fragment-specific. This pattern was scored as “cis.” In the other pattern of subclones, the up- and downstream PCRs were cleaved by enzymes of the opposite specificity and scored as “trans.”
Statistical Analysis. For different strains targeted with the same selectable marker, differences between strains in the number of hDNA and non-hDNA transformants at either the upstream or downstream locus were treated as categorical variables and analyzed by Fisher's two-tailed exact test. P values < 0.05 were considered statistically significant.
Results
Experimental System. We designed an experimental system that relies on restriction site polymorphisms to detect formation of heteroduplex between homologous regions of targeting DNA and the chromosome during gene targeting. To avoid mismatch repair of the resulting hDNA, the altered restriction sites were incorporated into 26-bp palindromic inserts that are poor substrates for mismatch repair (21).
After identifying transformants as isolated single colonies arising from targeted integration, we examined the regions of the flanking sequences containing the restriction site polymorphisms by PCR, digesting the PCR products with restriction endonucleases to distinguish chromosomal DNA from targeting DNA in the upstream and downstream flanking regions. Throughout this paper, “upstream” refers to the region 5′ to the ORF that contains the promoter of the relevant gene; “downstream” refers to the region 3′ to the ORF that contains the transcriptional terminator of that gene. Cleavage of the PCR product with one enzyme but not the other identifies that locus as containing exclusively chromosomal or exclusively targeting vector DNA. Partial cleavage with both enzymes indicates that the particular transformant gave rise to a colony with a mixed population of cells resulting from failure to repair hDNA formed during the targeting process (referred to as “sectoring”). Colonies showing hDNA formation in both the upstream and downstream flanking homology regions were further characterized by subclone analysis to determine whether the same or opposite strands of targeting DNA invaded the chromosome at the two ends, as shown in Fig. 1 (see Materials and Methods for details).
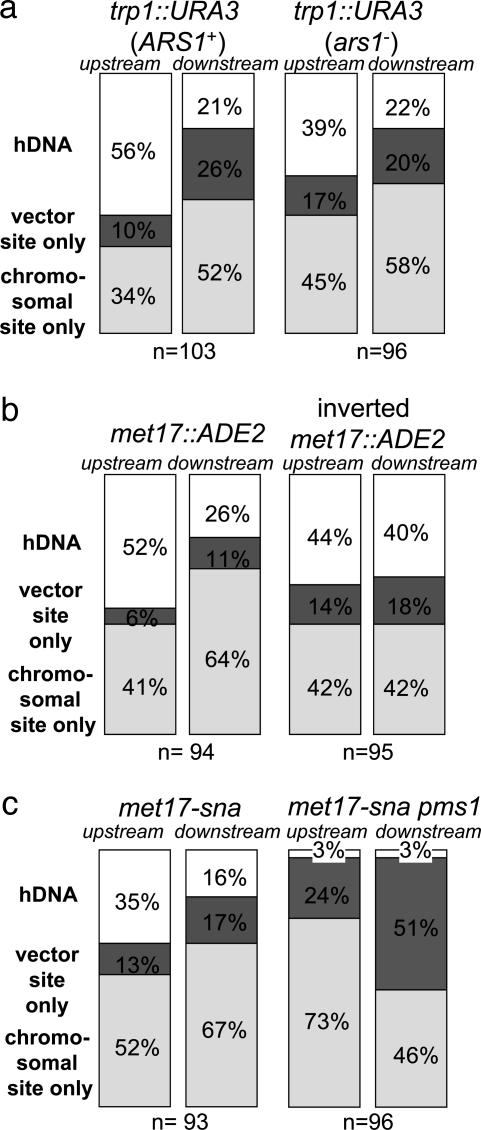
Targeted Gene Replacement Occurs by Invasion of Separate Strands at Each End. We first examined hDNA formation during targeted replacement of the chromosomal trp1::URA3 allele by TRP1 (Fig. 2a). Of 103 Trp+ Ura- transformants examined, 58 were sectored for the upstream hDNA marker (56%), and 22 were sectored downstream (21%) (Fig. 3a). Altogether, evidence of hDNA formation during gene targeting was identified in one or both flanking regions in 70 transformants (68%), although only 10 transformants (10%) showed hDNA formation in both flanking regions (Table 1). Subclone analysis of these transformants showed that nine were in the trans configuration, whereas only one was in the cis configuration (Table 1). These results are consistent with a model in which a different strand of targeting DNA invades the chromosome at each end (Fig. 1a), similar to what was found previously for targeted gene replacement in mouse cells (14).
Fig. 3.
hDNA formation during gene targeting. hDNA was analyzed by restriction analysis after gene targeting of each indicated strain. The number of transformants analyzed in each strain is indicated. Each stacked bar indicates the percentage of transformant colonies containing only the chromosome-specific (Bottom, chromosomal site only), only the targeting vector-specific (Middle, vector site only), or a mixture of both (Top, hDNA) restriction sites in the upstream or downstream flanking regions. (a) Gene replacement at the TRP1 locus. (b) Gene replacement at the MET17 locus. (c) Mutation correction by gene targeting at the MET17 locus.
Table 1. Summary of hDNA and subclone analysis.
| hDNA, %
|
||||
|---|---|---|---|---|
| Targeting DNA/targeted strain | Either end | Both ends | No. in trans | No. in cis |
| TRP1/trp1 :: URA3 (ARS1+) | 68 | 10 | 9 | 1 |
| TRP1-ars1-/trp1 :: URA3 (ars1-) | 53 | 7 | 7 | 0 |
| MET17/met17 :: ADE2 | 69 | 9 | 8 | 0 |
| MET17/inverted met17 :: ADE2 | 64 | 20 | 19 | 0 |
| MET17/met17-sna | 42 | 10 | 7 | 2 |
| MET17/met17-sna pms1 | 6 | 0 | 0 | 0 |
To confirm the generality of these results, we conducted a similar analysis at a second locus, this time replacing a chromosomal met17::ADE2 allele (where ADE2 is under control of the MET17 promoter) with MET17 (Fig. 2b). Although the hDNA markers were much nearer the ends of the targeting fragment in MET17, the results were remarkably similar to those at TRP1. Of the 94 Met+ Ade- transformants examined, 49 were sectored for the upstream flanking marker (52%), and 24 were sectored for the downstream flanking marker (26%) (Fig. 3b). Sixty-five transformants exhibited hDNA in one or both flanking regions (69%), and eight transformants (9%) showed evidence of hDNA in both flanking regions (Table 1). In this case, all eight double-sectored transformants were in the trans configuration (Table 1) indicating that, at least during gene replacement in wild-type yeast, gene targeting is normally initiated by different strands of DNA at the two ends of the molecule and not by assimilation of a single strand into hDNA.
Targeting of a Single Base Pair Mutation also Occurs by Separate Strand Invasions. To determine whether gene replacement with a selectable marker poses a barrier to gene targeting by single-strand invasion, we examined mutation correction by gene targeting using the met17-sna strain shown in Fig. 2b. In this case, the only difference between the MET17 sequence on the targeting DNA and the met17-sna allele on the chromosome is a single base pair insertion in an SnaBI site on the chromosomal allele, in addition to the palindromic insertions (8). Of 93 Met+ transformants shown to have replaced the met17-sna allele, 33 were sectored for the upstream flanking marker (35%), and 15 were sectored for the downstream flanking marker (16%) (Fig. 3c). Formation of hDNA was detected in at least one of the flanking regions in 39 transformants (42%), whereas nine transformants (10%) showed hDNA formation in both flanking regions (Table 1). Subclone analysis of these transformants showed seven and two in the trans and cis configurations, respectively (Table 1), suggesting that the mechanism of successful targeted integration does not differ significantly between gene replacement and mutation correction in wild-type cells (P = 0.47). These results do not exclude the possibility that single-strand invasion intermediates form in wild-type cells, but, if so, they are probably corrected to the chromosomal allele in most cases. In this way, our results are similar to those of Leung et al. (15), who found evidence for single-strand assimilation in mismatch repair-deficient pms1 mutants but not in wild-type cells.
PMS1 Prevents Gene Targeting by Assimilation of a Single Strand. To investigate the effects of mismatch repair, the gene targeting experiments at met17-sna were repeated in a strain deleted of PMS1, which encodes one of the four MutL homologs in Saccharomyces cerevisiae. We found a dramatic reduction in the number of transformants with hDNA in the pms1 strain, consistent with the formation, but not repair, of hDNA between a single strand of the MET17 targeting DNA and one strand of the chromosomal met17-sna locus (Fig. 1c). Replication of the unrepaired intermediate would yield only one daughter cell with the MET17 allele, resulting in an unsectored colony. Of 96 Met+ transformants shown to have replaced the met17-sna allele, only three were sectored for the upstream flanking marker (3%), three were sectored for the downstream flanking marker (3%) (Fig. 3c), and none of the transformants analyzed showed sectoring for both flanking markers. Along with the changes in hDNA formation, we also observed a >3-fold increase in the number of Met+ Ade- transformants in the pms1 strain (data not shown).
To create a MET17 strain by single-strand assimilation, the hDNA tract initiated by 3′ end invasion of the targeting DNA has to extend at least as far as the met17-sna mutation. The mutation in met17-sna is much closer to the downstream flanking marker than to the upstream flanking marker (Fig. 2b). Thus, if targeting occurs by single-strand assimilation without mismatch correction in the pms1 strain, we would expect the downstream flanking marker to be in hDNA with met17-sna more often than the upstream marker. Vector sequences were found at the downstream locus in 49 of 96 transformants (51%) but in only 23 of 96 (24%) at the upstream locus (P = 0.0002) (Fig. 3c). Transformants incorporating both flanking markers were rare (8 of 96, or 8%; data not shown), because this requires both that the hDNA tract be long (at least 2.2 kb) and that nuclease processing of the 5′ end of the assimilated strand be limited to <100 or 165 bp, depending on which 3′ end invaded (Fig. 2b). Altogether, the data from the pms1 mutant show that the mechanism of gene targeting of a point mutation is significantly altered in the absence of at least one component of the mismatch repair machinery in a way that is consistent with single-strand invasion/assimilation without mismatch correction (Fig. 1c).
Local DNA Dynamics Affect Interactions Between Targeting DNA and Targeted Locus. As shown in Fig. 3, hDNA formation at both TRP1 and MET17 was much greater in the upstream, promoter-proximal flanking region than in the downstream flanking region in wild-type cells. Both genes are transcribed in the same direction with respect to the centromere, so there were no clues as to whether the difference in hDNA formation up- and downstream at both loci was due to transcription or to some other factor, possibly the direction of replication fork movement through the locus or gross chromosome architecture.
To examine this question, we inverted the chromosomal met17::ADE2 allele on a 2.9-kb SpeI fragment containing the ADE2 ORF under control of the MET17 promoter and flanked by ≈600 bp of MET17 sequences on both sides (see Materials and Methods). Transformation was repeated by using the same MET17 targeting DNA as in the earlier experiments to replace the inverted met17::ADE2 allele. As shown in Fig. 3b, hDNA at the upstream flanking marker was essentially unchanged, with 42 transformants showing hDNA of 95 total Met+ Ade- transformants (44%), compared with 49 of 94 (52%) at the original met17::ADE2 locus (P = 0.31). In contrast, a small but significant increase in hDNA was observed at the downstream flanking region, with 38 sectors of 95 transformants (40%) compared with 24 of 94 (26%) in the original orientation (P = 0.044) (Fig. 3b). As a result, the difference in hDNA formation between the upstream and downstream flanking regions disappeared completely. Inversion of the met17::ADE2 locus did not greatly affect its transcription, because ADE2 mRNA levels in the inverted strain were 60% of those in the noninverted strain as determined by Northern blot analysis (data not shown). This outcome suggests that the difference between the two flanking regions in hDNA formation observed with the original met17::ADE2 allele was not caused by transcription. Finally, because of the higher rate of hDNA formation downstream with the inverted met17::ADE2 allele, the proportion of transformants showing hDNA in both flanking regions increased to 20% (Table 1). Subclone analysis of these transformants showed that all 19 were in the trans configuration (Table 1).
To determine whether the origin of DNA replication (ARS1) at the TRP1 locus had any effect on hDNA formation or the mechanism of targeted integration, we mutated the origin in a way that was previously shown to eliminate its ability to support plasmid replication (20). The same ars1 mutation was made in both the targeting fragment and the chromosome to avoid introducing additional heterologies between the two DNAs and to assure that neither the targeting DNA nor the targeted region was the site of replication initiation. ARS1 has previously been shown to be an active origin in the W303 yeast strain background used in our experiments (22), and its ability to support plasmid replication is not affected by the adjacent trp1::URA3 disruption (data not shown).
As shown in Fig. 3a, hDNA formation at the downstream flanking region was unaffected by the ars1 mutation. This is particularly noteworthy because the downstream hDNA marker at TRP1 is directly adjacent to the site of replication initiation at ARS1 (23). Thus, it was surprising to find a reduction in hDNA at the upstream flanking marker in the ars1 mutant, with 37 sectors of 96 Trp+ Ura- transformants (39%) compared with 58 of 103 (56%) with the wild-type ARS1 (P = 0.016). This result suggests that the presence of an active origin of DNA replication affects hDNA formation during gene targeting but, in this case, only at the upstream flanking marker distant from the origin and not at the downstream flanking marker located near the site of replication initiation. Furthermore, the ars1 mutation did not affect the mechanism of targeted integration: all seven transformants that showed hDNA formation in both the upstream and downstream flanking regions were in the trans configuration (Table 1).
Discussion
Several important differences between DSBR and gene targeting led us to investigate the mechanism of targeted gene replacement in yeast. Furthermore, conflicting data from yeast and mouse cells raised the possibility of a fundamental difference between these organisms that might explain the higher efficiency of homology-directed gene targeting in yeast. In particular, it was suggested that gene targeting in yeast might frequently occur by assimilation of a single strand of the targeting DNA into heteroduplex with the homologous chromosomal locus, followed by mismatch correction in favor of the targeting DNA, as shown in Fig. 1b (15). However, evidence for such a mechanism was found only in pms1 mutants, and the targeting constructs in that study differed from the targeted locus by just a few base pairs. Contrasting studies (e.g., 14) of gene replacement at a single locus in a mouse hybridoma cell line found little evidence for single-strand assimilation in mammalian cells.
Our studies focused on hDNA formation during gene targeting in wild-type yeast. To maximize the generality of our conclusions, hDNA formation was assayed at two different chromosomal loci, and both gene replacement and mutation correction by gene targeting were examined. Of 44 transformants where hDNA was detectable in both flanking regions in the gene replacement studies, only one of these was in the cis configuration (Table 1). Similarly, examination of mutation correction by gene targeting produced nine transformants wherein hDNA was detectable in both flanking regions, and only two of these were in the cis configuration (Table 1). No such transformants were identified in the corresponding pms1 mutant. Thus, gene targeting by single-strand assimilation appears not to be an important pathway in wild-type yeast, even in the absence of large heterologies between targeting and chromosomal DNA. Furthermore, it appears that gene replacement in mismatch repair-proficient mammals and yeast is initiated by invasion of the homologous chromosomal DNA by different strands of targeting DNA at the two ends, as shown in Fig. 1a.
In the course of our studies, we identified a notable difference in hDNA formation between the two flanking regions, with a greater level of hDNA detected in the upstream, promoter-proximal flanking region (Fig. 3) at both TRP1 and MET17. The differences in frequency of hDNA formation between the upstream and downstream loci could be due to differential end processing by nucleases or to different lengths of DNA engaged in hDNA during strand invasion. Alternatively, it is possible that hDNA forms efficiently at one or both ends of the targeting fragment but that the positions of strand cleavages required to connect both strands of the targeting DNA with the chromosomal locus vary in the upstream and downstream regions, resulting in the difference in frequency of retention of hDNA in the resulting transformants. To gain insight into the factors that influence formation of gene targeting intermediates, we first considered the possibility that some aspect of transcription initiation or transcription-related chromatin remodeling led to higher hDNA formation in the promoter-proximal flanking region. The stimulatory effects of transcription on recombination in general (24, 25) and on gene targeting in particular (26) have been appreciated for some time, suggesting transcription could be responsible for the observed disparity in hDNA formation. To examine this question, we inverted the met17::ADE2 locus on chromosome XII so that it is transcribed toward the centromere and repeated the transformation experiments. If the difference in hDNA in the upstream and downstream flanking regions was attributable to transcription alone, we predicted that the disparity would be retained when targeting the inverted locus, but in fact the difference disappeared (Fig. 3b) even though the inverted locus was still transcribed at 60% of the level of the noninverted locus. This result makes it extremely unlikely that transcription is responsible for the disparity observed when targeting the original met17::ADE2 locus.
A second possibility is that the disparity has to do with the effects of replication on gene targeting. A recent paper showed a dramatic effect of replication on the frequency of gene targeting in Schizosaccharomyces pombe, but the effect was limited to particular flanking sequences overlapping an active origin of replication (27). To test the effects of replication on hDNA formation during gene targeting, we mutated the ARS1 origin at the TRP1 locus on both the targeting DNA and on the chromosomal trp1::URA3 allele. hDNA formation at the downstream locus, in the vicinity of the ars1 mutation, was completely unaffected. Surprisingly, however, hDNA was reduced at the upstream locus (Fig. 3a). This result shows that hDNA formation at TRP1 is affected by the presence of the active replication origin. It is unclear whether the effect comes from binding of origin-specific proteins or from active initiation of DNA replication. We favor the latter hypothesis based on the fact that hDNA formation in the vicinity of ARS1, where origin-specific proteins would bind, was unaffected by the ars1 mutation.
Taken together, our studies lead to the firm conclusion that the early steps in gene targeting in wild-type yeast are analogous to those in DSBR, with the two ends of the targeting DNA separately engaging their homology regions in the chromosome as if each were one side of two separate double-strand breaks (Fig. 1a). From this point on, the analogy to DSBR breaks down because the opposite sides of the breaks are absent, and the only recombination/repair gene so far shown to be absolutely required for gene targeting in yeast is RAD52. The requirement for RAD52 in gene targeting in vertebrate cells is less strict (28, 29), although a defect in gene targeting is one of the few phenotypes of vertebrate RAD52-/- mutants (28, 29). Interestingly, gene targeting in mammalian cells is absolutely dependent on Ercc1 (30), which together with Xpf codes for a heterodimeric endonuclease. The requirement for the Ercc1 homolog in yeast, RAD10, is less stringent, although targeting frequencies are reduced 3- to 40-fold in rad10 and rad1 (Xpf homolog) mutants (10, 31–34). The peculiar genetic requirements of gene targeting are reminiscent of those for the single-strand annealing pathway for direct repeat recombination (35, 36), which suggests that a similar intermediate exists during gene targeting that requires the mediator and/or annealing functions of Rad52 to form and the endonuclease function of Rad1/10 to be processed. Although the details of such a mechanism remain unknown, it is clear from the changes in hDNA observed at the inverted MET17 locus and in the ars1 mutant at TRP1 that the formation and resolution of these intermediates are influenced a great deal by the chromosomal microenvironment of the targeted locus.
Acknowledgments
This work was supported by National Institutes of Health Grants GM54099 (to L.S.S.), T32 GM08224, and T32 CA09503 (to L.D.L.). We thank H. Klein and R. Rothstein (Columbia University) for strains and W. K. Holloman and B. Llorente for critical reading of the manuscript.
Author contributions: L.D.L. and L.S.S. designed the research; L.D.L. performed the research; L.D.L. and L.S.S. analyzed data; and L.D.L. and L.S.S. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: hDNA, heteroduplex DNA; DSBR, double-strand break repair.
References
- 1.Hinnen, A., Hicks, J. B. & Fink, G. R. (1978) Proc. Natl. Acad. Sci. USA 75, 1929-1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scherer, S. & Davis, R. W. (1979) Proc. Natl. Acad. Sci. USA 76, 4951-4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Orr-Weaver, T. L., Szostak, J. W. & Rothstein, R. J. (1981) Proc. Natl. Acad. Sci. USA 78, 6354-6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smithies, O., Gregg, R. G., Boggs, S. S., Koralewski, M. A. & Kucherlapati, R. S. (1985) Nature 317, 230-234. [DOI] [PubMed] [Google Scholar]
- 5.Thomas, K. R., Folger, K. R. & Capecchi, M. R. (1986) Cell 44, 419-428. [DOI] [PubMed] [Google Scholar]
- 6.Rothstein, R. J. (1983) Methods Enzymol. 101, 202-211. [DOI] [PubMed] [Google Scholar]
- 7.Thomas, K. R. & Capecchi, M. R. (1987) Cell 51, 503-512. [DOI] [PubMed] [Google Scholar]
- 8.Bartsch, S., Kang, L. E. & Symington, L. S. (2000) Mol. Cell. Biol. 20, 1194-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ira, G. & Haber, J. E. (2002) Mol. Cell. Biol. 22, 6384-6392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schiestl, R. H., Zhu, J. & Petes, T. D. (1994) Mol. Cell. Biol. 14, 4493-4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Symington, L. S. (2002) Microbiol. Mol. Biol. Rev. 66, 630-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mortensen, U. H., Bendixen, C., Sunjevaric, I. & Rothstein, R. (1996) Proc. Natl. Acad. Sci. USA 93, 10729-10734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hastings, P. J., McGill, C., Shafer, B. & Strathern, J. N. (1993) Genetics 135, 973-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li, J., Read, L. R. & Baker, M. D. (2001) Mol. Cell. Biol. 21, 501-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leung, W., Malkova, A. & Haber, J. E. (1997) Proc. Natl. Acad. Sci. USA 94, 6851-6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Negritto, M. T., Wu, X., Kuo, T., Chu, S. & Bailis, A. M. (1997) Mol. Cell. Biol. 17, 278-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thomas, B. J. & Rothstein, R. (1989) Genetics 123, 725-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sikorski, R. S. & Hieter, P. (1989) Genetics 122, 19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baudin, A., Ozier-Kalogeropoulos, O., Denouel, A., Lacroute, F. & Cullin, C. (1993) Nucleic Acids Res. 21, 3329-3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marahrens, Y. & Stillman, B. (1992) Science 255, 817-823. [DOI] [PubMed] [Google Scholar]
- 21.Nag, D. K., White, M. A. & Petes, T. D. (1989) Nature 340, 318-320. [DOI] [PubMed] [Google Scholar]
- 22.Liang, C., Weinreich, M. & Stillman, B. (1995) Cell 81, 667-676. [DOI] [PubMed] [Google Scholar]
- 23.Bielinsky, A. K. & Gerbi, S. A. (1998) Science 279, 95-98. [DOI] [PubMed] [Google Scholar]
- 24.Thomas, B. J. & Rothstein, R. (1989) Cell 56, 619-630. [DOI] [PubMed] [Google Scholar]
- 25.Saxe, D., Datta, A. & Jinks-Robertson, S. (2000) Mol. Cell. Biol. 20, 5404-5414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thyagarajan, B., Johnson, B. L. & Campbell, C. (1995) Nucleic Acids Res. 23, 2784-2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Segurado, M., Gomez, M. & Antequera, F. (2002) Mol. Cell 10, 907-916. [DOI] [PubMed] [Google Scholar]
- 28.Rijkers, T., Van Den Ouweland, J., Morolli, B., Rolink, A. G., Baarends, W. M., Van Sloun, P. P., Lohman, P. H. & Pastink, A. (1998) Mol. Cell. Biol. 18, 6423-6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamaguchi-Iwai, Y., Sonoda, E., Buerstedde, J. M., Bezzubova, O., Morrison, C., Takata, M., Shinohara, A. & Takeda, S. (1998) Mol. Cell. Biol. 18, 6430-6435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Niedernhofer, L. J., Essers, J., Weeda, G., Beverloo, B., de Wit, J., Muijtjens, M., Odijk, H., Hoeijmakers, J. H. & Kanaar, R. (2001) EMBO J. 20, 6540-6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schiestl, R. H. & Prakash, S. (1988) Mol. Cell. Biol. 8, 3619-3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schiestl, R. H. & Prakash, S. (1990) Mol. Cell. Biol. 10, 2485-2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saparbaev, M., Prakash, L. & Prakash, S. (1996) Genetics 142, 727-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Symington, L. S., Kang, L. E. & Moreau, S. (2000) Nucleic Acids Res. 28, 4649-4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fishman-Lobell, J. & Haber, J. E. (1992) Science 258, 480-484. [DOI] [PubMed] [Google Scholar]
- 36.Ivanov, E. L., Sugawara, N., Fishman-Lobell, J. & Haber, J. E. (1996) Genetics 142, 693-704. [DOI] [PMC free article] [PubMed] [Google Scholar]