ABSTRACT
The microbiota of the human gastrointestinal tract (GIT) may regularly be exposed to antibiotics, which are used to prevent and treat infectious diseases caused by bacteria and fungi. Bacterial communities of the gut retain a reservoir of antibiotic resistance (AR) genes, and antibiotic therapy thus positively selects for those microorganisms that harbor such genetic features, causing microbiota modulation. During the first months following birth, bifidobacteria represent some of the most dominant components of the human gut microbiota, although little is known about their AR gene complement (or resistome). In the current study, we assessed the resistome of the Bifidobacterium genus based on phenotypic and genotypic data of members that represent all currently recognized bifidobacterial (sub)species. Moreover, a comparison between the bifidobacterial resistome and gut metagenome data sets from adults and infants shows that the bifidobacterial community present at the first week following birth possesses a reduced AR arsenal compared to that present in the infant bifidobacterial population in subsequent weeks of the first year of life. Our findings reinforce the concept that the early infant gut microbiota is more susceptible to disturbances by antibiotic treatment than the gut microbiota developed at a later life stage.
IMPORTANCE The spread of resistance to antibiotics among bacterial communities has represented a major concern since their discovery in the last century. The risk of genetic transfer of resistance genes between microorganisms has been extensively investigated due to its relevance to human health. In contrast, there is only limited information available on antibiotic resistance among human gut commensal microorganisms such as bifidobacteria, which are widely exploited by the food industry as health-promoting microorganisms or probiotic ingredients. In the current study, we explored the occurrence of antibiotic resistance genes in the genomes of bifidobacteria and evaluated their genetic mobility to other human gut commensal microorganisms.
KEYWORDS: gut microbiomes, bifidobacteria, human gut, antibiotic resistance genes, resistomes
INTRODUCTION
The human gut microbiota plays an important role in health and disease of the host through its impact on immunology, nutrition, and pathogenesis (1, 2). The microbiota may regularly be exposed to a variety of antimicrobial agents, such as antibiotics, which are used in the treatment and prevention of bacterial infection in human and veterinary medicine (3). Antibiotic therapy may cause secondary effects such as distortion of the homeostasis of microbial gut consortia (4) and selection for antibiotic-resistant microorganisms. In fact, bacteria have counteracted the action of antibiotics through the acquisition of a specific genetic arsenal, also known as the resistome, which is involved in inactivation and/or removal of antibiotics. A large part of the resistome is contained within chromosomal DNA, although it may also be present on extrachromosomal replicons like plasmids and phages, which are transmissible to other members of the gut microbiota through horizontal gene transfer (HGT) events (5). Furthermore, antibiotic resistance may also be provided by a mutation in a gene encoding the antibiotic target, in which case the acquired resistance is not considered to be horizontally transferable (5). Antibiotic treatment selects for antimicrobial-resistant bacteria, where this selection is positively correlated with antibiotic usage (6). Furthermore, antibiotic resistance (AR) genes in gut commensal microorganisms are considered to be undesirable as they may lead to antibiotic resistance in human pathogens. Using HGT mechanisms, AR genes not only may be exchanged among members of the indigenous gut microbiota but also may be transferred to other bacteria that are just passing through the gastrointestinal tract (GIT), including several diet-associated bacteria (7). These studies have prompted the European Food Safety Authority (EFSA) to issue guidelines on the safety assessment of microorganisms used in food and feed production.
Bifidobacteria are common human gut microbiota members and are especially abundant during the first months following birth (2). Antibiotic therapy is commonly used to treat microbial infections in both infants and pregnant mothers and for antibiotic prophylaxis in preterm infants (8). Notably, infants without any exposure to antibiotics showed a higher percentage of Bifidobacteriaceae (8, 9). Thus, the occurrence of AR genes in bifidobacteria may increase their ecological fitness for gut persistence and colonization (10). Recently, the pan-genome, i.e., the full complement of genes of the Bifidobacterium genus has been reconstructed (11, 12), thereby providing a genomic data set, which will be important to identify the bifidobacterial resistome. Furthermore, we have evaluated the presence of AR genes in different strains belonging to the bifidobacterial (sub)species that are components of the qualified presumption of safety (QPS) list in accordance with the EFSA, such as Bifidobacterium bifidum, Bifidobacterium breve, Bifidobacterium adolescentis, Bifidobacterium longum subsp. longum, and Bifidobacterium animalis spp. (13, 14).
In the current study, we reconstructed the resistome of the Bifidobacterium genus based on both phenotypic and genotypic data, which together with the prediction of its genetic mobility allowed us to assess the potential of bifidobacterial AR genes to spread to the genomes of other gut bacteria. Furthermore, comparison of the, here identified, bifidobacterial resistome with those embedded in various microbiome data sets from infants and adults provides insights into the contribution of bifidobacteria to overall antibiotic resistance in the human gut microbial ecosystem.
RESULTS AND DISCUSSION
Assessment of antimicrobial susceptibility of bifidobacteria.
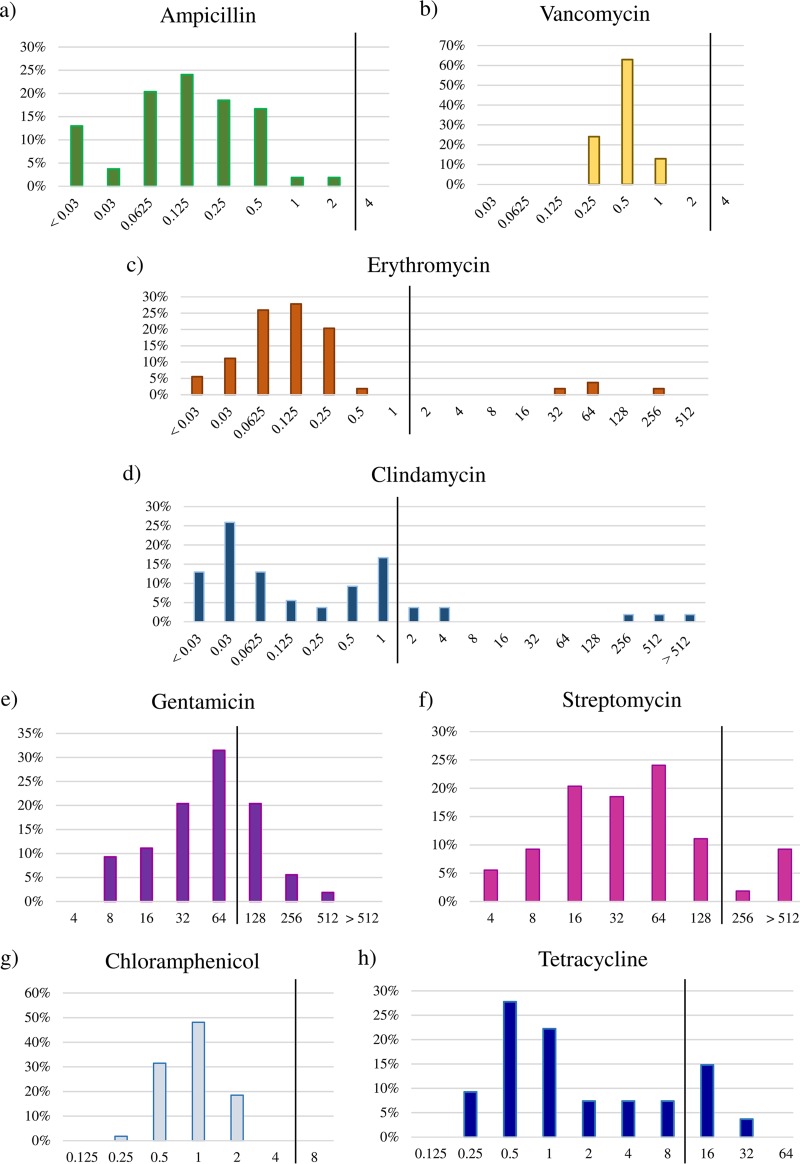
The susceptibility toward eight different antibiotics, i.e., ampicillin, vancomycin, gentamicin, streptomycin, erythromycin, tetracycline, chloramphenicol, and clindamycin, representing those indicated by EFSA (15, 51), was determined for 91 Bifidobacterium strains (Fig. 1 and Tables 1 and 2). As displayed in Fig. 1 and Table 1, we determined the breakpoint values for each antibiotic for the type strain of each of the currently known 54 bifidobacterial (sub)species. All type strains belonging to the 54 (sub)species of the Bifidobacterium genus showed a unimodal breakpoint value distribution for ampicillin (<0.03 to 2 μg/ml), vancomycin (0.25 to 1 μg/ml), gentamicin (8 to 512 μg/ml), and chloramphenicol (0.5 to 2 μg/ml) (Fig. 1a, b, e, and g). In contrast, for erythromycin, clindamycin, streptomycin, and tetracycline, a bimodal distribution with two or three different subpopulations (Fig. 1c, d, f, and h) was observed. In addition, we evaluated the susceptibility toward the same set of antibiotics at the intraspecies level (Table 2). Such analyses are crucial in order to understand the variability of antibiotic resistance within a particular taxon and to provide scientific support for the EFSA-suggested breakpoint values. Remarkably, the breakpoint values put forward by EFSA for the genus Bifidobacterium are based on values that were determined for a very small number of strains or for just a single strain (although proposed to apply to the whole genus) (15). Our analyses highlight a unimodal distribution of antibiotic resistance toward ampicillin, vancomycin, gentamicin, erythromycin, clindamycin, and chloramphenicol for all bifidobacterial species belonging to the QPS list. In contrast, we observed susceptibility variations to streptomycin and tetracycline at the intraspecific level for B. bifidum as well as B. breve and for B. bifidum, B. animalis, and B. adolescentis, respectively (Table 2). This information indicates that antibiotic resistance in bifidobacteria does not appear to follow a vertical route of evolution but may have been acquired through horizontal gene transfer, in a fashion similar to that previously observed for other gut commensal microorganisms (7).
FIG 1.
Microbiological breakpoint values. Distribution of breakpoint values for ampicillin (a), vancomycin (b), erythromycin (c), clindamycin (d), gentamicin (e) streptomycin (f), chloramphenicol (g), and tetracycline (h) in all type strains of the 54 (sub)species belonging to the Bifidobacterium genus. All bar plots report the antibiotic concentration values on the x axis (micrograms per milliliter) with the y axis showing the percentage of strains. The vertical black line represents the EFSA breakpoint value.
TABLE 1.
Antibiotic sensitivity of all type strains belonging to the Bifidobacterium genus
| Species | Resistance data (μg/ml) for: |
|||||||
|---|---|---|---|---|---|---|---|---|
| Ampicillin | Vancomycin | Gentamicin | Streptomycin | Erythromycin | Clindamycin | Tetracycline | Chloramphenicol | |
| B. actinocoloniiforme DSM 22766 | 0.0625 | 0.5 | 8 | 4 | 0.03 | 0.03 | 2 | 0.5 |
| B. adolescentis ATCC 15703 | 0.0625 | 0.5 | 128 | 32 | 0.0625 | 0.03 | 1 | 1 |
| B. aesculapii MRM 3/1 | 0.0625 | 0.5 | 64 | 16 | 0.0625 | <0.03 | 8 | 0.5 |
| B. angulatum LMG 11039 | 0.125 | 0.5 | 128 | 64 | 0.03 | 0.03 | 0.25 | 0.5 |
| B. animalis subsp. animalis LMG 10508 | 0.125 | 0.5 | 64 | 32 | 0.25 | 4 | 1 | 2 |
| B. animalis subsp. lactis DSM 10140 | 0.125 | 0.5 | 128 | 64 | 0.25 | 4 | 8 | 2 |
| B. asteroides LMG 10735 (PRL2011) | 0.0625 | 0.5 | 16 | 32 | 0.0625 | 0.0625 | 1 | 2 |
| B. biavatii DSM 23969 | 0.125 | 0.5 | 32 | 8 | 0.0625 | 0.03 | 0.25 | 0.5 |
| B. bifidum LMG 11041 | <0.06 | 0.25 | 8 | 8 | 0.125 | <0.06 | 0.5 | 0.5 |
| B. bohemicum DSM 22767 | 0.25 | 0.5 | 8 | 4 | 0.03 | <0.03 | 2 | 0.25 |
| B. bombi DSM 19703 | <0.03 | 1 | 8 | 4 | 0.0625 | 0.5 | 0.5 | 1 |
| B. boum LMG 10736 | <0.03 | 0.25 | 512 | 256 | 0.0625 | 0.125 | 32 | 0.5 |
| B. breve LMG 13208 | 0.0625 | 0.5 | 64 | 16 | 0.125 | 0.03 | 0.5 | 0.5 |
| B. callitrichos DSM 23973 | 0.0625 | 0.5 | 256 | 64 | 0.25 | 0.03 | 8 | 0.5 |
| B. catenulatum LMG 11043 | <0.03 | 0.5 | 128 | >512 | 0.0625 | 1 | 1 | 1 |
| B. choerinum LMG 10510 | 0.25 | 0.25 | 128 | 64 | 0.125 | 2 | 0.5 | 1 |
| B. coryneforme LMG 18911 | 0.25 | 0.5 | 32 | 16 | 0.0625 | 0.0625 | 4 | 2 |
| B. crudilactis LMG 23609 | 0.125 | 1 | 16 | 32 | 0.25 | 1 | 4 | 2 |
| B. cuniculi LMG 10738 | 0.5 | 0.5 | 32 | 32 | 0.125 | 1 | 2 | 1 |
| B. dentium LMG 11045 (Bd1) | 0.25 | 0.5 | 32 | 32 | 0.125 | 0.03 | 0.5 | 1 |
| B. eulemuris LMM E3 | 0.5 | 1 | 32 | 64 | 0.125 | 0.5 | 0.5 | 1 |
| B. gallicum LMG 11596 | 0.25 | 0.5 | 64 | 16 | <0.03 | 0.0625 | 0.5 | 0.5 |
| B. gallinarum LMG 11586 | 0.03 | 0.5 | 64 | >512 | <0.03 | 0.5 | 1 | 1 |
| B. hapali MRM 8.14 | 0.25 | 0.5 | 64 | 16 | 0.0625 | <0.03 | 0.5 | 1 |
| B. indicum LMG 11587 | 0.25 | 1 | 32 | 128 | 64 | 512 | 16 | 2 |
| B. kashiwanohense DSM 21854 | 0.5 | 0.5 | 128 | 64 | 32 | 256 | 0.5 | 1 |
| B. lemurum LMC13 | 0.5 | 1 | 64 | 32 | 0.03 | <0.03 | 0.5 | 1 |
| B. longum subsp. infantis ATCC 15697 | 0.125 | 0.5 | 8 | >512 | 0.125 | 0.0625 | 4 | 0.5 |
| B. longum subsp. longum LMG 13197 | 0.5 | 0.5 | 32 | 64 | 0.125 | <0.03 | 1 | 1 |
| B. longum subsp. suis LMG 21814 | <0.03 | 1 | 32 | >512 | 0.0625 | 0.03 | 16 | 1 |
| B. magnum LMG 11591 | <0.03 | 0.25 | 8 | 8 | <0.03 | 0.03 | 16 | 1 |
| B. merycicum LMG 11341 | 0.0625 | 0.5 | 64 | 64 | 0.125 | 0.03 | 16 | 1 |
| B. minimum LMG 11592 | 0.5 | 0.5 | 64 | 16 | 0.25 | 0.25 | 0.5 | 2 |
| B. mongoliense DSM 21395 | 0.25 | 1 | 16 | 8 | 0.25 | 0.0625 | 0.5 | 1 |
| B. moukalabense DSM 27321 | 0.125 | 0.5 | 128 | 128 | 0.125 | 1 | 0.5 | 0.5 |
| B. myosotis MRM 5.9 | 0.5 | 0.25 | 32 | 32 | 0.03 | <0.03 | 0.25 | 1 |
| B. pseudocatenulatum LMG 10505 | 0.0625 | 0.25 | 128 | 64 | 0.25 | 0.03 | 16 | 1 |
| B. pseudolongum subsp. globosum LMG 11569 | 0.125 | 0.5 | 128 | 128 | 0.0625 | 1 | 8 | 1 |
| B. pseudolongum subsp. pseudolongum LMG 11571 | 0.0625 | 0.25 | 64 | 128 | 0.125 | 0.125 | 2 | 1 |
| B. psychraerophilum LMG 21775 | 0.125 | 0.5 | 64 | 32 | 0.0625 | 1 | 4 | 2 |
| B. pullorum LMG 21816 | 1 | 0.25 | 32 | 16 | 64 | >1,024 | 32 | 2 |
| B. reuteri DSM 23975 | 0.125 | 0.5 | 32 | 16 | 0.25 | 0.5 | 0.25 | 0.5 |
| B. ruminantium LMG 21811 | 0.25 | 0.25 | 16 | 8 | 0.125 | 1 | 0.25 | 0.5 |
| B. saeculare LMG 14934 | 0.125 | 0.5 | 64 | >512 | 256 | 1 | 16 | 0.5 |
| B. saguini DSM 23967 | 0.5 | 0.5 | 64 | 16 | 0.25 | 1 | 1 | 1 |
| B. scardovii LMG 21589 | 0.5 | 0.5 | 64 | 16 | 0.25 | 0.25 | 1 | 1 |
| B. stellenboschense DSM 23968 | <0.03 | 0.5 | 16 | 16 | 0.0625 | 0.03 | 0.5 | 0.5 |
| B. stercoris DSM 24849 | 0.0625 | 0.5 | 64 | 64 | 0.25 | 0.03 | 16 | 1 |
| B. subtile LMG 11597 | 0.25 | 0.5 | 64 | 32 | 0.0625 | 0.5 | 1 | 1 |
| B. thermacidophilum subsp. porcinum LMG 21689 | 0.03 | 0.25 | 256 | 128 | 0.125 | 0.0625 | 1 | 0.5 |
| B. thermacidophilum subsp. thermacidophilum LMG 21395 | 0.125 | 0.25 | 16 | 64 | 0.125 | 0.125 | 0.5 | 2 |
| B. thermophilum JCM 1207 | 0.0625 | 0.25 | 256 | 128 | 0.125 | 0.0625 | 1 | 0.5 |
| B. tissieri MRM 5.18 | 2 | 0.25 | 128 | 64 | 0.03 | 0.03 | 1 | 1 |
| B. tsurumiense JCM 13495 | 0.125 | 0.5 | 128 | 64 | 0.5 | 2 | 1 | 1 |
TABLE 2.
Antibiotic sensitivity of different strains belonging to the QPS list
| Species | Strain | Resistance data (μg/ml) for: |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Ampicillin | Vancomycin | Gentamicin | Streptomycin | Erythromycin | Clindamycin | Tetracycline | Chloramphenicol | ||
| B. bifidum | PRL2010 | 0.0625 | 1 | 64 | 32 | 0.125 | <0.03 | 1 | 1 |
| LMG13195 | <0.03 | 0.5 | 64 | >1,024 | 0.0625 | 0.03 | 0.5 | 0.5 | |
| LMG11041 | <0.06 | 0.25 | 8 | 8 | 0.125 | <0.06 | 0.5 | 0.5 | |
| IPLA20017 | 0.03 | 0.5 | 32 | >1,024 | 0.125 | 0.0625 | 0.5 | 0.5 | |
| IPLA20015 | 0.0625 | 0.5 | 32 | >1,024 | 0.0625 | 0.0625 | 8 | 0.5 | |
| A8 | 0.03 | 1 | 64 | 8 | 0.0625 | 0.03 | 0.5 | 0.5 | |
| 156B | 0.0625 | 0.5 | 64 | 16 | 0.03 | 0.0625 | 1 | 1 | |
| 85B | 0.0625 | 0.5 | 32 | 16 | 0.03 | 0.0625 | 1 | 0.5 | |
| 324B | 0.0625 | 1 | 64 | 16 | 0.0625 | 0.0625 | 1 | 0.5 | |
| LMG13200 | 0.5 | 0.5 | 16 | 256 | 0.0625 | 0.25 | 8 | 0.5 | |
| LMG11583 | 0.5 | 0.5 | 32 | 16 | 0.03 | 0.25 | 16 | 1 | |
| LMG11582 | 0.5 | 0.5 | 32 | >1,024 | 0.125 | 0.25 | 16 | 0.5 | |
| B. breve | 689B | 0.5 | 0.5 | 32 | >1,024 | 0.125 | 0.25 | 0.5 | 0.5 |
| DSM20213 | 0.0625 | 0.5 | 64 | 16 | 0.125 | 0.03 | 0.5 | 0.5 | |
| 12L | 0.25 | 1 | 32 | 32 | 0.125 | 0.25 | 0.5 | 0.5 | |
| 2L | 0.25 | 1 | 16 | 32 | 0.125 | 0.25 | 0.25 | 0.5 | |
| D1-16 | 0.25 | 0.5 | 8 | >1,024 | 0.125 | <0.03 | 0.5 | 1 | |
| 31L | 0.5 | 1 | 32 | >1,024 | 0.125 | <0.03 | 0.5 | 0.5 | |
| UCC2003 | 0.125 | 1 | 64 | 32 | 0.25 | 0.5 | 0.125 | 1 | |
| B. animalis subsp. lactis | Bl12 | 0.125 | 0.5 | 128 | 64 | 0.25 | 4 | 8 | 2 |
| BLC1 | 0.125 | 0.5 | 128 | 64 | 0.25 | 4 | 8 | 2 | |
| DSM10140 | 0.125 | 0.5 | 128 | 64 | 0.25 | 4 | 8 | 2 | |
| 646 | 0.125 | 0.5 | 128 | 64 | 0.25 | 4 | 8 | 2 | |
| BB12 | 0.125 | 0.5 | 128 | 64 | 0.25 | 4 | 8 | 2 | |
| ADO11 | 0.125 | 0.5 | 128 | 64 | 0.25 | 4 | 8 | 2 | |
| B. animalis subsp. animalis | ATCC25527 | 0.125 | 0.5 | 64 | 32 | 0.25 | 4 | 1 | 2 |
| B. adolescentis | 22L | <0.03 | 0.25 | 32 | 32 | 0.5 | <0.03 | 0.5 | 2 |
| ATCC15703 | 0.0625 | 0.5 | 128 | 32 | 0.0625 | 0.03 | 1 | 1 | |
| LMG18897 | <0.03 | 0.25 | 16 | 32 | 0.25 | <0.03 | 0.25 | 2 | |
| LMG11579 | 0.25 | 0.25 | 32 | 32 | 1 | 1 | 16 | 1 | |
| LMG10733 | 0.0625 | 0.25 | 64 | 32 | 0.25 | <0.03 | 1 | 0.5 | |
| LMG10734 | 0.125 | 0.25 | 64 | 32 | 0.5 | <0.03 | 0.5 | 2 | |
| 42B | 0.0625 | 0.25 | 32 | 16 | 0.25 | <0.03 | 0.5 | 2 | |
| 70B | 0.0625 | 0.25 | 64 | 64 | 0.25 | 0.03 | 0.5 | 2 | |
| 487B | <0.03 | 0.25 | 32 | 32 | 0.5 | 0.125 | 0.5 | 2 | |
| 703B | 0.25 | 0.25 | 64 | 32 | 0.5 | <0.03 | 1 | 2 | |
| AD2-8 | 0.03 | 0.25 | 16 | 32 | 0.25 | <0.03 | 0.5 | 2 | |
| AL12-4 | 0.0625 | 0.25 | 32 | 16 | 0.5 | <0.03 | 1 | 2 | |
| AL46-2 | 1 | <0.03 | 32 | 32 | <0.03 | 0.125 | 16 | 0.5 | |
| AL46-7 | 0.0625 | 0.25 | 32 | 16 | 0.25 | 0.25 | 16 | 2 | |
| JCM15918 | 0.0625 | 0.5 | 64 | 64 | 0.25 | 0.03 | 16 | 1 | |
| B. longum subsp. longum | 296B | 0.125 | 0.5 | 32 | 64 | <0.03 | <0.03 | 1 | 2 |
| LMG13197 | 0.5 | 0.5 | 32 | 64 | 0.125 | <0.03 | 1 | 1 | |
| B2A2 | 0.125 | 0.25 | 32 | 64 | 0.0625 | <0.03 | 1 | 1 | |
Prediction of the bifidobacterial resistome.
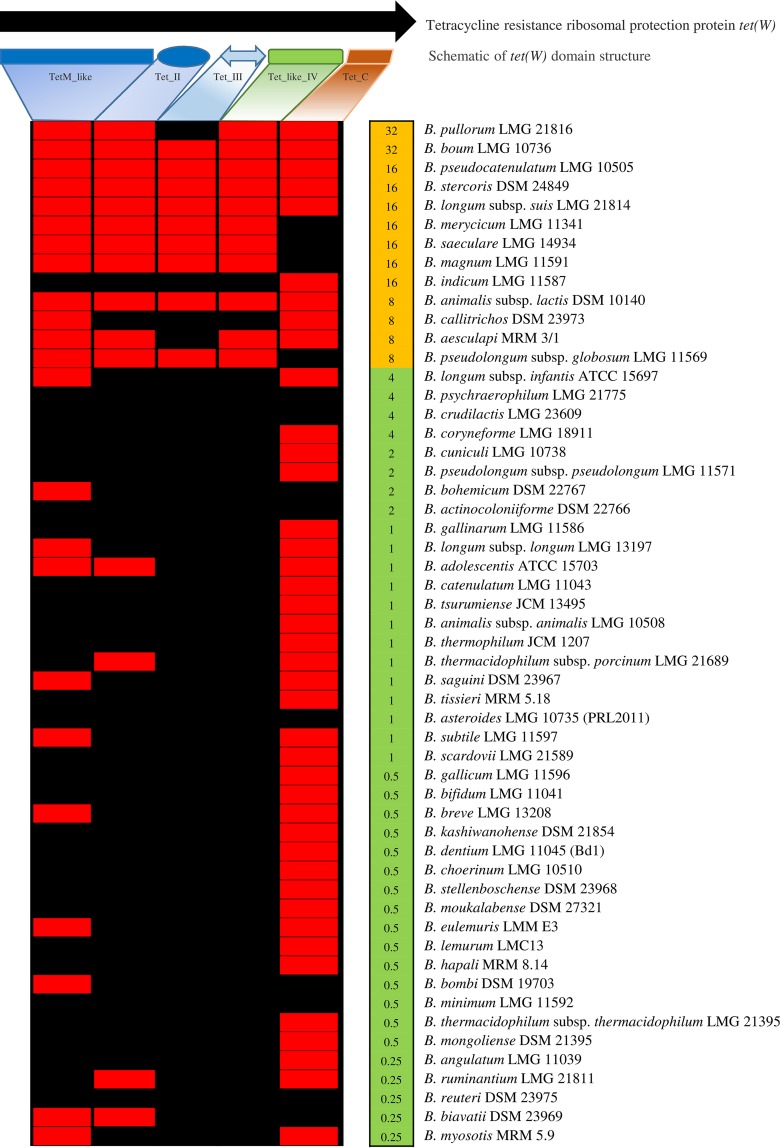
The genomes of 91 bifidobacterial strains that had been assessed for their susceptibility to antibiotics (see above) were screened for putative AR genes. This analysis was performed using the CARD AR gene database (16), to which two genes, predicted to encode aminoglycoside phosphotransferases (APH), had been added. Notably, the presence of these two genes in B. breve has previously been shown to be associated with resistance to gentamicin, streptomycin, and kanamycin (17). These in silico analyses revealed that the resistome of the genus Bifidobacterium is predicted to consist of 783 genes (see Table S1 in the supplemental material). Of the AR genes identified, 47% were shown to be conserved in the 54 (sub)species of the genus Bifidobacterium, thus representing bifidobacterial core genome sequences (12, 18). While 74% of these genes encode β-lactamases, 26% are involved in antimicrobial peptide resistance (Table S1). Interestingly, analysis of chromosomal sequences revealed the presence of a single copy of a predicted APH-encoding gene in all Bifidobacterium strains, Furthermore, in 20 bifidobacterial genomes, we identified two copies of such a putative APH-encoding gene, which may account for higher resistance toward streptomycin and gentamicin in these bifidobacterial strains (see Table S2 in the supplemental material). In contrast, tet(W) homologs, previously identified in Bifidobacterium animalis subsp. lactis (19, 20), are present in the genomes of 11 bifidobacterial species (Fig. 2), based on in silico tet(W) domain prediction, thus highlighting the fact that this AR gene represents a relatively broad species-specific genetic signature within the bifidobacterial pan-genome. Notably, in vitro-based MIC assays involving these strains revealed high resistance toward tetracycline (from 8 μg/ml to 32 μg/ml), consistent with the presence of the tet(W) gene in their genomes (Fig. 2). In addition, tetracycline resistance may be conferred by additional genetic features, since Bifidobacterium indicum LMG 11587 and Bifidobacterium callitrichos DSM 23973 encompass no less than three tet(W) domains (Fig. 2).
FIG 2.
Comparison of tet(W) gene identified in B. animalis subsp. lactis with the corresponding domains from all type strains of bifidobacteria. The top part of the figure depicts a schematic representation of the different domains identified in tet(W). The heat map displays an in silico prediction of domains encoded by tet(W) and the breakpoint value for tetracycline in all type strains belonging to the Bifidobacterium genus. The red color indicates gene presence, and the black color represents their absence.
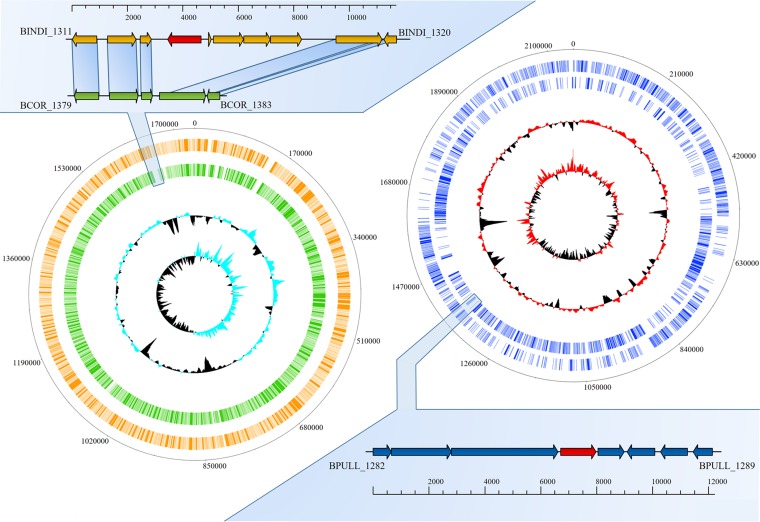
When the antibiotic susceptibility data were coupled to the predicted resistome information, we were able to detect good correspondence between phenotype and genotype. For instance, in B. indicum LMG 11587 and Bifidobacterium coryneforme LMG 18911, different susceptibilities to streptomycin, erythromycin, clindamycin and tetracycline were identified, which appeared to be supported by the genotypic data. Recently, Lugli et al. (12) highlighted that a very close genetic relatedness exists between B. indicum LMG11587 and B. coryneforme LMG18911 (average nucleotide identity [ANI] value of 98.13). Comparative genomic analysis involving the chromosomal sequences of these strains allowed the identification of genes that are absent in the genome of B. coryneforme LMG 18911 (Fig. 3). Interestingly, an in-depth functional analysis of the B. indicum LMG 11587 unique genes highlighted the presence of an open reading frame (ORF) predicted to encode a major facilitator superfamily (MFS) transporter that is classified as a member of the drug:H+-antiporter-3 (12 spanner) (DHA3) family, which may be responsible for the extrusion of erythromycin, streptomycin, and clindamycin from the (bifido)bacterial cell (Fig. 3). Similarly, Bifidobacterium pullorum LMG 21816 was shown to exhibit a high level of resistance against clindamycin (>1,024 μg/ml) compared to that identified for other type strains of the 54 (sub)species of the genus Bifidobacterium. Comparative genomics involving all type strains of the genus showed 138 protein-encoding genes that appeared to be unique to B. pullorum LMG21816. Notably, one of these genes (BPULL_1285) was predicted to encode an MFS transporter classified as a DHA3 family transporter. This putative multidrug transporter may thus be responsible for the noted resistance to clindamycin as also observed for B. coryneforme LMG 18911. Nonetheless, in five cases, Bifidobacterium catenulatum LMG 11043, Bifidobacterium gallinarum LMG 11586, Bifidobacterium longum subsp. infantis ATCC 15697, Bifidobacterium longum subsp. suis LMG 21814, and Bifidobacterium saeculare LMG 14934 (Table S1), the observed high resistance to streptomycin (>512 μg/ml) does not appear to be associated with the presence of a specific AR gene. This resistance may therefore either be due to a point mutation in a particular chromosomal gene causing innate resistance against streptomycin or be caused by high expression of a particular AR gene. Furthermore, based on the reconstruction of the genetic evolution of species of the genus Bifidobacterium based on their pan-genomes (11), we predicted the genetic origins of AR identified in bifidobacteria. Notably, 6% of the predicted bifidobacterial resistome appears to be acquired from other microbial genera such as Gardnerella and Lactobacillus, representing bacterial taxa that share the same environmental niche as bifidobacteria (Table S1). Nonetheless, we cannot exclude the possibility that these genes might be received independently from another common source. These findings therefore suggest that a sizeable portion of the genetic AR arsenal of bifidobacteria has been acquired by means of HGT events.
FIG 3.
Unique genetic loci identified in the genome of B. indicum LMG 11587 and B. pullorum LMG 21816. On the left, a circular genome atlas of B. indicum LMG 11587 (orange circle) and B. coryneforme LMG 18911 (green circle) are shown, while on the right, the same representation is proposed for the B. pullorum LMG 21816 genome (blue circles). Internal circles represent the G+C percent deviation followed by the GC skew (G−C/G+C). In the reported genetic maps, each arrow indicates an ORF where the red ones correspond to the major facilitator superfamily (MFS) identified in B. indicum LMG 11587 (orange arrows) and B. pullorum LMG 21816 (blue arrows).
Identification of putative mobile bifidobacterial AR genes.
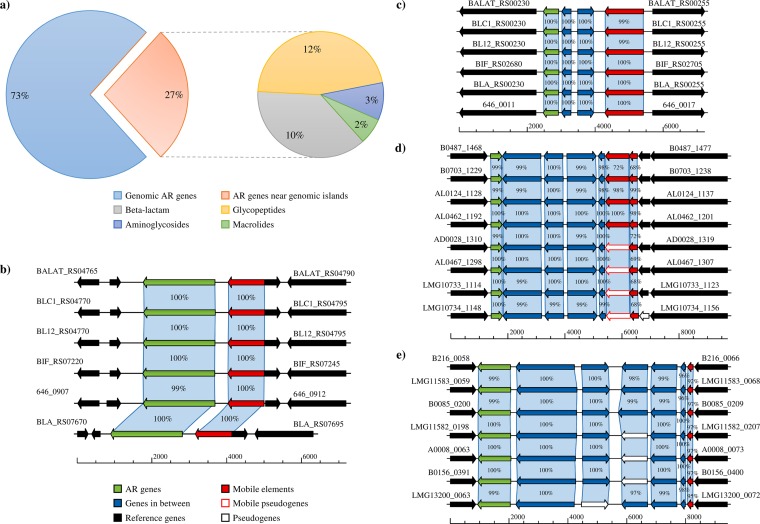
In order to evaluate the occurrence of bifidobacterial AR genes located on or in the proximity of mobile elements such as episomes, conjugative transposons, and prophages, we analyzed the flanking DNA sequences of the predicted AR genes (see Tables S1 and S3 in the supplemental material). Such genetic elements may be responsible for AR gene mobilization from and/or to other microorganisms. Homology-based analyses of the surrounding DNA regions of the predicted bifidobacterial AR genes were performed and combined with outputs of the software package COLOMBO (21). These analyses identified 208 AR genes placed within genomic regions that have putatively been acquired by HGT, thus representing the predicted horizontally acquired bifidobacterial resistome (HABR) (Fig. 4). Notably, HABR represents about 27% of the predicted bifidobacterial resistome, disregarding the proximity of the genes to transposable elements. Taken together, these results suggest that a substantial proportion of the bifidobacterial AR genes identified were acquired by HGT. Furthermore, in silico analysis disclosed that only seven of the AR genes identified are placed near a predicted prophage, while two are adjacent to putative plasmid replication genes.
FIG 4.
Distribution of mobile bifidobacterial AR genes. (a)The left pie chart represents chromosomal bifidobacterial AR genes that are (red) or are not (blue) predicted to be acquired by HGT, and the pie chart on the right displays the classification of such predicted HGT-acquired AR genes. (b to e) Bifidobacterial genomic regions containing putative AR genes (green arrows) located near mobile elements (red arrows) belonging to B. animalis subsp. lactis, B. adolescentis, and B. bifidum, respectively.
Remarkably, we noticed that the largest part of the predicted bifidobacterial HABR is directed toward glycopeptides (12%), followed by β-lactamase-encoding genes (10%) (Fig. 4a). Furthermore, the AR genes flanking prophage and plasmid DNA regions are represented by genes predicted to encode bleomycin, kanamycin, and bacitracin resistance (Table S1). Another intriguing bifidobacterial mobile AR gene predicted to be carried on a conjugative transposon is represented by the tet(W) gene, which is found flanking a mobile element in the genomes of all of the B. animalis subsp. lactis strains analyzed (Fig. 4b). Such findings confirm previous genomic data about this bifidobacterial taxon (19) and support the hypothesis that this resistance is due to a mobile tet(W) gene representing a common genetic feature of B. animalis subsp. lactis. Furthermore, we identified a putative mobile AR gene that encodes a predicted aminoglycoside protein with an APH domain for kanamycin resistance in all publicly available genomes of B. animalis subsp. lactis (Fig. 4c; see also Table S3 in the supplemental material). Yet another intriguing example is represented by analysis of the genomes of B. adolescentis, of which eight encompass an AR gene predicted to exert resistance against bleomycin, located within 4 kb from a putative mobile element (Fig. 4d and Table S3). Remarkably, seven strains of B. bifidum exhibit β-lactamase-encoding genes near truncated transposases (Fig. 4e and Table S3). Nevertheless, none of these transposase-encoding genes can be classified as a conjugal transposon, thus reducing the possibility of AR gene mobilization by HGT. These findings indicate that only a small fraction (1%) of the predicted resistome found in the genus Bifidobacterium resides in nearby mobile elements, which may facilitate AR gene transfer to other bacteria. The distribution of AR genes in bifidobacteria may thus be due to selective pressure imposed by extensive antibiotic use in their animal and human hosts in a fashion that is similar to what has previously been observed for other lactic acid bacteria (LAB) such as Lactobacillus (22), a phenomenon that is considered to represent microbe-host coevolution.
Metagenome analysis targeting bifidobacterial AR.
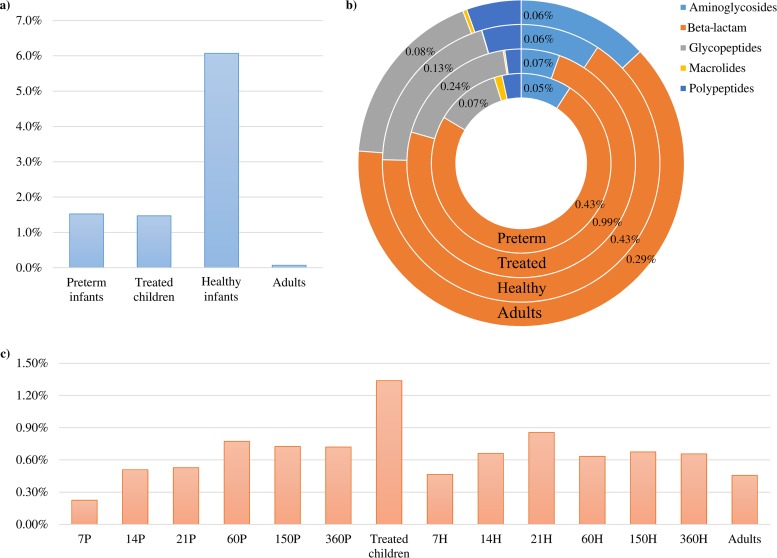
Bifidobacteria have predominantly been isolated from the mammalian GIT (23), where their functional contribution in terms of metabolism of various dietary carbohydrates and host glycans has been investigated (11, 18, 24–26). However, their contribution to the resistome of the mammalian gut microbiome has not been studied in any great detail. We therefore assessed the presence of AR-encoding bifidobacterial DNA sequences within two gut metagenome data sets from healthy human beings, including adult gut microbiomes (27) and infant gut microbiomes (https://www.broadinstitute.org/scientific-community/science/projects/microbiome-projects), which in both cases had been collected from U.S. citizens (see Table S4 in the supplemental material). In addition, we explored the bifidobacterial AR contribution to the resistomes of preterm infant gut microbiomes (https://www.broadinstitute.org/scientific-community/science/projects/microbiome-projects) and of 2- to 7-year-old children who had received intensive antibiotic treatment, i.e., a combination of β-lactams and macrolides (28). These metagenomic data sets were assayed for the presence of bifidobacteria by searching for bifidobacterial gene sequences (BGS), i.e., the presence of any of the combined genes collected from the 91 taxa analyzed. The minimum coverage of each gene included in the BGS collection was computed based on the metagenomic reads with 98% full-length identity. As displayed in Fig. 5, healthy infants showed a higher average percentage of metagenomic reads that correspond to BGS (6.1%, P < 0.001), ranging from 0.01% to 52%, than adults (0.1%), while preterm infants and children treated with antibiotics exhibited an average of 1.5%. This is consistent with previous findings supporting the ecological behavior of bifidobacteria as core gut microorganisms of the healthy human gut during the suckling stage (2, 23, 29–36). Furthermore, our analysis revealed reduced bifidobacterial populations in preterm infants and children extensively treated with antibiotics, which is consistent with the existing literature (37).
FIG 5.
Metagenomic abundance of bifidobacterial AR genes. (a) Average abundance of bifidobacterial reads retrieved in the metagenome samples of infants (healthy and preterm), children treated with antibiotics, and adults. (b) Abundance of different classes of bifidobacterial AR genes with respect to the total BGS identified in the samples. (c) Average abundance of bifidobacterial AR reads with respect to the total BGS retrieved in the metagenomes, where each number corresponds to the days following birth of healthy infants (H) or preterm infants (P).
The bifidobacterial AR genes collected from the genomic analysis allowed us to evaluate the contribution of the bifidobacterial resistome with respect to the total BGS identified in each sample. Interestingly, among the most frequently represented bifidobacterial AR genes in the metagenomic data sets from adult samples, an extensive repertoire of specific AR-encoding genes, such as those specifying β-lactamases, which confer resistance against penicillin, cephalosporin, carbapenem, and monobactam, is noteworthy, followed by genes that specify glycopeptide and aminoglycoside resistance (Fig. 5b). Such findings reinforce the notion that despite the lower abundance of bifidobacteria detected in the adult human gut, their functional contribution to the human gut microbiome may be important in terms of expanding the overall resistome of the human gut and thus affecting the maintenance of a gut climax following antibiotic therapies. Of note, the bifidobacterial AR-encoding gene distribution within the infant's microbiome was shown to be similar to that of adults with a preponderance of β-lactamase-encoding genes (Fig. 5b).
The availability of microbiomes from the Metagenome from Infant Gut project collected at different time points, i.e., 7, 14, 21, 60, 150, and 360 days, allowed us to evaluate the dynamics of bifidobacterial AR-encoding genes during the first year of life of healthy and preterm infants. Looking at the overall representation of bifidobacterial AR reads in the metagenomic data sets, a lower value is obtained for healthy and preterm infants in the first week of life, followed by an increase, which then stabilizes during a 60- to 360-day period, thus being comparable with an adult's profile (Fig. 5c). In contrast, in the metagenome data set of children extensively treated with antibiotics, we observed a higher abundance of bifidobacterial AR reads (1.34%, P < 0.006), suggesting the increased presence of bifidobacterial species that tolerate the administered dosage of antibiotics assumed during treatment (Fig. 5c). Furthermore, we observed a higher abundance in β-lactamase reads (0.99% of the bifidobacterial reads, P < 0.001) in the antibiotic-treated children with respect to those in the other three groups (ranging from 0.29% to 0.43%) (Fig. 5b), reflecting the antibiotic treatment provided. Accordingly, during the first days following birth, the infants' microbiota may be more sensitive to antibiotic treatment due to a very simple and fragile bifidobacterial community, which appears to be unable to cope with a high level of antibiotic administration. Thus, the particular antibiotic administered to the newborn may bring about an intestinal microbiota disturbance that can prevent or delay subsequent development of a normal microbiota (38).
In conclusion, bifidobacteria are dominant members of the infant gut microbiota, and their contribution to host health is well documented (24). Despite many reports investigating the susceptibility of human bifidobacterial species to various antibiotics, very little is known about their resistome. In this study, we performed a detailed assessment of the genetic traits that support antibiotic resistance in bifidobacteria. Notably, varying susceptibilities to antibiotics at the intraspecific level were identified for certain bifidobacterial species, an observation of relevance with respect to the scientific rationale for breakpoint values proposed by EFSA (15), as currently employed breakpoint values are based on MIC values determined for a single bifidobacterial strain. In addition, based on our analyses, the identification of higher MIC values with respect to breakpoint values proposed by EFSA does not always correspond with the occurrence of predicted mobile AR genes and thus does not necessarily pose any risk of genetic transferability. Interestingly, the resistome of bifidobacteria represents a substantial proportion of the predicted mobilome of the genus Bifidobacterium, thus supporting the hypothesis that the antibiotic era has somehow shaped the pan-genome of this group of commensal microorganisms. Notably, a similar trend has already been observed for other gut commensal microorganisms such as Lactobacillus (22). Acquisition of resistance to antibiotics represents a way used by bacteria to survive and thus to increase their ecological fitness (39). Characterization of the bifidobacterial resistome allowed us to obtain insights into the manner by which the bifidobacterial community contributes to the overall gut microbiota resistome. Our data show that the bifidobacterial communities in the infant gut possess a reduced AR arsenal compared to that present in the bifidobacterial gut microbiota of an older child. These data reinforce the notion that the infant gut microbiota, particular that present during the first weeks following birth, is more prone to perturbations following antibiotic therapy and may thus be highly susceptible to long-term disturbances, compared to the stable and more robust (adult) gut microbiota that develops subsequently.
MATERIALS AND METHODS
Bacterial strains.
All type strains belonging to the Bifidobacterium genus and several previously characterized strains belonging to species present on the qualified presumption of safety (QPS) list (11, 13, 19, 40–44) were used in this study (Tables 1 and 2). Cultures were grown under anaerobic conditions (2.99% H2, 17.01% CO2, and 80% N2) in a chamber (Concept 400; Ruskin) on De Man-Rogosa-Sharpe (MRS) broth (Scharlau Chemie, Barcelona, Spain) supplemented with 0.05% (wt/vol) l-cysteine hydrochloride and were incubated at 37°C. Prior to performance of the antibiotic susceptibility test, strains were precultivated (to allow adaptation) in the same medium used for the susceptibility test, based on the use of Iso-Sensitest (IST) broth (Oxoid).
Antibiotic susceptibility tests.
For selected Bifidobacterium strains, the MIC breakpoints (micrograms per milliliter) of eight antibiotics (ampicillin, vancomycin, gentamicin, streptomycin, erythromycin, clindamycin, tetracycline, and chloramphenicol) were determined using the broth microdilution method (MDIL) according to the ISO standard guidelines (15). All antibiotics were purchased from Sigma-Aldrich (Italy). Microplates were incubated under anaerobic conditions for 48 h at 37°C. Cell density was monitored by optical density measurements at 600 nm (OD600) using a plate reader (BioTek, VT, USA). The MIC breakpoint represents the highest concentration of a given antibiotic to which a particular bacterial strain is resistant.
Antibiotic resistance gene prediction.
The in silico proteome of 93 Bifidobacterium strains (Tables 1 and 2) was screened for enzymes that act as antibiotic inactivators using a custom script based on RapSearch2 software (45) and the database CARD (16). We decided not to include transporters in our analyses due to the low accuracy with which antibiotic transporters can be predicted.
Prediction of the mobile bifidobacterial AR genes.
The bifidobacterial strains used in this study were screened for genomic islands, evaluating the genes flanking the predicted AR genes in the range of 10 kb, using homology searches (46) and the software COLOMBO (21). Putative mobile elements such as episomes, conjugative transposons, and prophages were predicted through homology searches against in-house-generated databases (http://probiogenomics.unipr.it/sw/MobElemDB.zip), including genes retrieved from the National Center for Biotechnology Information (NCBI) database.
In silico analysis for resistome reconstruction.
All identified bifidobacterial AR genes were aligned with whole-genome sequencing (WGS) reads previously deposited at the NCBI (Sequence Read Archive [SRA] BioProject). This information was obtained from shotgun sequencing microbiome data sets of (fecal samples of) healthy adults from the Human Microbiome Project (PRJNA48479), healthy and preterm infants from the Metagenome from Infant Gut project (PRJNA63661), and children with high-frequency antibiotic treatment from the Child Gut Microbiome under Antibiotics project (PRJEB11685). Metagenomic data sets were filtered using the fastq-mcf script (https://expressionanalysis.github.io/ea-utils/) (minimum mean quality score, 20; window size, 5; quality threshold, 25; and minimum length, 80) to achieve high-quality reads exclusively. The resulting reads were aligned against the human genome using the Burrows-Wheeler Aligner program (47) (BWAMEM algorithm with trigger reseeding, 1.5; minimum seed length, 19; matching score, 1; mismatch penalty, 4; gap open penalty, 6; and gap extension penalty, 1) and further processed with the SAMtools software package (48) in order to remove human reads. The final mapping against putative bifidobacterial AR genes was performed using Bowtie 2 (49) through multiple-hit mapping and “very-sensitive” policy. The mapping was performed using a minimum score threshold function (–score-min C,−13,0) in order to limit reads of arbitrary length to two mismatches and retain those matches with at least 98% full-length identity. The software employed to calculate read counts corresponding to either bifidobacterial genes or bifidobacterium-specific AR genes was HTSeq (50) (running in union mode). The percentages of bifidobacterial genes for each sample were based on the total amount of filtered reads, while the percentages of bifidobacterial AR genes reported were based on the counts of reads mapped to bifidobacterial genes in each sample.
Statistical analysis.
SPSS software (IBM, Italy) was used to perform statistical analysis between groups by an analysis of variance (ANOVA) test.
Accession number(s).
The bifidobacterial sequences reported in this article have been deposited in the GenBank database under accession numbers MLZK00000000 and MLZL00000000. The versions described in this paper are MLZK01000000 and MLZL01000000.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by the EU Joint Programming Initiative, A Healthy Diet for a Healthy Life (JPI HDHL, http://www.healthydietforhealthylife.eu/) through MIUR and Science Foundation Ireland (SFI) to M.V. and D.V.S., respectively. We thank GenProbio srl for financial support of the Laboratory of Probiogenomics. S.D. is supported by Fondazione Caritro, Trento, Italy. L.M. is supported by Fondazione Cariparma, Parma, Italy. D.V.S. is a member of The APC Microbiome Institute funded by SFI (grant SFI/12/RC/2273).
We thank Paola Mattarelli for sharing bifidobacterial isolates.
We declare that we have no competing interests.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02894-16.
REFERENCES
- 1.Young VB. 2012. The intestinal microbiota in health and disease. Curr Opin Gastroenterol 28:63–69. doi: 10.1097/MOG.0b013e32834d61e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turroni F, Peano C, Pass DA, Foroni E, Severgnini M, Claesson MJ, Kerr C, Hourihane J, Murray D, Fuligni F, Gueimonde M, Margolles A, De Bellis G, O'Toole PW, van Sinderen D, Marchesi JR, Ventura M. 2012. Diversity of bifidobacteria within the infant gut microbiota. PLoS One 7:e36957. doi: 10.1371/journal.pone.0036957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marshall BM, Levy SB. 2011. Food animals and antimicrobials: impacts on human health. Clin Microbiol Rev 24:718–733. doi: 10.1128/CMR.00002-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Francino MP. 2015. Antibiotics and the human gut microbiome: dysbioses and accumulation of resistances. Front Microbiol 6:1543. doi: 10.3389/fmicb.2015.01543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ouwehand AC, Forssten S, Hibberd AA, Lyra A, Stahl B. 2016. Probiotic approach to prevent antibiotic resistance. Ann Med 48:246–255. doi: 10.3109/07853890.2016.1161232. [DOI] [PubMed] [Google Scholar]
- 6.Goossens H, Ferech M, Stichele RV, Elseviers M, Grp EP. 2005. Outpatient antibiotic use in Europe and association with resistance: a cross-national database study. Lancet 365:579–587. doi: 10.1016/S0140-6736(05)70799-6. [DOI] [PubMed] [Google Scholar]
- 7.Huddleston JR. 2014. Horizontal gene transfer in the human gastrointestinal tract: potential spread of antibiotic resistance genes. Infect Drug Resist 7:167–176. doi: 10.2147/IDR.S48820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arboleya S, Sanchez B, Milani C, Duranti S, Solis G, Fernandez N, de los Reyes-Gavilan CG, Ventura M, Margolles A, Gueimonde M. 2015. Intestinal microbiota development in preterm neonates and effect of perinatal antibiotics. J Pediatr 166:538–544. doi: 10.1016/j.jpeds.2014.09.041. [DOI] [PubMed] [Google Scholar]
- 9.Fouhy F, Guinane CM, Hussey S, Wall R, Ryan CA, Dempsey EM, Murphy B, Ross RP, Fitzgerald GF, Stanton C, Cotter PD. 2012. High-throughput sequencing reveals the incomplete, short-term recovery of infant gut microbiota following parenteral antibiotic treatment with ampicillin and gentamicin. Antimicrob Agents Chemother 56:5811–5820. doi: 10.1128/AAC.00789-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patel AR, Shah NP, Prajapati JB. 2012. Antibiotic resistance profile of lactic acid bacteria and their implications in food chain. World J Dairy Food Sci 7:202–211. [Google Scholar]
- 11.Milani C, Lugli GA, Duranti S, Turroni F, Bottacini F, Mangifesta M, Sanchez B, Viappiani A, Mancabelli L, Taminiau B, Delcenserie V, Barrangou R, Margolles A, van Sinderen D, Ventura M. 2014. Genomic encyclopedia of type strains of the genus Bifidobacterium. Appl Environ Microbiol 80:6290–6302. doi: 10.1128/AEM.02308-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lugli GA, Milani C, Turroni F, Duranti S, Ferrario C, Viappiani A, Mancabelli L, Mangifesta M, Taminiau B, Delcenserie V, van Sinderen D, Ventura M. 2014. Investigation of the evolutionary development of the genus Bifidobacterium by comparative genomics. Appl Environ Microbiol 80:6383–6394. doi: 10.1128/AEM.02004-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.European Food Safety Authority. 2015. Scientific opinion on the maintenance of the list of QPS biological agents intentionally added to food and feed (2013 update). EFSA J 11:3449. [Google Scholar]
- 14.Merenstein DJ, Tan TP, Molokin A, Smith KH, Roberts RF, Shara NM, Mete M, Sanders ME, Solano-Aguilar G. 2015. Safety of Bifidobacterium animalis subsp. lactis (B. lactis) strain BB-12-supplemented yogurt in healthy adults on antibiotics: a phase I safety study. Gut Microbes 6:66–77. doi: 10.1080/19490976.2015.1005484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.European Food Safety Authority. 2012. Guidance on the assessment of bacterial susceptibility to antimicrobials of human and veterinary importance. EFSA J 10:2740. [Google Scholar]
- 16.McArthur AG, Waglechner N, Nizam F, Yan A, Azad MA, Baylay AJ, Bhullar K, Canova MJ, De Pascale G, Ejim L, Kalan L, King AM, Koteva K, Morar M, Mulvey MR, O'Brien JS, Pawlowski AC, Piddock LJ, Spanogiannopoulos P, Sutherland AD, Tang I, Taylor PL, Thaker M, Wang W, Yan M, Yu T, Wright GD. 2013. The comprehensive antibiotic resistance database. Antimicrob Agents Chemother 57:3348–3357. doi: 10.1128/AAC.00419-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fouhy F, O'Connell Motherway M, Fitzgerald GF, Ross RP, Stanton C, van Sinderen D, Cotter PD. 2013. In silico assigned resistance genes confer Bifidobacterium with partial resistance to aminoglycosides but not to beta-lactams. PLoS One 8:e82653. doi: 10.1371/journal.pone.0082653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Milani C, Lugli GA, Duranti S, Turroni F, Mancabelli L, Ferrario C, Mangifesta M, Hevia A, Viappiani A, Scholz M, Arioli S, Sanchez B, Lane J, Ward DV, Hickey R, Mora D, Segata N, Margolles A, van Sinderen D, Ventura M. 2015. Bifidobacteria exhibit social behavior through carbohydrate resource sharing in the gut. Sci Rep 5:15782. doi: 10.1038/srep15782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Milani C, Duranti S, Lugli GA, Bottacini F, Strati F, Arioli S, Foroni E, Turroni F, van Sinderen D, Ventura M. 2013. Comparative genomics of Bifidobacterium animalis subsp. lactis reveals a strict monophyletic bifidobacterial taxon. Appl Environ Microbiol 79:4304–4315. doi: 10.1128/AEM.00984-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gueimonde M, Florez AB, van Hoek AH, Stuer-Lauridsen B, Stroman P, de los Reyes-Gavilan CG, Margolles A. 2010. Genetic basis of tetracycline resistance in Bifidobacterium animalis subsp. lactis. Appl Environ Microbiol 76:3364–3369. doi: 10.1128/AEM.03096-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Waack S, Keller O, Asper R, Brodag T, Damm C, Fricke WF, Surovcik K, Meinicke P, Merkl R. 2006. Score-based prediction of genomic islands in prokaryotic genomes using hidden Markov models. BMC Bioinformatics 7:142. doi: 10.1186/1471-2105-7-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gueimonde M, Sanchez B, de Los Reyes-Gavilán CG, Margolles A. 2013. Antibiotic resistance in probiotic bacteria. Front Microbiol 4:202. doi: 10.3389/fmicb.2013.00202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ventura M, Turroni F, Motherway MO, MacSharry J, van Sinderen D. 2012. Host-microbe interactions that facilitate gut colonization by commensal bifidobacteria. Trends Microbiol 20:467–476. doi: 10.1016/j.tim.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 24.Turroni F, Ventura M, Butto LF, Duranti S, O'Toole PW, Motherway MO, van Sinderen D. 2014. Molecular dialogue between the human gut microbiota and the host: a Lactobacillus and Bifidobacterium perspective. Cell Mol Life Sci 71:183–203. doi: 10.1007/s00018-013-1318-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bottacini F, Ventura M, van Sinderen D, O'Connell Motherway M. 2014. Diversity, ecology and intestinal function of bifidobacteria. Microb Cell Fact 13(Suppl 1):S4. doi: 10.1186/1475-2859-13-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Flint HJ, Scott KP, Duncan SH, Louis P, Forano E. 2012. Microbial degradation of complex carbohydrates in the gut. Gut Microbes 3:289–306. doi: 10.4161/gmic.19897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Human Microbiome Project Consortium. 2012. Structure, function and diversity of the healthy human microbiome. Nature 486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Korpela K, Salonen A, Virta LJ, Kekkonen RA, Forslund K, Bork P, de Vos WM. 2016. Intestinal microbiome is related to lifetime antibiotic use in Finnish pre-school children. Nat Commun 7:10410. doi: 10.1038/ncomms10410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Avershina E, Lundgard K, Sekelja M, Dotterud C, Storro O, Oien T, Johnsen R, Rudi K. 2016. Transition from infant- to adult-like gut microbiota. Environ Microbiol 18:2226–2236. doi: 10.1111/1462-2920.13248. [DOI] [PubMed] [Google Scholar]
- 30.Martin R, Makino H, Cetinyurek Yavuz A, Ben-Amor K, Roelofs M, Ishikawa E, Kubota H, Swinkels S, Sakai T, Oishi K, Kushiro A, Knol J. 2016. Early-life events, including mode of delivery and type of feeding, siblings and gender, shape the developing gut microbiota. PLoS One 11:e0158498. doi: 10.1371/journal.pone.0158498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Milani C, Hevia A, Foroni E, Duranti S, Turroni F, Lugli GA, Sanchez B, Martin R, Gueimonde M, van Sinderen D, Margolles A, Ventura M. 2013. Assessing the fecal microbiota: an optimized ion torrent 16S rRNA gene-based analysis protocol. PLoS One 8:e68739. doi: 10.1371/journal.pone.0068739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Milani C, Mancabelli L, Lugli GA, Duranti S, Turroni F, Ferrario C, Mangifesta M, Viappiani A, Ferretti P, Gorfer V, Tett A, Segata N, van Sinderen D, Ventura M. 2015. Exploring vertical transmission of bifidobacteria from mother to child. Appl Environ Microbiol 81:7078–7087. doi: 10.1128/AEM.02037-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nuriel-Ohayon M, Neuman H, Koren O. 2016. Microbial changes during pregnancy, birth, and infancy. Front Microbiol 7:1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roger LC, Costabile A, Holland DT, Hoyles L, McCartney AL. 2010. Examination of faecal Bifidobacterium populations in breast- and formula-fed infants during the first 18 months of life. Microbiology 156:3329–3341. doi: 10.1099/mic.0.043224-0. [DOI] [PubMed] [Google Scholar]
- 35.Turroni F, Duranti S, Bottacini F, Guglielmetti S, Van Sinderen D, Ventura M. 2014. Bifidobacterium bifidum as an example of a specialized human gut commensal. Front Microbiol 5:437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yassour M, Vatanen T, Siljander H, Hamalainen AM, Harkonen T, Ryhanen SJ, Franzosa EA, Vlamakis H, Huttenhower C, Gevers D, Lander ES, Knip M, DIABIMMUNE Study Group, Xavier RJ. 2016. Natural history of the infant gut microbiome and impact of antibiotic treatment on bacterial strain diversity and stability. Sci Transl Med 8:343ra381. doi: 10.1126/scitranslmed.aad0917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hussey S, Wall R, Gruffman E, O'Sullivan L, Ryan CA, Murphy B, Fitzgerald G, Stanton C, Ross RP. 2011. Parenteral antibiotics reduce bifidobacteria colonization and diversity in neonates. Int J Microbiol 2011:130574. doi: 10.1155/2011/130574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gibson MK, Crofts TS, Dantas G. 2015. Antibiotics and the developing infant gut microbiota and resistome. Curr Opin Microbiol 27:51–56. doi: 10.1016/j.mib.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Schaik W. 2015. The human gut resistome. Philos Trans R Soc Lond B Biol Sci 370:20140087. doi: 10.1098/rstb.2014.0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Duranti S, Milani C, Lugli GA, Mancabelli L, Turroni F, Ferrario C, Mangifesta M, Viappiani A, Sanchez B, Margolles A, van Sinderen D, Ventura M. 2016. Evaluation of genetic diversity among strains of the human gut commensal Bifidobacterium adolescentis. Sci Rep 6:23971. doi: 10.1038/srep23971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Duranti S, Milani C, Lugli GA, Turroni F, Mancabelli L, Sanchez B, Ferrario C, Viappiani A, Mangifesta M, Mancino W, Gueimonde M, Margolles A, van Sinderen D, Ventura M. 2015. Insights from genomes of representatives of the human gut commensal Bifidobacterium bifidum. Environ Microbiol 17:2515–2531. doi: 10.1111/1462-2920.12743. [DOI] [PubMed] [Google Scholar]
- 42.Bottacini F, O'Connell Motherway M, Kuczynski J, O'Connell KJ, Serafini F, Duranti S, Milani C, Turroni F, Lugli GA, Zomer A, Zhurina D, Riedel C, Ventura M, van Sinderen D. 2014. Comparative genomics of the Bifidobacterium breve taxon. BMC Genomics 15:170. doi: 10.1186/1471-2164-15-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O'Callaghan A, Bottacini F, O'Connell Motherway M, van Sinderen D. 2015. Pangenome analysis of Bifidobacterium longum and site-directed mutagenesis through by-pass of restriction-modification systems. BMC Genomics 16:832. doi: 10.1186/s12864-015-1968-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Milani C, Turroni F, Duranti S, Lugli GA, Mancabelli L, Ferrario C, van Sinderen D, Ventura M. 2015. Genomics of the genus Bifidobacterium reveals species-specific adaptation to the glycan-rich gut environment. Appl Environ Microbiol 82:980–991. doi: 10.1128/AEM.03500-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao Y, Tang H, Ye Y. 2012. RAPSearch2: a fast and memory-efficient protein similarity search tool for next-generation sequencing data. Bioinformatics 28:125–126. doi: 10.1093/bioinformatics/btr595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lugli GA, Milani C, Mancabelli L, van Sinderen D, Ventura M. 2016. MEGAnnotator: a user-friendly pipeline for microbial genomes assembly and annotation. FEMS Microbiol Lett 363:fnw049. doi: 10.1093/femsle/fnw049. [DOI] [PubMed] [Google Scholar]
- 47.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup . 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Anders S, Pyl PT, Huber W. 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.International Organization for Standardization (ISO). 2010. Milk and milk products—determination of the minimal inhibitory concentration (MIC) of antibiotics applicable to bifidobacteria and non-enterococcal lactic acid bacteria (LAB). ISO 10932:2010 (IDF 223:2010) ISO, Geneva, Switzerland. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.