ABSTRACT
Vibrio species are widely distributed in warm estuarine and coastal environments, and they can infect humans through the consumption of raw and mishandled contaminated seafood. In this study, we aimed to isolate and observe the distribution of enteropathogenic Vibrio spp. from environments of the southern coast of South Korea over a season cycle. A total of 10,983 isolates of Vibrio spp. were obtained from tidal water and mud samples over a 1-year period from five sampling sites along the southwest coast of South Korea. We found that Vibrio alginolyticus (n = 6,262) and Vibrio parahaemolyticus (n = 1,757) were ubiquitous in both tidal water and mud year round, whereas Vibrio cholerae (n = 24) and Vibrio vulnificus (n = 130) were seasonally specific to summer. While all V. cholerae isolates were nontoxigenic (non-O1 and non-O139), more than 88% of V. vulnificus isolates possessed the virulence factor elastolytic protease (encoded by vvp). Interestingly, V. parahaemolyticus, which was omnipresent in all seasons, contained the virulence factors thermostable direct hemolysin (encoded by tdh) and thermostable direct hemolysin-related hemolysin (encoded by trh) in larger amounts in June (29 trh-positive strains) and September (14 tdh-, 36 trh-, and 12 tdh- and trh-positive strains) than in December (4 trh-positive strains) and February (3 tdh-positive strains), and virulence factors were absent from isolates detected in April. To understand why virulence factors were detected only in the warm season and were absent in the cold season although the locations are static, long-term monitoring and particularly seasonal study are necessary.
IMPORTANCE The presence of enteropathogenic Vibrio species (Vibrio cholerae, Vibrio parahaemolyticus, and Vibrio vulnificus), which cause acute diarrheal infection, septicemia, and wound infections upon ingestion through food and water, is usually associated with temperature. The World Health Organization (WHO) has estimated that there are 1.4 to 4.3 million cases and 28,000 to 142,000 deaths per year worldwide caused by cholera disease. In South Korea alone, consumption is as much as 52.4 kg of fish and shellfish per year per capita. Our findings suggested that seasonally specific acceleration of these possible pathogenic Vibrio spp. may threaten seafood safety and increase the risk of illness in South Korea, where local people consume raw fish during warmer months.
KEYWORDS: Vibrio parahaemolyticus, Vibrio vulnificus, pathogens
INTRODUCTION
Vibrio species are omnipresent and widely distributed in aquatic environments all over the world (1). The occurrence of Vibrio spp. is commonly associated with temperature, especially a temperate climate. Generally, Vibrio species are periodically detectable in summer but less common in winter, whereas the Vibrio population variation is low in tropical and subtropical waters (1). Reports have shown a significant association between rising seawater temperature and an increase in the number of Vibrio infections, suggesting that global warming could be a factor in the emergence of Vibrio diseases in temperate areas due to its influence on resident bacterial communities (2). Over the past 25 years, marine-related illnesses along the east coast of the United States have risen steadily in correlation with El Niño occurrences (3). It has also been suggested that increases in seawater temperature, as a prospective consequence of climate change, may be responsible for Vibrio infection outbreaks in Israel, Denmark, Spain, Chile, and the United States (4). For example, the emergence of Vibrio infections in the Baltic Sea area was associated with increased sea surface temperature (4, 5). In Bangladesh, the risk of cholera was found to be 2 times higher 6 weeks after a 5°C increase in river, pond, and lake water temperature, and water temperature was found to have a direct correlation with cases of cholera (6).
The geographical distribution of Vibrio spp. in the environment is confined to warm and temperate regions at water temperatures ranging from 20°C to 30°C (7), which is the optimum growth temperature for these bacteria. Increases in surface seawater temperatures will undoubtedly increase human contact with Vibrio spp. and disease transmission (4). A study conducted in Helgoland Roads, North Sea, Germany, showed that the abundance of Vibrio spp. in seawater is higher in summer (3.4 × 104 cells/ml) and significantly lower in winter (5 × 102 cells/ml) (8). However, the seasonal distribution of Vibrio populations could be species specific. Böer et al. conducted a study at 10 recreational beaches along the German North Sea for 2 years and found that V. alginolyticus and V. parahaemolyticus were ubiquitous year round, whereas V. vulnificus was limited to the summer months (9).
In the past decade, unusual, incremental increases in seawater temperature along the shoreline have been linked to epidemic outbreaks of Vibrio-associated illness caused by V. parahaemolyticus in areas such as Chile, Peru, the United States, Europe, and Asia. Studies conducted all over the world have highlighted the environmental factors that affect the abundance and distribution of V. parahaemolyticus, such as water salinity, temperature, turbidity, and the levels of chlorophyll and organic matter in suspension (10). Although the mechanism underlying human infection by V. parahaemolyticus is not completely understood, there are two hemolysins that are commonly recognized as pathogenicity indicators: thermostable direct hemolysin (TDH), which is a pore-forming protein involved in bacterium invasion, and TDH-related hemolysin (TRH), which plays a significant role in virulence (10–12). Most of the clinical isolates of V. parahaemolyticus possess tdh and/or trh; however, a relatively low number of environmental isolates were found to carry these genes (10, 13). V. vulnificus is an opportunistic human pathogen. Although human infections are rare, V. vulnificus disease causes the highest hospitalization (91.3%) and mortality (34.4%) rates of all foodborne diseases in the United States (14–16). One of the possible virulence determinants in V. vulnificus, vvp, which encodes elastolytic protease, may be a causative agent for skin lesions (16–20). It is well known that epidemic cholera is caused mainly by toxigenic V. cholerae O1 and O139. The serotype O1 has been further divided into the classical (CL) and El Tor (ET) biotypes (21, 22). The pathogenicity of V. cholerae depends on a combination of properties, including the presence of cholera toxin (CT), encoded by ctxA, and the colonization and adhesion factor encoded by tcpA (22, 23). Studies have shown seasonal patterns for this species, with increased occurrence and incidence of wound infections at warmer temperatures (24).
In recent years, with the changing climate, specifically, rising air and sea temperatures and intensifying monsoons, the ecology of pathogenic bacteria that affect public health has been a great concern. More study on the ecology of enteropathogenic Vibrio species in the environment where human activities occur is especially important to those who rely on catching marine products to make a living and/or as a source of food (9). The current study focused on investigating the seasonal dynamics of enteropathogenic Vibrio species in the coastal tidal waters and mud along the southwestern coast of South Korea over a season cycle. The environmental factors that may affect the seasonal occurrence of Vibrio species and populations are also discussed.
RESULTS
Seasonal variation in Vibrio spp. with environmental factors.
The most probable number (MPN) of presumptive Vibrio spp. was found to differ significantly by month (P < 0.05) but not by sampling site (P > 0.05) (Fig. 1) as determined by analysis of variance (ANOVA). This shows that the MPN of presumptive Vibrio spp. in both habitats is affected by season and not location, although the total human population and major land uses differed at the sampling sites (see Table S1 in the supplemental material). Figure 1a shows that the average MPN of presumptive Vibrio spp. detected in tidal water in February was significantly different (post hoc, P < 0.05) from that in other months. The mean log10 MPN per milliliter of tidal water in February is 5.17, which is 3 logs higher than those in June (2.19) and September (2.45) and 4 logs higher than those in December (1.71) and April (1.39). The average MPN of presumptive Vibrio spp. detected in mud in February was significantly different (P < 0.05) from those in September, December, and April but not significantly different (P > 0.05) from that in June (Fig. 1b). The mean log10 MPNs per gram of mud in each season are 3.10 (June), 2.50 (September), 2.09 (December), 3.49 (February) and 2.70 (April). The overall average log10 MPN in mud is higher than that in tidal water. In addition, sampling month was found to have significant effect (P < 0.05) on temperature and pH in both tidal water and mud, as well as on salinity and biological oxygen demand (BOD) in tidal water (see Fig. S1 and S2 in the supplemental material). However, electric conductivity (EC) was found to be temporally specific in tidal water but spatially specific in mud (P < 0.05). Location had no significant effect (P > 0.05) on the tested parameters, except for EC in mud. Tidal water turbidity did not differ significantly according to season or location (P > 0.05) (see Table S4 in the supplemental material).
FIG 1.
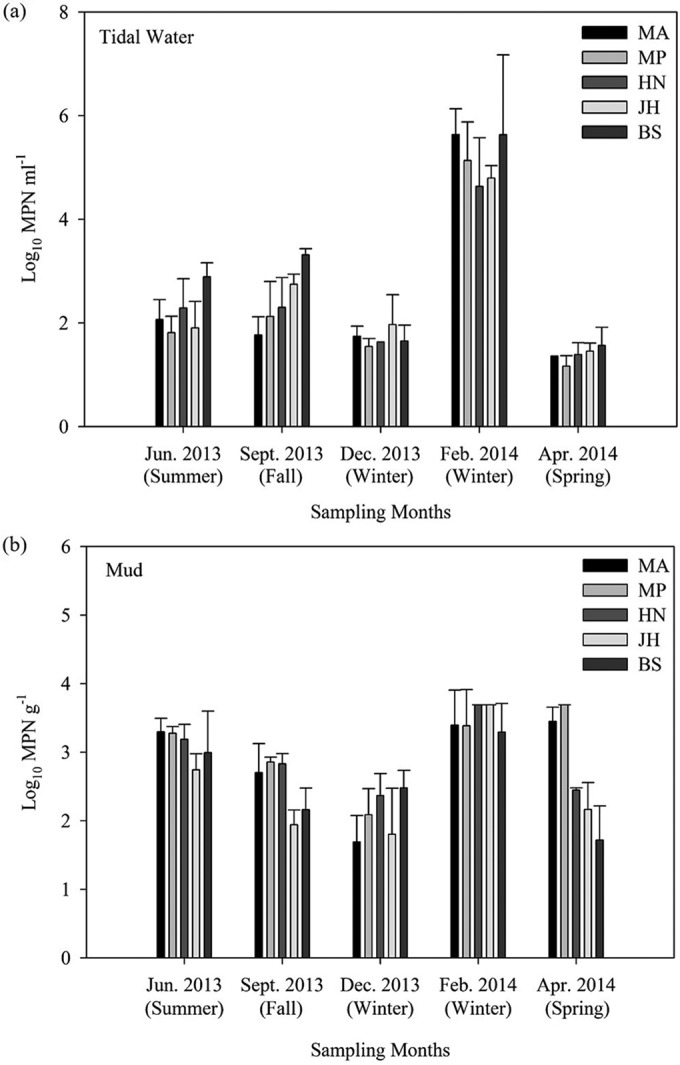
Distribution of presumptive Vibrio spp. (log10 MPN) in tidal water (a) and mud (b). Each bar in each month represents five sampling sites (MA, Muan; MP, Mokpo; HN, Haenam; JH, Jangheung; and BS, Boseong).
Principal-component analysis (PCA) was performed to investigate the correlation between the MPN of presumptive Vibrio spp. in the samples and environmental variables such as pH, temperature, salinity, EC, BOD, and turbidity according to sampling site and season (Fig. 2). The MPN in tidal water samples was found to be seasonally independent, whereas the MPN in mud samples was seasonally dependent, and the clusters were distinctly separated by the direction of the temperature projections. In tidal water (Fig. 2a), two major clusters of the variables were observed, a large cluster comprised of four sampling months (June, September, December, and April) and a small cluster comprised of a single month, February. The environmental variables distributed unevenly, with MPN and turbidity of tidal water having the longest projection score of near 1.0, which indicates that the variable is perfectly correlated. The differences in the variables in February compared to those in the other months were mostly affected by MPN, as observed by the direction of the projection arrow. MPN in tidal water was positively correlated with pH (r = 0.257, P = 0.215) and BOD (r = 0.120, P = 0.569) but negatively correlated with temperature (r = −0.318, P = 0.122), salinity (r = −0.224, P = 0.282), EC (r = −0.301, P = 0.144), and turbidity (r = −0.198, P = 0.343). In contrast, mud samples from April (spring), June (summer), and September (fall) were closely correlated but were separated from those collected in December (winter) and February (winter) by temperature (Fig. 2b). Although three clusters were observed in the mud samples, two small clusters comprised of variables from December and February, considered as one group, differed from those from April, June, and September by the projection arrow and the projection score of temperature. This indicates that the variables observed in winter were different from those in the other seasons, explained by temperature. In addition, the MPN in mud samples was positively correlated with pH (r = 0.290, P = 0.159) and temperature (r = 0.002, P = 0.991) but negatively correlated with EC (r = −0.106, P = 0.613).
FIG 2.
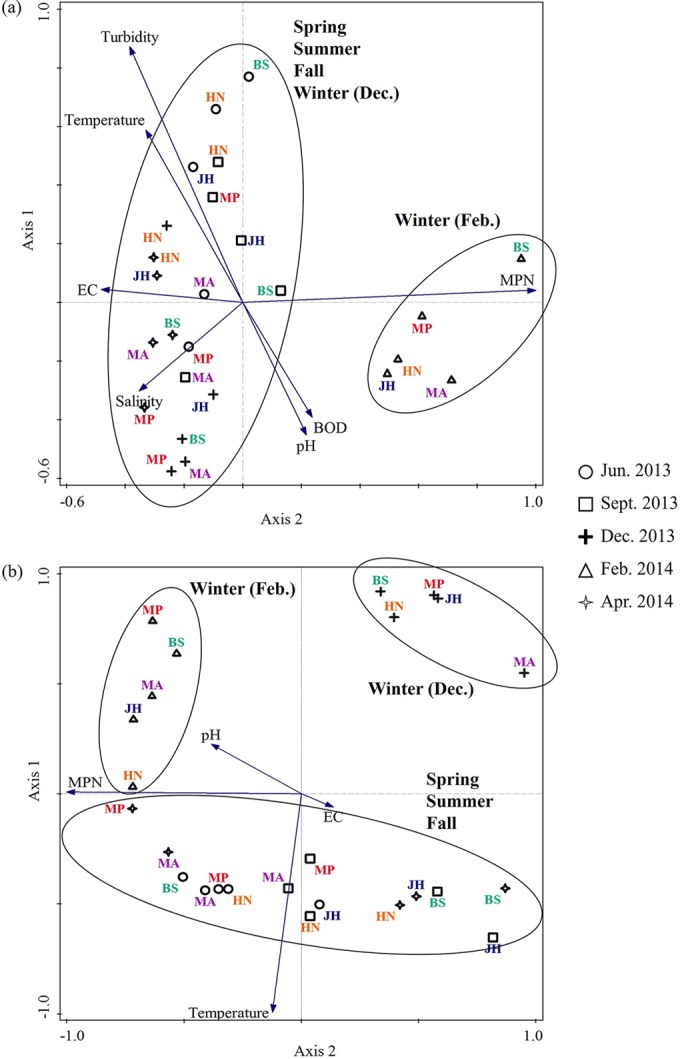
Principal-component analysis (PCA) projections of scores for correlation of the most probable number (MPN) and environmental parameters (BOD, EC, pH, salinity, temperature, and turbidity) from tidal water (a) and mud (b) in different seasons. A projection score near 1.0 indicates that the variables are perfectly correlated, whereas a score near −1.0 indicates that the variables are perfectly inversely correlated. Variables with vectors projected in the same plane could be considered positively correlated. Closely correlated variables are grouped in black ovals, indicated by months and sampling sites (MA, Muan; MP, Mokpo; HN, Haenam; JH, Jangheung; and BS, Boseong).
Relative abundance and genotypic diversity of enteropathogenic Vibrio spp.
Among the 11,772 phenotypically identified isolates of Vibrio spp., 10,983 isolates were genetically identified as Vibrio spp. by genus-specific 16S rRNA gene amplification, accounting for 93.7% and 92.9% of the presumptive Vibrio isolates from tidal water and mud, respectively (see Table S2 in the supplemental material). Among the enteropathogenic Vibrio sp. isolates identified, V. cholerae (n = 24) and V. vulnificus (n = 130) were found to be season specific, whereas V. alginolyticus (n = 6,262) and V. parahaemolyticus (n = 1,757) were season independent (Tables 1 and 2). V. cholerae was isolated in June only, with one strain from tidal water and 23 strains from mud samples. Thirty-four strains (2 and 32 from Haenam [HN] and Boseong [BS], respectively) of V. vulnificus were isolated in June and five strains (all from BS) were isolated in September from tidal water, whereas 91 strains (2, 59, 29, and 1 from Muan [MA], HN, Jangheung [JH], and BS, respectively) were isolated in June from mud samples. In contrast, V. alginolyticus and V. parahaemolyticus were isolated from both tidal water and mud samples collected in all sampling months and at all sites, except for tidal water samples collected in December and April from MA, MP, and HN, which did not contain V. parahaemolyticus. In general, in all seasons, the occurrence of V. parahaemolyticus was higher and more stable in mud samples than in tidal water. The occurrence of V. alginolyticus was higher than that of V. parahaemolyticus in both tidal water and mud. The environmental parameters were tested for correlation with the abundance of V. alginolyticus and V. parahaemolyticus. The log10 MPN of Vibrio spp. in tidal water (r2 = 0.172, P = 0.039) and temperature in mud (r2 = 0.178, P = 0.036) were found to have a significant linear relationship with V. alginolyticus abundance (Table 3). The log10 MPN of Vibrio spp. per milliliter of tidal water ranged from 1.17 to 5.64 (Fig. 1a), and the temperature of mud ranged from 3.0°C to 28.5°C (Fig. S2). However, the strength of the relationship is weak, as the coefficient of determination (r) values are less than 1. Despite this, none of the environmental variables were found to have a significant correlation with the abundance of V. parahaemolyticus in either tidal water or mud. Thus, the assumption can be made that changes in temperature may affect the number of V. alginolyticus bacteria in mud.
TABLE 1.
Vibrio species identification for the isolates collected from tidal water
| Sampling season | Vibrio species | No. (%) of isolates from tidal water ata: |
Total no. (%) of isolates | ||||
|---|---|---|---|---|---|---|---|
| MA | MP | HN | JH | BS | |||
| June 2013 | V. alginolyticus | 184 (76.0) | 177 (77.0) | 157 (71.4) | 159 (65.2) | 95 (40.6) | 772 (66.0) |
| V. cholerae | 0 | 0 | 0 | 0 | 1 (0.4) | 1 (0.1) | |
| V. parahaemolyticus | 10 (4.1) | 6 (2.6) | 19 (8.6) | 39 (16.0) | 18 (7.7) | 92 (7.9) | |
| V. vulnificus | 0 | 0 | 2 (0.9) | 0 | 32 (13.7) | 34 (2.9) | |
| Other Vibrio spp. | 48 (19.8) | 47 (20.4) | 42 (19.1) | 46 (18.9) | 88 (37.6) | 271 (23.2) | |
| September 2013 | V. alginolyticus | 68 (29.6) | 119 (51.5) | 112 (59.6) | 93 (41.5) | 120 (51.7) | 512 (46.3) |
| V. cholerae | 0 | 0 | 0 | 0 | 0 | 0 | |
| V. parahaemolyticus | 34 (14.8) | 50 (21.6) | 15 (8.0) | 54 (24.1) | 61 (26.3) | 214 (19.4) | |
| V. vulnificus | 0 | 0 | 0 | 0 | 5 (2.2) | 5 (0.5) | |
| Other Vibrio spp. | 128 (55.7) | 62 (26.8) | 61 (32.4) | 77 (34.4) | 46 (19.8) | 374 (33.9) | |
| December 2013 | V. alginolyticus | 119 (53.6) | 36 (41.4) | 212 (92.6) | 159 (70.7) | 51 (22.1) | 577 (58.0) |
| V. cholerae | 0 | 0 | 0 | 0 | 0 | 0 | |
| V. parahaemolyticus | 12 (5.4) | 18 (20.7) | 0 | 20 (8.9) | 90 (39.0) | 140 (14.1) | |
| V. vulnificus | 0 | 0 | 0 | 0 | 0 | 0 | |
| Other Vibrio spp. | 91 (41.0) | 33 (37.9) | 17 (7.4) | 46 (20.4) | 90 (39.0) | 277 (27.9) | |
| February 2014 | V. alginolyticus | 51 (54.3) | 17 (7.7) | 115 (69.3) | 42 (20.2) | 41 (31.1) | 266 (32.4) |
| V. cholerae | 0 | 0 | 0 | 0 | 0 | 0 | |
| V. parahaemolyticus | 7 (7.4) | 3 (1.4) | 5 (3.0) | 10 (4.8) | 10 (7.6) | 35 (4.3) | |
| V. vulnificus | 0 | 0 | 0 | 0 | 0 | 0 | |
| Other Vibrio spp. | 36 (38.3) | 201 (91.0) | 46 (27.7) | 156 (75.0) | 81 (61.4) | 520 (63.3) | |
| April 2014 | V. alginolyticus | 89 (46.1) | 236 (94.8) | 189 (88.7) | 204 (85.0) | 164 (67.8) | 882 (77.6) |
| V. cholerae | 0 | 0 | 0 | 0 | 0 | 0 | |
| V. parahaemolyticus | 0 | 0 | 0 | 1 (0.4) | 4 (1.7) | 5 (0.4) | |
| V. vulnificus | 0 | 0 | 0 | 0 | 0 | 0 | |
| Other Vibrio spp. | 104 (53.9) | 13 (5.2) | 24 (11.3) | 35 (14.6) | 74 (30.6) | 250 (22.0) | |
Total number of bacterial isolates grown on TCBS agar. All colonies grown on TCBS agar were selected, not exceeding 252 isolates for each sampling site. Locations: MA, Muan; MP, Mokpo; HN, Haenam; JH, Jangheung; BS, Boseong.
TABLE 2.
Vibrio species identification for the total isolates collected from mud samples
| Sampling season | Vibrio species | No. (%) of Vibrio isolates from mud ata: |
Total no. (%) of isolates | ||||
|---|---|---|---|---|---|---|---|
| MA | MP | HN | JH | BS | |||
| June 2013 | V. alginolyticus | 173 (68.7) | 170 (67.5) | 34 (13.5) | 79 (31.6) | 119 (47.8) | 575 (45.8) |
| V. cholerae | 2 (0.8) | 0 | 20 (7.9) | 1 (0.4) | 0 | 23 (1.8) | |
| V. parahaemolyticus | 28 (11.1) | 37 (14.7) | 67 (26.6) | 73 (29.2) | 77 (30.9) | 282 (22.5) | |
| V. vulnificus | 2 (0.8) | 0 | 59 (23.4) | 29 (11.6) | 1 (0.4) | 91 (7.3) | |
| Other Vibrio spp. | 47 (18.7) | 45 (17.9) | 72 (28.6) | 68 (27.2) | 52 (20.9) | 284 (22.6) | |
| September 2013 | V. alginolyticus | 106 (46.9) | 55 (25.7) | 82 (39.6) | 107 (52.2) | 114 (51.4) | 464 (43.2) |
| V. cholerae | 0 | 0 | 0 | 0 | 0 | 0 | |
| V. parahaemolyticus | 68 (30.1) | 98 (45.8) | 88 (42.5) | 54 (26.3) | 60 (27.0) | 368 (34.3) | |
| V. vulnificus | 0 | 0 | 0 | 0 | 0 | 0 | |
| Other Vibrio spp. | 52 (23.0) | 61 (28.5) | 37 (17.9) | 44 (21.5) | 48 (21.6) | 242 (22.5) | |
| December 2013 | V. alginolyticus | 200 (87.0) | 146 (70.9) | 104 (46.0) | 177 (82.7) | 94 (47.7) | 721 (67.2) |
| V. cholerae | 0 | 0 | 0 | 0 | 0 | 0 (0.0) | |
| V. parahaemolyticus | 6 (2.6) | 34 (16.5) | 79 (35.0) | 4 (1.9) | 69 (35.0) | 192 (17.9) | |
| V. vulnificus | 0 | 0 | 0 | 0 | 0 | 0 (0.0) | |
| Other Vibrio spp. | 24 (10.4) | 26 (12.6) | 43 (19.0) | 33 (15.4) | 34 (17.3) | 160 (14.9) | |
| February 2014 | V. alginolyticus | 137 (60.1) | 114 (60.3) | 172 (71.1) | 211 (84.1) | 138 (56.1) | 772 (66.8) |
| V. cholerae | 0 | 0 | 0 | 0 | 0 | 0 | |
| V. parahaemolyticus | 61 (26.8) | 28 (14.8) | 29 (12.0) | 12 (4.8) | 49 (19.9) | 179 (15.5) | |
| V. vulnificus | 0 | 0 | 0 | 0 | 0 | 0 | |
| Other Vibrio spp. | 30 (13.2) | 47 (24.9) | 41 (16.9) | 28 (11.2) | 59 (24.0) | 205 (17.7) | |
| April 2014 | V. alginolyticus | 107 (45.0) | 178 (77.7) | 153 (63.0) | 187 (76.3) | 96 (39.5) | 721 (60.2) |
| V. cholerae | 0 | 0 | 0 | 0 | 0 | 0 | |
| V. parahaemolyticus | 97 (40.8) | 16 (7.0) | 53 (21.8) | 13 (5.3) | 71 (29.2) | 250 (20.9) | |
| V. vulnificus | 0 | 0 | 0 | 0 | 0 | 0 | |
| Other Vibrio spp. | 34 (14.3) | 35 (15.3) | 37 (15.2) | 45 (18.4) | 76 (31.3) | 227 (18.9) | |
Total number of bacterial isolates grown on TCBS agar. All colonies grown on TCBS agar were selected, not exceeding 252 isolates for each sampling site. Locations: MA, Muan; MP, Mokpo; HN, Haenam; JH, Jangheung; BS, Boseong.
TABLE 3.
Statistical analysis of the relationship between the occurrences of ubiquitous V. alginolyticus and V. parahaemolyticus with different environmental parameters for tidal water and mud
| Environmental parameter | Linear regressiona |
|||
|---|---|---|---|---|
|
V. alginolyticus |
V. parahaemolyticus |
|||
| r2 value | P value | r2 value | P value | |
| Tidal water | ||||
| Log10 MPN | 0.172 | 0.039 | 0.016 | 0.550 |
| Temp (°C) | 0.074 | 0.189 | 0.001 | 0.882 |
| pH | 0.003 | 0.789 | 0.055 | 0.259 |
| Salinity (ppt) | 0.034 | 0.379 | 0.004 | 0.767 |
| BOD (mg liter−1) | 0.001 | 0.898 | 0.000 | 0.998 |
| Turbidity (NTUb) | 0.081 | 0.168 | 0.008 | 0.679 |
| Mud | ||||
| Log10 MPN | 0.003 | 0.807 | 0.000 | 0.974 |
| Temp (°C) | 0.178 | 0.036 | 0.099 | 0.126 |
| pH | 0.032 | 0.391 | 0.103 | 0.119 |
| EC (S m−1) | 0.010 | 0.635 | 0.013 | 0.593 |
Significance level, 0.05. Boldface indicates significant linear relationships.
NTU, nephelometric turbidity units.
Distribution of virulence genes among potential human-pathogenic strains of V. cholerae, V. parahaemolyticus, and V. vulnificus.
The distribution of virulence traits among the three potential human pathogens V. cholerae (n = 24), V. parahaemolyticus (n = 1,757), and V. vulnificus (n = 130) is summarized in Table 4. These virulence (ctxA), biotype (tcpA), and serogroup (rfb) genes were not detected in the 24 V. cholerae strains isolated from tidal water and mud. A possible virulence determinant, an elastolytic protease (vvp) gene of V. vulnificus strains that can cause infection in humans, was detected in most of the strains isolated. Among the 34 V. vulnificus isolates from tidal waters in June, 30 strains (88.2%) contained vvp. Five isolates from tidal water in September also contained vvp genes. Of the 91 V. vulnificus isolates from mud samples in June, 88 strains (96.7%) contained vvp. Although V. parahaemolyticus was isolated year round, pathogenic strains were detected mostly in June (29 strains with trh) and September (14 strains with tdh, 36 strains with trh, and 12 strains with tdh and trh). Of the 1,757 V. parahaemolyticus strains, 69 contained trh, including 41 and 28 strains from tidal water and mud, respectively. Among the 41 strains with trh from tidal water, 6, 34, and 1 were found in June, September, and December, respectively. Among the 28 strains with trh from mud, 23, 2, and 3 were found in June, September, and December, respectively. Most strains with trh were found in September (n = 34) in tidal water and in June (n = 23) in mud. Seventeen strains of V. parahaemolyticus possessed tdh, with 13 from tidal water and 4 from mud samples. Among the four strains with tdh from mud samples, one strain was found in September and three strains were found in February. Twelve strains of V. parahaemolyticus from tidal water in September contained both tdh and trh.
TABLE 4.
Distribution of virulence genes among isolates of V. cholerae, V. parahaemolyticus, and V. vulnificus
| Origin | Sampling season | No. (%) of positive strainsa |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
V. cholerae (n = 24) |
V. parahaemolyticus (n = 1,757) |
V. vulnificus (n = 130) |
||||||||
| ctxA | rfb O1 | rfb O139 | tcpA El Tor | tcpA classical | tdh | trh | tdh + trh | vvp | ||
| Tidal water | June 2013 | 0 | 0 | 0 | 0 | 0 | 0 | 6 (6.5) | 0 | 30 (88.2) |
| September 2013 | 0 | 0 | 0 | 0 | 0 | 13 (6.1) | 34 (15.9) | 12 (5.6) | 5 (100.0) | |
| December 2013 | 0 | 0 | 0 | 0 | 0 | 0 | 1 (0.7) | 0 | 0 | |
| February 2014 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| April 2014 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Mud | June 2013 | 0 | 0 | 0 | 0 | 0 | 0 | 23 (8.2) | 0 | 88 (96.7) |
| September 2013 | 0 | 0 | 0 | 0 | 0 | 1 (0.3) | 2 (0.5) | 0 | 0 | |
| December 2013 | 0 | 0 | 0 | 0 | 0 | 0 | 3 (1.6) | 0 | 0 | |
| February 2014 | 0 | 0 | 0 | 0 | 0 | 3 (1.7) | 0 | 0 | 0 | |
| April 2014 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
Presence of genes as determined by PCR. Genes: ctxA, cholera toxin (CT); rfb O1, serotype O1-specific rfb region; rfb O139, serotype O139-specific rfb region; tcpA El Tor, El Tor toxin-coregulated pilus (TCP); tcpA classical, classical TCP; tdh, thermostable direct hemolysin (TDH); trh, TDH-related hemolysin; vvp, elastolytic protease.
DISCUSSION
In this study, V. alginolyticus and V. parahaemolyticus were present year round, whereas V. vulnificus and V. cholerae were specific to the summer season. More interestingly, the virulence genes (tdh and trh) in V. parahaemolyticus were absent in April in both tidal water and mud samples and were present more frequently in June and September than in other months. The presence and seasonality of Vibrio spp. are similar to the results obtained in German coastal waters, where the presence of Vibrio spp. was significantly associated with temperature and salinity (2). It has been known that V. parahaemolyticus and V. alginolyticus are ubiquitous year round in the German North Sea, where salinity is high. Both species are known to respond strongly to increased water temperatures and seasonal cycles (2). In the central Wadden sea of northern Europe, V. alginolyticus and V. parahaemolyticus were omnipresent, whereas V. vulnificus was limited to summer months, suggesting that water temperature is the most important factor in the area (9).
In contrast, V. vulnificus was detected only in the summer months in the North Sea in Germany, whereas non-O1 and non-O139 V. cholerae strains occurred irregularly and did not follow any seasonal patterns (2). In French Mediterranean coastal lagoons, V. vulnificus was limited to summer in water, sediment, and shellfish samples (18). In a Danish marine environment, low numbers of V. vulnificus were present in water and sediment, with 0.8 to 19 CFU/liter from June to mid-September in water and 0.04 to 11 CFU/g from July to mid-November in sediment (25). In fact, V. vulnificus proliferates when the water temperature exceeds 18°C, usually in the range of 9 to 31°C and at low to moderate salinity (1 to 34 ppt) (18), and often the number dropped to nearly undetectable levels if the temperature dropped below 10°C (18, 19, 26). In this study, salinity and pH in tidal water were found to be significantly lower in summer (June). Possible reasons for this phenomenon might be freshwater discharge or submarine discharge due to the monsoon season in South Korea (27). Other than environmental factors, biological factors such as marine plankton, specifically zooplankton, show a stronger association with Vibrio species than phytoplankton (1). As many as 109 cells/g (wet weight) of Vibrio species were counted from plankton, which is higher than the Vibrio colony count from the surrounding water column (1). In the Chesapeake Bay, almost all the total viable count of plankton is composed of Vibrio species. When winter is over, vibrios released from the sediment attach to zooplankton and reproduce rapidly as the temperature increases (28). Other factors, such as V. vulnificus-positive fish, were found to be inversely correlated to salinity (Pearson's correlation coefficient r = −0.91077, P < 0.0001) and positively correlated to temperature (Pearson's correlation coefficient r = 0.62481, P < 0.0298) in the northern Gulf of Mexico (24). V. parahaemolyticus was frequently reported from tropical and subtropical oysters (99% positive in Brazil, 94% in India, 100% in the United States, and 71% in Taiwan) (10).
Most environmental V. parahaemolyticus strains are considered nonpathogenic because of the low frequencies of detection of tdh and trh. It has been shown that in general, only 1 to 2% of environmental V. parahaemolyticus strains contain tdh; however, this amount is sufficient to have a large impact on human health, particularly in tropical developing countries (29). In the highly populated regions along the coasts of South Carolina and Georgia in the United States, environmental isolates of V. parahaemolyticus containing both tdh and trh were detected at relatively low levels of 4.3% and 0.3%, respectively (12). In Galicia, Spain, the pattern of virulence genes observed in V. parahaemolyticus was that trh+ strains were most common in fall, when the seawater is warmer and less saline, and tdh+ strains were most common in the winter and spring (30). However, in contrast to what was observed in Spain, various studies of the distinct geographical distributions of the temperature-mediated affinity of tdh+ and trh+ strains have suggested that trh+ strains dominate in cold waters, whereas tdh+ strains dominate in warmer waters (31–34). Similar to our study, the year-round presence of V. alginolyticus and V. parahaemolyticus has been reported by others (2, 9). Non-O1/non-O139 V. cholerae and V. vulnificus, which were found in this study, showed season-specific prevalence and were found only in summer in tidal water and mud samples; however, in Germany, V. cholerae followed no seasonal pattern (2). V. parahaemolyticus strains containing trh were present across the summer and fall, with the highest occurrence in summer (6.5%) for mud samples and in fall (15.9%) for tidal water. Our results are similar to the findings in Galicia, Spain, where the prevalence of the trh gene peaked in fall, when the seawater temperature is higher (30). In contrast, the tdh gene was found in fall (6.1%) in tidal water and was rarely detected in mud samples. This result does not agree with the findings from other studies, where the tdh gene was found across the winter and spring in seawater in Galicia, Spain (30), and in the warmer waters in other places (31–34). V. parahaemolyticus strains containing trh and tdh genes also decreased to near zero in December and February and were absent in April, even though the temperature gradually increased. The vvp genes of V. vulnificus were detected in most strains isolated in June from both tidal water and mud samples and in September from tidal water, suggesting a higher possibility of causing diseases in humans during these times. In the United States, cases of oyster-associated human infections in warm-water months were equivalent to the amount of V. vulnificus cells present in seawater and shellfish (35).
In conclusion, Vibrio spp. isolated from southern South Korean coastal tidal water and mud samples showed species-specific seasonal patterns, with V. cholerae and V. vulnificus limited to summer. The other enteropathogenic species V. alginolyticus and V. parahaemolyticus were the dominant species in these areas year round. Interestingly, the MPN of Vibrio spp. in tidal water and temperature in mud showed a significant linear relationship with V. alginolyticus. Vibrio spp. were consistently more abundant in mud than in tidal water, suggesting that mud could serve as a reservoir for Vibrio spp., especially in winter. Moreover, the virulence traits of these enteropathogenic Vibrio spp. were restricted to warmer months, even though some species were present in all seasons. This year-round study suggests that the warmer season-specific virulence factors of Vibrio species and the risk to the human population of contagion by these pathogens might accelerate as the temperature increases due to global warming. Thus, it is necessary for long-term monitoring of these enteropathogenic Vibrio spp. to fully understand their distribution patterns in the region.
MATERIALS AND METHODS
Sampling and isolation of Vibrio strains from tidal water and mud samples.
Tidal water and mud samples were collected from five sites, Muan (MA), Mokpo (MP), Haenam (HN), Jangheung (JH), and Boseong (BS), located along the coast of southern South Korea (Fig. 3) in June (summer), September (fall), and December (winter) 2013 and in February (winter) and April (spring) 2014. The sampling locations, major land use and activities, and human populations are described in Table S1 in the supplemental material. Each sample was collected in triplicate from three points in the site that were approximately 10 m apart. Tidal water and mud samples were transported to the laboratory in sealed sterile plastic bottles and plastic bags on ice on the day of sampling. Approximately 2 liters of coastal sea surface tidal waters were collected, during either flood tide or ebb tide. Temperature, pH, salinity, electric conductivity (EC), and turbidity were measured in situ using a YSI 63 instrument (YSI Inc., Yellow Springs, OH) and a turbidimeter (TN-100; Thermo Scientific Eutech Products, Vernon Hills, IL). Biological oxygen demand (BOD) was measured with a YSI Pro 20-BOD probe (YSI Inc., Yellow Springs, OH) as previously described (36). A most-probable-number (MPN) estimate of Vibrio sp. numbers was done using standard MPN tables for a three-tube series in triplicate (15, 37). One milliliter of each tidal water sample was diluted using 9 ml of alkaline peptone water (APW), and 10−1 to 10−6 dilutions were made in 9 ml of APW in triplicates. After inoculation, all tubes were incubated overnight at 37°C. Vibrio strain isolation was conducted by filtering 1-ml, 10-ml, and 100-ml aliquots of tidal waters onto the surfaces of 0.45-μm-pore size membranes (Advantec, Tokyo, Japan). The membrane filters were incubated on thiosulfate-citrate-bile-sucrose (TCBS) (Difco, Detroit, MI) agar plates at 37°C for 18 to 24 h. Presumptive Vibrio isolates, which appeared as green and yellow colonies, were randomly selected, streaked, and incubated under the same conditions. All single colonies were enriched using APW and preserved at −70°C in TSG buffer (tryptic soy broth, 1% sodium chloride, and 24% glycerol) (38) in 96-well tissue culture test plates (SPL Life Sciences, Pucheon, South Korea) until use. For mud samples, about 10 cm of the upper layer of mud was removed, and mud samples underneath at about 5-cm depth (0.5 kg) were collected in duplicate. The temperature and EC of the mud sediment were measured in situ with ProCheck (Decagon Devices, Pullman, WA). The pH was measured using a pH Spear (Thermo Scientific Eutech Products) by suspending 10 g of air-dry mud soil into 10 ml of deionized water according to procedures described previously (39). For bacterial enrichment, 10 g of the mud samples was added to 40 ml of APW in a 50-ml plastic centrifuge tube (Corning Inc., Corning, NY) with shaking at 200 rpm on an orbital shaker (HB-203S; Hanbaek Scientific, Bucheon, South Korea) as previously described (40). After shaking for 2 h, the tubes were then centrifuged at 90 × g (lightly) for 10 min (Z 36 HK; Hermle, Wehingen, Germany). Subsequently, 1-ml, 10-ml, and 30-ml aliquots of the supernatant were filtered, and Vibrio strains were isolated as described above for the tidal water samples. For MPN enumeration, 10 g of each mud sample was added to 90 ml of APW, and 10−1 to 10−6 dilutions were made in 9 ml of APW in triplicates. The tubes were incubated overnight at 37°C (15, 37).
FIG 3.
Study area for five sampling points along the southern coastal region of South Korea: Muan (MA), Mokpo (MP), Haenam (HN), Jangheung (JH), and Boseong (BS). The map was generated using the Ocean Data View software (R. Schlitzer, 2015 [odv.awi.de]).
DNA extraction.
The isolated strains were cultivated by stamping on TCBS agar plates using a microplate replicator (Boekel Scientific, Feasterville, PA) and were grown overnight at 37°C. DNA was extracted as described previously (41). Briefly, single colonies were picked and suspended in 150 μl of 0.05 M NaOH in a 96-well PCR plate (Axygen Scientific, Union City, CA). The cells were heated at 95°C for 15 min in a thermal cycler (Mastercycler Gradient; Eppendorf, Hamburg, Germany) and then centrifuged for 10 min at 82 × g (Z400; Hermle, Wehingen, Germany).
Confirmation of genus (Vibrio) and identification of species.
All extracted DNA was tested by PCR to confirm the genus Vibrio and by multiplex PCR to identify the species using the primers listed in Table 5 and an A200 Gradient thermal cycler (LongGene, Hangzhou, China). Four Vibrio strains, V. alginolyticus KCTC 12696T, V. cholerae NCCP 11179, V. parahaemolyticus KCTC 2471, and V. vulnificus KCTC 2959T, were used as positive controls, and Escherichia coli NCCP 10004 and a nontemplate control were also used as negative controls. For genus identification, the amplification reaction mixtures contained 1× PCR buffer, 0.2 mM each deoxyribonucleoside triphosphate (dNTP), 1 U of nTaq-HOT polymerase (Enzynomics, Daejeon, South Korea), 10 pmol/μl forward and reverse primers (Macrogen, Daejeon, South Korea), and 1 μl of template in a final reaction volume of 10 μl. The program consisted of a 3-min initial denaturation step at 94°C, followed by 25 cycles at 94°C for 30 s, 57°C for 30 s, and 72°C for 1 min and a final elongation step at 72°C for 10 min. Amplified products were examined using a rapid automated gel electrophoresis system (QIAxcel Advanced System; Qiagen, Valencia, CA). Isolates identified as Vibrio were further tested by using multiplex PCR to determine the species. Amplification reaction mixtures contained 1× PCR buffer, 0.2 mM each dNTP, 0.1 U of nTaq-multi-HOT polymerase, 0.4 pmol/μl each primer (Macrogen, Daejeon, South Korea), and 1 μl of template in a final reaction volume of 10 μl. The PCR amplification program was as follows: a 3-min initial denaturation step, followed by 35 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 1 min and a final elongation step at 72°C for 7 min. Amplified products were examined using the QIAxcel Advanced System. Vibrio genus and species were determined by verifying the sizes of the amplified fragments (Table 5). Strains with species identified using the multiplex PCRs were further confirmed by 16S rRNA sequencing using universal primers (Macrogen, Daejeon, South Korea). Ten strains of each identified species for V. alginolyticus, V. cholerae, V. parahaemolyticus, and V. vulnificus were randomly selected from the stock cultures for sequencing (see Table S3 in the supplemental material).
TABLE 5.
Primers used in this study
| Species | Target genea | Primer | Sequence (5′ to 3′) | Size (bp) | Reference |
|---|---|---|---|---|---|
| Vibrio spp. | 16S rRNA | 567F | GGCGTAAAGCGCATGCAGGT | 120 | 44 |
| 680R | GAAATTCTACCCCCCTCTACAG | ||||
| V. alginolyticus | dnaJ | VM-Fb | CAGGTTTGYTGCACGGCGAAGA | 144 | 45 |
| V.al2-MmR | GATCGAAGTRCCRACACTMGGA | ||||
| V. cholerae | ctxA | CTX-F | GCAGTCAGGTGGTCTTATGC | 308 | 46 |
| CTX-R | CGTGCCTAACAAATCCCGTC | ||||
| dnaJ | VM-F | CAGGTTTGYTGCACGGCGAAGA | 375 | 45 | |
| VC-Rmm | AGCAGCTTATGACCAATACGCC | ||||
| rfb | O1-F | GTTTCACTGAACAGATGGG | 192 | 46 | |
| O1-R | GGTCATCTGTAAGTACAAC | ||||
| rfb | O139-F | AGCCTCTTTATTACGGGTGG | 449 | 46 | |
| O139-R | GTCAAACCCGATCGTAAAGG | ||||
| tcp | tcpA_72F | CACGATAAGAAAACCGGTCAAGAG | 22 | ||
| tcpA_477R | CGAAAGCACCTTCTTTCACGTTG | 451 (El Tor), 620 (classical) | |||
| tcpA_647R | TTACCAAATGCAACGCCGAATG | ||||
| V. parahaemolyticus | dnaJ | VM-F | CAGGTTTGYTGCACGGCGAAGA | 96 | 45 |
| VP-MmR | TGCGAAGAAAGGCTCATCAGAG | ||||
| tdh | tdh-F | TCCATCTGTCCCTTTTCCTGC | 278 | This study | |
| tdh-R | CGAACACAGCAGAATGACCG | ||||
| trh | trh_F | TACCTTTTCCTTCTCCAGGTTCGG | 122 | 47 | |
| trh_R | TCGTTTTATGTTTCGGTTTGTCCAGT | ||||
| V. vulnificus | dnaJ | VM-F | CAGGTTTGYTGCACGGCGAAGA | 412 | 45 |
| VV-Rmm | GTACGAAATTCTGACCGATCAA | ||||
| vvp | vvp_F | GACGTTCAAGCTGACGATGC | 598 | This study | |
| vvp_R | CACGCCCACTTGGTTAAACG |
Genes: dnaJ, housekeeping gene that encodes heat shock protein 40, for identification of Vibrio species; ctxA, cholera toxin; rfb, serotype O1/O139-specific rfb region; tcp, El Tor/classical toxin-coregulated pilus (TCP); tdh, thermostable direct hemolysin (TDH); trh, TDH-related hemolysin; vvp, elastolytic protease.
Universal forward primer for five Vibrio species.
Detection of virulence genes in V. cholerae, V. parahaemolyticus, and V. vulnificus.
Strains identified as V. cholerae (n = 24), V. parahaemolyticus (n = 1,757), and V. vulnificus (n = 130) were tested for the presence of virulence factors. Multiplex PCR was conducted using the primers listed in Table 5. V. cholerae NCCP 11179, V. parahaemolyticus DSM 25722, and V. vulnificus KCTC 2959T were used as positive controls. E. coli NCCP 10004 and a nontemplate control were included as negative controls. Amplification reaction mixtures contained 1× PCR buffer, 0.2 mM each dNTP, 1 U of nTaq-multi-HOT polymerase (Enzynomics, Daejeon, South Korea), 0.5 to 1 pmol/μl each primer (Macrogen, Daejeon, South Korea), and 1 μl of template in a final reaction volume of 20 μl. PCR amplifications were carried out as follows: a 10-min initial denaturation step, followed by 30 cycles of 94°C for 30 s, 51 to 60°C for a 30-s annealing step, and 72°C for 1 min and a final elongation step at 72°C for 7 min. Amplified products were examined by agarose gel electrophoresis (2%) and ethidium bromide staining.
Statistical analysis.
Principal-component analysis (PCA) was performed to examine the component distribution and correlation of the MPN of the presumptive Vibrio spp. with environmental parameters using CANOCO v5.0 (Microcomputer Power, Ithaca, NY). One-way ANOVA, Pearson's correlation, Spearman correlation, and linear regression were performed using SPSS Statistics v17.0 (SPSS Institute, Cary, NC) with a significance level of 0.05. One-way ANOVA was used to examine the mean difference between seasons and sites for all environmental parameters. A post hoc test was performed based on the least significant difference (LSD) at a significance level of 0.05. Pearson's correlation, Spearman correlation, and linear regression were used to investigate the correlations between the occurrence of omnipresent Vibrio spp. (V. alginolyticus and V. parahaemolyticus) and environmental parameters. By assuming that all variables are normally distributed, including those for the low-concentration samples, values below the limit of detection (LOD) were neglected (42, 43).
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Chang-Keun Kang, School of Earth Sciences and Environmental Engineering, Gwangju Institute of Science and Technology, Gwangju, South Korea, for reading the manuscript and giving us informative comments.
All of the authors of this study declare that there are no conflicts of interest.
This study was supported by the Environmental Health Action Program (RE201603079) funded by the Korea Ministry of Environment (MOE) and by the “Long-Term Change of Structure and Function in Marine Ecosystems of Korea” funded by the Ministry of Oceans and Fisheries, South Korea.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02680-16.
REFERENCES
- 1.Urakawa H, Rivera ING. 2006. Aquatic environments, p 175–189. In Thompson FL, Austin B, Swings J (ed), The biology of vibrios. ASM Press, Washington, DC. [Google Scholar]
- 2.Huehn S, Eichhorn C, Urmersbach S, Breidenbach J, Bechlars S, Bier N, Alter T, Bartelt E, Frank C, Oberheitmann B, Gunzer F, Brennholt N, Boer S, Appel B, Dieckmann R, Strauch E. 2014. Pathogenic vibrios in environmental, seafood and clinical sources in Germany. Int J Med Microbiol 304:843–850. doi: 10.1016/j.ijmm.2014.07.010. [DOI] [PubMed] [Google Scholar]
- 3.Harvell CD, Kim K, Burkholder JM, Colwell RR, Epstein PR, Grimes DJ, Hofmann EE, Lipp EK, Osterhaus AD, Overstreet RM, Porter JW, Smith GW, Vasta GR. 1999. Emerging marine diseases-climate links and anthropogenic factors. Science 285:1505–1510. doi: 10.1126/science.285.5433.1505. [DOI] [PubMed] [Google Scholar]
- 4.Baker-Austin C, Stockley L, Rangdale R, Martinez-Urtaza J. 2010. Environmental occurrence and clinical impact of Vibrio vulnificus and Vibrio parahaemolyticus: a European perspective. Environ Microbiol Rep 2:7–18. doi: 10.1111/j.1758-2229.2009.00096.x. [DOI] [PubMed] [Google Scholar]
- 5.Frank C, Littman M, Alpers K, Hallauer J. 2006. Vibrio vulnificus wound infections after contact with the Baltic Sea, Germany. Euro Surveill 11(33):pii=3024 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=3024. [DOI] [PubMed] [Google Scholar]
- 6.Huq A, Sack RB, Nizam A, Longini IM, Nair GB, Ali A, Morris JG Jr, Khan MN, Siddique AK, Yunus M, Albert MJ, Sack DA, Colwell RR. 2005. Critical factors influencing the occurrence of Vibrio cholerae in the environment of Bangladesh. Appl Environ Microbiol 71:4645–4654. doi: 10.1128/AEM.71.8.4645-4654.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tantillo GM, Fontanarosa M, Di Pinto A, Musti M. 2004. Updated perspectives on emerging vibrios associated with human infections. Lett Appl Microbiol 39:117–126. doi: 10.1111/j.1472-765X.2004.01568.x. [DOI] [PubMed] [Google Scholar]
- 8.Oberbeckmann S, Fuchs BM, Meiners M, Wichels A, Wiltshire KH, Gerdts G. 2012. Seasonal dynamics and modeling of a Vibrio community in coastal waters of the North Sea. Microb Ecol 63:543–551. doi: 10.1007/s00248-011-9990-9. [DOI] [PubMed] [Google Scholar]
- 9.Boer SI, Heinemeyer EA, Luden K, Erler R, Gerdts G, Janssen F, Brennholt N. 2013. Temporal and spatial distribution patterns of potentially pathogenic Vibrio spp. at recreational beaches of the German North Sea. Microb Ecol 65:1052–1067. doi: 10.1007/s00248-013-0221-4. [DOI] [PubMed] [Google Scholar]
- 10.Lopez-Hernandez KM, Pardio-Sedas VT, Lizarraga-Partida L, Williams Jde J, Martinez-Herrera D, Flores-Primo A, Uscanga-Serrano R, Rendon-Castro K. 2015. Environmental parameters influence on the dynamics of total and pathogenic Vibrio parahaemolyticus densities in Crassostrea virginica harvested from Mexico's Gulf coast. Mar Pollut Bull 91:317–329. doi: 10.1016/j.marpolbul.2014.11.015. [DOI] [PubMed] [Google Scholar]
- 11.Jones JL, Ludeke CH, Bowers JC, DeRosia-Banick K, Carey DH, Hastback W. 2014. Abundance of Vibrio cholerae, V. vulnificus, and V. parahaemolyticus in oysters (Crassostrea virginica) and clams (Mercenaria mercenaria) from Long Island sound. Appl Environ Microbiol 80:7667–7672. doi: 10.1128/AEM.02820-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gutierrez West CK, Klein SL, Lovell CR. 2013. High frequency of virulence factor genes tdh, trh, and tlh in Vibrio parahaemolyticus strains isolated from a pristine estuary. Appl Environ Microbiol 79:2247–2252. doi: 10.1128/AEM.03792-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leoni F, Talevi G, Masini L, Ottaviani D, Rocchegiani E. 2016. Trh (tdh−/trh+) gene analysis of clinical, environmental and food isolates of Vibrio parahaemolyticus as a tool for investigating pathogenicity. Int J Food Microbiol 225:43–53. doi: 10.1016/j.ijfoodmicro.2016.02.016. [DOI] [PubMed] [Google Scholar]
- 14.Givens CE, Bowers JC, DePaola A, Hollibaugh JT, Jones JL. 2014. Occurrence and distribution of Vibrio vulnificus and Vibrio parahaemolyticus—potential roles for fish, oyster, sediment and water. Lett Appl Microbiol 58:503–510. doi: 10.1111/lam.12226. [DOI] [PubMed] [Google Scholar]
- 15.Rodgers C, Parveen S, Chigbu P, Jacobs J, Rhodes M, Harter-Dennis J. 2014. Prevalence of Vibrio parahaemolyticus, and Vibrio vulnificus in blue crabs (Callinectes sapidus), seawater and sediments of the Maryland Coastal Bays. J Appl Microbiol 117:1198–1209. doi: 10.1111/jam.12608. [DOI] [PubMed] [Google Scholar]
- 16.Jones MK, Oliver JD. 2009. Vibrio vulnificus: disease and pathogenesis. Infect Immun 77:1723–1733. doi: 10.1128/IAI.01046-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cantet F, Hervio-Heath D, Caro A, Le Mennec C, Monteil C, Quemere C, Jolivet-Gougeon A, Colwell RR, Monfort P. 2013. Quantification of Vibrio parahaemolyticus, Vibrio vulnificus and Vibrio cholerae in French Mediterranean coastal lagoons. Res Microbiol 164:867–874. doi: 10.1016/j.resmic.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Strom MS, Paranjpye RN. 2000. Epidemiology and pathogenesis of Vibrio vulnificus. Microbes Infect 2:177–188. doi: 10.1016/S1286-4579(00)00270-7. [DOI] [PubMed] [Google Scholar]
- 19.Linkous DA, Oliver JD. 1999. Pathogenesis of Vibrio vulnificus. FEMS Microbiol Lett 174:207–214. doi: 10.1111/j.1574-6968.1999.tb13570.x. [DOI] [PubMed] [Google Scholar]
- 20.Miyoshi S. 2006. Vibrio vulnificus infection and metalloprotease. J Dermatol 33:589–595. doi: 10.1111/j.1346-8138.2006.00139.x. [DOI] [PubMed] [Google Scholar]
- 21.Alam M, Rashed SM, Mannan SB, Islam T, Lizarraga-Partida ML, Delgado G, Morales-Espinosa R, Mendez JL, Navarro A, Watanabe H, Ohnishi M, Hasan NA, Huq A, Sack RB, Colwell RR, Cravioto A. 2014. Occurrence in Mexico, 1998-2008, of Vibrio cholerae CTX+ El Tor carrying an additional truncated CTX prophage. Proc Natl Acad Sci U S A 111:9917–9922. doi: 10.1073/pnas.1323408111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rivera IN, Chun J, Huq A, Sack RB, Colwell RR. 2001. Genotypes associated with virulence in environmental isolates of Vibrio cholerae. Appl Environ Microbiol 67:2421–2429. doi: 10.1128/AEM.67.6.2421-2429.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li F, Du P, Li B, Ke C, Chen A, Chen J, Zhou H, Li J, Morris JG Jr, Kan B, Wang D. 2014. Distribution of virulence-associated genes and genetic relationships in non-O1/O139 Vibrio cholerae aquatic isolates from China. Appl Environ Microbiol 80:4987–4992. doi: 10.1128/AEM.01021-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tao Z, Larsen AM, Bullard SA, Wright AC, Arias CR. 2012. Prevalence and population structure of Vibrio vulnificus on fishes from the northern Gulf of Mexico. Appl Environ Microbiol 78:7611–7618. doi: 10.1128/AEM.01646-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoi L, Larsen JL, Dalsgaard I, Dalsgaard A. 1998. Occurrence of Vibrio vulnificus biotypes in Danish marine environments. Appl Environ Microbiol 64:7–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Banakar V, Constantin de Magny G, Jacobs J, Murtugudde R, Huq A, Wood RJ, Colwell RR. 2011. Temporal and spatial variability in the distribution of Vibrio vulnificus in the Chesapeake Bay: a hindcast study. Ecohealth 8:456–467. doi: 10.1007/s10393-011-0736-4. [DOI] [PubMed] [Google Scholar]
- 27.Park M-J, Oh HJ. 2004. Water masses and salinity in the Eastern Yellow Sea from winter to spring. Ocean Polar Res 26:65–75. doi: 10.4217/OPR.2004.26.1.065. [DOI] [Google Scholar]
- 28.Kaneko T, Colwell RR. 1973. Ecology of Vibrio parahaemolyticus in Chesapeake Bay. J Bacteriol 113:24–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cook DW, Bowers JC, DePaola A. 2002. Density of total and pathogenic (tdh+) Vibrio parahaemolyticus in Atlantic and Gulf coast molluscan shellfish at harvest. J Food Prot 65:1873–1880. [DOI] [PubMed] [Google Scholar]
- 30.Rodriguez-Castro A, Ansede-Bermejo J, Blanco-Abad V, Varela-Pet J, Garcia-Martin O, Martinez-Urtaza J. 2010. Prevalence and genetic diversity of pathogenic populations of Vibrio parahaemolyticus in coastal waters of Galicia, Spain. Environ Microbiol Rep 2:58–66. doi: 10.1111/j.1758-2229.2009.00064.x. [DOI] [PubMed] [Google Scholar]
- 31.Daniels NA, MacKinnon L, Bishop R, Altekruse S, Ray B, Hammond RM, Thompson S, Wilson S, Bean NH, Griffin PM, Slutsker L. 2000. Vibrio parahaemolyticus infections in the United States, 1973-1998. J Infect Dis 181:1661–1666. doi: 10.1086/315459. [DOI] [PubMed] [Google Scholar]
- 32.Gonzalez-Escalona N, Cachicas V, Acevedo C, Rioseco ML, Vergara JA, Cabello F, Romero J, Espejo RT. 2005. Vibrio parahaemolyticus diarrhea, Chile, 1998 and 2004. Emerg Infect Dis 11:129–131. doi: 10.3201/eid1101.040762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McLaughlin JB, DePaola A, Bopp CA, Martinek KA, Napolilli NP, Allison CG, Murray SL, Thompson EC, Bird MM, Middaugh JP. 2005. Outbreak of Vibrio parahaemolyticus gastroenteritis associated with Alaskan oysters. N Engl J Med 353:1463–1470. doi: 10.1056/NEJMoa051594. [DOI] [PubMed] [Google Scholar]
- 34.Martinez-Urtaza J, Lozano-Leon A, DePaola A, Ishibashi M, Shimada K, Nishibuchi M, Liebana E. 2004. Characterization of pathogenic Vibrio parahaemolyticus isolates from clinical sources in Spain and comparison with Asian and North American pandemic isolates. J Clin Microbiol 42:4672–4678. doi: 10.1128/JCM.42.10.4672-4678.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oliver JD. 2006. Vibrio vulnificus, p 349–366. In Thompson FL, Austin B, Swings J (ed), The biology of vibrios. ASM Press, Washington, DC. [Google Scholar]
- 36.Berho C, Guigues N, Togola A, Roy S, Fouillac A-M, Allan I. 2009. Evaluation of the field performance of emerging water quality monitoring tools. John Wiley & Sons, Hoboken, NJ, USA. [Google Scholar]
- 37.Blodgett R. October 2010. Bacteriological analytical manual, appendix 2. Most probable number from serial dilutions. http://www.fda.gov/Food/FoodScienceResearch/LaboratoryMethods/ucm109656.htm Accessed 20 September 2016.
- 38.Kaysner CA, DePaola A Jr. May 2004. Bacteriological analytical manual. Chapter 9, Vibrio. http://www.fda.gov/Food/FoodScienceResearch/LaboratoryMethods/ucm070830.htm Accessed 20 September 2016.
- 39.Sparks DL, Page AL, Helmke PA, Loeppert RH, Soltanpour PN, Tabatabai MA, Johnston CT, Sumner ME. 1996. Methods of soil analysis, part 3. Chemical methods. Soil Science Society of America, Inc., and American Society of Agronomy, Inc., Madison, WI, USA. [Google Scholar]
- 40.Kingsley MT, Bohlool BB. 1981. Release of Rhizobium spp. from tropical soils and recovery for immunofluorescence enumeration. Appl Environ Microbiol 42:241–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson LK, Brown MB, Carruthers EA, Ferguson JA, Dombek PE, Sadowsky MJ. 2004. Sample size, library composition, and genotypic diversity among natural populations of Escherichia coli from different animals influence accuracy of determining sources of fecal pollution. Appl Environ Microbiol 70:4478–4485. doi: 10.1128/AEM.70.8.4478-4485.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Armbruster DA, Pry T. 2008. Limit of blank, limit of detection and limit of quantitation. Clin Biochem Rev 29(Suppl 1):S49–S52. [PMC free article] [PubMed] [Google Scholar]
- 43.Shrivastava A, Gupta VB. 2011. Methods for the determination of limit of detection and limit of quantification of the analytical methods. Chron Young Sci 2:21–25. doi: 10.4103/2229-5186.79345. [DOI] [Google Scholar]
- 44.Tall A, Teillon A, Boisset C, Delesmont R, Touron-Bodilis A, Hervio-Heath D. 2012. Real-time PCR optimization to identify environmental Vibrio spp. strains. J Appl Microbiol 113:361–372. doi: 10.1111/j.1365-2672.2012.05350.x. [DOI] [PubMed] [Google Scholar]
- 45.Nhung PH, Ohkusu K, Miyasaka J, Sun XS, Ezaki T. 2007. Rapid and specific identification of 5 human pathogenic Vibrio species by multiplex polymerase chain reaction targeted to dnaJ gene. Diagn Microbiol Infect Dis 59:271–275. doi: 10.1016/j.diagmicrobio.2007.05.016. [DOI] [PubMed] [Google Scholar]
- 46.Hoshino K, Yamasaki S, Mukhopadhyay AK, Chakraborty S, Basu A, Bhattacharya SK, Nair GB, Shimada T, Takeda Y. 1998. Development and evaluation of a multiplex PCR assay for rapid detection of toxigenic Vibrio cholerae O1 and O139. FEMS Immunol Med Microbiol 20:201–207. doi: 10.1111/j.1574-695X.1998.tb01128.x. [DOI] [PubMed] [Google Scholar]
- 47.He P, Chen Z, Luo J, Wang H, Yan Y, Chen L, Gao W. 2014. Multiplex real-time PCR assay for detection of pathogenic Vibrio parahaemolyticus strains. Mol Cell Probes 28:246–250. doi: 10.1016/j.mcp.2014.06.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


