ABSTRACT
Lantibiotics are ribosomally synthesized and posttranslationally modified antimicrobial peptides that are characterized by the thioether cross-linked bisamino acids lanthionine (Lan) and methyllanthionine (MeLan). Duramycin contains 19 amino acids, including one Lan and two MeLans, an unusual lysinoalanine (Lal) bridge formed from the ε-amino group of lysine 19 and a serine residue at position 6, and an erythro-3-hydroxy-l-aspartic acid at position 15. These modifications are important for the interactions of duramycin with its biological target, phosphatidylethanolamine (PE). Based on the binding affinity and specificity for PE, duramycin has been investigated as a potential therapeutic, as a molecular probe to investigate the role and localization of PE in biological systems, and to block viral entry into mammalian cells. In this study, we identified the duramycin biosynthetic gene cluster by genome sequencing of Streptomyces cinnamoneus ATCC 12686 and investigated the dur biosynthetic machinery by heterologous expression in Escherichia coli. In addition, the analog duramycin C, containing six amino acid changes compared to duramycin, was successfully generated in E. coli. The substrate recognition motif of DurX, an α-ketoglutarate/iron(II)-dependent hydroxylase that carries out the hydroxylation of aspartate 15 of the precursor peptide DurA, was also investigated using mutagenesis of the DurA peptide. Both in vivo and in vitro results demonstrated that Gly16 is important for DurX activity.
IMPORTANCE Duramycin is a natural product produced by certain bacteria that binds to phosphatidylethanolamine (PE). Because PE is involved in many cellular processes, duramycin is an antibiotic that kills bacteria, but it has also been used as a molecular probe to detect PE and monitor its localization in mammalian cells and even whole organisms, and it was recently shown to display broad-spectrum inhibition of viral entry into host cells. In addition, the molecule has been evaluated as treatment for cystic fibrosis. We report here the genes that are involved in duramycin biosynthesis, and we produced duramycin by expressing those genes in Escherichia coli. We show that duramycin analogs can also be produced. The ability to access duramycin and analogs by production in E. coli opens opportunities to improve duramycin as an antibiotic, PE probe, antiviral, or cystic fibrosis therapeutic.
KEYWORDS: antibiotic, antiviral agents, biosynthesis, duramycin, phosphatidylethanolamine
INTRODUCTION
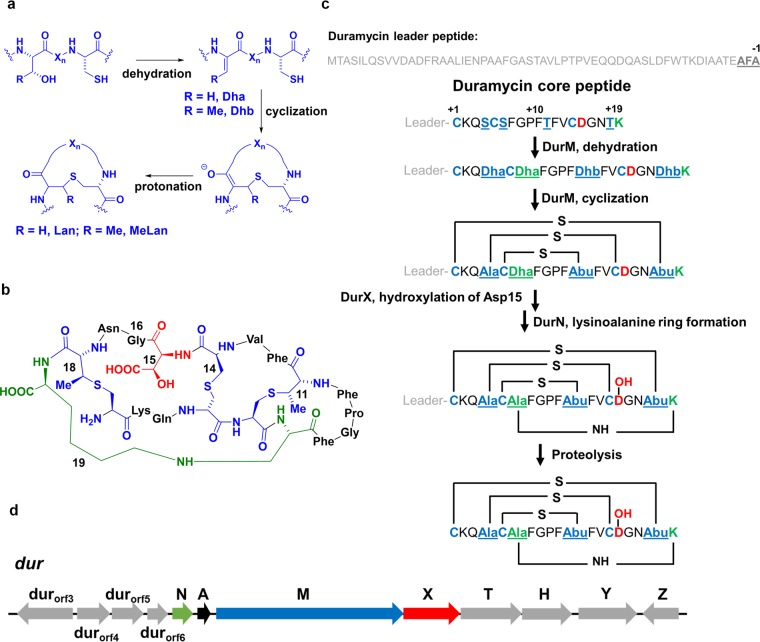
Lantibiotics are ribosomally synthesized and posttranslationally modified peptides (RiPPs) exhibiting antimicrobial activity. They are characterized by the thioether cross-linked amino acids lanthionine (Lan) and/or methyllanthionine (MeLan) (1), which are formed by dehydration of Ser and Thr residues, followed by intramolecular Michael-type addition of Cys thiols to the resulting dehydroalanine (Dha) and dehydrobutyrine (Dhb), respectively (2–4) (Fig. 1a). These posttranslational modifications take place in the core peptide region of the precursor peptide, whereas an N-terminal leader peptide is not modified but is critical for substrate recognition. Duramycin and its structurally closely related analogs cinnamycin, duramycin B, and duramycin C are produced by streptomycetes (5–10), whereas cinnamycin B was recently isolated from the actinomycete Actinomadura atramentaria NBRC 14695T (11). These peptides all contain 19 amino acids and exhibit a compact globular structure with one Lan and two MeLan cross-links (Fig. 1b; see also Fig. S1 in the supplemental material) (7–10). They also share an unusual lysinoalanine (Lal) cross-link formed between Lys19 and Ser6 and an erythro-3-hydroxy-l-aspartic acid resulting from the hydroxylation of Asp15 (Fig. 1b; see also Fig. S1). These compounds exhibit antimicrobial activity against Gram-positive bacteria (12) by binding to phosphatidylethanolamine (PE) (13). The compound recognizes the head group of PE with high affinity (14–16), in part because of the ionic interaction between the carboxylate of Asp15 and the ammonium group of PE and the additional binding energy arising from the β-hydroxyl group on Asp15. The interaction of duramycin with PE promotes chloride secretion in lung epithelial cells, which, in turn, causes mucus clearance from the lungs (17). Thus, duramycin entered phase II clinical trials for treatment of cystic fibrosis, a disorder characterized by disrupted chloride ion transport (17–19). In addition, recent studies have shown that duramycin can inhibit cell entry of viruses, such as Ebola, dengue, and West Nile viruses, that carry in their virions PE that is recognized by cellular phosphatidylserine receptors in the host cells (20). PE is highly abundant in biological membranes (21, 22) and is translocated or redistributed across the membrane bilayer during a number of important biological processes, including cell division and apoptosis (23). Based on the relatively low molecular weight, highly stable structure, and high binding affinity, duramycin-like lantibiotics have also been used as molecular probes for the localization of PE (24).
FIG 1.
(a) General biosynthetic route toward Lan and MeLan. (b) Structure of duramycin. Lan and MeLan are shown in blue, Lal is depicted in green, and the hydroxylated Asp is shown in red. (c) Posttranslational maturation of duramycin. The sequence of the DurA core and leader peptides is depicted, as well as the residues that form the cross-links. The putative sec cleavage sequence at the end of the leader peptide is shown by bold underlines. (d) Biosynthetic gene cluster for duramycin.
Characterization of the biosynthetic pathways toward duramycin could be beneficial to create recombinant libraries for pharmacological or diagnostic screening. In 2003, the cinnamycin biosynthetic gene cluster from Streptomyces cinnamoneus DSM 40005 was reported and used to produce cinnamycin (6). More recently, we characterized the biochemical processes involved in cinnamycin biosynthesis both in E. coli and in vitro (25). Although cinnamycin and duramycin differ by only a single amino acid (Fig. S1), duramycin is much more soluble and is more readily derivatized for diagnostic applications (23). In this study, we first identified the putative duramycin biosynthetic gene cluster from S. cinnamoneus ATCC 12686 by genome sequencing. We then investigated the biosynthetic machinery by heterologous production of duramycin in E. coli using a minimal functional cassette of biosynthetic genes (durA, durM, durN, and durX). Furthermore, using mutagenesis of the structural gene, we were able to generate duramycin C in E. coli.
RESULTS AND DISCUSSION
Analysis of the duramycin biosynthetic gene cluster from S. cinnamoneus ATCC 12686.
Based on the genome sequencing results of S. cinnamoneus ATCC 12686 and the known genes involved in the cinnamycin biosynthetic pathway (6), four open reading frames (ORFs) termed durA, durM, durN, and durX were identified in the duramycin biosynthetic gene cluster (Fig. 1d and Table 1). DurA is the precursor peptide consisting of a C-terminal 19-amino-acid core region that is transformed into duramycin by posttranslational modifications (PTMs) and a 59-amino-acid leader sequence (Fig. 1c). Similar to the cinnamycin precursor peptide CinA, but unlike most class II lantibiotics (1), DurA lacks a conserved GG/GA protease cleavage motif for leader peptide removal (26) and the cluster does not encode a specific protease. Instead, an AFA (AXA) motif that can be recognized by a type I signal peptidase is located between the leader sequence and core region, indicating that the general secretory (sec) pathway might be involved in the leader cleavage process (6, 27) (Fig. 1c). DurA shows 92% sequence identity to CinA, with 100% sequence coverage, which is consistent with the high structural homology between duramycin and cinnamycin. DurM exhibits high sequence similarity to class II lanthionine synthetases (generically called LanM), which catalyze dehydrations of Ser and Thr to Dha and Dhb, respectively, as well as the subsequent additions of Cys to the resulting dehydro amino acids. DurM shows 85% identity to CinM, with 100% coverage. DurX shows 89% identity to CinX, with 97% coverage. The latter has been biochemically characterized as a member of the non-heme Fe(II)-dependent family of enzymes. It catalyzes the hydroxylation of the β-carbon of Asp15 by utilizing α-ketoglutarate (α-KG) as a cofactor (25). The high sequence similarity between DurX and CinX suggests that DurX is responsible for the hydroxylation of Asp15 of duramycin. Another small gene in the duramycin gene cluster has 89% identity to cinorf7, with 97% coverage, indicating that like cinorf7, this gene is involved in Lal formation (25). Rather than continuing to call this set of orthologous genes lanorf7, we propose and use here the designation durN to indicate the involvement in Lal formation. In addition, a number of other putative genes with high sequence similarity to their counterparts in the cinnamycin biosynthetic pathways were also identified; these are presumably involved in secretion, self-resistance, and transcriptional regulation (6) (Fig. 1d and Table 1). For example, durorf4 encodes a helix-turn-helix DNA-binding motif which is similar to the pfam TETR family. durT and durH encode subunits of an ABC transporter. DurT appears to be the ATP-binding subunit showing 94% identity to CinT, while DurH is a putative integral membrane subunit of an ABC-2-type transporter that is commonly involved in drug efflux and resistance. durY encodes a protein that might possess an N-terminal signal peptide but otherwise has no significant homologies to known proteins. durZ encodes a member of the pfam UPF0029 family of proteins with unknown function. We further examined ca. 10 kb of up- and downstream sequences of the putative duramycin biosynthetic gene cluster, and no other homologs to cinnamycin biosynthetic genes could be identified (see Table S1 and Fig. S2 in the supplemental material). It is worth mentioning that by searching the whole genome sequence of S. cinnamoneus ATCC 12686, we could not identify a counterpart of the cinorf10 gene encoding a putative phospholipid N-methyltransferase that is present in the cinnamycin biosynthetic cluster (6). Cinorf10 was very recently shown to confer self-resistance by monomethylation of PE (47). We did not attempt to determine the boundary of the biosynthetic gene cluster of duramycin. However, compared to the homologs present in the cinnamycin biosynthetic pathway, the genes shown in Fig. 1d were anticipated to be sufficient for duramycin biosynthesis.
TABLE 1.
Proteins involved in duramycin biosynthesis
| Proteins | No. of residues | Putative annotation | Proposed function(s) in duramycin biosynthesis |
|---|---|---|---|
| DurA | 77 | Structural gene | Precursor peptide |
| DurM | 1,083 | Class II lanthipeptide synthetase | Dehydration of Ser/Thr and cyclization of Dha/Dhb with Cys |
| DurX | 328 | Non-heme Fe(II)-dependent hydroxylase | Hydroxylation of Asp15 |
| DurN | 119 | Hypothetical protein | Formation of lysinoalanine ring |
| Durorf6 | 119 | Hypothetical protein | Unknown |
| Durorf5 | 185 | Hypothetical protein | Unknown |
| Durorf4 | 190 | Putative TetR transcriptional regulator | Pathway regulation |
| Durorf3 | 288 | EamA-like transporter family/permease of the drug/metabolite transporter superfamily | Secretion and/or self-resistance |
| DurT | 352 | Putative ABC transporter | Secretion and/or self-resistance |
| DurH | 290 | Putative ABC transporter | Secretion and/or self-resistance |
| DurY | 340 | Hypothetical protein | Unknown |
| DurZ | 208 | Pfam UPF0029 family | Unknown |
Coexpression of DurA, DurM, DurN, and DurX in Escherichia coli.
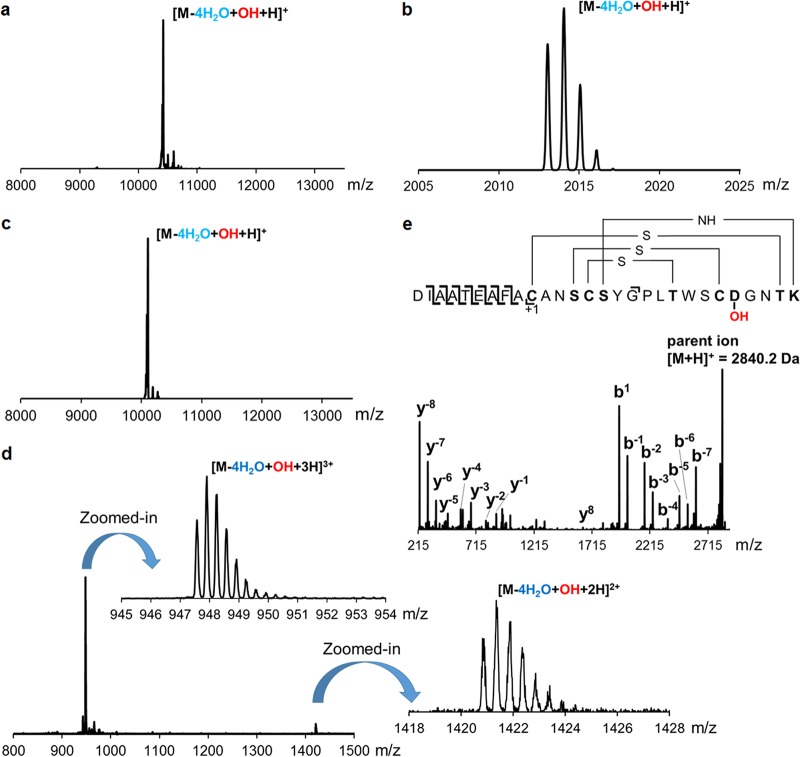
A number of recent studies from different laboratories have demonstrated the feasibility of reconstituting the posttranslational modifications toward lantibiotics in E. coli (25, 28–34). Similarly, N-terminal His6-DurA was coexpressed in this work with DurM in E. coli BL21(DE3). After metal affinity purification, analysis by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) showed that DurM-modified His6-DurA exhibited a mass difference compared to the unmodified peptide corresponding to four dehydrations (Fig. S3). The modified peptide was treated with iodoacetamide (IAA) to determine the presence of free thiols (35). Subsequent analysis by MALDI-TOF MS showed that acetamidomethyl adducts were not formed in comparison to a control with unmodified peptide, indicating the complete installation of all thioether cross-links by DurM (Fig. S3). Next, His6-DurA was coexpressed with DurM, DurN, and DurX in E. coli BL21(DE3). After purification, MALDI-TOF MS analysis revealed that the modified His6-DurA exhibited a mass corresponding to four dehydrations and one additional hydroxylation (Fig. 2a). These results demonstrated that DurM dehydrated all the available Ser and Thr residues in the DurA core peptide region as expected, and DurX hydroxylated Asp15 in E. coli. Since Lal formation would not result in a mass change, MS could not be used to verify its generation. However, the bioactivity of cinnamycin was abolished in a product containing all PTMs but lacking the Lal ring (25). Thus, we confirmed Lal formation by testing the bioactivity of the final mature compound. In order to generate mature duramycin, two steps of proteolytic cleavage of leader peptide were conducted. First, removal of most of the leader peptide was accomplished by treatment with Glu-C endoprotease, which leaves three residues (AFA) at the N terminus of the modified core peptide. Then the AFA-core fragment was treated with aminopeptidase. Since the first Cys residue in the DurA core peptide sequence is involved in a Lan cross-link, aminopeptidase was anticipated to not remove this residue (36–41), and thus, this treatment would remove only the N-terminal AFA residues and yield mature duramycin, which was indeed observed. The duramycin thus generated exhibited the same retention time as that of authentic duramycin in liquid chromatography (LC) (Fig. S4), and MALDI-TOF MS analysis demonstrated its correct mass (Fig. 2b). In addition, an agar diffusion growth inhibition assay against Bacillus subtilis ATCC 6633 showed that the duramycin generated in E. coli exhibited a zone of growth inhibition similar to that of authentic duramycin (Fig. 3). If DurM, DurX, or DurN was omitted from the coexpression system, the product produced was not active (Fig. S5). Collectively, these data confirmed that duramycin with all required posttranslational modifications correctly installed was indeed produced by coexpression in E. coli followed by proteolysis in vitro. Isothermal titration calorimetry (ITC) experiments were conducted next, and they showed that the binding interaction between PE and the generated duramycin was very similar to that of authentic duramycin (Fig. S6) (42, 43). These results confirmed successful generation of duramycin by heterologous expression of the minimal gene cassette of durA, durM, durN, and durX in E. coli.
FIG 2.
(a) MALDI-TOF mass spectrum of His6-DurA modified by DurM, DurN, and DurX (calculated m/z = 10,434.52 Da; observed m/z = 10,432.97 Da); (b) ESI-QTOF mass spectrum of His6-DurA modified by DurM, DurN, and DurX followed by Glu-C and aminopeptidase digestion (calculated m/z = 2,012.8852 Da; observed m/z = 2,012.9556 Da); (c) MALDI-TOF mass spectrum of His6-DurAduramycinC modified by DurM, DurN, and DurX (calculated m/z = 10,106.75 Da; observed m/z = 10,107.21 Da); (d) ESI mass spectrum of His6-DurAduramycinC modified by DurM, DurN, and DurX followed by Lys-C digestion (calculated m/z = 947.4123 Da for [M-4H2O+OH + 3H]3+ and observed m/z = 947.4321; calculated m/z = 1,420.6148 Da for [M-4H2O+OH + 2H]2+ and observed m/z = 1,420.6523 Da). (e) MS-MS fragmentation pattern of His6-DurAduramycinC modified by DurM, DurN, and DurX followed by Lys-C digestion.
FIG 3.
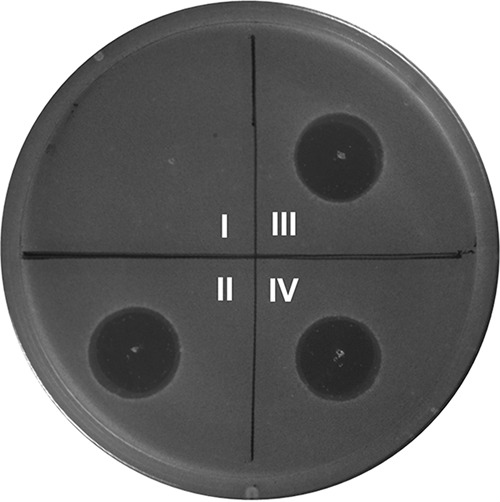
Antimicrobial agar diffusion growth inhibition assay of duramycin and duramycin C against Bacillus subtilis ATCC 6633. (I) Blank; (II) authentic duramycin; (III) duramycin produced by E. coli followed by in vitro proteolytic cleavage of leader peptide; (IV) duramycin C produced in E. coli followed by in vitro proteolytic removal of the leader peptide. Amount spotted for each assay: 100 pmol.
Heterologous production of duramycin C in E. coli.
The cinnamycin biosynthetic machinery has been used to make the variants duramycin A and B, but heterologous production of duramycin C was unsuccessful (6), possibly because it involves six simultaneous amino acid changes compared to cinnamycin. Duramycin C also differs at six positions compared to duramycin (Fig. 1; see also Fig. S1). With the successful establishment of a heterologous expression platform of duramycin in E. coli, we employed site-directed mutagenesis to generate a durA variant (durAduramycinC) encoding a precursor peptide containing the duramycin C core peptide sequence appended to the DurA leader sequence (i.e., DurA-K2A/Q3N/F7Y/F10L/F12W/V13S). His6-DurAduramycinC was first coexpressed with DurM in E. coli BL21(DE3). After purification, analysis by MALDI-TOF MS showed that DurM-modified His6-DurAduramycinC exhibited a mass corresponding to the desired four dehydrations (Fig. S7), indicating that DurM is substrate tolerant toward the duramycin C core peptide sequence. The completeness of installation of the three thioether cross-links was then verified by IAA assay (Fig. S7). Next, His6-DurAduramycinC was coexpressed with DurM, DurN, and DurX in E. coli BL21(DE3). After purification, analysis by MALDI-TOF MS showed that the modified His6-DurAduramycinC exhibited a mass corresponding to four dehydrations and one hydroxylation, indicating that all the required posttranslational modifications were accomplished in E. coli (Fig. 2c). However, another signal corresponding to four dehydrations without hydroxylation was also detected, suggesting that changing 6 out of 19 residues diminished the activity of DurX such that only a subset of the 4-fold-dehydrated species was hydroxylated (Fig. S8). Further optimization of expression conditions and change of expression vectors did not improve the modification by DurX, again suggesting that the effect might have been due to an unfavorable substrate sequence. Regardless, after proteolytic removal of the leader peptide using Glu-C and aminopeptidase and subsequent high-performance liquid chromatography (HPLC) purification, analysis by electrospray ionization–quadrupole time of flight mass spectrometry (ESI-QTOF MS) confirmed that a product corresponding to duramycin C was successfully generated (Fig. 2d). The correct formation of the Lan/MeLan cross-links was supported by MS/MS fragmentation (Fig. 2e). The duramycin C so produced was subjected to bioactivity and PE-binding assays as described above for duramycin (Fig. 3; see also Fig. S9), and the results strongly support the successful generation of duramycin C in E. coli. They also show that despite the six simultaneous amino acid changes compared to duramycin, duramycin C also recognizes PE.
Investigation of the recognition motif(s) for DurX.
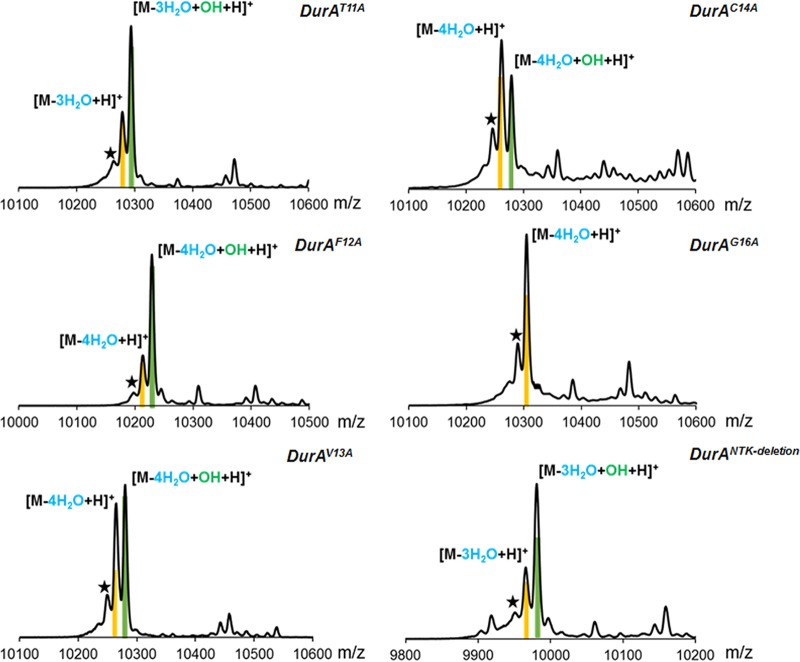
The observations discussed in the previous section suggest that DurM and DurN are quite permissive with respect to the sequence they require for catalysis. Conversely, the observed incomplete hydroxylation of the duramycin C core peptide sequence suggests that DurX has a preference for the native sequence. However, the six mutations are relatively far from Asp15 (Fig. S1). Since a sequence-tolerant Asp β-hydroxylase could be quite useful, we investigated the sequence specificity of DurX by performing alanine scanning on DurA for positions 11 to 16 (DurA mutations T11A, F12A, V13A, C14A, and G16A). In addition, another mutant was made in which the last three residues (NTK) were deleted. Previous studies have shown that for cinnamycin biosynthesis, CinM and CinX do not have a compulsory order of action in vitro, since CinX can hydroxylate CinM-modified CinA and vice versa (25). Thus, we first tested whether DurX could hydroxylate unmodified DurA. His6-DurA was coexpressed with DurX in E. coli BL21(DE3). After purification and Glu-C proteolysis, analysis by MALDI-TOF and ESI MS showed that only a limited amount of hydroxylated DurA could be observed (Fig. S10), indicating that unmodified DurA is not a favorable substrate for DurX. Next, His6-DurA was coexpressed with DurM and DurX in E. coli BL21(DE3). After purification and Glu-C proteolysis, analysis by MALDI-TOF MS demonstrated a single peak exhibiting a mass corresponding to four dehydrations and one hydroxylation, indicating that DurM-modified DurA is a good substrate for DurX and probably the native substrate. Then, the His6-DurA mutants were coexpressed with DurM in E. coli BL21(DE3). After purification, MALDI-TOF MS showed that DurM catalyzed the full dehydration of each mutant (four dehydrations for F12A, V13A, and C14A and three dehydrations for T11A and NTK deletion), illustrating its high substrate tolerance (Fig. S11). The products were also cyclized as shown by IAA assays (Fig. S12). Based on these results, we investigated the substrate specificity of DurX using a coexpression system encoding DurA variants DurM and DurX. After purification, MALDI-TOF MS analyses demonstrated that mutation of Thr11, Phe12, Val13, and Cys14 located N-terminally of Asp15, and Asn17, Thr18 and K19 located C-terminally of Asp15 did not abolish hydroxylation by DurX, although incomplete hydroxylation was observed for all of these mutants (Fig. 4). These findings are congruent with the observation for the core peptide of duramycin C in that the native sequence is the preferred substrate but partial activity is observed for each mutant. His6-DurX was also expressed in E. coli BL21(DE3) and purified by metal affinity chromatography. Incubation of His6-DurX with the His6-DurA mutants in vitro using conditions previously reported for CinX (25) and subsequent MALDI-TOF MS analysis provided results consistent with the coexpression data.
FIG 4.
MALDI-TOF mass spectra of DurM- and DurX-modified His6-DurA mutants in E. coli. Peaks corresponding to dehydrated products are highlighted in yellow, and peaks corresponding to dehydrated and hydroxylated products are indicated in green. The star indicates a peak resulting from laser-induced deamination, which occurs as a consequence of analyzing large peptides by MALDI-TOF MS in reflector mode.
We note that individual mutations at Thr11, Cys14, Thr18, and Lys19 each prevent formation of one of the four cross-links in duramycin. Yet all dehydrated peptides were partially hydroxylated, showing that none of the cross-links is absolutely required for recognition by DurX. However, a C-terminal Gly16 flanking Asp15 appears to be essential for DurX catalysis, since no hydroxylated species could be detected for the DurA-G16A variant (Fig. 4). Other members of the Fe(II)/α-KG-dependent enzyme family, among which are prolyl-4-hydroxylase and lysyl-5-hydroxylase, hydroxylate their substrates with a minimum chain length of six amino acids containing an Xaa-Pro/Lys-Gly sequence (44). Our results suggest that the recognition sequence for hydroxylation by DurX is similar (Xaa-Asp-Gly).
In summary, we describe the biosynthetic gene cluster for duramycin and show that its biosynthetic enzymes can produce the fully posttranslationally modified peptide in E. coli. The enzymes are sufficiently substrate tolerant that even a variant in which 6 out of 19 amino acids were changed was converted to a product that has high affinity for PE. Of the three proteins investigated (DurM, DurN, and DurX), the Asp hydroxylase DurX is least forgiving in that its activity toward analogs was decreased and that mutation of the Gly residue flanking Asp15 resulted in loss of activity in vitro and in E. coli. Collectively, these findings can be utilized for the design and application of variants of duramycin with improved properties for diagnostic or therapeutic applications (17, 20, 23).
MATERIALS AND METHODS
Materials.
All chemicals were purchased from Sigma-Aldrich, Roche Biosciences, or Fisher Scientific unless noted otherwise. All oligonucleotides were purchased from Integrated DNA Technologies and are shown in Table 2. Restriction endonucleases, DNA polymerases, deoxynucleoside triphosphate (dNTP) solutions, and a Gibson assembly cloning kit were purchased from New England BioLabs. Endoproteinases (Glu-C and Lys-C) were purchased from Roche Applied Science. Aminopeptidase was purchased from Sigma-Aldrich. Gel extraction, plasmid miniprep, and PCR purification kits were purchased from Qiagen. A genomic DNA isolation kit (UltraClean microbial DNA isolation kit) was purchased from Mo Bio Laboratories, Inc. Medium components for bacterial cultures were purchased from Fisher Scientific. Protein calibration standard I and peptide calibration standard II for MALDI-TOF MS were purchased from Bruker.
TABLE 2.
Primers used in the study
| Primer name | Sequence (5′-3′) |
|---|---|
| RSF-DurM-MCSII-up | ATATTAGTTAAGTATAAGAAGGAGATATACATATGGCAGATCTCAATTGGATATCGATGG |
| RSF-DurM-MCSII-dn | GCAGCGGTTTCTTTACCAGACTCGAGGGTACCTTACTGTTCGTGGTGCGACGGCGGGTCGA |
| RSF-DurA-MCSI-up | CAGGATCCGAATTCGAGCTCGGCGCGCCTGACCGCTTCGATTCTTCAGTCCGTCGTGGACG |
| RSF-DurA-MCSI-dn | TGCGGCCGCAAGCTTGTCGACCTGTTACTTGGTGTTGCCGTCACAC |
| PET-DurM-MCSII-up | ATATTAGTTAAGTATAAGAAGGAGATATACATATGGCAGATCTCAATTGGATATCGATGG |
| PET-DurM-MCSII-dn | GCAGCGGTTTCTTTACCAGACTCGAGGGTACCTTACTGTTCGTGGTGCGACGGCGGGTCGA |
| PET-DurA-MCSI-up | CAGGATCCGAATTCGAGCTCGGCGCGCCTGACCGCTTCGATTCTTCAGTCCGTCGTGGACG |
| PET-DurA-MCSI-dn | TGCGGCCGCAAGCTTGTCGACCTGTTACTTGGTGTTGCCGTCACAC |
| ACYC-DurX-MCSII-up | ATTAGTTAAGTATAAGAAGGAGATATACATATGGCCCTGAAGACCTGCGAGGAATTCCTG |
| ACYC-DurX-MCSII-dn | GCGTGGCCGGCCGATATCCAATTGAGATCTGCTCAGCCGTGGTGGCGGAGGGCGTCCATC |
| ACYC-DurN-MCSI-up | TTGTTTAACTTTAATAAGGAGATATACCATGAAAAGCGCCAAGGAACCGACGATCTACCA |
| ACYC-DurN-MCSI-dn | CTGGCTGTGGTGATGATGGTGATGGCTGCTGCCTCAGGACGTCAGCCGGGCGACCAACGT |
| RSF-DurX-MCSII-up | ATTAGTTAAGTATAAGAAGGAGATATACATATGGCCCTGAAGACCTGCGAGGAATTCCTG |
| RSF-DurX-MCSII-dn | GCGTGGCCGGCCGATATCCAATTGAGATCTGCTCAGCCGTGGTGGCGGAGGGCGTCCATC |
| RSF-DurN-MCSI-up | TGTTTAACTTTAATAAGGAGATATACCATGAAAAGCGCCAAGGAACCGACGATCTACCAG |
| RSF-DurN-MCSI-dn | GCGTGGCCGGCCGATATCCAATTGAGATCTGCTCAGCCGTGGTGGCGGAGGGCGTCCATC |
| RSF-DurAT11A-MCSI-dn | TGCGGCCGCAAGCTTGTCGACCTGTTACTTGGTGTTGCCGTCACACACGAAGGCGAACGGGCCGAAGCTGCAGCT |
| RSF-DurAF12A-MCSI-dn | AAGCTTGTCGACCTGCAGGCGCGCCGAGCTTTACTTGGTGTTGCCGTCACACACCGCGGT |
| RSF-DurAV13A-MCSI-dn | AAGCTTGTCGACCTGCAGGCGCGCCGAGCTTTACTTGGTGTTGCCGTCACACGCGAAGGT |
| RSF-DurAC14A-MCSI-dn | AAGCTTGTCGACCTGCAGGCGCGCCGAGCTTTACTTGGTGTTGCCGTCCGCCACGAAGGT |
| RSF-DurAG16A-MCSI-dn | TGCGGCCGCAAGCTTGTCGACCTGTTACTTGGTGTTGGCGTCACACACGAAGGTGAACGGGCCGAAGCTGCAGCT |
| RSF-DurANTK-deletion-MCSI-dn | TGCGGCCGCAAGCTTGTCGACCTGTTAGCCGTCACACACGAAGGTGAACGGGCCGAAGCTGCAGCT |
| His-DurX-MCSI-up | ACCATCATCACCACAGCCAGGATCCGAATTCGATGGCCCTGAAGACCTGCGAGGAATTC |
| His-DurX-MCSI-dn | AAGCTTGTCGACCTGCAGGCGCGCCGAGCTTCAGCCGTGGTGGCGGAGGGCGTCCATCCA |
| PET-DurAF12W, V13S-mcsI-up | GGATCCGAATTCGAGCTCGGCGCGCCTGACCGCTTCGATTCTTCAGTCCGTCGTGGACG |
| PET-DurAF12W, V13S-mcsI-dn | AGCATTATGCGGCCGCAAGCTTGTCGACCTGTTACTTGGTGTTGCCGTCACAGCTCCAGGTGAACGGGCCGAAGCT |
| PET-DurAK2A, Q3N, F12W, V13S-mcsI-up | AGGATCCGAATTCGAGCTCGGCGCGCCTGACCGCTTCGATTCTTCAGTCCGTCGTGGACG |
| PET-DurAK2A, Q3N, F12W, V13S-mcsI-dn | AGCATTATGCGGCCGCAAGCTTGTCGACCTGTTACTTGGTGTTGCCGTCACAGCTCCAGGTGAACGGGCCGAAGCTGCAGCTGTTCGCGCAGGCGAAGGCTTCCGTA |
| PET-DurAF7Y, F10L, K2A, Q3N, F12W, V13S-mcsI-up | CAGGATCCGAATTCGAGCTCGGCGCGCCTGACCGCTTCGATTCTTCAGTCCGTCGTGGACG |
| PET-DurAF7Y, F10L, K2A, Q3N, F12W, V13S-mcsI-dn | AGCATTATGCGGCCGCAAGCTTGTCGACCTGTTACTTGGTGTTGCCGTCACAGCTCCAGGTGAACGGGCCATAGCTGCAGCTGTTCGCGCA |
General methods.
For peptide residue numbering, positive residue numbers are used for amino acids in the core peptide counting forwards from the leader peptide cleavage site. Negative numbers are used for amino acids in the leader peptide counting backwards from the cleavage site. All PCRs were carried out on an automated thermocycler (C1000; Bio-Rad). Gibson assembly reaction solutions were made based on a published protocol (45). DNA sequencing was performed by the Biotechnology Center at the University of Illinois at Urbana-Champaign (UIUC) and ACGT, Inc. Matrix-assisted laser desorption–ionization time of flight mass spectrometry (MALDI-TOF MS) analyses were carried out at the Mass Spectrometry Laboratory at UIUC on an UltrafleXtreme mass spectrometer (Bruker Daltonics). For MALDI-TOF MS analysis, samples were desalted using ZipTip pipette tips containing C18 resin (Millipore) and spotted onto a MALDI target plate with a matrix solution containing 35 mg/ml of 2,5-dihydroxybenzoic acid (DHB) in 3:2 MeCN–H2O with 0.1% trifluoroacetic acid (TFA) or 15 mg/ml of sinapinic acid in 3:2 MeCN–H2O with 0.1% TFA. Peptides were desalted by C4 solid-phase extraction (SPE) and further purified by preparative reversed-phase high-performance liquid chromatography (RP-HPLC) on a Delta 600 instrument (Waters) equipped with a Phenomenex C18 column at a flow rate of 8 ml/min. For RP-HPLC, solvent A was 0.1% TFA in H2O and solvent B was 4:1 MeCN–H2O containing 0.086% TFA. An elution gradient from 0% solvent B to 100% solvent B over 45 min was used unless specified otherwise. Liquid chromatography-electrospray ionization tandem mass spectrometry (LC/ESI-QTOF-MS/MS) was carried out and processed using a Synapt ESI quadrupole TOF mass spectrometry system (Waters) equipped with an Acquity ultraperformance liquid chromatography (UPLC) system (Waters).
Strains and plasmids.
S. cinnamoneus ATCC 12686 (previously known as Streptoverticillium cinnamoneum forma azacoluta) and indicator strain Bacillus subtilis ATCC 6633 were obtained from the American Type Culture Collection (ATCC). E. coli DH5α and E. coli NEB Turbo were used as host for cloning and plasmid propagation, and E. coli BL21(DE3) was used as a host for expression of proteins and peptides. Coexpression vectors pRSFDuet-1, pACYCDuet-1, and pETDuet-1 were obtained from Novagen.
Sequencing and in silico analysis of S. cinnamoneus ATCC 12686 genome.
Total genomic DNA of S. cinnamoneus ATCC 12686 was isolated using a genomic DNA isolation kit (UltraClean microbial DNA isolation kit) and sequenced by Yun-juan Chang at the BioTechnology Center at UIUC using Illumina shotgun sequencing. The resulting sequence data were subsequently screened for putative open reading frames (ORFs) encoding peptides that contain the amino acid sequence of the duramycin core peptide (CKQSCSFGPFTFVCDGNTK). A small ORF (later named durA) was identified, encoding this sequence plus an N-terminal leader sequence (Fig. 1c). Prediction of other open reading frames was performed with FramePlot 4.0 (http://nocardia.nih.go.jp/fp4/), and functional annotation was based on BLASTP (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and Pfam (http://pfam.xfam.org/) searches. The Geneious software package (Biomatters, New Zealand) was used for the analysis and annotation of DNA and protein sequences. In addition to the putative ca. 11.5-kb duramycin biosynthetic gene cluster (dur genes [Fig. 1d and Table 2]) a number of flanking ORFs (orf1 and orf2 and orf8 to orf28; see Fig. S2 and Table S1 in the supplemental material) were identified.
Construction of pRSFDuet-1 derivative for coexpression of DurM and His6-DurA.
The gene encoding DurM harboring flanking sequences homologous to pRSFDuet-1 multiple-cloning site 2 was first amplified via PCR using genomic DNA of S. cinnamoneus ATCC 12686 as the template and primers RSF-DurM-mcsII-up and RSF-DurM-mcsII-dn (Table 2). Subsequently the PCR fragment was cloned into multiple-cloning site 2 of the pRSFDuet-1 vector (without a His tag) linearized by NdeI and KpnI using Gibson assembly to generate the pRSFDuet/DurM-2 vector. The gene encoding DurA harboring flanking sequences homologous to pRSFDuet-1 multiple-cloning site 1 (MCS1) were then amplified using primers RSF-DurA-mcsI-up and RSF-DurA-mcsI-dn (Table 2). Then the corresponding fragment was cloned into MCS1 of the pRSFDuet-1/DurM-2 vector (with His tag) linearized by AscI using Gibson assembly to generate pRSFDuet/His6-DurA/DurM.
Construction of pACYCDuet-1 derivative for coexpression of DurN and DurX.
The gene encoding DurX harboring flanking sequences homologous to pACYCDuet-1 multiple-cloning site 2 was first amplified using genomic DNA of S. cinnamoneus ATCC 12686 as the template and primers ACYC-DurX-mcsII-up and ACYC-DurX-mcsII-dn (Table 1). Subsequently the PCR fragment was cloned into the multiple-cloning site 2 of the pACYCDuet-1 vector (without a His tag) linearized by NdeI using Gibson assembly to generate pACYCDuet/DurX-2. The gene encoding DurN harboring flanking sequences homologous to pACYCDuet-1 multiple-cloning site 1 were then amplified using primers ACYC-DurN-mcsI-up and ACYC-DurN-mcsI-dn (Table 2). Then the corresponding fragment was cloned into MCS1 of the pACYCDuet/DurX-2 vector (without a His tag) linearized by NcoI using Gibson assembly to generate pACYCDuet/DurN/DurX.
Construction of pRSFDuet-1 derivatives for coexpression of DurM and His6-DurA-T11A, His6-DurA-F12A, His6-DurA-V13A, His6-DurA-C14A, His6-DurA-G16A, and His6-DurANTK-deletion.
Fragments of DurA variants harboring flanking sequences homologous to pRSFDuet-1 MCS1 were amplified using primers RSF-DurA-mcsI-up and corresponding RSF-DurA-variants-mcsI-dn (Table 2). Then the fragment was cloned into MCS1 of the pRSFDuet/DurM-2 vector (with a His tag) linearized by AscI using Gibson assembly to generate pRSFDuet/His6-DurA-variants/DurM.
Construction of pETDuet-1 derivatives for coexpression of DurM and DurAduramycinC.
A DNA fragment encoding DurA-F12W/V13S harboring flanking sequences homologous to pETDuet-1/DurM-2 multiple-cloning site 1 was first amplified using pRSFDuet/His-DurA/DurM as the template and primers PET-DurAF12W, V13S-mcsI-up and PET-DurAF12W, V13S-mcsI-dn (Table 2). Then the PCR fragment was cloned into multiple-cloning site 1 of the pETDuet/DurM-2 vector (with a His tag) linearized by AscI using Gibson assembly to generate pETDuet/His6-DurAF12W, V13S/DurM. Subsequently, the fragment DurAK2A, Q3N, F12W, V13S harboring flanking sequences homologous to pETDuet-1 multiple-cloning site 1 was first amplified using pETDuet/His6-DurAF12W, V13S/DurM as the template and primers PET-DurAK2A, Q3N, F12W, V13S-mcsI-up and PET-DurAK2A, Q3N, F12W, V13S-mcsI-dn (Table 2). Then the PCR fragment was cloned into multiple-cloning site 1 of the pETDuet/DurM-2 vector (with a His tag) linearized by AscI using Gibson assembly to generate pETDuet-1/His6-DurAK2A, Q3N, F12W, V13S/DurM. Finally, DurAduramycinC fragment harboring flanking sequences homologous to pETDuet/DurM-2 multiple-cloning site 1 was first amplified using pETDuet-1/His6-DurAK2A, Q3N, F12W, V13S/DurM as the template and primers PET-DurAF7Y, F10L, K2A, Q3N, F12W, V13S-mcsI-up and PET-DurAF7Y, F10L, K2A, Q3N, F12W, V13S-mcsI-dn (Table 2). Then the PCR fragment was cloned into multiple-cloning site 1 of the pETDuet/DurM-2 vector (with a His tag) linearized by AscI using Gibson assembly to generate pETDuet/His6-DurAduramycinC/DurM.
Construction of pRSFDuet-1 derivatives for coexpression of DurN and DurX.
The gene encoding DurX harboring flanking sequences homologous to pACYCDuet-1 multiple-cloning site 2 was first amplified using pACYCDuet/DurN/DurX as the template and primers RSF-DurX-mcsII-up and RSF-DurX-mcsII-dn (Table 2). Subsequently the PCR fragment was cloned into multiple-cloning site 2 of the pRSFDuet-1 vector (without a His tag) linearized by NdeI using Gibson assembly to generate pRSFDuet/DurX-2. The gene encoding DurN harboring flanking sequences homologous to pACYCDuet-1 multiple-cloning site 1 were then amplified using primers RSF-DurN-mcsI-up and RSF-DurN-mcsI-dn (Table 2). Then the corresponding fragment was cloned into MCS1 of the pRSFDuet/DurX-2 vector (without a His tag) linearized by NcoI using Gibson assembly to generate pRSFDuet/DurN/DurX.
Construction of pETDuet-1 derivatives for expression of DurX.
The gene encoding DurX harboring flanking sequences homologous to pETDuet-1 multiple-cloning site 1 was first amplified using genomic DNA of S. cinnamoneus ATCC 12686 as the template and primers His-DurX-mcsI-up and His-DurX-mcsI-dn (Table 2). Subsequently the PCR fragment was cloned into multiple-cloning site 1 of the pETDuet-1 vector (with a His tag) linearized by EcoRI using Gibson assembly to generate pETDuet/His6-DurX.
Heterologous production of modified His6-DurA and variants thereof in E. coli.
E. coli BL21(DE3) cells were transformed with pRSFDuet/His6-DurA (or DurA variants)/DurM and pACYCDuet/DurN/DurX and then plated on a Luria broth (LB) agar plate containing 50 mg/liter of kanamycin and 35 mg/liter of chloramphenicol. A single colony was picked and grown in 50 ml of LB with 50 mg/liter of kanamycin and 35 mg/liter of chloramphenicol at 37°C for 12 h, and the resulting culture was used to inoculate 8 liters of LB. Cells were cultured with shaking at 37°C until the optical density at 600 nm (OD600) reached 0.6 and cooled on ice for 30 min. Subsequently, isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 0.7 mM. The cells were cultured with shaking at 18°C for another 18 h before harvesting.
Heterologous expression of His6-DurX in E. coli BL21(DE3).
E. coli BL21(DE3) cells were transformed with pETDuet/His6-DurX and plated on an LB agar plate containing 100 mg/liter of ampicillin. A single colony was picked and grown in LB with 100 mg/liter of ampicillin at 37°C for 12 h, and the resulting culture was used to inoculate 2 liters of the same medium. Cells were cultured with shaking at 37°C until the OD600 reached 0.5 and cooled on ice for 30 min. Subsequently, IPTG was added to a final concentration of 0.2 mM. The cells were cultured with shaking at 18°C for another 18 h before harvesting.
Purification of modified His6-DurA and variants thereof.
The cell pellets were resuspended at room temperature in LanA start buffer (20 mM NaH2PO4 [pH 7.5 at 25°C], 500 mM NaCl, 0.5 mM imidazole, 20% glycerol) and lysed using a high-pressure homogenizer (Avestin, Inc.). The sample was centrifuged at 23,700 × g for 30 min, and the supernatant was kept. The pellets were then resuspended in LanA buffer 1 (6 M guanidine hydrochloride, 20 mM NaH2PO4 [pH 7.5 at 25°C], 500 mM NaCl, 0.5 mM imidazole) and lysed again. The insoluble portion was removed by centrifugation at 23,700 × g for 30 min, and the soluble portion was kept. Both soluble portions were passed through 0.45-μm syringe filters (Fisherbrand), and the His6-tagged modified peptides were purified by immobilized metal affinity chromatography (IMAC) as previously described (46). The eluted fractions were desalted by preparative RP-HPLC using a Waters Delta-pak C4 column (15 μm; 300 Å; 25 mm by 100 mm). The desalted peptides were lyophilized and stored at −20°C.
Purification of His6-DurX.
All steps were performed at 4°C in a cold room or on ice. The cell pellets were resuspended in buffer A (20 mM HEPES, 500 mM NaCl, 20% glycerol [pH 7.5 at 25°C]) and lysed using a high-pressure homogenizer (Avestin, Inc.). The sample was centrifuged at 23,700 × g for 30 min. The supernatant was passed through 0.45-μm syringe filters (Fisherbrand) and loaded onto a 5-ml HisTrap IMAC column precharged with Ni2+ and equilibrated with buffer A. The column was attached to an ÄKTA fast protein liquid chromatography (FPLC) system (GE Healthcare) and further washed with 25% buffer B (20 mM HEPES, 500 mM NaCl, 500 mM imidazole, 20% glycerol [pH 7.5 at 25°C]) in buffer A at a flow rate of 1.5 ml/min. Then the protein was eluted using a gradient of 25 to 100% buffer B. UV absorbance at 280 nm was monitored, and fractions were collected and analyzed by SDS-PAGE (Bio-Rad). The fractions containing the desired proteins were combined and the buffer exchanged to buffer A using a PD-10 desalting column (GE Healthcare). The protein was subsequently concentrated using an Amicon Ultra-15 centrifugal filter unit (Millipore). Protein aliquots were frozen in liquid nitrogen and stored at −80°C.
His6-DurX in vitro assay.
The His6-DurX in vitro assay was performed using the reported protocol for the CinX in vitro assay (25).
Proteolytic cleavage and purification of the DurA core peptide.
The modified DurA or DurAduramycinC precursor peptides were dissolved in 50 mM HEPES buffer to a concentration of 2 mg/ml at pH 7.5. To 5 ml of peptide solution 100 μl of 1 mg/ml Glu-C endoprotease was added, and the assay mixture was incubated for 12 h and then quenched with 0.1% TFA. The cleavage of the leader peptide was monitored by LC-ESI-QTOF MS. Purification of the digested core peptide was carried out using RP-HPLC as described in “General methods” above. The AFA-DurA core peptides thus obtained were dissolved in 50 mM HEPES to a concentration of 1 mg/ml at pH 7.5. To 5 ml of peptide solution 100 μl of 1-mg/ml aminopeptidase was added, and the assay mixture was incubated for 12 h and then quenched with 0.1% TFA. The removal of the AFA residues was monitored by LC-ESI-QTOF MS.
IAA assay to detect uncyclized Cys of duramycin and duramycin C.
An aliquot of duramycin/duramycin C was diluted to a final concentration of 1 mM in buffer containing 250 mM Tris-HCl, 25 mM iodoacetamide (IAA), and 13 mM tris-(2-carboxyethyl)phosphine, pH 9.0. The reaction mixture was incubated at room temperature for 60 min and analyzed by LC-ESI-QTOF MS.
LC-ESI-QTOF MS and MS/MS analyses.
A 5-μl volume of 100-μg/ml peptide was injected on a Waters Acquity UPLC system equipped with a BEH C8 column (1.7 μm, 100 mm by 1.0 mm; Waters). The column was preequilibrated in aqueous solvent. The solvents used for LC were as follows: solvent A, 0.1% formic acid in water, and solvent B, 0.1% formic acid in methanol. A solvent gradient of 3 to 97% B over 15 min was used, and the fractionated sample was directly subjected to ESI-QTOF MS analysis using a Waters Synapt mass spectrometer. The mass spectrometer was calibrated before any sample was injected. Data were acquired by ESI in positive mode with the capillary voltage set to 3.0 to 3.5 kV. The ionization source and desolvation gas were heated to 120°C and 300°C, respectively. Cone gas was set to 0 liters/h, and desolvation gas was set to 600 liters/h. The transfer collision energy was set to 4 V for both MS and MS/MS analyses. The trap collision energy was set to 6 V for MS analysis. For MS/MS analysis, a trap collision energy ramp ranging from 20 to 40 V was applied on multiply charged parent ions to achieve fragmentation. Suitable trap collision energy was determined by choosing the spectra where both fragment peaks and parent peak could be observed. [Glu1]-fibrinopeptide B (Sigma) was directly infused as lock mass with lock spray sampling if desired. The acquired spectra were processed using MaxEnt3 software and analyzed by Protein/Peptide Editor in BioLynx 4.1 (Waters). The MS/MS data of duramycin C were uploaded to the Global Natural Product Social Molecular Network (GNPS; MSV000080185).
Agar diffusion growth inhibition assay of duramycin and duramycin C.
Duramycin and duramycin C were obtained as described above by Glu-C cleavage of the posttranslationally modified DurA followed by aminopeptidase cleavage and HPLC purification. Peptides were dissolved in H2O to achieve a concentration of 500 μM. The peptide solutions were diluted to prepare a 10 μM solution. Agar plates were prepared by combining 20 ml of melted agar (cooled to 42°C for 5 min) with 200 μl of an overnight cell culture (OD600 = 2.3) of Bacillus subtilis ATCC 6633. The seeded agar was poured into a sterile 100-mm round petri dish (VWR) and allowed to solidify at 25°C for 30 min. A sample of 10 μl of 10 μM duramycin and duramycin C were directly spotted on the solidified agar. Plates were incubated at 30°C for 16 h, and the antimicrobial activity was determined by the presence or absence of zones of growth inhibition. A negative control was conducted using sterile H2O. A positive control was conducted using authentic duramycin purchased from Sigma-Aldrich.
ITC experiments.
Isothermal titration calorimetry (ITC) measurements for duramycin and duramycin C were performed using micelles composed of octyl-β-d-glucopyranoside (OG) as previously described (42). l-α-Phosphatidylethanolamine (1,2-diacyl-sn-glycero-3-phosphoethanolamine) from egg yolk (Sigma-Aldrich; 0.25 mg) and duramycin (Sigma-Aldrich; 25 or 50 μM) were dissolved in 36 mM OG solution, 10 mM Tris-HCl, and 0.1 M NaCl, pH 7.4. Then, the duramycin/OG solution (350 μl) was titrated with 1-μl aliquots of the PE/OG micelles (50 μl in total). ITC was performed with a Nano ITC titration calorimeter (TA Instruments). Solutions were degassed under vacuum before use. The calorimeter cell had a reaction volume of 350 μl. The titration pattern was analyzed in terms of a 1:1 peptide-lipid complex.
Accession number(s).
This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under accession number MOEP00000000.
Supplementary Material
ACKNOWLEDGMENTS
We thank Shan Huang (Fudan University, China) for help with cloning of the DurA variants during his stay at UIUC as an exchange student.
This work was supported by the National Institutes of Health (R37 GM058822).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02698-16.
REFERENCES
- 1.Chatterjee C, Paul M, Xie L, van der Donk WA. 2005. Biosynthesis and mode of action of lantibiotics. Chem Rev 105:633–684. doi: 10.1021/cr030105v. [DOI] [PubMed] [Google Scholar]
- 2.Willey JM, van der Donk WA. 2007. Lantibiotics: peptides of diverse structure and function. Annu Rev Microbiol 61:477–501. doi: 10.1146/annurev.micro.61.080706.093501. [DOI] [PubMed] [Google Scholar]
- 3.Bierbaum G, Sahl HG. 2009. Lantibiotics: mode of action, biosynthesis and bioengineering. Curr Pharm Biotechnol 10:2–18. doi: 10.2174/138920109787048616. [DOI] [PubMed] [Google Scholar]
- 4.Cotter PD, Hill C, Ross RP. 2005. Bacterial lantibiotics: strategies to improve therapeutic potential. Curr Protein Pept Sci 6:61–75. doi: 10.2174/1389203053027584. [DOI] [PubMed] [Google Scholar]
- 5.Kondo S, Sezaki M, Shimura M, Sato K, Hara T. 1964. Leucopeptin, a new peptide antibiotic. J Antibiot 17:262–263. [PubMed] [Google Scholar]
- 6.Widdick DA, Dodd HM, Barraille P, White J, Stein TH, Chater KF, Gasson MJ, Bibb MJ. 2003. Cloning and engineering of the cinnamycin biosynthetic gene cluster from Streptomyces cinnamoneus cinnamoneus DSM 40005. Proc Natl Acad Sci U S A 100:4316–4321. doi: 10.1073/pnas.0230516100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wakamiya T, Fukase K, Naruse N, Konishi M, Shiba T. 1988. Lanthiopeptin, a new peptide effective against herpes simplex virus: structural determination and comparison with Ro 09-0198, an immunopotentiating peptide. Tetrahedron Lett 29:4771–4772. doi: 10.1016/S0040-4039(00)80604-8. [DOI] [Google Scholar]
- 8.Fredenhagen A, Fendrich G, Marki F, Marki W, Gruner J, Raschdorf F, Peter HH. 1990. Duramycins B and C, two new lanthionine containing antibiotics as inhibitors of phospholipase A2. Structural revision of duramycin and cinnamycin. J Antibiot 43:1403–1412. [DOI] [PubMed] [Google Scholar]
- 9.Hayashi F, Nagashima K, Terui Y, Kawamura Y, Matsumoto K, Itazaki H. 1990. The structure of PA48009: the revised structure of duramycin. J Antibiot 43:1421–1430. doi: 10.7164/antibiotics.43.1421. [DOI] [PubMed] [Google Scholar]
- 10.Kessler H, Steuernagel S, Will M, Jung G, Kellner R, Gillessen D, Kamiyama T. 1988. The structure of the polycyclic nonadecapeptide Ro 09-0198. Helv Chim Acta 71:1924–1929. doi: 10.1002/hlca.19880710811. [DOI] [Google Scholar]
- 11.Kodani S, Komaki H, Ishimura S, Hemmi H, Ohnishi-Kameyama M. 2016. Isolation and structure determination of a new lantibiotic cinnamycin B from Actinomadura atramentaria based on genome mining. J Ind Microbiol Biotechnol 43:1159–1165. doi: 10.1007/s10295-016-1788-9. [DOI] [PubMed] [Google Scholar]
- 12.Lindenfelser LA, Pridham TG, Shotwell OL, Stodola FH. 1957. Antibiotics against plant disease. IV. Activity of duramycin against selected microorganisms. Antibiot Annu 5:241–247. [PubMed] [Google Scholar]
- 13.Märki F, Hanni E, Fredenhagen A, van Oostrum J. 1991. Mode of action of the lanthionine-containing peptide antibiotics duramycin, duramycin B and C, and cinnamycin as indirect inhibitors of phospholipase A2. Biochem Pharmacol 42:2027–2035. doi: 10.1016/0006-2952(91)90604-4. [DOI] [PubMed] [Google Scholar]
- 14.Zimmermann N, Freund S, Fredenhagen A, Jung G. 1993. Solution structures of the lantibiotics duramycin B and C Eur J Biochem 216:419–428. [DOI] [PubMed] [Google Scholar]
- 15.Seelig J. 2004. Thermodynamics of lipid-peptide interactions. Biochim Biophys Acta 1666:40–50. doi: 10.1016/j.bbamem.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 16.Hosoda K, Ohya M, Kohno T, Maeda T, Endo S, Wakamatsu K. 1996. Structure determination of an immunopotentiator peptide, cinnamycin, complexed with lysophosphatidylethanolamine by 1H-NMR. J Biochem 119:226–230. doi: 10.1093/oxfordjournals.jbchem.a021226. [DOI] [PubMed] [Google Scholar]
- 17.Oliynyk I, Varelogianni G, Roomans GM, Johannesson M. 2010. Effect of duramycin on chloride transport and intracellular calcium concentration in cystic fibrosis and non-cystic fibrosis epithelia. APMIS 118:982–990. doi: 10.1111/j.1600-0463.2010.02680.x. [DOI] [PubMed] [Google Scholar]
- 18.Grasemann H, Stehling F, Brunar H, Widmann R, Laliberte TW, Molina L, Doring G, Ratjen F. 2007. Inhalation of Moli1901 in patients with cystic fibrosis. Chest 131:1461–1466. doi: 10.1378/chest.06-2085. [DOI] [PubMed] [Google Scholar]
- 19.Jones AM, Helm JM. 2009. Emerging treatments in cystic fibrosis. Drugs 69:1903–1910. doi: 10.2165/11318500-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 20.Richard AS, Zhang A, Park SJ, Farzan M, Zong M, Choe H. 2015. Virion-associated phosphatidylethanolamine promotes TIM1-mediated infection by Ebola, dengue, and West Nile viruses. Proc Natl Acad Sci U S A 112:14682–14687. doi: 10.1073/pnas.1508095112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rouser G, Yamamoto A, Kritchevsky G. 1971. Cellular membranes. Structure and regulation of lipid class composition species differences, changes with age, and variations in some pathological states. Arch Intern Med 127:1105–1121. [DOI] [PubMed] [Google Scholar]
- 22.Spector AA, Yorek MA. 1985. Membrane lipid composition and cellular function. J Lipid Res 26:1015–1035. [PubMed] [Google Scholar]
- 23.Zhao M. 2011. Lantibiotics as probes for phosphatidylethanolamine. Amino Acids 41:1071–1079. doi: 10.1007/s00726-009-0386-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao M, Li Z, Bugenhagen S. 2008. 99mTc-labeled duramycin as a novel phosphatidylethanolamine-binding molecular probe. J Nucl Med 49:1345–1352. doi: 10.2967/jnumed.107.048603. [DOI] [PubMed] [Google Scholar]
- 25.Ökesli A, Cooper LE, Fogle EJ, van der Donk WA. 2011. Nine post-translational modifications during the biosynthesis of cinnamycin. J Am Chem Soc 133:13753–13760. doi: 10.1021/ja205783f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Håvarstein LS, Diep DB, Nes IF. 1995. A family of bacteriocin ABC transporters carry out proteolytic processing of their substrates concomitant with export. Mol Microbiol 16:229–240. doi: 10.1111/j.1365-2958.1995.tb02295.x. [DOI] [PubMed] [Google Scholar]
- 27.Carlos JL, Paetzel M, Brubaker G, Karla A, Ashwell CM, Lively MO, Cao G, Bullinger P, Dalbey RE. 2000. The role of the membrane-spanning domain of type I signal peptidases in substrate cleavage site selection. J Biol Chem 275:38813–38822. doi: 10.1074/jbc.M007093200. [DOI] [PubMed] [Google Scholar]
- 28.Caetano T, Krawczyk JM, Mosker E, Süssmuth RD, Mendo S. 2011. Heterologous expression, biosynthesis, and mutagenesis of type II lantibiotics from Bacillus licheniformis in Escherichia coli. Chem Biol 18:90–100. doi: 10.1016/j.chembiol.2010.11.010. [DOI] [PubMed] [Google Scholar]
- 29.Nagao J, Harada Y, Shioya K, Aso Y, Zendo T, Nakayama J, Sonomoto K. 2005. Lanthionine introduction into nukacin ISK-1 prepeptide by co-expression with modification enzyme NukM in Escherichia coli. Biochem Biophys Res Commun 336:507–513. doi: 10.1016/j.bbrc.2005.08.125. [DOI] [PubMed] [Google Scholar]
- 30.Shi Y, Yang X, Garg N, van der Donk WA. 2011. Production of lantipeptides in Escherichia coli. J Am Chem Soc 133:2338–2341. doi: 10.1021/ja109044r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garg N, Tang W, Goto Y, Nair SK, van der Donk WA. 2012. Lantibiotics from Geobacillus thermodenitrificans. Proc Natl Acad Sci U S A 109:5241–5246. doi: 10.1073/pnas.1116815109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang W, van der Donk WA. 2013. The sequence of the enterococcal cytolysin imparts unusual lanthionine stereochemistry. Nat Chem Biol 9:157–159. doi: 10.1038/nchembio.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin Y, Teng K, Huan L, Zhong J. 2011. Dissection of the bridging pattern of bovicin HJ50, a lantibiotic containing a characteristic disulfide bridge. Microbiol Res 166:146–154. doi: 10.1016/j.micres.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 34.Wang J, Ma H, Ge X, Zhang J, Teng K, Sun Z, Zhong J. 2014. Bovicin HJ50-like lantibiotics, a novel subgroup of lantibiotics featured by an indispensable disulfide bridge. PLoS One 9:e97121. doi: 10.1371/journal.pone.0097121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gomi T, Fujioka M. 1982. Inactivation of rat-liver S-adenosylhomocysteinase by iodoacetamide. Biochemistry 21:4171–4176. doi: 10.1021/bi00260a039. [DOI] [PubMed] [Google Scholar]
- 36.Velásquez JE, Zhang X, van der Donk WA. 2011. Biosynthesis of the antimicrobial peptide epilancin 15X and its unusual N-terminal lactate moiety. Chem Biol 18:857–867. doi: 10.1016/j.chembiol.2011.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao X, van der Donk WA. 2016. Structural characterization and bioactivity analysis of the two-component lantibiotic Flv system from a ruminant bacterium. Cell Chem Biol 23:246–256. doi: 10.1016/j.chembiol.2015.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bindman NA, van der Donk WA. 2013. A general method for fluorescent labeling of the N-termini of lanthipeptides and its application to visualize their cellular localization. J Am Chem Soc 135:10362–10371. doi: 10.1021/ja4010706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Majchrzykiewicz JA, Lubelski J, Moll GN, Kuipers A, Bijlsma JJ, Kuipers OP, Rink R. 2010. Production of a class II two-component lantibiotic of Streptococcus pneumoniae using the class I nisin synthetic machinery and leader sequence. Antimicrob Agents Chemother 54:1498–1505. doi: 10.1128/AAC.00883-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi Y, Bueno A, van der Donk WA. 2012. Heterologous production of the lantibiotic Ala(0)actagardine in Escherichia coli. Chem Commun (Cambridge) 48:10966–10968. doi: 10.1039/c2cc36336d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rink R, Arkema-Meter A, Baudoin I, Post E, Kuipers A, Nelemans SA, Akanbi MH, Moll GN. 2010. To protect peptide pharmaceuticals against peptidases. J Pharmacol Toxicol Methods 61:210–218. doi: 10.1016/j.vascn.2010.02.010. [DOI] [PubMed] [Google Scholar]
- 42.Machaidze G, Seelig J. 2003. Specific binding of cinnamycin (Ro 09-0198) to phosphatidylethanolamine. Comparison between micellar and membrane environments. Biochemistry 42:12570–12576. [DOI] [PubMed] [Google Scholar]
- 43.Iwamoto K, Hayakawa T, Murate M, Makino A, Ito K, Fujisawa T, Kobayashi T. 2007. Curvature-dependent recognition of ethanolamine phospholipids by duramycin and cinnamycin. Biophys J 93:1608–1619. doi: 10.1529/biophysj.106.101584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stenflo J, Holme E, Lindstedt S, Chandramouli N, Huang LH, Tam JP, Merrifield RB. 1989. Hydroxylation of aspartic acid in domains homologous to the epidermal growth factor precursor is catalyzed by a 2-oxoglutarate-dependent dioxygenase. Proc Natl Acad Sci U S A 86:444–447. doi: 10.1073/pnas.86.2.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA III, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 46.Li B, Cooper LE, van der Donk WA. 2009. In vitro studies of lantibiotic biosynthesis. Methods Enzymol 458:533–558. doi: 10.1016/S0076-6879(09)04821-6. [DOI] [PubMed] [Google Scholar]
- 47.O'Rourke S, Widdick D, Bibb M. 2016. A novel mechanism of immunity controls the onset of cinnamycin biosynthesis in Streptomyces cinnamoneus DSM 40646. J Ind Microbiol Biotechnol doi: 10.1007/s10295-016-1869-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.