Abstract
Two activation-dependent Abs to the integrin αL-subunit were used to study conformational rearrangement of αLβ2 on the cell surface. Activation lowered the concentration of Ca2+ required for maximal expression of each epitope. Each Ab requires the Ca2+-binding loop in the integrin genu and nearby species-specific residues in the thigh domain. Key thigh residues are shielded from Ab in the bent integrin conformation by the α-subunit calf-1 domain and the nearby bent β leg, suggesting that extension at the genu is required for epitope exposure. Activating stimuli and α/β I-like small molecule antagonists demonstrate that exposure of epitopes in the integrin α- and β-subunit legs is coordinate during integrin activation. A coordinating residue donated by the calf-1 domain is as important as Ca2+ for mAb binding. Together with inspection of the αV structure, this result suggests that the genu/calf-1 interface is maintained in integrin activation, and that extension occurs by a rearrangement at the thigh/genu interface.
Integrins are α/β heterodimeric adhesion molecules that transmit signals bidirectionally across the membrane to regulate ligand binding in the extracellular domains and signaling in the cytoplasm (1, 2). Recent crystal, NMR, and negative stain electron microscopic (EM) structures reveal a unifying mechanism of integrin activation (3-7). In the low affinity conformation, the headpiece of the integrin folds over its legs at the “genu” and faces down toward the membrane and extends upward in a switchblade-like opening upon activation (8).
Crystal and EM structures of integrin fragments reveal snap-shots of different conformational states; however, other methodologies are required to probe conformational movements on cell surfaces. A subset of mAbs to integrins recognize epitopes that become exposed on integrin extracellular domains upon activation by signals from inside the cell, as well as upon ligand binding. Epitope mapping has been instrumental in elucidating conformational rearrangement and movement between domains in the β-subunit (5, 9-11), and these rearrangements have been confirmed with structural studies (6, 7, 12, 13).
Much less is known about conformational rearrangement in the integrin α-subunit. The crystal structure of bent αVβ3 revealed a sharp bend between the αV thigh and calf-1 domains at the genu, a short, disulfide-bonded loop with a Ca2+-binding site (3). EM studies on αVβ3 suggest that in contrast to the flexible β-subunit, the α-subunit undergoes only a single change in the extended conformation (6). This change occurs at the genu between the α-subunit thigh and calf-1 domains, which docks into a distinctive, highly extended, rigid conformation. Abs mapping to the genu or nearby regions of the α-subunit could therefore be valuable probes of conformational change in the α leg for integrins on the cell surface.
NKI-L16 is a well characterized, Ca2+-dependent mAb that activates LFA-1 as well as reads out activation (14, 15). We have previously mapped the binding site of NKI-L16 to the C-terminal half of the αL extracellular region (16). AO3 is a mAb to the αL-subunit that induces activation of LFA-1 (P.S., J. M. Luk, and L.B.K., unpublished data). Here we show that NKI-L16 and AO3 bind to thigh domain residues, which are masked in the bent conformation, and recognize a Ca2+-bound, extended conformation at the genu. The results also suggest that rearrangements during α leg extension occur between the thigh and genu rather than between the genu and calf-1 domain.
Materials and Methods
Cell Lines and mAbs. HEK293T cells were grown in DMEM (Life Technologies) supplemented with 10% FBS, nonessential amino acids, 2 mM glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin. K562 transfectants expressing wild-type (WT) LFA-1 were described (17) and cultured in RPMI medium 1640/10% FBS supplemented with 4 μg/ml puromycin.
The murine mAbs TS1/18 (17), KIM127 (18), m24 (19) to human β2 (CD18), NKI-L16 (14), MHM24 (20) to human αL (CD11a), rat mAb M17/5 (21) to mouse αL, and the myeloma IgG1 X63 (American Type Culture Collection) were described. AO3 is a mouse anti-human αL mAb that was selected for its ability to activate LFA-1 (P.S., J. M. Luk, and L.B.K., unpublished data).
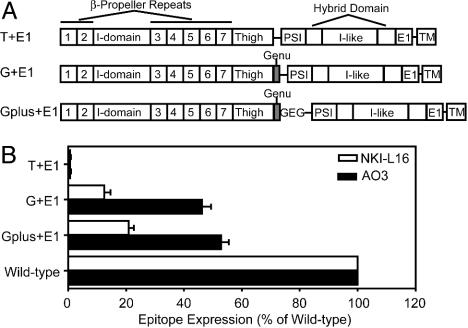
Plasmid Construction and Transient Transfection into 293T Cells. Single chain integrin LFA-1 constructs contained an N-terminal portion of the αL-subunit ending in residue N745 (“T+E1”) or residue E753 (“G+E1” and “Gplus+E1”) and a C-terminal portion of the β2-subunit containing residues 1-465 (Fig. 2 A). A linker (PRGGLENLYFQGG) containing a SacII site (bold) and a tobacco etch virus protease site (underlined) was inserted between α and β segments. Gplus+E1 construct contains an additional “GEG” sequence after E753 to mimic the calf-1 Ca2+-binding site. The αL and β2 segments were made by PCR. The linker was joined N-terminally to the β2 segment during PCR. Restriction enzyme EcoRV and SalI sites were incorporated 5′ of αL and 3′ of β2 fragments, respectively, by using the PCR primers. The SacII site shown above in the linker was incorporated in the 3′ αL and 5′ linker-β2 PCR primers. After digestion with EcoRV and SacII (αL segments), or SacII and SalI (β2 segment), a triple ligation was used to incorporate the fragments into the EcoRV and SalI sites of the pDisplay vector (Invitrogen) N-terminal to the transmembrane anchoring domain of platelet-derived growth factor receptor, which allows the target protein to be expressed and displayed on the cell surface. All PCR generated fragments were confirmed by DNA sequencing.
Fig. 2.
NKI-L16 and AO3 binding depends on the genu. (A) Schematic diagram of the single chain LFA-1 constructs. (B) Binding to single chain LFA-1 fragments measured by immunofluorescence flow cytometry. Expression was measured as specific mean immunofluorescence intensity and expressed as the percentage of specific mean immunofluorescence intensity of staining with MHM24 mAb to the αL I domain. Results are the mean ± SD of three independent experiments.
WT and chimeric human and mouse αL-subunit cDNAs were in the expression vector pCDNA3.1/hygro(-). Chimeric human and mouse αL-subunits were generated by overlap extension PCR (22, 23) by using a similar approach as described (9, 16). Chimeras were named according to the species origin of their segments. For example, h589m indicates that residues 1-589 are from the human αL-subunit and residues 590 to the C terminus are from the mouse αL-subunit. Amino acid sequence numbering was according to the mature human sequence (24). Human to mouse amino acid substitution and other mutants in this paper were generated by site-specific mutagenesis by using QuikChange (Stratagene). All mutations were confirmed by DNA sequencing.
The 293T cells were transiently transfected with WT or mutant integrin cDNAs by using calcium phosphate precipitation. In brief, 5 μg of single chain LFA-1 DNA or 2.5 μg of WT or mutant αL cDNA and 2.5 μg of β2 cDNA were used to cotransfect one 6-cm plate of 60-70% confluent cells. Cells were replated 1 d after transfection and harvested 2 d after transfection. Cells were detached from the plate with Hanks' balanced salt solution containing 10 mM EDTA and washed twice before flow cytometry.
Immunofluoresence Flow Cytometry. Cells were harvested and washed twice with Hepes-buffered saline (HBS) buffer (20 mM Hepes/150 mM NaCl, pH 7.4) and then incubated with primary Abs in 50 μl of HBS on ice for 30 min, or at 37°C for 30 min for KIM127 and m24 mAbs (18). The mAbs were used as purified IgG at 10 μg/ml or 1:200 dilutions of ascites. The control IgG X63 was used as 1:10 dilution of hybridoma supernatant. For Ca2+ dependence experiments, K562 transfectants expressing WT LFA-1 were harvested and pretreated with EDTA, followed by three washes with cation-depleted HBS. The cation-depleted HBS buffer was obtained by pretreating HBS buffer with Chelex 100 resin (Bio-Rad) following the manufacturer's instructions, and the Ca2+ concentration was determined by Oregon Green 488 Kit (Molecular Probes). After washing, cells were incubated with NKI-L16, AO3, or control mAb MHM24 in 50 μl of HBS in the presence of 1 mM Mg2+ and varying concentrations of Ca2+, with or without 1 μM XVA143, on ice for 30 min. For mAb binding under activating conditions, washed K562 cells expressing WT LFA-1 were incubated with primary mAbs in HBS containing appropriate concentrations of divalent cations and different stimuli for 30 min at 37°C. Cells were washed twice with HBS and incubated with FITC-conjugated goat anti-mouse IgG (Zymed) for 30 min on ice. After washing, cells were resuspended in cold HBS and analyzed on a FACScan (BD Biosciences).
Results
Dependence on Ca2+ and Activation of the NKI-L16 and AO3 Epitopes. The NKI-L16 epitope has been reported to be Ca2+- and activation-dependent (15, 25). However, the Ca2+ concentration optimal for probing activation with NKI-L16 has not been reported. Furthermore, the inter-relationships between the activation dependence and Ca2+ dependence of the NKI-L16 have not been investigated.
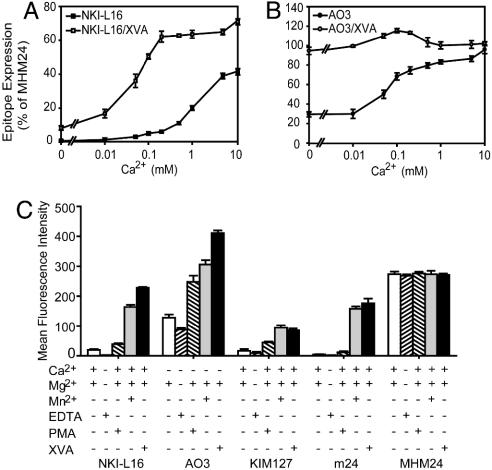
Under resting conditions, NKI-L16 did not bind to LFA-1 in Ca2+-depleted buffer (≤0.15 μM Ca2+) (Fig. 1A). Furthermore, 2 mM EDTA eliminated binding of NKI-L16 (Fig. 1C). Binding of NKI-L16 to resting LFA-1 was enhanced by increasing Ca2+ concentration in the range from 0.01 to 10 mM (Fig. 1 A). By contrast, AO3 mAb binds to resting LFA-1 in Ca2+-depleted buffer (Fig. 1B) and in the presence of 2 mM EDTA (Fig. 1C). However, higher Ca2+ concentrations enhanced AO3 binding (Fig. 1B).
Fig. 1.
Calcium and activation dependence of the NKI-L16 and AO3 epitopes. (A and B)Ca2+ dependence in the resting and extended conformations. K562 cells expressing WT αLβ2 were pretreated with EDTA and then washed twice with cation-depleted HBS. Binding of NKI-L16 and AO3 was measured in HBS containing 1 mM Mg2+ and varying concentrations of Ca2+, with or without 1 μM XVA143 to induce extension. (C) Preferential binding to activation epitopes. K562 transfectants pretreated as in A and B were incubated with primary mAbs in HBS in the presence of 0.2 mM Ca2+,1 mM Mg2+, 2 mM Mn2+, 1 μM PMA, and 1 μM XVA143 for 30 min at 37°C. Cells were washed and incubated with FITC-labeled second Ab for 15 min at 4°C. Epitope expression was measured as specific mean fluorescence intensity in C (after subtraction of control X63 intensity) or further quantitated as the percentage of fluorescence intensity with MHM24 mAb to the αL I domain in A and B. Results are the mean ± SD of three independent experiments.
We further examined the Ca2+ dependence of NKI-L16 and AO3 mAb binding in the presence of the α/β I-like allosteric antagonist XVA143, which is known to bind to the β2 I-like midas and to induce extension of αLβ2 as reported by mAbs to β2-subunit activation epitopes (26-28). XVA143 increased binding of NKI-L16 to LFA-1 at all Ca2+ concentrations (Fig. 1 A). Furthermore, low but significant binding of NKI-L16 to extended LFA-1 was observed even when Ca2+ was depleted. Half-maximal binding of NKI-L16 to extended LFA-1 was obtained at a Ca2+ concentration of 0.05 mM, compared to 1 mM in resting conformation (Fig. 1 A). XVA143 also enhanced binding of AO3 mAb to LFA-1 (Fig. 1B). As a control, binding of the extension-dependent mAb KIM127 was not affected by Ca2+ concentration in the presence and absence of XVA143 (data not shown).
Based on the above results, 0.2 mM Ca2+ was chosen as optimal to probe LFA-1 activation/extension with NKI-L16 mAb (Fig. 1 A), and 0 mM Ca2+ was chosen to probe activation/extension with AO3 mAb (Fig. 1B). At both of these Ca2+ concentrations and in the absence of other stimuli, LFA-1 is in the resting conformation, as confirmed by a lack of exposure of the KIM127 epitope in the β2 integrin-epidermal growth factor-like (I-EGF) 2 (9) and the m24 epitope in the β2 I-like domain (29) (Fig. 1C and data not shown).
We then tested the effect of the stimuli Mn2+ and phorbol 12-myristate 13-acetate (PMA), and for comparison XVA143, on exposure of the NKI-L16 and AO3 epitopes (Fig. 1C). Mn2+ markedly enhanced the NKI-L16 and AO3 epitopes in the αL-subunit, as it did the β-subunit KIM127 and m24 epitopes (Fig. 1C). PMA also reproducibly enhanced expression of the NKI-L16 and AO3 epitopes, with the latter showing the largest increase (Fig. 1C). PMA significantly increased the KIM127 epitope on I-EGF2 but had a much smaller effect on the m24 epitope on the I-like domain (Fig. 1C), as reported (9). By contrast, none of the activating conditions affect expression of constitutive epitopes as exemplified with mAb MHM24 to the αL I domain (Fig. 1C).
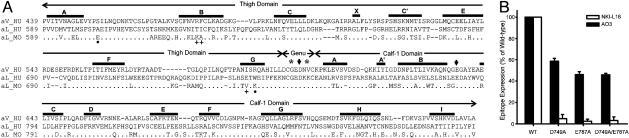
Requirement of the Genu of the αL-Subunit. The importance of the genu was demonstrated with single-chain integrins in which αL and β2 fragments were fused together with a linker and expressed on the cell surface with an artificial transmembrane domain (Fig. 2A). The NKI-L16 and AO3 mAbs react with fusion proteins in which the αL-subunit is truncated after residue 752 (G+E1 and Gplus+E1), but not after residue 745 (T+E1) (Fig. 2B). The sequence between residues 746 and 752 required for reactivity, CGEDKKC, corresponds to the disulfide-bonded loop of the genu. Reactivity was lesser than with WT, heterodimeric αLβ2, but this may result from lack of a Glu residue in the calf-1 domain that coordinates the genu Ca2+ (3), as suggested by greater reactivity with the Gplus+E1 construct that contains an additional Gly-Glu-Gly sequence following the genu (Fig. 2B) and the effect of mutating the corresponding calf-1 Glu in αL (see below).
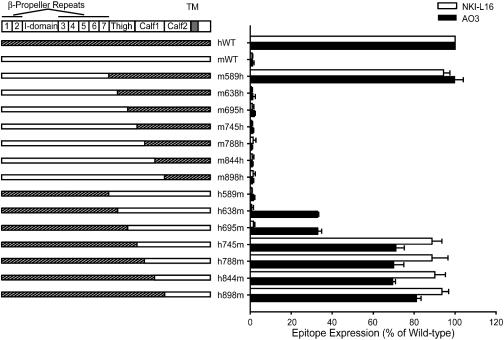
Epitope Mapping. Previous mapping to αL residues 443-1,063 (16) was refined with a series of human and mouse chimeric αL-subunit cDNAs (Fig. 3) that were used with human WT β2-subunit cDNA to cotransfect 293T cells. Immunostaining with anti-human-β2 mAb TS1/18, anti-human-αL I domain mAb MHM24 and anti-mouse-αL I domain mAb M17/5 showed that all chimeric αL/β2 complexes were expressed at comparable levels to the WT human and mouse αL complexes with human β2 (data not shown). The human αL segment 589-638 was absolutely required for binding by both NKI-L16 and AO3 mAbs, as shown by the presence of binding to chimera m589h and loss of binding to all chimeras in which the mouse-human junction was more C-terminal, such as m638h, m695h, and m745h (Fig. 3). Binding by mAb NKI-L16 also absolutely required the αL segment 695-745, as shown by binding to chimera h745 m and not to chimeras in which the human-mouse junction was more N-terminal, such as h695m, h638m, and h589m. Full reactivity by AO3 mAb also required residues in the 695-745 region, as shown by full reactivity with chimera h745m and partial reactivity with chimeras h695m and h638m.
Fig. 3.
Mapping of NKI-L16 and AO3 epitopes with αL chimeras. mAb reactivity was determined with chimeric human-mouse αL-subunits expressed with human β2 in 293T cells in the presence of 1 mM Ca2+ and 1 mM Mg2+ by using immunofluorescence flow cytometry. NKI-L16- and AO3-specific mean fluorescence intensity was determined as a ratio of that of TS1/18 mAb for each transfectant and expressed as a percentage of the ratio with WT αLβ2. Results are the mean ± SD of three independent experiments.
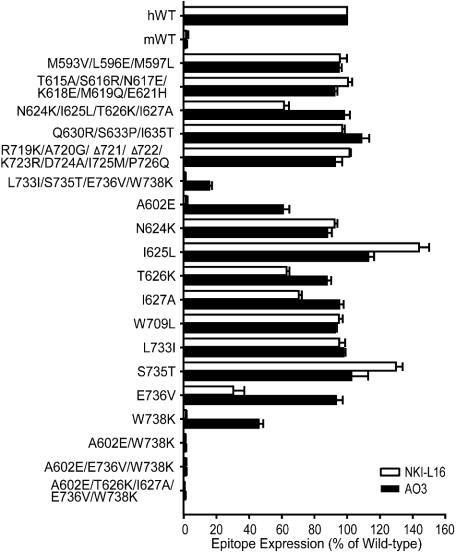
The regions 589-638 and 695-745 were subdivided into eight subregions with one or more substitutions each. Subsequently, single amino acid substitutions were made within any region that was found to affect mAb binding (Fig. 4). Five residues, A602, T626, I627, E736, and W738 affected NKI-L16 binding. Among these, A602 and W738 are each critical for epitope expression because substitution of either of these with the mouse residue abolished NKI-L16 binding (Fig. 4). The single substitution E736V also greatly decreased mAb binding, and the T626K and I627A substitutions had lesser but reproducible affects. Residues A602 and W738 were also part of the AO3 epitope; however, their substitution resulted in reduction rather than in elimination of binding (Fig. 4). The combined L733I/S735T/E736V/W738K substitution eliminated more AO3 binding than W738K alone; however, the individual substitutions L733I, S735T, and E736V did not show effects. Together with the complete elimination of AO3 binding by the h695m chimera and restoration by the h745m chimera, the results show that besides W738, other species-specific residues in the region between residues 695 and 745 contribute to the AO3 epitope. A602 and W738 are clearly the most important residues in the AO3 epitope, as shown by complete elimination of AO3 binding by the double A602E/W738K substitution (Fig. 4).
Fig. 4.
Fine mapping of mAb NKI-L16 and AO3 epitopes. WT αL or mutant αL containing multiple or single human-to-mouse amino acid substitutions was coexpressed with the human β2-subunit in 293T cells. Data are expressed as mean ± SD of three independent experiments, as in Fig. 3.
Ca2+-Coordinating Side Chains from both the Genu and Calf-1 Domain Are Critical for NKI-L16 and AO3 Binding. The coordinations to the genu Ca2+ in αV are formed by two backbone carbonyls in the genu, and Asp and Glu side chains in the genu and calf-1 domain, respectively. These latter residues are conserved as Asp-749 and Glu-787 in αL (Fig. 5A). Mutation of each of these residues abolished NKI-L16 binding and partially reduced AO3 binding (Fig. 5B). These results correspond almost exactly to the data on Ca2+ dependence for binding by the two mAbs (Fig. 1). The requirement for calf-1 residue Glu-787 suggests that the coordination by this residue is maintained in the extended integrin conformation.
Fig. 5.
Sequence alignment and effect of mutation of residues with side chains that coordinate the genu Ca2+ ion. (A) Sequence alignment. β-Strands are labeled. Residues with side chains and backbone carbonyls that coordinate the genu Ca2+ ion are marked above the sequence with arrows and asterisks, respectively. Residues in activation epitopes are marked below the sequence with filled circles for residues common to the NKI-L16 and AO3 epitopes and plus signs for additional residues in the NKI-L16 epitope. (B) Requirement for Ca2+-coordinating side chains for NKI-L16 and AO3 epitope expression. Mutants were tested for binding to NKI-L16 and AO3 mAbs as described in Fig. 3. Results are the mean ± SD of three independent experiments.
Discussion
To elucidate the basis for signal transduction between the integrin headpiece and cytoplasmic domains during integrin activation, we have mapped two activation epitopes to specific residues located close to the αL genu and examined the relationship between appearance in activation of these epitopes and epitopes in the β-subunit headpiece and leg. The data provide insights into integrin activation-based conformational change.
We separately investigated the dependence on activation and on Ca2+ of the NKI-L16 and AO3 epitopes. In all stimulation conditions (PMA, Mn2+, and XVA143) we tested, both NKI-L16 and AO3 mAbs showed enhanced binding to LFA-1, showing that their binding is activation dependent. The results agree with previous studies of NKI-L16 on lymphocytes, which reported that enhanced NKI-L16 binding was observed after PMA, IL-2 treatment, or after in vitro culture of resting lymphocyte (14, 15). Meanwhile, the Ca2+ concentration also plays an important role in epitope expression. As we show here, binding to resting LFA-1 by NKI-L16 is completely Ca2+ dependent. By contrast, Ca2+ increases binding of AO3 to resting LFA-1 but is not absolutely required. For both NKI-L16 and AO3, higher Ca2+ concentrations greatly enhanced activation-dependent epitope expression. We do not believe that Ca2+ favors the extended integrin conformation because structural studies suggest just the opposite, i.e., that Ca2+ favors the bent conformation (6). Ca2+ exerts this effect by binding to a site in the β I-like domain (30). Therefore, the Ca2+ dependence of the epitopes at the genu is likely to reflect a higher affinity of the Abs for the Ca2+-bound extended conformation compared to the Ca2+-free extended conformation.
Mutational studies show that the NKI-L16 and AO3 epitopes require species-specific residues on the face of the thigh domain that is innermost in the bent conformation and Ca2+-coordinating residues in the genu and calf-1 domain. The two most important species-specific residues in both epitopes, A602 and W738, are far apart in sequence. However, they are located close together in the thigh domain and near the genu (Fig. 6). The other, less important residues are nearby on the same face of the thigh domain in β-strands B and G (Figs. 5A and 6). There are no mouse-human substitutions in the genu; the genu may either be directly contacted by the NKI-L16 and AO3 mAbs and form part of the epitope or be required to maintain the structure of the portion of the thigh domain recognized by the mAbs. The role of Ca2+ in the NKI-L16 epitope has long been unclear. We have found that deletion of the disulfide-bonded genu loop abolishes both the NKI-L16 and AO3 epitopes. Furthermore, mutation of either of the two residues that contribute Ca2+-coordinating side chains, Asp-749 and Glu-787, almost completely eliminates NKI-L16 binding and partially eliminates AO3 binding, in excellent agreement with the Ca2+-dependence for binding of these Abs. These findings establish that it is the genu Ca2+-binding site that is responsible for the Ca2+ dependence of these epitopes.
Fig. 6.
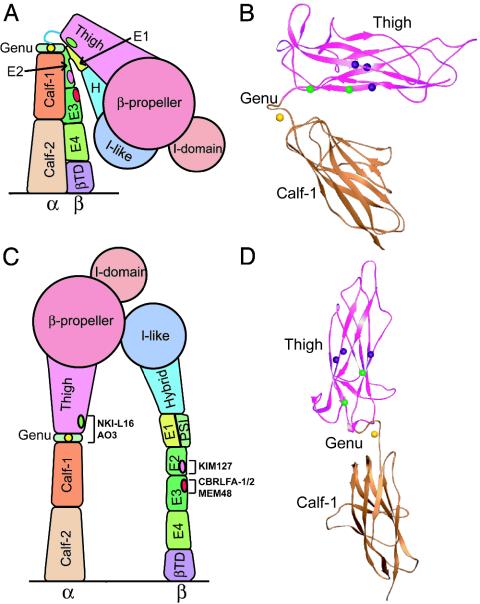
The bent and extended integrin conformations showing the location of the NKI-L16 and AO3 epitopes. (A and C) Cartoons based on crystal and EM structures of αVβ3 (3, 4, 6, 7) and NMR data on I-EGF domains 2 and 3 in β2 (5). (B and D) Ribbon diagrams of the thigh and calf-1 domains in the bent and extended conformations, respectively. The Cα atoms are shown as spheres for the αv residues corresponding (Fig. 5A) to the key NKI-L16 and AO3 epitope residues A602 and W738 (green) and additional NKI-L16 epitope residues T626, I627, and E736 (purple). The genu Ca2+ is shown as a gold sphere.
The mapping of the NKI-L16 and AO3 epitopes provides insights into structural rearrangements at the genu upon integrin activation. The conservation of the genu (Fig. 5A) and previous studies showing similar activation mechanisms in αVβ3 and αLβ2 (6, 27) justify presenting a model based on the detailed structure of αVβ3 and studies of activation epitopes in αLβ2. The NKI-L16 and AO3 epitopes are shielded from Ab recognition in the bent conformation because they are on the innermost face of the thigh domain that is shielded by the α-subunit calf-1 domain as well as the nearby I-EGF domains in the β-subunit (Fig. 6 A and B). Indeed, epitopes on I-EGF domains 2 and 3 in the LFA-1 β2-subunit that are shielded in the bent conformation and exposed in the extended conformation are nearby (Fig. 6 A and C). Use of a 25-Å radius probe to approximate an Fab fragment (data not shown) shows that αV residues equivalent to αL Glu-736 and Trp-738 (Fig. 5A) are completely shielded in the bent conformation and become exposed when the orientation at the genu is shifted to be similar to that of the extended αV leg seen in EM (6) (Fig. 6 C and D). Thus, the epitopes of NKI-L16 and AO3 are deeply buried in the bent conformation, and a switchblade-like opening would be required to make them accessible. There usually is overlap between Abs that bind better to activated than resting integrins and those that induce integrin activation; the common mechanism appears to be higher affinity or an absolute requirement for the active conformation and an equilibrium between different integrin conformational states (9). Indeed, besides activation dependence, NKI-L16 was also previously reported to increase LFA-1-mediated cell adhesion and aggregation (14, 15, 31-33); AO3 can also increase cell adhesion to ICAM-1 (P.S., J. M. Luk, and L.B.K., unpublished data). An equilibrium between the bent and extended conformations of LFA-1 on the cell surface may enable initial binding of activating mAbs and lead to activation of LFA-1.
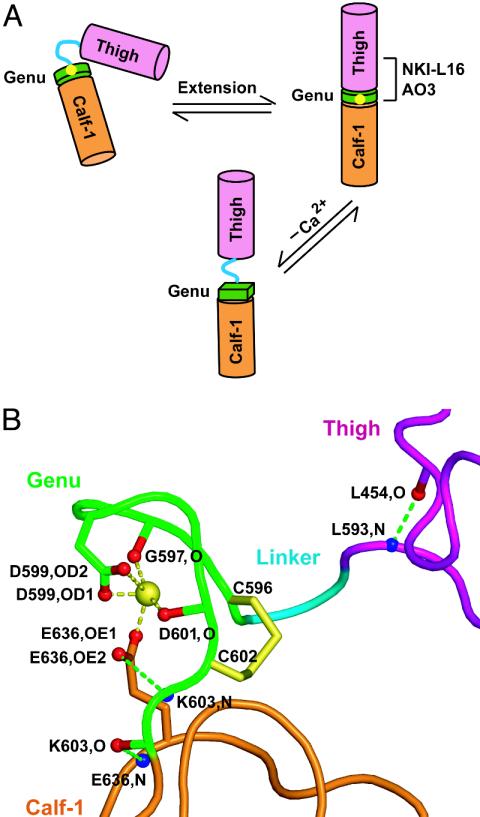
The Ca2+-coordinating residue αL Glu-787 in the calf-1 domain is required just as much as Ca2+ for the NKI-L16 and AO3 epitopes. This is most interesting because it strongly suggests that this coordination is maintained in the conversion of the bent conformation to the extended conformation. Therefore, the structural rearrangement at the genu must occur between the thigh domain and the genu rather than between the genu and the calf-1 domain (Fig. 7A). The αV structure is highly consistent with this finding (Fig. 7B). Thus, in the bent αV conformation, the Ca2+-coordinating residue Glu-636, which corresponds to αL calf-1 residue Glu-787, is at the center of a hydrogen bond network linking the genu to the calf-1 domain. αV Glu-636 coordinates both the Ca2+ and the genu residue 603 backbone with its side chain and also forms a backbone-backbone hydrogen bond to residue 603 (Fig. 7B). By contrast, there are no hydrogen bonds or Ca2+ coordinations from the thigh domain to the genu. Furthermore, αV residues Leu-594 and Asp-595 form no hydrogen bonds whatsoever. These residues link the last hydrogen-bonded residue in the thigh domain, Leu-593, to Cys-596, the first residue of the disulfide-bonded loop of the genu. Therefore, the structural and functional data strongly suggest that extension of the α-subunit leg occurs by a structural rearrangement involving linker residues 594-595 between the thigh domain and genu (Fig. 7A). Furthermore, the importance of the genu to the AO3 and NKI-L16 epitopes strongly suggests that there is a strong, stable interaction between the thigh and the genu in the extended conformation. These data are in agreement with EM observations that upon integrin extension, the α-subunit snaps into a well defined, rigid orientation between the thigh and calf-1 domains (6).
Fig. 7.
Genu rearrangement and structure. (A) Schematic diagram of genu rearrangement. (B) The structure of the αVβ3 genu (3). Backbones and carbon atom portions of side chains are pink (thigh), cyan (linker), green (genu), and orange (calf-1). The disulfide bond is yellow. Atoms involved in the hydrogen bonds nearest to the genu and in genu Ca2+ coordination are labeled and shown in red (oxygen) and blue (nitrogen). Ca2+ coordination and hydrogen bonds are yellow and green dashed lines, respectively. The figure was prepared with pymol (www.pymol.org; ref. 35).
Our findings further demonstrate that upon extension induced by XVA143, and extension/activation induced by PMA and Mn2+, straightening of the αL leg at the genu is coordinated with extension of the β2 leg. KIM127 acts as a probe of the extension of the β2 leg (5, 9) and m24 probes activation of the β2 I-like domain (19). In all conditions we have tested, NKI-L16 and AO3 are induced coordinately with the KIM127 epitope in the β2 leg. As previously reported, the m24 epitope is induced by Mn2+ but not PMA (9, 34). These results are consistent with PMA and Mn2+ each inducing extended integrin conformations but with Mn2+ favoring more the extended conformation with the open headpiece (28). The exposure of activation epitopes on both the αL leg and the β2 leg under the same conditions demonstrates coordinated straightening of the legs, in agreement with EM observations on αVβ3 (6). In summary, our studies on an activation epitope in the αL leg provide a fuller view of integrin activation that unifies findings on the α and β legs in diverse integrins and provides information on dynamic integrin structural rearrangements on the cell surface.
Acknowledgments
We thank Carl Figdor (Nijmegen Center for Molecular Life Sciences, Nijmegen, The Netherlands) for kindly providing the mAb NKI-L16 and Carl Figdor and Martin Humphries for reviewing the manuscript. This work was supported by National Institutes of Health Grant CA31798.
Abbreviations: EM, electron microscopy/microscopic; PMA, phorbol 12-myristate 13-acetate; I-EGF, integrin-epidermal growth factor-like; HBS, Hepes-buffered saline.
References
- 1.Springer, T. A. (1990) Nature 346, 425-433. [DOI] [PubMed] [Google Scholar]
- 2.Hynes, R. O. (2002) Cell 110, 673-687. [DOI] [PubMed] [Google Scholar]
- 3.Xiong, J.-P., Stehle, T., Diefenbach, B., Zhang, R., Dunker, R., Scott, D. L., Joachimiak, A., Goodman, S. L. & Arnaout, M. A. (2001) Science 294, 339-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xiong, J. P., Stehle, T., Zhang, R., Joachimiak, A., Frech, M., Goodman, S. L. & Arnaout, M. A. (2002) Science 296, 151-155. [DOI] [PubMed] [Google Scholar]
- 5.Beglova, N., Blacklow, S. C., Takagi, J. & Springer, T. A. (2002) Nat. Struct. Biol. 9, 282-287. [DOI] [PubMed] [Google Scholar]
- 6.Takagi, J., Petre, B. M., Walz, T. & Springer, T. A. (2002) Cell 110, 599-611. [DOI] [PubMed] [Google Scholar]
- 7.Xiao, T., Takagi, J., Wang, J.-h., Coller, B. S. & Springer, T. A. (2004) Nature, in press. [DOI] [PMC free article] [PubMed]
- 8.Takagi, J. & Springer, T. A. (2002) Immunol. Rev. 186, 141-163. [DOI] [PubMed] [Google Scholar]
- 9.Lu, C., Ferzly, M., Takagi, J. & Springer, T. A. (2001) J. Immunol. 166, 5629-5637. [DOI] [PubMed] [Google Scholar]
- 10.Mould, A. P., Askari, J. A., Barton, S., Kline, A. D., McEwan, P. A., Craig, S. E. & Humphries, M. J. (2002) J. Biol. Chem. 277, 19800-19805. [DOI] [PubMed] [Google Scholar]
- 11.Mould, A. P., Barton, S. J., Askari, J. A., McEwan, P. A., Buckley, P. A., Craig, S. E. & Humphries, M. J. (2003) J. Biol. Chem. 278, 17028-17035. [DOI] [PubMed] [Google Scholar]
- 12.Takagi, J., Strokovich, K., Springer, T. A. & Walz, T. (2003) EMBO J. 22, 4607-4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mould, A. P., Symonds, E. J., Buckley, P. A., Grossmann, J. G., McEwan, P. A., Barton, S. J., Askari, J. A., Craig, S. E., Bella, J. & Humphries, M. J. (2003) J. Biol. Chem. 278, 39993-39999. [DOI] [PubMed] [Google Scholar]
- 14.Keizer, G. D., Visser, W., Vliem, M. & Figdor, C. G. (1988) J. Immunol. 140, 1393-1400. [PubMed] [Google Scholar]
- 15.van Kooyk, Y., Weder, P., Hogervorst, F., Verhoeven, A. J., van Seventer, G., te Velde, A. A., Borst, J., Keizer, G. D. & Figdor, C. G. (1991) J. Cell Biol. 112, 345-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang, C. & Springer, T. A. (1995) J. Biol. Chem. 270, 19008-19016. [DOI] [PubMed] [Google Scholar]
- 17.Lu, C. & Springer, T. A. (1997) J. Immunol. 159, 268-278. [PubMed] [Google Scholar]
- 18.Robinson, M. K., Andrew, D., Rosen, H., Brown, D., Ortlepp, S., Stephens, P. & Butcher, E. C. (1992) J. Immunol. 148, 1080-1085. [PubMed] [Google Scholar]
- 19.Dransfield, I. & Hogg, N. (1989) EMBO J. 8, 3759-3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hildreth, J. E. K., Gotch, F. M., Hildreth, P. D. K. & McMichael, A. J. (1983) Eur. J. Immunol. 13, 202-208. [DOI] [PubMed] [Google Scholar]
- 21.Sanchez-Madrid, F., Davignon, D., Martz, E. & Springer, T. A. (1982) Cell. Immunol. 73, 1-11. [DOI] [PubMed] [Google Scholar]
- 22.Ho, S. N., Hunt, H. D., Horton, R. M., Pullen, J. K. & Pease, L. R. (1989) Gene 77, 51-59. [DOI] [PubMed] [Google Scholar]
- 23.Horton, R. M., Cai, Z., Ho, S. N. & Pease, L. R. (1990) BioTechniques 8, 528-535.2357375 [Google Scholar]
- 24.Kishimoto, T. K., O'Connor, K., Lee, A., Roberts, T. M. & Springer, T. A. (1987) Cell 48, 681-690. [DOI] [PubMed] [Google Scholar]
- 25.van Kooyk, Y., Weder, P., Heije, K. & Figdor, C. G. (1994) J. Cell Biol. 124, 1061-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Welzenbach, K., Hommel, U. & Weitz-Schmidt, G. (2002) J. Biol. Chem. 277, 10590-10598. [DOI] [PubMed] [Google Scholar]
- 27.Shimaoka, M., Salas, A., Yang, W., Weitz-Schmidt, G. & Springer, T. A. (2003) Immunity 19, 391-402. [DOI] [PubMed] [Google Scholar]
- 28.Salas, A., Shimaoka, M., Kogan, A. N., Harwood, C., von Andrian, U. H. & Springer, T. A. (2004) Immunity 20, 393-406. [DOI] [PubMed] [Google Scholar]
- 29.Lu, C., Shimaoka, M., Zang, Q., Takagi, J. & Springer, T. A. (2001) Proc. Natl. Acad. Sci. USA 98, 2393-2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen, J. F., Salas, A. & Springer, T. A. (2003) Nat. Struct. Biol. 10, 995-1001. [DOI] [PubMed] [Google Scholar]
- 31.Binnerts, M. E., van Kooyk, Y., Simmons, D. L. & Figdor, C. G. (1994) Eur. J. Immunol. 24, 2155-2160. [DOI] [PubMed] [Google Scholar]
- 32.van Kooyk, Y., van de Wiel-van Kemenade, P., Weder, P., Kuijpers, T. W. & Figdor, C. G. (1989) Nature 342, 811-813. [DOI] [PubMed] [Google Scholar]
- 33.van Kooyk, Y., Weder, P., Heije, K., De Waal Malefijt, R. & Figdor, C. G. (1993) Cell Adhes. Commun. 1, 21-32. [DOI] [PubMed] [Google Scholar]
- 34.Stewart, M. P., Cabanas, C. & Hogg, N. (1996) J. Immunol. 156, 1810-1817. [PubMed] [Google Scholar]
- 35.DeLano, W. L. (2002) pymol Molecular Graphics System (DeLano Scientific, San Carlos, CA).