ABSTRACT
From the years 2008 to 2014, a total of 1,155 water samples were collected (spring to fall) from 24 surface water sampling sites located in a mixed-used but predominantly agricultural (i.e., dairy livestock production) river basin in eastern Ontario, Canada. Water was analyzed for viable F-specific DNA (F-DNA) and F-specific RNA (F-RNA) (genogroup I [GI] to GIV) coliphage and a suite of molecularly detected viruses (norovirus [GI to GIV], torque teno virus [TTV], rotavirus, kobuvirus, adenovirus, astrovirus, hepatitis A, and hepatitis E). F-DNA and F-RNA coliphage were detected in 33 and 28% of the samples at maximum concentrations of 2,000 and 16,300 PFU · 100 ml−1, respectively. Animal TTV, human TTV, kobuvirus, astrovirus, and norovirus GIII were the most prevalent viruses, found in 23, 20, 13, 12, and 11% of samples, respectively. Viable F-DNA coliphage was found to be a modest positive indicator of molecularly detected TTV. F-RNA coliphage, unlike F-DNA coliphage, was a modest positive predictor of norovirus and rotavirus. There were, however, a number of significant negative associations among F-specific coliphage and viruses. F-DNA coliphage densities of >142 PFU · 100 ml−1 delineated conditions when ∼95% of water samples contained some type of virus. Kobuvirus was the virus most strongly related to detection of any other virus. Land use had some associations with virus/F-specific coliphage detection, but season and surface water flow were the variables that were most important for broadly delineating detection. Higher relative levels of detection of human viruses and human F-RNA coliphage were associated with higher relative degrees of upstream human land development in a catchment.
IMPORTANCE This study is one of the first, to our knowledge, to evaluate relationships among F-specific coliphages and a large suite of enteric viruses in mixed-use but agriculturally dominated surface waters in Canada. This study suggested that relationships between viable F-specific coliphages and molecularly detected viruses do exist, but they are not always positive. Caution should be employed if viable F-specific coliphages are to be used as indicators of virus presence in surface waters. This study elucidates relative effects of agriculture, wildlife, and human activity on virus and F-specific coliphage detection. Seasonal and meteorological attributes play a strong role in the detection of most virus and F-specific coliphage targets.
KEYWORDS: viruses, F-specific coliphages, pollution, indicators, surface water, microbial source tracking
INTRODUCTION
Viruses such as norovirus (NoV), rotavirus (RV), hepatitis A virus (HAV), hepatitis E virus (HEV), adenovirus (AdV), and astrovirus can be excreted in high numbers in human and/or animal waste (1). These viruses can contaminate surface waters via wastewater discharge, runoff and drainage from farming operations, and leakage from faulty septic systems (2–8). With ever-increasing anthropogenic stresses and pressures on surface water resources, identifying the driving factors that govern fecal pollution in open watershed systems and implementing management practices/strategies designed to ultimately reduce infection risks are becoming more crucial (9, 10). Important in this regard is the determination of the source of fecal pollution via biomarkers and host-specific pathogens (e.g., human, wildlife, and livestock), the seasonality of pathogen occurrence, and waterborne pathogen linkages to environmental/land use factors (11–14).
As it is not yet practical to ubiquitously test for the presence of all waterborne pathogens in a timely and tractable manner, water impairment guidelines are defined on the basis of levels of fecal indicator organisms, such as Escherichia coli, fecal coliforms, or enterococci (9). However, viruses often survive longer in water and can be more resistant to environmental stress than fecal indicator bacteria (FIB). Moreover, infectious viruses have been recovered at critical levels from surface waters that have met FIB impairment criteria (15). Therefore, for human infectious viruses, other types of indicators with preferred characteristics, depending on the application, will be required (9, 16–21).
F-specific coliphages have been identified as indicators of fecal contamination, as they primarily infect coliform bacteria present in the mammalian gut (9, 16, 18, 22–24). The F-specific coliphage group is composed of multiple groups of bacteriophages that infect bacteria via the F-specific pili (F-pili) and contain members with single-stranded DNA genomes (F-specific DNA [F-DNA] coliphages) or RNA genomes (F-specific RNA [F-RNA] coliphages). The enumeration and characterization of viable F-specific coliphage are relatively inexpensive and rapid. While F-RNA coliphages have been well characterized and have the added potential of discriminating between human and animal sources, much less is known about the distribution, survival, and ecology of F-DNA coliphages (17, 24–27). The F-DNA coliphages belong to the Inoviridae family and appear to have a higher genetic relatedness as a group than the F-RNA coliphages (27). F-RNA coliphages belong to the Leviviridae family, which is subdivided into the genera Levivirus and Allolevirus, and 4 serologically and phylogenetically separate genogroups (28, 29). F-RNA genogroups I (F-RNA GI) and II (F-RNA GII) belong to the Levivirus genus, while F-RNA genogroups III [F-RNA GIII] and IV [F-RNA GIV] are part of the Allolevirus genus. Genogroups I and IV are generally associated with animals [animal F-RNA] (but genogroup I has been isolated in sewage), and genogroups II and III are predominantly associated with humans [human F-RNA] but have been isolated from swine and chicken excrements (17, 28, 30–32). Different rates of survival at different temperatures for subgroups of F-specific coliphages have been reported in surface water (27, 30, 33, 34). In addition, the numbers and shedding frequency of F-specific coliphages shed by individual hosts can be highly variable, which can result in variability in detection and density in surface waters when fecal contamination occurs (35–37).
Adenoviruses (AdV) are gaining attention as a potential viral indicator as they are ubiquitous in sewage and surface water and are more resistant to disinfection from UV treatment than other viruses (19). AdV have a double-stranded DNA genome and display great diversity, with 7 species and 52 serotypes, where AdV types 40 and 41 are typically associated with gastroenteritis in children (38). Detection of AdV can be used for fecal source tracking in water, as human, porcine, bovine, and avian AdV are host specific (1, 39). AdV infections in animals are either asymptomatic or associated with mild enteric or respiratory diseases (1). Although many types of human AdV (HAdV) can be detected using cell culture, they vary with serotype and cell line and are laborious and time-consuming to measure in water (40). HAdV are readily detected and quantified in surface waters using PCR methods (40–43). With molecular detection techniques, it is possible to detect nucleic acids of many human-pathogenic viruses, such as NoV, HAV, HEV, and others that are difficult or impossible to cultivate, although no conclusions about virus infectivity can be made from these techniques. Molecular detection methods have opened up research opportunities to explore the potential utility of other viruses as indicators of fecal contamination.
Torque teno virus (TTV) is a potential viral indicator of fecal contamination, as it is frequently shed by humans and animals (44–47). TTV is a small virus classified in the Anelloviridae family with a high-genetic-diversity DNA genome (44, 45, 48). TTV is associated with persistent and transient infections, such as acute gastroenteritis, but is also excreted by healthy individuals, so it is not clear if TTV is part of the normal gut flora (19, 48–50). Some studies have demonstrated that TTV is very stable in the environment and more resistant to decontamination treatments than AdV (51, 52). However, as for several enteric viruses, there is no cell line supporting TTV replication in vitro.
Further information and data on the associations between indicator viruses and human-pathogenic viruses in impacted surface waters will be required before such viral indicator approaches are employed broadly. A positive correlation between the concentrations of F-RNA genogroup II and HAdV was reported in river water of an urban area in France, while a significant correlation was not detected between genomic copies of HAdV and culturable coliphages, F-specific coliphages, or FIB in urban rivers of California (42, 53). A meta-analysis revealed that F-specific coliphages can be good indicators for viral pathogens (21). When 5 pathogens and 8 commonly used indicators were compared, a positive correlation was only found for F-specific coliphages and AdV but not for other comparisons (21). The U.S. Environmental Protection Agency (USEPA) provides an expansive review of the use of coliphages as a fecal pollution indicator organism, but several studies therein found insignificant and negative coliphage-virus associations (54). Evidence was also provided therein supporting potential relationships between coliphages found in water and human health.
The primary objective of this study was to determine the degree of correlation among viable F-specific coliphages and a suite of molecularly detected human and animal enteric viruses (i.e., AdV, TTV, HAV, HEV, astrovirus, NoV genogroup I [GI] to GIV, rotavirus [RV], and kobuvirus) in surface water of several watersheds in a mixed-use, but predominantly agricultural river basin in eastern Ontario, Canada. A secondary objective was to determine the seasonality and environment/land use factors associated with these virus and F-specific coliphage targets.
RESULTS
Occurrence of F-coliphage and viruses in water.
F-DNA and F-RNA coliphage were detected in 33 and 28% of the water samples, respectively. The prevalence of F-DNA and F-RNA coliphage was greater than that of any specific virus (Table 1). The maximum numbers of F-DNA coliphage were 8-fold lower than the maximum numbers of F-RNA coliphages, where F-RNA GI was detected most frequently (21%), while F-RNA GIV was not detected in any water sample. Just over half of the samples (55%) exhibited detections for any virus. Human TTV and animal TTV were detected in 20 and 23% of the samples, respectively, while astrovirus, NoV GIII (often associated with bovines), and kobuvirus were detected in more than 10% of the samples and AdV 40/41 in 3% of the samples.
TABLE 1.
Summary of virus detections and maximum F-specific coliphage densities
| Coliphage or virusc | No. of detections | % detections | No. of nondetections | % nondetections | Maximum PFU · 100 ml−1a |
|---|---|---|---|---|---|
| F-DNA | 383 | 33 | 786 | 67 | 2,000 |
| F-RNA | 330 | 28 | 835 | 72 | 16,300 |
| Human F-RNA | 161 | 14 | 1,004 | 86 | 16,300 |
| Animal F-RNA | 248 | 21 | 917 | 79 | 925 |
| F-RNA GI | 247 | 21 | 918 | 79 | 16,300 |
| F-RNA GII | 118 | 10 | 1,047 | 90 | 925 |
| F-RNA GIII | 53 | 5 | 1,112 | 95 | 450 |
| F-RNA GIV | 0 | 0 | 1,165 | 100 | ND |
| Hepatitis E (HEV) | 20 | 2 | 1,135 | 98 | NA |
| Hepatitis A (HAV) | 18 | 2 | 1,014 | 98 | NA |
| Astrovirus | 83 | 12 | 638 | 88 | NA |
| Norovirus GI (NoV GI) | 48 | 4 | 1,107 | 96 | NA |
| Norovirus GII (NoV GII) | 57 | 5 | 1,098 | 95 | NA |
| Norovirus GIII (NoV GIII) | 89 | 11 | 755 | 89 | NA |
| Norovirus GIV (NoV GIV) | 59 | 7 | 785 | 93 | NA |
| Rotavirus (RV) | 23 | 2 | 1,132 | 98 | NA |
| Human torque teno virus (TTV) | 236 | 20 | 919 | 80 | NA |
| Animal TTV | 260 | 23 | 895 | 77 | NA |
| Adenovirus 40/41 (AdV 40/41) | 28 | 3 | 1,004 | 97 | NA |
| General adenoviruses (general AdV) | 22 | 2 | 1,010 | 98 | NA |
| Kobuvirus | 16 | 13 | 107 | 87 | NA |
| Any virusb | 634 | 55 | 521 | 45 | NA |
ND, not detected (below detection limit); NA, not applicable.
F-specific coliphage not included.
See Materials and Methods for virus group definitions.
Table 2 shows the levels of detection of any virus, animal virus, human virus, and F-specific coliphage in water samples for individual sample sites. For all the sample sites, the average percentages of detection of any virus, animal virus, human virus, F-RNA or F-DNA coliphage, F-DNA coliphage, human F-RNA coliphage, and animal F-RNA coliphage in water were 55, 36, 42, 53, 33, 15, and 79%, respectively. Some notable observations include site 12 on the mixed-use Little Castor River, which had the highest detection by percentage of all sites for any virus (67%) and human virus (52%), while the stream draining from a forested catchment with no human or agricultural upstream land uses (site 24) had the lowest percentages of any virus (35%) and animal virus (15%) detection. The percentages for F-RNA and F-DNA coliphage detection were greatest at site 14 and lowest at site 24 (forested catchment). Site 14 is located on a surface drainage network draining crop fields and is an area associated with a suite of municipal sewage treatment lagoons (the degree of hydrological connectivity of the drainage channel with the lagoons is unknown). The lowest percentages of human F-RNA coliphage detection were found at site 22 and site 24. Site 22 was located immediately downstream of a protected stream channel/riparian zone that received primarily agricultural drainage but, interestingly, has been a site subjected to septic system leakage inputs (6, 39, 55). The lowest animal F-RNA coliphage detection was at site 22. The highest level of animal F-RNA coliphage detection was observed at site 24 and site 10 (a site immediately downstream of a cow pasture where cattle had access to the water course [56]).
TABLE 2.
Sample site characteristics and detection of grouped viruses and F-specific coliphages at sampling sites
| Site | Site characteristics |
Virus detection |
F-specific coliphage detection |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Description | Upstream contributing area (km2) | Agriculture (% land) | Urban/developed (% land) | Forest (% land) | Wetland (% land) | Other (% land) | No. | Any virus (%) | Animal virus (%) | Human virus(%) | No. | F-RNA or F-DNA (%) | F-DNA (%) | Human F-RNA (%) | Animal F-RNA (%) | |
| 1 | WTP intake | 2,371 | 52 | 2 | 42 | 3 | 2 | 47 | 57.4 | 40.4 | 44.7 | 48 | 52.1 | 39.6 | 12.5 | 79.2 |
| 2 | Main river | 2,371 | 52 | 2 | 42 | 3 | 2 | 42 | 54.8 | 38.1 | 35.7 | 41 | 51.2 | 36.6 | 12.2 | 80.5 |
| 3 | Agricultural stream | <5 | 67 | 3 | 30 | 0 | 1 | 46 | 43.5 | 26.1 | 32.6 | 46 | 69.6 | 37.0 | 13.3a | 73.3a |
| 4 | Tributary | 724 | 51 | 5 | 39 | 2 | 3 | 45 | 53.3 | 37.8 | 44.4 | 46 | 52.2 | 32.6 | 19.6 | 80.4 |
| 5 | Tributary | 81 | 65 | 2 | 30 | 1 | 2 | 72 | 59.7 | 45.8 | 27.8 | 74 | 63.5 | 48.6 | 14.9 | 81.1 |
| 6 | Tributary | 176 | 54 | 1 | 43 | 1 | 1 | 72 | 55.6 | 36.1 | 36.1 | 75 | 53.3 | 36.0 | 16.0 | 85.3 |
| 7 | Main river | 1,216 | 48 | 1 | 47 | 4 | 1 | 46 | 54.3 | 32.6 | 47.8 | 46 | 58.7 | 37.0 | 21.7 | 80.4 |
| 8 | Main river | 1,413 | 49 | 1 | 46 | 3 | 1 | 46 | 56.5 | 37 | 50 | 49 | 61.2 | 34.7 | 19.1b | 70.2b |
| 9 | Tributary | 54 | 72 | 1 | 23 | 4 | 1 | 72 | 45.8 | 37.5 | 29.2 | 75 | 56.0 | 34.7 | 14.7 | 76.0 |
| 10 | Tributary | 68 | 75 | 2 | 19 | 3 | 1 | 43 | 62.8 | 44.2 | 51.2 | 43 | 48.8 | 32.6 | 16.3 | 88.4 |
| 11 | Main river | 1,548 | 51 | 1 | 44 | 3 | 1 | 45 | 62.2 | 35.6 | 46.7 | 45 | 51.1 | 31.1 | 24.4 | 80.0 |
| 12 | Tributary | 96 | 67 | 2 | 29 | 1 | 2 | 46 | 67.4 | 41.3 | 52.2 | 49 | 55.1 | 30.6 | 20.4 | 77.6 |
| 13 | Tributary | 88 | 66 | 2 | 30 | 1 | 2 | 45 | 60.0 | 44.4 | 44.4 | 46 | 56.5 | 34.8 | 10.9 | 84.8 |
| 14 | Agricultural stream | <5 | 69 | 2 | 25 | 0 | 5 | 21 | 57.1 | 42.9 | 38.1 | 20 | 85.0 | 50.0 | 45.0 | 80.0 |
| 15 | Agricultural stream | <5 | 90 | 0 | 10 | 0 | 0 | 39 | 64.1 | 43.6 | 51.3 | 39 | 53.8 | 41.0 | 5.1 | 74.4 |
| 16 | Tributary | 739 | 51 | 5 | 40 | 2 | 3 | 44 | 63.6 | 38.6 | 47.7 | 44 | 47.7 | 25.0 | 9.1 | 72.7 |
| 17 | Main river | 1,450 | 50 | 1 | 45 | 3 | 1 | 45 | 64.4 | 37.8 | 51.1 | 45 | 57.8 | 35.6 | 17.8 | 71.1 |
| 18 | Agricultural stream | <5 | 90 | 0 | 10 | 0 | 0 | 69 | 50.7 | 37.7 | 33.3 | 71 | 56.3 | 29.6 | 4.2 | 69.0 |
| 19 | Agricultural stream | <5 | 89 | 0 | 11 | 0 | 0 | 47 | 53.2 | 27.7 | 44.7 | 46 | 39.1 | 19.6 | 15.2 | 84.8 |
| 20 | Agricultural stream | <5 | 90 | 0 | 9 | 0 | 1 | 64 | 45.3 | 26.6 | 31.3 | 66 | 36.4 | 18.2 | 7.6 | 81.8 |
| 21 | Agricultural stream | <5 | 77 | 0 | 21 | 0 | 1 | 40 | 57.5 | 37.5 | 50 | 39 | 43.6 | 23.1 | 15.4 | 82.1 |
| 22 | Agricultural stream | <5 | 90 | 0 | 10 | 0 | 0 | 39 | 51.3 | 28.2 | 35.9 | 39 | 53.8 | 28.2 | 2.6 | 66.7 |
| 23 | Agricultural stream | <5 | 90 | 0 | 10 | 0 | 0 | 34 | 52.9 | 35.3 | 44.1 | 32 | 50.0 | 37.5 | 9.7c | 77.4c |
| 24 | Forest stream | <5 | 0 | 0 | 100 | 0 | 0 | 46 | 34.8 | 15.2 | 28.3 | 45 | 22.2 | 17.8 | 2.2 | 91.1 |
n = 45.
n = 47.
n = 31.
Associations between F-specific coliphage and viruses in water.
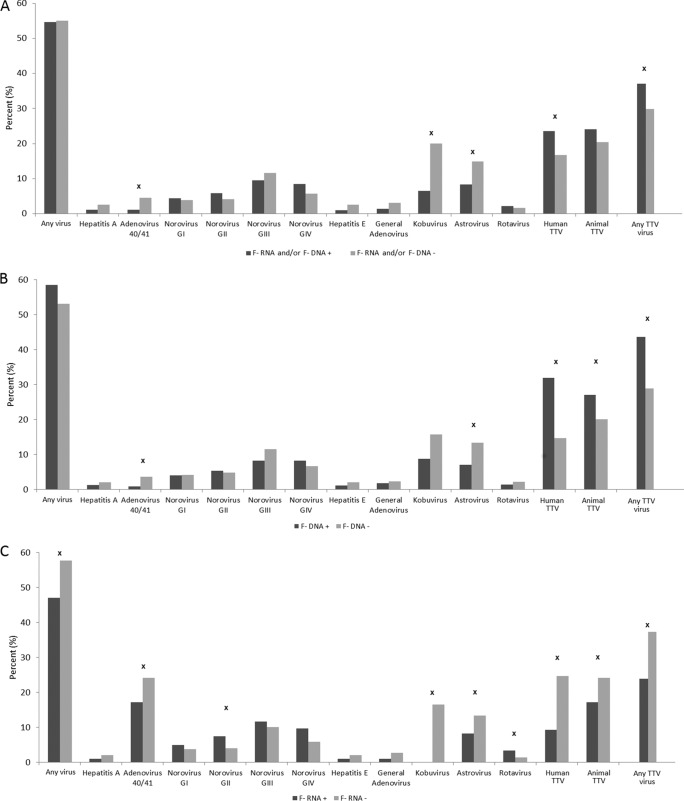
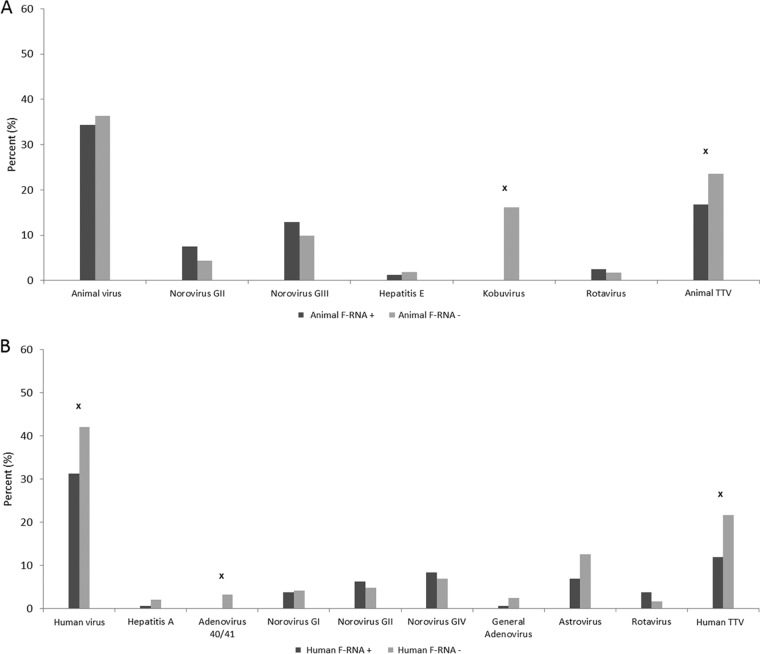
Fig. 2A shows significantly higher percentages of detection of kobuvirus, astrovirus, and AdV 40/41 when F-RNA coliphage and F-DNA coliphage were not detected in a sample. The reverse was true for human TTV and any TTV (there were higher detection percentages associated with F-RNA and/or F-DNA coliphage detection). For just F-DNA coliphage, these same relationships held true except for significantly higher levels of detection of animal TTV when F-DNA coliphage was detected in a water sample, and there was no significant difference in the levels of detection of kobuvirus with respect to F-DNA coliphage (Fig. 2B). For F-RNA coliphage only, significant detection associations were found for a vast array of viruses, but 7 out of the 9 significant associations indicated lower levels of virus detection when F-RNA coliphage was present in a water sample (Fig. 2C). Animal F-RNA coliphage associations with groups of viruses were only significant among kobuvirus and animal TTV, both negatively (Fig. 3A). Regarding human F-RNA coliphage detection, significant associations were found among human TTV, AdV 40/41, and human viruses; however, these detection relationships were all negative (Fig. 3B).
FIG 2.
The percentage of specific virus and virus group detections and nondetections in relation to detections and nondetections of F-DNA and/or F-RNA coliphages (A), F-DNA coliphages (B), and F-RNA coliphages (C). x = Fisher's exact test, P ≤ 0.05. + and − refer to F-coliphage detection and nondetection, respectively.
FIG 3.
The percentage of specific virus and virus group detections and nondetections in relation to detections and nondetections of animal F-RNA coliphages (A) and human F-RNA coliphages (B). x = Fisher's exact test, P ≤ 0.05. + and − refer to F-coliphage detection and nondetection, respectively.
Table 3 shows that a majority of the significant (P ≤ 0.05) associations among quantitative F-specific coliphage and virus detection/nondetection were negatively related. That is, F-specific coliphage average rank sums were lower when viruses were detected in a sample than when viruses were not detected in a sample. Positive associations, however, included (i) F-RNA coliphage versus NoV (GI, GII, and GIV) and RV, (ii) F-DNA coliphage versus any, human, or animal TTV, and (iii) F-DNA coliphage versus any, human, or animal virus.
TABLE 3.
Significant (P ≤ 0.05) Mann-Whitney U test results associated with F-specific coliphage densities in samples where a virus was or was not detecteda
| Virus grouping variable | Quantitative F-specific coliphage target | F-specific coliphage rank sum for virus detection group | F-specific coliphage rank sum for virus nondetection group | F-specific coliphage mean rank sum for virus detection group | F-specific coliphage mean rank sum for virus nondetection group | Positive association between virus and coliphage | U statistic of Mann-Whitney U test | Z adjusted | P value | Valid no. of virus detections | Valid no. of virus nondetections | Arithmetic mean of coliphage for virus detection groupa | Arithmetic mean of coliphage for virus nondetection groupa |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Astrovirus | F-DNA | 25,993 | 227,835 | 313 | 362 | No | 22,507 | −2.59 | 0.010 | 83 | 629 | 2 | 8 |
| Astrovirus | F-RNA | 26,533 | 225,872 | 320 | 360 | No | 23,047 | −2.03 | 0.042 | 83 | 627 | 58 | 72 |
| Norovirus GI | F-RNA | 37,478 | 609,475 | 658 | 564 | Yes | 25,735 | 2.63 | 0.009 | 57 | 1,080 | 447 | 28 |
| Norovirus GII | F-RNA GI | 36,225 | 610,728 | 636 | 565 | Yes | 26,988 | 2.21 | 0.027 | 57 | 1,080 | 440 | 24 |
| Norovirus GIV | F-RNA GIII | 26,134 | 320,394 | 443 | 414 | Yes | 21,243 | 2.26 | 0.024 | 59 | 773 | 0 | 1 |
| Rotavirus | F-RNA | 15,042 | 631,912 | 684 | 567 | Yes | 9,742 | 2.08 | 0.037 | 22 | 1,115 | 11 | 50 |
| Human TTV | F-DNA | 162,834 | 488,678 | 702 | 538 | Yes | 75,083 | 8.12 | 0.000 | 232 | 909 | 55 | 7 |
| Human TTV | F-RNA | 111,388 | 535,566 | 482 | 591 | No | 84,592 | −5.66 | 0.000 | 231 | 906 | 5 | 61 |
| Human TTV | F-RNA GI | 113,684 | 533,270 | 492 | 589 | No | 86,888 | −5.60 | 0.000 | 231 | 906 | 4 | 56 |
| Human TTV | F-RNA GII | 126,519 | 520,434 | 548 | 574 | No | 99,723 | −2.09 | 0.036 | 231 | 906 | 1 | 4 |
| Human TTV | F-RNA GIII | 127,619 | 519,335 | 552 | 573 | No | 100,823 | −2.35 | 0.019 | 231 | 906 | 0 | 1 |
| Animal TTV | F-DNA | 160,305 | 491,206 | 629 | 554 | Yes | 98,265 | 3.80 | 0.000 | 255 | 886 | 35 | 12 |
| Animal TTV | F-RNA | 136,154 | 510,800 | 540 | 577 | No | 104,276 | −1.98 | 0.048 | 252 | 885 | 26 | 56 |
| Adenovirus 40/41 | F-DNA | 11,246 | 508,444 | 402 | 513 | No | 10,840 | −2.38 | 0.017 | 28 | 991 | 11 | 19 |
| Adenovirus 40/41 | F-RNA | 11,682 | 503,938 | 417 | 511 | No | 11,276 | −2.07 | 0.039 | 28 | 987 | 13 | 56 |
| Any virus | F-DNA | 367,617 | 283,895 | 587 | 551 | Yes | 151,025 | 2.20 | 0.028 | 626 | 515 | 26 | 7 |
| Any virus | F-RNA | 339,859 | 307,094 | 546 | 596 | No | 146,106 | −3.21 | 0.001 | 622 | 515 | 54 | 45 |
| Any virus | F-RNA GI | 338,880 | 308,074 | 545 | 598 | No | 145,127 | −3.84 | 0.000 | 622 | 515 | 49 | 41 |
| Any TTV | F-DNA | 235,088 | 416,423 | 641 | 538 | Yes | 116,498 | 5.89 | 0.000 | 367 | 774 | 37 | 8 |
| Any TTV | F-RNA | 191,163 | 455,791 | 527 | 589 | No | 125,097 | −3.75 | 0.000 | 363 | 774 | 18 | 64 |
| Any TTV | F-RNA GI | 190,710 | 456,244 | 525 | 589 | No | 124,644 | −4.31 | 0.000 | 363 | 774 | 12 | 61 |
| Human virus | F-DNA | 281,884 | 369,628 | 610 | 544 | Yes | 138,768 | 3.97 | 0.000 | 462 | 679 | 34 | 6 |
| Human virus | F-RNA | 245,238 | 401,716 | 532 | 594 | No | 138,747 | −3.95 | 0.000 | 461 | 676 | 65 | 39 |
| Human virus | F-RNA GI | 246,422 | 400,531 | 535 | 593 | No | 139,931 | −4.11 | 0.000 | 461 | 676 | 63 | 33 |
| Human virus | F-RNA GII | 255,196 | 391,758 | 554 | 580 | No | 148,705 | −2.48 | 0.013 | 461 | 676 | 2 | 4 |
| Animal virus | F-DNA | 243,878 | 407,634 | 593 | 558 | Yes | 140,819 | 2.06 | 0.039 | 411 | 730 | 28 | 11 |
Mean trends may differ from rank sum trends. Concentrations are in PFU · 100 ml−1.
Classification and regression tree (CART) analyses were used to classify threshold F-DNA coliphage densities above which there would be a higher relative prevalence of viruses (groups). We focused this effort on F-DNA coliphage versus human virus, animal virus, and any virus. F-DNA coliphage, as already underscored, had the strongest positive associations with these virus groups, in relation to F-RNA coliphage. Table 4 indicates that a grand proportion of virus detections were associated with lower F-DNA threshold groupings (∼≤88 PFU · 100 ml−1 for human virus and ∼≤142 PFU · 100 ml−1 for animal and any virus), but the greatest classification percentages of virus-positive samples were above threshold values (percentages of virus positivity were 66, 87, and 95% for animal, human, and any virus, respectively), albeit the numbers of samples for these groups were small in relation to the total number of virus detections.
TABLE 4.
Classification of virus detections on the basis of F-DNA coliphage densities using CART analyses
| CART model | Dependent (target) variable | Classification criterion: F-DNA coliphage density (PFU · 100 ml−1) | No. of samples with: |
% virus detection | |
|---|---|---|---|---|---|
| Virus detection | Virus nondetection | ||||
| 1 | Human virus | >87.5 | 41 | 6 | 87 |
| Human virus | ≤87.5 | 421 | 673 | 39 | |
| 2 | Animal virus | >141.5 | 25 | 13 | 66 |
| Animal virus | ≤141.5 | 386 | 717 | 35 | |
| 3 | Any virus | >141.5 | 36 | 2 | 95 |
| Any virus | ≤141.5 | 590 | 513 | 54 | |
Intervirus associations in water.
Table 5 indicates that for HEV, there were a suite of significant Fisher's exact test results, primarily with NoV, but Phi coefficients, while all positive, were all low. Low Phi coefficients were also found for HAV. Astrovirus versus any virus had a modest Phi coefficient of 0.41. NoV GI and GII were associated with a suite of significant Fisher's exact test results; however, the associated Phi coefficients were predominantly positive but low. Interestingly, NoV GIII versus any virus had a modest positive Phi coefficient of 0.40. This could be important considering that NoV GIII is often associated with bovine fecal pollution, bovines being the dominant livestock in the region. Both animal TTV and human TTV versus any virus had modestly positive Phi coefficients (0.46 to 0.49). The strongest Phi coefficient belonged to kobuvirus versus any virus, at 0.51.
TABLE 5.
Significant Fisher's exact test results among virus targets, and associated Phi coefficients
| Virus vs virus | Phi coefficient | Fisher's exact test P value |
|---|---|---|
| Hepatitis E virus vs norovirus GI | 0.07 | 0.05 |
| Hepatitis E virus vs norovirus GIII | 0.24 | 0.00 |
| Hepatitis E virus vs norovirus GIV | 0.15 | 0.00 |
| Hepatitis E virus vs adenovirus 40/41 | 0.16 | 0.00 |
| Hepatitis E virus vs kobuvirus | 0.33 | 0.02 |
| Hepatitis E virus vs any virus | 0.12 | 0.00 |
| Hepatitis A virus vs astrovirus | 0.15 | 0.00 |
| Hepatitis A virus vs any virus | 0.12 | 0.00 |
| Astrovirus vs norovirus GIII | 0.10 | 0.02 |
| Astrovirus vs any virus | 0.41 | 0.00 |
| Norovirus GI vs norovirus GII | 0.11 | 0.00 |
| Norovirus GI vs norovirus GIII | 0.12 | 0.00 |
| Norovirus GI vs norovirus GIV | 0.12 | 0.00 |
| Norovirus GI vs human TTV | −0.09 | 0.00 |
| Norovirus GI vs any virus | 0.19 | 0.00 |
| Norovirus GII vs norovirus GIII | 0.10 | 0.01 |
| Norovirus GII vs animal TTV | −0.08 | 0.01 |
| Norovirus GII vs any virus | 0.21 | 0.00 |
| Norovirus GIII vs norovirus GIV | 0.27 | 0.00 |
| Norovirus GIII vs any virus | 0.40 | 0.00 |
| Norovirus GIV vs animal TTV | −0.07 | 0.02 |
| Norovirus GIV vs any virus | 0.32 | 0.00 |
| Rotavirus vs any virus | 0.13 | 0.00 |
| Human TTV vs animal TTV | 0.35 | 0.00 |
| Human TTV vs any virus | 0.46 | 0.00 |
| Animal TTV vs any virus | 0.49 | 0.00 |
| Adenovirus 40/41 vs general adenovirus | 0.31 | 0.00 |
| Adenovirus 40/41 vs any virus | 0.14 | 0.00 |
| General adenovirus vs any virus | 0.13 | 0.00 |
| Kobuvirus vs any virus | 0.51 | 0.00 |
Seasonality and environmental/land use associations with viruses and F-specific coliphage.
Fig. 4 shows the seasonal detection of F-RNA and F-DNA coliphage and viruses. Levels of detection were greatest for human F-RNA coliphage, animal F-RNA coliphage, F-RNA and/or F-DNA coliphage, and F-DNA coliphage in fall (24%), fall (38%), summer (62%), and summer (47% samples), respectively. For the statistically significant seasonal differences, levels of detection of HAV, NoV (GI, GII, and GIV), human TTV, animal TTV, AdV 40/41, and general AdV were greatest in summer (47%), summer (3%), summer (6, 7, and 12%, respectively), summer (25%), fall (27%), spring (8%), and spring (4% of samples), respectively. NoV GIII detection was greatest in spring (16% of samples), a time period when livestock manure is applied to land. Seasonality was not significant for the detection of any TTV, animal virus, HEV, astrovirus, RV, or kobuvirus.
FIG 4.
The percentage of various virus and F-specific coliphage detection and nondetection in samples grouped by season. Significant seasonality was determined via Fisher's exact test (3 seasons by presence/absence). x = P ≤ 0.05.
Table 6 documents exploratory data mining results for a suite of virus and F-specific coliphage targets. The independent variables used in this analysis are given in Table S1 and are exploratory variables that have been utilized consistently in other works for pathogen association (see, e.g., reference 13). The approach defines optimal independent variable conditions that classify dependent variables (virus and F-specific coliphage detection and nondetection) on the basis of independent variable criteria. Some noteworthy results are as follows: for F-DNA coliphage, most detections were associated with daily mean air temperature of >4.25°C and total rainfall on the day of sampling of ≤4.1 mm. The majority of the F-RNA coliphage as well as animal F-RNA coliphage detections occurred during fall, in relation to the other seasons. Human F-RNA coliphage were most strongly associated with a daily mean air temperature of ≤9.25°C and developed land upstream (5 km) of >1%; the developed land variable split condition suggests there was a positive association between human F-RNA coliphage and degree of urban/rural development for daily mean air temperatures of ≤9.25°C. Regarding human virus, a majority of the detections were associated with mean daily river discharge of ≤1.64 m3 · s−1 and developed land upstream (2 km) of >0.2%. The positive relationship between detection and human development for the discharge condition highlights positive-occurrence links between lower-discharge conditions and human land development. For animal virus, two independent variable criteria defined conditions associated with higher relative levels of virus detection; these were mean daily river discharge of >0.39 m3 · s−1 and total rainfall on the day of sampling of >0.30 mm, and a mean daily river discharge of ≤0.39 m3 · s−1. These are two contrasting situations, the first being higher relative discharge and rainfall inducing detection, and the second being lower-discharge conditions exclusively (animal virus presence is basically bimodal in the context of discharge). Some other notable virus-specific results include those for HEV, where greater HEV detection occurred when the total rainfall on the day of sampling was >0.60 mm and the electrical conductivity of water was >0.60 mS · cm−1 (higher salt content is usually linked to fertilizer-based nutrients in water in this area [13] under rainfall conditions). For NoV, the associations were different for each genogroup, where the detection of most NoV GI was associated with a daily mean air temperature of >5.90°C and a total rainfall on the day of sampling of ≤5.30 mm; proportionally higher levels of NoV GII detection occurred when the daily mean air temperature was >23.40°C. NoV GIII was associated with an oxidation-reduction potential of ≤145.10 mV or a combination of an oxidation-reduction potential of >145.10 mV and daily mean air temperature of ≤21.15°C, while NoV GIV detection was strongly associated with mean daily river discharge of ≤1.56 m3 · s−1 and pasture forage land upstream (10 km) of ≤35%.
TABLE 6.
CART classification tree results targeting detected F-specific coliphage and viruses using independent variables listed in Table 2
| Dependent (target) variable | CART classification criteria | No. of detections | No. of nondetections | Total samples | % positive | % negative | CART nodal class designationa | Classification accuracy (%) | Weighted classification accuracy of CART model (%) |
|---|---|---|---|---|---|---|---|---|---|
| F-DNA coliphage | RUS_MEANTEMP ≤4.25°C | 19 | 125 | 144 | 13 | 87 | − | 87 | 52 |
| F-DNA coliphage | RUS_MEANTEMP >4.25°C AND RUS_TOTALRAIN ≤4.10 mm | 344 | 519 | 863 | 40 | 60 | + | 40 | |
| F-DNA coliphage | RUS_MEANTEMP >4.25°C AND RUS_TOTALRAIN >4.10 mm | 20 | 142 | 162 | 12 | 88 | − | 88 | |
| F-RNA coliphage | SEASON = FALL | 157 | 190 | 347 | 45 | 55 | + | 45 | 67 |
| F-RNA coliphage | SEASON = SPRING, SUMMER AND RUS_MEANTEMP ≤22.90°C | 143 | 595 | 738 | 19 | 81 | − | 81 | |
| F-RNA coliphage | SEASON = SPRING, SUMMER AND RUS_MEANTEMP >22.90°C | 30 | 50 | 80 | 38 | 63 | + | 38 | |
| Human F-RNA | RUS_MEANTEMP >9.25°C | 75 | 716 | 791 | 9 | 91 | − | 91 | 76 |
| Human F-RNA | RUS_MEANTEMP ≤9.25°C AND DEVELP_5K ≤1% | 2 | 85 | 87 | 2 | 98 | − | 98 | |
| Human F-RNA | RUS_MEANTEMP ≤9.25°C AND DEVELP_5K >1% | 84 | 203 | 287 | 29 | 71 | + | 29 | |
| Animal F-RNA | SEASON = FALL | 132 | 215 | 347 | 38 | 62 | + | 38 | 69 |
| Animal F-RNA | SEASON = SPRING, SUMMER AND RUS_MEANTEMP ≤22.90°C | 91 | 647 | 738 | 12 | 88 | − | 88 | |
| Animal F-RNA | SEASON = SPRING, SUMMER AND RUS_MEANTEMP >22.90°C | 25 | 55 | 80 | 31 | 69 | + | 31 | |
| Any virus | DIS_RUS ≤0.39 m3 · s−1 | 56 | 0 | 56 | 100 | 0 | + | 100 | 62 |
| Any virus | DIS_RUS >0.39 m3 · s−1 AND TEMP ≤12.89°C | 146 | 230 | 376 | 39 | 61 | − | 61 | |
| Any virus | DIS_RUS >0.39 m3 · s−1 AND TEMP >12.89°C | 432 | 291 | 723 | 60 | 40 | + | 60 | |
| Human virus | DIS_RUS ≤1.64 m3 · s−1 AND DEVELP_2K ≤0.2% | 14 | 67 | 81 | 17 | 83 | − | 83 | 65 |
| Human virus | DIS_RUS ≤1.64 m3 · s−1 AND DEVELP_2K >0.2% | 276 | 215 | 491 | 56 | 44 | + | 56 | |
| Human virus | DIS_RUS >1.64 m3 · s−1 | 178 | 405 | 583 | 31 | 69 | − | 69 | |
| Animal virus | DIS_RUS ≤0.39 m3 · s−1 | 55 | 1 | 56 | 98 | 2 | + | 98 | 60 |
| Animal virus | DIS_RUS >0.39 m3 · s−1 AND RUS_TOTALRAIN ≤0.30 mm | 94 | 371 | 465 | 20 | 80 | − | 80 | |
| Animal virus | DIS_RUS >0.39 m3 · s−1 AND RUS_TOTALRAIN >0.30 mm | 268 | 366 | 634 | 42 | 58 | + | 42 | |
| Hepatitis E virus | RUS_TOTALRAIN ≤0.60 mm | 2 | 525 | 527 | 0 | 100 | − | 100 | 68 |
| Hepatitis E virus | RUS_TOTALRAIN >0.60 mm AND CONDUCTIVITY ≤0.60 mS · cm−1 | 0 | 246 | 246 | 0 | 100 | − | 100 | |
| Hepatitis E virus | RUS_TOTALRAIN >0.60 mm AND CONDUCTIVITY >0.60 mS · cm−1 | 18 | 364 | 382 | 5 | 95 | + | 5 | |
| Hepatitis A virus | RUS_MEANTEMP ≤11.50°C | 2 | 379 | 381 | 1 | 99 | − | 99 | 72 |
| Hepatitis A virus | RUS_MEANTEMP >11.50°C AND TOTPHO ≤0.08 mg · liter−1 | 15 | 287 | 302 | 5 | 95 | + | 5 | |
| Hepatitis A virus | RUS_MEANTEMP >11.50°C AND TOTPHO >0.08 mg · liter−1 | 1 | 348 | 349 | 0 | 100 | − | 100 | |
| Astrovirus | ORP ≤174.35 mV | 31 | 407 | 438 | 7 | 93 | − | 93 | 81 |
| Astrovirus | ORP >174.35 mV AND TEMP ≤11.13°C | 13 | 139 | 152 | 9 | 91 | − | 91 | |
| Astrovirus | ORP >174.35 mV AND TEMP >11.13°C | 39 | 92 | 131 | 30 | 70 | + | 30 | |
| Norovirus GI | RUS_MEANTEMP ≤5.90°C | 1 | 226 | 227 | 0 | 100 | − | 100 | 35 |
| Norovirus GI | RUS_MEANTEMP >5.90°C AND RUS_TOTALRAIN ≤5.30 mm | 46 | 750 | 796 | 6 | 94 | + | 6 | |
| Norovirus GI | RUS_MEANTEMP >5.90°C AND RUS_TOTALRAIN >5.30 mm | 1 | 131 | 132 | 1 | 99 | − | 99 | |
| Norovirus GII | RUS_MEANTEMP >23.40°C | 13 | 50 | 63 | 21 | 79 | + | 79 | 78 |
| Norovirus GII | RUS_MEANTEMP ≤23.40°C AND DIS_RUS ≤6.33 m3 · s−1 | 44 | 851 | 895 | 5 | 95 | − | 95 | |
| Norovirus GII | RUS_MEANTEMP ≤23.40°C AND DIS_RUS >6.33 m3 · s−1 | 0 | 197 | 197 | 0 | 100 | − | 0 | |
| Norovirus GIII | ORP ≤145.10 mV | 45 | 209 | 254 | 18 | 82 | + | 18 | 61 |
| Norovirus GIII | ORP >145.10 mV AND RUS_MEANTEMP ≤21.15°C | 21 | 451 | 472 | 4 | 96 | − | 96 | |
| Norovirus GIII | ORP >145.10 mV AND RUS_MEANTEMP >21.15°C | 23 | 95 | 118 | 19 | 81 | + | 19 | |
| Norovirus GIV | DIS_RUS ≤1.56 m3 · s−1 AND PASFOR_10K ≤35% | 51 | 222 | 273 | 19 | 81 | + | 19 | 73 |
| Norovirus GIV | DIS_RUS ≤1.56 m3 · s−1 AND PASFOR_10K >35% | 1 | 102 | 103 | 1 | 99 | − | 99 | |
| Norovirus GIV | DIS_RUS >1.56 m3 · s−1 | 7 | 461 | 468 | 1 | 99 | − | 99 | |
| Rotavirus | TOTKN ≤0.88 mg · liter−1 AND RUS_MEANTEMP ≤13.65°C | 11 | 343 | 354 | 3 | 97 | + | 3 | 50 |
| Rotavirus | TOTKN ≤0.88 mg · liter−1 AND RUS_MEANTEMP >13.65°C | 7 | 228 | 235 | 3 | 97 | + | 3 | |
| Rotavirus | TOTKN >0.88 mg · liter−1 | 5 | 561 | 566 | 1 | 99 | − | 99 | |
| Human TTV | RUS_MEANTEMP ≤20.40°C AND TEMP ≤16.06°C | 74 | 477 | 551 | 13 | 87 | − | 87 | 72 |
| Human TTV | RUS_MEANTEMP ≤20.40°C AND TEMP >16.06°C | 158 | 246 | 404 | 39 | 61 | + | 39 | |
| Human TTV | RUS_MEANTEMP >20.40°C | 4 | 196 | 200 | 2 | 98 | − | 98 | |
| Animal TTV | DIS_RUS ≤0.39 m3 · s−1 | 55 | 1 | 56 | 98 | 2 | + | 98 | 56 |
| Animal TTV | DIS_RUS >0.39 m3 · s−1 AND ORP ≤174.35 mV | 54 | 444 | 498 | 11 | 89 | − | 89 | |
| Animal TTV | DIS_RUS >0.39 m3 · s−1 AND ORP >174.35 mV | 151 | 450 | 601 | 25 | 75 | + | 25 | |
| Any TTV | DIS_RUS ≤0.49 m3 · s−1 | 82 | 18 | 100 | 82 | 18 | + | 82 | 61 |
| Any TTV | DIS_RUS >0.49 m3 · s−1 AND ORP ≤171.85 mV | 66 | 398 | 464 | 14 | 86 | − | 86 | |
| Any TTV | DIS_RUS >0.49 m3 · s−1 AND ORP >171.85 mV | 226 | 365 | 591 | 38 | 62 | + | 38 | |
| Adenovirus 40/41 | SEASON = SUMMER, FALL | 5 | 750 | 755 | 1 | 99 | − | 99 | 89 |
| Adenovirus 40/41 | SEASON = SPRING AND DIS_RUS ≤2.29 m3 · s−1 | 22 | 103 | 125 | 18 | 82 | + | 18 | |
| Adenovirus 40/41 | SEASON = SPRING AND DIS_RUS >2.29 m3 · s−1 | 1 | 151 | 152 | 1 | 99 | − | 99 | |
| Adenovirus | RUS_MEANTEMP ≤12.65°C | 4 | 413 | 417 | 1 | 99 | − | 99 | 50 |
| Adenovirus | RUS_MEANTEMP >12.65°C AND ORP ≤105.15 mV | 0 | 84 | 84 | 0 | 100 | − | 100 | |
| Adenovirus | RUS_MEANTEMP >12.65°C AND ORP >105.15 mV | 18 | 513 | 531 | 3 | 97 | + | 3 | |
| Kobuvirus | NITRATE ≤1.42 mg · liter−1 | 7 | 35 | 42 | 17 | 83 | + | 17 | 50 |
| Kobuvirus | NITRATE >1.42 mg · liter−1 AND TEMP ≤13.20°C | 1 | 46 | 47 | 2 | 98 | − | 98 | |
| Kobuvirus | NITRATE >1.42 mg · liter−1 AND TEMP >13.20°C | 8 | 26 | 34 | 24 | 76 | + | 24 |
+, classed as a detection group; −, classed as a nondetection group.
DISCUSSION
Detection and densities of F-specific coliphage and viruses in water and environmental/land use associations.
The detection of viruses in surface water varies greatly in the literature (54, 57). Payment and Locas summarize a large 20-year assessment of somatic and F-specific RNA coliphage and a suite of enteric viruses primarily in groundwater in Canada (58). Other studies focusing on surface water, such as those by Baggi et al. (59), Hot et al. (60), Skraber et al. (18), Westrell et al. (61), Espinosa et al. (62), Lodder et al. (63), and Viau et al. (64), examined collocated enteric virus and somatic and/or F-specific coliphage occurrence in fresh surface waters within a variety of impacted and lower-impacted regions throughout the world. But, to our knowledge, studies evaluating this broad suite of viral and F-specific coliphage genogroup targets over multiple years and surface waters from multiple (sub)watersheds have not been conducted for mixed-use predominantly agricultural regions. Untangling sources of fecal pollution in agro-ecosystems impacted by mixes of wildlife, livestock, and human fecal pollution is a rapidly emerging source water protection issue, and in parallel with new advances in molecular detection methods in combination with geospatial and statistical/data mining techniques, this work helps provide a Canadian baseline for future surveillance initiatives within an agricultural context.
F-DNA and F-RNA coliphages in this study were detected in 33 and 28% of the water samples, respectively. Wolf et al. found that 100% of river water samples likely impacted by both human and animal contamination were positive for F-RNA coliphage, with maximum numbers similar to those found herein (65). In this study, F-RNA coliphage GI was the most frequent and most abundant genogroup detected, which was expected, given that the samples were predominantly from agriculturally and wildlife-impacted surface waters (12). Moreover, increased rates of survival for F-RNA coliphage GI compared to those of other F-RNA coliphage subgroups at different temperatures may contribute to a greater prevalence or persistence of F-RNA coliphage GI in surface water (17, 30, 33, 66, 67). The stream systems studied had varied depth and substrates, which will contribute to there being a range in surface water temperatures affecting both persistence and viability. The lack of detection of F-RNA coliphage GIV in surface water found here is also in agreement with other studies (17, 65, 66).
Human and animal F-RNA coliphage detection levels were relatively low at site 22, an agricultural stream at the end of a cattle pasture area with a protected riparian buffer (55). Interestingly, this location has historically received highly transient fecal pollution inputs from leaking septic systems from homes immediately upstream, and from a relative framework, enhanced inputs of wildlife fecal matter resulting from riparian zone protection (e.g., Canada goose Bacteroidales markers [68]). Animal F-RNA was detected in relatively high quantities while human F-RNA was detected in relatively low quantities at site 24, a forested catchment area with no known upstream impact by humans or agriculture. This site did have some of the highest levels of detection of human and Canada goose Bacteroidales markers; however, as observed by Marti et al., the reasons for these observations are not fully clear (68). Frey et al. observed the presence of Campylobacter, Salmonella, and ruminant/bovine Bacteroidales markers in runoff from sediments in the area of study, especially around site 10 (impacted by pasturing cattle and direct cattle access to water) (56). In this study, at site 10, we observed some of the highest levels of detection of animal F-RNA coliphage, which is consistent with the Frey et al. ruminant/bovine marker observations. Site 12, located on a tributary with mixed land uses, had the greatest any virus detection, with associated detection levels of ∼20% human F-RNA and ∼78% animal F-RNA. For this site, few human, no pig, ∼20% ruminant, ∼7% muskrat, and no goose Bacteroidales markers were observed (68). The results of the site virus, F-specific coliphage, and source detections discussed here indicate that sources of viruses/coliphages are complex and vary in both time and space transiently; moreover, they are not always coherent and consistent with detections associated with other biomarkers and pathogens.
The detection results obtained in this study show that 55% of the samples analyzed had at least one virus detected, suggesting that there is a need to carry out more-refined microbial risk assessments (10). Human and animal TTV viruses were most frequently detected in 20% and 23% of the surface water samples, respectively. This is not surprising, since the TTV virus is highly prevalent in humans, yet its link with disease is not well defined. Some authors have reported that TTV could be part of the normal flora for both humans and animals (making it a potential source tracking marker, which would help explain its high prevalence in certain populations and in water observed in this study [69, 70]). Some recent studies have suggested that human TTV virus could be involved in cases of gastroenteritis and that TTVs could be responsible for pathologies in swine as well, particularly in coinfection with circovirus type 2 (50, 71–73). The presence of human TTV in water has been frequently observed by others. A recent study reported that human TTV was found in 38 to 100% of wastewaters (74).
Levels of detection of the main human-pathogenic viruses in the surface waters from this study varied from 2% for HAV and HEV up to 12% for astrovirus. Astrovirus is responsible for a considerable proportion of gastroenteritis cases in young children, the elderly, and the immunosuppressed (75–77). NoV GII, which is considered to be an etiologic agent frequently associated with cases of gastroenteritis, was found in only 5% of water samples, perhaps reflecting the agricultural impact in the watershed (as reflected also in the somewhat higher level of detection of NoV GIII, at ∼11%). Overall, the persistence of NoV in the environment, particularly in water, has often been observed, but there is in tandem high variability in terms of both prevalence and concentrations in surface and waste waters (78–80).
In this study, there were seasonal associations with the detection of F-DNA coliphages, where detection was highest in summer (when animal activity in the basin is greatest). Animal and human F-RNA coliphages were associated with the fall season. Cole et al. reported that the prevalence of F-DNA coliphages was significantly higher during the warmer months and early fall (17). A seasonal effect was reported for the survival of F-RNA coliphage in surface waters in California (67). F-DNA and F-RNA coliphages can die off faster at temperatures of 20°C or warmer than at 10°C or lower (27, 67). The presence or absence of a seasonal effect will be influenced by the climatic region in which the sample was taken (as well as the water body in terms of substrate and depth; see reference 81), as systemically higher maximum water temperatures in warmer climates may not be typical or sustained in a more northern climate. While overall, F-specific colipages were found in this study to be most strongly linked to seasonal attributes, the greatest number of human F-RNA coliphage detections were found via CART data mining analyses to be associated with both lower relative mean air temperature (i.e., cooler seasons) and higher relative degrees of developed land. This was the only CART-based F-specific coliphage finding to classify data on the basis of an urban/rural land development variable, with a 76% weighted classification accuracy. The CART result appears to be coherent with potential septic leakages that can occur in the area, as well as wastewater treatment plant effluents, which are typically discharged under higher-river-flow conditions in the cooler spring and fall seasons (6). Somewhat like human F-RNA coliphage, human virus associations with environment/land use indicated that most detections occurred where urban/rural development was relatively higher.
Environmental occurrences of and infections by NoV, AdV, RV, and astrovirus have been associated with cooler seasons (45, 82–84). In this work, we observed increased levels of detection of NoV GI, GII, and GIV in summer, while GIII was highest in spring, which could be related to agricultural practices, such as bovine manure applications on fields. Rotavirus was rarely observed, and astrovirus detections showed no seasonality.
The highest levels of animal virus detection were associated with higher relative water flow conditions and relatively higher daily rainfall, suggesting more spatially uniform hydrologically driven inputs (e.g., surface runoff, tile drainage, and in-stream mobilization of antecedent waters/sediment/stream detritus [55, 85]). HEV detection was also linked to relatively higher daily rainfalls, as well as higher relative water electrical conductivity values (i.e., salts). Wilkes et al. documented livestock Cryptosporidium and higher odds of bacterial pathogen occurrence in fall when stream/river discharge and nitrate (a salt that will increase electrical conductivity in water) concentrations in water were relatively higher (12). TTV was bimodal in detection with respect to relative river discharge (which might be suggestive of systemic hydrologically driven inputs from land to stream) and intrinsic inputs from wildlife/urban under nonflushing conditions (i.e., low-flow conditions where contaminants can accumulate).
Overall, seasonality, expressed through air and water temperature and season variables, was extremely important for delineating the detection of viruses and F-specific coliphage in water. Seasonality in pathogen/indicator prevalence in the study region is linked to seasonal application of livestock manures to land (spring and fall) and subsequent movement of fecal pollution to stream, wastewater treatment plant (WWTP) lagoon dumping (spring and fall) in the South Nation River proper, wildlife fecal inputs into water courses, and wetter fall conditions promoting water course fecal pollution (13, 86).
F-specific coliphage–virus relationships.
Associations between F-specific coliphage and viruses vary from modestly strong to negative (e.g., see references 21, 54, and 87). In this study, significant positive associations were found between the presence of F-DNA coliphages and human TTV, animal TTV, and any TTV. As TTV was the most frequently detected virus in the any virus group, the positive associations of F-DNA coliphages with the combined virus grouping were significantly influenced by TTV. In contrast, the presence of AdV 40/41, kobuvirus, astrovirus, human TTV, and any TTV (which includes human TTV as a subset) were associated with the absence of F-specific coliphage (F-RNA and F-DNA combined). The significance and number of the negative associations among the detection of viable F-specific coliphage and molecularly detected viruses were not entirely unexpected. Negative correlations might indicate that the viruses and F-specific coliphages had different fecal pollution sources and environmental reservoirs (54). Another factor could be that F-specific coliphages were detected by plaque assay and the viruses detected by molecular methods. Enteric viral genomes protected by a protein viral capsid can persist longer than some other infectious particles (42, 53). This also has bearing regarding differences in environmental persistence of the two microbial targets. Wu et al. reported that correlations between indicators and pathogens were strongest for conventional detection methods (odds ratio [OR], 2.36, positive association) and weakest for molecular detection methods (OR, 0.40) (21). In addition to the differences in the detection of infectious particles versus genomes, the absence of detection of F-specific coliphages and presence of other viruses may also be due to relative differences in viral and coliphage loads detected in the water samples. Fecal contamination from a single animal may also not be easily detected, since not all animals in a herd, for example, will carry and shed F-specific coliphages at equal levels; hence, inputs may be overwhelmingly diluted once entering the broader surface water environment (22, 88).
In our study, relatively small volumes of water were concentrated for the detection of viruses, while in many other surface water studies, larger volumes of water, up to 10 to 1,000 liters, are not uncommon (22, 65, 89). In the absence of significant fecal pollution inputs, the levels of F-specific coliphage may be well below the limit of detection. In such cases, perhaps an enrichment method for the detection of F-specific coliphages would provide more valuable information than direct enumeration (87). When Ballester et al. retested archived seawater sample concentrates prepared from large volumes (>100 liters of water) for the presence of coliphages, the rate of detection increased from 8% to 58% using a two-step enrichment method (87). However, with respect to sampling and laboratory management, processing larger volumes of water can be logistically cumbersome and would preclude, potentially, the identification of viral contamination in surface waters captured via the smaller-volume approaches defined in the current study, where over 1,150 samples from 24 surface water sampling sites were processed and analyzed. Such virus molecular detection approaches can be viewed as a preliminary screening for identifying where and when more-intensive monitoring or microbial risk assessments (e.g., viability) need to be performed, and potentially where coliphage could be used as an indicator of viral pollution in water.
Virus versus virus associations.
In this study, modestly strong associations were observed between the detection of astroviruses versus any virus, norovirus GIII versus any virus, animal and human TTV and any virus, as well as kobuvirus versus any virus. These virus groups were the most frequently detected viruses in the water samples. NoV GIII is associated with bovines, while kobuviruses are found in animals and humans (90, 91). Associations were not observed between TTV and kobuvirus. Information on correlations between TTV, kobuvirus, or NoV GIII and other pathogenic viruses is scarce (43). Diniz-Mendes et al. reported a lack of correlation between TTV and HAV in streams in a setting where both viruses were endemic (45).
TTV is being considered a suitable indicator of viral contamination in surface and drinking water broadly because it chronically infects humans; some studies have shown it is relatively abundant throughout the seasons, relative to human NoV and human AdV (for example, see references 45, 46, and 92–94). In this study, TTV was found to be positively associated with F-DNA coliphage and relatively abundant seasonally compared to other viruses.
Nevertheless, TTV results vary geographically, with reported human carriage rates as low as 10% to as high as 96% depending on the region/country (43, 47, 95, 96). In some cases, low concentrations of TTV DNA in sample water suggest that TTV may not necessarily be suitable as a single indicator of viral contamination of surface water (47). The limitations of TTV as a potential indicator virus were also raised by Verani et al. due to its low prevalence (97).
NoV GIII is associated with bovines, and very little information is currently available about its prevalence in Canadian cattle herds. However, the virus does seem to have the ability to transport and persist in the environment (98, 99), and in the South Nation River basin, where dairy livestock operations predominate, NoV GIII was observed in many surface water samples. NoV GIII had many significant positive associations with other viruses and virus groups. Hence, its potential as a virus indicator is promising. To our knowledge, the studies by Wilkes et al. (4, 39) are some of the first to report NoV GIII in an agriculture-dominated watershed setting.
Conclusion.
This study was one of the first surveillance attempts to assess associations and detections among viable F-DNA and F-RNA coliphage and a suite of molecularly detected viruses in surface waters of a mixed-use but predominantly agricultural region in Canada. Viable F-DNA coliphage was found to be a reasonable indicator of molecularly detected TTV. Stronger positive associations and codetection were found between F-DNA coliphage and TTV than between F-RNA coliphage and TTV. This has not been previously reported, to our knowledge. F-RNA coliphage, unlike F-DNA coliphage, was a reasonable predictor of NoV and RV, however. There was a large number of F-specific coliphage-versus-virus relationships that were negative, which suppresses the broad utility of F-specific coliphage as an indicator of virus occurrences in raw surface water, as documented in this study. Water sample size was relatively small to accommodate logistical constraints of this broader surveillance initiative, and therefore, we may have underestimated the presence of viruses in some circumstances where abundance may have been naturally lower. F-specific coliphages were also present at low levels in surface water samples, suggesting that alternate testing methods, such as a presence/absence approach using an enrichment method, may improve the frequency of detection. Notwithstanding, molecular methods for the detection of viruses in water surveillance initiatives, such as this, are desirable from a logistical and cost perspective; moreover, they provide for the detection of a vast array of viral targets to define fecal pollution sources and pollution hot spots. It can be seen in this way as a functional first-screening approach in terms of where to focus surface water sampling for future assessments of viability and for quantitative microbial risk assessments (10).
It was identified that F-DNA coliphage densities of >142 PFU · 100 ml−1 delineated conditions when ∼95% of the water samples contained some type of detected virus. This F-DNA coliphage density could be tested as a threshold value for virus detection in other surface waters to evaluate linkages with, for example, recreational virus exposure risks (10).
Generally speaking, the most important variables associated with virus and F-specific coliphage occurrence in surface water were season/temperature and surface water discharge. These factors are direct and indirect drivers of fecally derived pollution in this river basin. Seasonality is potentially linked to factors, such as wildlife activity, municipal wastewater lagoon discharge, septic leakages, land application of manure, and soil and hydrological conditions that promote off-field and instream transport processes. However, it was found that where human intervention was relatively lower, levels of detection of human viruses and human F-RNA coliphage were relatively lower (e.g., forest stream and small agricultural drainage systems hydrologically disconnected from human fecal pollution sources), and where human intervention was relatively more intensive, levels of detections of human viruses and human F-RNA coliphage were relatively higher.
MATERIALS AND METHODS
Study site and sample collection.
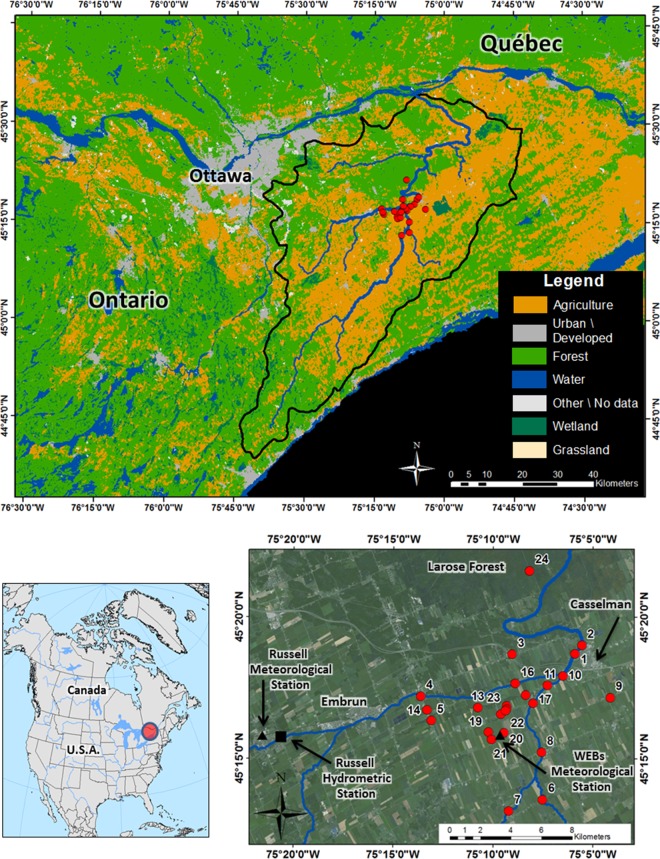
Water samples were collected in the South Nation River basin, in eastern Ontario, Canada, from spring 2008 to fall 2010 and spring 2013 to fall 2014 (Fig. 1). General basin land use is given in Table 2, where agriculture (dairy-based activities, including livestock cropping and land applications of manure in spring and fall), urban, forest, and wetlands occupied, respectively, 0 to 90, 0 to 5, 9 to 100, and 0 to 4% of the upstream contributing areas of water sample sites (using 2013 data). See two studies by Wilkes et al. for further details on the site characteristics and land use (13, 86). The maximum and minimum temperatures observed during the study period were 35°C (26 May 2010) and −35°C (17 December 2013). Total annual rainfall for the study period years ranged between 680 mm (2009) and 804 mm (2014) (climate data for Agriculture and Agri-Food Canada's WEBs station measured with Hobo weather station [Onset Computer Corporation, Bourne, MA] [Fig. 1]).
FIG 1.
Map of study site. Top, land cover of the South Nation river basin year 2013. This panel was adapted from that in reference 103. Bottom right, the location of the water sample sites. Bottom left, location of study area in North America.
Water sampling occurred on a biweekly basis at sites 1 to 24 (Fig. 1 and Table 2). A total of 1,155 site visits were made during this period, where a distinct 1 liter of sample water was collected for the molecular detection of viruses, and a separate 1 liter of sample water was collected for quantification of viable F-specific coliphages. Water samples were shipped overnight on ice to Agriculture and Agri-Food Canada's (AAFC's) research center in Lacombe, Alberta, Canada, for characterization of F-RNA and F-DNA coliphage, and to AAFC's research center in Saint-Hyacinthe, Quebec, Canada, for molecular detection of enteric viruses.
F-RNA and F-DNA coliphages.
Isolates were selected as described in reference 55. The RNA from confirmed F-RNA coliphage isolates was obtained from F-RNA coliphage suspensions that were thawed and diluted 1:50 in DNase/RNase-free water, boiled for 5 min, held on ice for 2 min, and then centrifuged at 14,000 × g for 10 s. The RNA extracts of F-RNA coliphages were genotyped into genogroups I through IV by real-time reverse transcription-PCR (RT-PCR). Real-time RT-PCRs were carried out with a QuantiTect multiplex no-ROX RT-PCR kit (Qiagen, Inc., Mississauga, Ontario, Canada) on a Stratagene MX3005P quantitative PCR (qPCR) thermocycler (Agilent), using conditions as described by Jones et al. (100). Genogroups I and IV were detected by a duplex assay using LV1 and GIV primers and probes, as described by Jones et al. (100), and genogroups II and III were detected in individual reactions using the primers and probes as described in Wolf et al. (65). Each 25-μl reaction mixture contained 200 nM each forward and reverse primers and probe, 0.25 μl of QuantiTect multiplex RT mixture, 0.03 μM of ROX reference dye, and 2.5 μl of RNA extract in 1× QuantiTect multiplex RT-PCR mastermix. RNA extracts that were positive for GI or GIV were assigned to F-RNA coliphage of animal origin, and those that were positive for GII or GIII were assigned to F-RNA coliphage of human origin for each water sample (taking into consideration that source definitions may not be absolute). The detection limits for viable phages were 5 liter−1, and the genome copies by RT-PCR were 120 liter−1.
Viruses.
Water samples were processed for the detection of HAV, HEV, astrovirus, NoV GI, GII, GIII, and GIV, RV, human TTV, animal TTV, AdV 40/41, general AdV, and kobuvirus. The samples were processed as described in reference 6. Feline calicivirus was added to each sample of water before any manipulation as an internal process control to monitor the concentration and extraction processes and to evaluate the presence of inhibitors; negative controls were also employed (6). For each detected virus, standard curves were made to estimate the amount of viral genomes, as described by Wilkes et al. (6).
For kobuvirus, which we describe in detail here (not described in the previous methods), the cDNA synthesis was carried out using SuperScript III (Invitrogen), according to the manufacturer's recommendations. Briefly, a 20-μl final volume containing 10 μl of RNA extract, 10 mM deoxynucleotide triphosphate (dNTP), 4 μl of 5× first-strand buffer, 10 pmol of the antisense primer, 40 U of RNaseOUT, and 200 U of SuperScript III. The reverse transcription was performed at 55°C for 1 h. PCRs were performed in a total volume of 50 μl using the HotStar Taq Plus master mix 2× kit (Qiagen), according to the manufacturer's recommendations, in an Mastercycler gradient PCR system (Eppendorf, Mississauga, Ontario, Canada). cDNA (10 μl) was used as the template. The PCR primers Univ-KOBU-F and Univ-KOBU-R were used at a final concentration of 500 nM each (90). They were designed for the conserved RNA-dependent RNA polymerase (RdRp) gene of kobuviruses and amplify a 216-bp-long PCR fragment. The PCR was conducted under the following conditions: 1 cycle at 95°C for 15 min and 40 cycles of 95°C for 30 s, 51°C for 90 s, and 72°C for 45 s, followed by a final elongation step of 72°C for 10 min. The amplified products were separated on a 2% agarose gel with amplicons visualized with ethidium bromide staining.
For this study, broad groupings of viruses were created based on their generalized host association (again taking into consideration that assignations may not be absolute) for microbial source tracking purposes (4). For defining animal virus, the grouped viruses were norovirus GII, norovirus GIII, animal TTV, kobuvirus, RV, and HEV. For human virus, the grouped viruses were HAV, astrovirus, norovirus GI, norovirus GII, norovirus GIV, RV, human TTV, AdV 40/41, and general AdV.
Statistical treatment.
Fisher's exact tests were used to examine the significance of contingency among the detection/nondetection of F-specific coliphage and detection nondetection of singular viruses (groups) (2 by 2 table) (R version 3.2.2; The Foundation for Statistical Computing, Vienna, Austria). For these same comparisons, Phi coefficients were calculated in R to discriminate the strength of the association between the detection/nondetection of viruses and the detection/nondetection of F-specific coliphage. A Phi coefficient of 1 is indicative of a strong positive correlation between categorical data, a Phi coefficient of −0.3 to 0.3 is indicative of no effective correlation, and a Phi coefficient of −1 is indicative of a strong negative correlation between categorical data. Fisher's exact tests were also used to test for seasonal significance among the singular virus (virus groups) and F-specific coliphage using a 3 by 2 (3 season and detection/nondetection) contingency table approach (R version 3.2.2, fisher.test function of stats package [101, 102]). To determine if quantitative F-specific coliphage distributions were significantly different among detection and nondetection groups of viruses, nonparametric Mann-Whitney U rank sum tests, using continuity corrections, were employed (Statistica version 12; StatSoft, Inc., Tulsa, OK). For all of the previous statistical treatments of the data, significance was defined as a P value of ≤0.05.
Data mining was conducted to explore associations among the detection/nondetection of microbiological targets (dependent variables) and a suite of independent land use (proportions of developed land, pasture and forage land, and agriculture land), physiographic (Strahler stream order [104]), and hydrological variables (e.g., stream flow) (Table S1). Classification and regression tree (CART) (Salford Predictive Modeler version 7.0 64-bit; Salford Systems, San Diego, CA) data mining was employed in classification tree mode, according to approaches outlined in reference 12. Also, we used CART in classification mode to evaluate threshold F-DNA densities associated with the detection and nondetection of viral targets. For CART analyses, we only present cross-validated tree models generated to a maximum of two levels (maximum of four classification groups) (4, 13).
Supplementary Material
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02763-16.
REFERENCES
- 1.Fong TT, Lipp EK. 2005. Enteric viruses of humans and animals in aquatic environments: health risks, detection, and potential water quality assessment tools. Microbiol Mol Biol Rev 69:357–371. doi: 10.1128/MMBR.69.2.357-371.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Irving LG, Smith FA. 1981. One-year survey of enteroviruses, adenoviruses, and reoviruses isolated from effluent at an activated-sludge purification plant. Appl Environ Microbiol 41:51–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Symonds EM, Griffin DW, Breitbart M. 2009. Eukaryotic viruses in wastewater samples from the United States. Appl Environ Microbiol 75:1402–1409. doi: 10.1128/AEM.01899-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilkes G, Brassard J, Edge TA, Gannon V, Gottschall N, Jokinen CC, Jones TH, Khan IU, Marti R, Sunohara MD, Topp E, Lapen DR. 2014. Long-term monitoring of waterborne pathogens and microbial source tracking markers in paired agricultural watersheds under controlled and conventional tile drainage management. Appl Environ Microbiol 80:3708–3720. doi: 10.1128/AEM.00254-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haack SK, Duris JW, Kolpin DW, Fogarty LR, Johnson HE, Gibson KE, Focazio M, Schwab KJ, Hubbard LE, Foreman WT. 2015. Genes indicative of zoonotic and swine pathogens are persistent in stream water and sediment following a swine manure spill. Appl Environ Microbiol 81:3430–3441. doi: 10.1128/AEM.04195-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilkes G, Brassard J, Edge TA, Gannon V, Jokinen CC, Jones TH, Neumann N, Pintar KD, Ruecker N, Schmidt PJ, Sunohara M, Topp E, Lapen DR. 2013. Bacteria, viruses, and parasites in an intermittent stream protected from and exposed to pasturing cattle: prevalence, densities, and quantitative microbial risk assessment. Water Res 47:6244–6257. doi: 10.1016/j.watres.2013.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeBorde DC, Woessner WW, Lauerman B, Ball PN. 1998. Virus occurrence and transport in a school septic system and unconfined aquifer. Ground Water 36:825–834. doi: 10.1111/j.1745-6584.1998.tb02201.x. [DOI] [Google Scholar]
- 8.Borchardt MA, Bradbury KR, Alexander EC, Kolberg RJ, Alexander SC, Archer JR, Braatz LA, Forest BM, Green JA, Spencer SK. 2011. Norovirus outbreak caused by a new septic system in a dolomite aquifer. Ground Water 49:85–97. doi: 10.1111/j.1745-6584.2010.00686.x. [DOI] [PubMed] [Google Scholar]
- 9.Yates MV. 2007. Classical indicators in the 21st century–far and beyond the coliform. Water Environ Res 79:279–286. doi: 10.2175/106143006X123085. [DOI] [PubMed] [Google Scholar]
- 10.Schmidt PJ. 2015. Norovirus dose-response: are currently available data informative enough to determine how susceptible humans are to infection from a single virus? Risk Anal 35:1364–1383. doi: 10.1111/risa.12323. [DOI] [PubMed] [Google Scholar]
- 11.Bernhard AE, Field KG. 2000. Identification of nonpoint sources of fecal pollution in coastal waters by using host-specific 16S ribosomal DNA genetic markers from fecal anaerobes. Appl Environ Microbiol 66:1587–1594. doi: 10.1128/AEM.66.4.1587-1594.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilkes G, Ruecker NJ, Neumann NF, Gannon VP, Jokinen C, Sunohara M, Topp E, Pintar KD, Edge TA, Lapen DR. 2013. Spatiotemporal analysis of Cryptosporidium species/genotypes and relationships with other zoonotic pathogens in surface water from mixed-use watersheds. Appl Environ Microbiol 79:434–448. doi: 10.1128/AEM.01924-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilkes G, Edge TA, Gannon VP, Jokinen C, Lyautey E, Neumann NF, Ruecker N, Scott A, Sunohara M, Topp E, Lapen DR. 2011. Associations among pathogenic bacteria, parasites, and environmental and land use factors in multiple mixed-use watersheds. Water Res 45:5807–5825. doi: 10.1016/j.watres.2011.06.021. [DOI] [PubMed] [Google Scholar]
- 14.Keeley A, Faulkner BR. 2008. Influence of land use and watershed characteristics on protozoa contamination in a potential drinking water resources reservoir. Water Res 42:2803–2813. doi: 10.1016/j.watres.2008.02.028. [DOI] [PubMed] [Google Scholar]
- 15.Pusch D, Oh D-Y, Wolf S, Dumke R, Schröter-Bobsin U, Höhne M, Röske I, Schreier E. 2005. Detection of enteric viruses and bacterial indicators in German environmental waters. Arch Virol 150:929–947. doi: 10.1007/s00705-004-0467-8. [DOI] [PubMed] [Google Scholar]
- 16.Allwood PB, Malik YS, Hedberg CW, Goyal SM. 2003. Survival of F-specific RNA coliphage, feline calicivirus, and Escherichia coli in water: a comparative study. Appl Environ Microbiol 69:5707–5710. doi: 10.1128/AEM.69.9.5707-5710.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cole D, Long SC, Sobsey MD. 2003. Evaluation of F+ RNA and DNA coliphages as source-specific indicators of fecal contamination in surface waters. Appl Environ Microbiol 69:6507–6514. doi: 10.1128/AEM.69.11.6507-6514.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Skraber S, Gassilloud B, Gantzer C. 2004. Comparison of coliforms and coliphages as tools for assessment of viral contamination in river water. Appl Environ Microbiol 70:3644–3649. doi: 10.1128/AEM.70.6.3644-3649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin J, Ganesh A. 2013. Water quality indicators: bacteria, coliphages, enteric viruses. Int J Environ Health Res 23:484–506. doi: 10.1080/09603123.2013.769201. [DOI] [PubMed] [Google Scholar]
- 20.Savichtcheva O, Okabe S. 2006. Alternative indicators of fecal pollution: relations with pathogens and conventional indicators, current methodologies for direct pathogen monitoring and future application perspectives. Water Res 40:2463–2476. doi: 10.1016/j.watres.2006.04.040. [DOI] [PubMed] [Google Scholar]
- 21.Wu J, Long SC, Das D, Dorner SM. 2011. Are microbial indicators and pathogens correlated? A statistical analysis of 40 years of research. J Water Health 9:265–278. [DOI] [PubMed] [Google Scholar]
- 22.Havelaar AH, van Olphen M, Drost YC. 1993. F-specific RNA bacteriophages are adequate model organisms for enteric viruses in fresh water. Appl Environ Microbiol 59:2956–2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grabow W. 2004. Bacteriophages: update on application as models for viruses in water. Water SA 27:251–268. [Google Scholar]
- 24.Vinjé J, Oudejans SJ, Stewart JR, Sobsey MD, Long SC. 2004. Molecular detection and genotyping of male-specific coliphages by reverse transcription-PCR and reverse line blot hybridization. Appl Environ Microbiol 70:5996–6004. doi: 10.1128/AEM.70.10.5996-6004.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Havelaar AH, Furuse K, Hogeboom WM. 1986. Bacteriophages and indicator bacteria in human and animal faeces. J Appl Bacteriol 60:255–262. doi: 10.1111/j.1365-2672.1986.tb01081.x. [DOI] [PubMed] [Google Scholar]
- 26.Hsu F-C, Shieh YSC, Van Duin J, Beekwilder MJ, Sobsey MD. 1995. Genotyping male-specific RNA coliphages by hybridization with oligonucleotide probes. Appl Environ Microbiol 61:3960–3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Long SC, Sobsey MD. 2004. A comparison of the survival of F+RNA and F+DNA coliphages in lake water microcosms. J Water Health 2:15–22. [PubMed] [Google Scholar]
- 28.Furuse K. 1987. Distribution of coliphages in the environment: general considerations, vol 87 John Wiley & Sons, New York, NY. [Google Scholar]
- 29.Bollback JP, Huelsenbeck JP. 2001. Phylogeny, genome evolution, and host specificity of single-stranded RNA bacteriophage (family Leviviridae). J Mol Evol 52:117–128. doi: 10.1007/s002390010140. [DOI] [PubMed] [Google Scholar]
- 30.Schaper M, Jofre J, Uys M, Grabow WOK. 2002. Distribution of genotypes of F-specific RNA bacteriophages in human and non-human sources of faecal pollution in South Africa and Spain. J Appl Microbiol 92:657–667. doi: 10.1046/j.1365-2672.2002.01600.x. [DOI] [PubMed] [Google Scholar]
- 31.Scott TM, Rose JB, Jenkins TM, Farrah SR, Lukasik J. 2002. Microbial source tracking: current methodology and future directions. Appl Environ Microbiol 68:5796–5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sundram A, Jumanlal N, Ehlers MM. 2006. Genotyping of F-RNA coliphages isolated from wastewater and river water samples. Water SA 32:65–70. [Google Scholar]
- 33.Brion GM, Meschke JS, Sobsey MD. 2002. F-specific RNA coliphages: occurrence, types, and survival in natural waters. Water Res 36:2419–2425. doi: 10.1016/S0043-1354(01)00547-4. [DOI] [PubMed] [Google Scholar]
- 34.Yang Y, Griffiths MW. 2013. Comparative persistence of subgroups of F-specific RNA phages in river water. Appl Environ Microbiol 79:4564–4567. doi: 10.1128/AEM.00612-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Calci KR, Burkhardt W III, Watkins WD, Rippey SR. 1998. Occurrence of male-specific bacteriophage in feral and domestic animal wastes, human feces, and human-associated wastewaters. Appl Environ Microbiol 64:5027–5029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Havelaar AH, Pot-Hogeboom WM, Furuse K, Pot R, Hormann MP. 1990. F-specific RNA bacteriophages and sensitive host strains in faeces and wastewater of human and animal origin. J Appl Bacteriol 69:30–37. doi: 10.1111/j.1365-2672.1990.tb02908.x. [DOI] [PubMed] [Google Scholar]
- 37.Jones TH, Johns MW. 2009. Improved detection of F-specific RNA coliphages in fecal material by extraction and polyethylene glycol precipitation. Appl Environ Microbiol 75:6142–6146. doi: 10.1128/AEM.00436-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gironés R, Bofill-Mas S, Cook N. 2013. Virus indicators for food and water, p 483–509. In Cook N. (ed), Viruses in food and water: risks, surveillance and control. Woodhead Publishing Ltd., Cambridge, United Kingdom. [Google Scholar]
- 39.Wilkes G, Brassard J, Edge T, Gannon V, Jokinen C, Jones TH, Marti R, Neumann N, Ruecker N, Sunohara M, Topp E, Lapen DR. 2013. Coherence among different microbial source tracking markers in a small agricultural stream with and without livestock exclusion practices. Appl Environ Microbiol 79:6207–6219. doi: 10.1128/AEM.01626-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang SC. 2006. Human adenoviruses in water: occurrence and health implications: a critical review. Environ Sci Technol 40:7132–7140. doi: 10.1021/es060892o. [DOI] [PubMed] [Google Scholar]
- 41.Albinana-Gimenez N, Miagostovich MP, Calgua B, Huguet JM, Matia L, Girones R. 2009. Analysis of adenoviruses and polyomaviruses quantified by qPCR as indicators of water quality in source and drinking-water treatment plants. Water Res 43:2011–2019. doi: 10.1016/j.watres.2009.01.025. [DOI] [PubMed] [Google Scholar]
- 42.Choi S, Jiang SC. 2005. Real-time PCR quantification of human adenoviruses in urban rivers indicates genome prevalence but low infectivity. Appl Environ Microbiol 71:7426–7433. doi: 10.1128/AEM.71.11.7426-7433.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haramoto E, Kitajima M, Katayama H, Ohgaki S. 2010. Real-time PCR detection of adenoviruses, polyomaviruses, and torque teno viruses in river water in Japan. Water Res 44:1747–1752. doi: 10.1016/j.watres.2009.11.043. [DOI] [PubMed] [Google Scholar]
- 44.Brassard J, Gagné M-J, Lamoureux L, Inglis G, Leblanc D, Houde A. 2008. Molecular detection of bovine and porcine Torque teno virus in plasma and feces. Vet Microbiol 126:271–276. doi: 10.1016/j.vetmic.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 45.Diniz-Mendes L, de Paula VS, Luz SLB, Niel C. 2008. High prevalence of human Torque teno virus in streams crossing the city of Manaus, Brazilian Amazon. J Appl Microbiol 105:51–58. doi: 10.1111/j.1365-2672.2007.03720.x. [DOI] [PubMed] [Google Scholar]
- 46.Griffin JS, Plummer JD, Long SC. 2008. Torque teno virus: an improved indicator for viral pathogens in drinking waters. Virol J 5:112. doi: 10.1186/1743-422X-5-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hamza IA, Jurzik L, Uberla K, Wilhelm M. 2011. Evaluation of pepper mild mottle virus, human picobirnavirus and Torque teno virus as indicators of fecal contamination in river water. Water Res 45:1358–1368. doi: 10.1016/j.watres.2010.10.021. [DOI] [PubMed] [Google Scholar]
- 48.Bendinelli M, Pistello M, Maggi F, Fornai C, Freer G, Vatteroni ML. 2001. Molecular properties, biology, and clinical implications of TT virus, a recently identified widespread infectious agent of humans. Clin Microbiol Rev 14:98–113. doi: 10.1128/CMR.14.1.98-113.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Irshad M, Joshi YK, Sharma Y, Dhar I. 2006. Transfusion transmitted virus: a review on its molecular characteristics and role in medicine. World J Gastroenterol 12:5122–5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brassard J, Gagne MJ, Leblanc D, Poitras E, Houde A, Boras VF, Inglis GD. 2015. Association of age and gender with Torque teno virus detection in stools from diarrheic and non-diarrheic people. J Clin Virol 72:55–59. doi: 10.1016/j.jcv.2015.08.020. [DOI] [PubMed] [Google Scholar]
- 51.Carducci A, Morici P, Pizzi F, Battistini R, Rovini E, Verani M. 2008. Study of the viral removal efficiency in a urban wastewater treatment plant. Water Sci Technol 58:893–897. doi: 10.2166/wst.2008.437. [DOI] [PubMed] [Google Scholar]
- 52.Haramoto E, Katayama H, Ohgaki S. 2008. Quantification and genotyping of Torque teno virus at a wastewater treatment plant in Japan. Appl Environ Microbiol AEM 1 74:7434–7436. doi: 10.1128/AEM.01605-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ogorzaly L, Tissier A, Bertrand I, Maul A, Gantzer C. 2009. Relationship between F-specific RNA phage genogroups, faecal pollution indicators and human adenoviruses in river water. Water Res 43:1257–1264. doi: 10.1016/j.watres.2008.12.011. [DOI] [PubMed] [Google Scholar]
- 54.USEPA. 2015. Review of coliphages as possible indicators of fecal contamination for ambient water quality. 820-R-15-098. Office of Water, U.S. Environmental Protection Agency, Washington, DC. [Google Scholar]
- 55.Sunohara MD, Topp E, Wilkes G, Gottschall N, Neumann N, Ruecker N, Jones TH, Edge TA, Marti R, Lapen DR. 2012. Impact of riparian zone protection from cattle on nutrient, bacteria, F5 coliphage, Cryptosporidium, and Giardia loading of an intermittent stream. J Environ Qual 41:1301–1314. doi: 10.2134/jeq2011.0407. [DOI] [PubMed] [Google Scholar]
- 56.Frey SK, Gottschall N, Wilkes G, Gregoire DS, Topp E, Pintar KD, Sunohara M, Marti R, Lapen DR. 2015. Rainfall-induced runoff from exposed streambed sediments: an important source of water pollution. J Environ Qual 44:236–247. doi: 10.2134/jeq2014.03.0122. [DOI] [PubMed] [Google Scholar]
- 57.Tran NH, Gin KY, Ngo HH. 2015. Fecal pollution source tracking toolbox for identification, evaluation and characterization of fecal contamination in receiving urban surface waters and groundwater. Sci Total Environ 538:38–57. doi: 10.1016/j.scitotenv.2015.07.155. [DOI] [PubMed] [Google Scholar]
- 58.Payment P, Locas A. 2011. Pathogens in water: value and limits of correlation with microbial indicators. Ground Water 49:4–11. doi: 10.1111/j.1745-6584.2010.00710.x. [DOI] [PubMed] [Google Scholar]
- 59.Baggi F, Demarta A, Peduzzi R. 2001. Persistence of viral pathogens and bacteriophages during sewage treatment: lack of correlation with indicator bacteria. Res Microbiol 152:743–751. doi: 10.1016/S0923-2508(01)01255-4. [DOI] [PubMed] [Google Scholar]
- 60.Hot D, Legeay O, Jacques J, Gantzer C, Caudrelier Y, Guyard K, Lange M, Andreoletti L. 2003. Detection of somatic phages, infectious enteroviruses and enterovirus genomes as indicators of human enteric viral pollution in surface water. Water Res 37:4703–4710. doi: 10.1016/S0043-1354(03)00439-1. [DOI] [PubMed] [Google Scholar]
- 61.Westrell T, Teunis P, van den Berg H, Lodder W, Ketelaars H, Stenström TA, de Roda Husman AM. 2006. Short-and long-term variations of norovirus concentrations in the Meuse River during a 2-year study period. Water Res 40:2613–2620. doi: 10.1016/j.watres.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 62.Espinosa AC, Arias CF, Sánchez-Colón S, Mazari-Hiriart M. 2009. Comparative study of enteric viruses, coliphages and indicator bacteria for evaluating water quality in a tropical high-altitude system. Environ Health 8:1. doi: 10.1186/1476-069X-8-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lodder W, van den Berg H, Rutjes S, de Roda Husman A. 2010. Presence of enteric viruses in source waters for drinking water production in The Netherlands. Appl Environ Microbiol 76:5965–5971. doi: 10.1128/AEM.00245-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Viau EJ, Lee D, Boehm AB. 2011. Swimmer risk of gastrointestinal illness from exposure to tropical coastal waters impacted by terrestrial dry-weather runoff. Environ Sci Technol 45:7158–7165. doi: 10.1021/es200984b. [DOI] [PubMed] [Google Scholar]
- 65.Wolf S, Hewitt J, Rivera-Aban M, Greening GE. 2008. Detection and characterization of F+ RNA bacteriophages in water and shellfish: application of a multiplex real-time reverse transcription PCR. J Virol Methods 149:123–128. doi: 10.1016/j.jviromet.2007.12.012. [DOI] [PubMed] [Google Scholar]
- 66.Hartard C, Rivet R, Banas S, Gantzer C. 2015. Occurrence of and sequence variation among F-specific RNA bacteriophage subgroups in feces and wastewater of urban and animal origins. Appl Environ Microbiol 81:6505–6515. doi: 10.1128/AEM.01905-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ravva SV, Sarreal CZ. 2016. Persistence of F-specific RNA coliphages in surface waters from a produce production region along the central coast of California. PLoS One 11:e0146623. doi: 10.1371/journal.pone.0146623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Marti R, Gannon VP, Jokinen C, Lanthier M, Lapen DR, Neumann NF, Ruecker NJ, Scott A, Wilkes G, Zhang Y, Topp E. 2013. Quantitative multi-year elucidation of fecal sources of waterborne pathogen contamination in the South Nation River basin using Bacteroidales microbial source tracking markers. Water Res 47:2315–2324. doi: 10.1016/j.watres.2013.02.009. [DOI] [PubMed] [Google Scholar]
- 69.Virgin HW, Wherry EJ, Ahmed R. 2009. Redefining chronic viral infection. Cell 138:30–50. doi: 10.1016/j.cell.2009.06.036. [DOI] [PubMed] [Google Scholar]
- 70.Simmonds P, Prescott L, Logue C, Davidson F, Thomas A, Ludlam C. 1999. TT virus—part of the normal human flora? J Infect Dis 180:1748–1749. doi: 10.1086/315103. [DOI] [PubMed] [Google Scholar]
- 71.Lee Y, Lin C-M, Jeng C-R, Chang H-W, Chang C-C, Pang VF. 2015. The pathogenic role of torque teno sus virus 1 and 2 and their correlations with various viral pathogens and host immunocytes in wasting pigs. Vet Microbiol 180:186–195. doi: 10.1016/j.vetmic.2015.08.027. [DOI] [PubMed] [Google Scholar]
- 72.Liu J, Guo L, Zhang L, Wei Y, Huang L, Wu H, Liu C. 2013. Three new emerging subgroups of Torque teno sus viruses (TTSuVs) and co-infection of TTSuVs with porcine circovirus type 2 in China. Virol J 10:189. doi: 10.1186/1743-422X-10-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kekarainen T, Sibila M, Segales J. 2006. Prevalence of swine Torque teno virus in post-weaning multisystemic wasting syndrome (PMWS)-affected and non-PMWS-affected pigs in Spain. J Gen Virol 87:833–837. doi: 10.1099/vir.0.81586-0. [DOI] [PubMed] [Google Scholar]
- 74.Charest A, Plummer J, Long S, Carducci A, Verani M, Sidhu J. 2015. Global occurrence of Torque teno virus in water systems. J Water Health 13:777–789. doi: 10.2166/wh.2015.254. [DOI] [PubMed] [Google Scholar]
- 75.Bosch A, Pinto RM, Guix S. 2014. Human astroviruses. Clin Microbiol Rev 27:1048–1074. doi: 10.1128/CMR.00013-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Coppo P, Scieux C, Ferchal F, Clauvel J, Lassoued K. 2000. Astrovirus enteritis in a chronic lymphocytic leukemia patient treated with fludarabine monophosphate. Ann Hematol 79:43–45. doi: 10.1007/s002770050008. [DOI] [PubMed] [Google Scholar]
- 77.Jarchow-Macdonald AA, Halley S, Chandler D, Gunson R, Shepherd SJ, Parcell BJ. 2015. First report of an astrovirus type 5 gastroenteritis outbreak in a residential elderly care home identified by sequencing. J Clin Virol 73:115–119. doi: 10.1016/j.jcv.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 78.Petterson SR, Stenström TA, Ottoson J. 2016. A theoretical approach to using faecal indicator data to model norovirus concentration in surface water for QMRA: Glomma River, Norway. Water Res 91:31–37. doi: 10.1016/j.watres.2015.12.037. [DOI] [PubMed] [Google Scholar]
- 79.Pérez-Sautu U, Sano D, Guix S, Kasimir G, Pinto RM, Bosch A. 2012. Human norovirus occurrence and diversity in the Llobregat River catchment, Spain. Environ Microbiol 14:494–502. doi: 10.1111/j.1462-2920.2011.02642.x. [DOI] [PubMed] [Google Scholar]
- 80.Nordgren J, Matussek A, Mattsson A, Svensson L, Lindgren PE. 2009. Prevalence of norovirus and factors influencing virus concentrations during one year in a full-scale wastewater treatment plant. Water Res 43:1117–1125. doi: 10.1016/j.watres.2008.11.053. [DOI] [PubMed] [Google Scholar]
- 81.Sinokrot BA, Stefan HG. 1993. Stream temperature dynamics: measurements and modeling. Water Resour Res 29:2299–2312. doi: 10.1029/93WR00540. [DOI] [Google Scholar]
- 82.Mounts AW, Ando T, Koopmans M, Bresee JS, Noel J, Glass RI. 2000. Cold weather seasonality of gastroenteritis associated with Norwalk-like viruses. J Infect Dis 181(Suppl 2):S284–S287. [DOI] [PubMed] [Google Scholar]
- 83.Dey SK, Hoq I, Okitsu S, Hayakawa S, Ushijima H. 2013. Prevalence, seasonality, and peak age of infection of enteric adenoviruses in Japan, 1995–2009. Epidemiol Infect 141:958–960. doi: 10.1017/S0950268812001586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Arena C, Amoros JP, Vaillant V, Ambert-Balay K, Chikhi-Brachet R, Jourdan-Da Silva N, Varesi L, Arrighi J, Souty C, Blanchon T, Falchi A, Hanslik T. 2014. Acute diarrhea in adults consulting a general practitioner in France during winter: incidence, clinical characteristics, management and risk factors. BMC Infect Dis 14:574. doi: 10.1186/s12879-014-0574-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Corsi SR, Borchardt M, Spencer S, Hughes PE, Baldwin AK. 2014. Human and bovine viruses in the Milwaukee River watershed: hydrologically relevant representation and relations with environmental variables. Sci Total Environ 490:849–860. doi: 10.1016/j.scitotenv.2014.05.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wilkes G, Edge T, Gannon V, Jokinen C, Lyautey E, Medeiros D, Neumann N, Ruecker N, Topp E, Lapen DR. 2009. Seasonal relationships among indicator bacteria, pathogenic bacteria, Cryptosporidium oocysts, Giardia cysts, and hydrological indices for surface waters within an agricultural landscape. Water Res 43:2209–2223. doi: 10.1016/j.watres.2009.01.033. [DOI] [PubMed] [Google Scholar]
- 87.Ballester NA, Fontaine JH, Margolin AB. 2005. Occurrence and correlations between coliphages and anthropogenic viruses in the Massachusetts Bay using enrichment and ICC-nPCR. J Water Health 3:59–68. [PubMed] [Google Scholar]
- 88.Long SC, El-Khoury SS, Oudejans SJ, Sobsey MD, Vinjé J. 2005. Assessment of sources and diversity of male-specific coliphages for source tracking. Environ Eng Sci 22:367–377. doi: 10.1089/ees.2005.22.367. [DOI] [Google Scholar]
- 89.Jiang S, Noble R, Chu W. 2001. Human adenoviruses and coliphages in urban runoff-impacted coastal waters of Southern California. Appl Environ Microbiol 67:179–184. doi: 10.1128/AEM.67.1.179-184.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Reuter G, Boldizsar A, Pankovics P. 2009. Complete nucleotide and amino acid sequences and genetic organization of porcine kobuvirus, a member of a new species in the genus Kobuvirus, family Picornaviridae. Arch Virol 154:101–108. doi: 10.1007/s00705-008-0288-2. [DOI] [PubMed] [Google Scholar]
- 91.Di Felice E, Mauroy A, Dal Pozzo F, Thiry D, Ceci C, Di Martino B, Marsilio F, Thiry E. 2016. Bovine noroviruses: a missing component of calf diarrhoea diagnosis. Vet J 207:53–62. doi: 10.1016/j.tvjl.2015.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bibby K, Viau E, Peccia J. 2011. Viral metagenome analysis to guide human pathogen monitoring in environmental samples. Lett Appl Microbiol 52:386–392. doi: 10.1111/j.1472-765X.2011.03014.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Haramoto E, Katayama H, Oguma K, Koibuchi Y, Furumai H, Ohgaki S. 2006. Effects of rainfall on the occurrence of human adenoviruses, total coliforms, and Escherichia coli in seawater. Water Sci Technol 54:225–230. doi: 10.2166/wst.2006.473. [DOI] [PubMed] [Google Scholar]
- 94.Haramoto E, Katayama H, Oguma K, Yamashita H, Tajima A, Nakajima H, Ohgaki S. 2006. Seasonal profiles of human noroviruses and indicator bacteria in a wastewater treatment plant in Tokyo, Japan. Water Sci Technol 54:301–308. doi: 10.2166/wst.2006.888. [DOI] [PubMed] [Google Scholar]
- 95.Abe K, Inami T, Asano K, Miyoshi C, Masaki N, Hayashi S, Ishikawa K-i, Takebe Y, Win KM, El-Zayadi AR, Han KH, Zhang DY. 1999. TT virus infection is widespread in the general populations from different geographic regions. J Clin Microbiol 37:2703–2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Desai SM, Muerhoff AS, Leary TP, Erker JC, Simons JN, Chalmers ML, Birkenmeyer LG, Pilot-Matias TJ, Mushahwar IK. 1999. Prevalence of TT virus infection in US blood donors and populations at risk for acquiring parenterally transmitted viruses. J Infect Dis 179:1242–1244. doi: 10.1086/314735. [DOI] [PubMed] [Google Scholar]
- 97.Verani M, Casini B, Battistini R, Pizzi F, Rovini E, Carducci A. 2006. One-year monthly monitoring of Torque teno virus (TTV) in river water in Italy. Water Sci Technol 54:191–195. doi: 10.2166/wst.2006.468. [DOI] [PubMed] [Google Scholar]
- 98.Costantini V, Loisy F, Joens L, Le Guyader FS, Saif LJ. 2006. Human and animal enteric caliciviruses in oysters from different coastal regions of the United States. Appl Environ Microbiol 72:1800–1809. doi: 10.1128/AEM.72.3.1800-1809.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zakhour M, Maalouf H, Di Bartolo I, Haugarreau L, Le Guyader FS, Ruvoën-Clouet N, Le Saux J-C, Ruggeri FM, Pommepuy M, Le Pendu J. 2010. Bovine norovirus: carbohydrate ligand, environmental contamination, and potential cross-species transmission via oysters. Appl Environ Microbiol 76:6404–6411. doi: 10.1128/AEM.00671-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jones T, Houde A, Poitras E, Ward P, Johns M. 2009. Development and evaluation of a multiplexed real-time TaqMan RT-PCR assay with a sample process control for detection of F-specific RNA coliphage genogroups I and IV. Food Environ Virol 1:57–65. doi: 10.1007/s12560-009-9008-7. [DOI] [Google Scholar]
- 101.Mehta CR, Patel NR. 1986. Algorithm 643: FEXACT: a FORTRAN subroutine for Fisher's exact test on unordered r×c contingency tables. ACM Trans Math Softw 12:154–161. doi: 10.1145/6497.214326. [DOI] [Google Scholar]
- 102.Clarkson DB, Fan Y-A, Joe H. 1993. A remark on algorithm 643: FEXACT: an algorithm for performing Fisher's exact test in r×c contingency tables. ACM Trans Math Softw 19:484–488. doi: 10.1145/168173.168412. [DOI] [Google Scholar]
- 103.Lapen DR, Schmidt PJ, Thomas JL, Edge TA, Flemming C, Keithlin J, Neumann N, Pollari F, Ruecker N, Simhon A, Topp E, Wilkes G, Pintar KDM. 2016. Towards a more accurate quantitative assessment of seasonal Cryptosporidium infection risks in surface waters using species and genotype information. Water Res 105:625–637. doi: 10.1016/j.watres.2016.08.023. [DOI] [PubMed] [Google Scholar]
- 104.Strahler AN. 1952. Hypsometric (area-altitude) analysis of erosional topography. Geol Soc Am Bull 63:1117–1142. doi: 10.1130/0016-7606(1952)63[1117:HAAOET]2.0.CO;2. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.