ABSTRACT
Understanding the mechanisms behind the typicity of regional wines inevitably brings attention to microorganisms associated with their production. Oenococcus oeni is the main bacterial species involved in wine and cider making. It develops after the yeast-driven alcoholic fermentation and performs the malolactic fermentation, which improves the taste and aromatic complexity of most wines. Here, we have evaluated the diversity and specificity of O. oeni strains in six regions. A total of 235 wines and ciders were collected during spontaneous malolactic fermentations and used to isolate 3,212 bacterial colonies. They were typed by multilocus variable analysis, which disclosed a total of 514 O. oeni strains. Their phylogenetic relationships were evaluated by a second typing method based on single nucleotide polymorphism (SNP) analysis. Taken together, the results indicate that each region holds a high diversity of strains that constitute a unique population. However, strains present in each region belong to diverse phylogenetic groups, and the same groups can be detected in different regions, indicating that strains are not genetically adapted to regions. In contrast, greater strain identity was seen for cider, white wine, or red wine of Burgundy, suggesting that genetic adaptation to these products occurred.
IMPORTANCE This study reports the isolation, genotyping, and geographic distribution analysis of the largest collection of O. oeni strains performed to date. It reveals that there is very high diversity of strains in each region, the majority of them being detected in a single region. The study also reports the development of an SNP genotyping method that is useful for analyzing the distribution of O. oeni phylogroups. The results show that strains are not genetically adapted to regions but to specific types of wines. They reveal new phylogroups of strains, particularly two phylogroups associated with white wines and red wines of Burgundy. Taken together, the results shed light on the diversity and specificity of wild strains of O. oeni, which is crucial for understanding their real contribution to the unique properties of wines.
KEYWORDS: Oenococcus oeni, biogeography, wine, lactic acid bacteria
INTRODUCTION
The biogeography of microbial populations aims to understand the diversity of microorganisms at the local, regional, continental, and environmental scales, in order to understand their distribution and factors that contribute to it (1, 2). Some microorganisms have ubiquitous distribution, while others present specific biogeographic patterns, which are more influenced by environmental differences between habitats and separations due to geographical barriers rather than to geographical distances (1, 3–6). Biogeography studies have a particular implication in oenology since they address the concept of “terroir,” which refers to a geographic area characterized mainly by its climate, soil, and human factors that contribute to produce typical wines. The main question is whether the microorganisms of soil, grapes, and wine can be associated with particular regions and considered a component of the terroir that contributes to the typicality of wine. From a microbial perspective, this has come to mean the presence of genetically unique geographically distinct populations of organisms.
A complex microbial consortium is associated with grapes and wine; it includes molds, yeasts, and bacteria. The main contributors and best-known species involved in the alcoholic fermentation (AF) and malolactic fermentation (MLF) are the yeast Saccharomyces cerevisiae and the lactic acid bacteria Oenococcus oeni, respectively. It has been recently shown that the fungal and bacterial grape microbiotas are influenced by the vineyard environmental conditions, suggesting that there is a nonrandom microbial fingerprint associated with terroir (7, 8). Ecological studies based on global sampling of S. cerevisiae from diverse origins suggest that different strain populations are associated with different foods and beverages such as wine, spirits, beer, or bread, while geographic origin explains only 28% of variability (9, 10). In contrast, larger sample sizes from fewer locations provide evidence for a regional delineation of S. cerevisiae populations associated with vines and the wines produced from them (11). A direct correlation was established between the origin of yeasts that conduct AF in New Zealand and the chemical composition of wines, suggesting that microbial populations are important for the regional identity of wine (12).
Contrary to S. cerevisiae, little is known about the biogeography of O. oeni. The species was first described in 1967 (13) and reclassified in 1995 (14). It is a fastidious bacterium that is rarely detected in the environment and requires a rich medium for growth. In contrast to its environmental scarcity, O. oeni is the best-adapted species to develop in wine and cider, thanks to its tolerance to low pH and ethanol. While numerous lactic acid bacteria (LAB) species are present in grape must, O. oeni becomes generally the only detectable species after AF (15). During MLF, it reduces the acid taste of wine by converting malic acid into lactic acid and carbon dioxide. O. oeni also produces or degrades a variety of aromatic compounds, thus contributing to the aromatic complexity of wine (15–19). Numerous studies based on various molecular methods have revealed that there is a huge diversity of strains performing MLF in wine (20–22). Strain diversity is important not only in regions but also in wine estates: up to 10 different genotypes have been detected simultaneously during MLF (21, 23–25), with one or more genotypes being predominant during all or part of MLF (21, 26). Moreover, strains can persist in a cellar over several consecutive vintages (21).
Population structure analyses based on multilocus sequence typing (MLST) of strains from diverse wines or ciders and geographic origins have revealed that the O. oeni species consist of two major genetic groups of strains, named A and B, and possibly a third group, C (27, 28). All group A strains were isolated from wine, while group B strains were isolated from wine and cider. Interestingly, some strains from specific products or geographic areas such as Champagne, Chile, and South Africa formed distinct subgroups (28). Comparative genomics has confirmed the distribution of strains in groups A, B, and C and also revealed genetic properties that can be linked with adaptation to wine, such as exopolysaccharide biosynthesis, sugar and amino acid transport, and metabolism (29–32). It is suggested that O. oeni strains were domesticated to cider and wine, with some strains possibly being further domesticated to specific wines such as champagne (31).
Recent studies based on small numbers of strains collected in a few regions have shown that regional strains may belong to different genetic groups (A and B) and are able to ferment local wines more or less efficiently (33–36). Here, with the aim of investigating the biogeography of O. oeni, we have analyzed 2,997 O. oeni isolates from 235 wines and ciders produced in five French regions and in Lebanon. O. oeni strains were genotyped by multilocus variable-number tandem-repeat analysis (MLVA) as recently reported (37) to get insight on the diversity of the species over the regions, wines, and ciders that were studied. In order to assign them to the genetic groups A or B, we have developed a strategy based on single nucleotide polymorphism (SNP) genotyping using the Sequenom MassArray iPLEX platform (38). The results shed light on the huge diversity of this important enological species and its dissemination over regional fermented products.
RESULTS
O. oeni strain collection.
O. oeni strains were isolated from 201 red wines and 25 white wines collected during MLF in 69 wineries of the Aquitaine, Burgundy, Val de Loire, and Languedoc-Roussillon regions of France as well as several locations in Lebanon. Nine ciders from Brittany were also analyzed in order to include cider strains in the panel. As expected, LAB populations were around 2 × 107 CFU/ml in most samples, with lower levels (about 5 × 106 CFU/ml) in ciders and Burgundy wines. To investigate the diversity of the O. oeni strains that predominate during MLF, 10 to 15 colonies per sample were analyzed in agreement with previous works (24, 34). The total sampling included 3,212 bacterial isolates. To our knowledge this is the largest sampling of wine bacteria reported to date. A PCR screening identified 2,997 colonies (93.3%) as O. oeni isolates, confirming that it is the best-adapted species for MLF in wine and cider (Table 1). Non-O. oeni isolates were detected in all regions, but mainly in ciders and in Burgundy wines, in which they accounted for 23% and 7.5% of all isolates, respectively. They were identified by 16S rRNA gene sequencing. In Burgundy, it was mainly Pediococcus damnosus, a species often detected in wine and associated with the spoilage known as the “ropy” character (39). In cider, the main species detected were Zymomonas mobilis and Lactobacillus paracollinoides, which are frequently reported in this product (40, 41). The 2,997 O. oeni isolates were further analyzed by MLVA according to Claisse and Lonvaud-Funel (37). This revealed 514 different genotypes, which were considered 514 different strains: 489 from wines and 25 from ciders (Table 1).
TABLE 1.
Collection of O. oeni strains isolated from wines and ciders
| Region | No. of samples | No. of LAB isolates | No. of O. oeni isolates | No. of complete MLVA genotypes | No. of incomplete MLVA genotypes | No. of O. oeni strainsa |
|---|---|---|---|---|---|---|
| Aquitaine | 80 | 1,125 | 1,072 | 912 | 160 | 200 |
| Burgundy | 70 | 895 | 837 | 631 | 206 | 142 |
| Languedoc-Roussillon | 36 | 534 | 514 | 379 | 135 | 134 |
| Val de Loire | 8 | 120 | 117 | 91 | 26 | 29 |
| Lebanon | 32 | 403 | 353 | 339 | 14 | 57 |
| Brittany (cider) | 9 | 135 | 104 | 59 | 45 | 25 |
| Total | 235 | 3,212 | 2,997 | 2,411 | 586 | 514b |
Each variable-number tandem-repeat profile was considered to represent a different strain.
Strains present in different regions are counted only once in the total.
Relative abundance of isolates and strains.
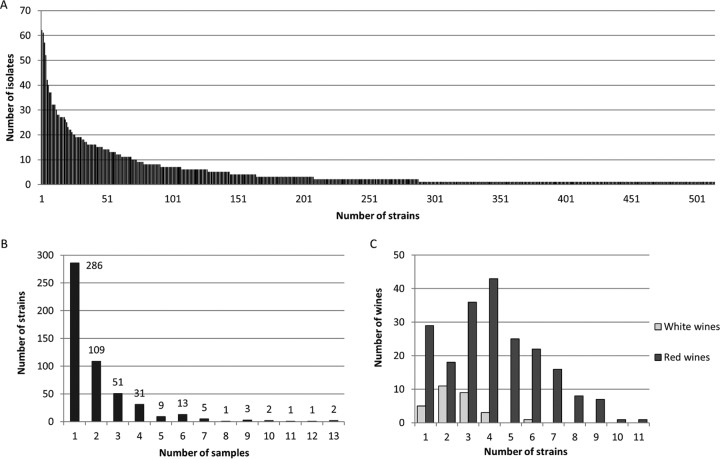
Figure 1 shows the number of isolates obtained for each strain. It reveals that the majority of strains (306, i.e., 59.6%) were isolated only once or twice (Fig. 1A). Only 19 isolates (3.7%) were counted more than 25 times and up to 62 times for the most frequent. The same trend was observed in each region (data not shown). In addition, the isolates of the same strain were generally obtained from only one or two samples and rarely from more than three (Fig. 1B). This suggests that there is a huge diversity of O. oeni strains in each region and that no strain is predominant. This conclusion is also supported by the relatively high number of strains identified in each sample, although there was a slight difference between red and white wines: there were 1 to 11 strains in red wines (average, 4.23) and 1 to 4 in white wines (average, 2.46) (Fig. 1C). In addition, some isolates carrying the same MLVA genotypes as those of three commercial starters were detected: one isolate of the two strains CiNe and L31 in two red wines of Lebanon and 25 isolates of strain Lalvin VP41 in six wines of Aquitaine and Burgundy (not shown). This was unexpected because the wines that were analyzed were not inoculated with commercial starters to perform MLF, but it is possible that the starters were used in the wineries during previous vintages. However, this limited number of starters further confirms that there is not wide dissemination of commercial strains in the wine environment.
FIG 1.
Frequency of isolates and strains of O. oeni. The distribution of 2,997 isolates and 514 strains of O. oeni was examined to determine the number of isolates obtained from each strain (A), the number of samples in which a same strain was detected (B), and the number of strains detected in each sample of white or red wine (C).
Diversity of strains in regions, wines, and ciders.
When we looked at the distribution of strains, there was a clear distinction between ciders and wines. While 228 strains were detected at least in 2 and up to 13 different samples, none was detected in both cider and wine (Fig. 2A). It is unlikely that this situation results from a geographical separation because ciders were collected just a few dozen kilometers from the wine region Val de Loire. In contrast, it is possible that strains are better adapted to either cider or wine. Similarly, the vast majority of wine strains were detected in either red or white wine but not in both, suggesting that strains are better adapted to a single type of wine (Fig. 2A). The same trend was observed for rosé wine strains, although it only concerns very few strains.
FIG 2.
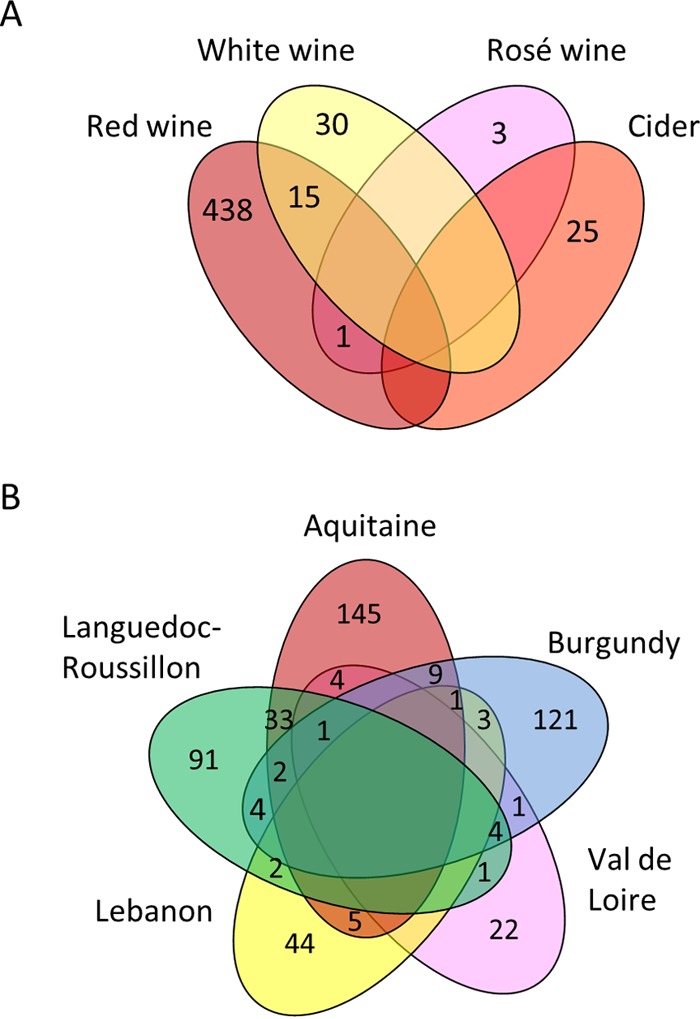
Venn diagrams denoting the numbers of unique and shared strains in different wines and ciders (A) and wine-production regions (B).
According to Table 2, 89% of the wine strains (435 strains in total) were detected in only one region. Figure 2B shows their detailed regional distribution. Besides the 435 “region-specific strains,” 62 strains were detected in two regions, 3 in three regions, and only 1 in four regions. Aquitaine and Languedoc-Roussillon share together a total of 36 strains and many fewer with Burgundy, although a similar sampling was performed in all three regions and they are almost equally distant. In addition, the geographic distance does not appear as a hindrance to the spread of strains since 11 of the 57 strains isolated from Lebanon were also detected in at least one of the French regions.
TABLE 2.
O. oeni populations in each region
| Region | No. of strains | Region-specific strains (no. [%])a | Estimated maximum no. of strainsb | Shannon diversity index | Pielou diversity index |
|---|---|---|---|---|---|
| Aquitaine | 200 | 150 (75) | 410 | 4.57 | 0.86 |
| Burgundy | 142 | 124 (87.3) | 300 | 4.19 | 0.84 |
| Languedoc-Roussillon | 134 | 93 (69.4) | 350 | 4.28 | 0.87 |
| Val de Loire | 29 | 22 (75.8) | NDc | ND | ND |
| Lebanon | 57 | 46 (80.7) | 123 | 3.17 | 0.82 |
| Brittany (cider) | 25 | 25 (100) | ND | ND | ND |
Strains detected in only one region.
Determined using EstimateS with 95% upper and lower limits (46).
ND, not determined.
The population diversity in each region was estimated by rarefaction analyses and diversity indexes (Table 2). Comparable populations were found in Aquitaine, Languedoc-Roussillon, and Burgundy, with an estimated maximum number of strains on the order of several hundreds. The Pielou and Shannon diversity indexes were close to 1 and 4.5, respectively, for all three regions. These are considered high values in environmental studies and suggest that the populations are complex, with no or few dominating strains. This also confirms the quality of samplings carried out in those regions, since it appears that the maximum diversity was almost reached. A quite different situation was observed in Lebanon, where the maximum population was about three times lower and where the values of diversity indexes were lower, indicating a less homogeneous population with more dominant strains. Not enough samples were collected from the Val de Loire and Brittany regions to perform reliable diversity analyses.
Development of a genotyping method based on SNP analysis.
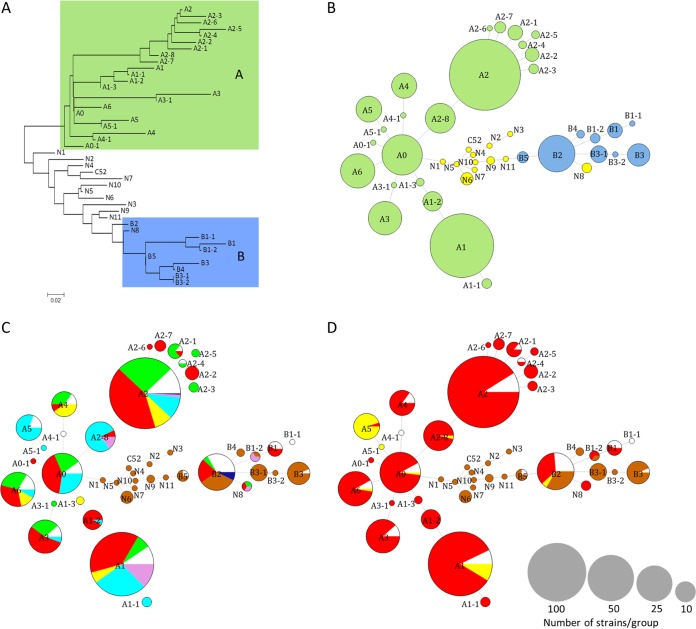
We have developed a genotyping method based on SNP analysis using the Sequenom MassArray iPLEX platform (38) in order to classify the strains according to their genetic group (A or B) or subgroups, A phylogenetic tree based on the 50 O. oeni genomes available in databases was used to delineate 11 groups of phylogenetically related strains (see Fig. S1 in the supplemental material). They were named groups A and B, according to previous studies (27, 31), and subgroups A1 to A6 and B1 to B3 (Fig. S1). A comparative genomic analysis revealed 11 to 1,695 SNPs specific for each of the 11 groups (see Table S1 in the supplemental material). A total of 40 SNPs were manually selected, with 2 to 6 SNPs specific for each group of strains, except for subgroup B2 for which no SNP was retained (see Materials and Methods). Concatenation of the 40 selected SNPs generated 11 sequence types (STs), corresponding to each of the 10 groups, plus strain C52 (group N), which does not belong to either group A or group B (28, 31) (see Table S2 in the supplemental material).
The 40 SNPs were determined for each of the 514 strains isolated in this work and for 63 “control” strains characterized in a previous study (28). SNP data analysis revealed that 466 of the 577 strains possessed SNP combinations corresponding to the 11 predefined STs (Table S2). The remaining 111 strains (19.2%) had variant SNP combinations corresponding to 32 new STs (Table S2). Ninety-three of them differ from the 11 predefined STs at only one or two SNP positions and were attributed to new subgroups A or B (Fig. 3A). The 18 other strains (15 from cider and 3 from wines of Aquitaine, Val de Loire, and Languedoc-Roussillon) showed hybrid combinations of SNPs that were attributed to the group N or newly defined subgroups N1 to N11, along with the previously described strain C52 (28). A tree based on the alignment of all 43 STs showed that these 18 new STs group together, apart from the STs of group A and B strains (Fig. 3A).
FIG 3.
Distribution of strains in phylogroups. (A) A neighbor-joining tree was constructed using the 43 different concatenated sequences of SNP identified by analyzing 577 O. oeni strains. (B to D) Minimum spanning trees represent the distribution of strains in the genetic groups and subgroups and are colorized according to their groups of affiliation (B), their region of origin (C), and their wine or cider of origin (D). The size of the circles is proportional to the number of strains belonging to the phylogroup: maximum 148 for A2 and minimum 1 for the smallest.
Distribution of strains in phylogroups.
To analyze the distribution of the 577 strains, their STs were used to construct minimum spanning trees (MSTs). Figure 3B shows that the vast majority of strains (466/577, 84.2%) belong to group A and only 12.6% (73/577) belong to group B. This distribution is in agreement with previous reports (27, 28). Subgroups A2 and A1 are by far the largest. They contain, respectively, 148 and 116 strains, which represents 45.7% of all strains. It is likely that they contain strains that could be separated into various subgroups, but the SNPs analyzed in this study are not sufficiently informative for this.
When looking at the distribution of strains according to their region of origin, it appeared that each region contained strains from different subgroups (Fig. 3C). For example, strains of Aquitaine belong to 16 subgroups spanning both groups A and B. This also implies that most of the subgroups of significant size (more than 5 strains) contained strains from different regions. Interestingly subgroups A5 and A2-8 contain, respectively, 17 and 28 strains that come almost exclusively from Burgundy. These results show that all regions were colonized by strains of different genetic origins, and there are few or no genetic groups that are specific to a particular region.
The distribution of strains according to the beverage from which they were isolated (red wine, white wine, or cider) shows a quite different picture (Fig. 3D). First, all cider strains are found in the subgroups B and N, which separates them from almost all wine strains. Only the subgroup B2 contains a combination of wine and cider strains (38 in total), but it is possible that analyzing SNPs different from the 40 selected would separate them. Second, white wine strains were distributed in very few subgroups (mainly A5 and A1). Although many fewer white wine than red wine strains were analyzed (25 and 464, respectively), this low dispersion suggests that strains found in white wines actually have unique genetic characteristics. This is particularly evident for group A5, which contains a large majority of strains from white wines of Burgundy (17/21) and four other strains isolated for white wine of Champagne (control strains). Interestingly, another group consists almost exclusively of Burgundy strains, but only strains isolated from red wine (subgroup A2-8). It is remarkable that strains of this region form two genetic groups associated with two types of wines, i.e., red and white.
DISCUSSION
The growing interest of consumers for traditional foods produced under geographical indications has raised questions about the contribution of microorganisms to the unique properties of regional wines (42). In the context of winemaking, the key issue is to determine whether microorganisms can contribute and be considered a real component of the terroir (43). Here we have considered this issue by determining whether there are genetically unique populations of O. oeni in distinct geographic regions. This species is present in the environment along with many other LAB, but it is barely detectable in environmental samples since it is slow growing and unable to compete with other LAB except in wine and cider, thanks to its tolerance to low pH and ethanol. Its preferred environment is recreated each year during the short period starting from AF to the end of MLF. It is not certain that strains that become predominant during MLF are really representative of the diversity of the species, but at least they represent the diversity that is industrially relevant and possibly involved in the unique regional properties of the resulting wines. Our analysis of 3,212 isolates collected during MLF of 235 wines and ciders from several regions of France and Lebanon brings a new picture of the biodiversity and regional distribution of O. oeni strains.
When we look at the overall O. oeni population, whichever the region, a first conclusion is that this species population is highly diverse, with a huge number of strains. By using the MLVA genotyping method (37), we have discriminated at least 514 different genotypes among the 3,212 isolates. We have assumed that they likely represent 514 different strains because MLVA has the highest Simpson index value for discrimination (DI = 0.994) in comparison to those for the other two most popular typing methods for O. oeni: pulsed-field gel electrophoresis (DI = 0.989) and multilocus sequence typing (DI = 0.965) (44). Not only is this a high number of strains, but also the number of strains is clearly underestimated since genotyping failed for 19.5% of the isolates. In these cases, one or two of the five PCR amplicons expected using MLVA were not obtained despite several attempts, probably because the corresponding DNA regions were missing in the target genomes. This high diversity is reflected in regions. Here we have estimated that the total number of predominant strains (with populations high enough to be detected) during MLF in Aquitaine, Burgundy, and Languedoc-Roussillon is close to 400, which is already high and underestimated if we consider that only the predominant ones and those that develop well in wine have been counted. A second insight is that the O. oeni population is not dominated by a few strains since none has been detected a great number of times or in many samples. On the contrary, there is a huge diversity of strains, which are likely present in wine at more or less similar population levels. We can conclude that there is no “invasion” of the oenological environment by a few strains that would be particularly well adapted to all kinds of wines. This is of importance for winemakers because this means that the use of commercial starter strains, which are inoculated in wine at a high population level in order to manage MLF, does not constitute a risk to the biodiversity, indigenous strains remaining predominant in the environment. Another interesting insight is that the vast majority of wine strains belong to the phylogroup A. The SNP genotyping procedure that we have developed for analyzing the 514 strains was much more efficient than MLVA for typing the strains, but it has revealed their phylogenetic distribution. It has confirmed the disparity of the distribution of wine strains in phylogroups A and B (84.2% and 12.6%, respectively). This had been previously suggested by analyzing selected strains using MLST (27, 28) or comparative genomics (31, 32). An opposite situation has been observed for cider strains. They are associated with the phylogroup B, except for a single strain that forms a third phylogroup, C (28). Here, we have confirmed that there is no cider strain from the phylogroup A. Most of them belong to the phylogroup B, but, interestingly, we have also detected 18 strains that could not be attributed to either A or B. Their SNP genotypes suggest that they form at least one new phylogroup, possibly with the single group C strain reported previously (28). Does this distribution of wine strains mean that group A strains are better adapted to wine? It is possible because 14 of the 15 commercial starters, i.e., strains that are normally selected for their exceptional capacity to survive in wine and perform MLF, that have been analyzed to date belong to the phylogroup A (29, 31, 32). However, it would be inexact to consider that all group B strains are not adapted to wine. A few of them have been recently isolated from wines of South Catalonia in Spain and have proved to be efficient in performing MLF (33). It is also tempting to speculate that O. oeni had first developed in the environment on overripe fruits containing a low level of alcohol produced by contaminant yeasts and then it has become adapted to human-made environments containing increasing levels of alcohol: up to 6 to 8% in cider and 15 to 16% in wine. This domestication process has been suggested by Campbell-Sills et al. (31). A further domestication of wine strains to specific types of wines such as champagne has also been suggested. Here, this possibility is reinforced with the identification of a subgroup A5, which contains 17 strains isolated almost exclusively from white wines, predominantly from Burgundy, and a second phylogroup A2-8 with 28 strains coming almost exclusively from red wines of Burgundy. A genomic and phenotypic characterization of these strains is in progress to determine if they are actually adapted to these wines, which would be a real case of recent domestication.
When looking at the biodiversity in the regions, our results show that each region holds a vast majority of strains that are not detected in other regions: from 69.4% to 87.5% in Aquitaine, Burgundy, and Languedoc-Roussillon where the largest samplings have been made. Other authors have also reported a vast majority of “unique” strains in some regions of Portugal or Spain (26, 45). In our case, this situation is partly due to the high diversity of strains, with a large number of them (286 strains) being detected in a single sample and in a single region. However, if we consider only those strains that have been isolated in two or more samples (228 strains), the vast majority (158 strains) also come from a single region. As suggested in other studies, this can lead to the conclusion that the strains identified in a region are specific to that region. However, we assume that regional strains cannot be considered “specific” to regions because they are not “genetically” specific. In fact, our SNP genotyping has shown that each region holds a patchwork of strains that belong to different genetic groups, mainly from subgroup A but also in some cases from subgroup B. Conversely, strains that are genetically close and assigned to the same phylogroup are detected in several regions. There is no doubt that new strains and phylogroups appear first in a single region and are virtually specific to this region until they spread to other places. This is probably what happens with the A5 and A2-8 subgroups described above. It is very likely that strains from these subgroups have developed relatively recently in the two types of wines produced in Burgundy because they contain a vast majority of strains isolated from the wines of this region. However, these strains can spread in the environment and sometimes develop in wines of other regions. For instance, 4 of the 21 strains of group A5 have been isolated from champagne (31).
In conclusion, the biogeography of O. oeni provides new insights on the contribution of indigenous strains to the properties of regional wines. In agreement with previous studies suggesting that microorganisms of the vineyard (7) and wine yeasts (12) are components of the terroir, we can assume that O. oeni is also part of the terroir because each region holds a different combination of strains and a majority of unique strains. However, the strains are not genetically specific to the regions and a nonnegligible fraction of them is present in diverse regions. Therefore, it is unlikely that regional strains can shape all of the wines of a region similarly, providing them with distinguishable properties. The different strains present in each region may be responsible for the production of wines with different properties. However, at the level of each wine estate, it is possible that O. oeni strains contribute to the production of specific wines because these strains may persist in the estates for several years (21). This study has also revealed another specific situation concerning some types of wines such as the white and red wines produced in Burgundy. The discovery of the genetic groups A5 and A2.8 confirms the existence of strains which are genetically adapted to specific types of wines and probably provide them with specific properties.
MATERIALS AND METHODS
Sampling and strain collection.
The bacterial strains analyzed in this work were isolated from 235 wines and ciders collected during malolactic fermentation from 74 vineyards distributed in four major wine-producing regions of France (Aquitaine, Burgundy, Languedoc-Roussillon, and Val de Loire), different wine-producing areas of Lebanon (mainly the Beqaa Valley), and one cider-producing region (Brittany). Samplings were performed during vintages 2011 in Lebanon (32 wines); 2012 in Aquitaine (69 wines), Burgundy (59 wines), and Languedoc-Roussillon (36 wines); and 2013 in Aquitaine (11 wines), Burgundy (11 wines), Val de Loire (8 wines), and Brittany (9 ciders). All of the 514 new strains reported here were deposited in the Biological Resources Center CRB OENO (ISVV, Villenave d'Ornon, France). Representative strains are available upon request. All other bacteria used in this work were obtained from the CRB OENO.
Isolation and storage of bacterial strains and cell lysates.
Dilutions of wine and cider samples were plated on a grape juice medium containing 250 ml·liter−1 commercial red grape juice, 5 g·liter−1 yeast extract, 1 ml·liter−1 Tween 80, 15 g·liter−1 agar, and 100 mg·liter−1 pimaricin, adjusted to pH 4.8. Plates were incubated anaerobically (AnaeroGen; Oxoid) for 7 to 10 days at 25°C. Ten to 15 colonies were randomly selected from each sample and inoculated individually in 1-ml volumes of liquid grape juice medium. After 7 days of incubation, an aliquot of each culture was preserved at −80°C in 30% glycerol for subsequent isolation of bacteria. Another aliquot of 200 μl was centrifuged at 10,000 × g. for 5 min. The cell pellet was resuspended in 200 μl of sterile water, and cells were lysed by freezing at −20°C and melting at room temperature. Cell lysates were kept at −20°C until use.
MLVA genotyping.
Bacterial colonies were genotyped by multilocus variable-number tandem-repeat analysis (MLVA) specific to O. oeni. The MLVA was performed as described by Claisse and Lonvaud-Funel (37). Briefly, for each colony two multiplex PCRs were performed using 2 or 3 pairs of labeled primers in order to amplify 5 regions of tandem repeats (TRs). The two reaction mixtures of 10 μl contained 1 μl of cell lysate, 5 μl of 2× Qiagen multiplex PCR master mix, and 5 pmol of each primer targeting tandem repeat regions 1 and 2 (TR1 and TR2) for the first reaction (M1) or 2.5 pmol of TR3 and TR4 primers and 5 pmol of TR5 primers for the second reaction (M2). Both PCRs were performed under the same conditions in a thermocycler (T_100; Bio-Rad, France) with the following program: 95°C for 15 min, followed by 30 cycles of 30 s at 94°C, 90 s at 62°C, and 90 s at 72°C, and a final step of 30 min at 60°C. Then PCR products M1 and M2 were diluted 40 and 60 times, respectively, and pooled. Next, 2 μl of the mixture was added to 9 μl of highly deionized (HI-DI) formamide (Applied Biosystems) and analyzed by MWG Eurofins Operon (Cochin Institute, France).
The genotyping results were processed with GenMarker (SoftGenetics) software in which a specific MLVA panel has been incorporated in order to automatically determine the number of repetitions of each TR. The combination of the number of repetitions of TR1 to TR5 represents the digital profile of a colony. All of the MLVA profiles were then integrated in a database of BioNumerics v5.1 (Applied Maths, Belgium) software, and a number was affiliated with each different profile to facilitate the analysis. A minimum spanning tree was then calculated by ranking the variables of each TR and profile number by category (calculate minimum spanning tree, coefficient: categorical).
16S rRNA gene analysis.
Colony PCRs were performed in 20-μl reaction mixtures containing 10 pmol each of universal primers BSF8 (5′-AGAGTTTGATCCTGGCTCAG-3′) and BSR1541 (5′-AAGGAGGTGATCCAGCCGCA-3′), 4 μl of Taq 5× MasterMix (New England BioLabs), and 2 μl of cell lysate. The PCR program was 95°C for 5 min, 30 cycles of 30 s at 95°C, 30 s at 55°C, and 30 s at 72°C, and a final step of 10 min at 72°C. PCR products were sequenced on both DNA strands (MWG Operon, Ebersberg, Germany), and species identification was achieved using BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
Pielou and Shannon-Weaver diversity indexes.
Shannon-Weaver and Pielou diversity indexes and rarefaction curves were calculated using EstimateS 9.1.0 software (46). These two indexes are complementary and make it possible to assess the diversity of O. oeni strains and the evenness of their distribution across the regions studied.
Classification of strains in phylogroups using SNP genotyping.
A method for strain classification by SNP genotyping was developed to assign the newly identified strains of O. oeni to phylogenetic groups A and B and their respective subgroups previously reported in Campbell-Sills et al. (31). The 49 genomes reported by Campbell-Sills et al. (31) were mapped against the genome of strain PSU-1 and revealed a whole set of 6,120 SNPs that discriminate strains from the different subgroups of A and B strains with 100% accuracy (Table S1). A manual curation and selection process was performed to select 40 SNPs that could be amplified and sequenced by the Sequenom strategy. In order to amplify their genomic regions, we designed multiplex PCRs of primers with the software Suite 1.0 Assay Design (Sequenom Inc., San Diego, CA). Genotyping of the collected strains was performed using the iPLEX GOLD kit in the MassArray facility (Sequenom). The extension products were spotted onto a SpectroCHIP and analyzed by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF). The Sequenom analysis was performed using TYPER 4.0 (Sequenom). The assignment of alleles was done in real time using SpectroCALLER, and then the results were displayed on SpectroACQUIRE (Sequenom).
The genotyping results for the 40 SNPs for each strain were concatenated into a single sequence of 40 bp. The sequence alignments and phylogenetic analysis were performed with MEGA software 6.0.5 (47) with 1,000 bootstraps using the neighbor-joining distance calculation with the Kimura 2 parameter. The data were also included in BioNumerics v5.1 software (Applied Maths, Belgium). A similarity matrix was then calculated with the neighbor-joining clustering parameter, with a 100% open gap penalty for pairwise alignment, and an MST was built from this matrix.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to M.-C. Colosio, E. Vinsonneau, P. Cottereau, and V. Gerbaux from the Institut Français du Vin, S. Becquet from the Syndicat des Vignerons Bio d'Aquitaine, R. Bauduin from the Institut Français de Production du Cidre, and V. Pladeau from SudVinBio and many wine estates for providing wine and cider samples.
This study was supported in parts by the European Commission (FP7-SME project Wildwine, grant agreement no. 315065) and the French Ministry of Agriculture (project LevainsBio CASDAR AAP-2012 no. 1220).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02322-16.
REFERENCES
- 1.Green J, Bohannan BJM. 2006. Spatial scaling of microbial biodiversity. Trends Ecol Evol 21:501–507. doi: 10.1016/j.tree.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 2.Ramette A, Tiedje JM. 2007. Biogeography: an emerging cornerstone for understanding prokaryotic diversity, ecology, and evolution. Microb Ecol 53:197–207. doi: 10.1007/s00248-005-5010-2. [DOI] [PubMed] [Google Scholar]
- 3.Whitaker RJ, Grogan DW, Taylor JW. 2003. Geographic barriers isolate endemic populations of hyperthermophilic Archaea. Science 301:976–978. doi: 10.1126/science.1086909. [DOI] [PubMed] [Google Scholar]
- 4.Horner-Devine MC, Lage M, Hughes JB, Bohannan BJM. 2004. A taxa–area relationship for bacteria. Nature 432:750–753. doi: 10.1038/nature03073. [DOI] [PubMed] [Google Scholar]
- 5.Martiny JBH, Bohannan BJM, Brown JH, Colwell RK, Fuhrman JA, Green JL, Horner-Devine MC, Kane M, Krumins JA, Kuske CR, Morin PJ, Naeem S, Øvreås L, Reysenbach A-L, Smith VH, Staley JT. 2006. Microbial biogeography: putting microorganisms on the map. Nat Rev Microbiol 4:102–112. doi: 10.1038/nrmicro1341. [DOI] [PubMed] [Google Scholar]
- 6.Nemergut DR, Costello EK, Hamady M, Lozupone C, Jiang L, Schmidt SK, Fierer N, Townsend AR, Cleveland CC, Stanish L, Knight R. 2011. Global patterns in the biogeography of bacterial taxa. Environ Microbiol 13:135–144. doi: 10.1111/j.1462-2920.2010.02315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bokulich NA, Thorngate JH, Richardson PM, Mills DA. 2014. Microbial biogeography of wine grapes is conditioned by cultivar, vintage, and climate. Proc Natl Acad Sci U S A 111:E139–E148. doi: 10.1073/pnas.1317377110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zarraonaindia I, Owens SM, Weisenhorn P, West K, Hampton-Marcell J, Lax S, Bokulich NA, Mills DA, Martin G, Taghavi S, van der Lelie D, Gilbert JA. 2015. The soil microbiome influences grapevine-associated microbiota. mBio 6:e02527-14. doi: 10.1128/mBio.02527-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fay JC, Benavides JA. 2005. Evidence for domesticated and wild populations of Saccharomyces cerevisiae. PLoS Genet 1:e5. doi: 10.1371/journal.pgen.0010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Legras J-L, Merdinoglu D, Cornuet J-M, Karst F. 2007. Bread, beer and wine: Saccharomyces cerevisiae diversity reflects human history. Mol Ecol 16:2091–2102. doi: 10.1111/j.1365-294X.2007.03266.x. [DOI] [PubMed] [Google Scholar]
- 11.Knight S, Goddard MR. 2015. Quantifying separation and similarity in a Saccharomyces cerevisiae metapopulation. ISME J 9:361–370. doi: 10.1038/ismej.2014.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Knight S, Klaere S, Fedrizzi B, Goddard MR. 2015. Regional microbial signatures positively correlate with differential wine phenotypes: evidence for a microbial aspect to terroir. Sci Rep 5:14233. doi: 10.1038/srep14233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garvie EI. 1967. Leuconostoc oenos sp. nov. J Gen Microbiol 48:431–438. doi: 10.1099/00221287-48-3-431. [DOI] [PubMed] [Google Scholar]
- 14.Dicks LMT, Dellaglio F, Collins MD. 1995. Proposal to reclassify Leuconostoc oenos as Oenococcus oeni [corrig.] gen. nov., comb. nov. Int J Syst Bacteriol 45:395–397. doi: 10.1099/00207713-45-2-395. [DOI] [PubMed] [Google Scholar]
- 15.Fleet GH, Lafon-Lafourcade S, Ribereau-Gayon P. 1984. Evolution of yeasts and lactic acid bacteria during fermentation and storage of Bordeaux wines. Appl Environ Microbiol 48:1034–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lonvaud-Funel A. 1999. Lactic acid bacteria in the quality improvement and depreciation of wine. Antonie Van Leeuwenhoek 76:317–331. doi: 10.1023/A:1002088931106. [DOI] [PubMed] [Google Scholar]
- 17.Bae S, Fleet GH, Heard GM. 2006. Lactic acid bacteria associated with wine grapes from several Australian vineyards. J Appl Microbiol 100:712–727. doi: 10.1111/j.1365-2672.2006.02890.x. [DOI] [PubMed] [Google Scholar]
- 18.Barata A, Malfeito-Ferreira M, Loureiro V. 2012. The microbial ecology of wine grape berries. Int J Food Microbiol 153:243–259. doi: 10.1016/j.ijfoodmicro.2011.11.025. [DOI] [PubMed] [Google Scholar]
- 19.Sumby KM, Jiranek V, Grbin PR. 2013. Ester synthesis and hydrolysis in an aqueous environment, and strain specific changes during malolactic fermentation in wine with Oenococcus oeni. Food Chem 141:1673–1680. doi: 10.1016/j.foodchem.2013.03.087. [DOI] [PubMed] [Google Scholar]
- 20.Kelly WJ, Huang CM, Asmundson RV. 1993. Comparison of Leuconostoc oenos strains by pulsed-field gel electrophoresis. Appl Environ Microbiol 59:3969–3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reguant C, Bordons A. 2003. Typification of Oenococcus oeni strains by multiplex RAPD-PCR and study of population dynamics during malolactic fermentation. J Appl Microbiol 95:344–353. doi: 10.1046/j.1365-2672.2003.01985.x. [DOI] [PubMed] [Google Scholar]
- 22.Larisika M, Claus H, König H. 2008. Pulsed-field gel electrophoresis for the discrimination of Oenococcus oeni isolates from different wine-growing regions in Germany. Int J Food Microbiol 123:171–176. doi: 10.1016/j.ijfoodmicro.2007.11.081. [DOI] [PubMed] [Google Scholar]
- 23.López I, Tenorio C, Zarazaga M, Dizy M, Torres C, Ruiz-Larrea F. 2007. Evidence of mixed wild populations of Oenococcus oeni strains during wine spontaneous malolactic fermentations. Eur Food Res Technol 226:215–223. doi: 10.1007/s00217-006-0529-0. [DOI] [Google Scholar]
- 24.Cappello MS, Zapparoli G, Stefani D, Logrieco A. 2010. Molecular and biochemical diversity of Oenococcus oeni strains isolated during spontaneous malolactic fermentation of Malvasia Nera wine. Syst Appl Microbiol 33:461–467. doi: 10.1016/j.syapm.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 25.González-Arenzana L, Pérez-Martín F, Palop ML, Seseña S, Santamaría P, López R, López-Alfaro I. 2015. Genomic diversity of Oenococcus oeni populations from Castilla La Mancha and La Rioja Tempranillo red wines. Food Microbiol 49:82–94. doi: 10.1016/j.fm.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 26.González-Arenzana L, Santamaría P, López R, López-Alfaro I. 2013. Indigenous lactic acid bacteria communities in alcoholic and malolactic fermentations of Tempranillo wines elaborated in ten wineries of La Rioja (Spain). Food Res Int 50:438–445. doi: 10.1016/j.foodres.2012.11.008. [DOI] [Google Scholar]
- 27.Bilhère E, Lucas PM, Claisse O, Lonvaud-Funel A. 2009. Multilocus sequence typing of Oenococcus oeni: detection of two subpopulations shaped by intergenic recombination. Appl Environ Microbiol 75:1291–1300. doi: 10.1128/AEM.02563-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bridier J, Claisse O, Coton M, Coton E, Lonvaud-Funel A. 2010. Evidence of distinct populations and specific subpopulations within the species Oenococcus oeni. Appl Environ Microbiol 76:7754–7764. doi: 10.1128/AEM.01544-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Borneman AR, McCarthy JM, Chambers PJ, Bartowsky EJ. 2012. Comparative analysis of the Oenococcus oeni pan genome reveals genetic diversity in industrially-relevant pathways. BMC Genomics 13:373. doi: 10.1186/1471-2164-13-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dimopoulou M, Vuillemin M, Campbell-Sills H, Lucas PM, Ballestra P, Miot-Sertier C, Favier M, Coulon J, Moine V, Doco T, Roques M, Williams P, Petrel M, Gontier E, Moulis C, Remaud-Simeon M, Dols-Lafargue M. 2014. Exopolysaccharide (EPS) synthesis by Oenococcus oeni: from genes to phenotypes. PLoS One 9:e98898. doi: 10.1371/journal.pone.0098898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Campbell-Sills H, Khoury ME, Favier M, Romano A, Biasioli F, Spano G, Sherman DJ, Bouchez O, Coton E, Coton M, Okada S, Tanaka N, Dols-Lafargue M, Lucas PM. 2015. Phylogenomic analysis of Oenococcus oeni reveals specific domestication of strains to cider and wines. Genome Biol Evol 7:1506–1518. doi: 10.1093/gbe/evv084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sternes PR, Borneman AR. 2016. Consensus pan-genome assembly of the specialised wine bacterium Oenococcus oeni. BMC Genomics 17:308. doi: 10.1186/s12864-016-2604-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bordas M, Araque I, Alegret JO, El Khoury M, Lucas P, Rozès N, Reguant C, Bordons A. 2013. Isolation, selection, and characterization of highly ethanol-tolerant strains of Oenococcus oeni from south Catalonia. Int Microbiol Off J Span Soc Microbiol 16:113–123. [DOI] [PubMed] [Google Scholar]
- 34.González-Arenzana L, Santamaría P, López R, López-Alfaro I. 2014. Oenococcus oeni strain typification by combination of multilocus sequence typing and pulsed field gel electrophoresis analysis. Food Microbiol 38:295–302. doi: 10.1016/j.fm.2013.07.014. [DOI] [PubMed] [Google Scholar]
- 35.Garofalo C, El Khoury M, Lucas P, Bely M, Russo P, Spano G, Capozzi V. 2015. Autochthonous starter cultures and indigenous grape variety for regional wine production. J Appl Microbiol 118:1395–1408. doi: 10.1111/jam.12789. [DOI] [PubMed] [Google Scholar]
- 36.Wang C, García-Fernández D, Mas A, Esteve-Zarzoso B. 2015. Fungal diversity in grape must and wine fermentation assessed by massive sequencing, quantitative PCR and DGGE. Front Microbiol 6:1156. doi: 10.3389/fmicb.2015.01156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Claisse O, Lonvaud-Funel A. 2014. Multiplex variable number of tandem repeats for Oenococcus oeni and applications. Food Microbiol 38:80–86. doi: 10.1016/j.fm.2013.08.012. [DOI] [PubMed] [Google Scholar]
- 38.Gabriel S, Ziaugra L, Tabbaa D. 2001. SNP genotyping using the Sequenom MassArray iPLEX platform. Curr Protoc Hum Genet Chapter 2:Unit 2.12. doi: 10.1002/0471142905.hg0212s60. [DOI] [PubMed] [Google Scholar]
- 39.Dols-Lafargue M, Lee HY, Le Marrec C, Heyraud A, Chambat G, Lonvaud-Funel A. 2008. Characterization of gtf, a glucosyltransferase gene in the genomes of Pediococcus parvulus and Oenococcus oeni, two bacterial species commonly found in wine. Appl Environ Microbiol 74:4079–4090. doi: 10.1128/AEM.00673-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coton E, Coton M. 2003. Microbiological origin of “framboisé” in French ciders. J Inst Brew 109:299–304. doi: 10.1002/j.2050-0416.2003.tb00601.x. [DOI] [Google Scholar]
- 41.Ladero V, Coton M, Fernández M, Buron N, Martín MC, Guichard H, Coton E, Alvarez MA. 2011. Biogenic amines content in Spanish and French natural ciders: application of qPCR for quantitative detection of biogenic amine-producers. Food Microbiol 28:554–561. doi: 10.1016/j.fm.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 42.Capozzi V, Spano G. 2011. Food microbial biodiversity and “microbes of protected origin”. Front Microbiol 2:237. doi: 10.3389/fmicb.2011.00237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bokulich NA, Collins TS, Masarweh C, Allen G, Heymann H, Ebeler SE, Mills DA. 2016. Associations among wine grape microbiome, metabolome, and fermentation behavior suggest microbial contribution to regional wine characteristics. mBio 7:e00631-16. doi: 10.1128/mBio.00631-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Claisse O, Lonvaud-Funel A. 2012. Development of a multilocus variable number of tandem repeat typing method for Oenococcus oeni. Food Microbiol 30:340–347. doi: 10.1016/j.fm.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 45.Ana P, Marques AJD. 2011. Genomic diversity of Oenococcus oeni from different winemaking regions of Portugal. Int Microbiol Off J Span Soc Microbiol 14:155–162. [DOI] [PubMed] [Google Scholar]
- 46.Colwell RK, Elsensohn JE. 2014. EstimateS turns 20: statistical estimation of species richness and shared species from samples, with non-parametric extrapolation. Ecography 37:609–613. doi: 10.1111/ecog.00814. [DOI] [Google Scholar]
- 47.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.