ABSTRACT
Lactobacillus paracasei DG is a bacterial strain with recognized probiotic properties and is used in commercial probiotic products. However, the mechanisms underlying its probiotic properties are mainly unknown. In this study, we tested the hypothesis that the ability of strain DG to interact with the host is at least partly associated with its ability to synthesize a surface-associated exopolysaccharide (EPS). Comparative genomics revealed the presence of putative EPS gene clusters in the DG genome; accordingly, EPS was isolated from the surface of the bacterium. A sample of the pure EPS from strain DG (DG-EPS), upon nuclear magnetic resonance (NMR) and chemical analyses, was shown to be a novel branched hetero-EPS with a repeat unit composed of l-rhamnose, d-galactose, and N-acetyl-d-galactosamine in a ratio of 4:1:1. Subsequently, we demonstrated that DG-EPS displays immunostimulating properties by enhancing the gene expression of the proinflammatory cytokines tumor necrosis factor alpha (TNF-α) and interleukin 6 (IL-6), and particularly that of the chemokines IL-8 and CCL20, in the human monocytic cell line THP-1. In contrast, the expression of the cyclooxygenase enzyme COX-2 was not affected. In conclusion, DG-EPS is a bacterial macromolecule with the ability to boost the immune system either as a secreted molecule released from the bacterium or as a capsular envelope on the bacterial cell wall. This study provides additional information about the mechanisms supporting the cross talk between L. paracasei DG and the host.
IMPORTANCE The consumption of food products and supplements called probiotics (i.e., containing live microbial cells) to potentially prevent or treat specific diseases is constantly gaining popularity. The lack of knowledge on the precise mechanisms supporting their potential health-promoting properties, however, greatly limits a more appropriate use of each single probiotic strain. In this context, we studied a well-known probiotic, Lactobacillus paracasei DG, in order to identify the constitutive molecules that can explain the documented health-promoting properties of this bacterium. We found a novel polysaccharide molecule, named DG-EPS, that is secreted by and covers the bacterium. We demonstrated that this molecule, which has a chemical structure never identified before, has immunostimulatory properties and therefore may contribute to the ability of the probiotic L. paracasei DG to interact with the immune system.
KEYWORDS: Capsular EPS, probiotic, Enterolactis, THP-1, MAMPs, immunostimulation, Lactobacillus casei DG, Lactobacillus paracasei
INTRODUCTION
Strains of Lactobacillus paracasei are Gram-positive, non-spore-forming bacteria that are common inhabitants of the human intestinal tract. Specific strains of L. paracasei are found naturally in a number of fermented food products, and they have traditionally been used in the production of fermented milks and cheeses. Recently, specific strains of L. paracasei have been used in probiotic dietary supplements, including the strain L. paracasei DG (commercially known as Lactobacillus casei DG [Enterolactis]). A range of health-promoting properties have been assigned to L. paracasei DG, including the improvement of ulcerative colitis (1), a reduction in the side effects associated with therapies for the eradication of Helicobacter pylori (2), and treatment of small intestinal bacterial overgrowth (3). It has also been shown that L. paracasei DG is able to modulate the levels of fecal Clostridiales bacteria and butyrate in healthy adults (4). Despite the significant clinical evidence for the health benefits associated with the consumption of L. paracasei DG, the molecular mechanisms underlying these health effects are still unknown.
A number of mechanisms have been proposed for the probiotic effect. One of the most studied mechanisms relates to the ability of probiotic bacteria to antagonize pathogenic organisms either by the excretion of antimicrobial agents (5) or by the displacement of pathogenic organisms through the competitive occupancy of adhesion sites (6). In addition, a number of reports suggest that health benefits result from stimulation of the immune system by components presented at the surfaces of probiotic strains (7, 8). Both in vivo and in vitro experiments have demonstrated that the polysaccharides present at the surfaces of the bacteria, referred to either as capsule or as exopolysaccharides (EPSs), can play roles both in the displacement of pathogenic organisms and in the stimulation of the immune system (9, 10).
In an attempt to understand the molecular mechanisms underlying the probiotic activity of L. paracasei DG, we undertook a study to identify EPS molecules in L. paracasei DG. We isolated, purified, and characterized a novel EPS molecule (which we named DG-EPS) and studied its ability to mediate the probiotic properties of L. paracasei DG by monitoring adhesion on a Caco-2 cell layer and immunostimulatory activity on Caco-2 enterocyte-like and THP-1 macrophage-like cell models.
RESULTS
Identification of putative genetic clusters for exopolysaccharide biosynthesis.
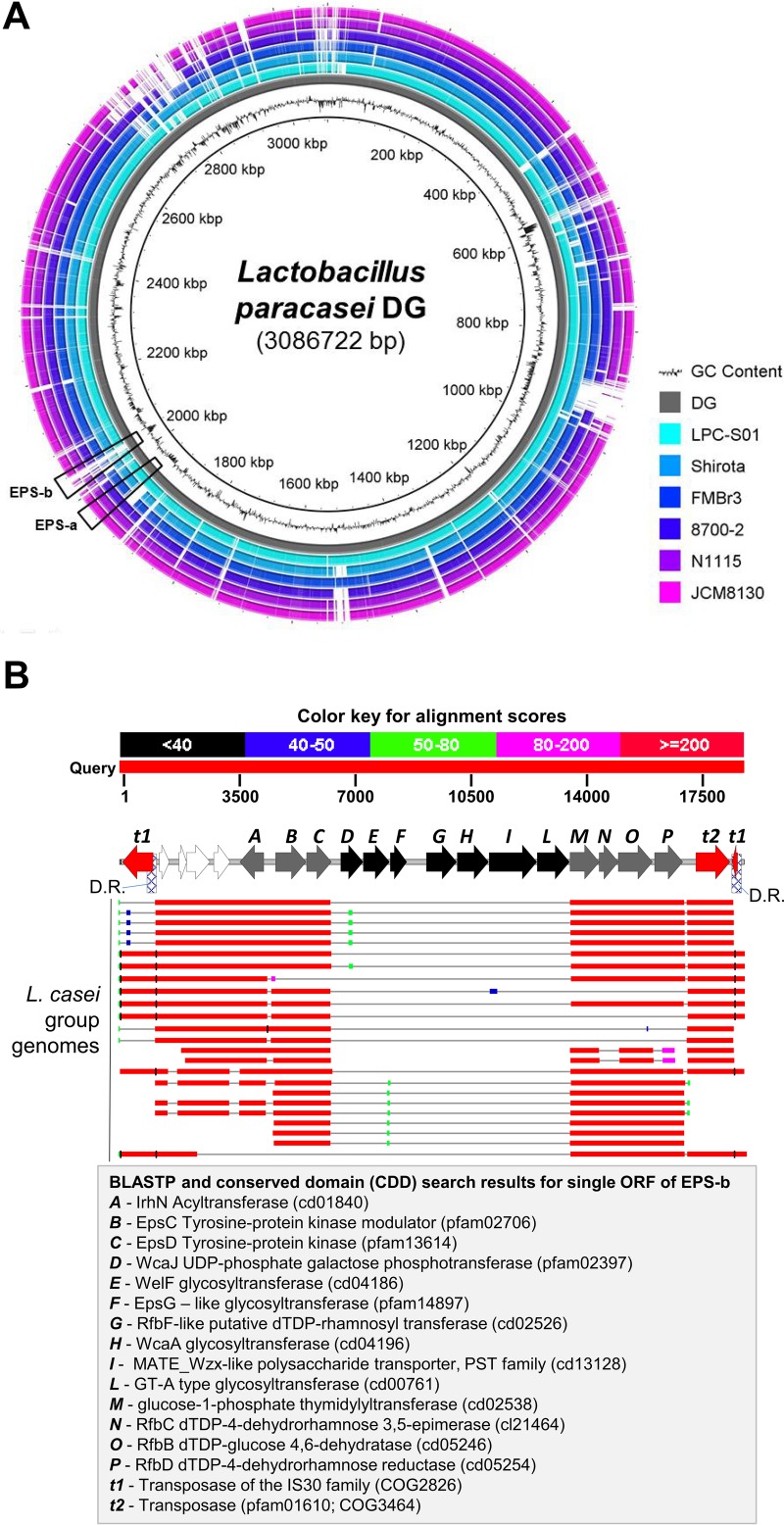
In light of the potential importance of EPS molecules in the cross talk between probiotic bacteria and the host (9–11), we performed in silico analyses to identify putative EPS operons in the draft genome of the probiotic strain L. paracasei DG. Specifically, comparative analysis with other genomes of the same species led to the identification in the genome of strain DG of two different regions encoding open reading frames (ORFs) putatively involved in the biosynthesis of EPS molecules (Fig. 1A, regions EPS-a and EPS-b). Notably, whereas the EPS-a region is common to all L. paracasei genomes investigated, EPS-b is a 13-kb region coding for several putative glycosyltransferases and includes a region of about 7 kb in the center of the cluster for which a BLASTN search found no match with other sequences in GenBank (Fig. 1B). The GC content of the 7-kb region is much lower (36%) than the average GC content of the whole genome of DG (approximately 46%), supporting the idea of the acquisition of these genes by horizontal gene transfer from a phylogenetically unrelated host.
FIG 1.
Comparative genomic analysis of Lactobacillus paracasei DG and other L. paracasei strains with complete genome sequences. (A) Circular genome atlas of L. paracasei DG (reference genome) and six other publicly available L. paracasei genomes. Highlighted in the atlas are the two putative exopolysaccharide (EPS) regions of strain DG. (B) In silico-predicted functional organization of the EPS-b region of L. paracasei DG and BLASTN search results for the EPS-b region. A map of the putative EPS gene cluster is shown above a graphic representation of the BLASTN output for the genomes of 24 L. casei group strains. Arrows in the map represent ORFs. Open arrows, ORFs outside the putative EPS operon; shaded arrows, ORFs for which homologous sequences were found in GenBank; filled arrows, ORFs that do not share significant homology with other sequences in GenBank. D.R., direct repeat sequence.
The identification of putative EPS operons in the genome of L. paracasei DG, along with the formation of a pellet much weaker than those of other L. paracasei strains after centrifugation (12), was considered an indication of the probable ability of strain DG to produce EPS molecules. Consequently, we undertook experiments aimed at the extraction and purification of the EPS fraction from broth cultures of DG.
Production and isolation of the EPS from L. paracasei DG.
A culture of L. paracasei DG was grown in a chemically defined medium (CDM). Although growth was slower than that observed in more-conventional media, such as de Man-Rogosa-Sharpe (MRS) broth, CDM was chosen because it does not contain contaminating polysaccharides, which interfere with the characterization of bacterial polysaccharides by nuclear magnetic resonance (NMR) (13). In order to isolate a polysaccharide sample suitable for characterization, it was necessary to grow L. paracasei DG for 3 days, at which point the cell biomass was separated from the fermentation liquors by centrifugation. High-purity EPS was isolated from the supernatant by fractional precipitation of material. The addition of 1 volume of ethanol released small amounts of an EPS material contaminated with proteins (typically 20 to 25 mg from a 500-ml batch fermentation). The addition of a second volume of ethanol also precipitated a relatively small amount of EPS (20 to 25 mg), but with much greater purity, rendering it suitable for characterization by NMR. Because the yields of EPS were low, and in order to determine if additional material was being retained with the biomass, various methods were used in an effort to recover capsular material bound to the cells. Stirring a suspension of the cells overnight in an aqueous solution of sodium hydroxide (1 M) and then precipitating crude polysaccharide by adding 2 volumes of ethanol yielded a significant amount of material, which included both polysaccharide and protein. The same approach was adopted for the isolation of extracellular polysaccharide molecules from strain L. paracasei LPC-S01, whose genome has been reported to harbor two distinct putative EPS operons (14), one of which is identical to strain DG EPS-a. However, we failed to obtain significant amounts of EPS from LPC-S01.
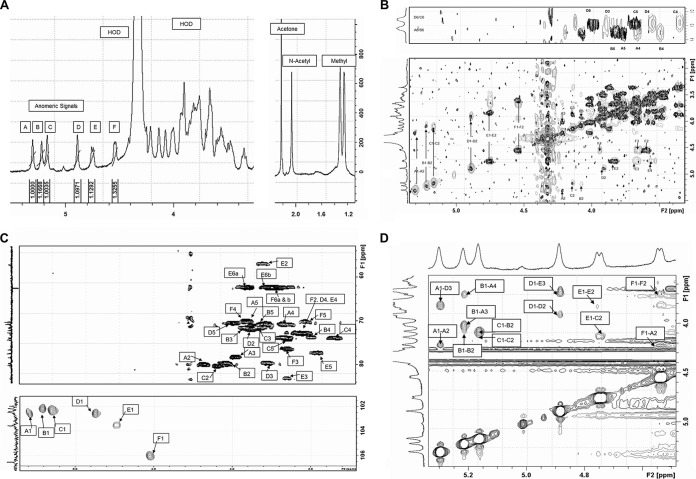
The purity of the EPSs released into the supernatant was established by examination of a 1H NMR spectrum of the sample (Fig. 2A). The low-field region of the spectrum contained six anomeric signals. Within the error of the experiments, the peak area integrals for each of the anomeric signals were identical, suggesting that a single EPS with a repeating unit containing six monosaccharides had been isolated. In addition to the anomeric signals, a single resonance with an integral height of 3 was visible at a chemical shift of 2.05 ppm, which is indicative of the presence of an N-acetylhexosamine, and a further two sets of overlapping doublets, each integrating to six protons, were present at 1.25 and 1.32 ppm; these signals indicated that the repeat unit contained four 6-deoxyhexoses.
FIG 2.
NMR analysis of the EPS isolated from Lactobacillus paracasei DG. (A) Selected regions of the 1H-NMR spectrum of DG-EPS recorded at 70°C in D2O using acetone as an internal standard. Anomeric (H-1) resonances are labeled A to F in order of decreasing chemical shift. (B) Selected regions of overlaid COSY (black contours) and TOCSY (gray contours) spectra for DG-EPS recorded at 70°C. Top, 13C-methyl region; bottom, 13C region for anomeric and ring protons. The letters A to F designate individual sugars, and numbers 1 to 6 indicate the C/H ring position. (C) Selected regions of the HSQC spectrum of DG-EPS. (Top) Locations of the individual rings and H6 protons and carbons; (bottom) locations of the anomeric protons and carbons. The spectrum was recorded in D2O at 70°C. (D) Anomeric region of a ROESY spectrum recorded for DG-EPS. Inter- and intraresidue NOEs from the anomeric hydrogens to ring protons are individually labeled.
Monomer analysis, linkage analysis, and determination of the absolute configuration of the monosaccharides in the repeating-unit structure.
Gas chromatography–mass spectrometry (GC-MS) analysis of the alditol acetates generated during monomer analysis of the EPS revealed the presence of 1,2,3,4,5-penta-O-acetyl-l-rhamnitol, 1,2,3,4,5,6-hexa-O-acetyl-d-galactitol, and 2-acetamido-1,3,4,5,6-penta-O-acetyl-2-deoxy-d-galactitol at a ratio of 4:1:1. Monomer analysis revealed the presence of rhamnose, galactose, and N-acetylgalactosamine in the repeating unit.
The methylated alditol acetates generated during linkage analysis included a 1,5-di-O-acetyl-2,3,4,6-tetra-O-methylhexitol, which confirms that the galactose is present in its pyranose form as a terminal sugar; a 1,3,5-tri-O-acetyl-2-(acetylmethylamino)-2-deoxy-4,6-di-O-methylgalacitol, which confirms that the N-acetylgalactosamine is present in its pyranose form as a 1,3-linked monosaccharide; two 1,2,5-tri-O-acetyl-3,4-di-O-methyl-6-deoxyhexitols, which indicate that two of the rhamnose monomers are 1,2-linked; a 1,3,5-tri-O-acetyl-6-deoxy-2,4-di-O-methylhexitol, which indicates that one of the rhamnose monomers is 1,3-linked; and finally, a 1,2,3,5-tetra-O-acetyl-6-deoxy-4-O-methylhexitol, suggesting that the final rhamnose is a 1,2,3-linked rhamnose present as a bridging point in the repeating unit.
Conversion of the monomers to mixtures of their epimeric 2-butyl glycosides confirmed that all the rhamnose monomers were of l-absolute configuration while both the galactose and N-acetylgalactosamine were of d-absolute configuration.
Determination of the sequence of the monomers in the repeat unit using 1D and 2D NMR.
The chemical shifts of each of the protons and carbons in the repeat unit were determined through the inspection of a series of 1-dimensional (1D) and 2D NMR spectra and are listed in Table 1. Analysis of the 1H-1H correlation spectroscopy (COSY) spectrum (Fig. 2B, black contours) in combination with the 1H-1H total correlation spectroscopy (TOCSY) spectrum (Fig. 2B, gray contours) allowed the scalar coupling within the individual sugars (labeled A to F in order of decreasing chemical shift [Fig. 2A]) to be tracked from the anomeric protons from H-1 to H-6 in the rhamnose sugars (A, B, C, and D) and from H-1 to H-4 in the galactose and N-acetylgalactosamine monomers (Fig. 2B).
TABLE 1.
Chemical shifts of protons and carbons in the repeat unit of L. paracasei DG-EPS determined through the inspection of a series of 1D and 2D NMR spectra
| Position | Chemical shift (ppm) at the indicated positiona |
|||||
|---|---|---|---|---|---|---|
| A | B | C | D | E | F | |
| H-1 (C-1) | 5.30 (102.6) | 5.21 (102.2) | 5.15 (102.4) | 4.87 (102.5) | 4.72 (103.5) | 4.53 (106.0) |
| H-2 (C-2) | 4.21 (79.8) | 4.07 (79.6) | 4.13 (80.3) | 3.90 (71.6) | 3.82 (57.1) | 3.59 (72.5) |
| H-3 (C-3) | 4.01 (78.2) | 3.91 (72.0) | 3.86 (71.3) | 3.79 (79.5) | 3.65 (83.1) | 3.63 (76.5) |
| H-4 (C-4) | 3.67 (70.4) | 3.49 (73.6) | 3.35 (73.7) | 3.55 (72.9) | 3.54 (72.6) | 3.93 (70.1) |
| H-5 (C-5) | 3.82 (70.6) | 3.77 (70.6) | 3.66 (73.8) | 4.02 (70.5) | 3.45 (77.3) | 3.53 (70.0) |
| H-6 (C-6) | 1.32 (18.6) | 1.32 (18.1) | 1.25 (18.0) | 1.25 (17.9) | 3.91/3.75 (62.4) | 3.75 (62.3) |
The 13C chemical shifts for the N-acetyl group are 23.5 ppm (CH3) and 175.6 ppm (CO), and the corresponding proton methyl resonance occurs at 2.05 ppm.
A heteronuclear single quantum coherence (HSQC) spectrum was used to correlate ring carbons with their attached protons (Fig. 2C), and the distinctive position of C-2 and identification of H-2 of the N-acetylgalactosamine confirmed, by observation of scalar coupling to H-1 resonance at 4.72 on the COSY spectrum, that the anomeric resonance at 4.72 ppm (E in Fig. 2A) was that of the N-acetylgalactosamine. The remaining anomeric resonance at 4.53 ppm must therefore belong to the terminal galactose.
Through inspection of the chemical shifts of the rhamnose ring carbons, it was possible to identify points of linkage by locating those carbons whose chemical shifts had moved toward low-field positions (above 78 ppm) compared to the values normally associated with unsubstituted ring positions (less than 74 ppm for rhamnose sugars). This identified A as a 2,3-linked rhamnose (C-2, δ 79.8 ppm; C-3, δ 78.2 ppm), B as a 2-linked rhamnose (C-2, δ 79.6 ppm), C as a 2-linked rhamnose (C-2, δ 80.3 ppm), and D as a 3-linked rhamnose (C-3, δ 79.5 ppm). The results of the linkage analysis already showed that the N-acetylgalactosamine (E) was 1,3-linked, and this was confirmed by the high chemical shift of C-3 in E (δ 83.1 ppm). Finally, the chemical shifts of the carbons in residue F are in agreement with the conclusion that this is a terminal galactose monomer.
The anomeric configuration of the monosaccharides was determined by measuring the 1JC1-H1 coupling constants that were visible on a coupled HSQC spectrum (not shown). Residues A to D had 1JC1-H1 coupling constants of 177 Hz, 172 Hz, 175 Hz, and 174 Hz, respectively. The fact that these coupling constants are >170 Hz indicates that the rhamnose residues are alpha-linked, while the sizes of the 1JC1-H1 coupling constants for E (164 Hz) and F (157 Hz) identify these two resonances as beta-linked monomers.
Finally, the sequence of the sugars in the repeating unit was established through inspection of both a 1H-13C heteronuclear multiple bond correlation (HMBC) spectrum (not shown) and a 1H-1H rotating frame nuclear Overhauser effect spectroscopy (ROESY) spectrum (Fig. 2D). Interresidue scalar coupling observed on the HMBC spectrum included coupling between A H-1 and D C-3, indicating that A is linked to the 3-position of D; coupling between C H-1 and B C-2, indicating that C is linked to the 2-position of B; coupling between D H-1 and E C-3, indicating that D is linked to the 3-position of E; and coupling between F H-1 and A C-2, indicating that F is linked to the 2-position of A. On the ROESY spectrum, interresidue nuclear Overhauser effects (NOEs) were observed between A H-1 and D H-3, confirming the A(1-3)D linkage; between B H-1 and A H-3 (strong) and between B H-1 and A H-4 (moderate), showing that B is linked to the 3-position of A; between C H-1 and B H-2, confirming the C(1-2)B linkage; between D H-1 and E H-3, identifying a D(1-3)E linkage; between E H-1 and C H-2, identifying the E(1-2)C linkage; and also between F H-1 and A H-2, confirming the F(1-2)A linkage.
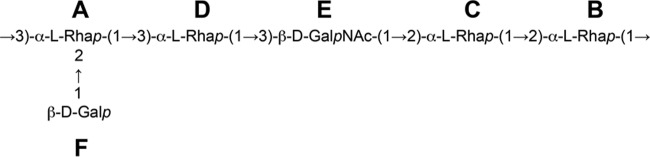
The combined results of the chemical and NMR analyses of the EPS isolated from L. paracasei DG indicate that DG-EPS is a novel heteropolysaccharide that has the repeating-unit structure reported in Fig. 3.
FIG 3.
Repeating unit structure of DG-EPS, the heteropolysaccharide isolated from Lactobacillus paracasei DG.
In the next set of experiments, we studied the possible involvement of the characterized EPS molecule in mediating the cross talk between L. paracasei DG and the host.
Experiments in an enterocyte-like Caco-2 cell model.
In order to investigate the ability of DG-EPS to mediate the bacterium's interaction with the host, we first used the Caco-2 cell line, which is considered a valuable in vitro tool for studying the mechanisms underlying interactions between bacterial cells and the human gut (7, 15, 16).
In a previous publication, L. paracasei DG was shown to be moderately adhesive on a Caco-2 cell layer (14). In this study, the potential involvement of DG-EPS was assessed by testing the adhesion ability of strain DG after the removal of the EPS molecule (“naked” DG cells [nDG], prepared by phosphate-buffered saline [PBS] washes and mild sonication) and upon preincubation of Caco-2 cells with purified EPS. Several studies have demonstrated that bacterial EPS molecules may either promote adhesion (17) or, in contrast, reduce adherence by masking cell wall adhesins (18). Nonetheless, in our experiments, purified EPS was unable to affect the adhesion properties of strain DG. In fact, the adhesion of L. paracasei DG was not significantly different from that of nDG and was quite modest (about 300 bacteria per 100 Caco-2 cells), as reported previously (14); in addition, adhesion ability was not affected by coincubation of the bacterial cells with purified EPS (data not shown).
Afterwards, Caco-2 cells were employed to investigate the effect of DG-EPS on the activation of the transcription factor NF-κB. We recently showed that L. paracasei DG possesses an evident ability to reduce NF-κB activation in Caco-2 cells at baseline and upon stimulation with the proinflammatory cytokine interleukin 1β (IL-1β) (14), as determined through a reporter system obtained by transfecting Caco-2 cells with a luciferase reporter vector (6, 7). Here we used the same immunological model to test the EPS macromolecule isolated from strain DG. We found that the purified EPS molecule, in contrast to the whole bacterial cells and their exhausted broth (14), was not able to affect NF-κB activation either at baseline or in the presence of the proinflammatory stimulus IL-1β (data not shown). Other L. paracasei strains under study (namely, strains LPC-S01 and Shirota) displayed the same ability as strain DG to reduce NF-κB activation in Caco-2 cells (14), further suggesting that DG-EPS does not contribute to this specific immunomodulatory effect.
The DG-EPS molecule triggers the expression of chemokines and cytokines by THP-1 cells.
We exploited the THP-1 macrophage model to test the immunomodulatory properties of L. paracasei DG and its isolated EPS. We used real-time quantitative PCR (qRT-PCR) to quantify the gene expression of tumor necrosis factor alpha (TNF-α), IL-6, chemokine (C-C motif) ligand 20 (CCL20), the chemokine IL-8, and cyclooxygenase 2 (COX-2). We tested three concentrations of the purified DG-EPS molecule (0.1, 1, and 10 μg ml−1). We also performed the same experiments in the presence of 1 μg ml−1 of the proinflammatory stimulus lipopolysaccharide (LPS). We found that the purified DG-EPS can stimulate the expression of all the genes under study, except for COX-2, in a concentration-dependent manner (Fig. 4). In particular, the chemokines IL-8 and CCL20 were induced approximately 70-fold by 10 μg ml−1 DG-EPS, whereas the proinflammatory cytokines TNF-α and IL-6 were induced 26- and 39-fold, respectively. A similar stimulatory profile was observed with bacterial cells of strain DG (multiplicity of infection [MOI], 50), even if to a lower extent (see Fig. S1 in the supplemental material). In addition, when EPS was removed by PBS washes and mild sonication, the bacterial cells of strain DG lost their ability to stimulate the expression of the TNF-α, IL-8, and CCL20 genes; the addition of 1 μg ml−1 purified EPS partially reconstituted the stimulatory capacity of the cells (Fig. S1).
FIG 4.
Gene expression analysis by qRT-PCR in THP-1 human macrophages after 4 h of stimulation with the purified DG-EPS molecule (0.1, 1, and 10 μg ml−1), with or without the addition of LPS (1 μg ml−1). Expression levels of TNF-α, IL-6, IL-8, CCL20, and COX-2 are shown as the fold change in induction (FOI) relative to expression by the control (unstimulated macrophages), which was set at a value of 1. Data are means of results from three independent experiments ± standard deviations. Asterisks indicate statistically significant differences (according to a two-tailed unpaired Student t test) from results for unstimulated (left y axis) or LPS-stimulated (right y axis) THP-1 cells. *, P < 0.05; **, P < 0.01.
As expected, the stimulation of THP-1 cells with LPS led to marked overexpression of all genes tested, particularly the IL-6, IL-8, and CCL20 genes. However, the addition of DG-EPS did not significantly affect the LPS-associated induction of any of the genes tested (Fig. 4).
DISCUSSION
The use of probiotic and, in general, food-grade bacteria as modulators of host immune responses appears to be a promising strategy for the prevention and management of inflammatory conditions at the gut mucosa level. With this aim, the identification of microbial cell components mediating the interaction with host cells is of pivotal importance. The enormous biodiversity of the microbial world provides a virtually inexhaustible source of molecular stimuli for the immune system, which has been only marginally explored to date. Particularly, bacterial cell wall components and secreted molecules are key ligands that can interact with host receptors and activate various signaling pathways, thus triggering the final probiotic effect. Among the most studied probiotics, lactic acid bacteria (LAB) possess a cell wall that is typically composed of a thick peptidoglycan layer decorated with proteins, teichoic acids, and polysaccharides (19), which can act as microbe-associated molecular patterns (MAMPs). EPSs, apart from their technological properties and industrial applications, have been found to contribute to several health-promoting features of probiotics, such as antitumor, antiulcer, immunomodulatory, antiviral, and cholesterol-lowering activities (20, 21), but their mechanisms of action have not been clearly established and are probably diverse and complex.
Thanks to the genomic analysis of L. paracasei strain DG, we could identify two gene clusters putatively coding for EPS biosynthesis. Interestingly, a GenBank search revealed that one of these regions, EPS-b, is unprecedented. Since we failed to obtain knockout mutants for EPS-b, the link between this gene cluster and the production of DG-EPS, although probable, is not definitely demonstrated. Nonetheless, the identification of EPS gene clusters in the genome of DG inspired us to investigate biologically the potential presence of EPS molecules on the outer surface of the bacterium.
After purification of the exopolysaccharidic cell fraction of strain DG, we characterized the EPS repeating unit by means of chromatographic methods and NMR spectroscopy, establishing that it possesses a novel structure. The repeating-unit structures of a number of different strains of the Lactobacillus casei group of species (i.e., L. casei, L. paracasei, and Lactobacillus rhamnosus) have been published previously (22–28), and the repeating-unit structure reported here is different. Moreover, it is also different from the published structures of the EPSs characterized for a range of LAB (20, 29). A feature of the repeating-unit structure of the EPS reported here and those of a number of probiotic bacteria is the presence of rhamnose in the polymeric backbone (20); nonetheless, the very high content of rhamnose (accounting for two-thirds of the sugar moieties in molarity) in DG-EPS is, to the best of our knowledge, unique in LAB.
The ability of EPSs isolated from LAB to trigger immunomodulatory responses has been observed in several studies (11, 30–33); nonetheless, the definition of a common immunological outcome for LAB EPSs is a difficult (and potentially impracticable) task, because their broad structural diversity may influence recognition by immune system receptors (30). Therefore, any EPS molecule with a different monosaccharide sequence may represent a novel MAMP. In this context, we undertook an in vitro immunological characterization of DG-EPS. The initial experiments clearly demonstrated that the previously reported ability of strain DG to modulate NF-κB activation in Caco-2 enterocytes (14) is independent of the EPS.
After this preliminary immunological investigation on epithelial cells, we performed experiments with macrophages, which are cells more properly belonging to the immune system. Macrophages, like monocytes and dendritic cells (DCs), are antigen-presenting cells (APCs) responsible for the detection of microorganisms and involved in their clearance through phagocytosis and the production of proinflammatory cytokines. Specifically, in our study, we used the human leukemia monocytic cell line THP-1, which, upon differentiation with phorbol myristate acetate (PMA), expresses a phenotype with similarities to human peripheral blood mononuclear cell (PBMC) monocyte-derived macrophages (34). With this model, we found that DG-EPS displays immunostimulatory properties by enhancing the gene expression of the proinflammatory cytokines TNF-α and IL-6 and, particularly, the chemokines IL-8 and CCL20. In contrast, the expression level of the cyclooxygenase enzyme COX-2 was not affected. TNF-α and IL-6 are among the first cytokines to be produced by APCs during the innate immune response to bacteria. TNF-α and IL-6 have pleiotropic effects, such as induction of the acute-phase response and activation of macrophages, and, also, together with IL-8, play crucial roles as chemoattractants for neutrophils, immature dendritic cells, natural killer cells, and activated T cells (35, 36). Furthermore, CCL20 is a chemokine that attracts immature DCs (37); it has been shown to be upregulated by TNF-α (38) and also to be elicited by probiotics such as L. rhamnosus GG (39).
In accordance with the results of our study, the EPS isolated from L. rhamnosus KL37 was shown in a previous study to display a proinflammatory profile by inducing the production of TNF-α, IL-6, and IL-12 in mouse peritoneal macrophages (30). Similarly, in a different study, the EPS from L. rhamnosus RW-9595M, which is a rhamnose-rich heteropolysaccharide (rhamnose-glucose-galactose at a 4:2:1 molar ratio) like DG-EPS, elicited TNF-α, IL-6, and IL-12p40 production by both the murine RAW 264.7 macrophage-like cell line and PBMCs (40). The ability to stimulate the production of proinflammatory cytokines by APCs, therefore, might be a common property of the (rhamnose-rich) hetero-exopolysaccharides produced by the lactobacilli of the L. casei group.
Notably, in our experiments, the levels of induction of proinflammatory cytokines and chemokines were much lower upon stimulation with DG-EPS than with LPS, even when DG-EPS was used at a 10-times-higher concentration. In addition, when we used EPS or DG cells in combination with LPS, we did not observe any additive effect over the induction levels of cytokines triggered by LPS. Accordingly, it was proposed that probiotics can moderately stimulate the synthesis of proinflammatory cytokines in the absence of an inflammatory response but may have suppressive effects or no effect when an inflammatory response has already been triggered (41). In this context, probiotics, though inoffensive during infection-derived inflammations, act as mild boosters of the innate immunity that may contribute to a more efficient and faster immune response against potential infectious agents.
Several studies failed to assign evident immunostimulatory abilities to the EPSs, and it was proposed that the EPSs anchored on the bacterial outer surface are immunologically inert molecules that keep cell wall-associated MAMPs from direct contact with immune cell receptors (18, 42, 43). However, in our study, the purified EPS produced by L. paracasei DG displayed immunostimulatory activity, particularly in terms of chemokine expression. Thus, the EPS from L. paracasei DG, rather than being an inert molecule, can be considered a bacterial product that can boost the immune system either as a secreted molecule released from the bacterium or, plausibly, as a capsular envelope on the bacterial cell wall. Our results concerning the immunostimulatory abilities of DG-EPS derive, however, from a preliminary in vitro study, and further investigations are needed in order to decipher the potential role of DG-EPS in the in vivo setting, where the final response depends on the combination of signals coming from diverse epithelial and immune cells in an environmental context populated by numerous diverse commensal microbes.
In conclusion, our study provides additional information about the well-characterized probiotic strain L. paracasei DG by demonstrating that it produces a unique rhamnose-rich hetero-exopolysaccharide, named DG-EPS, possessing immunostimulatory properties. DG-EPS may represent a new molecule for potential nutraceutical and pharmaceutical applications.
MATERIALS AND METHODS
Unless otherwise stated, all reagents were purchased from Sigma-Aldrich Company Ltd. (Poole, Dorset, UK) and were used as supplied.
Identification of the putative EPS gene cluster.
The draft genome sequence of L. paracasei DG was obtained through Ion Torrent PGM (Life Technologies, Germany) as described previously (44). The raw sequence data were assembled using MIRA, version 3.9 (http://www.chevreux.org/projects_mira.html), applying the default parameters recommended for Ion Torrent data processing. Initial automated annotation of the genome was performed using RAST, combined with BLASTX. The results of the gene finder program were combined manually with data from BLASTP analysis against a nonredundant protein database provided by the National Center for Biotechnology Information (NCBI). The draft genome sequence of DG was compared with other L. paracasei genome sequences by means of the BLAST Ring Image Generator (BRIG) (45). Functional annotation of the EPS-b region was carried out by combining the results of BLASTN, BLASTP, and the “CD-search” of the Conserved Domain Database (CDD), available at the NCBI website (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi).
EPS isolation and purification.
Lactobacillus paracasei strain DG (deposited at the Collection Nationale de Culture de Microorganismes of the Pasteur Institute under code CNCM I-1572) was grown at 37°C in de Man-Rogosa-Sharpe (MRS) broth (Difco Laboratories Inc., Detroit, MI) for 24 h. This culture was used to inoculate the chemically defined medium (CDM) (Table 2). The multistep extraction and purification of EPS was performed from about 1 liter of CDM supplemented with 2% glucose. After growth at 37°C for 48 h, cells were collected by centrifugation at 12,000 × g for 15 min at 4°C (Avant J-26 XPI system; Beckman Coulter Ltd., High Wycombe, UK) and were separated from the exhausted medium. The two fractions were then treated separately. An equal volume of absolute ethanol was added to the exhausted medium, and the mixture was stored at 4°C for 48 h. After storage, it was centrifuged at 25,000 × g for 35 min at 4°C. The pellet obtained (fraction S1) was dissolved in deionized water (about 20 to 50 ml), whereas the supernatant was added to a second volume of ethanol, and this mixture was stored at 4°C for 48 h. Subsequently, the centrifugation step was repeated, and the pellet (fraction S2) was dissolved in deionized water as described above. For cell fractions, the pellet was washed with phosphate-buffered saline (PBS) to remove polysaccharide impurities, treated with 1 M sodium hydroxide, and stirred overnight at 4°C. Afterwards, it was centrifuged again at 12,000 × g and 4°C for 15 min in order to remove sodium hydroxide. Crude EPS was precipitated by the addition of an equal volume of chilled absolute ethanol; this was stored for 48 h at 4°C and was then centrifuged at 25,000 × g and 4°C for 35 min. The recovered pellet (fraction C1) was redissolved in deionized water (about 20 ml). The resulting supernatant was then added to a second volume of absolute ethanol and was again incubated for 48 h at 4°C. After another centrifugation, as described above, a second precipitated fraction (C2) was recovered and was dissolved in deionized water. Small neutral sugars and proteins were then removed by dialysis (with cellulose acetate membranes [cutoff, 100 kDa]) of the extracted fractions for 72 h at 4°C, against three changes of deionized water per day. After 3 days, the contents of the dialysis membrane were collected and were lyophilized in a freeze-dryer (Northern Scientific, York, UK). The dry mass of EPS was then determined. The EPS preparations were tested for the presence of contaminating bacterial DNA through real-time quantitative PCR (q-PCR) with two primer pairs: universal primers targeting the 16S rRNA gene (EUB) and DG strain-specific primers targeting the welF gene (14). This analysis revealed the presence of 10 to 63 ng ml−1 in the 1-mg ml−1 stock solutions of EPS, corresponding to an overall maximum concentration of 0.6 ng ml−1 of DNA incubated with THP-1 cells when the highest concentration of EPS (10 μg ml−1) was used in immunological experiments.
TABLE 2.
Chemically defined medium used to cultivate L. paracasei DG for extraction of the exopolysaccharide
| Component | Concn (g liter−1) |
|---|---|
| Solution 1 | |
| (NH4)2SO4 | 2 |
| MgSO4·7H2O | 0.15 |
| MnSO4·4H2O | 0.02 |
| Solution 2 | |
| Adenine | 0.005 |
| Pyridoxal | 0.002 |
| Nicotinic acid | 0.001 |
| Ca2+–d-pantothenate | 0.001 |
| Riboflavin | 0.001 |
| Thiamine | 0.001 |
| Vitamin B12 | 0.000001 |
| Biotin | 0.00001 |
| p-Aminobenzoic acid | 0.000005 |
| Folic acid | 0.00001 |
| Solution 4a | |
| Guanine | 0.005 |
| Xanthine | 0.005 |
| Uracil | 0.005 |
| Solution 5, K2HPO4 | 4.56 |
| Solution 6 | |
| Sodium acetate | 0.05 |
| Sodium citrate | 0.02 |
| KH2PO4 | 0.01 |
| NaCl | 0.002 |
| CaCl2 | 0.002 |
| Solution 7 | |
| Tween 80 | 1 |
| Tween 20 | 1 |
| Glycerol | 1 |
| Glucose | 20 |
| Casamino Acids | 10 |
Dissolved in 1 M NaOH. An equal volume of 1 M HCl was added to the medium to neutralize the pH.
Determination of monomer composition and linkage analysis.
To determine the monomer composition of the polysaccharide, the EPS (3 mg) was hydrolyzed by treatment with 2 M trifluoroacetic acid (TFA) at 120°C for 2 h; the released monosaccharides were subsequently derivatized to form alditol acetates, which were analyzed by gas chromatography–mass spectrometry (GC-MS). To derivatize the monomers, 10 mg NaBH4 was added to the dried monosaccharides, which were resuspended in 1 ml Milli-Q water, and the mixture was incubated at 40°C for 2 h. After evaporation of the solution, 1 ml glacial acetic acid was added to the residue, and this was again evaporated to dryness. Subsequently, 3 ml methanol was added and then evaporated in order to remove the borate complex and to produce the methylated sugar alditols. Then 2 ml pyridine and 2 ml acetic anhydride were added; the acetylation reaction ran at 100°C for 2 h. At the end of the reaction, the solution was evaporated and the acetylated monomers resuspended in water. Extraction with chloroform was performed to collect the organic phase, containing the alditol acetate sugars. Any trace of water was removed by adding anhydrous sodium sulfate and storing the sample for 30 min at 4°C. Sodium sulfate was removed by filtration on filter paper and chloroform by evaporation. The resulting residue was resuspended in acetone. The GC-MS analysis was performed on an Agilent (Santa Clara, CA, USA) 7890A GC system coupled to an Agilent 5675c quadrupole MS. The samples were eluted from an HP-5 column (length, 30 m; inside diameter, 0.25 mm; film thickness, 0.25 μm) with helium as the carrier (9 lb/in2; flow rate, 1 ml min−1) and the following temperature program: start temperature, 150°C; hold time, 4 min; final column temperature, 250°C, reached via a rising gradient of 4°C min−1). The ratios of the different sugars were determined by examination of the relative responses of the different alditol acetates with reference to those determined for a standard mixture of alditol acetates. As reported previously (46), the integral area for amino sugars was low, because they had undergone thermal decomposition during analysis. The final monomer ratio, for the amino sugar, was taken from integration of the nuclear magnetic resonance (NMR) peak integrals for the respective anomeric and H-2 protons. The absolute configurations of monosaccharides were determined by conversion to their 2-butyl glycosides using the procedure described by Gerwig et al. (47). To determine the linkage pattern of the EPS, the sample was permethylated using the procedures described by Ciucanu and Kerek (48). The permethylated polysaccharide was then hydrolyzed (2 M TFA at 120°C for 2 h), and the methylated monosaccharides were converted to methylated alditol acetates. The identities of the methylated alditol acetates were determined by analysis of their individual mass spectrum fragmentation patterns generated during GC-MS analysis. The GC-MS analyses were performed on the same instrumentation as the monomer analysis but used the following temperature program: start temperature, 155°C; hold time, 1 min; final column temperature, 195°C, reached via a rising gradient of 0.75°C min−1.
NMR analysis of the EPS from L. paracasei.
NMR spectra were recorded for EPS samples that were dissolved (10 to 20 mg ml−1) directly in D2O (Goss Scientific Instruments Ltd., Essex, UK). The spectra were recorded at a probe temperature of 70°C on a Bruker Avance 500.13-MHz 1H (125.75-MHz 13C) spectrometer (Bruker-Biospin, Coventry, UK) operating with Z-field gradients where appropriate and using Bruker's pulse programs. Chemical shifts are expressed in parts per million relative to internal acetone (δ 2.225 for 1H and δ 31.55 for 13C). Spectra recorded included a 2D gradient-selected double quantum filtered correlation spectrum (gs-DQF-COSY) recorded in magnitude mode at 70°C, total correlation spectroscopy (TOCSY) spectra recorded with variable mixing times (60, 90, and 120 ms), 1H-13C heteronuclear single quantum coherence (HSQC) spectra (decoupled and coupled), a heteronuclear multiple bond correlation (HMBC) spectrum, and finally, a rotating frame nuclear Overhauser effect (ROESY) spectrum. The 2D spectra were recorded with 256 experiments of 1,024 data points. The ROESY spectrum was recorded using a Bruker pulse sequence and 256 experiments of 1,024 data points using a mixing time of 200 ms. For the majority of spectra, time domain data were multiplied by phase-shifted (squared) sine-bell functions. After zero filling and Fourier transformation, data sets of 1,024 by 1,024 points were obtained.
Bacterial adhesion to Caco-2 cells.
The adhesion of L. paracasei strains to a Caco-2 (ATCC HTB-37) cell layer was assessed as described previously (14). In brief, for adhesion experiments, fully differentiated Caco-2 cells were used (i.e., 15 days after confluence). EPS (100 μg ml−1) was incubated with a monolayer of Caco-2 cells for 1 h at 37°C. Finally, monolayers were examined microscopically (magnification, ×400) under oil immersion after Giemsa staining (15). All experiments were performed in duplicate.
NF-κB activation by exopolysaccharides.
The activation of nuclear factor κB (NF-κB) was studied by means of a recombinant Caco-2 cell line stably transfected with vector pNiFty2-Luc (InvivoGen, Labogen, Rho, Italy) as described in detail elsewhere (49, 50). In brief, recombinant Caco-2 cell monolayers (approximately 3 × 105 cells/well), cultivated in the presence of 50 μg ml−1 Zeocin, were washed with 0.1 M Tris-HCl buffer (pH 8.0) and then suspended in fresh Dulbecco's modified Eagle medium (DMEM) containing 100 mM HEPES (pH 7.4), together with 0.1 ml of L. paracasei DG-EPS, corresponding to a final concentration of 100 μg ml−1. The stimulation was conducted by adding 10 ng ml−1 of interleukin 1β (IL-1β). After incubation at 37°C for 4 h, the samples were treated, and bioluminescence was measured as described by Stuknyte et al. (50). Two independent experiments were conducted in triplicate for each condition.
Activation of the THP-1 human macrophage cell line: cell culture, growth conditions, and stimulation protocol.
The monocytic THP-1 cell line was purchased from the American Type Culture Collection (Manassas, VA, USA). THP-1 cells were originally cultured from the peripheral blood of a 1-year child with acute monocytic leukemia (51). These are nonadherent cells, which can be differentiated into macrophage-like cells through a protein kinase C-mediated, reactive oxygen species (ROS)-dependent signaling pathway (52) by treatment with phorbol myristate acetate (PMA). The normal growth medium for THP-1 cells consisted of RPMI 1640 medium (Lonza, Basel, Switzerland) supplemented with 10% (vol/vol) fetal bovine serum (FBS) (Gibco-BRL, Life Technologies, Milan, Italy), 2 mM l-glutamine, 100 U ml−1 penicillin, and 100 μg ml−1 streptomycin (Sigma-Aldrich). Cells were seeded at a density of 5 × 105/well in 24-well plates and were incubated at 37°C under a humidified atmosphere of 95% air and 5% CO2. Differentiation was induced by the addition of PMA (Sigma-Aldrich) to the cellular medium at a final concentration of 100 nM and was allowed to proceed for 24 h. Afterwards, cells were washed once with sterile PBS buffer to remove all nonadherent cells, and fresh complete medium was added. Bacteria were used at a multiplicity of infection (MOI) of 50, EPS at final concentrations of 0.1, 1, and 10 μg ml−1, and lipopolysaccharide (LPS) from Salmonella enterica (Sigma-Aldrich) at a final concentration of 1 μg ml−1. An untreated sample, consisting only of RPMI 1640 medium with 10% (vol/vol) FBS, was used as a control.
Preparation of RNA and qRT-PCR.
After the incubation of THP-1 cells at 37°C for 4 h, the supernatant was carefully removed from each well, and the total cellular RNA was isolated from the adherent cells with the Total RNA Blood and Cultured Cells kit (GeneAid, New Taipei City, Taiwan). Afterwards, traces of DNA were removed by treatment with DNase enzyme (Sigma-Aldrich) according to the manufacturer's instructions. RNA concentration and purity were determined with a Take3 Multi-Volume plate reader (BioTek, Lucerne, Switzerland), and reverse transcription to cDNA was performed with the iScript Select cDNA synthesis kit (Bio-Rad Laboratories, Hercules, CA), using the following thermal cycle: 5 min at 25°C, 30 min at 42°C, and 5 min at 85°C. Real-time quantitative reverse transcription-PCR (qRT-PCR) was carried out in order to measure the mRNA expression levels of cytokines by means of the SYBR green technology using SsoFast EvaGreen Supermix (Bio-Rad) on a Bio-Rad CFX96 system according to the manufacturer's instructions. The primers were as follows: GAPDH forward, 5′-GGGAAGGTGAAGGTCGGAGT-3′; GAPDH reverse, 5′-TCAGCCTTGACGGTGCCATG-3′; IL-6 forward, 5′-CGGTACATCCTCGACGGCAT; IL-6 reverse, 5′-TCACCAGGCAAGTCTCCTCAT-3′; IL-8 forward, 5′-TGTGGTATCCAAGAATCAGTGAA-3′; IL-8 reverse, 5′-TATGTTCTGGATATTTCATGGTACA-3′; CCL20 forward, 5′-CTGCTTGATGTCAGTGCTG; CCL20 reverse, 5′-CACCCAAGTCTGTTTTGG-3′; TNF-α forward, 5′-TCAGCTCCACGCCATT-3′; TNF-α reverse, 5′-CCCAGGCAGTCAGATCAT-3′; COX-2 forward, 5′-CCCTTGGGTGTCAAAGGTAA; COX-2 reverse, 5′-TGAAAAGGCGCAGTTTACG-3′. All primers were designed previously, and their specificity was assessed with melting curves during amplification and by 1% agarose gels (53). Quantitative PCR was carried out according to the following cycle: an initial hold at 95°C for 30 s, followed by 39 cycles at 95°C for 2 s and 60°C (for TNF-α and cyclooxygenase COX-2) or 58.2°C (for IL-6, IL-8, and CCL20) for 5 s. Gene expression was normalized to the reference glyceraldehyde-3-phosphate dehydrogenase (gapdh) gene. The amount of template cDNA used for each sample was 15 ng. All results regarding cytokine mRNA expression levels are reported as the fold induction (FOI) relative to expression by the control (unstimulated THP-1 cells), to which we attributed an FOI of 1. Statistically significant differences have been determined by an unpaired Student t test with a two-tailed distribution.
Accession number(s).
The DNA sequence of the EPS-b region has been deposited in the EMBL database under accession number LT629195.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge the financial support of Sofar S.p.A. (research contract 16693 with the University of Milan). Part of the study was financially supported by the “Tesoretto Giovani 2014” grant (MAGIC-MAMPs project) of the University of Milan.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02702-16.
REFERENCES
- 1.D'Inca R, Barollo M, Scarpa M, Grillo AR, Brun P, Vettorato MG, Castagliuolo I, Sturniolo GC. 2011. Rectal administration of Lactobacillus casei DG modifies flora composition and Toll-like receptor expression in colonic mucosa of patients with mild ulcerative colitis. Dig Dis Sci 56:1178–1187. doi: 10.1007/s10620-010-1384-1. [DOI] [PubMed] [Google Scholar]
- 2.Tursi A, Brandimarte G, Giorgetti GM, Modeo ME. 2004. Effect of Lactobacillus casei supplementation on the effectiveness and tolerability of a new second-line 10-day quadruple therapy after failure of a first attempt to cure Helicobacter pylori infection. Med Sci Monit 10:CR662–CR666. [PubMed] [Google Scholar]
- 3.Rosania R, Giorgio F, Principi M, Amoruso A, Monno R, Di Leo A, Ierardi E. 2013. Effect of probiotic or prebiotic supplementation on antibiotic therapy in the small intestinal bacterial overgrowth: a comparative evaluation. Curr Clin Pharmacol 8:169–172. doi: 10.2174/15748847113089990048. [DOI] [PubMed] [Google Scholar]
- 4.Ferrario C, Taverniti V, Milani C, Fiore W, Laureati M, De Noni I, Stuknyte M, Chouaia B, Riso P, Guglielmetti S. 2014. Modulation of fecal Clostridiales bacteria and butyrate by probiotic intervention with Lactobacillus paracasei DG varies among healthy adults. J Nutr 144:1787–1796. doi: 10.3945/jn.114.197723. [DOI] [PubMed] [Google Scholar]
- 5.Spinler JK, Taweechotipatr M, Rognerud CL, Ou CN, Tumwasorn S, Versalovic J. 2008. Human-derived probiotic Lactobacillus reuteri demonstrate antimicrobial activities targeting diverse enteric bacterial pathogens. Anaerobe 14:166–171. doi: 10.1016/j.anaerobe.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guglielmetti S, Taverniti V, Minuzzo M, Arioli S, Zanoni I, Stuknyte M, Granucci F, Karp M, Mora D. 2010. A dairy bacterium displays in vitro probiotic properties for the pharyngeal mucosa by antagonizing group A streptococci and modulating the immune response. Infect Immun 78:4734–4743. doi: 10.1128/IAI.00559-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taverniti V, Stuknyte M, Minuzzo M, Arioli S, De Noni I, Scabiosi C, Cordova ZM, Junttila I, Hamalainen S, Turpeinen H, Mora D, Karp M, Pesu M, Guglielmetti S. 2013. S-layer protein mediates the stimulatory effect of Lactobacillus helveticus MIMLh5 on innate immunity. Appl Environ Microbiol 79:1221–1231. doi: 10.1128/AEM.03056-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guglielmetti S, Zanoni I, Balzaretti S, Miriani M, Taverniti V, De Noni I, Presti I, Stuknyte M, Scarafoni A, Arioli S, Iametti S, Bonomi F, Mora D, Karp M, Granucci F. 2014. Murein lytic enzyme TgaA of Bifidobacterium bifidum MIMBb75 modulates dendritic cell maturation through its cysteine- and histidine-dependent amidohydrolase/peptidase (CHAP) amidase domain. Appl Environ Microbiol 80:5170–5177. doi: 10.1128/AEM.00761-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hidalgo-Cantabrana C, Lopez P, Gueimonde M, de Los Reyes-Gavilan CG, Suarez A, Margolles A, Ruas-Madiedo P. 2012. Immune modulation capability of exopolysaccharides synthesised by lactic acid bacteria and bifidobacteria. Probiotics Antimicrob Proteins 4:227–237. doi: 10.1007/s12602-012-9110-2. [DOI] [PubMed] [Google Scholar]
- 10.Ruas-Madiedo P, Gueimonde M, de los Reyes-Gavilan CG, Salminen S. 2006. Effect of exopolysaccharide isolated from “viili” on the adhesion of probiotics and pathogens to intestinal mucus. J Dairy Sci 89:2355–2358. doi: 10.3168/jds.S0022-0302(06)72307-4. [DOI] [PubMed] [Google Scholar]
- 11.Liu CF, Tseng KC, Chiang SS, Lee BH, Hsu WH, Pan TM. 2011. Immunomodulatory and antioxidant potential of Lactobacillus exopolysaccharides. J Sci Food Agric 91:2284–2291. [DOI] [PubMed] [Google Scholar]
- 12.Polak-Berecka M, Wasko A, Paduch R, Skrzypek T, Sroka-Bartnicka A. 2014. The effect of cell surface components on adhesion ability of Lactobacillus rhamnosus. Antonie Van Leeuwenhoek 106:751–762. doi: 10.1007/s10482-014-0245-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alhudhud M, Humphreys P, Laws A. 2014. Development of a growth medium suitable for exopolysaccharide production and structural characterisation by Bifidobacterium animalis ssp. lactis AD011. J Microbiol Methods 100:93–98. doi: 10.1016/j.mimet.2014.02.021. [DOI] [PubMed] [Google Scholar]
- 14.Balzaretti S, Taverniti V, Rondini G, Marcolegio G, Minuzzo M, Remagni MC, Fiore W, Arioli S, Guglielmetti S. 2015. The vaginal isolate Lactobacillus paracasei LPC-S01 (DSM 26760) is suitable for oral administration. Front Microbiol 6:952. doi: 10.3389/fmicb.2015.00952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guglielmetti S, Tamagnini I, Minuzzo M, Arioli S, Parini C, Comelli E, Mora D. 2009. Study of the adhesion of Bifidobacterium bifidum MIMBb75 to human intestinal cell lines. Curr Microbiol 59:167–172. doi: 10.1007/s00284-009-9415-x. [DOI] [PubMed] [Google Scholar]
- 16.Taverniti V, Guglielmetti S. 2011. The immunomodulatory properties of probiotic microorganisms beyond their viability (ghost probiotics: proposal of paraprobiotic concept). Genes Nutr 6:261–274. doi: 10.1007/s12263-011-0218-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanmani P, Satish Kumar R, Yuvaraj N, Paari KA, Pattukumar V, Arul V. 2013. Probiotics and its functionally valuable products—a review. Crit Rev Food Sci Nutr 53:641–658. doi: 10.1080/10408398.2011.553752. [DOI] [PubMed] [Google Scholar]
- 18.Dertli E, Mayer MJ, Narbad A. 2015. Impact of the exopolysaccharide layer on biofilms, adhesion and resistance to stress in Lactobacillus johnsonii FI9785. BMC Microbiol 15:8. doi: 10.1186/s12866-015-0347-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lebeer S, Vanderleyden J, De Keersmaecker SC. 2010. Host interactions of probiotic bacterial surface molecules: comparison with commensals and pathogens. Nat Rev Microbiol 8:171–184. doi: 10.1038/nrmicro2297. [DOI] [PubMed] [Google Scholar]
- 20.De Vuyst L, Degeest B. 1999. Heteropolysaccharides from lactic acid bacteria. FEMS Microbiol Rev 23:153–177. doi: 10.1111/j.1574-6976.1999.tb00395.x. [DOI] [PubMed] [Google Scholar]
- 21.Ryan PM, Ross RP, Fitzgerald GF, Caplice NM, Stanton C. 2015. Sugar-coated: exopolysaccharide producing lactic acid bacteria for food and human health applications. Food Funct 6:679–693. doi: 10.1039/C4FO00529E. [DOI] [PubMed] [Google Scholar]
- 22.Kojic M, Vujcic M, Banina A, Cocconcelli P, Cerning J, Topisirovic L. 1992. Analysis of exopolysaccharide production by Lactobacillus casei CG11, isolated from cheese. Appl Environ Microbiol 58:4086–4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Calsteren MR, Pau-Roblot C, Begin A, Roy D. 2002. Structure determination of the exopolysaccharide produced by Lactobacillus rhamnosus strains RW-9595M and R. Biochem J 363:7–17. doi: 10.1042/0264-6021:3630007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Landersjo C, Yang Z, Huttunen E, Widmalm G. 2002. Structural studies of the exopolysaccharide produced by Lactobacillus rhamnosus strain GG (ATCC 53103). Biomacromolecules 3:880–884. doi: 10.1021/bm020040q. [DOI] [PubMed] [Google Scholar]
- 25.Robijn GW, Wienk HL, van den Berg DJ, Haas H, Kamerling JP, Vliegenthart JF. 1996. Structural studies of the exopolysaccharide produced by Lactobacillus paracasei 34-1. Carbohydr Res 285:129–139. [DOI] [PubMed] [Google Scholar]
- 26.Vanhaverbeke C, Bosso C, Colin-Morel P, Gey C, Gamar-Nourani L, Blondeau K, Simonet JM, Heyraud A. 1998. Structure of an extracellular polysaccharide produced by Lactobacillus rhamnosus strain C83. Carbohydr Res 314:211–220. doi: 10.1016/S0008-6215(98)00297-3. [DOI] [PubMed] [Google Scholar]
- 27.Gorska S, Schwarzer M, Jachymek W, Srutkova D, Brzozowska E, Kozakova H, Gamian A. 2014. Distinct immunomodulation of bone marrow-derived dendritic cell responses to Lactobacillus plantarum WCFS1 by two different polysaccharides isolated from Lactobacillus rhamnosus LOCK 0900. Appl Environ Microbiol 80:6506–6516. doi: 10.1128/AEM.02104-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gorska-Fraczek S, Sandstrom C, Kenne L, Rybka J, Strus M, Heczko P, Gamian A. 2011. Structural studies of the exopolysaccharide consisting of a nonasaccharide repeating unit isolated from Lactobacillus rhamnosus KL37B. Carbohydr Res 346:2926–2932. doi: 10.1016/j.carres.2011.10.024. [DOI] [PubMed] [Google Scholar]
- 29.Harutoshi T. 2013. Exopolysaccharides of lactic acid bacteria for food and colon health applications, chapter 22 In Kongo JM. (ed), Lactic acid bacteria—R & D for food, health and livestock purposes. InTech doi: 10.5772/50839. [DOI] [Google Scholar]
- 30.Ciszek-Lenda M, Nowak B, Srottek M, Gamian A, Marcinkiewicz J. 2011. Immunoregulatory potential of exopolysaccharide from Lactobacillus rhamnosus KL37: effects on the production of inflammatory mediators by mouse macrophages. Int J Exp Pathol 92:382–391. doi: 10.1111/j.1365-2613.2011.00788.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao K, Wang C, Liu L, Dou X, Liu J, Yuan L, Zhang W, Wang H. 14 May 2015. Immunomodulation and signaling mechanism of Lactobacillus rhamnosus GG and its components on porcine intestinal epithelial cells stimulated by lipopolysaccharide. J Microbiol Immunol Infect doi: 10.1016/j.jmii.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 32.Patten DA, Laws AP. 2015. Lactobacillus-produced exopolysaccharides and their potential health benefits: a review. Benef Microbes 6:457–471. doi: 10.3920/BM2014.0117. [DOI] [PubMed] [Google Scholar]
- 33.Vinderola G, Perdigon G, Duarte J, Farnworth E, Matar C. 2006. Effects of the oral administration of the exopolysaccharide produced by Lactobacillus kefiranofaciens on the gut mucosal immunity. Cytokine 36:254–260. doi: 10.1016/j.cyto.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 34.Chanput W, Mes JJ, Wichers HJ. 2014. THP-1 cell line: an in vitro cell model for immune modulation approach. Int Immunopharmacol 23:37–45. doi: 10.1016/j.intimp.2014.08.002. [DOI] [PubMed] [Google Scholar]
- 35.Miettinen M, Lehtonen A, Julkunen I, Matikainen S. 2000. Lactobacilli and streptococci activate NF-κB and STAT signaling pathways in human macrophages. J Immunol 164:3733–3740. doi: 10.4049/jimmunol.164.7.3733. [DOI] [PubMed] [Google Scholar]
- 36.Habil NY. 2015. Probiotic induce macrophage cytokine production via activation of STAT-3 pathway. Automat Control Intellig Syst 3:1–7. [Google Scholar]
- 37.Zlotnik A, Yoshie O. 2000. Chemokines: a new classification system and their role in immunity. Immunity 12:121–127. doi: 10.1016/S1074-7613(00)80165-X. [DOI] [PubMed] [Google Scholar]
- 38.Harant H, Eldershaw SA, Lindley IJ. 2001. Human macrophage inflammatory protein-3α/CCL20/LARC/Exodus/SCYA20 is transcriptionally upregulated by tumor necrosis factor-alpha via a non-standard NF-κB site. FEBS Lett 509:439–445. doi: 10.1016/S0014-5793(01)03138-6. [DOI] [PubMed] [Google Scholar]
- 39.Veckman V, Miettinen M, Matikainen S, Lande R, Giacomini E, Coccia EM, Julkunen I. 2003. Lactobacilli and streptococci induce inflammatory chemokine production in human macrophages that stimulates Th1 cell chemotaxis. J Leukoc Biol 74:395–402. doi: 10.1189/jlb.0402212. [DOI] [PubMed] [Google Scholar]
- 40.Chabot S, Yu H-L, Léséleuc LD, Cloutier D, Van Calsteren M-R, Lessard M, Roy D, Lacroix M, Oth D. 2001. Exopolysaccharides from Lactobacillus rhamnosus RW-9595M stimulate TNF. Lait 81:683–697. doi: 10.1051/lait:2001157. [DOI] [Google Scholar]
- 41.Jorjao AL, de Oliveira FE, Leao MV, Carvalho CA, Jorge AO, de Oliveira LD. 2015. Live and heat-killed Lactobacillus rhamnosus ATCC 7469 may induce modulatory cytokines profiles on macrophages RAW 264.7. ScientificWorldJournal 2015:716749. doi: 10.1155/2015/716749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lebeer S, Claes IJ, Verhoeven TL, Vanderleyden J, De Keersmaecker SC. 2011. Exopolysaccharides of Lactobacillus rhamnosus GG form a protective shield against innate immune factors in the intestine. Microb Biotechnol 4:368–374. doi: 10.1111/j.1751-7915.2010.00199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yasuda E, Serata M, Sako T. 2008. Suppressive effect on activation of macrophages by Lactobacillus casei strain Shirota genes determining the synthesis of cell wall-associated polysaccharides. Appl Environ Microbiol 74:4746–4755. doi: 10.1128/AEM.00412-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guglielmetti S, Balzaretti S, Taverniti V, Miriani M, Milani C, Scarafoni A, Corona S, Ciranna A, Arioli S, Santala V, Iametti S, Bonomi F, Ventura M, Mora D, Karp M. 2014. TgaA, a VirB1-like component belonging to a putative type IV secretion system of Bifidobacterium bifidum MIMBb75. Appl Environ Microbiol 80:5161–5169. doi: 10.1128/AEM.01413-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. 2011. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Patten DA, Leivers S, Chadha MJ, Maqsood M, Humphreys PN, Laws AP, Collett A. 2014. The structure and immunomodulatory activity on intestinal epithelial cells of the EPSs isolated from Lactobacillus helveticus sp. Rosyjski and Lactobacillus acidophilus sp. 5e2. Carbohydr Res 384:119–127. doi: 10.1016/j.carres.2013.12.008. [DOI] [PubMed] [Google Scholar]
- 47.Gerwig GJ, Kamerling JP, Vliegenthart JFG. 1979. Determination of the absolute configuration of mono-saccharides in complex carbohydrates by capillary G.L.C. Carbohydr Res 77:1–7. doi: 10.1016/S0008-6215(00)83788-X. [DOI] [PubMed] [Google Scholar]
- 48.Ciucanu I, Kerek F. 1984. A simple and rapid method for the permethylation of carbohydrates. Carbohydr Res 131:209–217. [Google Scholar]
- 49.Guglielmetti S, Taverniti V, Minuzzo M, Arioli S, Stuknyte M, Karp M, Mora D. 2010. Oral bacteria as potential probiotics for the pharyngeal mucosa. Appl Environ Microbiol 76:3948–3958. doi: 10.1128/AEM.00109-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stuknyte M, De Noni I, Guglielmetti S, Minuzzo M, Mora D. 2011. Potential immunomodulatory activity of bovine casein hydrolysates produced after digestion with proteinases of lactic acid bacteria. Int Dairy J 21:763–769. doi: 10.1016/j.idairyj.2011.04.011. [DOI] [Google Scholar]
- 51.Tsuchiya S, Yamabe M, Yamaguchi Y, Kobayashi Y, Konno T, Tada K. 1980. Establishment and characterization of a human acute monocytic leukemia cell line (THP-1). Int J Cancer 26:171–176. doi: 10.1002/ijc.2910260208. [DOI] [PubMed] [Google Scholar]
- 52.Traore K, Trush MA, George M Jr, Spannhake EW, Anderson W, Asseffa A. 2005. Signal transduction of phorbol 12-myristate 13-acetate (PMA)-induced growth inhibition of human monocytic leukemia THP-1 cells is reactive oxygen dependent. Leuk Res 29:863–879. doi: 10.1016/j.leukres.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 53.Taverniti V, Minuzzo M, Arioli S, Junttila I, Hamalainen S, Turpeinen H, Mora D, Karp M, Pesu M, Guglielmetti S. 2012. In vitro functional and immunomodulatory properties of the Lactobacillus helveticus MIMLh5-Streptococcus salivarius ST3 association that are relevant to the development of a pharyngeal probiotic product. Appl Environ Microbiol 78:4209–4216. doi: 10.1128/AEM.00325-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.