ABSTRACT
More than 75 “species-level” phylotypes of spirochete bacteria belonging to the genus Treponema reside within the human oral cavity. The majority of these oral treponeme phylotypes correspond to as-yet-uncultivated taxa or strains of uncertain standing in taxonomy. Here, we analyze phylogenetic and taxonomic relationships between oral treponeme strains using a multilocus sequence analysis (MLSA) scheme based on the highly conserved 16S rRNA, pyrH, recA, and flaA genes. We utilized this MLSA scheme to analyze genetic data from a curated collection of oral treponeme strains (n = 71) of diverse geographical origins. This comprises phylogroup 1 (n = 23) and phylogroup 2 (n = 48) treponeme strains, including all relevant American Type Culture Collection reference strains. The taxonomy of all strains was confirmed or inferred via the analysis of ca. 1,450-bp 16S rRNA gene sequences using a combination of bioinformatic and phylogenetic approaches. Taxonomic and phylogenetic relationships between the respective treponeme strains were further investigated by analyzing individual and concatenated flaA (1,074-nucleotide [nt]), recA (1,377-nt), and pyrH (696-nt) gene sequence data sets. Our data confirmed the species differentiation between Treponema denticola (n = 41) and Treponema putidum (n = 7) strains. Notably, our results clearly supported the differentiation of the 23 phylogroup 1 treponeme strains into five distinct “species-level” phylotypes. These respectively corresponded to “Treponema vincentii” (n = 11), Treponema medium (n = 1), “Treponema sinensis” (Treponema sp. IA; n = 4), Treponema sp. IB (n = 3), and Treponema sp. IC (n = 4). In conclusion, our MLSA-based approach can be used to effectively discriminate oral treponeme taxa, confirm taxonomic assignment, and enable the delineation of species boundaries with high confidence.
IMPORTANCE Periodontal diseases are caused by persistent polymicrobial biofilm infections of the gums and underlying tooth-supporting structures and have a complex and variable etiology. Although Treponema denticola is strongly associated with periodontal diseases, the etiological roles of other treponeme species/phylotypes are less well defined. This is due to a paucity of formal species descriptions and a poor understanding of genetic relationships between oral treponeme taxa. Our study directly addresses these issues. It represents one of the most comprehensive analyses of oral treponeme strains performed to date, including isolates from North America, Europe, and Asia. We envisage that our results will greatly facilitate future metagenomic efforts aimed at characterizing the clinical distributions of oral treponeme species/phylotypes, helping investigators to establish a more detailed understanding of their etiological roles in periodontal diseases and other infectious diseases. Our results are also directly relevant to various polymicrobial tissue infections in animals, which also involve treponeme populations.
KEYWORDS: dentistry, periodontitis, spirochete, oral microbiome, infection, DNA sequencing, MLSA, Treponema, bacterial genome, phylogeny, soft tissue infection, taxonomy
INTRODUCTION
Oral treponeme bacteria (genus Treponema) constitute a small but significant part of the “normal” human oral microbiota (1). This genus of spirochete bacteria shares a distinctive helical or flat-wave cell morphology, and many species are highly motile in liquid or semisolid environments, locomoting via a corkscrew-like motion (2–4). They typically occupy anaerobic niches, especially within dental plaque located in the gingival crevice: the narrow gap between the gingivae (gums) and the base of the crown of the tooth (5, 6). Their proportions are often highly elevated within diseased periodontal sites, and clinical studies have consistently associated them with various forms of periodontal disease, especially chronic periodontitis (7–12). High levels of treponeme taxa are also frequently detected within endodontic infections and oral-implant infections (e.g., peri-implantitis) (13–17).
Nine species of oral treponemes have been formally characterized in the scientific literature (the reporting year is indicated in parentheses): Treponema denticola (1993) (18); Treponema pectinovorum (1983) (19); Treponema socranskii (1998; three subspecies: socranskii, paredis, and buccale) (20); Treponema maltophilum (1996) (21); Treponema medium (1997) (22), whose description was amended in 2003 (23); Treponema amylovorum (1997) (24); Treponema lecithinolyticum (1999) (25); Treponema parvum (2001) (26); and Treponema putidum (2004) (27). “Treponema vincentii” is also generally considered to constitute an oral treponeme species, having been studied and reported in the scientific literature for more than 60 years (28–31). However, in the absence of a recent systematic characterization, its precise taxonomic definition remains obscure (9, 30).
Dewhirst et al. proposed a taxonomic framework for the classification of oral treponemes, based on shared 16S rRNA gene homology (32), which has become widely adopted by the scientific community. This places oral treponeme taxa within 10 different “phylogroups” (phylogenetic clusters) that respectively share more than 90% 16S rRNA gene sequence similarity (over a 500-bp region). At this time, more than 75 Treponema “species-level” phylotypes have been identified in the human oral cavity using 16S rRNA gene sequence-based approaches (1, 33). Many phylotypes have been assigned human oral taxon (HOT) codes, as part of the Human Oral Microbiome Database (HOMD) classification scheme (1). However, roughly three-quarters of identified oral treponeme phylotypes remain to be cultivated and formally described.
Placed within phylogroup 2, clinical investigations have consistently associated the presence or abundance of T. denticola cells with the occurrence and severity of periodontal disease (10–12, 34, 35). This phylogroup also contains T. putidum, a poorly studied treponeme that has been detected within periodontitis, acute necrotizing ulcerative gingivitis (ANUG), and endodontic lesions (27, 36). Notably, phylogroup 2 treponemes share high levels of phylogenetic similarity with Treponema pallidum, Treponema phagedenis, and Treponema pedis (31, 37), as well as many other isolates and as-yet-uncultivated taxa commonly found in polymicrobial tissue infections in livestock animals (38, 39).
The species T. medium and T. vincentii both belong to phylogroup 1. Consensus results from previous molecular clinical investigations have indicated that treponeme taxa belonging to phylogroup 1 are particularly prevalent, abundant and diverse within human subgingival plaque (33, 40). Furthermore, several phylogroup 1 taxa and species have previously been associated with periodontal disease (12, 33, 41, 42).
Complete genome sequences for T. putidum OMZ 758T (ATCC 700334T) and Treponema sp. strain OMZ 838 (“Treponema sinensis” [ATCC 700772]) have recently been published (43, 44). Furthermore, in as-yet-unpublished work, researchers working under the auspices of the Human Microbiome Project (45) have recently deposited genome sequence data for the type strain of T. medium (ATCC 700293, NCBI Genome BioProject no. 169490, sequenced by the Broad Institute), as well as for two strains of T. vincentii (ATCC 35580, NCBI Genome BioProject no. 34095, sequenced by the J. Craig Venter Institute; and F0403/N9, NCBI Genome BioProject no. 169493, sequenced by the Broad Institute). However, due to the relative paucity of genetic and genomic data for treponeme isolates belonging to phylogroup 1, the unambiguous identification and classification of taxa within this group remains highly problematic.
Multilocus sequence analysis (MLSA) and related multilocus sequence typing (MLST) approaches have previously been successfully used to delineate boundaries between closely related spirochete species, subspecies, and component strains. This has included Borrelia (46, 47), Brachyspira (48), and Leptospira (49) spp. and treponemes associated with digital dermatitis in cloven-hoofed animals (38, 50). Phylogenetic analysis of the 16S rRNA gene and 16S-23S rRNA intergenic spacer regions have also been commonly used for the analysis of Treponema spp. within animal foot and soft tissue lesions (see, for example, references 51 and 52).
Here, we used a four-gene MLSA approach to delineate genetic similarities and differences between oral treponeme reference strains and clinical isolates belonging to phylogroups 1 and 2. Our data suggest that several isolates previously classified as T. vincentii and T. medium should be placed within other taxonomic groupings, some of which most probably constitute novel species.
RESULTS
Obtaining gene sequences data sets from phylogroup 1 and 2 oral treponeme strains.
A total of 71 human oral treponeme strains belonging to oral phylogroups 1 and 2 were included in our MLSA (Table 1). This included T. denticola (n = 41), T. putidum (n = 7), T. medium (n = 1), and T. vincentii (n = 11), as well as treponeme strains of uncertain standing in taxonomy (n = 11). These treponeme strains were originally isolated from various (diseased) oral sites, predominantly periodontitis lesions, from individuals who lived in North America, Europe, or Asia. This included all relevant ATCC (and DSMZ) oral treponeme reference strains. Additional details about the origins of these strains are summarized in Table S1 in the supplemental material.
TABLE 1.
Summary of oral treponeme strains included in MLSA
| Phylogroup and species (no. of strains) | Strains |
|---|---|
| Phylogroup 1 | |
| T. vincentii (11) | ATCC 35580 (OMZ 293, LA-1), ATCC 700013 (OMZ 779, N9, F0403), ATCC 700765 (OMZ 800, DSM16788), ATCC 700774 (OMZ 860), OMZ 801, OMZ 802, OMZ 858, OMZ 859, OMZ 861, OMZ 862, OMZ 863 |
| T. medium (1) | ATCC 700293T (ex. G7201, OMZ 824) |
| T. sinensis IA (4) | ATCC 700772 (OMZ 838, DSM 16789), OMZ 855, OMZ 856, OMZ 857 |
| Treponema sp. IB (3) | OMZ 305 (T. vincentii Ritz A), OMZ 805, ATCC 700767 (OMZ 806, DSM 16787) |
| Treponema sp. IC (4) | ATCC 700766 (OMZ 804, MH1F1), OMZ 803, OMZ 899, OMZ 906 |
| Phylogroup 2 | |
| T. denticola (41) | ATCC 35405T (strain a, OMZ 661, DSM 14222), ATCC 35404 (strain c, TD-4, OMZ 663), ATCC 33521 (strain 11, OMZ 662), ATCC 33520 (strain W), ATCC 700771 (OMZ 834), ATCC 700768 (OMZ 830), CD-1 (OMZ 294), S2, OTK, OKA3, OT2B, MS25, GM-1, ST10 (OMZ 829), NY531, NY535, NY545, NY553, F0402, OMZ 823, OMZ 845, OMZ 849, OMZ 850, OMZ 852, OMZ 853, OMZ 854, OMZ 898, OMZ 905, OMZ 908, OMZ 910, H1-T (HMS-570), SP37 (HMS-569), SP44, SP33, SP32, SP23, AL-2 (HMS-575), ASLM (HMS-574), US-Trep (HMS-573), MYR-T (HMS-572), H-22 (HMS-571) |
| T. putidum (7) | ATCC 700334T (OMZ 758), OMZ 730, OMZ 835, OMZ 844, OMZ 846, OMZ 847, OMZ 848 |
Four genes were selected for analysis in each oral treponeme strain: the 16S rRNA gene (rrs), as well as the flaA, recA, and pyrH coding DNA sequences (CDSs). The recA gene encodes the main DNA homologous recombination and repair protein in bacteria, recombinase A (53). The pyrH gene encodes uridylate kinase (UMP kinase), which plays a key role in nucleotide metabolism in bacteria (54). The flaA gene encodes the FlaA flagellar outer sheath protein, which coats the core of the (periplasmic) flagellar filament, which is comprised of the FlaB protein (55). The 16S rRNA gene is present in two identical copies (rrsA and rrsB) in Treponema taxa (37). The others are single-copy CDSs, which correspond to the following three loci in the genome-sequenced T. denticola ATCC 35405T strain: pyrH (TDE2085; 696 bp), flaA (TDE1712; 1,050 bp), and recA (TDE0872; 1,245 bp) (37). These three CDSs were selected based on the results obtained from our previous phylogenetic analysis of 20 T. denticola strains (56).
We were able to obtain full-length gene sequence data for flaA, recA, and pyrH from 36 oral phylogroup 1 and 2 strains by searching the publically accessible nucleotide sequence depositories. Using these data, various sets of PCR primers were designed to amplify full-length flaA, recA, and pyrH gene sequences from phylogroup 1 and 2 oral treponeme strains (summarized in Table 2). In addition, a total of 35 phylogroup 1 and 2 oral treponeme strains from our curated collection were cultivated, and full-length gene sequences were successfully obtained for these strains. The primer sequences and PCR conditions used are summarized in Table 2. Near-full-length (∼1,450-bp) regions of the 16S rRNA gene from each cultivated treponeme strain were also obtained. The accession codes for all gene sequences used in this study are summarized in Table S2 in the supplemental material.
TABLE 2.
Primers used for PCR amplification of target gene sequences
| Gene | Primer | Sequence (5′–3′) | Tma (range in °C) | Description (source or reference) |
|---|---|---|---|---|
| 16S rRNA | TPU1 | AGAGTTTGATCMTGGCTCAG | 56–53 | All treponeme strains (33) |
| C90 | GTTACGACTTCACCCTCCT | |||
| pyrH | TvpyrH-F | ATGGTACGGGTCTTATCGGTAG | 55–49 | All phylogroup 1 treponemes and primers are based on T. vincentii ATCC 35580 (this study) |
| TvpyrH-R | TTAACCTATCGTTGTGCCTTTAA | |||
| pyrH-F | ATGGTAACTGTTTTGTCGGT | 54–47 | All phylogroup 2 treponemes and primers are based on T. denticola ATCC 35405 (56) | |
| pyrH-R | TTAGCCGATTACCGTTCCTT | |||
| recA | TvrecA-F | ATGGCAAAAACAAAATCAGAAA | 55–52 | All phylogroup 1 treponemes and primers are based on T. vincentii ATCC 35580 (this study) |
| TvrecA-R | TTAAAAGAGCTCGTTGTCGC | |||
| recA-F1 | GTGGCAAAAGCAAAAAACGAAG | 55–52 | Most phylogroup 2 treponemes except T. denticola ATCC 700771, ATCC 700768, and MS25 and primers are based on T. denticola ATCC 35405 (56) | |
| recA-R1 | TTAAAAAAGACTGTCGTCCGCC | |||
| recA-F2 | TTCATATTGGCCGCATTTG | 54–47 | T. denticola ATCC 700768 and MS25 and primers are based on regions 117 bp upstream and 69 bp downstream of recA gene on T. denticola ATCC 35405 chromosome (56) | |
| recA-R2 | TTGTGTACTCATAATGCCGCTC | |||
| recA-F1 | GTGGCAAAAGCAAAAAACGAAG | 55–49 | T. denticola ATCC 700771 and primers are as described above (56) | |
| recA-R2 | TTGTGTACTCATAATGCCGCTC | |||
| flaA | TvflaA-F | ATGAAAAGAACAAGCATACTTGTAG | 55–49 | All phylogroup 1 treponemes except Treponema sp. IB and primers are based on T. vincentii ATCC 35580 (this study) |
| TvflaA-R | CTATTGCTTTGTATTTTCGGC | |||
| TvflaA-F | ATGAAAAGAACAAGCATACTTGTAG | 55–49 | Treponema sp. IB treponemes and the 305flaA-R primer are based on the flaA gene extracted from genome sequence data for Treponema sp. strain OMZ 305 (this study) | |
| 305flaA-R | TTAGTTTGCAGCCTCTGTTG | |||
| TDE1712-F | ATGAAAAAAACATTTATACTTGTTG | 52–46 | All phylogroup 2 treponemes and primers are based on T. denticola ATCC 35405 (56) | |
| TDE1712-R | TTATTGTTGGTTCTTTTCGG |
Touchdown PCRs were performed, with the annealing temperature decreased by ca. 0.5 to 1°C every cycle for the first six cycles (according to the range of temperatures indicated in the Tm column), thereafter using the lowest temperature within the range shown for the remaining 27 cycles.
Assignment of taxonomy and phylogeny of oral treponeme strains based on the 16S rRNA gene.
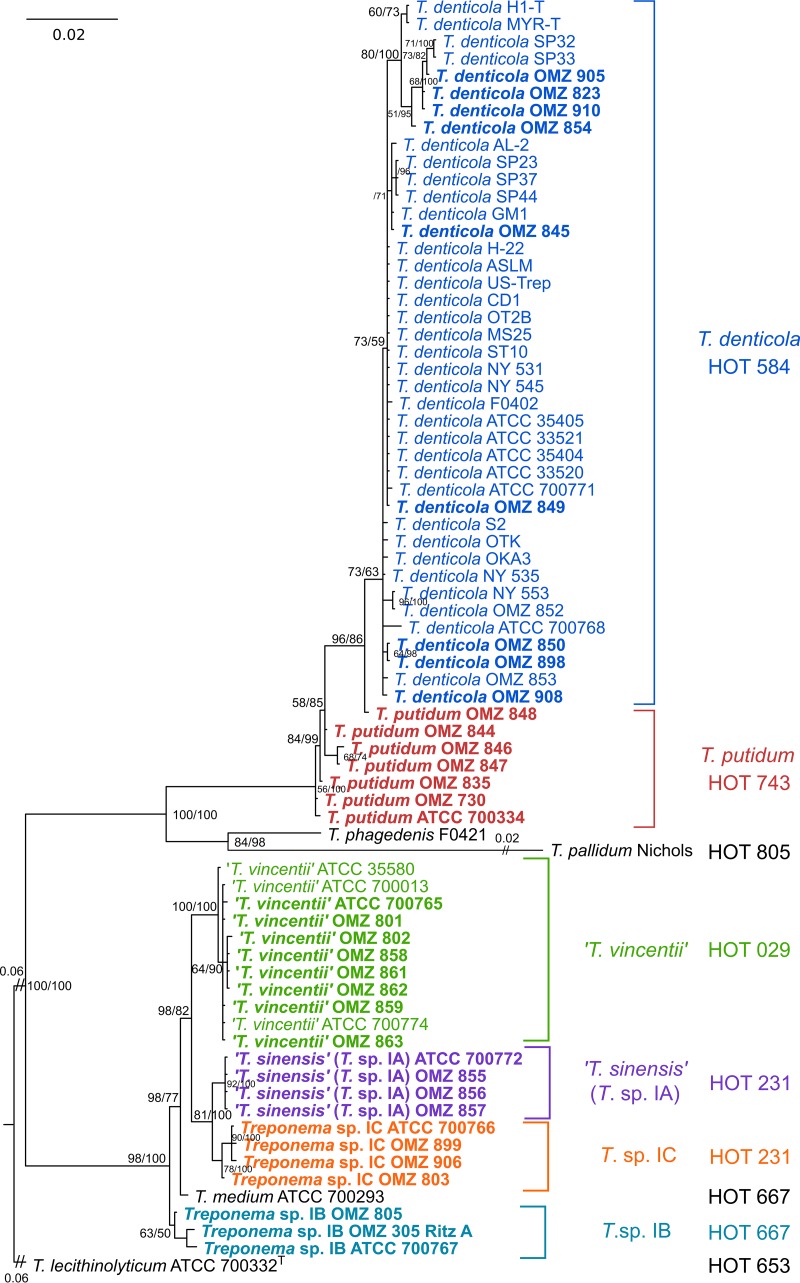
We first assigned taxonomy for each oral treponeme strain (n = 71) to the species or phylotype level by analyzing the 16S rRNA gene sequence data sets using a maximum-likelihood (ML) phylogenetic approach. This included 23 phylogroup 1 strains and 48 phylogroup 2 strains (listed in Table 1). T. phagedenis F0421 and T. pallidum subsp. pallidum strain Nichols were included for comparative purposes, and T. lecithinolyticum ATCC 700332T (which belongs to phylogroup 4) was used as an outgroup. The GTR+I+G nucleotide substitution model was determined to be optimal for the analysis of 16S rRNA data sets according to jModelTest (57). The ML tree generated is shown in Fig. 1. For clarity, the respective treponeme species (or proposed species) are indicated with different colors. The corresponding human oral taxon (HOT) numbers, as defined by the HOMD (58), are also indicated for each strain.
FIG 1.
ML phylogenetic tree of oral treponeme 16S rRNA genes. The maximum clade credibility tree topology is supported by bootstrapping for 1,000 replicates (first number) and Bayesian posterior probabilities (second number) separated with a forward slash “/” symbol in percentages; only values over 40% are shown beside the branch nodes. Sequences generated in this study are in boldface type. A scale bar indicates the nucleotide changes per position. Excessively long branches were trimmed in proportion to the scale bar (indicated by “//” on the branches). The respective oral treponeme species (phylogroups) are indicated with different color shadings. The corresponding HOT numbers are also indicated as follows: T. denticola (HOT 584), dark blue; T. putidum (HOT 743), red; T. vincentii (HOT 029), light green; Treponema sp. IA (HOT 231), purple; Treponema sp. IB (HOT 667), light blue; and Treponema sp. IC (HOT 231), orange.
It may be seen in the phylogram (Fig. 1) that the 41 T. denticola (HOT 584; dark blue) and 7 T. putidum (HOT 743; red) strains were well separated from each other based on their 16S rRNA gene sequences. Both of these phylogroup 2 species were well separated from T. pallidum (HOT 805), T. phagedenis, and the phylogroup 1 treponeme strains. This was as expected and is in good accordance to analogous phylogenetic analyses of oral treponeme populations reported in previous studies (1, 27, 31–33). Six of the seven T. putidum strains, including the ATCC 700334T strain, clustered together. The OMZ 848 strain appeared to be slightly divergent, being more closely related to T. denticola strains. It should be noted that our analysis includes every T. putidum strain that has been characterized and reported in the literature (27).
The phylogroup 1 treponeme strains formed four well-separated clades with good bootstrap support. The T. medium type strain (ATCC 700293, ex. [formerly] G7201 [22, 23]) formed a single taxon branch. All 11 T. vincentii strains formed a single clade (HOT 029; light green in Fig. 1). This clade included four American Type Culture Collection (ATCC) reference strains: ATCC 35580 (LA-1), 700013 (F0403, N9), 700774 (OMZ 860), and 700765 (OMZ 800). The three other clades were designated Treponema sp. IA (purple), Treponema sp. IB (light blue), and Treponema sp. IC (orange), respectively. The IA and IC phylotypes both corresponded to HOT 231, whereas the IB phylotypes corresponded to HOT 667, which also contains T. medium ATCC 700293T.
It has been proposed (C. Wyss, unpublished data) that Treponema sp. strain ATCC 700772 (OMZ 838), whose full genome sequence has recently been reported (44), as well as three other Treponema strains—OMZ 855, OMZ 856, and OMZ 857—correspond to a new species that has tentatively been named Treponema sinensis (to reflect the strain's Chinese origins) (59). These four T. sinensis strains correspond to Treponema sp. IA and are well separated from the other phylotypes and species. Furthermore, since the respective T. medium, T. vincentii, T. sinensis (IA), and Treponema sp. IB and IC taxa are all well segregated into distinct phylogenetic lineages in the ML tree in Fig. 1, it appears to be valid to refer to these respective clades or single branch taxa as “species-level” phylotypes.
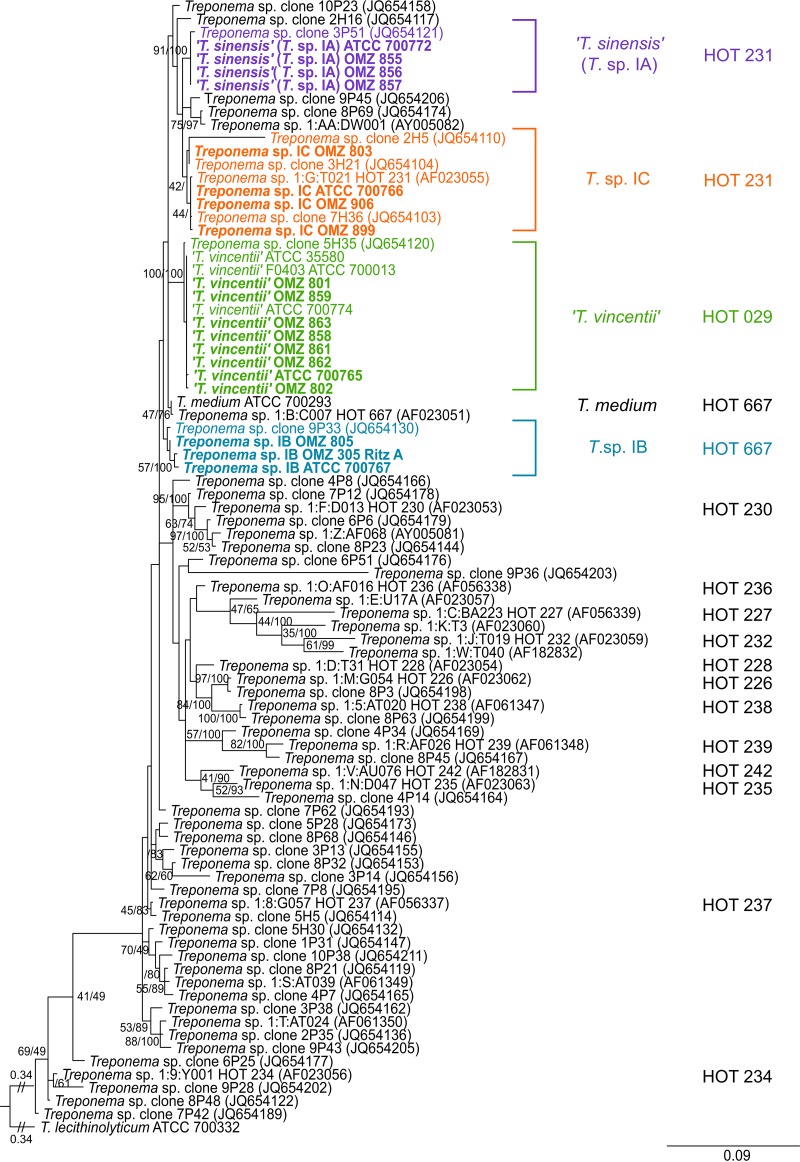
To further investigate taxonomic and phylogenetic relationships between phylogroup 1 treponeme strains, we performed a further ML phylogenetic analysis incorporating an additional set of n = 41 16S rRNA gene sequences, which were previously obtained from a plasmid library of (human) oral treponeme 16S rRNA gene amplicons (33). These were representatives of 265 unique cloned amplicon sequences based on a 99% similarity cutoff. This plasmid library was constructed in an analogous manner to the procedure employed for this study, using DNA extracted from pooled subgingival plaque samples obtained from a cohort of periodontitis patients (n = 10) and periodontitis-free individuals (n = 10). The section of the ML phylogram generated, which contains all oral phylogroup taxa is shown in Fig. 2. The corresponding oral treponeme phylotypes that have been previously defined by Dewhirst et al. (32) and You et al. (33) are indicated for comparative purposes.
FIG 2.
ML phylogenetic tree of 16S rRNA genes from phylogroup 1 oral treponemes. Refer to Fig. 1 legend for explanatory details.
It may be seen that the overall topology (branching pattern) of the ML tree generated from the 16S rRNA data set that included all the additional (uncultivated) treponeme phylotypes was very similar to that of the ML tree shown in Fig. 1. Specifically, the phylogenetic arrangements and strain compositions of the five respective species or “species-level” phylotypes—T. vincentii (HOT 029), T. medium (HOT 667), Treponema sp. IB (HOT 667), T. sinensis (Treponema sp. IA; HOT 231), and Treponema sp. IC (HOT 231)—were well preserved after the addition of the uncultivated oral treponeme taxa. The T. vincentii clade contained the Treponema sp. clone 5H35 (GenBank accession no. JQ654120) (33). The T. sinensis (Treponema sp. IA) clade contained the Treponema sp. clone 3P51 (JQ654121) (33). The Treponema sp. IC clade contained the “group 01 cluster G (I:G:T21)” (AF023055) (32), and the Treponema sp. clones 2H5 (JQ654110), 3H21 (JQ654104), and 7H36 (JQ654103) (33). The T. medium clade contained the “group 01 cluster B (I:B:C7)” (AF023051) (32), and the Treponema sp. IB clade contained the Treponema sp. clone 9P33 (JQ654130) (33). The Treponema sp. clones 10P23 (JQ654158), 2H16 (JQ654117), 9P45 (JQ654206), and 8P69 (JQ654174) and the “group 01 cluster AA (1:AA:DW001)” (AY005082) (32) were closely related to, but distinct from, the T. sinensis and Treponema sp. IC clades.
Phylogenetic analyses of individual flaA, recA, and pyrH genes.
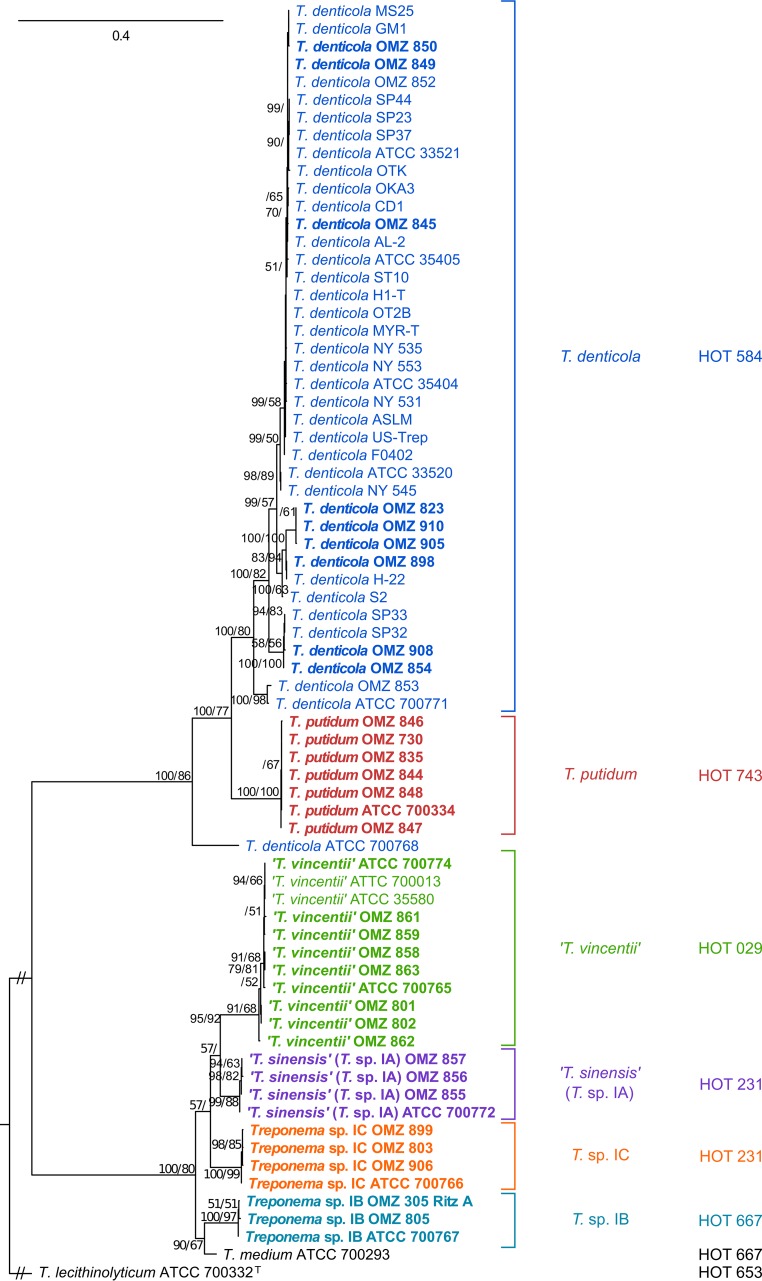
Individual ML trees were calculated for the flaA, recA, and pyrH genes to investigate phylogenetic relationships between oral treponeme strains at three different genetic loci. The optimal nucleotide substitution models were determined to be GTR+G for flaA, and GTR+I+G for recA and pyrH according to jModelTest (57). In the flaA (Fig. 3) and pyrH (see Fig. S1B in the supplemental material) ML phylogenetic trees, the seven Treponema putidum strains were closely clustered and well separated from T. denticola strains. Interestingly, T. denticola ATCC 700768 was more closely related to T. putidum in the pyrH ML tree and was an outlier in the flaA ML tree. The recA ML tree (see Fig. S1A in the supplemental material) had a different topology, with the seven T. putidum strains forming three clades that were intertwined with T. denticola strains. The branching patterns for the T. denticola strains exhibited considerable variation between the three respective single gene ML trees.
FIG 3.
ML phylogenetic tree of individual flaA genes. Refer to Fig. 1 legend for explanatory details.
For the phylogroup 1 oral treponemes, there was consistency in the branching patterns observed in the respective flaA, recA, and pyrH ML trees (Fig. 3 and see Fig. S1 in the supplemental material). In each tree, there were four clusters that respectively contained the T. vincentii, T. sinensis (Treponema sp. IA), Treponema sp. IB, and Treponema sp. IC taxa. The topologies were analogous to those present in the 16S rRNA ML tree, with only two minor differences. The Treponema sp. IB OMZ 305 (Ritz A) strain was distinct from the other Treponema sp. IB taxa and formed a clade with the T. medium type strain (ATCC 700293, ex. G7201) in the recA ML tree (see Discussion for the details of further analyses). The T. vincentii ATCC 700013 (F0403, N9) strain differed from the other T. vincentii strains in the pyrH ML tree by being more closely related to the other phylogroup 1 strains outside the T. vincentii clade. The phylogeny of these three genetic loci consistently showed that T. medium ATCC 700293T was closely related to the Treponema sp. IB taxa.
Phylogenetic analyses of concatenated two-gene and three-gene sequence data sets.
ML phylogenetic analysis was next performed on three different combinations of two-gene concatenations (i.e., flaA-recA [2,360 bp], flaA-pyrH [1,667 bp], and recA-pyrH [2,017 bp]) in order to determine which pair of protein-coding genes may be most effective for the delineation of phylogroup 1 and 2 treponeme strains. The best nucleotide substitution model for flaA-recA and recA-pyrH was GTR+I+G, with GTR+G being optimal for flaA-pyrH. The three ML phylogenetic trees generated are shown, respectively, in Fig. S2A to C in the supplemental material. In the ML trees generated from all three of the two-gene combinations, the 23 phylogroup 1 taxa showed the same clustering patterns, with T. vincentii, Treponema sp. IA, Treponema sp. IB, and Treponema sp. IC taxa each forming individual clades. T. medium ATCC 700293T formed a single branch, which was most closely related to the Treponema sp. IB clade. The 42 phylogroup 2 strains showed very similar clustering patterns in all three of the two-gene ML trees, which were analogous to those present in the individual flaA and pyrH ML trees. T. denticola ATCC 700768 was more closely related to T. putidum in the recA-pyrH ML tree and was an outlier in the flaA-recA and flaA-pyrH ML trees.
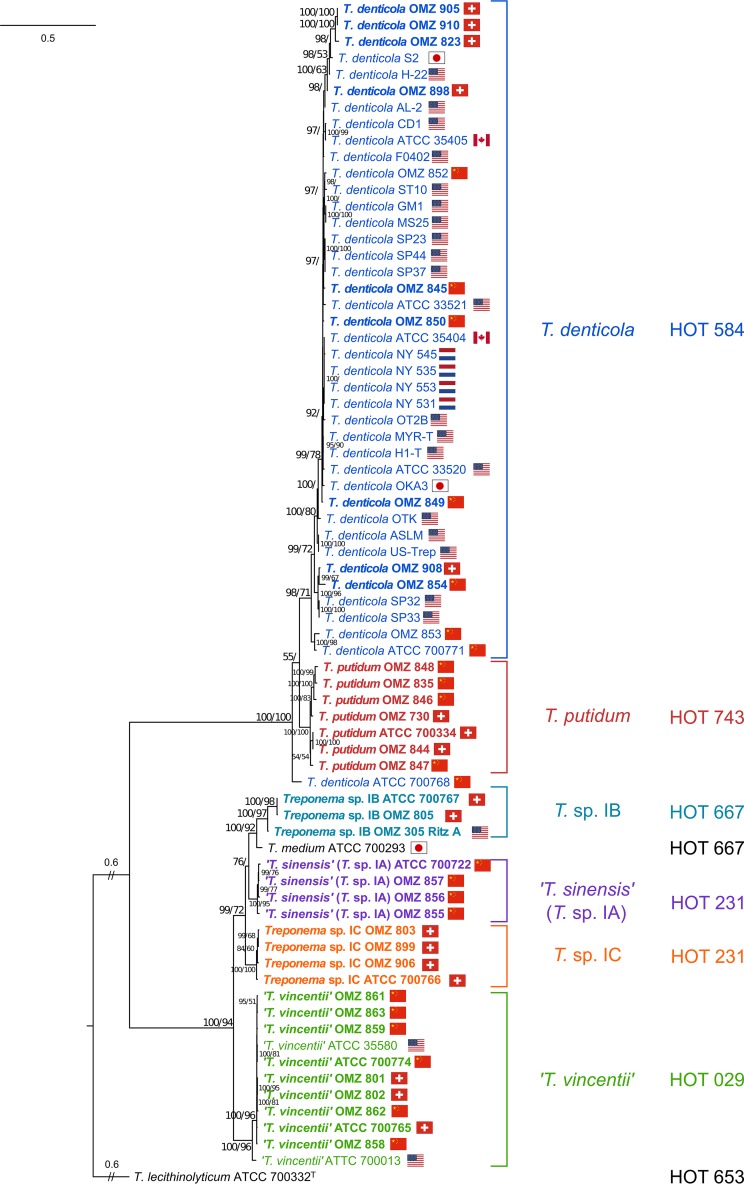
The optimum nucleotide substitution model for the ML phylogenetic analysis of the concatenated flaA-recA-pyrH three-gene data set (3,022 bp) was determined to be GTR+I+G. The phylogenetic analysis revealed a very clear distinction between phylogroups 1 and 2 and also well-defined clades within each phylogroup, all with high bootstrap and Bayesian posterior probability support values (Fig. 4). The concatenated three-gene ML tree had a very similar overall topology as the recA-pyrH ML tree, but with generally higher statistical support for the branches. As noted above, T. denticola ATCC 700768 was a notable outlier in the three-gene ML tree, being more closely related to the T. putidum strains. All 11 of the T. vincentii strains formed a separate clade, with the ATCC 700013 strain appearing somewhat distinct from the others. The respective T. sinensis and Treponema sp. IC taxa formed very clearly demarcated clades, with very high levels of statistical support. The distinction between T. medium ATCC 700293 and the three Treponema sp. IB taxa was less well defined, especially regarding the phylogenetic position of the Treponema sp. IB Ritz A (OMZ 305) strain.
FIG 4.
ML phylogenetic tree constructed from concatenated flaA-recA-pyrH gene data sets. Refer to Fig. 1 legend for explanatory details. The geographical origins of the respective clinical treponeme isolates are indicated with the corresponding countries' flags.
Bioinformatic analysis of 16S rRNA, flaA, recA, and pyrH gene sequences.
The respective lengths of the 16S rRNA gene sequences obtained were highly consistent with the taxonomic assignment (Table 3). All of the T. denticola strains had 16S rRNA genes 1,497 bp in length, making them distinct from the T. putidum strains, which encoded 16S rRNA genes that were 1,496 bp in length. All of the phylogroup 1 treponeme strains had 16S rRNA genes 1,502 bp in length. There were only relatively minor differences in the mean G+C content of the 16S rRNA genes present in the respective species/phylotypes, ranging from ca. 51.9 to 52.5% (Table 3).
TABLE 3.
Average gene length and G+C content of oral treponeme 16S rRNA, flaA, pyrH, and recA genes analyzed
| Species (no. of strains) and parameter | Mean gene length and G+C content |
|||
|---|---|---|---|---|
| 16S rRNA | flaA | recA | pyrH | |
| T. vincentii (11) | ||||
| Mean gene length in bp (range) | 1,502 (1,502) | 1,062 (1,062) | 1,326 (1,326) | 687 (687) |
| Mean gene G+C content (%) ± SD | 51.9 ± 0.03 | 45.4 ± 0.07 | 50.5 ± 0.10 | 47.5 ± 0.76 |
| T. medium (1) | ||||
| Mean gene length in bp (range) | 1,502 (1,502) | 1,068 (1,068) | 1,362 (1,362) | 687 (687) |
| Mean gene G+C content (%) ± SD | 52.3 | 45.3 | 50.8 | 45.0 |
| T. sinensis (4) | ||||
| Mean gene length in bp (range) | 1,502 (1,502) | 1,059 (1,059) | 1,338 (1,338) | 687 (687) |
| Mean gene G+C content (%) ± SD | 52.5 ± 0.00 | 44.7 ± 0.13 | 50.7 ± 0.19 | 46.3 ± 0.14 |
| Treponema sp. IB (3) | ||||
| Mean gene length in bp (range) | 1,502 (1,502) | 1,068 (1,068) | 1,362 (1,359−1,368) | 687 (687) |
| Mean gene G+C content (%) ± SD | 52.4 ± 0.15 | 46.5 ± 0.12 | 51.3 ± 0.17 | 46.7 ± 0 |
| Treponema sp. IC (4) | ||||
| Mean gene length in bp (range) | 1,502 (1,502) | 1,064 (1,062−1,065) | 1,348 (1,347−1,350) | 687 (687) |
| Mean gene G+C content (%) ± SD | 52.3 ± 0.00 | 45.4 ± 0.13 | 50.6 ± 0.17 | 46.0 ± 0.25 |
| T. denticola (41) | ||||
| Mean gene length in bp (range) | 1,497 (1,497) | 1,050 (1,050) | 1,245 (1,245) | 696 (696) |
| Mean gene G+C content (%) ± SD | 52.4 ± 0.09 | 40.6 ± 0.40 | 45.6 ± 0.44 | 41.8 ± 0.38 |
| T. putidum (7) | ||||
| Mean gene length in bp (range) | 1,496 (1,496) | 1,050 (1,050) | 1,245 (1,245) | 696 (696) |
| Mean gene G+C content (%) ± SD | 52.3 ± 0.18 | 39.8 ± 0.05 | 45.0 ± 0.61 | 40.1 ± 0.04 |
The lengths of the pyrH gene sequences (obtained from databases or determined experimentally here) were strictly conserved within the phylogroup 1 and phylogroup 2 strains, respectively (Table 3). The pyrH gene was 696 bp in length for all phylogroup 2 (T. denticola and T. putidum) strains and was 687 bp in all phylogroup 1 taxa (T. vincentii, T. medium, T. sinensis [Treponema sp. IA], Treponema sp. IB, and Treponema sp. IC). The flaA genes in all phylogroup 2 strains were 1,050 bp in length. However, this was one to four codons shorter than the corresponding flaA genes in phylogroup 1 strains, which were different in each of the species or phylotypes: T. vincentii (1,062 bp), T. medium (1,068 bp), T. sinensis (1,059 bp), Treponema sp. IB (1,068 bp), and Treponema sp. IC (1,062 to 1,065 bp). Similarly, the length of recA gene was identical within all phylogroup 2 strains (1,245 bp) but varied among the phylogroup 1 phylotypes. The recA gene sequences within the phylogroup 1 strains were notably longer (1,326 to 1,368 bp) than those from phylogroup 2. The respective recA gene lengths were consistent within T. vincentii (1,326 bp) and T. sinensis (1,338 bp) phylotypes but varied slightly within the Treponema sp. IB (1,359 to 1,368 bp) and Treponema sp. IC (1347 to 1350 bp) phylotypes. The recA gene in T. medium (1,362 bp) was distinct from the other phylotypes.
There were notable trends in the respective G+C contents of the flaA, recA, and pyrH genes. The respective G+C contents of the individual flaA, recA, or pyrH genes within the respective treponeme phylotypes or species were very similar (± 0-0.76%). However, the respective G+C contents of the flaA, recA, and pyrH genes in the oral phylogroup 1 treponemes were all ca. 4 to 7% higher than the corresponding genes in T. denticola and T. putidum. For example, the mean G+C content for the pyrH gene in T. denticola strains was 41.8 ± 0.38%, whereas in T. vincentii it was 47.5 ± 0.76%. In addition, in all strains studied, the respective G+C content of the recA gene was ca. 4 to 5% higher than that of the corresponding flaA or pyrH genes (Table 3).
Nucleotide diversity, polymorphism, and evolutionary conservation of codon sites.
The respective levels of nucleotide diversity (π) and the number of polymorphic sites were calculated for all four genes at the phylogroup level and at the species/phylotype level. The evolutionary conservation of codon sites was calculated for the flaA, pyrH, and recA genes (since the 16S rRNA gene does not encode a protein). The results are summarized in Table 4 (see also Table S3 in the supplemental material).
TABLE 4.
Summary of polymorphic sites, nucleotide diversity per site, global rate ratios, and positively/negatively selected codon sitesa
| Species (no. of strains) | Gene | No. of nt | No. of codons | No. (%) of polymorphic sites | Mean nucleotide diversity (π) ± SD | Mean dN/dS (global ω; 95% CI) | No. of sites (%) |
|
|---|---|---|---|---|---|---|---|---|
| Negatively selected | Positively selected | |||||||
| T. vincentii (11) | flaA | 1,062 | 353 | 19 (1.79) | 0.006 ± 0.0010 | 0.103 (0.032–0.239) | 1 (0.28) | 0 |
| recA | 1,326 | 441 | 24 (1.71) | 0.004 ± 0.0005 | 0.060 (0.019–0.139) | 0 | 0 | |
| pyrH | 687 | 228 | 106 (14.60) | 0.031 ± 0.0194 | 0.067 (0.042–0.102) | 0 | 0 | |
| 16S | 1,450 | NA | 6 (0.41) | 0.001 ± 0.0004 | NA | NA | NA | |
| T. medium (1) | flaA | 1,068 | 355 | NA | NA | NA | NA | NA |
| recA | 1,362 | 453 | NA | NA | NA | NA | NA | |
| pyrH | 687 | 228 | NA | NA | NA | NA | NA | |
| 16S | 1,450 | NA | NA | NA | NA | NA | NA | |
| T. sinensis (4) | flaA | 1,059 | 352 | 5 (0.47) | 0.003 ± 0.0008 | 0.058 (0.003–0.257) | 0 | 0 |
| recA | 1,338 | 445 | 50 (3.57) | 0.019 ± 0.0066 | 0.052 (0.019–0.112) | 0 | 0 | |
| pyrH | 687 | 228 | 23 (3.17) | 0.017 ± 0.0051 | 0.183 (0.078–0.357) | 0 | 0 | |
| 16S | 1,450 | NA | 0 (0.00) | 0.000 ± 0.0000 | NA | NA | NA | |
| Treponema sp. IB (3) | flaA | 1,068 | 355 | 5 (0.47) | 0.003 ± 0.0010 | 0.074 (0.004–0.328) | 0 | 0 |
| recA | 1,368 | 455 | 124 (8.85) | 0.061 ± 0.0282 | 0.052 (0.019–0.112) | 0 | 0 | |
| pyrH | 687 | 228 | 0 (0.00) | 0.000 ± 0.0000 | 0.001 (0.00–10000) | 0 | 0 | |
| 16S | 1,450 | NA | 6 (0.41) | 0.003 ± 0.0008 | NA | NA | NA | |
| Treponema sp. IC (4) | flaA | 1,065 | 354 | 6 (0.56) | 0.003 ± 0.0006 | 0.148 (0.025–0.456) | 0 | 0 |
| recA | 1,350 | 449 | 40 (2.86) | 0.016 ± 0.0043 | 0.108 (0.052–0.195) | 0 | 0 | |
| pyrH | 687 | 228 | 10 (1.38) | 0.009 ± 0.0021 | 0.061 (0.102–0.190) | 1 (0.44) | 0 | |
| 16S | 1,450 | NA | 4 (0.28) | 0.001 ± 0.0006 | NA | NA | NA | |
| T. denticola (41) | flaA | 1,050 | 349 | 218 (20.76) | 0.029 ± 0.0070 | 0.121 (0.097–0.149) | 13 (3.71) | 0 |
| recA | 1,245 | 414 | 185 (13.20) | 0.035 ± 0.0030 | 0.094 (0.076–0.115) | 58 (13.98) | 1 | |
| pyrH | 696 | 231 | 149 (20.52) | 0.031 ± 0.0067 | 0.082 (0.061–0.106) | 31 (13.36) | 0 | |
| 16S | 1,450 | NA | 25 (1.72) | 0.003 ± 0.0005 | NA | NA | NA | |
| T. putidum (7) | flaA | 1,050 | 349 | 2 (0.19) | 0.001 ± 0.0002 | 0.00 (0.00–0.263) | 0 | 0 |
| recA | 1,245 | 414 | 154 (10.99) | 0.058 ± 0.0064 | 0.122 (0.091–0.160) | 9 (2.17) | 0 | |
| pyrH | 696 | 231 | 3 (0.41) | 0.001 ± 0.0004 | NA | 0 | 0 | |
| 16S | 1,450 | NA | 20 (1.38) | 0.005 ± 0.0016 | NA | NA | NA | |
NA, not applicable; nt, nucleotides; 95% CI, 95% confidence interval.
The proportions of polymorphic sites were equivalent in the 16S rRNA gene sequences from the respective sets of oral phylogroup 1 strains (2.52%) and oral phylogroup 2 strains (2.74%). Within the respective species (phylotypes) there was considerable variation in the numbers (%) of 16S rRNA polymorphic sites present. The numbers of polymorphic sites present within the 41 strains of T. denticola (1.72%) were broadly comparable to those present in the 7 T. putidum strains (1.38%). The proportions of polymorphic sites were considerably lower in the respective phylogroup 1 species, ranging from 0.00% across the 4 T. sinensis strains to 0.41% across the 11 T. vincentii strains. The number of 16S rRNA polymorphic sites was ca. 11% across all the treponeme strains analyzed, reflecting the considerable phylogenetic distance between phylogroups 1 and 2.
The overall proportions of polymorphic sites in the respective flaA, pyrH, and recA genes were broadly equivalent in the two respective phylogroups, ranging from ca. 19 to 25%. However, this masked considerable differences in the degree of gene polymorphism in these three respective genes within the respective species or phylotypes. For example, 10.99% of sites in the recA genes from T. putidum were polymorphic, which is considerably higher than levels present in flaA (0.19%) or pyrH (0.41%). An analogous situation was observed for the Treponema sp. IB taxa. However, in T. vincentii the levels of nucleotide polymorphisms were much higher in the pyrH gene (14.6%) than in the flaA (1.79%) or recA genes (1.71%). The numbers of polymorphic sites in the respective flaA, pyrH, and recA genes ranged from ca. 39 to 46% across all of the treponeme strains analyzed. The levels of nucleotide diversity (π) for the respective flaA, pyrH, and recA genes were slightly higher in the phylogroup 1 strains (0.078 to 0.098) than in the phylogroup 2 strains (0.042 to 0.064), which probably reflects the imbalance in the strains analyzed.
Mean global omega (ω) values, which are equivalent to the ratio of nonsynonymous (dN) to synonymous (dS) mutations (i.e., dN/dS) for each codon site, were calculated for the flaA, recA, and pyrH genes within the respective sets of oral phylogroup 1 and 2 strains to gauge the evolutionary pressures. These are reported with the corresponding 95% confidence intervals (CI). The ω values were fairly similar for the corresponding flaA and pyrH genes in oral phylogroup 1 and 2 strains. However, the ω values for the recA gene were higher in phylogroup 1 strains (ω = 0.300; 95% CI = 0.276 to 0.326), compared to phylogroup 2 strains (ω = 0.103; 95% CI = 0.087 to 0.121). The means ω values for the flaA and pyrH genes in all the strains included (n = 71) was calculated to be <0.12, whereas the mean ω value for recA gene was 0.21. This indicates that there was a strong purifying selective pressure to conserve the function of these three proteins, although recA was under slightly less selective pressure than flaA and pyrH.
The respective numbers of negatively and positively selected codon sites were also calculated for the flaA, recA, and pyrH genes the at the phylogroup level. Negative (purifying) selection indicates the conservation of gene sequence and function, while positive selection indicates evolutionary pressure for gene sequence change and adaptation. There were similar proportions of negatively selected codon sites in the flaA and pyrH genes within both phylogroup 1 and phylogroup 2 strains (ca. 7% and ca. 13%, respectively). For the recA gene, there were more negatively selected codon sites within the phylogroup 2 strains (17.9%) compared to phylogroup 1 (11.9%). Notably, there was a single positively selected site in the recA gene in T. denticola. This corresponded to the amino acid residue Val-8 (see Fig. S3 in the supplemental material). The specific role of the Val-8 in the biological activities of the RecA protein remains obscure (53).
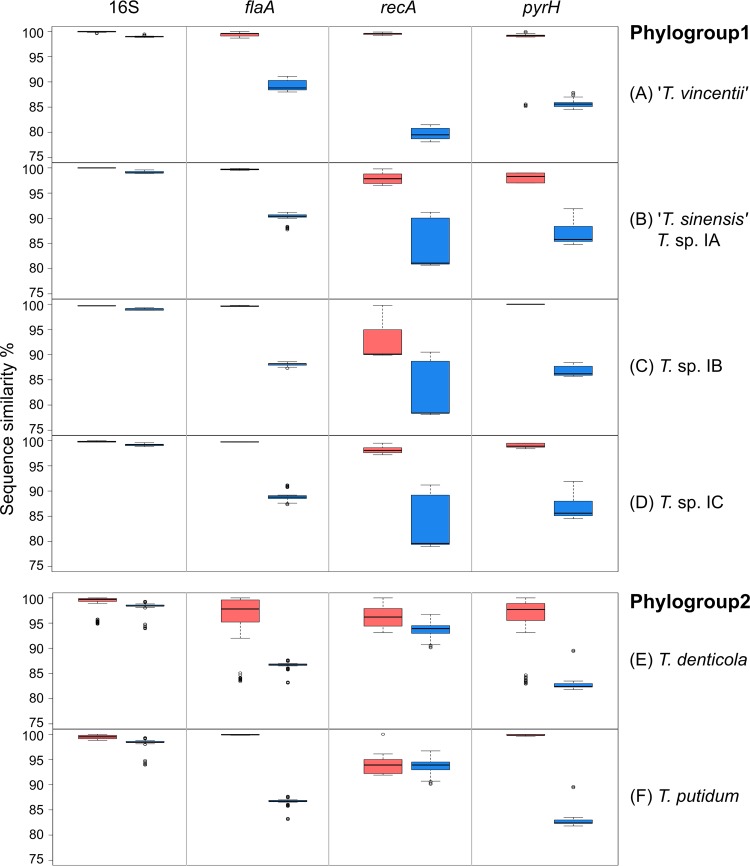
Intraspecific and interspecific sequence similarity.
The intraspecific and interspecific sequence similarities for each of the four genes (16S rRNA, flaA, recA, and pyrH) were calculated for each of the respective treponeme species/phylotypes. These are summarized in Fig. 5 and Fig. S4 in the supplemental material. Here, we defined intraspecific gene sequence similarity as that which occurs between strains belonging to the same species or phylotype. Correspondingly, interspecific gene sequence similarity was defined as that which occurred between taxa belonging to different species or phylotypes. In accordance with the phylogenetic analysis described above, T. medium ATCC 700293T was treated as a unique taxon. It should be carefully noted that gene sequence similarity does not directly correlate with phylogenetic distances due to their different calculation/estimation methods.
FIG 5.
Intra- and interspecific sequence similarity (%) for individual treponeme phylogroups. The respective levels of intra- and interspecific sequence similarity (in %) for the 16S rRNA, flaA, recA, and pyrH genes are shown for T. vincentii (A), T. sinensis (Treponema sp. IA; panel B), Treponema sp. IB (C), Treponema sp. IC (D), T. denticola (E), and T. putidum (F). Interspecific gene sequence similarity is indicated by blue shading, and intraspecific gene sequence is indicated by pink shading. The boxes indicates the lower/upper quartiles and the median, and the whiskers indicate the maximum and minimum values (Q3 + 1.5 IQR and Q1 − 1.5 IQR), excluding the outliers, which are indicated as open circles.
The average intraspecific sequence similarities for each of the four genes, within each of the six respective species/phylotypes are shown in Fig. S4 in the supplemental material. Each species/phylotype is represented with the same color scheme used in the ML phylograms shown in Fig. 1 to 4, and error bars are excluded, to aid visual comparison. As expected, the 16S rRNA gene showed the lowest levels of sequence variation (or highest similarity) within each of the respective species/phylotypes. The ranges of intraspecific sequence similarities were very small, especially for the phylogroup 1 species (99.5 to 100%). The 11 T. vincentii taxa shared 99.8 to 100% similarity, the four T. sinensis taxa had identical 16S rRNA sequences, and the three Treponema sp. IB and four Treponema sp. IC strains shared 99.5 to 99.6% and 99.7 to 99.9% 16S rRNA gene sequence similarities, respectively. There were higher levels of 16S rRNA gene sequence variation within the T. denticola and T. putidum taxa; these variations ranged from 98.9 to 100.0% and from 98.7 to 99.9%, respectively. These data are consistent with those presented in Table 5 and Table S4 in the supplemental material, as well as with the general topology of the 16S rRNA ML tree in Fig. 1.
TABLE 5.
Interspecific 16S rRNA gene sequence similarities between species or phylotypes
| Phylogroup and species | % gene sequence similarities (range) |
|||
|---|---|---|---|---|
| T. vincentii | T. sinensis (Treponema sp. IA) | Treponema sp. IB | Treponema sp. IC | |
| Phylogroup 1 | ||||
| T. medium | 98.9 (98.8–99.0) | 98.9 | 99.3 (99.2–99.6) | 99.0 (99.0–99.1) |
| T. vincentii | 98.6 (98.5–98.7) | 98.7 (98.4–99.0) | 98.5 (98.3–98.8) | |
| T. sinensis | 98.9 (98.8–99.0) | 99.3 (99.2–99.5) | ||
| Treponema sp. IB | 99.0 (98.8–99.1) | |||
| T. putidum | ||||
| Phylogroup 2 | ||||
| T. denticola | 98.4 (97.8–99.3) | |||
The respective levels of intraspecific sequence similarity for the flaA, recA, and pyrH genes varied considerably across the six treponeme species/phylotypes. The general patterns of gene sequence similarity/variation mirrored those reported above and summarized in Table 5. Overall, the recA gene showed the highest levels of sequence variation across the six species/phylotypes, followed by pyrH and then flaA.
The ranges of intraspecific and interspecific gene sequence similarity for the 16S rRNA, flaA, recA, and pyrH genes, for each of the six species/phylotypes are shown using a standard box-plot format in Fig. 5 and Fig. S5 in the supplemental material. Such plots can be used to establish “cutoff” points to help delineate species or other taxonomic groupings. The data have been analyzed and presented in three different ways. Figure 5A to D shows the interspecific and intraspecific gene sequence similarity relationships for phylogroup 1 treponemes only. Figure 5E and F shows the equivalent data for phylogroup 2 treponemes only. Figure S5 in the supplemental material shows the interspecific and intraspecific gene sequence similarity relationships for both phylogroup 1 and 2 treponemes.
As seen in Fig. 5 and Fig. S5 in the supplemental material, the “cutoff” between T. denticola and T. putidum taxa may be placed at ca. 99% 16S rRNA gene sequence similarity. The “species” boundaries appear to be tighter for phylogroup 1 oral treponemes, since it requires a similarity cutoff greater than ca. 99.5% to separate the respective species or phylotypes based on their 16S rRNA sequences (Fig. 5 and see Fig. S5).
For the flaA, recA, and pyrH genes, gene sequence similarity levels greater than ca. 90% are generally sufficient to separate the respective treponeme species/phylotypes. Of the three CDSs, the recA gene had the widest range of intraspecific sequence similarities (visualized by larger pink Q1 to Q3 boxes in Fig. 5 and Fig. S5 in the supplemental material). Thus, it can effectively distinguish different Treponema strains within the same species/phylotypes. However, the intra-/interspecific boundaries for the recA gene were often ambiguous (visualized by overlap between the pink and blue boxes in Fig. 5 and Fig. S5), that would hinder taxonomic assignation based on these criteria alone. In this regard, the flaA and pyrH gene sequences may offer better discrimination between the species/phylotypes. It should also be noted that the majority of the outliers (indicated with open circles in Fig. 5 and Fig. S5) correspond to T. denticola ATCC 700768, which is highly phylogenetically distinct from the other T. denticola strains.
DISCUSSION
Taxa belonging to the genus Treponema represent a significant proportion of the bacterial diversity present within the human oral cavity. Of the 75 oral treponeme species or “species-level” phylotypes currently listed in the web-accessible HOMD database (www.homd.org) (58), 15 correspond to oral phylogroup 1, and 5 correspond to phylogroup 2. The vast majority of phylogroup 2 taxa commonly identified in the oral cavity (e.g., via 16S rRNA gene amplicon sequencing-based approaches) correspond to T. denticola (HOT 584), with T. putidum (HOT 743) or other phylotypes encountered much less frequently (see, for example, references 1, 32, 33, and 40). There is a large body of evidence linking T. denticola with periodontal disease, with many putative pathological mechanisms having been identified (see, for example, references 10, 11, 34, 35, and 60). However, our current knowledge of the respective distributions of phylogroup 1 treponeme taxa within the oral cavity is far less well understood. This is greatly complicated by the lack of formal species descriptions for previously isolated/cultivated phylogroup 1 phylotypes and strains. Thus, many of the ca. 200- to 500-bp 16S rRNA gene reads commonly generated from next-generation DNA sequencing analyses cannot be accurately assigned to specific treponeme species (or putative species). This prevents us from obtaining a more detailed picture of the “T. vincentii-like” and “T. medium-like” taxa present within specific oral niches or infected clinical sites, greatly hampering our understanding of disease etiology. The four-gene multilocus genetic analysis described here was primarily aimed at addressing this important issue. An additional objective was to analyze the phylogenetic differences between T. denticola and T. putidum strains, which share many phenotypic characteristics and have high levels of 16S rRNA gene sequence similarity (27).
In our previous seven-gene MLSA of human oral T. denticola strains (56), our results indicated that the flaA, recA, and pyrH genes exhibited the highest levels of gene sequence diversity. Consequently, we selected these three genes for the phylogenetic analysis of a large and comprehensive collection of oral treponeme strains belonging to phylogroups 1 (n = 23) and 2 (n = 48). Taxonomy was assigned based on 16S rRNA gene phylogeny (Fig. 1). To facilitate comparisons with previous studies, we directly correlated the results from our phylogenetic analyses with both the HOMD taxonomic framework, as well as the oral treponeme taxonomic framework originally established by Paster and coworkers (1, 32, 40, 61). Our results supported the division of the (n = 23) “T. vincentii-like” and “T. medium-like” phylogroup 1 treponeme strains into five “species-level” groupings: T. vincentii (HOT 029), T. sinensis (Treponema sp. IA; HOT 231), Treponema sp. IC (HOT 231), T. medium (HOT 667), and Treponema sp. IB (HOT 667). This phylogeny-based taxonomic assignment was still strongly supported when 16S rRNA data from relevant as-yet-uncultivated oral taxa were added (Fig. 2).
Our data strongly support the long-held (though not yet officially recognized) designation of T. vincentii as a distinct “species” of oral treponeme, since in both phylogenetic (Fig. 1 to 4) and sequence similarity-based (Fig. 5) analyses the component strains (n = 11) consistently grouped together. The gene lengths (Table 3) and nucleotide compositions (Table 4) were also highly conserved in component T. vincentii strains and were different from those of the other four “species-level” groups. It may be noted that Correia et al. previously reported the 16S rRNA gene sequences of the T. vincentii OMZ 858, 860, 861, and 862 strains as part of their genetic analysis of the subtilisin operon in oral treponemes (62). Importantly, our data clearly indicate that the Ritz A (OMZ 305) strain, which was originally classified as being T. vincentii (63), does not belong within this species group. It belongs within the Treponema sp. IB group. It should be noted that the Ritz A strain has previously been included in a number of investigations as a representative T. vincentii strain (e.g., see references 64 to 67). Consequently, any previous “species-specific” generalizations involving the Ritz A strain should be carefully reappraised.
Our data are also consistent with the taxonomic position of T. medium as a distinct species. However, it should be noted that there is a paucity of T. medium strains reported in the literature. To the best of our knowledge, the properties of only one T. medium strain (ATCC 700293; ex. G7201) have been reported (22, 23). Thus, in the absence of data from additional strains, the precise nature of the relationship between T. medium and Treponema sp. IB taxa (ATCC 700767 = OMZ 806, Ritz A = OMZ 305, and OMZ 805) remains somewhat ill defined. As noted above, T. sinensis (Treponema sp. IA) has been proposed to constitute a distinct putative “species” of oral treponeme (Wyss, unpublished). With the exception of the recent publication of the genome sequence of the OMZ 838 (ATCC 700772) strain (44), there is a dearth of genetic or biological information about this proposed species. The properties of the Treponema sp. IC strains have previously been reported. Riviere et al. previously noted that the Treponema sp. IC strain OMZ 804 (ATCC 700766) could not be amplified by either “T. vincentii-specific” or “T. medium-specific” primer sets. In addition, the results from these authors' arbitrarily primed PCR analysis of genomic DNA indicated that the OMZ 804 strain was distinct from the T. vincentii ATCC 33580, T. vincentii 802, T. medium G7201, and Treponema sp. IB OMZ 805 strains (68). These observations are consistent with our MLSA.
A wide range of MLSA or MLST studies have previously utilized recA and/or pyrH genes to delineate species (subspecies) boundaries within diverse bacterial taxa, such as Vibrio spp. (69), Streptococcus spp. (70), and Achromobacter spp. (71). Of note, the pyrH gene had the highest resolution for discriminating the diversity of Vibrio cholerae and Vibrio mimicus species (72), and the recA gene was most effective for differentiating Achromobacter taxa (71). Genes encoding components of the flagellum have previously been included in a number of MLSA and (multilocus) bacterial typing systems, e.g., in Campylobacter spp. (73) and Clostridium difficile (74). The flaB2 gene has previously been used to discriminate treponemes associated with animal hoof/foot infections (38).
Clegg et al. (50) recently designed and employed an MLST system based on seven housekeeping genes for the discrimination and phylogenetic analysis of 121 treponeme strains isolated from digital dermatitis (DD) lesions of cloven-hoofed (farm) animals. These researchers used a PCR-based strategy to analyze ca. 500- to 600-bp fragments of the groEL, recA, glpK, adk, gdh, pyrG, and rplB genes and included data from the genome sequenced T. sinensis OMZ 838 and T. medium ATCC 700293 strains for references. These authors noted that both of these human oral strains were highly distinct from all the (nonoral) animal strains analyzed. To further probe this issue, we directly compared our human oral treponeme recA gene data with corresponding recA gene data from the 121 animal (nonoral) treponeme isolates reported by Clegg et al. via an ML phylogenetic analysis of the entire data set. The resultant ML tree is shown in Fig. S6 in the supplemental material. This clearly indicated that the human and animal treponemes had no direct phylogenetic overlap, with the very notable exception of the Treponema sp. IB Ritz A (OMZ 305) strain. This single human oral strain clustered along with 31 phylogroup I DD strains of uncertain standing in taxonomy, which corresponded to the recA “allele 1” defined in the study by Clegg et al. (50). However, it should be noted that this represents a single gene locus of this strain, which may not be wholly representative of the phylogenic origins of the rest of its genome. The implications of this phylogenetic relationship remain to be determined.
Regions of the FlaB and/or FlaA (flagellin) proteins of phylogroup 1 and 2 treponeme strains may be surface exposed, thereby becoming antigenic, and may be glycosylated (64, 67, 68, 75, 76). This phenotypic property is responsible for the ability of several strains of T. denticola, T. putidum, T. medium, T. vincentii, and “T. vincentii-like” species to cross-react with monoclonal antibodies (e.g., H9-2) raised against T. pallidum. This led to the definition of the term pathogen-related oral spirochete (PROS), a term that was discontinued due to its somewhat confusing and poorly defined nature (68). Many of the strains we analyzed here were previously identified as “PROS-positive” strains (27, 64, 67, 68).
Since the (antigenic) FlaA protein may become intimately associated with the host tissues, it may be hypothesized that the flaA gene could be under different selective pressures compared to genes encoding intracellular “housekeeping” proteins, such as RecA and PyrH. However, our findings here do not support this hypothesis. The overall topologies (branching patterns) of the ML trees constructed from the flaA (Fig. 3), recA (see Fig. S1A in the supplemental material), and pyrH (Fig. S1B) single gene data sets were all highly congruent with one another. Although there were differences in the relative numbers of polymorphic sites within these three protein-coding genes, all exhibited a strong stabilizing selection (Table 4). This is consistent with our previous findings for T. denticola strains (56). There was no evidence of genetic recombination for the flaA and pyrH genes. However, one possible recombination site was identified in the recA gene of the phylogroup 2 strains (Val-8), which is where a polymorphic site is located (see Fig. S3 in the supplemental material). Based on the functions and predicted tertiary structure of the RecA protein (53), the biological reasons underlying this variation remain obscure.
The results from the ML phylogenetic analysis of the concatenated flaA-recA-pyrH data sets clearly separated T. putidum from T. denticola (Fig. 4), with the T. denticola ATCC 700768 (OMZ 830) strain being a notable outlier. This is consistent with a previous report (27) which stated that this strain had a notably different phenotype from the other T. denticola and T. putidum strains analyzed. However, the phylogenetic distinction between T. putidum and T. denticola strains is not always clear-cut for the individual recA and pyrH genes (see Fig. S1 in the supplemental material), suggesting that these two species may share a certain amount of overlap in their genome compositions. It should also be noted that several of the T. putidum and T. denticola strains studied here were originally isolated from ANUG lesions in Chinese individuals (27, 59), as opposed to periodontitis lesions (see Table S1 in the supplemental material). It remains to be seen whether there are any significant genomic differences in treponeme taxa that inhabit ANUG versus periodontitis lesions or play etiological roles in these two oral infectious diseases.
We were curious to investigate whether there were any correlations between the phylogenies of the concatenated flaA-recA-pyrH gene sequences and the geographical origins of these clinical treponeme isolates. Based on the phylogenetic branching patterns (which may be visualized by the respective distributions of the country flags in Fig. 4), we tentatively speculate that there may be some genomic similarities for certain sets of oral treponeme strains isolated from the same geographical region. For example, several of the T. putidum strains isolated from Switzerland (ATCC 700334 and OMZ 844) and China (OMZ 848, OMZ 835, and OMZ 846) cluster together. The two Swiss Treponema sp. IB strains (OMZ 805 and ATCC 700767) cluster together. The four T. sinensis strains, all isolated in China, share high levels of phylogenetic similarity. Also, the two Chinese T. vincentii strains OMZ 861 and 863 cluster closely together. However, there is also evidence to support the hypothesis that there are treponeme strain lineages that may have a “global” distribution, as evidenced by phylogenetically closely related strains being isolated from geographically distinct locations. For example, the T. vincentii ATCC 35580 (USA) and ATCC 700774 (China) strains cluster together, as do the T. vincentii OMZ 801 (Switzerland), OMZ 802 (Switzerland), and OMZ 862 (China) strains. These speculative conclusions are consistent with the findings from our previous seven-gene MLSA analysis of 20 T. denticola isolates (56). However, we are cautious with these correlations, since although these three respective protein-coding genes are widely separated on the chromosome (37, 44), they only represent a very small snapshot of the entire genome composition.
Conclusions.
Taken together, the results from our multilocus phylogenetic and sequence-similarity-based analyses of oral treponeme strains clearly indicate that the 16S rRNA, flaA, recA, and pyrH genes can be used to effectively characterize and discriminate phylogroup 1 and 2 taxa spanning multiple species. Our data confirm the taxonomic positions of T. vincentii, T. medium, and T. putidum and defines three new “species-level” phylotypes: T. sinensis (Treponema sp. IA), Treponema sp. IB, and Treponema sp. IC. We envisage that results from future whole-genome-based analyses and systematic polyphasic taxonomic studies will support the oral treponeme “species” definitions proposed here. Our MLSA-based approach may be easily applied for the systematic characterization and phylogenetic analysis of treponemes from other phylogroups, as well as those from other ecological systems.
MATERIALS AND METHODS
Strain culture.
Details of the oral treponeme strains used in this study are summarized in Table 1 and see Table S1 in the supplemental material. Strains of Treponema denticola, Treponema putidum, Treponema vincentii, and Treponema medium, as well as other Treponema strains of uncertain taxonomic standing, were either purchased from the ATCC or were kindly provided by Chris Wyss, Georgios Belibasakis, and R. Gmür (University of Zurich, Switzerland); E. Peter Greenberg (University of Washington, USA); and Barry McBride (University of British Columbia, Canada). All isolates were stored at −70°C and were cultivated anaerobically in TYGVS medium supplemented with thiamine pyrophosphate (Sigma-Aldrich, USA), volatile fatty acids (Sigma-Aldrich, USA), and 10% rabbit serum (Gibco, USA) as described previously (22, 77). Strains were incubated anaerobically (Thermo Scientific Forma anaerobic system; gas mix: 85% nitrogen, 10% hydrogen, 5% carbon dioxide) at 37°C for 5 to 7 days until active growth became apparent. The cells were harvested by centrifugation (14,000 rpm; 5 min) and washed with 5 ml of phosphate-buffered saline prior to genomic DNA extraction using a QIAamp DNA mini-extraction kit (Qiagen, Germany) according to the manufacturer's Gram-negative protocol.
Primer design and PCR gene amplification.
All primer sets were designed using Omiga 2.0 (Oxford Molecular) and are listed in Table 2. For phylogroup 1 strains, the respective sets of PCR primers for amplifying the flaA, recA, and pyrH genes were designed based on the sequences of the corresponding genes present in the genomes of T. vincentii ATCC 35580 (accession no. PRJNA34095), T. vincentii F0403 (accession no. PRJNA169493), and T. medium ATCC 700293 (accession no. PRJNA169490), as well as preliminary genome sequence data for Treponema sp. IB OMZ 305 (Y. Chan et al., unpublished data). See Table S2 in the supplemental material for additional details. For phylogroup 2 strains, PCR primers targeting the flaA (TDE1712), recA (TDE0872), and pyrH (TDE2085) genes were designed based on the genome sequence of the type strain Treponema denticola ATCC 35405 (37). Respective “touchdown” PCRs were performed on a Veriti 96-Well Fast thermal cycler (Applied Biosystems, USA). PCRs (50 μl) contained 10 μl of 5× Go Taq Flexi buffer (Promega, USA), 5 μl of 7.5 mM MgCl2 (Invitrogen, USA), 2 μl of deoxynucleoside triphosphates (2.5 mM each; Invitrogen), 2 μl (10 μM) of forward and reverse primers (Integrated and Technologies, USA), 1 μl of purified genomic DNA (ca. 50 ng/μl), 1 μl of GoTaq DNA polymerase (5 U/μl; Promega), and 27 μl of sterile water. The PCR thermal cycling program consisted of the following: (i) an initial denaturation (95°C, 2 min); (ii) six cycles of denaturation (95°C, 30 s), annealing (temperatures as indicated in the Tm column in Table 2, 30 s) decreasing by ca. 0.5 to 1°C every cycle, and extension (72°C, 90 s); (iii) 27 cycles of denaturation (95°C, 30 s), annealing (lowest temperature of the range indicated in Table 2, 30 s), and extension (72°C, 90 s); and (iv) a final extension (72°C, 8 min). The PCR products were examined by using 1% agarose gel electrophoresis in Tris-borate-EDTA buffer and stained with ethidium bromide.
Clone library construction.
PCR products were gel purified using a QIAquick gel extraction kit (Qiagen) according to the manufacturer's instructions. The purified PCR products were cloned into pCR2.1-TOPO vectors by using a TA cloning kit (Invitrogen). Ligation mixtures were transformed into Escherichia coli DH10B competent cells and plated on Luria-Bertani (LB) agar plates supplemented with kanamycin (50 μg/ml) and X-Gal (5-bromo-4-chloro-indolyl-β-d-galactopyranoside; 20 μg/ml). Plates were incubated overnight at 37°C for blue-white screening. Five to six white colonies from each plate were picked and inoculated in LB medium. Plasmid DNA was extracted using the QIAprep spin miniprep kit (Qiagen) and commercially sequenced in both directions using the M13 universal forward and reverse primers.
Phylogenetic analysis.
Sequence data were edited and assembled using OMIGA 2.0 (78) and MEGA 7.0.14 (79). Multiple sequence alignments were performed using MUSCLE (80) and manually edited using BioEdit v.7.1.3 (81). The ranges of intraspecific and interspecific sequence similarities for each gene (flaA, recA, pyrH, and 16S rRNA) among T. putidum, T. denticola, and Treponema phylogroup 1 strains were calculated using the sequence identity matrix implemented in BioEdit and plotted in R using the default boxplot code. The G+C content and the numbers and locations of polymorphic sites were determined using DnaSP5 (82). The aligned sequences of the three genes were concatenated (flaA-recA-pyrH) using SeaView (83).
A maximum-likelihood (ML) approach was utilized to analyze the individual (single-gene) and the concatenated (two- or three-gene) sequence data sets. The best-fit substitution model and gamma rate heterogeneity for the individual gene and concatenated gene data sets were determined using Akaike's information criterion implemented in jModelTest (57). ML trees for single gene and multiple genes concatenation were generated using the Genetic Algorithm for Rapid Likelihood Inference (GARLI) (84) and Phylogenetic Analysis Using Parsimony (PAUP) (85), with branch support calculated from 1,000 bootstrap replicates, and Bayesian posterior probabilities were calculated using MrBayes v3.4 (86) on a maximum clade credibility tree topology selected by SumTrees using DendroPy 4.1.0 (87).
Detection of recombination and natural selection.
A codon-based approach implemented in DataMonkey (88) was used to analyze selection pressures. Genetic Algorithm Recombination Detection (GARD) was used to identify putative recombination breakpoints within genes. Single-likelihood ancestor counting was used to calculate the average global nonsynonymous (dN) and synonymous (dS) nucleotide substitution rate ratios (ω = dN/dS), as well as to test the selection of variable codon sites based on the most appropriate nucleotide substitution model and tree topology, with a critical P value of 0.05.
Accession number(s).
The sequences generated in this study were submitted to NCBI GenBank with the following accession numbers (see Table S2 in the supplemental material): 16S rRNA gene (KT192142 to KT192159, KP101520 to KP101532, and JN244657 to JN244660), flagellar filament outer layer protein (flaA) gene (KU877111 to KU877130, KP101533 to KP101547, and KP144319), recombinase A (recA) gene (KU877131 to KU877148, KP101548 to KP101564, and KP144320), and uridylate kinase (pyrH) gene (KU877149 to KU877166, KP101565 to KP101581, and KP144321).
Supplementary Material
ACKNOWLEDGMENTS
We sincerely thank Chris Wyss for sharing his unpublished experimental data and for invaluable discussions and guidance regarding oral treponeme biology and taxonomy. We thank Chris Wyss, Georgios Belibasakis, R. Gmür, E. Peter Greenberg, and Barry McBride for generously supplying many of the oral treponeme isolates included in this study. We thank Simon Clegg for sharing animal treponeme recA gene data. We also thank Joyce Yau of the Centralized Research Laboratories of the Faculty of Dentistry for technical assistance.
This study was financially supported by the Research Grants Council of Hong Kong through a GRF grant (grant 17105115) to R.M.W. Support from the Infection and Immunology Strategic Research Theme (SRT) of the University of Hong Kong is also acknowledged.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02499-16.
REFERENCES
- 1.Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, Lakshmanan A, Wade WG. 2010. The human oral microbiome. J Bacteriol 192:5002–5017. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnson RC. 1977. The spirochetes. Annu Rev Microbiol 31:89–106. doi: 10.1146/annurev.mi.31.100177.000513. [DOI] [PubMed] [Google Scholar]
- 3.Charon NW, Goldstein SF. 2002. Genetics of motility and chemotaxis of a fascinating group of bacteria: the spirochetes. Annu Rev Genet 36:47–73. doi: 10.1146/annurev.genet.36.041602.134359. [DOI] [PubMed] [Google Scholar]
- 4.Fenno JC, McBride BC. 1998. Virulence factors of oral treponemes. Anaerobe 4:1–17. doi: 10.1006/anae.1997.0131. [DOI] [PubMed] [Google Scholar]
- 5.Rosebury T. 1962. The indigenous spirochetes, p 186–221. In Rosebury T. (ed), Microorganisms indigenous to man. McGraw-Hill, New York, NY. [Google Scholar]
- 6.Listgarten MA. 1976. Structure of the microbial flora associated with periodontal health and disease in man: a light and electron microscopic study. J Periodontol 47:1–18. doi: 10.1902/jop.1976.47.1.1. [DOI] [PubMed] [Google Scholar]
- 7.Loesche WJ. 1988. The role of spirochetes in periodontal disease. Adv Dent Res 2:275–283. [DOI] [PubMed] [Google Scholar]
- 8.Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL Jr. 1998. Microbial complexes in subgingival plaque. J Clin Periodontol 25:134–144. doi: 10.1111/j.1600-051X.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 9.Chan EC, McLaughlin R. 2000. Taxonomy and virulence of oral spirochetes. Oral Microbiol Immunol 15:1–9. doi: 10.1034/j.1399-302x.2000.150101.x. [DOI] [PubMed] [Google Scholar]
- 10.Sela MN. 2001. Role of Treponema denticola in periodontal diseases. Crit Rev Oral Biol Med 12:399–413. doi: 10.1177/10454411010120050301. [DOI] [PubMed] [Google Scholar]
- 11.Ellen RP, Galimanas VB. 2005. Spirochetes at the forefront of periodontal infections. Periodontol 2000 38:13–32. doi: 10.1111/j.1600-0757.2005.00108.x. [DOI] [PubMed] [Google Scholar]
- 12.Perez-Chaparro PJ, Goncalves C, Figueiredo LC, Faveri M, Lobao E, Tamashiro N, Duarte P, Feres M. 2014. Newly identified pathogens associated with periodontitis: a systematic review. J Dent Res 93:846–858. doi: 10.1177/0022034514542468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sakamoto M, Siqueira JF Jr, Rocas IN, Benno Y. 2009. Diversity of spirochetes in endodontic infections. J Clin Microbiol 47:1352–1357. doi: 10.1128/JCM.02016-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maruyama N, Maruyama F, Takeuchi Y, Aikawa C, Izumi Y, Nakagawa I. 2014. Intraindividual variation in core microbiota in peri-implantitis and periodontitis. Sci Rep 4:6602. doi: 10.1038/srep06602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leite FR, Nascimento GG, Demarco FF, Gomes BP, Pucci CR, Martinho FC. 2015. Prevalence of Treponema species detected in endodontic infections: systematic review and meta-regression analysis. J Endod 41:579–587. doi: 10.1016/j.joen.2015.01.020. [DOI] [PubMed] [Google Scholar]
- 16.Belibasakis GN, Mir-Mari J, Sahrmann P, Sanz-Martin I, Schmidlin PR, Jung RE. 2016. Clinical association of Spirochaetes and Synergistetes with peri-implantitis. Clin Oral Implants Res 27:656–661. doi: 10.1111/clr.12690. [DOI] [PubMed] [Google Scholar]
- 17.Zhuang LF, Watt RM, Mattheos N, Si MS, Lai HC, Lang NP. 2016. Periodontal and peri-implant microbiota in patients with healthy and inflamed periodontal and peri-implant tissues. Clin Oral Implants Res 27:13–21. doi: 10.1111/clr.12508. [DOI] [PubMed] [Google Scholar]
- 18.Chan EC, Siboo R, Keng T, Psarra N, Hurley R, Cheng SL, Iugovaz I. 1993. Treponema denticola (ex Brumpt 1925) sp. nov., nom. rev., and identification of new spirochete isolates from periodontal pockets. Int J Syst Bacteriol 43:196–203. doi: 10.1099/00207713-43-2-196. [DOI] [PubMed] [Google Scholar]
- 19.Smibert RM, Burmeister JA. 1983. Treponema pectinovorum sp. nov. isolated from humans with periodontitis. Int J Syst Bacteriol 33:852–856. [Google Scholar]
- 20.Smibert RM, Johnson JL, Ranney RR. 1984. Treponema socranskii sp. nov., Treponema socranskii subsp. socranskii subsp. nov., Treponema socranskii subsp. buccale subsp. nov, and Treponema socranskii subsp. paredis subsp. nov. isolated from the human periodontia. Int J Syst Bacteriol 34:457–462. [Google Scholar]
- 21.Wyss C, Choi BK, Schupbach P, Guggenheim B, Gobel UB. 1996. Treponema maltophilum sp. nov., a small oral spirochete isolated from human periodontal lesions. Int J Syst Bacteriol 46:745–752. doi: 10.1099/00207713-46-3-745. [DOI] [PubMed] [Google Scholar]
- 22.Umemoto T, Nakazawa F, Hoshino E, Okada K, Fukunaga M, Namikawa I. 1997. Treponema medium sp. nov., isolated from human subgingival dental plaque. Int J Syst Bacteriol 47:67–72. doi: 10.1099/00207713-47-1-67. [DOI] [PubMed] [Google Scholar]
- 23.Nakazawa F, Hoshino E, Fukunaga M, Jinno T, Asai Y, Yamamoto H, Ogawa T. 2003. Amended biochemical characteristics and phylogenetic position of Treponema medium. Oral Microbiol Immunol 18:127–130. doi: 10.1034/j.1399-302X.2003.00015.x. [DOI] [PubMed] [Google Scholar]
- 24.Wyss C, Choi BK, Schupbach P, Guggenheim B, Gobel UB. 1997. Treponema amylovorum sp. nov., a saccharolytic spirochete of medium size isolated from an advanced human periodontal lesion. Int J Syst Bacteriol 47:842–845. doi: 10.1099/00207713-47-3-842. [DOI] [PubMed] [Google Scholar]
- 25.Wyss C, Choi BK, Schupbach P, Moter A, Guggenheim B, Gobel UB. 1999. Treponema lecithinolyticum sp. nov., a small saccharolytic spirochaete with phospholipase A and C activities associated with periodontal diseases. Int J Syst Bacteriol 49(Pt 4):1329–1339. doi: 10.1099/00207713-49-4-1329. [DOI] [PubMed] [Google Scholar]
- 26.Wyss C, Dewhirst FE, Gmur R, Thurnheer T, Xue Y, Schupbach P, Guggenheim B, Paster BJ. 2001. Treponema parvum sp. nov., a small, glucuronic, or galacturonic acid-dependent oral spirochaete from lesions of human periodontitis and acute necrotizing ulcerative gingivitis. Int J Syst Evol Microbiol 51:955–962. doi: 10.1099/00207713-51-3-955. [DOI] [PubMed] [Google Scholar]
- 27.Wyss C, Moter A, Choi BK, Dewhirst FE, Xue Y, Schupbach P, Gobel UB, Paster BJ, Guggenheim B. 2004. Treponema putidum sp. nov., a medium-sized proteolytic spirochaete isolated from lesions of human periodontitis and acute necrotizing ulcerative gingivitis. Int J Syst Evol Microbiol 54:1117–1122. doi: 10.1099/ijs.0.02806-0. [DOI] [PubMed] [Google Scholar]
- 28.Klein HS. 1946. What role do Treponema vincentii play in certain diseases of the gingiva? Acta Odontol Scand 7:92–96. doi: 10.3109/00016354609040426. [DOI] [PubMed] [Google Scholar]
- 29.Hampp EG, Scott DB, Wyckoff RW. 1948. Morphologic characteristics of certain cultured strains of oral spirochetes and treponema pallidum as revealed by the electron microscope. J Bacteriol 56:755–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smibert RM. 1984. Genus III. Treponema Schaudinn 1905, 1728 AL, p 49–57. In Krieg NR, Holt JG (ed), Bergey's manual of systematic bacteriology, vol 1 Williams & Wilkins, Baltimore, MD. [Google Scholar]
- 31.Olsen I, Paster BJ, Dewhirst FE. 2000. Taxonomy of spirochetes. Anaerobe 6:39–57. doi: 10.1006/anae.1999.0319. [DOI] [Google Scholar]
- 32.Dewhirst FE, Tamer MA, Ericson RE, Lau CN, Levanos VA, Boches SK, Galvin JL, Paster BJ. 2000. The diversity of periodontal spirochetes by 16S rRNA analysis. Oral Microbiol Immunol 15:196–202. doi: 10.1034/j.1399-302x.2000.150308.x. [DOI] [PubMed] [Google Scholar]
- 33.You M, Mo S, Leung WK, Watt RM. 2013. Comparative analysis of oral treponemes associated with periodontal health and disease. BMC Infect Dis 13:174. doi: 10.1186/1471-2334-13-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ishihara K. 2010. Virulence factors of Treponema denticola. Periodontol 2000 54:117–135. doi: 10.1111/j.1600-0757.2009.00345.x. [DOI] [PubMed] [Google Scholar]
- 35.Dashper SG, Seers CA, Tan KH, Reynolds EC. 2011. Virulence factors of the oral spirochete Treponema denticola. J Dent Res 90:691–703. doi: 10.1177/0022034510385242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rocas IN, Siqueira JF Jr. 2005. Occurrence of two newly named oral treponemes—Treponema parvum and Treponema putidum—in primary endodontic infections. Oral Microbiol Immunol 20:372–375. doi: 10.1111/j.1399-302X.2005.00238.x. [DOI] [PubMed] [Google Scholar]
- 37.Seshadri R, Myers GS, Tettelin H, Eisen JA, Heidelberg JF, Dodson RJ, Davidsen TM, DeBoy RT, Fouts DE, Haft DH, Selengut J, Ren Q, Brinkac LM, Madupu R, Kolonay J, Durkin SA, Daugherty SC, Shetty J, Shvartsbeyn A, Gebregeorgis E, Geer K, Tsegaye G, Malek J, Ayodeji B, Shatsman S, McLeod MP, Smajs D, Howell JK, Pal S, Amin A, Vashisth P, McNeill TZ, Xiang Q, Sodergren E, Baca E, Weinstock GM, Norris SJ, Fraser CM, Paulsen IT. 2004. Comparison of the genome of the oral pathogen Treponema denticola with other spirochete genomes. Proc Natl Acad Sci U S A 101:5646–5651. doi: 10.1073/pnas.0307639101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Evans NJ, Brown JM, Demirkan I, Murray RD, Vink WD, Blowey RW, Hart CA, Carter S. 2008. Three unique groups of spirochetes isolated from digital dermatitis lesions in UK cattle. Vet Microbiol 130:141–150. doi: 10.1016/j.vetmic.2007.12.019. [DOI] [PubMed] [Google Scholar]
- 39.Klitgaard K, Boye M, Capion N, Jensen TK. 2008. Evidence of multiple Treponema phylotypes involved in bovine digital dermatitis as shown by 16S rRNA gene analysis and fluorescence in situ hybridization. J Clin Microbiol 46:3012–3020. doi: 10.1128/JCM.00670-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paster BJ, Boches SK, Galvin JL, Ericson RE, Lau CN, Levanos VA, Sahasrabudhe A, Dewhirst FE. 2001. Bacterial diversity in human subgingival plaque. J Bacteriol 183:3770–3783. doi: 10.1128/JB.183.12.3770-3783.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Griffen AL, Beall CJ, Campbell JH, Firestone ND, Kumar PS, Yang ZK, Podar M, Leys EJ. 2012. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J 6:1176–1185. doi: 10.1038/ismej.2011.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mason MR, Nagaraja HN, Camerlengo T, Joshi V, Kumar PS. 2013. Deep sequencing identifies ethnicity-specific bacterial signatures in the oral microbiome. PLoS One 8:e77287. doi: 10.1371/journal.pone.0077287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lacap-Bugler DC, Jiang J, Huo YB, Chan Y, Leung FC, Watt RM. 2014. Complete genome sequence of the oral spirochete bacterium Treponema putidum strain OMZ 758T (ATCC 700334T). Genome Announc 2:e01076-14. doi: 10.1128/genomeA.01076-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chan Y, Ma AP, Lacap-Bugler DC, Huo YB, Keung Leung W, Leung FC, Watt RM. 2014. Complete genome sequence for Treponema sp. OMZ 838 (ATCC 700772, DSM 16789), isolated from a necrotizing ulcerative gingivitis lesion. Genome Announc 2:e01333-14. doi: 10.1128/genomeA.01333-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Group NHW, Peterson J, Garges S, Giovanni M, McInnes P, Wang L, Schloss JA, Bonazzi V, McEwen JE, Wetterstrand KA, Deal C, Baker CC, Di Francesco V, Howcroft TK, Karp RW, Lunsford RD, Wellington CR, Belachew T, Wright M, Giblin C, David H, Mills M, Salomon R, Mullins C, Akolkar B, Begg L, Davis C, Grandison L, Humble M, Khalsa J, Little AR, Peavy H, Pontzer C, Portnoy M, Sayre MH, Starke-Reed P, Zakhari S, Read J, Watson B, Guyer M. 2009. The NIH human microbiome project. Genome Res 19:2317–2323. doi: 10.1101/gr.096651.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gomez-Diaz E, Boulinier T, Sertour N, Cornet M, Ferquel E, McCoy KD. 2011. Genetic structure of marine Borrelia garinii and population admixture with the terrestrial cycle of Lyme borreliosis. Environ Microbiol 13:2453–2467. doi: 10.1111/j.1462-2920.2011.02515.x. [DOI] [PubMed] [Google Scholar]
- 47.Margos G, Gatewood AG, Aanensen DM, Hanincova K, Terekhova D, Vollmer SA, Cornet M, Piesman J, Donaghy M, Bormane A, Hurn MA, Feil EJ, Fish D, Casjens S, Wormser GP, Schwartz I, Kurtenbach K. 2008. MLST of housekeeping genes captures geographic population structure and suggests a European origin of Borrelia burgdorferi. Proc Natl Acad Sci U S A 105:8730–8735. doi: 10.1073/pnas.0800323105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rasback T, Johansson KE, Jansson DS, Fellstrom C, Alikhani MY, La T, Dunn DS, Hampson DJ. 2007. Development of a multilocus sequence typing scheme for intestinal spirochaetes within the genus Brachyspira. Microbiology 153:4074–4087. doi: 10.1099/mic.0.2007/008540-0. [DOI] [PubMed] [Google Scholar]
- 49.Ahmed A, Thaipadungpanit J, Boonsilp S, Wuthiekanun V, Nalam K, Spratt BG, Aanensen DM, Smythe LD, Ahmed N, Feil EJ, Hartskeerl RA, Peacock SJ. 2011. Comparison of two multilocus sequence based genotyping schemes for Leptospira species. PLoS Negl Trop Dis 5:e1374. doi: 10.1371/journal.pntd.0001374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Clegg SR, Carter SD, Birtles RJ, Brown JM, Hart CA, Evans NJ. 2016. Multilocus sequence typing of pathogenic treponemes isolated from cloven hoofed animals and their comparison to human isolates. Appl Environ Microbiol 82:4523–4536. doi: 10.1128/AEM.00025-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Karlsson F, Svartstrom O, Belak K, Fellstrom C, Pringle M. 2013. Occurrence of Treponema spp. in porcine skin ulcers and gingiva. Vet Microbiol 165:402–409. doi: 10.1016/j.vetmic.2013.03.031. [DOI] [PubMed] [Google Scholar]
- 52.Stamm LV, Bergen HL, Walker RL. 2002. Molecular typing of papillomatous digital dermatitis-associated Treponema isolates based on analysis of 16S-23S ribosomal DNA intergenic spacer regions. J Clin Microbiol 40:3463–3469. doi: 10.1128/JCM.40.9.3463-3469.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cox MM. 2007. Motoring along with the bacterial RecA protein. Nat Rev Mol Cell Biol 8:127–138. doi: 10.1038/nrm2099. [DOI] [PubMed] [Google Scholar]
- 54.Serina L, Blondin C, Krin E, Sismeiro O, Danchin A, Sakamoto H, Gilles AM, Barzu O. 1995. Escherichia coli UMP-kinase, a member of the aspartokinase family, is a hexamer regulated by guanine nucleotides and UTP. Biochemistry 34:5066–5074. doi: 10.1021/bi00015a018. [DOI] [PubMed] [Google Scholar]
- 55.Limberger RJ. 2004. The periplasmic flagellum of spirochetes. J Mol Microbiol Biotechnol 7:30–40. doi: 10.1159/000077867. [DOI] [PubMed] [Google Scholar]
- 56.Mo S, You M, Su YC, Lacap-Bugler DC, Huo YB, Smith GJ, Leung WK, Watt RM. 2013. Multilocus sequence analysis of Treponema denticola strains of diverse origin. BMC Microbiol 13:24. doi: 10.1186/1471-2180-13-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Posada D. 2008. jModelTest: phylogenetic model averaging. Mol Biol Evol 25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- 58.Chen T, Yu WH, Izard J, Baranova OV, Lakshmanan A, Dewhirst FE. 2010. The Human Oral Microbiome Database: a web accessible resource for investigating oral microbe taxonomic and genomic information. Database (Oxford) 2010:baq013. doi: 10.1093/database/baq013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gmur R, Wyss C, Xue Y, Thurnheer T, Guggenheim B. 2004. Gingival crevice microbiota from Chinese patients with gingivitis or necrotizing ulcerative gingivitis. Eur J Oral Sci 112:33–41. doi: 10.1111/j.0909-8836.2004.00103.x. [DOI] [PubMed] [Google Scholar]
- 60.Fenno JC. 2012. Treponema denticola interactions with host proteins. J Oral Microbiol 2012:4. doi: 10.3402/jom.v4i0.9929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Choi BK, Paster BJ, Dewhirst FE, Gobel UB. 1994. Diversity of cultivable and uncultivable oral spirochetes from a patient with severe destructive periodontitis. Infect Immun 62:1889–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Correia FF, Plummer AR, Ellen RP, Wyss C, Boches SK, Galvin JL, Paster BJ, Dewhirst FE. 2003. Two paralogous families of a two-gene subtilisin operon are widely distributed in oral treponemes. J Bacteriol 185:6860–6869. doi: 10.1128/JB.185.23.6860-6869.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reijntjens FM, Mikx FH, Wolters-Lutgerhorst JM, Maltha JC. 1986. Adherence of oral treponemes and their effect on morphological damage and detachment of epithelial cells in vitro. Infect Immun 51:642–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Choi BK, Wyss C, Gobel UB. 1996. Phylogenetic analysis of pathogen-related oral spirochetes. J Clin Microbiol 34:1922–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Heuner K, Meltzer U, Choi BK, Gobel UB. 2001. Outer sheath associated proteins of the oral spirochete Treponema maltophilum. FEMS Microbiol Lett 197:187–193. doi: 10.1016/S0378-1097(01)00104-5. [DOI] [PubMed] [Google Scholar]
- 66.Heuner K, Bergmann I, Heckenbach K, Gobel UB. 2001. Proteolytic activity among various oral Treponema species and cloning of a prtP-like gene of Treponema socranskii subsp. socranskii. FEMS Microbiol Lett 201:169–176. doi: 10.1016/S0378-1097(01)00264-6. [DOI] [PubMed] [Google Scholar]
- 67.Wyss C. 1998. Flagellins, but not endoflagellar sheath proteins, of Treponema pallidum and of pathogen-related oral spirochetes are glycosylated. Infect Immun 66:5751–5754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Riviere GR, Smith KS, Willis SG, Riviere KH. 1999. Phenotypic and genotypic heterogeneity among cultivable pathogen-related oral spirochetes and Treponema vincentii. J Clin Microbiol 37:3676–3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Thompson FL, Gevers D, Thompson CC, Dawyndt P, Naser S, Hoste B, Munn CB, Swings J. 2005. Phylogeny and molecular identification of vibrios on the basis of multilocus sequence analysis. Appl Environ Microbiol 71:5107–5115. doi: 10.1128/AEM.71.9.5107-5115.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yu J, Sun Z, Liu W, Xi X, Song Y, Xu H, Lv Q, Bao Q, Menghe B, Sun T. 2015. Multilocus sequence typing of Streptococcus thermophilus from naturally fermented dairy foods in China and Mongolia. BMC Microbiol 15:236. doi: 10.1186/s12866-015-0551-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gomila M, Prince-Manzano C, Svensson-Stadler L, Busquets A, Erhard M, Martinez DL, Lalucat J, Moore ER. 2014. Genotypic and phenotypic applications for the differentiation and species-level identification of Achromobacter for clinical diagnoses. PLoS One 9:e114356. doi: 10.1371/journal.pone.0114356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Thompson CC, Thompson FL, Vicente AC. 2008. Identification of Vibrio cholerae and Vibrio mimicus by multilocus sequence analysis (MLSA). Int J Syst Evol Microbiol 58:617–621. doi: 10.1099/ijs.0.65461-0. [DOI] [PubMed] [Google Scholar]
- 73.Korczak BM, Zurfluh M, Emler S, Kuhn-Oertli J, Kuhnert P. 2009. Multiplex strategy for multilocus sequence typing, fla typing, and genetic determination of antimicrobial resistance of Campylobacter jejuni and Campylobacter coli isolates collected in Switzerland. J Clin Microbiol 47:1996–2007. doi: 10.1128/JCM.00237-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lemee L, Bourgeois I, Ruffin E, Collignon A, Lemeland JF, Pons JL. 2005. Multilocus sequence analysis and comparative evolution of virulence-associated genes and housekeeping genes of Clostridium difficile. Microbiology 151:3171–3180. doi: 10.1099/mic.0.28155-0. [DOI] [PubMed] [Google Scholar]
- 75.Cockayne A, Sanger R, Ivic A, Strugnell RA, MacDougall JH, Russell RR, Penn CW. 1989. Antigenic and structural analysis of Treponema denticola. J Gen Microbiol 135:3209–3218. [DOI] [PubMed] [Google Scholar]
- 76.Ruby JD, Li H, Kuramitsu H, Norris SJ, Goldstein SF, Buttle KF, Charon NW. 1997. Relationship of Treponema denticola periplasmic flagella to irregular cell morphology. J Bacteriol 179:1628–1635. doi: 10.1128/jb.179.5.1628-1635.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fenno JC. 2005. Laboratory maintenance of Treponema denticola. Curr Protoc Microbiol Chapter 12:Unit 12B 11. [DOI] [PubMed] [Google Scholar]
- 78.Kramer JA. 2001. Omiga: a PC-based sequence analysis tool. Mol Biotechnol 19:97–106. doi: 10.1385/MB:19:1:097. [DOI] [PubMed] [Google Scholar]
- 79.Kumar S, Nei M, Dudley J, Tamura K. 2008. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform 9:299–306. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98. [Google Scholar]
- 82.Librado P, Rozas J. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 83.Galtier N, Gouy M, Gautier C. 1996. SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput Appl Biosci 12:543–548. [DOI] [PubMed] [Google Scholar]
- 84.Zwickl DJ. 2006. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. PhD thesis University of Texas, Austin, TX. [Google Scholar]
- 85.Swofford DL. 2002. PAUP* phylogenetic analysis using parsimony (* and other methods), v4. Sinauer Associates, Sunderland, MA. [Google Scholar]
- 86.Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 87.Sukumaran J, Holder MT. 2010. DendroPy: a Python library for phylogenetic computing. Bioinformatics 26:1569–1571. doi: 10.1093/bioinformatics/btq228. [DOI] [PubMed] [Google Scholar]
- 88.Pond SL, Frost SD. 2005. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics 21:2531–2533. doi: 10.1093/bioinformatics/bti320. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.