ABSTRACT
Biological soil crusts (biocrusts) are slow-growing, phototroph-based microbial assemblages that develop on the topsoils of drylands. Biocrusts help maintain soil fertility and reduce erosion. Because their loss through human activities has negative ecological and environmental health consequences, biocrust restoration is of interest. Active soil inoculation with biocrust microorganisms can be an important tool in this endeavor. We present a culture-independent, two-step process to grow multispecies biocrusts in open greenhouse nursery facilities, based on the inoculation of local soils with local biocrust remnants and incubation under seminatural conditions that maintain the essence of the habitat but lessen its harshness. In each of four U.S. Southwest sites, we tested and deployed combinations of factors that maximized growth (gauged as chlorophyll a content) while minimizing microbial community shifts (assessed by 16S rRNA sequencing and bioinformatics), particularly for crust-forming cyanobacteria. Generally, doubling the frequency of natural wetting events, a 60% reduction in sunlight, and inoculation by slurry were optimal. Nutrient addition effects were site specific. In 4 months, our approach yielded crusts of high inoculum quality reared on local soil exposed to locally matched climates, acclimated to desiccation, and containing communities minimally shifted in composition from local ones. Our inoculum contained abundant crust-forming cyanobacteria and no significant numbers of allochthonous phototrophs, and it was sufficient to treat ca. 6,000 m2 of degraded dryland soils at 1 to 5% of the typical crust biomass concentration, having started from a natural crust remnant as small as 6 to 30 cm2.
IMPORTANCE Soil surface crusts can protect dryland soils from erosion, but they are often negatively impacted by human activities. Their degradation causes a loss of fertility, increased production of fugitive dust and intensity of dust storms with associated traffic problems, and provokes general public health hazards. Our results constitute an advance in the quest to actively restore biological soil covers by providing a means to obtain high-quality inoculum within a reasonable time (a few months), thereby allowing land managers to recover essential, but damaged, ecosystem services in a sustainable, self-perpetuating way as provided by biocrust communities.
KEYWORDS: biological soil crusts, soil restoration, cyanobacteria, 16S rRNA, erosion control, degraded soils, drylands, soil microbiome
INTRODUCTION
Drylands are characterized by sparsely vegetated soils that are subject to aeolian erosion. Because drylands occupy approximately 45% of the Earth's terrestrial surface and are home to more than 38% of the global population (1, 2), consequences of dryland soil degradation can have global impacts (2). Soil surface crusts, both physical and biological, can protect dryland soil surfaces from erosion (3), but they are often negatively impacted by human activities, such as agriculture, construction, trampling by cattle, off-road vehicle use, or military training (4). The degradation of surface cover results not only in the loss of local soil fertility but also in the increased production of fugitive dust and the intensity of dust storms (5) since unconsolidated surfaces are prone to become sources of particulate pollution by entrainment under otherwise inconsequential windy conditions. Associated problems include reduced visibility, traffic accidents, and road closures (4), as well as more general public health hazards caused by the dispersal of microbial pathogens and toxins (6) and by the chronic impact of particulate pollutants on the respiratory tract (7). Cities such as Phoenix, AZ, USA, and its metropolitan area consistently rank in the worst 5% for particulate pollution according to the American Lung Association (6). Effects can reach far from the dust source. Increased dust deposition on the Rocky Mountains is known to promote earlier snowmelt, modifying hydrological patterns in the Upper Colorado River Basin, an important water supply in several U.S. and Mexican states (8). For these reasons, there is a broad societal interest in stabilizing dryland soils to protect not only the functioning of ecosystems but also human populations that reside within arid land communities.
A potential avenue for soil surface restoration consists of regenerating biological soil crust (biocrust) cover. Biocrusts are complex, topsoil microbial assemblages that develop on the primary production of soil cyanobacteria, microalgae (sometimes in algal symbioses), or mosses and that support a large diversity of heterotrophic bacteria (9), archaea (10), and fungi (11). Considered to be a “mantle of fertility” in arid lands (12), biocrusts provide essential goods and services; they stabilize soils and thus reduce rates of wind erosion and dust particle production (13), can influence soil temperature (14, 15), contribute significantly to soil C and N inputs into the ecosystem (16), increase the lixiviation of micronutrients (17), control soil hydrological dynamics (18), and are thought to provide good conditions for plant germination and establishment (19).
Yet, several land uses, such as agriculture (20), livestock grazing (21–23), and recreation (24), negatively affect biocrusts. Recovery is a slow process, given that biological activity in these organosedimentary assemblages is highly limited by water and restricted to short periods of time after precipitation events (25) and has been estimated to be at least decades (3). However, there is also evidence that active intervention can accelerate recovery times (26, 27). For lichen- and moss-dominated biocrusts, recovery can be enhanced simply by providing increasing moisture and nutrient availability to existing remnant populations (28) or by additional inoculation with cultivated biomass (29). Active inoculation of bare soils with biocrust microorganisms can be an important tool in this enterprise (3). Cultivated cyanobacteria, for example, can be grown in the laboratory or in outdoor racetrack facilities and then inoculated to fix unconsolidated soil particles in degraded drylands (21, 30–36). The results of such attempts have been difficult to assess because of the limited amount of inoculum produced (19). These cultivation approaches present the additional shortcoming of yielding an inoculum acclimated to optimal, nutrient-replete conditions and one that is of low fitness in the soil, particularly if standard strains are used that may prove suboptimal for the local climate and edaphic properties of the site. Because there is clear evidence of microbial biogeographical patterns of distribution in biocrust microorganisms (37–39), the active inoculation of allochthonous strains may also bring about the risk of unintended introduction of invasive species. When using open-air facilities, the production of inoculum with a high content on nonterrestrial, water, or airborne contaminant phototrophs is virtually unavoidable, making it necessary to (i) restrict the growth of airborne, noncrust cyanobacteria by imposing recurrent desiccation and (ii) carry out microbiological quality control monitoring of the microbial composition of the products.
We present here an alternative approach to the production of biocrust inoculum that is based on the enhancement of natural populations of remnant biocrust organisms on native soils in controlled, but seminatural, greenhouse facilities (biocrust nurseries). The main goal of our approach was to produce abundant biomass for inoculation that is of high fitness and low ecological risk in that the resulting microbes are of local origin, designed to match the community composition of the original biocrusts, and acclimated to the edaphic and climatic conditions of the target area. To meet our goal, we developed methodologies for the greenhouse rearing of biocrusts from local remnants and probed variations in factors that would optimize yields without causing major shifts in microbial community composition. We used a two-step approach applied to several settings with two geographically and climatically distinct biocrusts and two soil types in each. First, we screened six factors that can logically and potentially enhance the growth of biocrust communities, while minimizing microbial community composition shifts. We then validated our results in a large-scale incubation experiment that yielded enough biomass to be relevant in restoration efforts in the field.
RESULTS
The period and conditions of incubation during the first set of experiments were sufficient to allow robust growth in most, but not all, treatments. In some cases, growth yield was well beyond the initial levels (INIT; red lines in Fig. 1) and close to the levels originally observed in the mature field biocrusts used as inocula (INOC; blue lines in Fig. 1). Screening models applied to this data set determined the main factors that resulted in significantly higher growth at each site; water and light were important in all sites and nutrients were relevant only in the hot desert sites. We fitted reduced linear models to the data involving relevant factors. All models were statistically significant according to one-way analysis of variance (ANOVA) results (F ≥ 3.86; P < 0.05; see Table S1 in the supplemental material). Analyses of estimated effects revealed that a high watering frequency and a low light intensity promoted the growth of biocrust biomass in all sites (least-squares [LS] means tests, P < 0.05; Table 1). Similarly, the addition of nutrients enhanced the yield of biocrust growth in hot desert sites: P plus N in Fort Bliss (FB) but only P in Jornada (J) (LS means tests, P < 0.05; Table 1).
FIG 1.
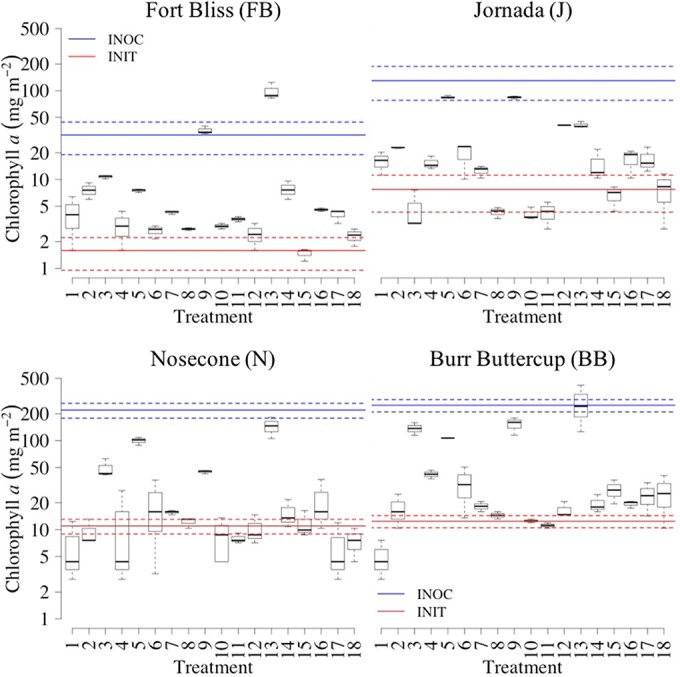
Boxplots for the final phototrophic biomass (as aerial chl a content) obtained after greenhouse incubation of native soils from 4 sites (each panel shows a site) inoculated with natural biocrusts from their respective site under 18 different treatments. Boxes denote lower and upper quartiles (with median values depicted as black solid lines), and whiskers denote lower and upper extremes (n = 3). Blue lines indicate the chl a content of field biocrust samples used as inoculum (INOC), and red lines indicate initial chl a content in the inoculated soils (INIT) (color solid lines indicate mean, and color dashed lines indicate standard deviations of n = 3).
TABLE 1.
Results of linear models for the effect of selected factors, as obtained after the preliminary screening process for each of the four sites, on chlorophyll a and Bray-Curtis dissimilarity index as an estimate of community composition shift based on bacterial phyla and cyanobacteriaa
| Dependent variable | Fort Bliss |
Jornada |
Nosecone |
Burr Buttercup |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Factor (level) | df | F | P | Factor (level) | df | F | P | Factor (level) | df | F | P | Factor (level) | df | F | P | |
| Chl a content | Water (+) | 1 | 4.14 | 0.039 | Water (+) | 1 | 11.03 | 0.005 | Light (−) | 1 | 8.99 | 0.009 | Water (+) | 1 | 10.84 | 0.005 |
| Light (−) | 1 | 3.97 | 0.042 | Light (−) | 1 | 5.56 | 0.034 | Water (+) | 1 | 6.24 | 0.024 | Light (−) | 1 | 9.92 | 0.006 | |
| Nutrients (P+N) | 2 | 3.87 | 0.047 | Nutrients (P) | 2 | 3.94 | 0.041 | |||||||||
| BC (bacterial phyla) | Inoculum (S) | 1 | 6.41 | 0.024 | Inoculum (S) | 1 | 5.49 | 0.041 | Water (+) | 1 | 6.16 | 0.027 | Nutrients (P+N) | 2 | 11.78 | 0.001 |
| Nutrients (P) | 2 | 4.81 | 0.026 | Nutrients (P) | 2 | 3.97 | 0.040 | Light (−) | 1 | 5.95 | 0.029 | Inoculum (S) | 1 | 10.43 | 0.008 | |
| Nutrients (N) | 2 | 3.88 | 0.047 | Calcium (+) | 1 | 8.83 | 0.012 | |||||||||
| Water (+) | 1 | 7.81 | 0.017 | |||||||||||||
| Light (−) | 1 | 6.63 | 0.023 | |||||||||||||
| BC (cyanobacteria) | Inoculum (S) | 1 | 3.99 | 0.048 | Water (+) | 1 | 6.65 | 0.021 | Inoculum (S) | 1 | 9.65 | 0.008 | ||||
| Calcium (−) | 1 | 5.85 | 0.029 | Light (−) | 1 | 6.18 | 0.025 | Water (+) | 1 | 6.07 | 0.028 | |||||
| Nutrients (P+N) | 2 | 4.84 | 0.026 | |||||||||||||
In parentheses, levels of factors that maximized the production of biomass (chlorophyll a [Chl a]) or minimized changes in community composition (Bray-Curtis [BC] based on bacterial phyla or cyanobacteria) according to LS means tests (P ≤ 0.05) (P, addition of phosphorus; N, addition of nitrogen; S, slurry-like inoculum).
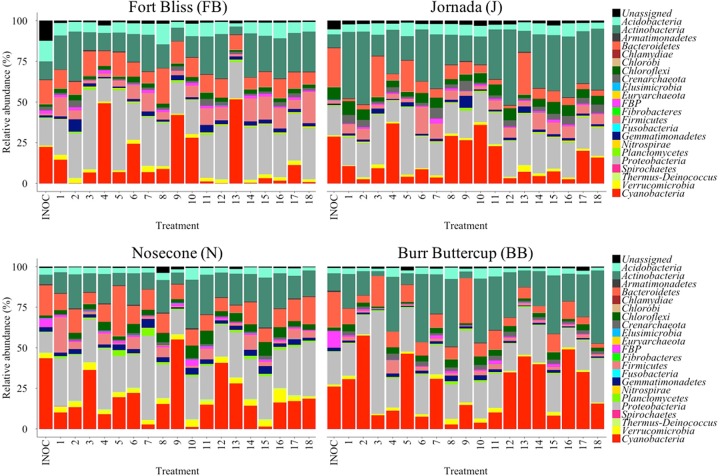
Figure 2 shows community composition at the level of bacterial phyla for each treatment as well as that of the respective biocrusts used as inoculum (INOC). In general, no major differences were conspicuous at this level of phylogenetic resolution. As is typical for biocrusts, cyanobacteria were the dominant phototrophs (11, 40), although there were small contributions by diatoms and streptophytes (detected through plastid 16S rRNA sequences, which fall in the cyanobacterial phylum). Cyanobacteria were important overall in all treatments and inoculum communities, along with Proteobacteria, Actinobacteria, Firmicutes, and Bacteriodetes; this overall distribution is also quite typical for biocrusts (9, 41). Bray-Curtis distances in community composition, calculated between each treatment and its respective inoculum, varied between 0.19 and 0.57 (see Fig. S1 in the supplemental material). Screening models applied to this treatment data set teased apart the factors that resulted in significantly minimal community composition deviations. While there was more site dependence regarding this than we had found for overall growth, nutrients and inoculum type emerged as important drivers. In cold sites, other factors (water and light) played an important role. Upon fitting reduced linear models to the data involving the relevant factors, all models were statistically significant according to one-way ANOVA results (F ≥ 3.27; P < 0.05; Table S1). Analyses of estimated effects showed that for inoculum type, slurries resulted in minimal shifts (LS means tests, P < 0.05; Table 1). For nutrients, the type of nutrient (N, P, or N plus P) was dependent on site (LS means tests, P < 0.05; Table 1). Whenever light was of relevance, the shaded treatment resulted in less shifts; whenever watering was of relevance, the highest level resulted in more stable communities (LS means tests, P < 0.05; Table 1).
FIG 2.
Endpoint bacterial community composition by phylum for each of the treatments in the fractional factorial experiments. Each panel corresponds to a different site. Data are averages of three independent determinations (biological replicates). Also included are the community composition determined for the biocrust samples used as inoculum (INOC; n = 3, technical replicates).
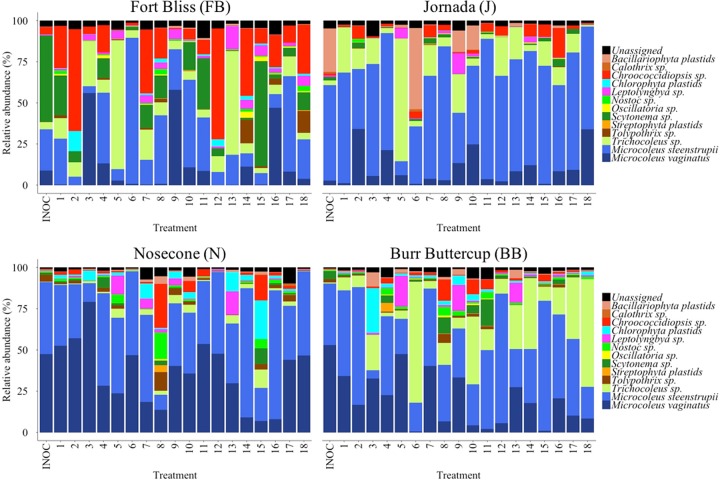
In Fig. 3, we show a similar community composition shift analysis but for cyanobacteria. In this case, the taxonomic resolution is much higher since the biocrust cyanobacteria are much better known in terms of diversity, and so the sensitivity to change is much enhanced as would be desirable for this functionally important group. The cyanobacterial community composition matched our expectations in terms of major cyanobacterial taxa for filamentous (42, 43) and heterocystous forms (44) as well as their biogeographical distribution, which is known to be heavily determined by average temperatures (38). Algal plastids were, as expected, minor components in most sites, but the presence of significant contributions of diatoms in the J inoculum was an unusual trait. Generally speaking, we can observe some differences among and between treatments and their inoculum communities, particularly in the FB and Burr Buttercup (BB) sites. The treatment-elicited shifts in community composition were generally larger than those seen in bacterial phyla and also more variable among treatments: Bray-Curtis dissimilarity distances calculated between each treatment and its respective inoculum varied between 0.15 and 0.99 (Fig. S1). In some treatments, relatively minor components of the biocrust microbial community become important, as is the case in FB, J, and BB samples, where we observed a high relative abundance of Trichocoleus in various treatments (Fig. 3). Here again, initial screening models teased apart, from the treatments, the main factors that minimized community composition shifts. We did not detect any significant factor in the FB site, but in the J site, inoculum and calcium were important. In the cold deserts, light and water were determinant, and nutrients and inoculum played a role in BB samples only. Results of one-way ANOVA tests after fitting reduced linear models with the relevant factors showed that models were statistically significant (F ≥ 5.77; P < 0.05; Table S1). As with bacterial phyla, analyses of the estimated effects revealed that slurries resulted in minimal shifts as did low light (LS means tests, P < 0.05; Table 1). The highest level of watering resulted in more stable communities (LS means tests, P < 0.05; Table 1). For nutrients, the addition of P plus N in BB resulted in minimal changes (LS means tests, P < 0.05; Table 1).
FIG 3.
Endpoint cyanobacterial community composition by major clades for each of the treatments in the fractional factorial experiments. Each panel corresponds to a different site. Data are averages of three independent determinations (biological replicates). Also included are the community composition determined for the biocrust samples used as inoculum (INOC; n = 3, technical replicates).
To test the predicted ability of the small-scale fractional factorial experiments for large-scale inoculum production, we applied incubation conditions deemed optimal for each site in a second greenhouse incubation trial. Here we used slurries, low light levels, the highest watering frequency, and added P to all sites. In the FB and BB samples, we also added N. Those were all factors that (i) maximized or were neutral to yield and (ii) minimized or were neutral to community shifts for bacteria and cyanobacteria in all sites. Because calcium additions had contradictory effects (promoting versus preventing community shifts) in different sites, we excluded this factor from the final formulation. Under these potentially optimal conditions, we observed a consistent and significant increase in chlorophyll a (chl a) content in all cases according to Wilcoxon's W signed-rank test results (W ≥ 25.08; P < 0.05) (Fig. 4), the magnitude of which varied between 1 and 2 orders from initial levels (INIT; red lines in Fig. 4). Final yields matched or exceeded the phototroph biomass contained in the biocrusts used as inocula (INOC; blue lines in Fig. 4). Microbial community structures at the phylum resolution remained rather constant in all treatments (Fig. 5), with low Bray-Curtis distances between the inoculum and the final and with differences in compositions never significant according to null model results (standardized effect sizes [SES] ≥ −4.35; P < 0.05) (Table 2). For cyanobacterial composition, however, community composition shifts during growth remained insignificant in 3 out of 4 sites (SES ≥ −1.98; P < 0.05) (Table 2; Fig. 6). Although noticeable in all sites, the development of populations of N-fixing Nostoc sp. and diatoms in FB samples resulted in significant deviations in community composition there (SES = −0.40; P > 0.05) (Table 2; Fig. 6). Pioneer crust-forming cyanobacteria (Microcoleus vaginatus, Microcoleus steenstrupii) (43) retained significant proportions of the final biomass in all sites (18%, 29%, 51%, and 47% for FB, J, Nosecone (N), and BB, respectively) in the greenhouse-grown biomass (Fig. 6). Nostoc, Tolypothrix, Leptolyngbya, and diatoms (through plastid 16S rRNA sequences) grew in samples cultivated in the greenhouse, whereas their relative abundances were not so important in the initial biocrust, especially in FB, J, and BB sites (Fig. 6).
FIG 4.
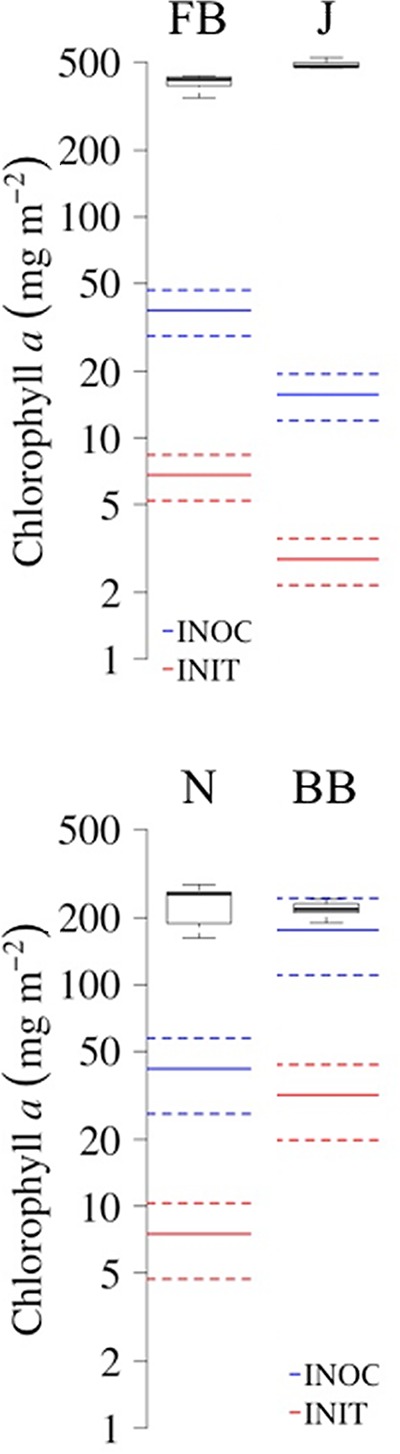
Boxplots showing aerial chl a contents at the end of the greenhouse incubation period for the second set of experiments. Boxes denote lower and upper quartiles (with median values depicted as black solid lines), and whiskers denote lower and upper extremes (n = 5). Blue lines indicate the chl a content of field biocrust samples used as inoculum (INOC), and red lines indicate the initial chl a content in the inoculated soils (INIT) (color solid lines indicate mean, and color dashed lines indicate standard deviations of n = 5) (FB, Fort Bliss; J, Jornada; N, Nosecone; BB, Burr Buttercup).
FIG 5.
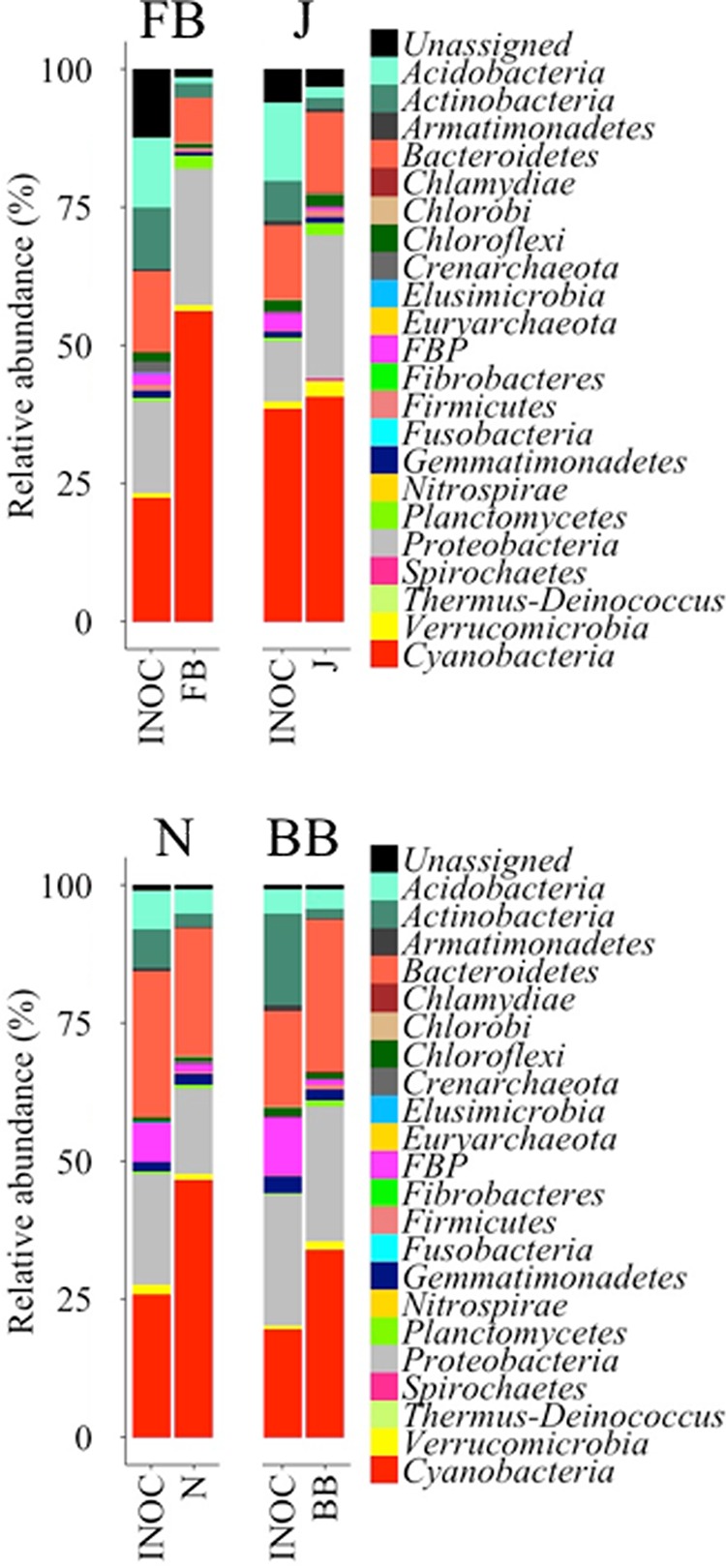
Endpoint bacterial community composition by phylum at the end of the greenhouse incubation period for the second set of experiments that were optimized according to previous results versus that obtained for the field biocrusts used as inoculum. Each panel corresponds to one site. Data are averages from five technical replicates (INOC, inoculum biocrust samples) or five biological replicates (cultivated biocrusts) (FB, Fort Bliss; J, Jornada; N, Nosecone; BB, Burr Buttercup).
TABLE 2.
Summary statistics of null models calculated with randomized microbial community data of bacterial phyla and cyanobacteria to assess the similarity between inoculum and nursery-reared biocrusts during the optimized, second set of experiments, according to sitea
| Site | Bacterial phyla |
Cyanobacteria |
||||
|---|---|---|---|---|---|---|
| BC | SES | P | BC | SES | P | |
| Fort Bliss | 0.47 | −4.35 | 0.000 | 0.73 | −0.40 | 0.345 |
| Jornada | 0.23 | −5.31 | 0.000 | 0.49 | −1.98 | 0.040 |
| Nosecone | 0.21 | −5.53 | 0.000 | 0.39 | −2.50 | 0.015 |
| Burr Buttercup | 0.39 | −4.69 | 0.000 | 0.48 | −1.99 | 0.034 |
BC, Bray-Curtis dissimilarity index, as an estimate of community composition shift based on bacterial phyla and cyanobacteria; SES, standardized effect size.
FIG 6.
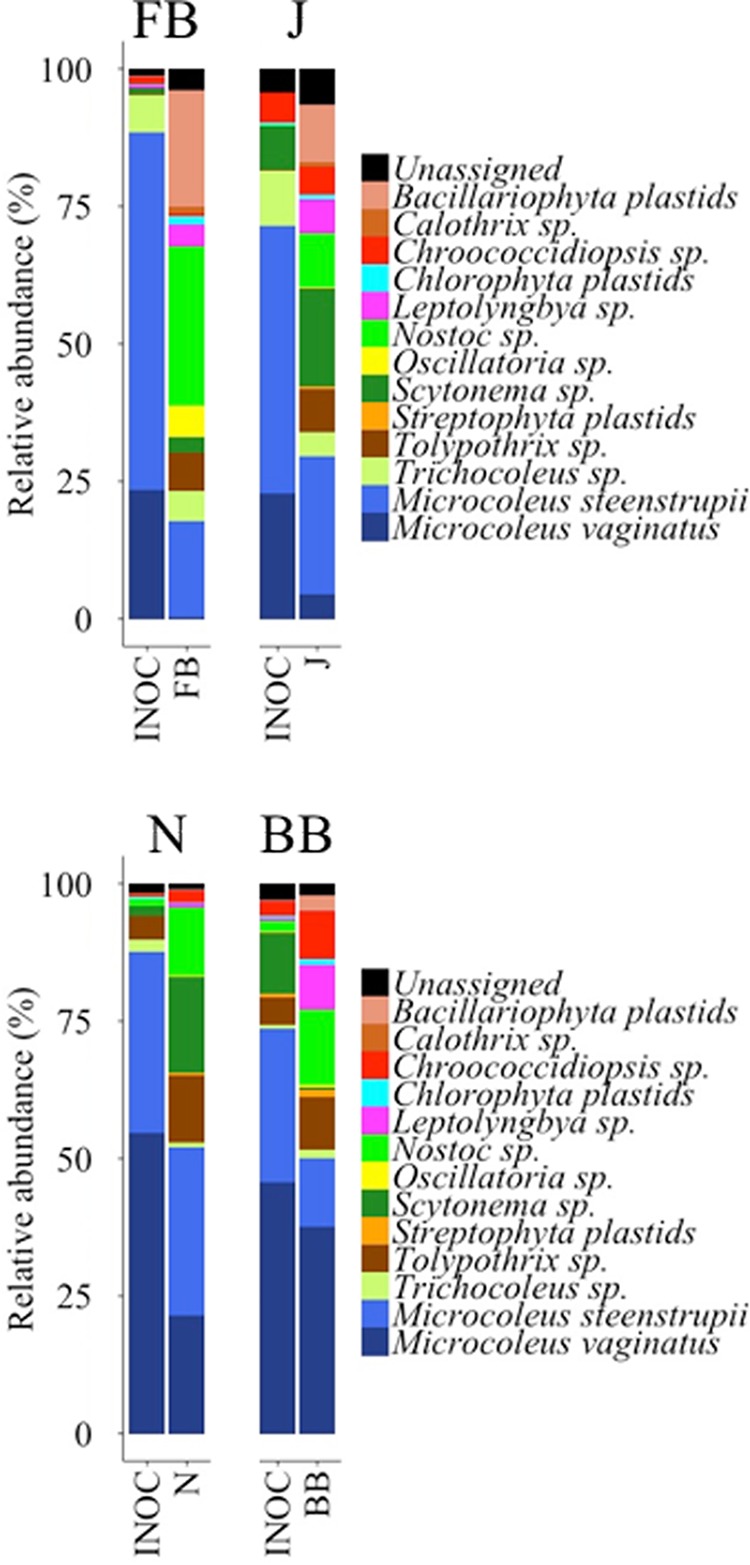
Endpoint cyanobacterial community composition by clades at the end of the greenhouse incubation period for the second set of experiments that were optimized according to previous results versus that obtained for the field biocrusts used as inoculum. Each panel corresponds to one site. Data are averages from five technical replicates (INOC, inoculum biocrust samples) or five biological replicates (cultivated biocrusts) (FB, Fort Bliss; J, Jornada; N, Nosecone; BB, Burr Buttercup).
DISCUSSION
Our work clearly shows that it is feasible to produce large amounts of biocrust biomass from low levels of natural inoculum within relatively short incubation times by using controlled, but seminatural, greenhouse facilities (biocrust nurseries). Our procedure was based on a two-step approach, whereby the incubation conditions leading to an optimal outcome with respect to maximal biomass development and minimal shifts in community structure are determined in a fractional factorial experiment first (3 months) to then proceed with a focused scale-up based on the optimized conditions (1 to 2 months). In our case, we found that watering crusts at a frequency double that experienced in the original sites and decreasing the intensity of sunlight by 60% yielded the best results in all of the sites and soil types tested. The addition of nutrients had a positive effect on some but not all sites. The procedures resulted in yield factors close to 30-fold (1.75 m2 of biocrust inoculum out of 0.06 m2 of natural biocrust remnant), which in turn can be used to inoculate ca. 6,000 m2 at a 5% inoculum level with quality-controlled, pedigreed, drought-acclimated inoculum.
After the first set of experiments, we observed that optimally nursed biocrusts attained or exceeded the biomass concentrations typical of field-collected mature communities. This was even in the presence of recurrent, full-scale cycles of desiccation and wetting designed to mimic the naturally pulsed nature of growth in biocrusts (25) and to avoid allochthonous contamination by nonterrestrial forms in our open system, a problem that cannot be avoided if open containers with the constant presence of liquid media are used. Separate controls show that open soil incubations with recurrent desiccation did not develop even incipient crusts for up to 6 months (unpublished results). However, not all incubation conditions resulted in such positive outcomes, and several treatments resulted consistently in either poor growth or even in loss of inoculum biomass (Fig. 1). Across different crust types, incubations under enhanced watering regimes (equivalent to double the natural rainfall averages of origin) and decreased light stress consistently resulted in high growth rates. These results are in line with what may have been surmised from the literature: rainfall frequency and light intensity are among the most important factors that contribute to the growth and activity of biocrusts (32, 34, 45–47). Moreover, some field observations indicate that shaded, wet, and relatively cool conditions promote biocrust growth and development (47, 48). This makes physiological sense in that photosynthetic microorganisms in these assemblages respond rapidly to hydration (49) but require sufficiently long periods of hydration to turn on the specific sets of genes involved in nutrient uptake, chl a, and ATP synthesis, as well as DNA repair after drought episodes (25) before net growth can occur. With respect to light intensity being a stress factor, this is also in line with preliminary findings in our laboratory (32) and elsewhere (50), and it is in agreement with the known physiological adaptive responses of cyanobacteria to photochemical stress in biocrusts, where the production of microbial sunscreens (14) or the phototactic responses of motile microorganisms (51) are important for microbial fitness. Contrary to some of the literature reports (52), we could not find statistical support for inoculum type playing a role in the biomass yield eventually attained. Finally, nutrient additions seemed to be important for biomass yield in some samples but not in all. Literature reports also give a somewhat contradictory picture regarding nutrient effects on biocrust development, one that includes some negative (32, 53, 54) and some positive effects (52). It stands to reason that differences in soil nutrient content may determine whether nutrient amendments will be needed to obtain high yields or not. In our case, the sites that responded to nutrient amendments (FB and J) were those with low soil nutrient levels (see Table S2 in the supplemental material). While the number of sites is too small to close this case, this explanation seems quite viable. The same lack of generality can be found in the literature with respect to micronutrient effects (Ca and/or metals) (32, 55), even though in our case they never determined yield. In summary, our results suggest that shading and increasing moisture availability are factors to be included generically in attempts to optimize biocrust nursery operations and that nutrient amendments should be evaluated on a case-by-case basis.
As the physiologically taxing conditions typical of biocrusts (UV irradiance, recurrent wetting and drying cycles, long periods of drought, exposure to extreme temperatures, abrasion by saltating soil particles, or lack of mobile nutrients) are relaxed in a nursery facility in order to allow for faster microbial growth, one also runs the risk of opening a window of opportunity for adventitious microbes that would not have been fit under natural conditions and will not survive placement in the wild. These are expected to be inherently fast-growing, weed-like forms that are kept in check in the field but enriched under nursery conditions, not unlike what we observed with Trichocoleus sp. or with diatoms in some of our treatments (Fig. 3). The enrichment of Trichocoleus in some treatments is consistent with some of our own unpublished data, showing that this clade is apparently better adapted to more eutrophic, fast-growth conditions than other crust cyanobacteria. Large amounts of such weedy organisms would produce an inoculum of low quality and fitness. For this reason, we implemented the following second constraint to our protocols: the monitoring of microbial community structure during incubations to ensure minimal deviations from local composition in the field. The issue of inoculum quality has in fact received very little attention in the literature even in open systems, but it is important in terms of the prevention of unintended inoculation of field sites with invasive microbes. It is, in fact, the case that populations of biocrust microbes, from lichens (56) to filamentous (38) and heterocystous cyanobacteria (37) to chemolithotrophic bacteria and archaea (10, 39), show clear biogeographical patterns of dominance and distribution. Therefore, it is important to develop site-specific inocula and to ensure that community composition shifts are minimized. Finally, only a few microbial types are considered to be pioneers capable of initiating biocrust formation (43). These are mostly some rope-forming cyanobacterial taxa in the form genus Microcoleus, which can colonize bare soils, hold soil particles together, and allow the establishment of other members in the assemblage (41), such as the N2-fixing cyanobacteria (Nostoc, Scytonema, and Tolypothrix), surface lichens, and mosses. The quality of a nursery-raised inoculum in terms of its potential to promote further biocrust formation in the wild will heavily hinge on the presence of robust and viable populations of such pioneer species.
The incubation protocols tested here were apparently moderate enough to maintain bacterial community composition rather invariant when gauged with the rough phylogenetic resolution of bacterial phyla. By determining the best combination of factors that minimized changes in the microbial community structure while allowing optimal growth, we succeeded in obtaining inoculum from each of the sites that had no significant difference in bacterial phyla composition from its respective initial inoculum (Fig. 5; Table 2). It was very useful to determine factors that minimize community shifts during our initial experiments. Fortunately, factors that promoted growth, such as high watering frequency and low light exposure, did not promote cyanobacterial community shifts (Fig. 6; Table 2). Importantly, the slurry method of inoculation was effective in preserving the community structure of biocrust biomass grown ex situ. This may be because it ensures the dispersal of motile (i.e., Microcoleus) and sessile species (Nostoc and Scytonema in their vegetative stage) alike, whereas mosaic inoculation would unduly favor motile species. By applying the optimal combination of factors, we observed that cyanobacterial communities in most cases can be maintained within compositional stasis (Table 2) and with a high content of crust-forming Microcoleus-like phylotypes (Fig. 6). However, the fact that the FB communities notably shifted in the nursery reminds us that community monitoring must remain a requirement in the quality control of inoculum production. Even these FB nursery-reared biocrusts, however, could be the basis for useful inoculation as the vast majority of phylotypes were detected originally in the field site and biocrust-forming Microcoleus-like phylotypes still made up more than 18% of the final community. It may be interesting to speculate what steps one could take when and if very serious deviations in community structure were to be discovered upon monitoring during incubations. An obvious approach would be to restart the experiment while imposing a range of conditions that resembles more the conditions typically seen in the field sites of origin. The logic behind this is that the natural local populations must be well adapted and are likely selected by those climatic and edaphic conditions. The more one deviates from those selective conditions in order to obtain increased growth, the more likely it is that alternative soil bacteria will outgrow the original types. Precisely such “niche separation” is what will result in the lower fitness of the new community when returned to the field. Different factors can, however, promote the growth of fast-growing, weed-like forms. Seasonality can be one of them, although we performed all of the experiments in both sets during the same season to minimize potential problems.
While the biocrusts that we produced are potentially of high inoculum quality, in that they have been reared on local soil substrates and exposed to locally matched climates, are acclimated to recurrent wetting and drying, are enriched to contain microbial communities that are minimally shifted in composition from local communities, and contain abundant crust-forming organisms, the question of inoculum quality ultimately must be tested experimentally in field inoculation experiments. Long-term experiments monitoring viability and effectiveness in the use (and fate) of nursery-reared biocrusts in field restoration are under way at all of our sites.
In summary, this work proposes a two-step process to obtain a high-quality biocrust biomass ex situ. We have demonstrated that it is possible to find an optimal combination of factors to allow biocrust biomass increases by orders of magnitude in nursery settings and within reasonable time frames for land managers, while preserving the quality and potential fitness of the communities to serve as inoculum in large-scale restoration efforts. Active restoration programs can markedly enhance the recovery of biological soil crusts in degraded dryland soils compared to no action (26), and the methodology proposed in this work should find use in such attempts.
MATERIALS AND METHODS
Study sites and sampling procedures.
The biocrust communities of two geographical locations were chosen. Soil and inoculum biocrust samples from hot deserts were from Fort Bliss military base (northern El Paso, TX, USA) and Jornada Basin Long Term Ecological Research Station (northeastern Las Cruces, NM, USA), whereas those from cold deserts were taken at Hill Air Force Base-Utah Test and Training Range (western Salt Lake City, UT, USA). In each type of desert, we took samples from sandy and silty soil sites, designated Fort Bliss (FB; lat 32.431069°, long −105.984151°) and Jornada (J; lat 32.545580°, long −106.723240°) in the hot deserts and Nosecone (N; lat 41.104198°, long −113.023194°) and Burr Buttercup (BB; lat 41.104211°, long −113.008204°) in the cold deserts. The maximum average temperatures in January are 13°C and 3°C, respectively, for the warm and cold locations, whereas the maximum average temperatures in July are 36°C and 34°C, respectively. Mean average precipitation is 250 mm (∼42-year period) in the hot locations and 200 mm (∼97-year period) in the cold locations. Biocrust samples were taken down to 1 cm deep by means of a dough steel scraper. Bulk soil to be used as growth substrate in the nursery was collected 10 cm below the surface. All samples were transported to greenhouse facilities within 2 days of collection. Bulk soil was then dried, sieved using a 0.45-mm metallic sieve, and stored dry; biocrusts were dried and then stored. Experiments involving hot desert sites were set up in a greenhouse facility at Arizona State University (ASU; Tempe, AZ, USA; 350 m above sea level), whereas those involving cold desert samples were carried out in a greenhouse facility at Northern Arizona University (NAU; Flagstaff, AZ, USA; 2,100 m above sea level), which has a much cooler climate so as to match temperature ranges as much as possible. Both greenhouse facilities have regular borosilicate glass panes, which block the UV-B portion of solar radiation but not its UV-A, thus providing an UV environment that is less harsh than the field but not free of stress, noting that phototrophs are especially sensitive to the oxygen-mediated, photosensitized effects of UV-A (57).
Experimental design.
In a first set of experiments, we performed a fractional factorial experiment on each of the four sites, with three replicates per treatment to test the effects of six main factors (water frequency, light intensity, type of inoculum, nutrients, calcium, and essential metals) on the growth and composition of biocrusts. In the case of composition, some replicates were lost and, as a consequence, 6 out of 56 treatments had only duplicates and one treatment had a single determination. A fractional factorial experiment, widely used in health sciences, is a carefully chosen fraction of a full factorial design, exploiting the sparsity-of-effects principle to reveal information about the most important features of the problem studied while using a fraction of the effort and cost of a full factorial design but still with a suitable power resolution (58). The use of a fractional factorial experiment helps to save space and time when several factors are tested in comparison to a full factorial experiment. The 18-treatment design used in this study is shown in Table 3. This first set of experiments was run on 15- by 15- by 5-cm transparent plastic containers (0.02 m2), filled to 4 cm with bulk soil and inoculated with the field biocrust samples to yield a final dilution of 5% (surface to surface) of the original. These experiments ran for 137 days during the fall/winter of 2013 in appropriate greenhouse nursery settings (Fig. 7A, B, and C). The water frequency factor had two levels: a high frequency (+, where crusts samples were watered every 3 days for hot desert sites and every 2 days for cold desert sites) and a low frequency (−, where crusts were watered every 9 and 4 days, respectively). The frequency of watering per location was based on local rainfall records after calculating average rainfall event frequencies. In each watering event, crust samples received an amount of water through mist emitters designed to attain ca. 80% of the water holding capacity of the soil and were allowed to dry naturally thereafter. The light intensity (illumination) factor also had two levels: a high light intensity (+, exposed to full greenhouse sunlight) and a low light intensity (−, crusts were covered with a black cloth that blocked approximately 60% of sunlight). The inoculum factor consisted of two types: mosaic (M), where 15 discrete fragments of appropriate biocrust, 0.4 cm in diameter and 1 cm deep, were directly transplanted on top of the bare soil, in a mosaic pattern, and slurry (S), where 15 discrete fragments of biocrust, 0.4 cm in diameter and 1 cm deep, were slurried and then spread over the bare soil. The nutrient factor had three levels: P (addition of a mix of KH2PO4 and K2HPO4 to a final concentration of 75 μg P g soil−1), N (addition of NH4NO3 to a final concentration of 150 μg N g soil−1), and P plus N (addition of both P and N); all nutrients were prepared in fresh, autoclaved, double-distilled water and added as a unique pulse on day 1 of the experiments. The calcium factor had two levels: a high content of calcium (+, addition of Ca as calcium carbonate pellets to a final concentration of approximately 40 μg Ca g soil−1) and a low content of calcium (−, no addition of Ca). Finally, the trace metal factor had two levels: a high content in trace metals (+, addition of the trace metal solution of the BG-11 medium [59] to a final concentration of 2 μg metal solution g soil−1) and a low content in essential metals (−, no addition of this metal solution); the metal solution was prepared in fresh, autoclaved, double-distilled water and added as a unique pulse on day 1 of the experiments.
TABLE 3.
Treatments for greenhouse incubations used in the fractional factorial design experiments (first phase experiments), which were combinations of independent factorsa
| Treatment | Watering | Light | Nutrient(s) | Inoculum | Calcium | Metals |
|---|---|---|---|---|---|---|
| 1 | + | + | P+N | M | + | + |
| 2 | + | + | N | S | + | − |
| 3 | + | − | N | M | − | + |
| 4 | + | + | P | M | − | − |
| 5 | + | − | P | S | + | + |
| 6 | − | − | N | M | − | + |
| 7 | − | − | P+N | S | − | − |
| 8 | − | + | P | S | − | + |
| 9 | + | − | P | M | + | − |
| 10 | − | + | P | M | − | − |
| 11 | − | + | P+N | M | + | + |
| 12 | + | + | N | S | − | + |
| 13 | + | − | P+N | S | − | − |
| 14 | − | − | P+N | S | − | − |
| 15 | − | + | N | S | + | − |
| 16 | − | − | P | S | + | + |
| 17 | + | + | P+N | M | + | + |
| 18 | − | − | N | M | + | − |
P, addition of phosphorus; N, addition of nitrogen; M, mosaic-like inoculum; S, slurry-like inoculum; high (+) or low (−) watering; ambient (+) or shaded (−) illumination; and additions (+) or not (−) of calcium and metals.
FIG 7.
General aspect of the microbial nursery facilities. Initial fractional factorial experiment (first phase experiments) at the Arizona State University (ASU) greenhouse (A), with a detailed view of Fort Bliss (B) and Jornada (C) soil incubations; plastic containers are 15 by 15 cm, and greenhouse benches are 2.74 by 0.91 m. Large-scale incubations (second phase experiments) in the Northern Arizona University (NAU) nursery (D), with top views of the final biocrusts produced for Nosecone (E) and Burr Buttercup (F) samples; plastic containers are 86 by 14 cm.
To test the validity of the results obtained in the fractional factorial experiment, we ran a second set of experiments for 120 days during the spring/summer of 2014. This second set involved only the factors and levels that were found to maximize the growth of the biocrust and minimize changes in the microbial community structure in the first phase. These experiments used five 86- by 40- by 12-cm transparent plastic containers (0.35 m2), filled to 1 cm with soil and inoculated with the appropriate amount of biocrust sample at a dilution factor (surface to surface) of 18% of the field crusts used as inocula. Both bulk soil and biocrust samples were newly collected at each site for the second set of experiments. Inoculated biocrusts were incubated in the appropriate greenhouse facility, either Tempe or Flagstaff, AZ, USA (Fig. 7D, E, and F).
Microbiological response variables.
Aerial content in chlorophyll a (chl a) was determined in the initial field biocrust samples that were used as inocula and at the end of the experimental incubations as a proxy for autotrophic biomass in each replicate. Seven 0.4-cm-diameter cores of biocrust, 1 cm deep, were randomly taken in each replicate or field biocrust sample, mixed, and extracted in 95% ethanol at 4°C in the dark for 24 h. Extracts were then centrifuged (5,000 rpm for 5 min at 4°C), and chl a concentrations were quantified according to references 60 and 61 in a Shimadzu UV-1601 spectrophotometer (Shimadzu Corporation, Kyoto, Japan).
To determine the microbial community structure of biocrust samples, we used high-throughput 16S rRNA gene amplicon sequencing. At the end of each experiment, seven 0.4-cm-diameter core samples of biocrusts, 1 cm deep, were randomly taken in each replicate and in all of the inoculum biocrust samples and stored at −80°C until further processing. In each replicate, or in each biocrust sample used as inoculum, the seven cores were pooled in composite samples, and whole community DNA was then extracted using the PowerSoil DNA isolation kit (Mo Bio, Carlsbad, CA, USA) according to manufacturer's recommendations. The V4 region of the 16S rRNA gene was amplified using the barcoded primer set 515F/806R (62). PCR conditions were as follows: 3 min at 94°C followed by 35 cycles of 45 s at 94°C, 60 s at 50°C, and 90 s at 72°C and a final elongation for 10 min at 72°C. PCR amplifications for each sample were done in triplicate and then pooled and quantified by using the Quant-iT PicoGreen double-stranded DNA assay kit (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA). A total of 240 ng of DNA per sample was pooled and then cleaned using the QIAquick PCR purification kit (Qiagen, Valencia, CA, USA). The DNA concentration of the PCR pooled library was quantified by the Illumina library quantification kit ABI Prism (Kapa Biosystems, Wilmington, MA, USA). The PCR pooled library was diluted to a final concentration of 4 nM and denatured before being mixed with 30% (vol/vol) of 4 pM denatured Phi X viral DNA. Finally, the PCR pooled library and Phi X mixture was loaded in the MiSeq Illumina sequencer cartridge, and the run was performed using chemistry version 2 (2 × 150 paired end) following the recommendations of the manufacturer (Illumina, San Diego, CA, USA).
All of the paired-end reads obtained were assembled with PANDAseq (63). Sequences between 250 and 290 bp were further used for downstream analyses in QIIME (64) to remove barcodes and low-quality reads, to pick operational taxonomic units (OTUs), assign taxonomy, align multiple sequences, build phylogenetic trees, and define an OTU table. OTUs were defined with a threshold of 97% sequence similarity and clustered using UCLUST (65). Each OTU was initially taxonomically assigned by using the Ribosomal Database Project (RDP) classifier (66). Representative sequences of each OTU were then aligned against the Greengenes database core reference alignment (67). Additionally, all cyanobacterial OTUs were subject to individual scrutiny against our own biocrust database in order to produce correct taxonomic assignments at this low level of resolution, for which representative sequences of each OTU were placed in a reference tree to determine their positions within known clades. Sequences were then aligned using the Guidance2 server and MAFFT7 (68, 69). The tree was built using RAxML 8 (70) through bootstrap and maximum likelihood workflow on the CIPRES cluster (71). OTUs were aligned to the reference alignment using PaPaRa (72) and then placed on the reference tree using the RAxML 8 evolutionary placement algorithm (70). The placed sequences were visualized by using the iTOL 3 server (73). Once all of the OTUs were defined and taxonomically assigned, an abundance table with all of the OTUs and samples was built and then used in the analyses described below.
Statistical analyses.
A screening model was applied to the data of the fractional factorial experiments to identify which factors maximized the growth of biocrusts and minimized shifts in community structure. Results of linear models fitted after initial screening were tested via one-way analysis of variance (ANOVA), and model statistical significance was determined by Fisher's F test value (74). The estimated effects of factor levels were tested by least-squares (LS) means tests (74). Diagnostic plots were used to assess potential deviations from homoscedasticity and normality of residuals in these models. Overall phototroph growth was assessed as the difference between final and initial aerial concentrations in chl a. Shifts in community structure were gauged pairwise by using the Bray-Curtis dissimilarity index as an estimator of the taxonomic distance between treatments and the biocrust inoculum (0 indicates that two samples have the same composition, whereas 1 indicates that two samples do not share any taxon). Experimental design and data analyses were performed in JMP Pro 12 (SAS Institute, Cary, NC, USA). Bray-Curtis dissimilarity indices were calculated using the vegan package (75) written in R language (76). A P value of 0.05 was set as the significance threshold for all of these statistical analyses.
Differences in the mean aerial chl a content between the biocrust samples grown in the greenhouse and the initial values in the second set of experiments were tested by using Wilcoxon's W signed-rank tests (74). Here, we were interested not just in gauging relative deviations from the original composition but also in testing whether those changes were statistically significant. For this, we tested the similarity of microbial community structures through Bray-Curtis dissimilarity indices between the biocrust samples grown in the greenhouse and the inoculum (INOC) by generating abundance matrices of random communities that were then used to build a null distribution model to which the observed Bray-Curtis dissimilarity index value was compared (77). If we consider a matrix with taxa in rows, sites in columns, and abundances as entries, we maintained the richness of each row (i.e., row sums are fixed) and we set abundances among columns to be equiprobable (i.e., all sites had the same average number of entries) to build the null model (78). This fixed rows-equiprobable columns null distribution model retains taxa frequencies, i.e., rare taxa remain rare and common taxa remain common (79). The use of this null distribution model is recommended because it has a low probability of type I errors (78). Statistical significance was then assessed by comparing observed Bray-Curtis dissimilarity indices to the distribution of distances calculated after the randomization process (77). To avoid any directional bias associated with the decrease in variance in expected values with increasing species richness, we calculated standardized effect sizes (SES) according to reference 77. All of these statistical analyses were completed in R (76) using the vegan (75) and picante packages (80) written in R language. A P value of 0.05 was set as the significant threshold for all of these statistical analyses.
Accession number(s).
Raw sequence data were submitted to NCBI and are publicly available under BioProject number PRJNA343817.
Supplementary Material
ACKNOWLEDGMENTS
We are indebted to Javier Seoane Pinilla for his assistance with the statistical analyses and to Juan Manuel Maldonado Ortiz for help with bioinformatics. We thank Anita J. Antoninka and Matthew A. Bowker for help with the bionursery at Northern Arizona University (Flagstaff, AZ, USA).
This study was supported by a Strategic Environmental Research and Development Grant (SERDP) (W912HQ-13-C-0035-P00005 RC-2329) of the U.S. Department of Defense.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02179-16.
REFERENCES
- 1.Prăvălie R. 2016. Drylands extent and environmental issues: a global approach. Earth Sci Rev 161:259–278. doi: 10.1016/j.earscirev.2016.08.003. [DOI] [Google Scholar]
- 2.Huang J, Yu H, Guan X, Wang G, Guo R. 2016. Accelerated dryland expansion under climate change. Nat Clim Chang 6:166–172. [Google Scholar]
- 3.Belnap J. 2003. The world at your feet: desert biological soil crusts. Front Ecol Environ 1:181–189. [Google Scholar]
- 4.Belnap J, Walker BJ, Munson SM, Gill RA. 2014. Controls on sediment production in two U.S. deserts. Aeolian Res 14:15–24. doi: 10.1016/j.aeolia.2014.03.007. [DOI] [Google Scholar]
- 5.Field JP, Belnap J, Breshears DD, Neff JC, Okin GS, Whicker JJ, Painter TH, Ravi S, Reheis MC, Reynolds RL. 2010. The ecology of dust. Front Ecol Environ 8:423–430. [Google Scholar]
- 6.Griffin DW. 2007. Atmospheric movement of microorganisms in clouds of desert dust and implications for human health. Clin Microbiol Rev 20:459–477. doi: 10.1128/CMR.00039-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Powell JT, Chatziefthimiou AD, Banack SA, Cox PA, Metcalf JS. 2015. Desert crust microorganisms, their environment, and human health. J Arid Environ 112:127–133. doi: 10.1016/j.jaridenv.2013.11.004. [DOI] [Google Scholar]
- 8.Painter TH, Skiles SM, Deems JS, Bryant AC, Landry CC. 2012. Dust radiative forcing in snow of the Upper Colorado River Basin. 1. A 6 year record of energy balance, radiation, and dust concentrations. Wat Resour Res 48:W07521. doi: 10.1029/2012WR011985. [DOI] [Google Scholar]
- 9.Nunes da Rocha U, Cadillo-Quiroz H, Karaoz U, Rajeev L, Klitgord N, Dunn S, Truong V, Buenrostro M, Bowen BP, Garcia-Pichel F, Mukhopadhyay A, Northen TR, Brodie EL. 2015. Isolation of a significant fraction of non-phototroph diversity from a desert biological soil crust. Front Microbiol 6:277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soule T, Anderson IJ, Johnson SL, Bates ST, Garcia-Pichel F. 2009. Archaeal populations in biological soil crusts from arid lands in North America. Soil Biol Biochem 41:2069–2074. doi: 10.1016/j.soilbio.2009.07.023. [DOI] [Google Scholar]
- 11.Davies LO, Schäfer H, Marshall S, Bramke I, Oliver RG, Bending GD. 2013. Light structures phototroph, bacterial and fungal communities at the soil surface. PLoS One 8:e69048. doi: 10.1371/journal.pone.0069048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia-Pichel F, Johnson S, Youngkin D, Belnap J. 2003. Small-scale vertical distribution of bacterial biomass and diversity in biological soil crusts from arid lands in the Colorado Plateau. Microb Ecol 46:312–321. doi: 10.1007/s00248-003-1004-0. [DOI] [PubMed] [Google Scholar]
- 13.Belnap J, Phillips S, Herrick J, Johansen J. 2007. Wind erodibility of soils at Fort Irwin, California (Mojave Desert), USA, before and after trampling disturbance: implications for land management. Earth Surf Process Landforms 32:75–84. doi: 10.1002/esp.1372. [DOI] [Google Scholar]
- 14.Couradeau E, Karaoz U, Lim HC, Nunes U, Northen T, Brodie E, Garcia-Pichel F. 2016. Bacteria increase arid-land soil surface temperature through the production of sunscreens. Nat Commun 7:10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiao B, Wang H, Fan J, Fischer T, Veste M. 2013. Biological soil crusts decrease soil temperature in summer and increase soil temperature in winter in semiarid environment. Ecol Eng 58:52–56. doi: 10.1016/j.ecoleng.2013.06.009. [DOI] [Google Scholar]
- 16.Strauss SL, Day TA, Garcia-Pichel F. 2012. Nitrogen cycling in desert biological soil crusts across biogeographic regions in the Southwestern United States. Biogeochemistry 108:171–182. doi: 10.1007/s10533-011-9587-x. [DOI] [Google Scholar]
- 17.Beraldi-Campesi H, Hartnett HE, Anbar A, Gordon GW, Garcia-Pichel F. 2009. Effect of biological soil crusts on soil elemental concentrations: implications for biogeochemistry and as traceable biosignatures of ancient life on land. Geobiology 7:348–359. doi: 10.1111/j.1472-4669.2009.00204.x. [DOI] [PubMed] [Google Scholar]
- 18.Chamizo S, Cantón Y, Rodríguez-Caballero E, Domingo F. 2016. Biocrusts positively affect the soil water balance in semiarid ecosystems. Ecohydrology 9:1208–1221. doi: 10.1002/eco.1719. [DOI] [Google Scholar]
- 19.Lan S, Zhang Q, Wu L, Liu Y, Zhang D, Hu C. 2014. Artificially accelerating the reversal of desertification: cyanobacterial inoculation facilitates the succession of vegetation communities. Environ Sci Technol 48:307–315. doi: 10.1021/es403785j. [DOI] [PubMed] [Google Scholar]
- 20.Zaady E, Arbel S, Barkai D, Sarig S. 2013. Long-term impact of agricultural practices on biological soil crusts and their hydrological processes in a semiarid landscape. J Arid Environ 90:5–11. doi: 10.1016/j.jaridenv.2012.10.021. [DOI] [Google Scholar]
- 21.Williams WJ, Eldridge DJ, Alchin BM. 2008. Grazing and drought reduce cyanobacterial soil crusts in an Australian Acacia woodland. J Arid Environ 72:1064–1075. doi: 10.1016/j.jaridenv.2007.11.017. [DOI] [Google Scholar]
- 22.Thomas AD. 2012. Impact of grazing intensity on seasonal variations in soil organic carbon and soil CO2 efflux in two semiarid grasslands in Southern Botswana. Philos Trans R Soc Lond B Biol Sci 367:3076–3086. doi: 10.1098/rstb.2012.0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hagemann M, Henneberg M, Felde VJ, Drahorad SL, Berkowicz SM, Felix-Henningsen P, Kaplan A. 2015. Cyanobacterial diversity in biological soil crusts along a precipitation gradient, Northwest Negev Desert, Israel. Microb Ecol 70:219–230. doi: 10.1007/s00248-014-0533-z. [DOI] [PubMed] [Google Scholar]
- 24.Ferrenberg S, Reed SC, Belnap J. 2015. Climate change and physical disturbance cause similar community shifts in biological soil crusts. Proc Natl Acad Sci U S A 112:12116–12121. doi: 10.1073/pnas.1509150112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rajeev L, da Rocha UN, Klitgord N, Luning EG, Fortney J, Axen SD, Shih PM, Bouskill NJ, Bowen BP, Kerfeld CA, Garcia-Pichel F, Brodie EL, Northen TR, Mukhopadhyay A. 2013. Dynamic cyanobacterial response to hydration and dehydration in a desert biological soil crust. ISME J 7:2178–2191. doi: 10.1038/ismej.2013.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li XR, Xiao HL, Zhang JG, Wang XP. 2004. Long-term ecosystem effects of sand-binding vegetation in the Tengger Desert, Northern China. Restor Ecol 12:376–390. doi: 10.1111/j.1061-2971.2004.00313.x. [DOI] [Google Scholar]
- 27.Read C, Duncan D, Vesk P, Elith J. 2011. Surprisingly fast recovery of biological soil crusts following livestock removal in southern Australia. J Veg Sci 22:905–916. doi: 10.1111/j.1654-1103.2011.01296.x. [DOI] [Google Scholar]
- 28.Bowker MA, Belnap J, Davidson DW, Goldstein H. 2006. Correlates of biological soil crust abundance across a continuum of spatial scales: support for a hierarchical conceptual model. J Appl Ecol 43:152–163. doi: 10.1111/j.1365-2664.2006.01122.x. [DOI] [Google Scholar]
- 29.Antoninka A, Bowker MA, Reed SC, Doherty K. 2016. Production of greenhouse-grown biocrust mosses and associated cyanobacteria to rehabilitate dryland soil function. Restor Ecol 24:324–335. doi: 10.1111/rec.12311. [DOI] [Google Scholar]
- 30.Xu Y, Rossi F, Colica G, Deng S, De Philippis R, Chen L. 2013. Use of cyanobacterial polysaccharides to promote shrub performances in desert soils: a potential approach for the restoration of desertified areas. Biol Fertil Soils 49:143–152. doi: 10.1007/s00374-012-0707-0. [DOI] [Google Scholar]
- 31.Acea M, Prieto-Fernández A, Diz-Cid N. 2003. Cyanobacterial inoculation of heated soils: effect on microorganisms of C and N cycles and on chemical composition in soil surface. Soil Biol Biochem 35:513–524. doi: 10.1016/S0038-0717(03)00005-1. [DOI] [Google Scholar]
- 32.Bu C, Wu S, Yang Y, Zheng M. 2014. Identification of factors influencing the restoration of cyanobacteria-dominated biological soil crusts. PLoS One 9:e90049. doi: 10.1371/journal.pone.0090049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buttars SM, St. Clair LL, Johansen JR, Sray JC, Payne MC, Webb BL, Terry RE, Pendleton BK, Warren SD. 1998. Pelletized cyanobacterial soil amendments: laboratory testing for survival, escapability, and nitrogen fixation. Arid Soil Res Rehabil 12:165–178. [Google Scholar]
- 34.Chen L, Xie Z, Hu C, Li D, Wang G, Liu Y. 2006. Man-made desert algal crusts as affected by environmental factors in Inner Mongolia, China. J Arid Environ 67:521–527. doi: 10.1016/j.jaridenv.2006.02.018. [DOI] [Google Scholar]
- 35.Wang W, Liu Y, Li D, Hu C, Rao B. 2009. Feasibility of cyanobacterial inoculation for biological soil crusts formation in desert area. Soil Biol Biochem 41:926–929. doi: 10.1016/j.soilbio.2008.07.001. [DOI] [Google Scholar]
- 36.Zhang B, Zhang Y, Su Y, Wang J, Zhang J. 2013. Responses of microalgal-microbial biomass and enzyme activities of biological soil crusts to moisture and inoculated Microcoleus vaginatus gradients. Arid Land Res Manag 27:216–230. doi: 10.1080/15324982.2012.754514. [DOI] [Google Scholar]
- 37.Nagy ML, Pérez A, Garcia-Pichel F. 2005. The prokaryotic diversity of biological soil crusts in the Sonoran Desert (Organ Pipe Cactus National Monument, AZ). FEMS Microbiol Ecol 54:233–245. doi: 10.1016/j.femsec.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 38.Garcia-Pichel F, Loza V, Marusenko Y, Mateo P, Potrafka RM. 2013. Temperature drives the continental-scale distribution of key microbes in topsoil communities. Science 340:1574–1577. doi: 10.1126/science.1236404. [DOI] [PubMed] [Google Scholar]
- 39.Marusenko Y, Bates ST, Anderson I, Johnson SL, Soule T, Garcia-Pichel F. 2013. Ammonia-oxidizing archaea and bacteria are structured by geography in biological soil crusts across North American arid lands. Ecol Process 2:9. doi: 10.1186/2192-1709-2-9. [DOI] [Google Scholar]
- 40.Steven B, Gallegos-Graves LV, Belnap J, Kuske CR. 2013. Dryland soil microbial communities display spatial biogeographic patterns associated with soil depth and soil parent material. FEMS Microbiol Ecol 86:101–113. doi: 10.1111/1574-6941.12143. [DOI] [PubMed] [Google Scholar]
- 41.Gundlapally SR, Garcia-Pichel F. 2006. The community and phylogenetic diversity of biological soil crusts in the Colorado Plateau studied by molecular fingerprinting and intensive cultivation. Microb Ecol 52:345–357. doi: 10.1007/s00248-006-9011-6. [DOI] [PubMed] [Google Scholar]
- 42.Garcia-Pichel F, López-Cortés A, Nübel U. 2001. Phylogenetic and morphological diversity of cyanobacteria in soil desert crusts from the Colorado Plateau. Appl Environ Microbiol 67:1902–1910. doi: 10.1128/AEM.67.4.1902-1910.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garcia-Pichel F, Wojciechowski MF. 2009. The evolution of a capacity to build supra-cellular ropes enabled filamentous cyanobacteria to colonize highly erodible substrates. PLoS One 4:e7801. doi: 10.1371/journal.pone.0007801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yeager CM, Kuske CR, Carney TD, Johnson SL, Ticknor LO, Belnap J, Brodie EL, Berkeley L. 2012. Response of biological soil crust diazotrophs to season, altered summer precipitation, and year-round increased temperature in an arid grassland of the Colorado Plateau, USA. Front Microbiol 3:358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bowker MA, Soliveres S, Maestre FT. 2010. Competition increases with abiotic stress and regulates the diversity of biological soil crusts. J Ecol 98:551–560. doi: 10.1111/j.1365-2745.2010.01647.x. [DOI] [Google Scholar]
- 46.Kidron GJ, Vonshak A, Dor I, Barinova S, Abeliovich A. 2010. Properties and spatial distribution of microbiotic crusts in the Negev Desert, Israel. Catena 82:92–101. doi: 10.1016/j.catena.2010.05.006. [DOI] [Google Scholar]
- 47.Kidron GJ, Vonshak A. 2012. The use of microbiotic crusts as biomarkers for ponding, subsurface flow and soil moisture content and duration. Geoderma 181–182:56–64. [Google Scholar]
- 48.Bowker MA. 2007. Biological soil crust rehabilitation in theory and practice: an underexploited opportunity. Restor Ecol 15:13–23. doi: 10.1111/j.1526-100X.2006.00185.x. [DOI] [Google Scholar]
- 49.Garcia-Pichel F, Belnap J. 1996. Microenvironments and microscale productivity of cyanobacterial desert crusts. J Phycol 32:774–782. doi: 10.1111/j.0022-3646.1996.00774.x. [DOI] [Google Scholar]
- 50.Chen L, Liu Y, Song L. 2002. The function of exopolysaccharides of Microcoleus vaginatus in the formation of desert soil. Acta Hydrobiol Sin 26:155–159. [Google Scholar]
- 51.Garcia-Pichel F, Pringault O. 2001. Cyanobacteria track water in desert soils. Nature 413:380–381. doi: 10.1038/35096640. [DOI] [PubMed] [Google Scholar]
- 52.Maestre FT, Martín N, Díez B, López-Poma R, Santos F, Luque I, Cortina J. 2006. Watering, fertilization, and slurry inoculation promote recovery of biological crust function in degraded soils. Microb Ecol 52:365–377. doi: 10.1007/s00248-006-9017-0. [DOI] [PubMed] [Google Scholar]
- 53.Belnap J, Phillips SL, Flint S, Money J, Caldwell M. 2008. Global change and biological soil crusts: effects of ultraviolet augmentation under altered precipitation regimes and nitrogen additions. Glob Chang Biol 14:670–686. doi: 10.1111/j.1365-2486.2007.01509.x. [DOI] [Google Scholar]
- 54.Delgado-Baquerizo M, Morillas L, Maestre FT, Gallardo A. 2013. Biocrusts control the nitrogen dynamics and microbial functional diversity of semi-arid soils in response to nutrient additions. Plant Soil 372:643–654. doi: 10.1007/s11104-013-1779-9. [DOI] [Google Scholar]
- 55.Bowker M, Belnap J, Davidson DW, Phillips SL. 2005. Evidence for micronutrient limitation of biological soil crusts: importance to arid-lands restoration. Ecol Appl 15:1941–1951. doi: 10.1890/04-1959. [DOI] [Google Scholar]
- 56.Bowker M, Belnap J, Büdel B, Sannier C, Pietrasiak N, Eldridge DJ, Rivera-Aguilar V. 2016. Controls on distribution patterns of biological soil crusts at micro- to global scales, p 173–198. In Weber B, Büdel B, Belnap J (ed), Biological soil crusts: an organizing principle in drylands. Springer Nature, Berlin, Germany. [Google Scholar]
- 57.Garcia-Pichel F. 1998. Solar ultraviolet and the evolutionary history of cyanobacteria. Orig Life Evol Biosph 28:321–347. doi: 10.1023/A:1006545303412. [DOI] [PubMed] [Google Scholar]
- 58.Box G, Hunter J, Hunter W. 2005. Statistics for experimenters: design, innovation and discovery, 2nd ed John Wiley and Sons, Hoboken, NJ. [Google Scholar]
- 59.Stanier R, Kunisawa R, Mandel M, Cohen-Bazire G. 1971. Purification and properties of unicellular blue-green algae (order Chroococcales). Bacteriol Rev 35:171–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wintermans J, de Mots A. 1965. Spectrophotometric characteristics of chlorophyll a and b and their phaeophytins in ethanol. Biochim Biophys Acta 109:448–453. doi: 10.1016/0926-6585(65)90170-6. [DOI] [PubMed] [Google Scholar]
- 61.Ritchie RJ. 2006. Consistent sets of spectrophotometric chlorophyll equations for acetone, methanol and ethanol solvents. Photosynth Res 89:27–41. doi: 10.1007/s11120-006-9065-9. [DOI] [PubMed] [Google Scholar]
- 62.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R. 2012. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Masella AP, Bartram AK, Truszkowski JM, Brown DG, Neufeld JD. 2012. PANDAseq: paired-end assembler for Illumina sequences. BMC Bioinformatics 13:31. doi: 10.1186/1471-2105-13-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, Mcdonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 66.Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P. 2012. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sela I, Ashkenazy H, Katoh K, Pupko T. 2015. GUIDANCE2: accurate detection of unreliable alignment regions accounting for the uncertainty of multiple parameters. Nucleic Acids Res 43:W7–W14. doi: 10.1093/nar/gkv318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees, p 1–8. In Proceedings of the Gateway Computing Environments Workshop (GCE) IEEE, New Orleans, LA. [Google Scholar]
- 72.Berger SA, Stamatakis A. 2011. Aligning short reads to reference alignments and trees. Bioinformatics 27:2068–2075. doi: 10.1093/bioinformatics/btr320. [DOI] [PubMed] [Google Scholar]
- 73.Letunic I, Bork P. 2007. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23:127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- 74.Zar JH. 1998. Biostatistical analysis, 4th ed Prentice Hall, New York, NY. [Google Scholar]
- 75.Oksanen J, Guillaume Blanchet F, Kindt R, Legendre P, Minchin P, O'Hara R, Simpson G, Solymos P, Stevens M, Wagner H, R Development Core Team . 2015. “vegan”: community ecology package. R package version 2.3-4. The R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 76.R Development Core Team. 2016. R: a language and environment for statistical computing. The R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 77.Swenson N. 2014. Functional and phylogenetic ecology in R. Springer-Verlag, New York, NY. [Google Scholar]
- 78.Gotelli NJ. 2008. Null model analysis of species co-occurrence patterns. Ecology 81:2606–2621. [Google Scholar]
- 79.Maestre FT, Martínez I, Escolar C, Escudero A. 2009. On the relationship between abiotic stress and co-occurrence patterns: an assessment at the community level using soil lichen communities and multiple stress gradients. Oikos 118:1015–1022. doi: 10.1111/j.1600-0706.2009.17362.x. [DOI] [Google Scholar]
- 80.Kembel S, Ackerly D, Blomberg S, Cornwell W, Cowan P, Helmus M, Morlon H, Webb C, R Development Core Team . 2015. “picante”: R tool for integrating phylogenies and ecology. R package version 1.6-2. The R Foundation for Statistical Computing, Vienna, Austria. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.