ABSTRACT
The APOBEC3 (A3) enzymes, A3G and A3F, are coordinately expressed in CD4+ T cells and can become coencapsidated into HIV-1 virions, primarily in the absence of the viral infectivity factor (Vif). A3F and A3G are deoxycytidine deaminases that inhibit HIV-1 replication by inducing guanine-to-adenine hypermutation through deamination of cytosine to form uracil in minus-strand DNA. The effect of the simultaneous presence of both A3G and A3F on HIV-1 restriction ability is not clear. Here, we used a single-cycle infectivity assay and biochemical analyses to determine if coencapsidated A3G and A3F differ in their restriction capacity from A3G or A3F alone. Proviral DNA sequencing demonstrated that compared to each A3 enzyme alone, A3G and A3F, when combined, had a coordinate effect on hypermutation. Using size exclusion chromatography, rotational anisotropy, and in vitro deamination assays, we demonstrate that A3F promotes A3G deamination activity by forming an A3F/G hetero-oligomer in the absence of RNA which is more efficient at deaminating cytosines. Further, A3F caused the accumulation of shorter reverse transcripts due to decreasing reverse transcriptase efficiency, which would leave single-stranded minus-strand DNA exposed for longer periods of time, enabling more deamination events to occur. Although A3G and A3F are known to function alongside each other, these data provide evidence for an A3F/G hetero-oligomeric A3 with unique properties compared to each individual counterpart.
IMPORTANCE The APOBEC3 enzymes APOBEC3F and APOBEC3G act as a barrier to HIV-1 replication in the absence of the HIV-1 Vif protein. After APOBEC3 enzymes are encapsidated into virions, they deaminate cytosines in minus-strand DNA, which forms promutagenic uracils that induce transition mutations or proviral DNA degradation. Even in the presence of Vif, footprints of APOBEC3-catalyzed deaminations are found, demonstrating that APOBEC3s still have discernible activity against HIV-1 in infected individuals. We undertook a study to better understand the activity of coexpressed APOBEC3F and APOBEC3G. The data demonstrate that an APOBEC3F/APOBEC3G hetero-oligomer can form that has unique properties compared to each APOBEC3 alone. This hetero-oligomer has increased efficiency of virus hypermutation, raising the idea that we still may not fully realize the antiviral mechanisms of endogenous APOBEC3 enzymes. Hetero-oligomerization may be a mechanism to increase their antiviral activity in the presence of Vif.
KEYWORDS: APOBEC3, DNA-protein interactions, HIV, deaminase, mutagenesis, oligomerization, processivity, protein-protein interactions, reverse transcriptase
INTRODUCTION
APOBEC3 (A3) enzymes are a family of deoxycytidine deaminases that can act as host restriction factors for HIV-1 (referred to as HIV) (1). To restrict HIV replication, A3 enzymes must first become encapsidated in HIV virions from a producer cell (2–4). In the next cell that is infected, the HIV genomic RNA is reverse transcribed to form the minus-strand DNA. During this time, single-stranded minus-strand DNA is vulnerable to A3 deaminations of cytosine that form uracil (2, 3, 5). The reverse transcriptase is forced to use uracil as a template during plus-strand DNA synthesis, which induces C/G→T/A mutations and can functionally inactivate the virus or lead to its degradation through DNA repair pathways that excise uracil (6). The effect of A3 enzymes on HIV replication in infected individuals is severely dampened by the viral infectivity factor (Vif) that interacts with A3 enzymes, recruits an E3 ubiquitin ligase complex, and induces A3 ubiquitination and degradation in the proteasome (7–12). From the seven human A3 enzymes, there are four, A3D, A3F, A3G, and A3H, that can inhibit HIV replication in the absence of Vif (13). These four enzymes are coordinately expressed in CD4+ T cells and induced upon T cell activation (14, 15).
However, even in the presence of Vif, there is evidence of A3 deaminations inducing hypermutation of integrated proviral genomes (16–23). There are two ways in which this occurs. In one route, the A3s are encapsidated in the presence of Vif, but at a much smaller amount (24). Additionally, viruses can adapt to have a less fit Vif that is less efficient at inducing A3 degradation (25). From these proviral DNA sequences, G→A mutations have been attributed to specific A3s through identification of their preferred sequence contexts surrounding the mutations by both cellular and in vitro studies (2, 3, 5, 26–29). The sequence contexts showed that A3G uniquely prefers to deaminate the 3′C in 5′CC motifs, resulting in a GG→GA mutation signature (5, 29). A3D, A3F, and A3H all deaminate in 5′TC motifs, resulting in a GA→AA mutation signature (26–28). More detailed analysis of the signatures revealed that A3G is most active at a 5′CCC motif, and A3D can be differentiated from A3F and A3H by examining a larger surrounding sequence 3′ of the cytosine (30). Studies that sequenced integrated proviral genomes have shown that proviral DNAs contain mutations in multiple sequence contexts, suggesting that multiple A3s can mutate the same genome (16–23). However, what was unable to be concluded from these studies was if the mutations induced from multiple A3s occurred in a single round of replication or multiple rounds of replication. If they occurred in a single round of replication, then the A3s would need to be coencapsidated into HIV virions. Recently, it was found that A3G is primarily encapsidated into virions by binding nonspecifically to cellular RNAs (31). The only requirement for being virion encapsidated was that HIV Gag was also bound to the same RNA (32). In this model, the encapsidation is not competitive and multiple A3s should be coencapsidated. However, a computational study determined from sequence analysis of proviral DNA that A3G and A3F rarely comutate the same genome (33). This endpoint analysis is in contrast to cellular studies that have identified that A3 enzymes are able to be coencapsidated, and some studies have found that there is a synergistic effect where more mutations or more viral restriction than expected by an additive relationship are induced (26, 30, 34). Despite this synergistic effect being identified, a mechanism for how multiple A3 enzymes can result in more than additive mutations or virus restriction has not been determined.
Based on previous findings, there are two possible mechanisms by which coencapsidated A3s could result in higher levels of mutations. One mechanism is to slow down the reverse transcriptase to keep the minus-strand DNA single stranded for longer to allow A3s more time to scan the DNA for their preferred deamination motif (35–37). This is thought to be accomplished by A3 enzymes binding to the template and blocking the progression of reverse transcriptase. A second mechanism is to increase the processivity of the A3 enzyme (38, 39). The processivity is the ability of an enzyme to deaminate multiple cytosines in a single enzyme-substrate encounter. A3 enzyme processivity is mediated by facilitated diffusion (1). Facilitated diffusion is a term that describes Brownian motion-driven diffusion and results in enzymes moving along the DNA phosphate backbone by sliding, diffusing within the charged domain of the DNA by a mechanism termed jumping, and moving between DNA segments through a doubly bound state termed intersegmental transfer (40, 41). The DNA scanning enables the A3 enzyme to search and find the cytosine among the nontarget nucleotides and enables the continuation of the search after a deamination without completely dissociating from the DNA. The sliding movements cover small distances, up to approximately 20 nucleotides (nt) (38, 39). For A3G, the average sliding distance is 9 nt (42). The jumping and intersegmental transfer movements enable the enzyme to move distally, e.g., 100 nt or more (38, 43).
We initiated this study to investigate if coexpressed A3F and A3G have an additive or synergistic ability to restrict HIV replication. We focused on A3F and A3G, since these enzymes are most commonly expressed together in the human population and are active against HIV (14, 15). While A3H haplotypes II, V, and VII are highly restrictive of HIV, they occur less frequently in the overall population, i.e., haplotype II, 26.5%; haplotype V, 20%; and haplotype VII, 0.9% (44, 45). A3D is much less active against HIV than A3F and A3G (46). In the present work, the data show that A3F and A3G are able to induce a combined increase in mutations in HIV DNA and synergistic decrease in HIV infectivity in single-cycle infectivity assays. The mechanism for this cooperation was 2-fold. First, A3F and A3G can hetero-oligomerize in the absence of RNA, and this A3F/G hetero-oligomer has improved processivity. Second, the A3F/G hetero-oligomer can decrease the efficiency of reverse transcriptase, which provides increased time for A3-catalyzed deaminations. Together, the data suggest that A3F and A3G are able to function together with distinct and improved properties from the individual enzymes.
RESULTS
A3F and A3G can hetero-oligomerize in an RNA-independent manner.
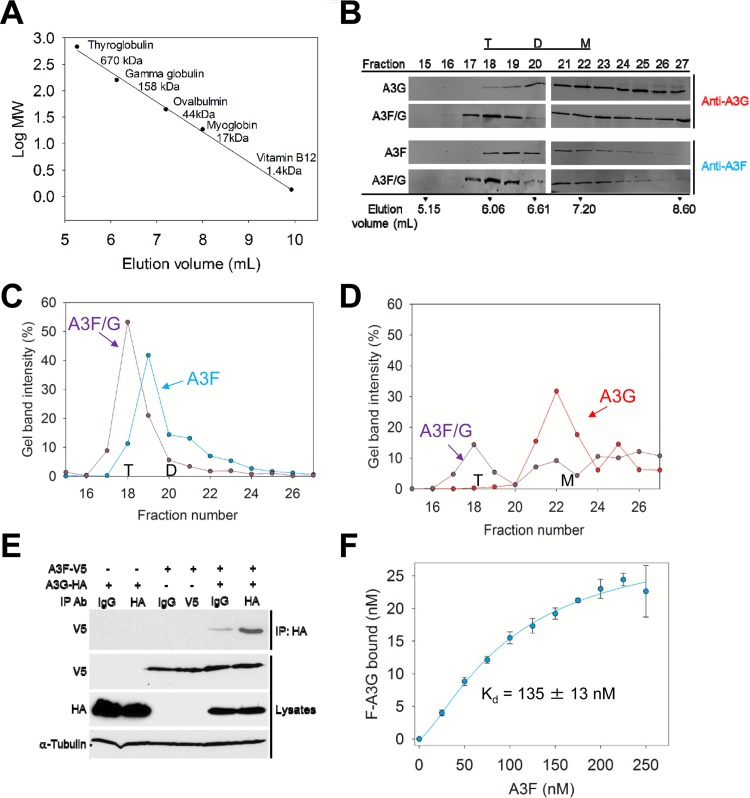
A3F and A3G have been shown to hetero-oligomerize on RNA in an RNA-dependent manner (47). However, A3F and A3G can also form homo-oligomers in the absence of RNA (38, 48, 49). Here, we examined if A3F and A3G could also form hetero-oligomers in the absence of RNA. We used A3F and A3G enzymes purified in the presence of RNase A to remove the bound cellular RNA that causes the enzymes to form higher-order oligomers, i.e., >650 kDa (43, 50). Based on comparison to a calibration curve, size exclusion chromatography (SEC) confirmed the absence of higher-order oligomers (Fig. 1A and B). For SEC, we used a relatively low concentration of enzymes (1 μM) and detected the enzymes by quantitative immunoblotting to avoid potential artifacts caused by high concentrations of A3G and A3F. A monomer of A3G or A3F is predicted to be 46.4 or 45 kDa, respectively, based on the amino acid sequences. Consistent with previous reports, A3G was present in both monomer and dimer fractions (Fig. 1B, fraction 20, dimers, 101 kDa; fraction 22, monomers, 48 kDa). A3F was present in trimer and dimer fractions (Fig. 1B, fraction 19, trimers, 153 kDa; fraction 20, dimers, 101 kDa) (48).
FIG 1.
A3F and A3G hetero-oligomerize. (A to D) Size exclusion chromatography was conducted with a 10-ml G200 Superdex column. (A) A calibration curve was used to calculate the molecular weights (MW) and oligomerization states of the enzymes. (B) The A3G, A3F, and A3F/G SEC experiments used quantitative immunoblotting to detect A3G or A3F with antibodies to the native proteins. The integrated band intensities calculated using LiCor/Odyssey software were used to generate chromatograms. The T, D, and M notations indicate peak fractions for tetramers, dimers, and monomers, respectively. (C) The integrated band intensities for A3F indicated that A3F alone was primarily a trimer (fraction 19, 153 kDa) with a minority of dimers (fraction 20, 101 kDa). The A3F in the A3F/G combined run was primarily a tetramer (fraction 18, 207 kDa). (D) The integrated band intensities for A3G indicated that A3G alone was primarily a monomer (fraction 22, 46 kDa) with a minority of dimers (fraction 20, 101 kDa). The A3G in the A3F/G combined run maintained a population of monomers (fraction 22) but also was able to fractionate with the peak fraction corresponding to tetramers (fraction 18, 207 kDa). (E) Coimmunoprecipitation of A3F-V5 with A3G-HA. A3G-HA and A3F-V5 were transfected alone or in combination. The immunoprecipitation of cell lysates used either anti-HA antibody or rabbit IgG (mock), and samples were immunoblotted with antibodies against α-tubulin, HA, and V5. Cell lysates show the expression of α-tubulin, HA, and V5. (F) Steady-state fluorescence depolarization was used to measure the rotational anisotropy of F-A3G interacting with A3F. Rotational anisotropy was normalized to fraction F-A3G bound. An apparent Kd was calculated by regression analysis of the saturation curve from three independent experiments. A sigmoidal fit was chosen by least-squares analysis and resulted in an apparent Kd of 135 ± 13 nM.
To examine whether A3F and A3G can hetero-oligomerize, we mixed together an equal amount of each enzyme before loading onto the column. The A3F/G blots show the SEC profile for the combined run where either A3G (anti-A3G) or A3F (anti-A3F) was detected (Fig. 1B). These bands were quantified and plotted for comparison to SEC of each enzyme alone (Fig. 1C and D). For A3F, the peak fraction of trimers (fraction 19) shifted to a peak fraction of tetramers (fraction 18, 207 kDa) in the presence of A3G (Fig. 1B and C). Additionally, the dimer peak (fraction 20) of A3F was decreased (Fig. 1B and C). These data suggest that A3F, which is predominantly a trimer, can interact with one molecule of A3G to form a tetrameric A3F/G hetero-oligomer. To investigate this further, we analyzed the A3G SEC data. A3G alone predominantly fractionated as a monomer (Fig. 1B and D, fraction 22). In the presence of A3F, and concomitant with A3F and A3G forming a hetero-oligomer, the A3G peak fraction shifted to the tetrameric fraction (Fig. 1B and D, fraction 18, 207 kDa). However, a significant amount of A3G monomers are still present in the presence of A3F, supporting the conclusion that there are fewer A3G molecules than A3F molecules in the A3F/G hetero-oligomer (Fig. 1B). Together with the A3F SEC and immunoblotting, the data support that a trimer of A3F interacts with a monomer of A3G and that this complex is a protein-protein interaction and not mediated by RNA.
To determine if this interaction would be relevant in cells, we used two additional approaches. In one experiment, we conducted a coimmunoprecipitation (co-IP) to determine if A3G-HA expressed in 293T cells could immunoprecipitate A3F-V5. Cells transiently expressing A3G-HA, A3F-V5, or A3G-HA and A3F-V5 were lysed, incubated with RNase A, and then incubated with anti-HA antibody or normal rabbit IgG (mock) and Sepharose A beads. Even in the absence of RNA, A3G-HA was able to immunoprecipitate A3F-V5, indicating that A3F and A3G associate through a protein-protein interaction in cells (Fig. 1E). In another experiment, we quantified the strength of the interaction between A3F and A3G using fluorescence depolarization to measure the rotational anisotropy of fluorescein-labeled A3G (F-A3G) in the presence of increasing amounts of A3F. As A3F formed a complex with F-A3G, the anisotropy increased and reached saturation where all of the F-A3G was bound. From this saturation curve, an apparent dissociation constant (Kd) of 135 nM was calculated by fitting the curve to a Sigmoidal equation by regression analysis (Fig. 1F). These data indicated that A3F and A3G interacted with high affinity and further support that they could interact in cells. That the binding curve best fit a Sigmoidal equation rather than a rectangular hyperbola by least-squares analysis further supports that multiple A3F molecules are binding to A3G, consistent with the SEC data (Fig. 1B to D).
A3F and A3G coexpression results in higher levels of HIV restriction.
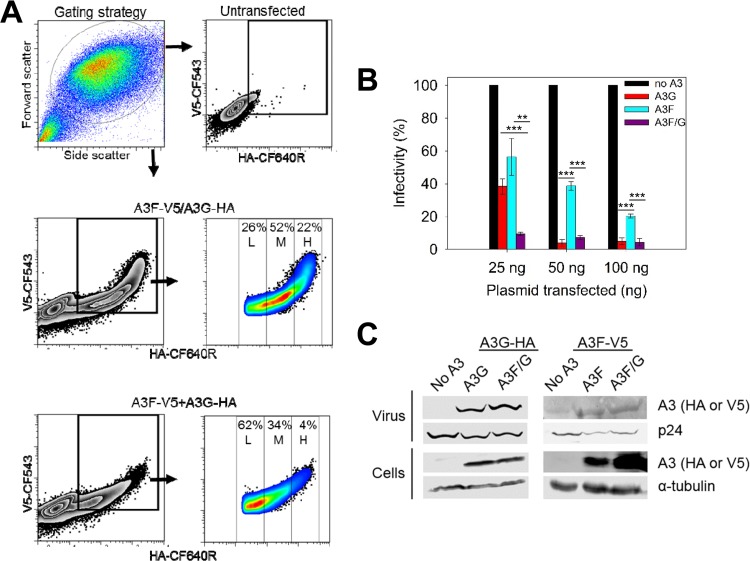
To determine if the A3F/G hetero-oligomer influences the ability of the A3 enzymes to restrict HIV replication, we conducted single-cycle infectivity assays. To model cellular conditions where A3F and A3G are coexpressed, we used a plasmid that has two transcription units, enabling the combined expression of both A3F-V5 and A3G-HA on a single-cell basis. Flow cytometry to detect the V5 and hemagglutinin (HA) tags demonstrated a distinct population of cells that expressed both A3F-V5 and A3G-HA when A3F-V5 and A3G-HA were expressed from the same plasmid (Fig. 2A). The population of cells expressing both A3F-V5 and A3G-HA showed a normal distribution of cells, with low (L; 26%), medium (M; 52%), and high (H; 22%) coexpression on a single-cell basis (Fig. 2A). In contrast, a cotransfection strategy using separate plasmids expressing A3F-V5 and A3G-HA showed a skewed distribution of the cell population with a predominantly small amount of cells coexpressing A3F-V5 and A3G-HA (Fig. 2A, 62% L, 34% M, and 4% H). As a result, the virions generated from cell populations with a normal distribution of A3F-V5 and A3G-HA coexpression will be more likely to encapsidate more homogenous levels of each A3 enzyme across the population than the virions generated from cell populations with a skewed distribution of A3F-V5 and A3G-HA coexpression. Thus, our experiments used only A3F-V5 and A3G-HA that were coexpressed from one plasmid.
FIG 2.
Coexpressed A3F and A3G enhances the restriction of HIV replication. (A) Flow cytometry was used to detect A3G-HA or A3F-V5 in individual cells after transient transfection using fluorescently labeled anti-HA or anti-V5 antibodies. Cells were either transfected with one pVIVO2 vector expressing both A3G-HA and A3F-V5 or individual pVIVO2 vectors each expressing A3G-HA or A3F-V5. (B) HIV Δvif infectivity was measured by eGFP expression in 293T cells infected with HIV Δvif that was produced in the absence or presence of A3G-HA (A3G) or A3F-V5 (A3F) or coexpressed A3F-V5 and A3G-HA (A3F/G). Results normalized to the no-A3 condition are shown with the standard deviations (SD) of the means calculated from at least three independent experiments. Designations for significant difference of values were a P value of ≤0.001 (***), ≤0.01(**), or ≤0.05 (*). (C) Immunoblotting of HA and V5 tags was used to detect A3 enzymes expressed in cells and encapsidated into HIV Δvif virions. The cell lysate and virion loading controls were α-tubulin and p24, respectively.
Using increasing amounts of A3 plasmid for A3G-HA (A3G), A3F-V5 (A3F), and coexpressed A3G-HA/A3F-V5 (A3F/G), we observed that the maximum amount of restriction occurred at a lower level of transfected plasmid when A3F and A3G were coexpressed (Fig. 2B). Specifically, only 25 ng of the A3F/G plasmid was required for restriction of HIV compared to 50 ng of A3G plasmid or 100 ng of A3F plasmid. A3F-mediated viral restriction was consistently less than that of A3G, as previously reported (26, 38, 46, 51). However, most notable was that for the 25-ng plasmid transfection experiment, the combined expression of A3F and A3G resulted in 4- to 6-fold greater restriction than either A3G or A3F alone (Fig. 2B, 25 ng). At 50 ng and 100 ng of transfection plasmid, the restriction of A3G was not significantly different from the combined restriction of A3F/G (Fig. 2B, 50 ng and 100 ng). The infectivity data demonstrate that at lower levels of A3G and A3F expression, when their restriction effect is not saturated, their combined action is better than their individual action in inhibiting HIV infectivity. This effect is similar to previous reports (26, 34), although the reason for this enhancement of restriction has not been previously investigated.
To investigate if the enhanced effect was a result of higher encapsidation levels of A3G or A3F, we analyzed cell lysates and virions by immunoblotting (Fig. 2C). Since the A3s have different tags they were blotted with different antibodies, and as a result we are unable to compare A3G to A3F. However, we can determine if the same amount of A3G or A3F is encapsidated under the different expression conditions. The virus blot showed that the same amount of A3G was encapsidated in both the A3G and A3F/G experiments (Fig. 2C). Similarly, A3F was encapsidated at the same level whether it was expressed alone or with A3G (Fig. 2C). These results indicate that the enhanced restriction was not due to higher levels of enzymes being encapsidated and suggested that coencapsidated A3G and A3F were able to synergistically restrict HIV replication.
Enhanced mutagenesis of HIV Δvif induced by coencapsidated A3F and A3G.
Recently it was shown that A3F and A3G are indeed able to be coencapsidated into HIV virions (30). To identify if one or both A3s were increasing the level of mutagenesis when coencapsidated, we sequenced a portion of the proviral DNA. We used proviral DNA from the 25-ng A3 plasmid transfection experiment to avoid analyzing proviral DNA that may be saturated with mutations and PCR amplified the protease region for DNA sequencing. The mutation frequencies showed that A3G mutated the proviral DNA more than A3F (A3G, 7.5 mutations/kb; A3F, 1.6 mutations/kb) (Table 1). When A3G and A3F were coexpressed, the mutation frequency was 14 mutations/kb (Table 1). If coencapsidated A3G and A3F were acting independently of each other, the mutation frequency would be expected to be the sum of each mutation frequency alone (estimated 9.1 mutations/kb). That the mutation frequency when A3G and A3F were combined was 1.5-fold more than the sum of each mutation frequency alone suggests that A3G and A3F can coordinate to restrict HIV replication. When 50 ng of plasmid was transfected, we observed that for the A3F/G condition, the mutations were 2-fold greater than the sum of the mutations of A3G and A3F alone (data not shown). At 100 ng of plasmid, there was no increase in mutations in the A3F/G condition compared to A3G and A3F alone, presumably because the restriction was reaching saturation, which would make it difficult to differentiate between the conditions (Fig. 2B, 100 ng, and data not shown).
TABLE 1.
Analysis of A3-induced mutagenesis of protease DNA from integrated HIV Δvif
| A3 enzyme | No. of: |
||||||
|---|---|---|---|---|---|---|---|
| Base pairs sequenced | G→A mutations (total) | GG→AG mutations (total) | GA→AA mutations (total) | Deaminations per kb | GG→AG mutations per kb | GA→AA mutations per kb | |
| A3G | 8,775 | 66 | 60 | 6 | 7.5 | 6.8 | 0.7 |
| A3F | 10,179 | 16 | 1 | 15 | 1.6 | 0.1 | 1.5 |
| A3F/G | 9,126 | 129 | 104 | 24 | 14 | 11 | 2.6 |
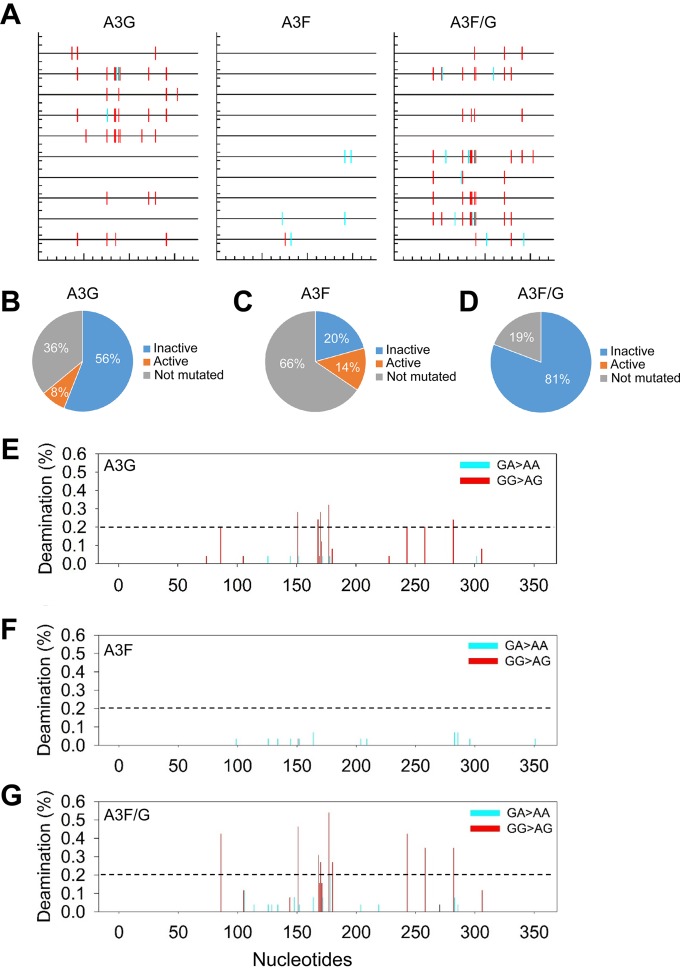
Since A3G causes mutations in the plus-strand DNA predominantly at GG sites and A3F predominantly at GA sites, sequence analysis can determine which A3 was inducing the increases in mutational load (5, 26). Consistent with previous observations, we found that A3G alone mutated GG sites for 91% of the mutations (6.8 mutations/kb) and mutated GA sites for 9% of the mutations (0.7 mutations/kb) (Table 1). A3F predominantly mutated GA sites (94% of GA sites, 1.5 mutations/kb) but could also mutate GG sites at a low level (6% of mutations, 0.1 mutations/kb) (Table 1). This meant that we could use the sequence context to determine which A3 was inducing increases in the mutational load. For the 25-ng transfection condition, the combined mutation frequency (14 mutations/kb) was composed of 11 mutations/kb at GG sites (A3G induced) and 2.6 mutations/kb at GA sites (A3F induced) (Table 1). This is a 1.6-fold or 1.7-fold increase in GG mutations (6.8 mutations/kb increased to 11 mutations/kb) or GA mutations (1.5 mutations/kb increased to 2.6 mutations/kb), respectively (Table 1). When A3F and A3G were coexpressed, we recovered both GG and GA mutations in the same clone more frequently than when each A3 was expressed alone, indicating that indeed both A3F and A3G were coencapsidated (Fig. 3A). Specifically, for the A3F/G cotransfection (25 ng), 52% of mutated clones were comutated (Fig. 3A). Of those not comutated, the mutations recovered contained only A3G-induced mutations at GG sites, consistent with there being a low mutation efficiency of A3F (Fig. 3A and Table 1). For the 50-ng and 100-ng transfections, 75% of mutated clones were comutated (data not shown).
FIG 3.
Coexpressed A3F and A3G comutate the same HIV proviral genome. (A) Spectral plot generated by Hypermut (86) was generated using representative samples from the 25-ng A3G, A3F, and A3F/G infectivity experiments. Across the protease gene, GG→AG (expected A3G-induced mutation) changes are shown by a red line, and GA→AA (expected A3F-induced mutations) changes are shown by a cyan line. (B to D) Individual analysis of each protease clone for A3G (B), A3F (C), and A3F/G (D) enabled determination of the percentage of clones that would result in a mutated and inactive (blue) or mutated and active (orange) protease. The percentage of clones that were not mutated is shown in gray. (E to G) Mutational spectra for all clones shows the percentage of clones with a mutation at a particular site in the protease gene for A3G (E), A3F (F), and A3F/G (G). The GG→GA (red) and GA→AA (cyan) mutations are distinguished by color and demonstrate the specificity of the deamination targets for A3G and A3F.
Despite only a 1.5-fold increase in mutagenesis, there was a 4-fold (A3G) to 6-fold (A3F) increase in restriction ability when the two enzymes were coexpressed in virus producer cells and coencapsidated in virions (Table 1 and Fig. 2B). This inconsistency may be due to the ability of A3G to induce the formation of numerous stop codons in plus-strand DNA, which would be able to completely ablate virus replication with only a single mutation (5). Stop codons arise from deamination of cytosines in minus-strand DNA Trp codons, e.g., 5′CCA. When the minus-strand DNA is deaminated to 5′UCA or 5′CUA, the sequence becomes 5′TGA or 5′TAG in plus-strand DNA and results in a stop codon. Within the 351-nt-long protease region that we sequenced, there was one Trp codon that was a hot spot for mutagenesis. A3G alone induced stop codon formation in 32% of the clones, A3F alone induced stop codon formation in 6.9% of the clones, and coencapsidated A3F/A3G induced stop codon formation in 50% of the clones. There are also numerous other A3-induced missense mutations that can inactivate protease (38, 39, 52). The inactivation of protease as a result of missense mutations was inferred from results from an extensive protease mutagenesis study conducted by Loeb et al. (53). Taking into account both stop codons and inactivation by missense mutations, we found that A3G alone inactivated 56% of mutated protease and left 8% of mutated protease active (Fig. 3B). Due to the low A3 plasmid transfection level used in the experiment (25 ng), we recovered protease clones that were not mutated 36% of the time (Fig. 3B). In contrast, A3F inactivated only 20% of mutated protease and left 14% mutated and active and 66% not mutated (Fig. 3C). When A3F and A3G were coexpressed, there were no protease clones that were mutated and active (Fig. 3D). Eighty-one percent of protease clones were mutated and inactivated, and 19% were left unmutated. This means that when A3F and A3G are coexpressed they are able to more efficiently inactivate the proviral DNA. Thus, although the combined effect of A3F/G on mutations was at most 2-fold, this enabled a larger decrease in infectivity (4- to 6-fold) and further supports that coencapsidation of A3G and A3F enhances HIV restriction from each A3 alone (Fig. 2B and Table 1).
The mutations per clone were plotted to visualize the mutations across the sequenced protease region of the integrated provirus. In the 351-nt clone, there are distinct hotspots in which A3G induces mutations (Fig. 3E). The hotspots are deaminated in at least 20% of clones, and there are 8 of these sites at positions 86, 151, 168, 170, 177, 243, 258, and 282. Under the combined A3F/G condition, the same hotspots are deaminated in more clones, but no new hotspots were identified (Fig. 3E and G, see hashed reference line). The increases in deamination at each of the sites ranged from 1.3-fold (nucleotide position 168) to 2.1-fold (nucleotide positions 86 and 243), suggesting that deamination activity across the protease gene was increased similarly (Fig. 3E and G). For A3F, there were no distinct hotspots due to the low level of induced mutagenesis (Fig. 3F). Increases in deamination for A3F under the A3F/G condition occurred at new sites or sites deaminated when A3F was expressed alone (Fig. 3F and G, new sites 106, 114, 129, 148, and 178). However, in accordance with the overall mutation frequency, the increases were not high. The higher mutation frequency of A3F under the A3F/G condition was primarily due to new site mutations. Together, the data indicate that for A3G and A3F, coexpression increases their mutagenic activity, but for A3G there is no change in the deamination hotspots, in contrast to A3F, which is able to mutate more new sites during minus-strand DNA synthesis. Since A3G and A3F can form an A3F/G hetero-oligomer (Fig. 1), we interpreted the mutation data to suggest that the A3F/G hetero-oligomer is working to increase deamination activity (observed for A3G) and accessibility to single-stranded DNA (ssDNA) (observed for A3F).
A3G, when part of an A3F/G hetero-oligomer, has an improved ability to jump over RNA/DNA hybrids.
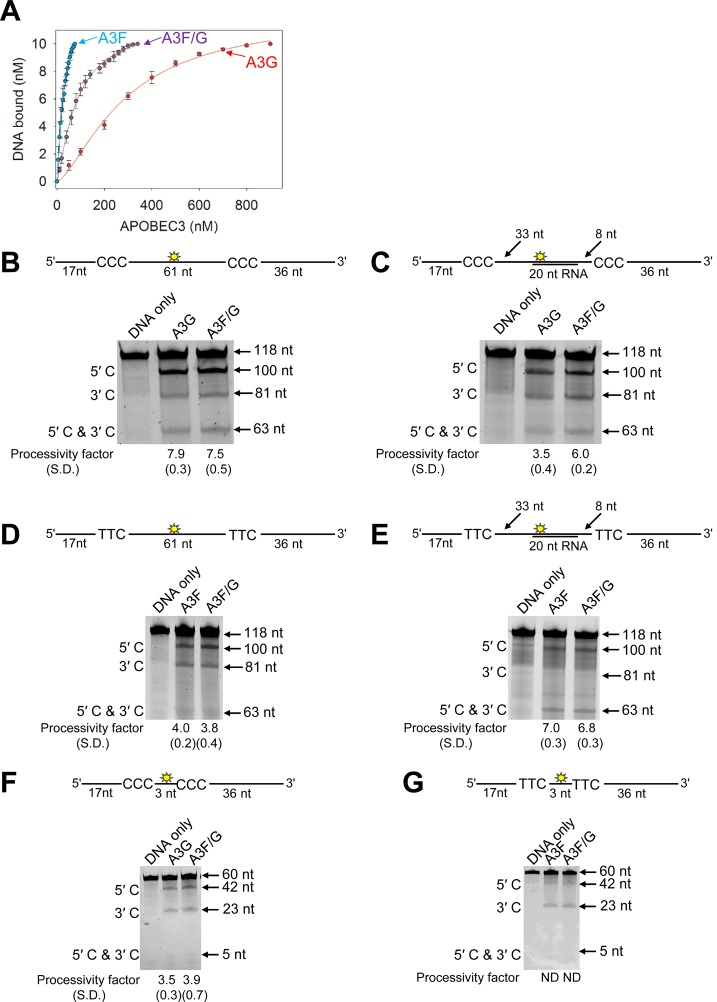
To test our hypothesis that the A3F/G hetero-oligomer can increase the activity of A3G, we conducted in vitro deamination and binding assays on synthetic oligonucleotides. Using fluorescence depolarization, we found the A3F/G hetero-oligomer bound fluorescein-labeled ssDNA with an apparent Kd that was unique from that of either A3F or A3G alone (Fig. 4A). The A3F/G hetero-oligomer was formed by preincubating equal amounts of A3F and A3G. The A3F/G hetero-oligomer bound ssDNA with an apparent Kd of 114 nM, which indicates 3-fold less affinity than A3F and 2.5-fold more affinity than A3G alone. Since the binding experiment was conducted under steady state, the apparent Kd of the A3F/G hetero-oligomer is an average of all possible associations, i.e., A3F/G, A3G, and A3F. However, the data support the conclusion that the A3F/G hetero-oligomer may have biochemical properties distinct from those of A3G and A3F.
FIG 4.
Biochemical properties of the A3F/G hetero-oligomer are distinct from A3G and A3F. (A) Fluorescence depolarization was used to detect changes in rotational anisotropy of an F-labeled 118-nt ssDNA upon titration of A3G or A3F or an A3F/G hetero-oligomer into the solution. The rotational anisotropy was normalized to fraction F-ssDNA bound and analyzed by regression analysis. The binding curves fit to a sigmoidal binding curve as determined by least-squares analysis. The apparent Kd values were calculated to be 286 ± 17 nM for A3G, 39 ± 6 nM for A3F, and 114 ± 20 nM for A3F/G. Error bars represent the standard deviations of the means from three independent experiments. (B to G) Processivity of A3G, A3F, or A3F/G was tested on ssDNA substrates that contain a fluorescein-labeled deoxythymidine (yellow star) between two 5′CCC (for A3G) or 5′TTC (for A3F) deamination motifs separated by different distances. (B) Deamination of a 118-nt ssDNA substrate with two 5′CCC deamination motifs spaced 61 nt apart. Single deaminations of the 5′C and 3′C are detected as the appearance of labeled 100- and 81-nt fragments, respectively; double deamination of both C residues on the same molecule results in a 63-nt labeled fragment. (C) Deamination of the same substrate shown in panel B but with a 20-nt cRNA annealed between the two 5′CCC motifs. (D) Deamination of the same substrate shown in panel B, but with two 5′TTC motifs. (E) Deamination of the same substrate shown in panel D, but with a 20-nt cRNA annealed between the two 5′TTC motifs. (F) Deamination of a 60-nt ssDNA substrate with two 5′CCC motifs spaced 3 nt apart. Single deaminations of the 5′C and 3′C are detected as the appearance of labeled 42- and 23-nt fragments, respectively; double deamination of both C residues on the same molecule results in a 5-nt labeled fragment. (G) Deamination of the same substrate shown in panel F, but with two 5′TTC motifs. The notation ND means not able to be determined due the absence of a detectable 5′C and 3′C band. The measurements of enzyme processivity (processivity factor) and the SD are shown below the gel. All values are calculated from three independent experiments.
Previously we have shown that the processive scanning mechanism of the A3 enzymes that is used to search for the preferred deamination motifs in the DNA substrate can be a predictor of mutagenic potential (38). Therefore, we hypothesized that since one A3G molecule appeared to interact with three molecules of A3F (Fig. 1A to D), A3F may be able to influence the processive scanning mechanism of A3G. A3F is able to jump larger distances than A3G (38). Jumping movements are required for overcoming in a processive manner RNA/DNA hybrid obstacles left on minus-strand DNA by RNase H activity (1). To examine the processivity and scanning mechanism of A3, the reactions were conducted under single-hit conditions where an ssDNA is encountered by only one enzyme during the course of the reaction (54). To characterize processivity and scanning movement, we used an ssDNA with either two CCC motifs (for A3G activity) or two TTC motifs (for A3F activity) separated by differing distances. Under our reaction conditions, we observed no deamination of CCC motifs by A3F or TTC motifs by A3G (data not shown). Thus, we examined how the A3F/G hetero-oligomer could improve activity of either A3G or A3F by using the relevant substrate. To form the hetero-oligomer, we preincubated A3F and A3G together, added the enzymes to the reaction buffer, and started the reaction by the addition of ssDNA. We did not subsequently purify the A3F/G hetero-oligomer from unbound A3F or A3G to simulate cellular conditions, where there is likely to be an “average” population of A3F, A3G, and the A3F/G hetero-oligomer.
To examine jumping, we used deamination motifs that were distantly spaced (61 nt). On the CCC motif containing ssDNA, A3G and the A3F/G hetero-oligomer had the same processivity factor of approximately 8 (Fig. 4B). This processivity factor is calculated using the integrated gel band intensity of the 5′C and 3′C deamination band and means that the enzyme is 8-fold more likely to make a processive deamination than a nonprocessive deamination (see Materials and Methods). To challenge the jumping ability of A3G and the A3F/G hetero-oligomer, we annealed a complementary 20-nt RNA between the two CCC motifs (Fig. 4C, sketch). Since A3G and A3F cannot bind to this RNA/DNA hybrid, the enzyme complex must jump over the obstacle to undergo processive deamination of both CCC motifs (38). A3G had a characteristic decrease in processivity due to the obstacle (2.5-fold) (Fig. 4B and C) (38, 55). When in complex with A3F, the A3G processivity decreased only 1.3-fold (Fig. 4B and C). Thus, for A3G, being in complex with A3F (A3F/G) can improve the jumping ability. We also conducted the equivalent experiment for A3F but used the TTC motif containing ssDNA. For A3F alone, the processivity is enhanced by annealing a cRNA molecule, presumably due to the annealed portion enabling the TTC sites to be juxtaposed at an optimal distance for A3F jumping movements, as previously observed (Fig. 4D and E) (38). There was no change in the A3F processivity factor on either DNA substrate in the presence of A3G (Fig. 4D and E, A3F or A3F/G).
To examine processive sliding movements, we used an ssDNA with closely spaced deamination motifs (3 nt apart). On the CCC-containing ssDNA we found no change in the A3G processivity in the presence of A3F (Fig. 4F). A3F is not able to processively slide to deaminate closely spaced TTC motifs, thus, on the substrate with the TTC motifs separated by 3 nt, we observed no deamination of both the 5′C and 3′C (Fig. 4G). Despite A3G being able to slide, there was no 5′C and 3′C band detected for the A3F/G hetero-oligomer on the TTC containing ssDNA (Fig. 4G). Altogether, the data demonstrate that A3F influences the processive jumping movements of A3G when they form a hetero-oligomer, but A3F processivity is not influenced by A3G. This is consistent with there being more A3F molecules in the A3F/G hetero-oligomer that could exert more of an effect on A3G than vice versa (Fig. 1). These data are also consistent with the A3F/G hetero-oligomer having 1.7-fold higher specific activity on CCC motif ssDNA than A3G alone and no effect on specific activity on TTC motif ssDNA (data not shown).
A3F and A3F/G decrease reverse transcripts more efficiently than A3G.
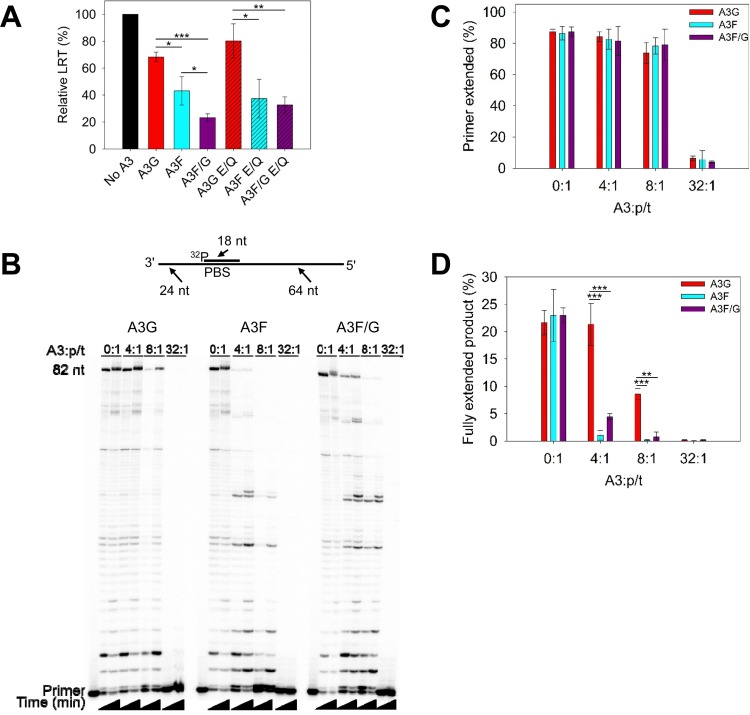
Despite A3F-induced mutations being increased in the presence of A3G (Table 1), the reason for this did not appear to be due to an increase in the processive search mechanism of A3F. Another mechanism by which A3-induced mutations can be increased is if reverse transcription was slowed down, which would leave minus-strand DNA single stranded for a longer time (56–58). It is known that both A3G and A3F can decrease reverse transcriptase efficiency, which results in less late reverse transcript (LRT) formation and less proviral integration (36, 37, 59–68). We hypothesized that the A3F/G hetero-oligomer would inhibit reverse transcriptase more efficiently, and this would allow more time to search for and deaminate TC motifs in minus-strand DNA.
To investigate this, we conducted experiments that quantified the LRT in infected 293T cells and in vitro primer extension of HIV reverse transcriptase from a primer binding site (PBS) primer in the presence and absence of A3s. The relative LRT formed in the presence of the A3s was decreased for all conditions with A3G, A3F, or A3F/G. Consistent with previous findings, A3F inhibited LRT formation more than A3G (Fig. 5A, 1.6-fold difference), and the inhibition of LRT formation was independent from deamination ability (Fig. 5A, catalytic E→Q mutants) (60, 65). The A3F/G hetero-oligomer suppressed LRT formation approximately 2-fold more than A3F alone. These data were consistent with a primer extension assay in which purified HIV reverse transcriptase was used to extend the 18-nt PBS primer from a 106-nt template that had a nucleotide sequence matching the corresponding region of the HIV genome (Fig. 5B, sketch). For the A3F/G condition, equimolar amounts of A3F and A3G were preincubated before being added to the reaction. All reactions had the same total moles of A3 for each titration. By titrating increasing amounts of A3 into the reaction mixture, we observed an increasing amount of inhibition of DNA polymerization (Fig. 5B). Analysis of the extended primer showed that each A3 inhibited total primer extension similarly (Fig. 5C). However, visual inspection of the gel for the 4:1, 8:1, and 32:1 ratios of A3 to primer/template (p/t) conditions demonstrated that A3F and the A3F/G hetero-oligomer can inhibit the formation of full-length products (82 nt) at a 4:1 molar ratio, whereas at least 2-fold more A3G is required for the same inhibition (Fig. 5B and D). Altogether, the data indicate that the presence of either A3F or the A3F/G hetero-oligomer can slow down reverse transcription. This may enable the A3 enzymes more time to deaminate the minus-strand DNA.
FIG 5.
A3 enzymes can decrease reverse transcriptase efficiency. (A) Quantification of late reverse transcripts (LRT) by qPCR demonstrated that A3G, A3F, and A3F/G and their catalytic mutants can decrease LRT relative abundance. The E/Q notation means an E259Q mutation for A3G or an E251Q mutation for A3F. Error bars represent the standard deviations of the means from three independent experiments. (B) An 18-nt 32P-labeled RNA primer containing a sequence complementary to the HIV PBS was annealed to a 106-nt RNA containing PBS (sketch). Complete extension of the primer results in a product of 82 nt (sketch). The p/t was used at a concentration of 10 nM. Primer extension by reverse transcriptase (480 nM) in the absence (0:1) or presence (4:1, 8:1, and 32:1) of increasing amounts of A3G, A3F, or A3F/G relative to the p/t concentration is shown. Reaction mixtures were sampled at 2.5 and 60 min. (C) Quantification of primer extension (in percent) from gels shown in panel B for no A3 or for A3G, A3F, and A3F/G at 60 min. (D) Quantification of fully extended 82-nt product (in percent) for no A3 or for A3G, A3F, and A3F/G at 60 min. (C and D) Error bars represent the standard deviations of the means from three independent experiments. (A, C, and D) Designations for significant difference of values were a P value of ≤0.001 (***), ≤0.01 (**), or ≤0.05 (*).
DISCUSSION
A3 enzymes are coexpressed in CD4+ T cells and have been shown to coordinately restrict HIV replication (14, 15, 47, 69). However, the mechanism of this coordinate restriction is not clear. For A3G and A3F, it has been proposed that each contributes individually to HIV mutagenesis due to not being coencapsidated (33), that A3F and A3G enzymes are coencapsidated but act independently of one another (26, 30), and that A3 enzymes can enhance each other's activity (26, 34). Thus, we undertook a study of A3F and A3G to examine how they may coordinately restrict HIV infectivity when expressed in the same HIV producer cell. We observed that A3F and A3G can form an A3F/G hetero-oligomer in cells and in vitro in the absence of an RNA intermediate and inferred that they can frequently coencapsidate into HIV particles due to 52 to 75% of proviruses being comutated. Overall, our data show that the A3F/G hetero-oligomer has enhanced ability to decrease the infectivity of HIV through increased mutagenic activity, resulting from unique biochemical properties of the A3F/G hetero-oligomer compared to the individual enzymes.
A3F/G oligomerization.
We determined that A3F and A3G hetero-oligomerize using several methods. Using uniquely tagged versions of A3F and A3G, we conducted a co-IP similar to what was done originally by Wiegand et al., except that we added RNase A to the cell lysates (47). Even in the absence of RNA, A3G-HA could immunoprecipitate A3F-V5, indicating that they hetero-oligomerized in cells (Fig. 1E). The complex has a high-affinity association in the nanomolar range and is stable enough to be identified through SEC (Fig. 1A to D and F). We previously determined by SEC that A3F was approximately 158 kDa in the peak fraction, which would be a trimer or tetramer (3.5-fold the molecular weight of a monomer) (38). However, using a similar method and column but with quantitative immunoblotting to better visualize the bands, we identified the A3F peak fraction more exactly to be 153 kDa, which confirmed that the molecular weight is closer to that of a trimer than a tetramer. The oligomerization of A3F does not appear to be concentration dependent, since it formed these trimers at protein concentrations in the low micromolar range (1 μM A3F was used for SEC) (Fig. 1B and C). In contrast, the oligomerization of A3G is concentration dependent. In the absence of RNA at low concentrations, A3G is primarily a monomer, and at higher concentrations it forms predominantly dimers (Fig. 1) (24, 38, 48, 49). This dynamic nature of A3G oligomerization may facilitate the ability to oligomerize with A3F (Fig. 1). Conversely, the stable oligomer of A3F may inhibit multiple A3G molecules from binding. With 50% amino acid similarity between A3F and A3G, it is reasonable to assume that they oligomerize similarly. However, making a Y131A/W132A mutant in A3F, which is equivalent to the dimer disruption mutant F126A/W127A of A3G, did not disrupt A3F oligomerization (data not shown), suggesting that A3F oligomerizes in a manner unique from that of A3G. A crystal structure of the rhesus A3G N-terminal domain (NTD) shows that A3G (NTD) dimerizes using helix 6 and loop 7, where F126/W127 are located, confirming previous mutational studies (48, 70, 71). Further, this dimerization of rhesus A3G NTD is distinct from any of the existing A3F dimer models that have resulted from A3F C-terminal domain structures (72–75). It remains to be determined how full-length A3F oligomerizes in solution. However, since A3F does not tetramerize by itself (Fig. 1B and C), this suggests that for a tetramer to form, a different association interface must be used. Since the A3F/G oligomer formed in cell lysates, it is likely to also form in virions where the apparent concentration of the components is estimated to be in the high micromolar range due to the restricted volume of the capsid (Fig. 1E) (76).
A3G and A3F cooperate to restrict HIV replication.
All studies to date on A3G and A3F coencapsidation have found different results. Most recently, Desimmie et al. showed on a single-virion level that A3G and A3F are indeed coencapsidated (30). In the experimental system, the authors transfected an HIV plasmid and separate plasmids for each A3G and A3F. This resulted in ∼50% coencapsidation (30). The coencapsidation in our study is implied by our data, in which we recovered proviruses that had mutations at both GG and GA, representing mutation sites due to A3G and A3F deamination, respectively. However, we observed that 52 to 75% of the proviruses were commutated depending on the transfection conditions. Under the same approximated restriction conditions, we recovered 1.5-fold more comutated viral genomes than Desimmie et al. (30) (75% in this study versus 49.6% in theirs). In contrast to our study, those authors found that coencapsidated A3G and A3F had an additive, not cooperative, effect (30). This may be due to the different A3 expression strategy. Desimmie et al. used separate plasmids to express the two A3s, whereas our study used one plasmid to express A3G and A3F, thereby ensuring each transfected cell had both enzymes expressed, which may have resulted in a more defined transfection population (Fig. 2A) (30). Further, we titrated our expression plasmids to avoid saturation of the mutations that may obscure any cooperative effects (Fig. 2B). Interestingly, Krisko et al. found that in a BLT humanized mouse model, consistent HIV Δvif restriction required the combined antiviral activities of A3G and A3F and that A3F was contributing through a deamination-independent mechanism (34). Despite more consistent HIV restriction in the presence of both A3F and A3G, the mutations were predominated by GG→GA, and GA→AA represented only ∼1% of mutations (34). These data are in agreement with our observation that A3G deamination activity is being enhanced by A3F and that A3F deamination activity was low (Fig. 3E to G). Our studies are in further agreement that A3F has a more robust activity to inhibit reverse transcriptase polymerization than A3G when acting alone or in the presence of A3G (Fig. 5). However, data from Krisko et al. are in contrast to another humanized mouse model study in which HIV proviral DNA was found to contain mutations that resulted from only A3G deamination or predominantly A3F deamination, suggesting that A3G and A3F act individually (77). In the earliest study to combine A3G and A3F plasmids in the producer cell, Liddament et al. showed a synergistic 2.7-fold increase in the mutations caused by A3G and A3F coexpression compared to each expressed alone, but only an additive effect on infectivity was found (26). Thus, the authors concluded that A3G and A3F have both an independent and synergistic (which they termed “dependent”) ability to induce mutagenesis of proviral DNA (26). Although we observed a synergistic effect for infectivity (4- to 6-fold) and a lesser effect on mutagenesis (∼2-fold), the conclusions of Liddament et al. are in agreement with ours (26).
Independent and dependent modes of A3F and A3G restriction.
As discussed already, previous studies on A3F and A3G coencapsidation and corestriction are largely complementary to our work. However, our study identified possible mechanisms that can enable coencapsidated A3F and A3G to act cooperatively. Although A3F and A3G were identified to be coexpressed in CD4+ T cells and hetero-oligomerize in an early study, the role of RNA in mediating the hetero-oligomerization was not clear (47). Defining if their interaction was mediated by an RNA intermediate is important to identify because it provides information on whether they act independently while coencapsidated, such as A3F inhibiting reverse transcriptase polymerization while A3G deaminates the minus-strand DNA, or if they are dependent on each other through forming a hetero-oligomer.
That we identified A3F and A3G to form a hetero-oligomer in the absence of RNA in vitro and in cells (Fig. 1) led us to investigate a dependent mechanism of cooperation. The SEC indicated that there were more molecules of A3F in the hetero-oligomer than molecules of A3G (Fig. 1B to D). These data imply that if equal amounts of A3G and A3F are coencapsidated, there would be a population of free A3G molecules and A3F/G hetero-oligomers and a minority of free A3F molecules. We tested if the A3F/G hetero-oligomer had properties that could enhance HIV restriction and found that A3G in the A3F/G hetero-oligomer was better able to processively scan ssDNA with an RNA/DNA hybrid region than A3G alone (Fig. 4B and C). The processive scanning mechanism of an A3 has been shown to be a determinant of restriction efficiency (38).
The A3F/G hetero-oligomer was able to slow down the polymerization efficiency of reverse transcriptase (Fig. 5B to D). In the primer extension assay, we kept the total moles of enzyme equal for experiments with A3G, A3F, and A3F/G. Thus, under the A3F/G condition, there is half the amount of A3F as there is under the A3F-alone condition. Despite this, the A3F/G hetero-oligomer could inhibit synthesis of full-length product as efficiently as A3F, further supporting the conclusion that the A3F/G hetero-oligomer has distinct biochemical properties and functionality. None of the conditions resulted in the inhibition of primer initiation, consistent with other studies that were focused on A3G (Fig. 5C) (35, 65, 66). However, A3G, A3F, and A3F/G could inhibit synthesis of full-length product (Fig. 5D). The analyses of inhibition of full-length product formation are consistent with cellular data and demonstrate that the ability of A3F to inhibit reverse transcriptase is greater than that of A3G (Fig. 5A and D). This may be due to A3F binding ssDNA with higher affinity than A3G (Fig. 4A). Consistent with the A3G model that oligomers on the ssDNA block reverse transcriptase progression, both A3F and the A3F/G hetero-oligomer form trimers and tetramers, respectively, in solution and bind ssDNA cooperatively, indicating that they further oligomerize on ssDNA and suggesting a mechanism of action similar to that of A3G (Fig. 4A) (36). The slower reverse transcription that occurred in vitro and in cells in the presence of the A3s could provide more time to deaminate minus-strand DNA, resulting in an independent mechanism of synergy where one A3 oligomer is inhibiting the reverse transcriptase and another is deaminating the DNA (Fig. 5A and B). This may facilitate increased deamination of A3F in the A3F/G hetero-oligomer. The A3F/G hetero-oligomer bound ssDNA with approximately 2-fold less affinity than A3F alone (Fig. 4A). The high affinity of A3F for ssDNA has been suggested to inhibit its movement on DNA, adding to the inefficiency with which it searches the substrate for deamination motifs (38). The higher Kd of the A3F/G hetero-oligomer may increase the ability of A3F in the A3F/G hetero-oligomer to search the minus-strand DNA before it becomes double stranded (38). In combination with the decrease in reverse transcriptase efficiency imposed by A3 enzymes, this provides an explanation for the increased number of A3F-induced mutations when A3F and A3G are coexpressed (Fig. 3 and 5 and Table 1).
Conclusions.
Overall, the data support that A3F and A3G in cells can form a hetero-oligomer with properties unique from the individual enzymes. The extent to which A3F and A3G oligomerize with each other in HIV-infected CD4+ T cells could not be determined due to the lack of a high-sensitivity antibody for A3F. However, it is likely that these interactions identified here in cell culture and in vitro contribute to the hypermutation identified in proviral genomes of HIV-infected people, since the higher mutagenic activity could enable more efficient mutagenesis despite the presence of Vif (78). It is also possible that other A3s hetero-oligomerize and synergistically restrict HIV. Altogether our data identify a mechanism that may contribute to enhanced activity of A3s and raises the idea that we still may not fully realize all of the antiviral mechanisms of endogenous A3 enzymes.
MATERIALS AND METHODS
Protein expression and purification.
Recombinant baculovirus production for expression of glutathione S-transferase (GST)-A3G, GST-A3F, and GST-nucleocapsid protein in Sf9 cells was carried out using the transfer vector pAcG2T(BD Biosciences), as previously described (38, 39, 43). Sf9 cells were infected with recombinant virus at a multiplicity of infection (MOI) of 1 for GST-A3G and GST-NC and an MOI of 2 for GST-A3F. After 72 h of infection, recombinant baculovirus-infected Sf9 cells were harvested for protein purification. A3G and NC were purified in the presence of RNase A and the GST tag cleaved on the affinity column, as previously described (48). A3F was purified in the presence of RNase A and the GST tag cleaved in solution, as previously described (38, 43). Proteins were estimated to be approximately 95% pure by SDS-PAGE.
The Escherichia coli strain containing the plasmids to express HIV reverse transcriptase and HIV protease was provided by Stuart Le Grice (National Cancer Institute). Expression of the HIV reverse transcriptase and protease were carried out as previously described (79). Cell lysates produced using sonication were clarified by centrifugation and then purified using a HisTrap FF crude column (GE Healthcare) and HiTrap heparin HP (GE Healthcare) as described previously (79).
Expression of A3G and A3F in 293T cells.
To express both A3F-V5 and A3G-HA on a single-cell basis, we used an expression plasmid, pVIVO2 (Invivogen), with two transcription units in a single vector. A3G and A3F were PCR amplified from pcDNA3.1 vectors previously used in the laboratory with primers that contained a C-terminal 1× HA-tag sequence (for A3G) or C-terminal V5 tag sequence (for A3F) (38). A3F-V5 was cloned using an XbaI site in MCS1. A3G-HA was cloned using an EcoRI site in MCS2. Using this cloning strategy, plasmids containing both A3F-V5 and A3G-HA (pVIVO2 A3F-V5/A3G-HA), A3F-V5 alone (pVIVO2 A3F-V5), and A3G-HA alone (pVIVO2 A3G-HA) were constructed. These constructs were also used as a template for site-directed mutagenesis to create catalytic mutants of A3F and A3G that resulted in pVIVO2 A3F-V5 E251Q/A3G-HA E259Q, pVIVO2 A3F-V5 E251Q, and pVIVO2 A3G-HA E259Q. Primer sequences are available upon request.
SEC.
To determine the oligomerization state of the A3 enzymes, we used size exclusion chromatography (SEC). A 10-ml Superdex 200 (GE Healthcare) resin bed contained in a column with a 0.5-cm diameter and 16-cm height was used. The running buffer used contained 50 mM Tris, pH 8.0, 200 mM NaCl, and 1 mM dithiothreitol (DTT). The molecular masses and oligomerization states were calculated from the standard curve obtained by using a Bio-Rad gel filtration standard set. For A3G or A3F alone, a total of 15 μg of the purified A3 enzyme was loaded on the column. For the combined run of A3F and A3G, 15 μg of each purified enzyme was preincubated at 21°C for 3 min before loading onto the column. Proteins in fractions were detected by Western blot analysis by using anti-A3G (ApoC17 rabbit antiserum; number 10082; NIH AIDS Reagent Program) and anti-A3F (number GTX47211; GeneTex) antibodies.
Steady-state rotational anisotropy assays.
To measure protein-protein and protein-ssDNA binding interactions, we used steady-state fluorescence depolarization (rotational anisotropy), where one of the binding partners was fluorescein (F) labeled. Data were collected using a QuantaMaster QM-4 spectrofluorometer (Photon Technology International) with a dual emission channel. Measurements were made at 21°C. Samples were excited with vertically polarized light at 495 nm (6-nm band pass), and vertical and horizontal emissions were measured at 520 nm (6-nm band pass). Apparent dissociation constants (Kd) were obtained by fitting to a sigmoidal curve using Sigma Plot 11.2 software.
For binding to ssDNA, we measured the ability of A3F alone, A3G alone, and A3F/A3G to bind to a 118-nt F-labeled ssDNA, as described previously (38). Reactions were performed in a 60-μl total volume, which contained F-labeled ssDNA (10 nM) in RT buffer (50 mM Tris, pH 7.5, 40 mM KCl, 10 mM MgCl2, 1 mM DTT) and A3G (0 to 900 nM), A3F (0 to 75 nM), or A3F/A3G in combination (0 to 340 nM). For A3F/A3G binding in combination, the enzymes were first mixed at an equimolar ratio and preincubated at 21°C for 3 min before titration into the reaction mixture.
To measure protein-protein binding, A3G was F labeled using the fluorescein-EX protein labeling kit (Life Technologies) and used as the binding substrate for A3F. This assay was carried out in a 60-μl total volume that contained 25 nM F-labeled A3G in the presence of RT buffer and increasing amounts of A3F (0 to 250 nM).
Co-IP assay.
Coimmunoprecipitation (co-IP) assays were conducted as described previously (80). Briefly, the 293T cells (2.5 × 106 cells per 75-cm2 flask) were transfected with 1 μg of total DNA. Equal amounts of each plasmid, pVIVO2 A3G-HA, pVIVO2-A3F-V5, or pVIVO2 A3F-V5/A3G-HA, were used to transfect the cells. GeneJuice transfection reagent (EMD Millipore) was used according to the manufacturer's instructions. At 64 h posttransfection, the cells were washed with phosphate-buffered saline and lysed in co-IP buffer (50 mM Tris-Cl, pH 7.4, 1% Nonidet P-40, 0.1% sodium deoxycholate, 10% glycerol, 150 mM NaCl) supplemented with RNase A (20 μg/ml; Roche) and EDTA-free protease inhibitor (Roche). Clarified supernatants were precleared with protein A-agarose-conjugated normal rabbit IgG (2 μg; Santa Cruz Biotechnology) in the presence of RNase A (20 μg/ml; Roche). One-half of the precleared supernatant was then incubated with protein A-agarose-conjugated polyclonal rabbit anti-HA antibody (2 μg; Sigma), and the other half (mock) was incubated with protein A-agarose-conjugated normal rabbit IgG (2 μg; Santa Cruz Biotechnology) at 4°C for 2 h. Resin was washed and the samples were then resuspended in Laemmli sample buffer and prepared for SDS-PAGE.
After SDS-PAGE, proteins were transferred to a nitrocellulose membrane. For detection of A3G-HA and A3F-V5 in cell lysates, the nitrocellulose membrane was probed with polyclonal rabbit HA (1:1,000; Sigma) and monoclonal mouse V5 antibodies (1:1,000, Sigma), respectively. For the loading control, monoclonal mouse anti-α-tubulin (1:1,000; Sigma) was used. HA- and rabbit IgG-immunoprecipitated lysates were probed with anti-V5 mouse monoclonal antibodies. After incubation with horse radish peroxidase (HRP)-conjugated secondary antibodies, the blots were visualized with X-ray film using Super Signal West Pico chemiluminescence substrate (Thermo-Scientific).
Intracellular detection of A3F-V5 and A3G-HA in 293T cells by flow cytometry.
Single-cell suspensions of untransfected 293T cells and A3F-V5- and A3G-HA-transfected 293T cells were prepared in phosphate-buffered saline containing 2% fetal bovine serum (FBS) and 0.05% sodium azide. Intracellular staining was performed by following an established procedure (81). Briefly, transfected and untransfected cells were fixed using BD Cytofix/Cytoperm buffer containing 4.2% paraformaldehyde. Fixed cells were permeabilized with 1× BD Perm/Wash buffer containing saponin and FBS. Cells were stained with rabbit anti-V5-CF543 and rabbit anti-HA-CF640R antibodies (Biotium Inc.) in BD Perm/Wash buffer for 30 min at 4°C. After three washes with BD Perm/Wash buffer, cells were resuspended in phosphate-buffered saline containing 2% FBS and 0.05% sodium azide. Data were acquired by flow cytometry on a FACSCalibur using CellQuest software (BD Biosciences) and analyzed with FlowJo software (TreeStar).
Single-cycle infectivity assay.
In order to generate vesicular stomatitis virus glycoprotein (VSV-G)-pseudotyped HIV Δvif NL4-3 viruses, 3 × 105 293T cells were transfected using GeneJuice (EMD Millipore) transfection reagent as previously described (38). Cells were transfected with 500 ng of pNL4-3 HIV Δvif, which expresses an enhanced green fluorescent protein (eGFP) reporter gene (82), and 200 ng of pMDG, which expresses the VSV-G protein (83, 84), in the presence of empty pVIVO2, pVIVO2 A3F-V5/A3G-HA (A3F/A3G combined), pVIVO2 A3F-V5 (A3F-V5 alone), or pVIVO2 A3G-HA (A3G-HA alone). A titration of A3 expression vector was used (25, 50, and 100 ng). We used empty pVIVO2 vector to achieve a total of 800 ng DNA. Sixteen hours after the transfection, the medium (Dulbecco's modified Eagle's medium [DMEM] with 10% FBS) was replaced after washing the cells with phosphate-buffered saline. Virus-containing supernatants were collected 48 h after the medium change and filtered through 0.45-μm polyvinylidene difluoride (PVDF) syringe filters. Virus was quantified by a p24 enzyme-linked immunosorbent assay (QuickTiter lentivirus titer kit [Cell Biolabs Inc.] or HIV-1 p24 enzyme-linked immunosorbent assay kit [XpressBio]). Target 293T cells were infected with virus by spinoculation at 800 × g for 1 h in the presence of 8 μg/ml of Polybrene (85). Infection levels in 293T cells were determined by flow cytometry by detecting eGFP fluorescence at 48 h postinfection and normalized to HIVΔvif infections in the absence of A3 enzymes (38). Statistical significance of results was determined using an unpaired t test.
Quantitative immunoblotting.
The 293T cells expressing A3G-HA and A3F-V5 from single-cycle infectivity assays were detected using anti-HA and anti-V5 antibodies (Sigma) in cell lysates (40 μg total protein) and virions. Virions were prepared for immunoblotting by concentration using Retro-X concentrator (Clontech), and 20 μl of concentrated virus was used. Loading controls for cell lysates (α-tubulin; Sigma) and virions (p24; number 3537; NIH AIDS Reagent Program) were detected using mouse monoclonal antibodies. Proteins of interest and loading controls were detected in parallel on the same gel by using the Licor/Odyssey system (IRDye 680-labeled goat anti-rabbit secondary antibody and IRDye 800-labeled goat anti-mouse secondary antibody).
Sequencing of integrated proviral DNA.
After 48 h of infection, total DNA from infected 293T cells was extracted using DNAzol reagent (Life Technologies). DNA was treated with DpnI (New England BioLabs) for 1 h at 37°C to remove possible contaminating plasmid DNA, and the protease (prot) (nt 2280 to 2631) sequences were amplified by PCR using Q5 polymerase (New England BioLabs). Primers have been published previously (38). PCR products were purified and cloned with the CloneJET PCR cloning kit (Thermo). DNA was sequenced with kit-specific primers and carried out at the National Research Council of Canada (Saskatoon, Canada). Analysis of the sequence context of the mutations was done using Original Hypermut (86).
In vitro deamination assays.
All of the ssDNA substrates used in this study were synthesized from Tri-Link Biotechnologies and have been published previously (38). In brief, substrates all contain two deamination motifs, either 5′CCC for A3G or 5′TTC for A3F, and a fluorescein-labeled thymidine between the motifs. Reactions were carried out under single-hit conditions, i.e., <15% substrate usage, to ensure that a single ssDNA substrate was interacting with at most a single enzyme (54). Under these conditions, a processivity factor can be determined. The processivity factor is a ratio of the quantified total number of deaminations occurring at two sites on the same ssDNA substrate with a calculated theoretical value of deaminations that would occur at these two sites if the deamination event were not processive (see reference 43). For A3F deamination reactions, 100 nM A3F was incubated with 100 nM ssDNA. For A3G, 30 nM A3G was incubated with 100 nM substrate. Due to differences in specific activity on different substrates, sample times ranged from 1 to 30 min after incubation at 37°C in RT buffer. The time points where only 10 to 11% of the substrate was deaminated were considered in processivity factor determination. For experiments using A3F and A3G in combination, we first preincubated the enzymes at an equimolar ratio for 3 min at 21°C before addition to the reaction mixture. Reactions were stopped using phenol and chloroform extractions. The ssDNA was then treated with uracil DNA glycosylase (New England BioLabs) and heated under alkaline conditions to induce DNA breakage at deaminated motifs. The DNA fragments were then resolved on a 10% or 20% (vol/vol) denaturing polyacrylamide gel depending on the expected sizes. Gel pictures were obtained using a Typhoon Trio multipurpose scanner (GE Healthcare), and gel band intensities were quantified by ImageQuant software (GE Healthcare).
Quantification of LRT formation during HIV replication.
For quantifying late reverse transcripts (LRT) by quantitative real-time PCR (qPCR), we followed the method of Belanger et al. (59). HIV Δvif-infected 293T cells were harvested 8 h postinfection, and total DNA was extracted and then treated with DpnI. For each reaction, 0.9 pmol/ml of each primer (as listed in reference 59), 0.25 pmol/ml of the probe (as listed in reference 59), and 10 ng of template DNA were used in a 20-μl reaction volume. Reactions were performed in triplicate with TaqMan Gene Expression master mix (Applied Biosystems). qPCR cycling conditions were 10 min at 95°C, followed by 40 cycles of 15 s at 95°C and 1 min at 60°C. qPCR was carried out using a StepOnePlus real-time PCR system (Applied Biosystems). The copy numbers in each sample were normalized for DNA input using human RNase P copy number assay (number 4403326; Applied Biosystems). Relative LRT quantitation was calculated by following the comparative CT (threshold cycle) method (ΔΔCT method).
Primer extension assay.
An RNA template encompassing the primer binding site (PBS) and upstream region near the 5′ end of the HIV genome (nt 571 to 674) was generated as described previously (35). An 18-nt 32P-labeled RNA primer to mimic tRNALys,3 primer was heat annealed to the 106-nt template RNA containing the PBS as described previously (35). Reactions were conducted in the presence of 10 nM primer/template, RT buffer, 200 μM deoxynucleoside triphosphates (dNTPs), 175 nM nucleocapsid, and 480 nM reverse transcriptase in the absence or presence of A3G alone, A3F alone, or A3F and A3G in combination. Each A3 alone was used at concentrations of 40, 80, and 320 nM. For A3F and A3G in combination, an equimolar ratio was preincubated for 3 min at 21°C and then added to reactions to achieve a final concentration of 40, 80, or 320 nM enzyme complex. Reaction mixtures were preincubated at 37°C for 1 min before the addition of dNTPs, which started the reaction. A negative control was used which contained all reaction components except reverse transcriptase to ensure there was no contaminating polymerase activity. Reactions were stopped by adding a 5-fold excess of 20 mM EDTA and 95% formamide. Primer extension was visualized by resolving samples on a 16% denaturing 8 M urea polyacrylamide gel. Gel band intensities were measured by phosphorimaging with a Bio-Rad FX scanner. The integrated gel band intensities of all bands in a lane were calculated with ImageQuant software (GE Healthcare) and used to determine the relative amounts of extended and unextended primers as well as fully extended product (82 nt). Statistical significance of primer extension assay results was determined using an unpaired t test.
REFERENCES
- 1.Feng Y, Baig TT, Love RP, Chelico L. 2014. Suppression of APOBEC3-mediated restriction of HIV-1 by Vif. Front Microbiol 5:450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, Trono D. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424:99–103. doi: 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- 3.Zhang H, Yang B, Pomerantz RJ, Zhang C, Arunachalam SC, Gao L. 2003. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature 424:94–98. doi: 10.1038/nature01707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sheehy AM, Gaddis NC, Choi JD, Malim MH. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- 5.Yu Q, Konig R, Pillai S, Chiles K, Kearney M, Palmer S, Richman D, Coffin JM, Landau NR. 2004. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat Struct Mol Biol 11:435–442. doi: 10.1038/nsmb758. [DOI] [PubMed] [Google Scholar]
- 6.Harris RS, Dudley JP. 2015. APOBECs and virus restriction. Virology 479-480:131–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mariani R, Chen D, Schrofelbauer B, Navarro F, Konig R, Bollman B, Munk C, Nymark-McMahon H, Landau NR. 2003. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 114:21–31. doi: 10.1016/S0092-8674(03)00515-4. [DOI] [PubMed] [Google Scholar]
- 8.Marin M, Rose KM, Kozak SL, Kabat D. 2003. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat Med 9:1398–1403. doi: 10.1038/nm946. [DOI] [PubMed] [Google Scholar]
- 9.Sheehy AM, Gaddis NC, Malim MH. 2003. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat Med 9:1404–1407. doi: 10.1038/nm945. [DOI] [PubMed] [Google Scholar]
- 10.Stopak K, de Noronha C, Yonemoto W, Greene WC. 2003. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol Cell 12:591–601. doi: 10.1016/S1097-2765(03)00353-8. [DOI] [PubMed] [Google Scholar]
- 11.Yu X, Yu Y, Liu B, Luo K, Kong W, Mao P, Yu XF. 2003. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302:1056–1060. doi: 10.1126/science.1089591. [DOI] [PubMed] [Google Scholar]
- 12.Jager S, Kim DY, Hultquist JF, Shindo K, LaRue RS, Kwon E, Li M, Anderson BD, Yen L, Stanley D, Mahon C, Kane J, Franks-Skiba K, Cimermancic P, Burlingame A, Sali A, Craik CS, Harris RS, Gross JD, Krogan NJ. 2012. Vif hijacks CBF-beta to degrade APOBEC3G and promote HIV-1 infection. Nature 481:371–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hultquist JF, Lengyel JA, Refsland EW, LaRue RS, Lackey L, Brown WL, Harris RS. 2011. Human and rhesus APOBEC3D, APOBEC3F, APOBEC3G, and APOBEC3H demonstrate a conserved capacity to restrict Vif-deficient HIV-1. J Virol 85:11220–11234. doi: 10.1128/JVI.05238-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Refsland EW, Stenglein MD, Shindo K, Albin JS, Brown WL, Harris RS. 2010. Quantitative profiling of the full APOBEC3 mRNA repertoire in lymphocytes and tissues: implications for HIV-1 restriction. Nucleic Acids Res 38:4274–4284. doi: 10.1093/nar/gkq174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koning FA, Newman EN, Kim EY, Kunstman KJ, Wolinsky SM, Malim MH. 2009. Defining APOBEC3 expression patterns in human tissues and hematopoietic cell subsets. J Virol 83:9474–9485. doi: 10.1128/JVI.01089-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Janini M, Rogers M, Birx DR, McCutchan FE. 2001. Human immunodeficiency virus type 1 DNA sequences genetically damaged by hypermutation are often abundant in patient peripheral blood mononuclear cells and may be generated during near-simultaneous infection and activation of CD4(+) T cells. J Virol 75:7973–7986. doi: 10.1128/JVI.75.17.7973-7986.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vartanian JP, Henry M, Wain-Hobson S. 2002. Sustained G→A hypermutation during reverse transcription of an entire human immunodeficiency virus type 1 strain Vau group O genome. J Gen Virol 83:801–805. doi: 10.1099/0022-1317-83-4-801. [DOI] [PubMed] [Google Scholar]
- 18.Vartanian JP, Meyerhans A, Asjo B, Wain-Hobson S. 1991. Selection, recombination, and G→A hypermutation of human immunodeficiency virus type 1 genomes. J Virol 65:1779–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vartanian JP, Meyerhans A, Sala M, Wain-Hobson S. 1994. G→A hypermutation of the human immunodeficiency virus type 1 genome: evidence for dCTP pool imbalance during reverse transcription. Proc Natl Acad Sci U S A 91:3092–3096. doi: 10.1073/pnas.91.8.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pathak VK, Temin HM. 1990. Broad spectrum of in vivo forward mutations, hypermutations, and mutational hotspots in a retroviral shuttle vector after a single replication cycle: substitutions, frameshifts, and hypermutations. Proc Natl Acad Sci U S A 87:6019–6023. doi: 10.1073/pnas.87.16.6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kieffer TL, Kwon P, Nettles RE, Han Y, Ray SC, Siliciano RF. 2005. G→A hypermutation in protease and reverse transcriptase regions of human immunodeficiency virus type 1 residing in resting CD4+ T cells in vivo. J Virol 79:1975–1980. doi: 10.1128/JVI.79.3.1975-1980.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pace C, Keller J, Nolan D, James I, Gaudieri S, Moore C, Mallal S. 2006. Population level analysis of human immunodeficiency virus type 1 hypermutation and its relationship with APOBEC3G and vif genetic variation. J Virol 80:9259–9269. doi: 10.1128/JVI.00888-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Piantadosi A, Humes D, Chohan B, McClelland RS, Overbaugh J. 2009. Analysis of the percentage of human immunodeficiency virus type 1 sequences that are hypermutated and markers of disease progression in a longitudinal cohort, including one individual with a partially defective Vif. J Virol 83:7805–7814. doi: 10.1128/JVI.00280-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nowarski R, Britan-Rosich E, Shiloach T, Kotler M. 2008. Hypermutation by intersegmental transfer of APOBEC3G cytidine deaminase. Nat Struct Mol Biol 15:1059–1066. doi: 10.1038/nsmb.1495. [DOI] [PubMed] [Google Scholar]
- 25.Simon V, Zennou V, Murray D, Huang Y, Ho DD, Bieniasz PD. 2005. Natural variation in Vif: differential impact on APOBEC3G/3F and a potential role in HIV-1 diversification. PLoS Pathog 1:e6. doi: 10.1371/journal.ppat.0010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liddament MT, Brown WL, Schumacher AJ, Harris RS. 2004. APOBEC3F properties and hypermutation preferences indicate activity against HIV-1 in vivo. Curr Biol 14:1385–1391. doi: 10.1016/j.cub.2004.06.050. [DOI] [PubMed] [Google Scholar]
- 27.Dang Y, Wang X, Esselman WJ, Zheng YH. 2006. Identification of APOBEC3DE as another antiretroviral factor from the human APOBEC family. J Virol 80:10522–10533. doi: 10.1128/JVI.01123-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harari A, Ooms M, Mulder LC, Simon V. 2009. Polymorphisms and splice variants influence the antiretroviral activity of human APOBEC3H. J Virol 83:295–303. doi: 10.1128/JVI.01665-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harris RS, Bishop KN, Sheehy AM, Craig HM, Petersen-Mahrt SK, Watt IN, Neuberger MS, Malim MH. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113:803–809. doi: 10.1016/S0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- 30.Desimmie BA, Burdick RC, Izumi T, Doi H, Shao W, Alvord WG, Sato K, Koyanagi Y, Jones S, Wilson E, Hill S, Maldarelli F, Hu WS, Pathak VK. 2016. APOBEC3 proteins can copackage and comutate HIV-1 genomes. Nucleic Acids Res 44:7848–7865. doi: 10.1093/nar/gkw653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Apolonia L, Schulz R, Curk T, Rocha P, Swanson CM, Schaller T, Ule J, Malim MH. 2015. Promiscuous RNA binding ensures effective encapsidation of APOBEC3 proteins by HIV-1. PLoS Pathog 11:e1004609. doi: 10.1371/journal.ppat.1004609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.York A, Kutluay SB, Errando M, Bieniasz PD. 2016. The RNA binding specificity of human APOBEC3 proteins resembles that of HIV-1 nucleocapsid. PLoS Pathog 12:e1005833. doi: 10.1371/journal.ppat.1005833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ebrahimi D, Anwar F, Davenport MP. 2012. APOBEC3G and APOBEC3F rarely co-mutate the same HIV genome. Retrovirology 9:113. doi: 10.1186/1742-4690-9-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krisko JF, Begum N, Baker CE, Foster JL, Garcia JV. 2016. APOBEC3G and APOBEC3F act in concert to extinguish HIV-1 replication. J Virol 90:4681–4695. doi: 10.1128/JVI.03275-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adolph MB, Webb J, Chelico L. 2013. Retroviral restriction factor APOBEC3G delays the initiation of DNA synthesis by HIV-1 reverse transcriptase. PLoS One 8:e64196. doi: 10.1371/journal.pone.0064196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chaurasiya KR, McCauley MJ, Wang W, Qualley DF, Wu T, Kitamura S, Geertsema H, Chan DS, Hertz A, Iwatani Y, Levin JG, Musier-Forsyth K, Rouzina I, Williams MC. 2014. Oligomerization transforms human APOBEC3G from an efficient enzyme to a slowly dissociating nucleic acid-binding protein. Nat Chem 6:28–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gillick K, Pollpeter D, Phalora P, Kim EY, Wolinsky SM, Malim MH. 2013. Suppression of HIV-1 infection by APOBEC3 proteins in primary human CD4(+) T cells is associated with inhibition of processive reverse transcription as well as excessive cytidine deamination. J Virol 87:1508–1517. doi: 10.1128/JVI.02587-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ara A, Love RP, Chelico L. 2014. Different mutagenic potential of HIV-1 restriction factors APOBEC3G and APOBEC3F is determined by distinct single-stranded DNA scanning mechanisms. PLoS Pathog 10:e1004024. doi: 10.1371/journal.ppat.1004024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feng Y, Chelico L. 2011. Intensity of deoxycytidine deamination of HIV-1 proviral DNA by the retroviral restriction factor APOBEC3G is mediated by the noncatalytic domain. J Biol Chem 286:11415–11426. doi: 10.1074/jbc.M110.199604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berg OG, Winter RB, von Hippel PH. 1981. Diffusion-driven mechanisms of protein translocation on Nucleic acids. 1. Models and theory. Biochemistry 20:6929–6948. [DOI] [PubMed] [Google Scholar]
- 41.von Hippel PH, Berg OG. 1989. Facilitated target location in biological systems. J Biol Chem 264:675–678. [PubMed] [Google Scholar]
- 42.Senavirathne G, Jaszczur M, Auerbach PA, Upton TG, Chelico L, Goodman MF, Rueda D. 2012. Single-stranded DNA scanning and deamination by APOBEC3G cytidine deaminase at single molecule resolution. J Biol Chem 287:15826–15835. doi: 10.1074/jbc.M112.342790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chelico L, Pham P, Calabrese P, Goodman MF. 2006. APOBEC3G DNA deaminase acts processively 3′→5′ on single-stranded DNA. Nat Struct Mol Biol 13:392–399. doi: 10.1038/nsmb1086. [DOI] [PubMed] [Google Scholar]
- 44.OhAinle M, Kerns JA, Li MM, Malik HS, Emerman M. 2008. Antiretroelement activity of APOBEC3H was lost twice in recent human evolution. Cell Host Microbe 4:249–259. doi: 10.1016/j.chom.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang X, Abudu A, Son S, Dang Y, Venta PJ, Zheng YH. 2011. Analysis of human APOBEC3H haplotypes and anti-human immunodeficiency virus type 1 activity. J Virol 85:3142–3152. doi: 10.1128/JVI.02049-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chaipan C, Smith JL, Hu WS, Pathak VK. 2013. APOBEC3G restricts HIV-1 to a greater extent than APOBEC3F and APOBEC3DE in human primary CD4+ T cells and macrophages. J Virol 87:444–453. doi: 10.1128/JVI.00676-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wiegand HL, Doehle BP, Bogerd HP, Cullen BR. 2004. A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. EMBO J 23:2451–2458. doi: 10.1038/sj.emboj.7600246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chelico L, Prochnow C, Erie DA, Chen XS, Goodman MF. 2010. Structural model for deoxycytidine deamination mechanisms of the HIV-1 inactivation enzyme APOBEC3G. J Biol Chem 285:16195–16205. doi: 10.1074/jbc.M110.107987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bennett RP, Salter JD, Liu X, Wedekind JE, Smith HC. 2008. APOBEC3G subunits self-associate via the C-terminal deaminase domain. J Biol Chem 283:33329–33336. doi: 10.1074/jbc.M803726200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang X, Dolan PT, Dang Y, Zheng YH. 2007. Biochemical differentiation of APOBEC3F and APOBEC3G proteins associated with HIV-1 life cycle. J Biol Chem 282:1585–1594. doi: 10.1074/jbc.M610150200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zennou V, Bieniasz PD. 2006. Comparative analysis of the antiretroviral activity of APOBEC3G and APOBEC3F from primates. Virology 349:31–40. doi: 10.1016/j.virol.2005.12.035. [DOI] [PubMed] [Google Scholar]
- 52.Love RP, Xu H, Chelico L. 2012. Biochemical analysis of hypermutation by the deoxycytidine deaminase APOBEC3A. J Biol Chem 287:30812–30822. doi: 10.1074/jbc.M112.393181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Loeb DD, Swanstrom R, Everitt L, Manchester M, Stamper SE, Hutchison CA III. 1989. Complete mutagenesis of the HIV-1 protease. Nature 340:397–400. doi: 10.1038/340397a0. [DOI] [PubMed] [Google Scholar]
- 54.Creighton S, Bloom LB, Goodman MF. 1995. Gel fidelity assay measuring nucleotide misinsertion, exonucleolytic proofreading, and lesion bypass efficiencies. Methods Enzymol 262:232–256. doi: 10.1016/0076-6879(95)62021-4. [DOI] [PubMed] [Google Scholar]
- 55.Feng Y, Love RP, Chelico L. 2013. HIV-1 viral infectivity factor (Vif) alters processive single-stranded DNA scanning of the retroviral restriction factor APOBEC3G. J Biol Chem 288:6083–6094. doi: 10.1074/jbc.M112.421875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Suspene R, Rusniok C, Vartanian JP, Wain-Hobson S. 2006. Twin gradients in APOBEC3 edited HIV-1 DNA reflect the dynamics of lentiviral replication. Nucleic Acids Res 34:4677–4684. doi: 10.1093/nar/gkl555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wurtzer S, Goubard A, Mammano F, Saragosti S, Lecossier D, Hance AJ, Clavel F. 2006. Functional central polypurine tract provides downstream protection of the human immunodeficiency virus type 1 genome from editing by APOBEC3G and APOBEC3B. J Virol 80:3679–3683. doi: 10.1128/JVI.80.7.3679-3683.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hu C, Saenz DT, Fadel HJ, Walker W, Peretz M, Poeschla EM. 2010. The HIV-1 central polypurine tract functions as a second line of defense against APOBEC3G/F. J Virol 84:11981–11993. doi: 10.1128/JVI.00723-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Belanger K, Savoie M, Rosales Gerpe MC, Couture JF, Langlois MA. 2013. Binding of RNA by APOBEC3G controls deamination-independent restriction of retroviruses. Nucleic Acids Res 41:7438–7452. doi: 10.1093/nar/gkt527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Holmes RK, Koning FA, Bishop KN, Malim MH. 2007. APOBEC3F can inhibit the accumulation of HIV-1 reverse transcription products in the absence of hypermutation. Comparisons with APOBEC3G. J Biol Chem 282:2587–2595. [DOI] [PubMed] [Google Scholar]
- 61.Luo K, Wang T, Liu B, Tian C, Xiao Z, Kappes J, Yu XF. 2007. Cytidine deaminases APOBEC3G and APOBEC3F interact with human immunodeficiency virus type 1 integrase and inhibit proviral DNA formation. J Virol 81:7238–7248. doi: 10.1128/JVI.02584-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mbisa JL, Bu W, Pathak VK. 2010. APOBEC3F and APOBEC3G inhibit HIV-1 DNA integration by different mechanisms. J Virol 84:5250–5259. doi: 10.1128/JVI.02358-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Newman EN, Holmes RK, Craig HM, Klein KC, Lingappa JR, Malim MH, Sheehy AM. 2005. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Curr Biol 15:166–170. doi: 10.1016/j.cub.2004.12.068. [DOI] [PubMed] [Google Scholar]
- 64.Bishop KN, Holmes RK, Malim MH. 2006. Antiviral potency of APOBEC proteins does not correlate with cytidine deamination. J Virol 80:8450–8458. doi: 10.1128/JVI.00839-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bishop KN, Verma M, Kim EY, Wolinsky SM, Malim MH. 2008. APOBEC3G inhibits elongation of HIV-1 reverse transcripts. PLoS Pathog 4:e1000231. doi: 10.1371/journal.ppat.1000231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Iwatani Y, Chan DS, Wang F, Maynard KS, Sugiura W, Gronenborn AM, Rouzina I, Williams MC, Musier-Forsyth K, Levin JG. 2007. Deaminase-independent inhibition of HIV-1 reverse transcription by APOBEC3G. Nucleic Acids Res 35:7096–7108. doi: 10.1093/nar/gkm750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mbisa JL, Barr R, Thomas JA, Vandegraaff N, Dorweiler IJ, Svarovskaia ES, Brown WL, Mansky LM, Gorelick RJ, Harris RS, Engelman A, Pathak VK. 2007. Human immunodeficiency virus type 1 cDNAs produced in the presence of APOBEC3G exhibit defects in plus-strand DNA transfer and integration. J Virol 81:7099–7110. doi: 10.1128/JVI.00272-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shindo K, Takaori-Kondo A, Kobayashi M, Abudu A, Fukunaga K, Uchiyama T. 2003. The enzymatic activity of CEM15/Apobec-3G is essential for the regulation of the infectivity of HIV-1 virion but not a sole determinant of its antiviral activity. J Biol Chem 278:44412–44416. doi: 10.1074/jbc.C300376200. [DOI] [PubMed] [Google Scholar]
- 69.Refsland EW, Hultquist JF, Harris RS. 2012. Endogenous origins of HIV-1 G-to-A hypermutation and restriction in the nonpermissive T cell line CEM2n. PLoS Pathog 8:e1002800. doi: 10.1371/journal.ppat.1002800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xiao X, Li SX, Yang H, Chen XS. 2016. Crystal structures of APOBEC3G N-domain alone and its complex with DNA. Nat Commun 7:12193. doi: 10.1038/ncomms12193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huthoff H, Autore F, Gallois-Montbrun S, Fraternali F, Malim MH. 2009. RNA-dependent oligomerization of APOBEC3G is required for restriction of HIV-1. PLoS Pathog 5:e1000330. doi: 10.1371/journal.ppat.1000330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nakashima M, Ode H, Kawamura T, Kitamura S, Naganawa Y, Awazu H, Tsuzuki S, Matsuoka K, Nemoto M, Hachiya A, Sugiura W, Yokomaku Y, Watanabe N, Iwatani Y. 2015. Structural insights into HIV-1 Vif-APOBEC3F interaction. J Virol 90:1034–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bohn MF, Shandilya SM, Albin JS, Kouno T, Anderson BD, McDougle RM, Carpenter MA, Rathore A, Evans L, Davis AN, Zhang J, Lu Y, Somasundaran M, Matsuo H, Harris RS, Schiffer CA. 2013. Crystal structure of the DNA cytosine deaminase APOBEC3F: the catalytically active and HIV-1 Vif-binding domain. Structure 21:1042–1050. doi: 10.1016/j.str.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shaban NM, Shi K, Li M, Aihara H, Harris RS. 2016. 1.92 Angstrom zinc-free APOBEC3F catalytic domain crystal structure. J Mol Biol 428:2307–2316. doi: 10.1016/j.jmb.2016.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Siu KK, Sultana A, Azimi FC, Lee JE. 2013. Structural determinants of HIV-1 Vif susceptibility and DNA binding in APOBEC3F. Nat Commun 4:2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Briggs JA, Simon MN, Gross I, Krausslich HG, Fuller SD, Vogt VM, Johnson MC. 2004. The stoichiometry of Gag protein in HIV-1. Nat Struct Mol Biol 11:672–675. doi: 10.1038/nsmb785. [DOI] [PubMed] [Google Scholar]
- 77.Sato K, Izumi T, Misawa N, Kobayashi T, Yamashita Y, Ohmichi M, Ito M, Takaori-Kondo A, Koyanagi Y. 2010. Remarkable lethal G-to-A mutations in vif-proficient HIV-1 provirus by individual APOBEC3 proteins in humanized mice. J Virol 84:9546–9556. doi: 10.1128/JVI.00823-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Albin JS, Harris RS. 2010. Interactions of host APOBEC3 restriction factors with HIV-1 in vivo: implications for therapeutics. Expert Rev Mol Med 12:e4. doi: 10.1017/S1462399409001343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Le Grice SF, Gruninger-Leitch F. 1990. Rapid purification of homodimer and heterodimer HIV-1 reverse transcriptase by metal chelate affinity chromatography. Eur J Biochem 187:307–314. doi: 10.1111/j.1432-1033.1990.tb15306.x. [DOI] [PubMed] [Google Scholar]
- 80.Baig TT, Feng Y, Chelico L. 2014. Determinants of efficient degradation of APOBEC3 restriction factors by HIV-1 Vif. J Virol 88:14380–14395. doi: 10.1128/JVI.02484-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ahmed KA, Wang L, Griebel P, Mousseau DD, Xiang J. 2015. Differential expression of mannose-6-phosphate receptor regulates T cell contraction. J Leukoc Biol 98:313–318. doi: 10.1189/jlb.2HI0215-049RR. [DOI] [PubMed] [Google Scholar]
- 82.Zhang H, Zhou Y, Alcock C, Kiefer T, Monie D, Siliciano J, Li Q, Pham P, Cofrancesco J, Persaud D, Siliciano RF. 2004. Novel single-cell-level phenotypic assay for residual drug susceptibility and reduced replication capacity of drug-resistant human immunodeficiency virus type 1. J Virol 78:1718–1729. doi: 10.1128/JVI.78.4.1718-1729.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Langlois MA, Beale RC, Conticello SG, Neuberger MS. 2005. Mutational comparison of the single-domained APOBEC3C and double-domained APOBEC3F/G anti-retroviral cytidine deaminases provides insight into their DNA target site specificities. Nucleic Acids Res 33:1913–1923. doi: 10.1093/nar/gki343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Naldini L, Blomer U, Gallay P, Ory D, Mulligan R, Gage FH, Verma IM, Trono D. 1996. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 85.O'Doherty U, Swiggard WJ, Malim MH. 2000. Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. J Virol 74:10074–10080. doi: 10.1128/JVI.74.21.10074-10080.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rose PP, Korber BT. 2000. Detecting hypermutations in viral sequences with an emphasis on G→A hypermutation. Bioinformatics 16:400–401. doi: 10.1093/bioinformatics/16.4.400. [DOI] [PubMed] [Google Scholar]