ABSTRACT
The J subgroup of avian leukosis virus (ALV-J) infects domestic chickens, jungle fowl, and turkeys. This virus enters the host cell through a receptor encoded by the tvj locus and identified as Na+/H+ exchanger 1. The resistance to avian leukosis virus subgroup J in a great majority of galliform species has been explained by deletions or substitutions of the critical tryptophan 38 in the first extracellular loop of Na+/H+ exchanger 1. Because there are concerns of transspecies virus transmission, we studied natural polymorphisms and susceptibility/resistance in wild galliforms and found the presence of tryptophan 38 in four species of New World quails. The embryo fibroblasts of New World quails are susceptible to infection with avian leukosis virus subgroup J, and the cloned Na+/H+ exchanger 1 confers susceptibility on the otherwise resistant host. New World quails are also susceptible to new avian leukosis virus subgroup J variants but resistant to subgroups A and B and weakly susceptible to subgroups C and D of avian sarcoma/leukosis virus due to obvious defects of the respective receptors. Our results suggest that the avian leukosis virus subgroup J could be transmitted to New World quails and establish a natural reservoir of circulating virus with a potential for further evolution.
IMPORTANCE Since its spread in broiler chickens in China and Southeast Asia in 2000, ALV-J remains a major enzootic challenge for the poultry industry. Although the virus diversifies rapidly in the poultry, its spillover and circulation in wild bird species has been prevented by the resistance of most species to ALV-J. It is, nevertheless, important to understand the evolution of the virus and its potential host range in wild birds. Because resistance to avian retroviruses is due particularly to receptor incompatibility, we studied Na+/H+ exchanger 1, the receptor for ALV-J. In New World quails, we found a receptor compatible with virus entry, and we confirmed the susceptibilities of four New World quail species in vitro. We propose that a prospective molecular epidemiology study be conducted to identify species with the potential to become reservoirs for ALV-J.
KEYWORDS: ALV-J, antiretroviral resistance, Na+/H+ exchanger, New World quail, retroviral receptor
INTRODUCTION
The range of hosts susceptible to a given retrovirus results from the process of coevolution between the viral envelope glycoproteins and host cell receptors. Selection forces imposed by the retrovirus drive the positive selection of receptors toward variants with decreased or even abrogated binding to retroviral envelopes. Vice versa, the highly error prone replication of retroviruses enables the rapid evolution of new strains with the capacity to bind to the variant receptors or even to quite new cell surface molecules. The results of these processes acting over evolutionary time are visible particularly in avian sarcoma/leukosis viruses (ASLVs), murine leukemia viruses (MLV), and feline leukemia viruses, where closely related virus subgroups differ in envelope glycoprotein sequences and infect different ranges of host species through different receptors (1–4).
The impact of the host-virus arms race on retroviral receptors has been much less studied than retroviral envelopes. It can be nicely documented by interspecific polymorphisms in receptors for retroviruses circulating naturally in feral populations. For example, multiple variants of xenotropic and polytropic MLV (XP-MLV) use the same receptor, mouse XPR1 (5) or its orthologs in other species, but show different ranges of rodent hosts as a result of diversification of both retroviral envelopes and host cell XPR1 receptors. Correlation of host range virus variants with receptor polymorphisms enabled the identification of amino acids critical for virus entry within two extracellular loops (ECL) of XPR1 (6). Interestingly, highly restrictive XPR1 alleles with disabling mutations of ECL3 and ECL4, either K496QE or Q579E, were also found in chickens and several other fowl and raptor species (7). At least three restrictive XPR1 alleles have single- or 5-amino-acid deletions in the critical region of ECL4. The positive selection at codon 496 of ECL3, geographic overlaps, and possible contact between XP-MLV-infected rodents suggest that transmission of the virus drove the parallel selection of virus-resistant receptor orthologs (7).
The second example of receptor polymorphism is the CD4 alleles, either permissive or resistant to human immunodeficiency virus type 1 (HIV-1) infection, that have been described for various primate species. Single amino acid differences at position 39 of CD4 distinguish species with different susceptibilities to HIV-1 isolates from early and late stages of human infection. The N39 alleles of humans and chimpanzees are functional receptors for laboratory-adapted (mostly CXCR4-tropic) and chronic-stage HIV-1 isolates, as well as for CCR5-tropic HIV-1 variants isolated from infected individuals soon after virus acquisition. In contrast, I39N CD4 alleles from rhesus and pig-tailed macaques are suboptimal receptors for early-stage HIV-1 isolates and efficiently support infection with laboratory-adapted isolates only (8). A wide screen of CD4 alleles in multiple primate species revealed strong positive selection against N39 (9, 10) and identified three species of New World owl monkeys compatible with early-stage HIV-1 isolates (10). At least in one captive colony of Spix's owl monkey, the CD4 is polymorphic, with the N39 allele prevailing over I39, suggesting that CD4 diversification is still in progress (10). The diversification of CD4 alleles has obviously been exerted by circulating simian immunodeficiency viruses and appears as preexisting resistance to a newly emerging HIV-1 strain.
Avian leukosis virus subgroup J (ALV-J) is a separate envelope subgroup of ASLVs (11), which arose by recombination of exogenous and endogenous counterparts (12) and utilizes Na+/H+ exchanger type 1 (NHE1) as the cell surface receptor (13). Originally identified in chronic myelocytomatosis in commercial meat breeds of domestic chicken in the United Kingdom, ALV-J diversified into new strains that also induced erythroblastosis, hemangiomas, and cholangiomas in layer chicks (14). This transition in pathogenesis coincided with the global spread of new variants of ALV-J, particularly in China and other Asian countries (see reference 15 for a review). Rigorous control of chicken breeds resulted in the successful elimination of ALV-J in Europe and the United States, with the last American outbreak of the PDRC-59831 strain (16) occurring in 2007, but there are concerns about the geographic spread of new Asian variants and their reintroduction. Furthermore, there are reports of ALV-J isolates from wild passeriform and anseriform bird species in Northeast China (17, 18).
The range of bird species susceptible to ALV-J infection remains to be explored systematically. According to very limited in vitro screens, only domestic chickens, jungle fowls, and turkeys support efficient ALV-J replication (19, 20). The resistant species, including quail, pheasants, Guinea fowl, and chukar, have been shown to carry NHE1 receptor-disabling mutations or deletions of W38 in ECL1 (18). In contrast, there are no polymorphisms of the NHE1 amino acid sequence in chickens of various genetically distant breeds (21). More-detailed knowledge of the host range of ALV-J and interspecific polymorphisms in NHE1 is desirable from the point of view of virus-host coevolution, because susceptible species might host a natural reservoir of circulating infectious virus. Hence, we studied the NHE1 receptor polymorphisms in galliform species and found functional receptors in New World quails. We confirmed the susceptibility of four New World quail species to ALV-J in vitro and validated the W38-based screen for prospective molecular epidemiology studies.
RESULTS
NHE1 sequences of New World quails encode W38, a key determinant of susceptibility to ALV-J.
In our previous study (20), we identified W38 of chicken NHE1 as a critical amino acid distinguishing between resistant and susceptible galliform species. In order to find new susceptible species in addition to the domestic chicken, red jungle fowl, and turkey, we analyzed multiple galliform species, including New World quails, for the presence of a W38 homolog as a marker of susceptibility. The amino acid sequences were deduced from the cDNA sequences obtained from embryo fibroblasts of four New World quail species, California quail, Gambel's quail, northern bobwhite, and mountain quail. The amino acid sequence of the putative ECL1 of NHE1 is shown in Fig. 1, aligned with the corresponding sequence of chicken. All four New World quail species contain the tryptophan residue homologous with W38 of chicken NHE1 (chNHE1), suggesting that these NHE1 forms might be compatible with ALV-J. Within ECL1, the New World quails exhibit several differences from chNHE1. Some polymorphisms are specific for New World quails; some are shared with other nonchicken galliform species; and some are specific for individual species or genera (Fig. 1). The rest of the NHE1 amino acid sequence was found to be well conserved between New World quails and chickens, except for three residues in putative transmembrane region 1 and two residues in the putative ECL4. In conclusion, our results confirmed the accumulation of polymorphisms within the N-terminal part of ECL1 of otherwise well-conserved NHE1 sequences and suggested that New World quails might be susceptible to ALV-J infection.
FIG 1.
Polymorphisms of NHE1 amino acid sequences from New World quails. The deduced amino acid sequences of ECL1 and adjacent parts of transmembrane regions M1 and M2, corresponding to chNHE1 amino acids 23 to 104, are aligned and compared. The borders between ECL1 and putative transmembrane domains M1 and M2 are shown. The W38 residue is indicated by a vertical arrow. Amino acids matching the chNHE1 sequence are shown on a gray background.
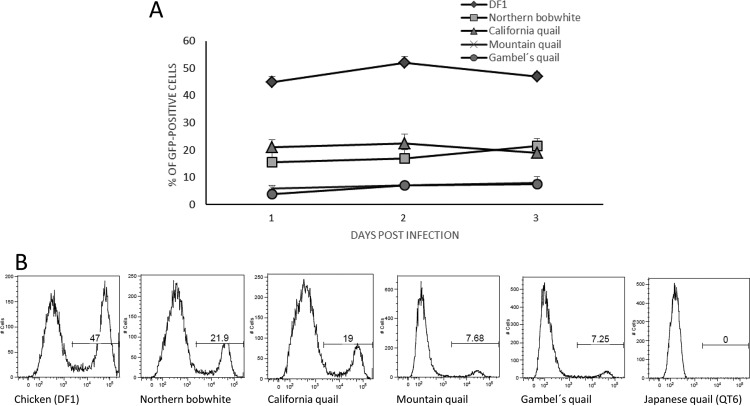
New World quails are susceptible to ALV-J.
The susceptibility of New World quail cells was tested by infection of cultured embryo fibroblasts with a recombinant reporter virus of subgroup J, RCASBP(J)GFP. The spread of infection was monitored by quantification of green fluorescent protein (GFP)-positive cells. Embryo fibroblasts from all four New World quail species displayed fewer GFP-positive cells than chicken DF-1 cells (Fig. 2A). RCASBP(J)GFP infected ca 20% of northern bobwhite and California quail cells, which is less than half of the proportion of GFP-positive DF-1 chicken cells. Mountain quail and Gambel's quail embryo fibroblasts were even less susceptible, with only ca. 5% of the cells infected on day 1, and the virus spread slowly; ca. 8% of cells were infected on day 3. The GFP-negative and GFP-positive cells are clearly distinct, as shown by the presence of two separated peaks in the fluorescence-activated cell sorter (FACS) histograms (Fig. 2B). These data confirmed that galliform species with NHE1 containing W38 are susceptible to infection with ALV-J. New World quails, however, are less susceptible than chickens, probably due to amino acid substitutions in ECL1 or nonreceptor postentry restriction of ALV-J.
FIG 2.
Time course of infection of New World quail embryo fibroblasts with ALV-J. Embryo fibroblasts were infected at a multiplicity of infection of 10 with replication-competent ALV-J encoding the GFP reporter proteins RCASBP(J)GFP. (A) The percentages of GFP-positive cells were determined by FACS analysis on the indicated days postinfection. The relative GFP fluorescence was plotted against the cell count, and the percentages of GFP-positive cells area are shown. Data are means for three parallel dishes. Error bars show standard deviations wherever they are large enough to be visible. (B) FACS histograms of RCASBP(J)GFP-infected chicken DF-1 cells, Japanese quail QT6 cells, and northern bobwhite, California quail, mountain quail, and Gambel's quail embryo fibroblasts at 3 days postinfection.
Because of the very slow increase of GFP-positive cells in RCASBP(J)GFP-infected cells, we tested the propagation of replication-competent virus in New World quail cells. Supernatants of New World cell cultures were sampled 3 days postinfection and were transferred to chicken embryonic fibroblasts (CEFs). GFP-positive CEFs indicated the transfer of infectious virus from New World quail cells. The titer of RCASBP(J)GFP virus produced by New World quail cells was very low, ca. 103 per ml (data not shown), which corresponds to the low number of infected cells and the slow kinetics of infection.
New World quail NHE1 confers susceptibility to ALV-J.
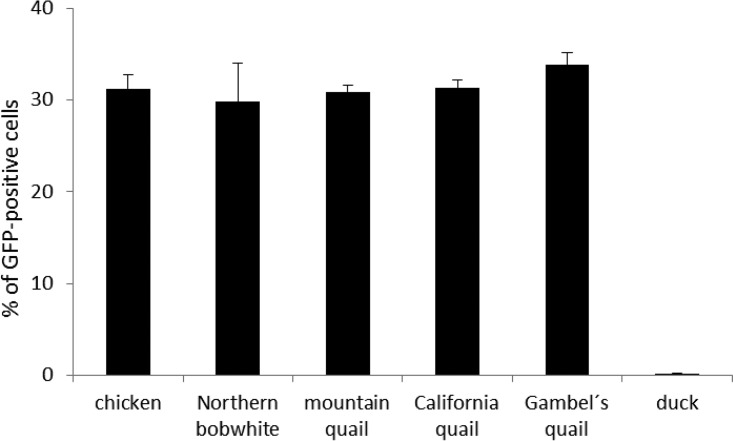
Having proven that New World quails are susceptible to ALV-J in vitro, we tested the capacity of their NHE1 genes to confer this susceptibility on otherwise resistant Japanese quail QT6 cells. We prepared expression vectors encoding hybrid NHE1 receptors with ECL1 of all four New World quails examined and the rest of NHE1 of chicken origin, transfected them into QT6 cells, and analyzed the subsequent RCASBP(J)GFP infection by flow cytometry. The efficiency of transfection with individual NHE1 expression vectors was normalized by the coexpression of a fluorescent tdTomato reporter from the same construct. We observed that ca. 30% of the transfected tdTomato-positive cells were GFP positive on the second day after transfection of all New World quail NHE1 expression vectors, a level comparable to that with chicken NHE1 (Fig. 3). This result corroborates the susceptibility of New World quails to ALV-J infection and shows that NHE1 orthologs of New World quails encode functional ALV-J receptors. However, the differences in susceptibility to ALV-J between northern bobwhite and California quail, on the one hand, and mountain quail and Gambel's quail, on the other, were not reproduced in this experiment. This, together with the comparable efficiency of chNHE1, indicates that the amino acid mismatches observed among the ECL1 sequences of New World quail NHE1 are not the cause of species-specific differences in susceptibility to ALV-J. Rather, some nonreceptor restriction mechanisms present in New World quails might be in play. We also cannot exclude the effect of polymorphisms present either in other ECLs or in membrane regions, which have not been tested in our experiment.
FIG 3.
Expression of cloned wild-type chNHE1 and its hybrid variants with ECL1 of New World quails, but not duck, confers susceptibility to ALV-J on QT6 cells. QT6 cells were transfected with NHE1 expression constructs and were infected with RCASBP(J)GFP on the next day. The percentage of GFP-positive cells was analyzed by flow cytometry 2 days postinfection. The RCASBP(J)GFP susceptibility conferred on QT6 cells by NHE1 hybrid variants containing ECL1 derived from New World quail NHE1 may be compared to that conferred by chicken NHE1 or hybrid NHE1 containing ECL1 from duck by observing the percentages of efficiently transfected tdTomato-positive cells that were GFP positive. Results were calculated as the averages for three parallel wells ± standard deviations.
NHE1 expression in New World quails.
Considering the lower efficiency of ALV-J infection in New World quails, we followed the possibility of reduced NHE1 expression in these species. We therefore examined the levels of NHE1 mRNA in cultured embryo fibroblasts of four New World quail species and compared them with those in chicken cells. qPCR analysis showed that NHE1 mRNA levels are comparable in chicken, northern bobwhite, Gambel's quail, and California quail (Fig. 4). In mountain quail, the NHE1 mRNA level is significantly lower, but still reasonably high compared to the level of the GAPDH housekeeping gene. These results do not suggest that the low infection efficiency in New World quails could be explained by insufficient expression of NHE1.
FIG 4.
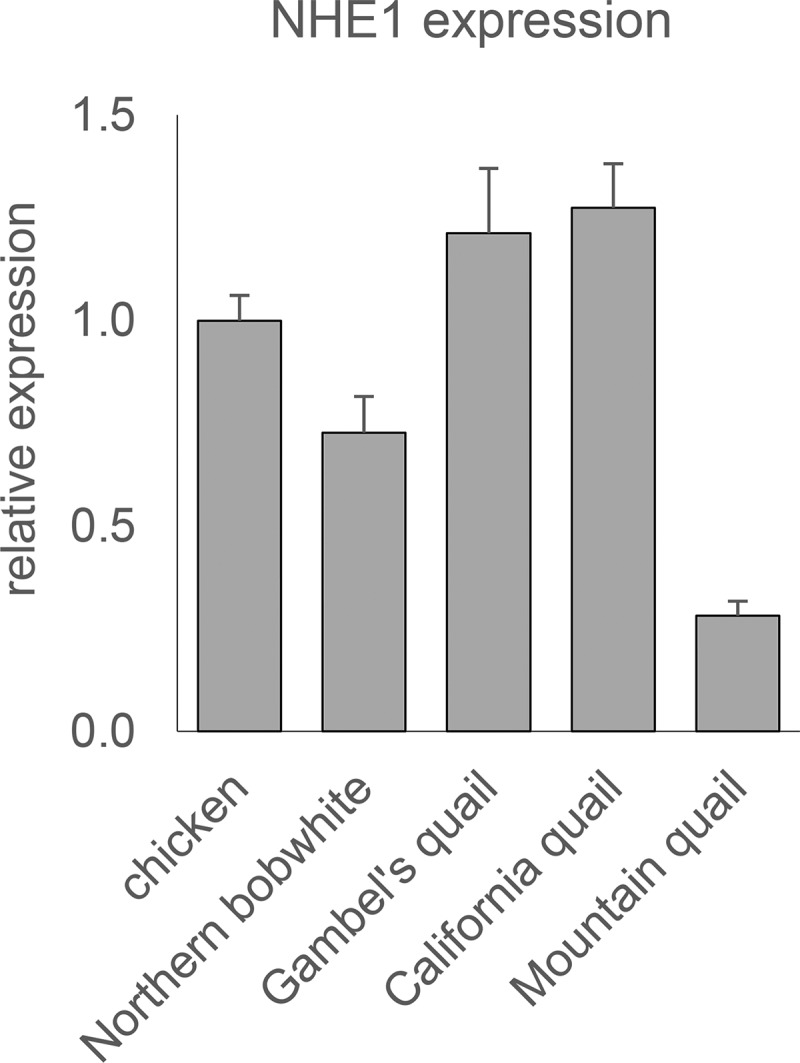
Comparison of NHE1 expression in chicken and New World quails. Cultured embryo fibroblasts were analyzed using qRT-PCR, and the resulting mRNA levels were normalized to those for the GAPDH housekeeping control. Results are shown as fold expression relative to the chNHE1 mRNA level. Error bars indicate standard errors for three parallel samples in one experiment.
Susceptibility/resistance of New World quail species to avian sarcoma/leukosis viruses of other subgroups.
In order to further analyze the susceptibility of New World quails to the ASLV complex, we infected cultured embryo fibroblasts with RCAS vectors of subgroups A to D. In general, New World quails were completely resistant to subgroups A and B and only marginally susceptible to subgroups C and D (Table 1). Subgroup C was the most efficient at infecting New World quails; the proportion of GFP-positive cells reached >10% in mountain quail and Gambel's quail 4 days after infection. In sharp contrast, chicken DF-1 cells were fully susceptible to the subgroups examined, reaching almost-complete infection with subgroups B, C, and D on the second day after infection.
TABLE 1.
Host ranges of ASLV subgroups A to Da in New World quails
| Host | % GFP-positive cells 2, 4 days after infectionb with the following virus: |
|||
|---|---|---|---|---|
| RCASBP(A)GFP | RCASBP(B)GFP | RCASBP(C)GFP | RCASBP(D)GFP | |
| Chicken (DF1) | 45.4, 68.8 | 96.9, 99.7 | 99.9, 100 | 99.6, 99.5 |
| Northern bobwhite | <0.05, <0.05 | <0.05, <0.05 | 0.39, 0.47 | 0.06, 0.09 |
| Mountain quail | <0.05, <0.05 | <0.05, <0.05 | 9.7, 12.3 | 0.21, 0.10 |
| California quail | <0.05, <0.05 | <0.05, <0.05 | <0.05, <0.05 | 0.08, 0.10 |
| Gambel's quail | <0.05, <0.05 | <0.05, <0.05 | 2.93, 10.87 | <0.05, 0.07 |
RCASBP(A)GFP enters the host cells through the Tva receptor, RCASBP(B)GFP and RCASBP(D)GFP through the Tvb receptor, and RCASBP(C)GFP through the Tvc receptor.
A value of 0.05% for GFP-positive cells represents the natural autofluorescence of mock-infected cells. All results are averages for two parallel infections. The standard deviation was calculated for each average value and reached only low values between ±0.016 and 0.029 (not shown).
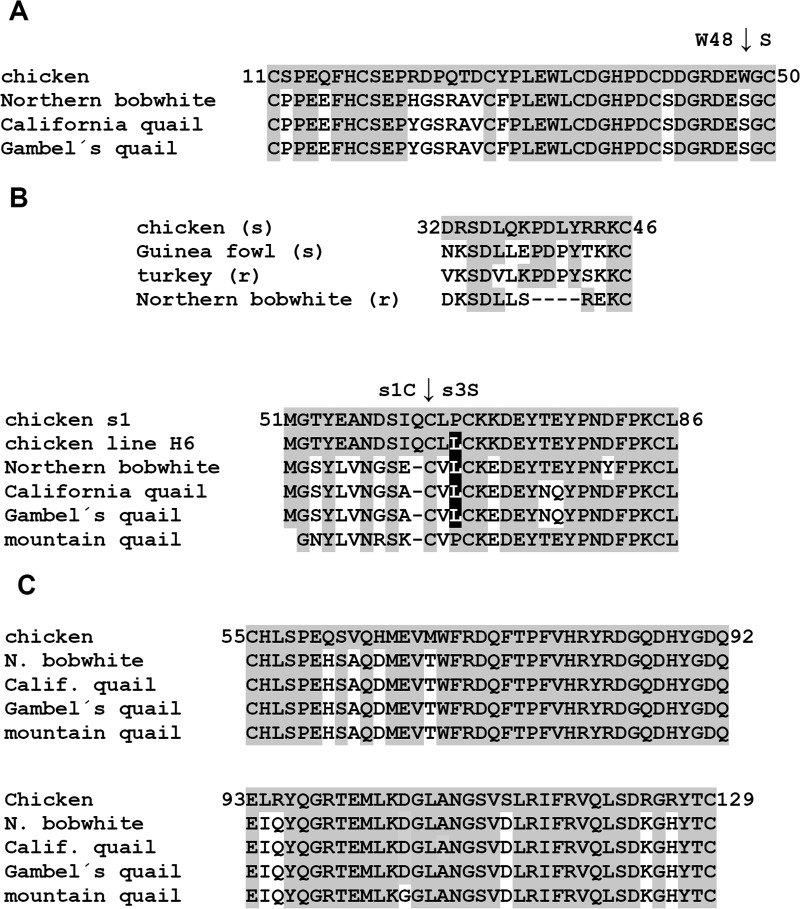
Because of the evolutionary distance between chicken and New World quails, we can assume that the divergence of tva, tvb, and tvc loci could explain this resistance. Therefore, we deduced the amino acid sequences of Tva, Tvb, and Tvc from the cDNAs of New World quails and compared them with chicken orthologs. An alignment of partial chicken ortholog sequences with the Tva (subgroup A receptor) sequences of three New World quail species (Fig. 5A) shows multiple amino acid substitutions within and around the low-density lipoprotein receptor-related motif, which is the major virus interaction determinant (22, 23). Particularly, the W48S substitution alone abrogates virus infectivity in chicken cells (22), in accordance with the complete resistance of New World quails to A-subgroup RCASBP(A)GFP. The tva sequence of mountain quail could not be analyzed, because we failed to amplify it. The Tvb (receptor for subgroups B and D) sequence is highly divergent from the chicken ortholog, with many polymorphisms common to all species of New World quails and several species-specific polymorphisms. The resistance of New World quails to RCASBP(B)GFP could be explained by a 4-amino acid deletion within the N-terminal peptide (amino acids 32 to 46) (Fig. 5B) of Tvb that was found in a draft genome assembly of northern bobwhite (24). This N-terminal peptide is sufficient for virus entry, and even single amino acid deletions abrogate its receptor capacity (25). Although the critical C62 residue defining the s1 allele (26) is present in all New World quails (Fig. 5B), low conservation around this site could contribute to the observed resistance of all four species to subgroup B and their marginal susceptibility to RCASBP(D)GFP. One of the substitutions present in three New World quail species, P64L, is also found in the inbred chicken line H6 (Fig. 5B). The Tvc (subgroup C receptor) amino acid sequence is well conserved between chicken and New World quails (Fig. 5C). There are only 10 substitutions within the IgV domain, the main determinant of subgroup C virus interaction, and the two residues that were described as critical for virus binding (27, 28), W69 and Y127, are conserved in New World quails. This is in accordance with their reduced but still significant susceptibility to RCASBP(C)GFP. We did not find any particular polymorphism unequivocally correlated with the complete resistance of California quail. In summary, New World quails turned out to be mostly resistant to ASLV subgroups A to D. Multiple changes in the Tva, Tvb, and Tvc receptors were found, corresponding to receptor-inactivating mutations described previously for resistant chicken lines (23, 26, 27). We can infer that these changes are responsible for the resistance or strongly reduced susceptibility of New World quails.
FIG 5.
Partial Tva, Tvb, and Tvc amino acid sequences of New World quails aligned to the chicken orthologs. Amino acids matching the chicken sequence are shown on a gray background. (A) Low-density lipoprotein receptor motif (amino acid residues 11 to 50) of Tva. The critical W48 residue mutated to S in New World quails is indicated by a vertical arrow. (B) (Top) Amino acid sequence of the N-terminal Tvb peptide sufficient for virus entry (amino acids 32 to 46). Chicken, guinea fowl (both sensitive to subgroup B), turkey, and northern bobwhite (both resistant to subgroup B) sequences are aligned. (Bottom) Amino acid sequence of New World quail Tvb around the polymorphic site C62S distinguishing the s1 and s3 alleles. A Tvb variant of the inbred chicken line H6 shows the presence of a P64L substitution (on a black background). (C) Amino acid sequence of the IgV domain of Tvc, delimited by C residues 55 and 129.
Susceptibility of New World quails to new ALV subgroup J variants.
Taking our findings together, we demonstrated that New World quail cells can be infected with ALV-J in accordance with the conservation of NHE1 W38, the amino acid critical for the entry of the virus into chicken cells. It is therefore interesting to inquire whether New World quails could be infected with subgroup J variants circulating in domestic chicken breeds. We answered this question by infecting cultured embryo fibroblasts with recombinant reporter RCAS vectors equipped with the subgroup J envelopes of two recently described Chinese isolates, ZB110604-5 and WB11016j, and two American isolates, ADOL7501 and PDRC-59831. The respective RCAS vectors were constructed and employed recently (21). Susceptibility was quantified as the percentage of cells that were GFP positive 2 days after infection. We observed that all species were susceptible to all four isolates (Table 2). However, RCASBP(JPDRC)GFP and RCASBP(JWB)GFP were much more effective than RCASBP(J)GFP with env from the prototypic strain HPRS103 (Fig. 2); RCASBP(JZB)GFP, in contrast, infected cells of all four New World quail species only marginally. The lower susceptibility of mountain quail and Gambel's quail to prototypic RCASBP(J)GFP was recapitulated only in part and not for all strains (Table 2).
TABLE 2.
Susceptibilities of New World quails to new J subgroup variants
| Host | % GFP-positive cells 2 days after infectiona |
|||
|---|---|---|---|---|
| RCASBP(JPDRC)GFP | RCASBP(JADOL)GFP | RCASBP(JWB)GFP | RCASBP(JZB)GFP | |
| Northern bobwhite | 36.6 | 19.1 | 41.1 | 1.9 |
| Mountain quail | 12.9 | 15.1 | 30.6 | 0.9 |
| California quail | 33.8 | 15.9 | 36.9 | 2.1 |
| Gambel's quail | 15.3 | 14.5 | 31.0 | 1.0 |
All results are averages for two parallel infections. The standard deviation was calculated for each value and reached only low values between ±0.027 and 1.24 (not shown).
Molecular evolution of NHE1 in galliform birds.
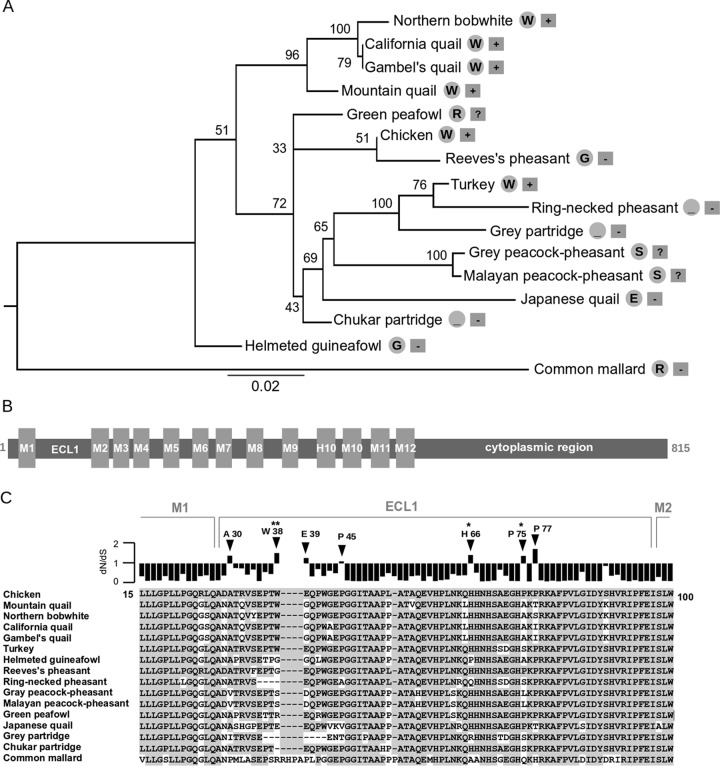
Since the ECL1 region of the NHE1 gene determines the host susceptibility to ALV-J, we further analyzed its evolution in galliform birds examined previously (20, 21) and in this study. We looked at the signature of positive selection acting on this region. Maximum likelihood (ML) phylogenetic analysis of partial NHE1 coding sequences produced the expected tree (Fig. 6A), supporting the orthologous character of amplified sequences. Some branches of the tree are not fully resolved or display low statistical support because of the short region of NHE1 sequence available and the small number of orthologous sequences examined. The topology corroborates previously published data that place guinea fowl and New World quails, belonging to the families Numididae and Odontophoridae, respectively, outside of the monophyletic Phasianidae. Within the family Phasianidae, the branching pattern is mostly in accordance with those of phylogenetic trees based on mitochondrial sequences, nuclear sequences, or CR1 insertions (29–31). Our topology does not support the monophyly of the subfamily Phasianinae, because Reeves's pheasant groups with chicken, and ring-necked pheasant groups with turkey. Also, the close relationship between peafowl and peacock-pheasant (29) is not recovered by our analysis.
FIG 6.
Molecular evolution of NHE1 in galliform birds. (A) ML tree of NHE1 partial nucleotide sequences of galliform species with the common mallard as an outlier. Nonparametric bootstrap supports are given next to the corresponding nodes. For each species, the critical residue or deletion at position 38 relative to the chicken sequence, taken as a reference, is shown in a gray circle on the right. Information about the sensitivity of the species to ALV-J infection is shown in a gray rectangle on the far right (+, susceptible; −, resistant; ?, unknown). Bar, substitutions per nucleotide. (B) Domain structure of the NHE1 protein. Membrane regions (M1 to M12, H10), extracellular loop 1 (ECL1), and the C-terminal cytoplasmic domain are shown. Annotation is based on the human NHE1 ortholog. (C) Amino acid sequence alignment and positive-selection analysis of the ECL1 region. Residues identical to the chicken sequence are shown on a gray background. Ratios of nonsynonymous to synonymous substitution rates (dN/dS) for each alignment position are shown above the alignment. Residues with positive-selection signatures (dN/dS, >1) are indicated by arrowheads. Residues with a posterior probability for positive selection of >0.95 or >0.99 are marked with one or two asterisks, respectively. Positively selected sites were predicted by the REL method.
The presence of positively selected residues in the ECL1 region of avian NHE1 (Fig. 6B) was examined using the random effect likelihood (REL) method. The ratio of nonsynonymous to synonymous substitution rates (dN/dS)—usually used to describe the presence of natural selection in coding sequences—was estimated for each position in an ECL1 alignment (Fig. 6C). Residues A30, W38, E39, P45, H66, P75, and P77, numbered according to the chicken NHE1 coding sequence, show dN/dS ratios of >1, indicating the presence of positive selection. For three of these residues, H66, P75, and W38, the occurrence of positive selection is statistically significant. Interestingly, the highest posterior probability of positive selection (>99%) was detected for W38. In contrast to the clear functional importance of W38, the polymorphisms at positively selected sites 66 and 75 are less probably linked to receptor functions, because they do not correlate with the host's resistance/susceptibility status. Particularly, either P75 or S75 can occur in ALV-J-susceptible chicken and turkey. The rest of the ECL1 region displays low dN/dS values, suggesting a strong purifying selection. Further analysis of additional avian NHE1 orthologs would be needed to identify specific lineages where positive selection occurred.
DISCUSSION
Avian sarcoma/leukosis viruses and galliform birds are a useful example of the host-virus arms race. We have focused on the host side, i.e., the receptors for virus entry, and described the sequence variation in the ALV-J-specific receptor, NHE1, of New World quails. Our finding that W38 is present in ECL1 of the NHE1 receptor suggested that New World quails are susceptible to ALV-J, and this was directly confirmed by infecting New World quail embryo fibroblasts with a recombinant retroviral vector of subgroup J specificity. Our results not only identified New World quails susceptible to ALV-J but also validated our bioprospective approach based on the detection of W38 in galliform species (20, 21). Future surveys of galliform species might reveal new NHE1 alleles with the ability to serve as functional receptors for ALV-J. So far, all species carrying W38 in the ECL1 of NHE1 turned out to be susceptible to ALV-J. However, we cannot exclude the possibility of imperfect correlation between W38 positivity and susceptibility to ALV-J due to another disabling mutation. Actually, the most frequent link between genetic and phenotypic variation is RNA splicing, and splicing defects of the tva receptor gene have already been found in in an ASLV subgroup A-resistant chicken line (32). The perfect analogy of receptor-disabling mutations and deletions in the critical ECL regions of XPR1 (7) and NHE1 (20; also this work) suggests that this is a common mechanism leading to host resistance to a virus.
Our finding of W38 in NHE1 of New World quails is based on embryo fibroblasts derived from several individuals. We did not observe any intraspecific polymorphisms within the region of ECL1 examined, which suggests that all individuals were homozygous. We cannot, however, exclude the presence of ΔW38 alleles in wild populations and the eventual low frequency of ALV-J-resistant individuals. Such a population structure was described for the CCR5 coreceptor of HIV/simian immunodeficiency virus (SIV). In the Caucasian population, 1% of individuals are homozygous for the CCR5Δ32 allele, abrogating cell surface display, and hence are highly resistant to CCR5-using HIV isolates (33, 34). A similar in-frame deletion allele, CCR5Δ24, exists in a majority of red-capped mangabeys (Cercocebus torquatus), and SIVRCM, as an exception among SIV strains, enters cells through CCR2b (35). The same allele rarely occurs in certain populations of sooty mangabeys (Cercocebus atys), but no homozygous animals were found (35, 36). Another example of this parallel evolution is the frameshift mutation CCR5Δ2 in sooty mangabeys. Accordingly, SIVSMM has extended coreceptor usage and employs CXCR6, GPR15, and GPR1 molecules for cell entry (37).
The susceptibility of New World quails to ALV-J is profoundly lower than that of chicken or turkey. This difference could be explained by several amino acid substitutions found in ECL1; however, some of these substitutions were identical with the sequence of turkey. Furthermore, the difference between chicken and New World quails was neither reproduced in the experimental induction of susceptibility after ectopic expression of NHE1 alleles (Fig. 3) nor correlated with different levels of NHE1 expression (Fig. 4). We can speculate that New World quails are protected by some host factors that restrict retrovirus replication; however, there is no evidence of such factors in birds so far. Experiments with other ASLV subgroups did not suggest any explanation, as the resistance of New World quails to subgroups A and B and their low susceptibility to subgroups C and D mostly correlated with restrictive receptors Tva, Tvb, and Tvc (Table 1; Fig. 5).
It is a question whether New World quails could naturally acquire ALV-J from either domestic poultry or wild birds. ALV-J has not been documented in the Americas since the last outbreak in 2007 (16), and reintroduction of the new Chinese ALV-J variants would require complicated transduction via migratory birds, most probably wild species of ducks. North American and Asian duck populations share wintering grounds, and at least two reports indicate the presence of ALV-J in ducks and partridges (17, 18). Avian leukosis viruses of other subgroups can infect ducks and establish persistent infections in these heterologous hosts (38, 39). On the other hand, we reported that NHE1 receptors of duck species are restrictive, and in vitro infection of duck embryo fibroblasts with ALV-J was inefficient (21). However, the evolution of the virus and its gradual adaptation to new hosts are well documented; human immunodeficiency virus, koala endogenous retrovirus, and X-MLV are good examples of naturally occurring transspecies or even transclass transmission, which led to disease in a new host and started further diversification of the virus. Among the four species of New World quails examined, northern bobwhite is the best candidate to host a natural reservoir of circulating infectious virus, because it lives in high-density populations and tolerates agricultural disturbances.
Considering the arms race between ASLVs and galliform hosts, it was important that we documented positive selection at several amino acid residues, particularly W38, within the otherwise negatively selected ECL1 of NHE1. This finding further corroborated the importance of W38 for receptor competence and suggested the possible virus-driven selection of this amino acid. Again, there is a similarity to XPR1, where two amino acid residues within the critical ECL3 are under positive selection in mice and birds (7). What virus left behind the remarkable variation of resistant receptor alleles remains open to speculation. This question seems to be answered in the case of S3/S1 alleles of the tvb gene, where the occurrence of the S1 allele has been attributed to the selection imposed by RAV-0 endogenous retrovirus (26). There is, however, little information on the diversity of ASLVs and their distribution in nonchicken galliform birds. Dimcheff et al. (40) detected gag sequences similar to exogenous ASLVs in all species of galliforms examined, including northern bobwhite. Phylogenetic analysis based on these gag sequences placed ASLVs of bobwhite closer to the ASLVs of grouses and pheasants than to Gallus ASLVs, including the exogenous ALV-J HPRS103 (40). The gag sequences of bobwhite displayed only limited similarity (42%) to the gag of ev/J, the endogenous predecessor of ALV-J, and contain a stop codon (40). Therefore, the ASLVs in bobwhite are probably replication defective even though they are actively transcribed (41). The ASLV env sequences of bobwhite have not been studied yet, and the relative endogenous ASLVs in grouses and ptarmigans are heavily deleted (41). In conclusion, the New World quails contain ASLVs that could have triggered the evolution of retrovirus restriction factors and restrictive receptor alleles. The subgroup identity of New World quail ASLVs is, however, unclear, and ev/J probably did not exist here, which left New World quails susceptible to the recent ALV-J.
In the present study, we concentrated on the receptor part of host-pathogen coevolution. We suggest that W38 of NHE1 is a residue under positive selection imposed by a virus pathogen, probably a retrovirus of J specificity. It can be assumed that positive selection also works on env sequences of ALV-J. Further studies on the molecular evolution of ALV-J isolates and changes in the infection efficiency are therefore warranted.
MATERIALS AND METHODS
Preparation of embryo fibroblasts and cell culture.
Embryonated eggs of chicken and four New World quail species—northern bobwhite (Colinus virginianus), California quail (Callipepla californica), Gambel's quail (Callipepla gambellii), and mountain quail (Oreortyx pictus)—were obtained from fancy fowl breeders in the Czech Republic, and embryo fibroblasts were prepared in the middle of incubation, i.e., on day 11 (California and Gambel's quails) or 12 (northern bobwhite and mountain quail). The procedure for the preparation of chicken embryo fibroblasts has been described previously (42). All embryo fibroblasts, as well as the permanent chicken cell line DF-1 (43) and the Japanese quail tumor cell line QT6 (44), were grown in a mixture of 2 parts Dulbecco's modified Eagle's medium and 1 part F-12 medium supplemented with 8% fetal calf serum, 2% chicken serum, and 1× antibiotic-antimycotic solution (Sigma) under a 5% CO2 atmosphere at 37°C.
Analysis of the NHE1, tva, tvb, and tvc sequences from New World quail species.
Total RNAs from cultured embryo fibroblasts of four New World quail species were prepared using the TRI reagent (Sigma). cDNA was reverse transcribed from 1 μg total RNA with ProtoScript II reverse transcriptase (New England BioLabs) and oligo(dT)15 primers (Promega). Then we amplified the NHE1 coding sequence, comprising all predicted transmembrane domains and extra- and intracellular loops (13), using forward primer NWQTVJ1L (5′-CTACGAGCCGTCCCCTGG-3′; complementary to the sequence encoding 2 amino acids of the N-terminal intracytoplasmic tail and part of transmembrane region 1) and reverse primer chTVJR6 (5′-CAGGAACTGCGTGTGGATCTC-3′; complementary to nucleotides 1537 to 1557 of chNHE1). PCR conditions were as follows: 98°C for 180 s; 39 cycles of 98°C for 10 s, 67.6°C for 30 s, and 72°C for 100 s; and terminal extension at 72°C for 7 min with recombinant Taq (rTaq) polymerase (TaKaRa). The resulting PCR products of 1,533 bp were treated with ExoSAP-IT reagent (USB) and were then directly sequenced from primer chTVJ5′RACE1 (5′-TCATCAGGCAGGCCAGCAGGAT-3′; complementary to nucleotides 301 to 322 of chNHE1) and from primer chTVJ6R using a BigDye Terminator cycle sequencing kit (version 3.1; Perkin-Elmer Applied Biosystems).
cDNAs for the tva sequence, encompassing the low-density lipoprotein receptor-related motif, were amplified by rTaq polymerase (TaKaRa) using primers NWQTVA4 (5′-CATGGCGCGGCTGCTGGC-3′) and NWQTVA10 (5′-GATTCCTGGATGGCAGAGAG-3′) with the following PCR conditions: 98°C for 180 s; 40 cycles of 98°C for 10 s, 65°C for 30 s, and 72°C for 30 s; and terminal extension at 72°C for 5 min. The resulting 306-bp product was analyzed. cDNAs for the tvb sequence, encompassing the critical C62 residue defining the s1 allele, were amplified by Ex Taq HS polymerase (TaKaRa) using primers NWQTVB3 (5′-CGGACAAGTCAGATCTC-3′) and TVB4 (5′-CCCAGGCACTTGGGAAA-3′) with the following PCR conditions: 98°C for 180 s; 35 cycles of 98°C for 10 s, 58°C for 30 s, and 72°C for 20 s; and terminal extension at 72°C for 5 min. The resulting 136-bp product was analyzed. cDNAs for the tvc sequence, encompassing the IgV domain, were amplified by Ex Taq HS polymerase (TaKaRa) using forward primer Btf6 (5′-ATCCGAATTCCATGGAGACGATGTTTTTTGGCTG-3′) and reverse primer IG3R (5′-CCAGGAGAAGGAGTCCGCATTC-3′) with the following PCR conditions: 94°C for 60 s; 30 cycles of 94°C for 10 s, 58°C for 30 s, and 68°C for 45 s; and terminal extension at 68°C for 5 min.
Virus propagation and susceptibility assay for New World quail embryo fibroblasts.
Infectious virus was produced in DF-1 cells transfected with RCASBP(A)GFP, RCASBP(B)GFP, RCASBP(C)GFP, RCASBP(D)GFP (42), RCASBP(J)GFP, RCASBP(JADOL)GFP, RCASBP(JPDRC)GFP, RCASBP(JZB)GFP, or RCASBP(JWB)GFP (21) plasmid DNA. Virus stocks were harvested on day 9 or 10 posttransfection. The cell supernatants were cleared of debris by centrifugation at 2,000 × g for 10 min at 10°C, and aliquoted viral stocks were stored at −80°C. The virus titer was determined by terminal dilution and subsequent infection of DF-1 cells; it reached 106 infection units (IU) per ml. Susceptibility to ALV-J was assessed by RCAS(J)GFP virus spread as described by Reinišová et al. (45). Briefly, New World quail embryo fibroblasts, DF-1 cells, or QT6 cells were seeded at a density of 5 × 104 per well in a 24-well plate and were infected with RCASBP(J)GFP virus at a multiplicity of infection of 10 the day after seeding. The percentage of GFP-positive cells was quantitated by fluorescence-activated cell sorting (FACS) using an LSR II analyzer (Becton, Dickinson) on days 1, 2, 3, and 4 postinfection (p.i.). The cells were trypsinized, washed in phosphate-buffered saline (PBS), and resuspended in Hoechst solution (Sigma) before analysis.
Construction of NHE1 expression vectors and transfection experiments.
We constructed New World quail-specific NHE1 expression vectors based on the pVitrotdT-tvj vector with chNHE1 constructed previously (20). The ECL1 cassettes of all four New World quail species and that of domestic duck, corresponding to the ApaI-BamHI fragment of chNHE1, were synthesized (Integrated DNA Technologies) and cloned into ApaI-BamHI (New England BioLabs)-linearized pVitrotdT-tvj. The expression constructs with chicken and New World quail NHE1 were transfected into QT6 cells. A total of 5 × 104 cells were seeded onto a 24-well plate in the cultivation medium without antibiotics. Transfection was performed 12 h later by a mixture of 1 μl of X-tremeGENE HP DNA transfection reagent (Roche) and 0.5 μg plasmid in Opti-MEM (Gibco BRL) per well. The cells were used for RCAS(J)GFP infection and a virus spread assay 24 h after transfection. The tdTomato fluorescent protein was used as a marker of successfully transfected cells, and infection efficiency was expressed as the percentage of GFP-positive cells among the tdTomato-positive cells as measured by FACS analysis (see above).
Quantitative RT-PCR.
Total RNA was isolated from 2 × 106 cultured embryo fibroblasts using RNAzol RT (Molecular Research Center, Inc.). cDNA was reverse transcribed from 1 μg total RNA with ProtoScript II reverse transcriptase (New England BioLabs) and oligo(dT)15 primers (Promega). Gene expression was then evaluated by relative quantitative reverse transcription-PCR (qRT-PCR) using the Mesa Green qPCR MasterMix Plus for SYBR assay (Eurogentec) in a C1000 Touch thermal cycler (Bio-Rad). Signals were normalized to the corresponding glyceraldehyde-3-phosphate dehydrogenase (GAPDH) housekeeping gene controls. The relative quantification of the target genes was carried out using the ΔΔCT method. All reactions were run in triplicate, and the average threshold cycles (CTs) were used for quantitation. The following primers were used: chNHE1-FW (5′-GCATCGACTACTCGCACGTC-3′), chNHE1-RV (5′-GCTCTCGGGAACCACTTTGG-3′), chGAPDH-FW (5′-CATCGTGCACCACCAACTG-3′), and chGAPDH-RV (5′-CGCTGGGATGATGTTCTGG-3′). Cycling conditions were as follows: 40 cycles of 95°C for 15 s, 55°C for 20 s, and 72°C for 30 s. The negative controls contained water instead of a template. Data analysis was conducted with the aid of CFX Manager software, version 3.1 (Bio-Rad).
Phylogenetic inference and positive selection analysis.
NHE1 nucleotide coding sequences of the following bird species, obtained in previous studies (20, 21) or this study, were used: chicken (NCBI Nucleotide database accession number DQ256198.1), turkey (DQ883630.1), mountain quail (KX823320), northern bobwhite (KX823321), California quail (KX823322), Gambel's quail (KX823323), helmeted guineafowl (KX823324), Reeves′s pheasant (KX823325), ring-necked pheasant (KX823326), gray peacock-pheasant (KX823327), Malayan peacock-pheasant (KX823328), green peafowl (KX823329), Japanese quail (KX823330), gray partridge (KX823331), chukar partridge (KX823332), and common mallard (KX823333). The sequences were aligned using a codon-based MUSCLE algorithm implemented in MEGA 6 software (46) and were checked manually. The maximum likelihood phylogeny of NHE1 was constructed using PhyML software, version 3.1 (47). A Kimura 2-parameter (K80) model with gamma distribution (4 categories) of rates among sites was used as a substitution model. The starting tree was generated by the BioNJ method. Nonparametric bootstrap analysis was performed with 1,000 replicates.
The random effect likelihood (REL) method implemented in the Datamonkey Web server (48) was used to detect positively selected sites in the ECL1 region of avian NHE1. Bayes empirical Bayes (BEB) positive-selection analysis (49), implemented in PAML, version 4.7 (50), was performed under an M2a site model and was used as a validation of the REL method. Trimmed alignment of a 318-nucleotide coding sequence containing the ECL1 region was used for the analyses.
Accession number(s).
The NHE1 sequences determined in this study were deposited in GenBank under accession numbers KX823320 (mountain quail), KX823321 (northern bobwhite), KX823322 (California quail), and KX823323 (Gambel's quail).
ACKNOWLEDGMENTS
This work was supported by grant 13-30983S from the Czech Science Foundation. We also acknowledge the institutional support by projects RVO 68378050 and NPU I LO1419.
We thank Zbyněk Laube, Pavel Doubek, and Jaroslav Huf for providing us with embryonated eggs of New World quails. We thank Lenka Mikušová and Marie Hejnarová for excellent technical assistance.
REFERENCES
- 1.Weiss RA. 1992. Cellular receptors and viral glycoproteins involved in retrovirus entry, p 1–108. In Levy JA. (ed), Retroviridae, vol 2 Plenum Press, New York, NY. [Google Scholar]
- 2.Barnard RJO, Elleder D, Young JAT. 2006. Avian sarcoma and leukosis virus-receptor interactions: from classical genetics to novel insights into virus-cell membrane vision. Virology 344:25–29. doi: 10.1016/j.virol.2005.09.021. [DOI] [PubMed] [Google Scholar]
- 3.Kozak CA. 2011. Naturally occurring polymorphisms of the mouse gammaretrovirus receptors CAT-1 and XPR1 alter virus tropism and pathogenicity. Adv Virol 2011:975801. doi: 10.1155/2011/975801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Watanabe S, Kawamura M, Odahara Y, Anai Y, Ochi H, Nakagawa S, Endo Y, Tsujimoto H, Nishigaki K. 2013. Phylogenetic and structural diversity in the feline leukemia virus env gene. PLoS One 8:e61009. doi: 10.1371/journal.pone.0061009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tailor CS, Nouri A, Lee CG, Kozak C, Kabat D. 1999. Cloning and characterization of a cell surface receptor for xenotropic and polytropic murine leukemia viruses. Proc Natl Acad Sci U S A 96:927–932. doi: 10.1073/pnas.96.3.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yan Y, Liu Q, Kozak C. 2009. Six host range variants of the xenotropic/polytropic gammaretroviruses define determinants for entry in the XPR1 cell surface receptor. Retrovirology 6:87. doi: 10.1186/1742-4690-6-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martin C, Buckler-White A, Wollenberg K, Kozak CA. 2013. The avian XPR1 gammaretrovirus receptor is under positive selection and is disabled in bird species in contact with virus-infected wild mice. J Virol 87:10094–10104. doi: 10.1128/JVI.01327-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Humes D, Emery S, Laws E, Overbaugh J. 2012. A species-specific amino acid difference in the macaque CD4 receptor restricts replication by global circulating HIV-1 variants representing viruses from recent infection. J Virol 86:12472–12483. doi: 10.1128/JVI.02176-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meyerson NR, Rowley PA, Swan CH, Le DT, Wilkerson GK, Sawyer SL. 2014. Positive selection of primate genes that promote HIV-1 replication. Virology 454–455:291–298. doi: 10.1016/j.virol.2014.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meyerson NR, Sharma A, Wilkerson GK, Overbaugh J, Sawyer SL. 2015. Identification of owl monkey CD4 receptors broadly compatible with early-stage HIV-1 isolates. J Virol 89:8611–8622. doi: 10.1128/JVI.00890-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Payne LN, Brown SR, Bumstead N, Howes K, Frazier JA, Thouless ME. 1991. A novel subgroup of exogenous avian leukosis virus in chickens. J Gen Virol 72:801–807. doi: 10.1099/0022-1317-72-4-801. [DOI] [PubMed] [Google Scholar]
- 12.Sacco MA, Howes K, Smith LP, Nair VK. 2004. Assessing the roles of endogenous retrovirus EAV-HP in avian leukosis virus subgroup J emergence and tolerance. J Virol 78:10525–10535. doi: 10.1128/JVI.78.19.10525-10535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chai N, Bates P. 2006. Na+/H+ exchanger type 1 is a receptor for pathogenic subgroup J avian leukosis virus. Proc Natl Acad Sci U S A 103:5531–5536. doi: 10.1073/pnas.0509785103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Venugopal K, Howes K, Flannery DMJ, Payne LN. 2000. Isolation of acutely transforming subgroup J avian leukosis viruses that induce erythroblastosis and myelocytomatosis. Avian Pathol 29:497–503. doi: 10.1080/030794500750047252. [DOI] [PubMed] [Google Scholar]
- 15.Payne LN, Nair V. 2012. The long view: 40 years of avian leukosis research. Avian Pathol 41:11–19. doi: 10.1080/03079457.2011.646237. [DOI] [PubMed] [Google Scholar]
- 16.Malhotra S, Justice J, Lee N, Li Y, Zavala G, Ruano M, Morgan R, Beemon K. 2015. Complete genome sequence of an American avian leukosis virus subgroup J isolate that causes hemangiomas and myeloid leukosis. Genome Announc 3:e01586-14. doi: 10.1128/genomeA.01586-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang L, Zeng X, Hua Y, Gao Q, Fan Z, Chai H, Wang Q, Qi X, Wang Y, Gao H, Gao Y, Wang X. 2014. Genetic diversity and phylogenetic analysis of glycoprotein gp85 of avian leukosis virus subgroup J wild-bird isolates from Northeast China. Arch Virol 159:1821–1826. doi: 10.1007/s00705-014-2004-8. [DOI] [PubMed] [Google Scholar]
- 18.Zeng X, Liu L, Hao R, Han C. 2014. Detection and molecular characterization of J subgroup avian leukosis virus in wild ducks in China. PLoS One 9:e94980. doi: 10.1371/journal.pone.0094980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Payne LN, Howes K, Gillespie AM, Smith LM. 1992. Host range of Rous sarcoma virus pseudotype RSV(HPRS-103) in 12 avian species: support for a new avian retrovirus envelope subgroup, designated J. J Gen Virol 73:2995–2997. doi: 10.1099/0022-1317-73-11-2995. [DOI] [PubMed] [Google Scholar]
- 20.Kučerová D, Plachý J, Reinišová M, Šenigl F, Trejbalová K, Geryk J, Hejnar J. 2013. Nonconserved tryptophan 38 of the cell surface receptor for subgroup J avian leukosis virus discriminates sensitive from resistant avian species. J Virol 87:8399–8407. doi: 10.1128/JVI.03180-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reinišová M, Plachý J, Kučerová D, Šenigl F, Vinkler M, Hejnar J. 2016. Genetic diversity of NHE1, receptor for subgroup J avian leukosis virus, in domestic chicken and wild anseriform species. PLoS One 11:e0150589. doi: 10.1371/journal.pone.0150589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zingler K, Bélanger C, Peters R, Agard E, Young JAT. 1995. Identification and characterization of the viral interaction determinant of the subgroup A avian leukosis virus receptor. J Virol 69:4261–4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zingler K, Young JAT. 1996. Residue Trp-48 of Tva is critical for viral entry but not for high-affinity binding to the SU glycoprotein of subgroup A avian leukosis and sarcoma viruses. J Virol 70:7510–7516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Halley YA, Dowd SE, Decker JE, Seabury PM, Bhattarai E, Johnson CD, Rollins D, Tizard IR, Brightsmith DJ, Peterson MJ, Taylor JF, Seabury CM. 2014. A draft de novo genome assembly for the northern bobwhite (Colinus virginianus) reveals evidence for a rapid decline in effective population size beginning in the Late Pleistocene. PLoS One 9:e90240. doi: 10.1371/journal.pone.0090240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knauss DJ, Young JAT. 2002. A fifteen-amino-acid TVB peptide serves as a minimal soluble receptor for subgroup B avian leukosis and sarcoma viruses. J Virol 76:5404–5410. doi: 10.1128/JVI.76.11.5404-5410.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adkins HB, Brojatsch J, Young JAT. 2000. Identification and characterization of a shared TNFR-related receptor for subgroup B, D, and E avian leukosis viruses reveal cysteine residues required specifically for subgroup E viral entry. J Virol 74:3572–3578. doi: 10.1128/JVI.74.8.3572-3578.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elleder D, Stepanets V, Melder DC, Šenigl F, Geryk J, Pajer P, Plachý J, Hejnar J, Svoboda J, Federspiel MJ. 2005. The receptor for the subgroup C avian sarcoma and leukosis viruses, Tvc, is related to mammalian butyrophilins, members of the immunoglobulin superfamily. J Virol 79:10408–10419. doi: 10.1128/JVI.79.16.10408-10419.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Munguia A, Federspiel MJ. 2008. Efficient subgroup C avian sarcoma and leukosis virus receptor activity requires the IgV domain of the Tvc receptor and proper display on the cell membrane. J Virol 82:11419–11428. doi: 10.1128/JVI.01408-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dimcheff DE, Drovetski SV, Mindell DP. 2002. Phylogeny of Tetraoninae and other galliform birds using mitochondrial 12S and ND2 genes. Mol Phylogenet Evol 24:203–215. doi: 10.1016/S1055-7903(02)00230-0. [DOI] [PubMed] [Google Scholar]
- 30.Smith EJ, Shi L, Tu Z. 2005. Gallus gallus agrecan gene-based phylogenetic analysis of selected avian groups. Genetica 124:23–32. doi: 10.1007/s10709-004-5184-4. [DOI] [PubMed] [Google Scholar]
- 31.Kaiser VB, van Tuinen M, Ellegren H. 2007. Insertion events of CR1 retrotransposable elements elucidate the phylogenetic branching order in Galliform birds. Mol Biol Evol 24:338–347. doi: 10.1093/molbev/msl164. [DOI] [PubMed] [Google Scholar]
- 32.Reinišová M, Plachý J, Trejbalová K, Šenigl F, Kučerová D, Geryk J, Svoboda J, Hejnar J. 2012. Intronic deletions that disrupt mRNA splicing of the tva receptor gene result in decreased susceptibility to infection by avian sarcoma and leukosis virus subgroup A. J Virol 86:2021–2030. doi: 10.1128/JVI.05771-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu R, Paxton WA, Choe S, Ceradini D, Martin SR, Horuk R, MacDonald ME, Stuhlmann H, Koup RA, Landau NR. 1996. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell 86:367–377. doi: 10.1016/S0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 34.Samson M, Libert F, Doranz BJ, Rucker J, Liesnard C, Farber CM, Saragosti S, Lapoumeroulie C, Cognaux J, Forceille C, Muyldermans G, Verhofstede C, Burtonboy G, Georges M, Imai T, Rana S, Yi Y, Smyth RJ, Collman RG, Doms RW, Vassart G, Parmentier M. 1996. Resistance to HIV-1 infection in Caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature 382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 35.Chen Z, Kwon D, Jin Z, Monard S, Telfer P, Jones MS, Lu CY, Aguilar RF, Ho DD, Marx PA. 1998. Natural infection of a homozygous delta24 CCR5 red-capped mangabey with an R2b-tropic simian immunodeficiency virus. J Exp Med 188:2057–2065. doi: 10.1084/jem.188.11.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palacios E, Digilio L, McClure HM, Chen Z, Marx PA, Goldsmith MA, Grant RM. 1998. Parallel evolution of CCR5-null phenotypes in humans and in a natural host of simian immunodeficiency virus. Curr Biol 8:943–946. doi: 10.1016/S0960-9822(07)00378-8. [DOI] [PubMed] [Google Scholar]
- 37.Riddick NE, Hermann EA, Loftin LM, Elliott ST, Wey WC, Cervasi B, Taaffe J, Engram JC, Li B, Else JG, Li Y, Hahn BH, Derdeyn CA, Sodora DL, Apetrei C, Paiardini M, Silvestri G, Collman RG. 2010. A novel CCR5 mutation common in sooty mangabeys reveals SIVsmm infection of CCR5-null natural hosts and efficient alternative coreceptor use in vivo. PLoS Pathog 6:e1001064. doi: 10.1371/journal.ppat.1001064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nehyba J, Svoboda J, Karakoz I, Geryk J, Hejnar J. 1990. Ducks: a new experimental host system for studying persistent infection with avian leukaemia retroviruses. J Gen Virol 71:1937–1945. doi: 10.1099/0022-1317-71-9-1937. [DOI] [PubMed] [Google Scholar]
- 39.Stepanets V, Vernerová Z, Vilhelmová M, Geryk J, Hejnar J, Svoboda J. 2001. Amyloid A amyloidosis in non-infected and avian leukosis virus-C persistently infected inbred ducks. Avian Pathol 30:33–42. doi: 10.1080/03079450020023177. [DOI] [PubMed] [Google Scholar]
- 40.Dimcheff DE, Drovetski SV, Krishnan M, Mindell DP. 2000. Cospeciation and horizontal transmission of avian sarcoma and leukosis virus gag genes in galliform birds. J Virol 74:3984–3995. doi: 10.1128/JVI.74.9.3984-3995.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dimcheff DE, Krishnan M, Mindell DP. 2001. Evolution and characterization of tetraonine endogenous retrovirus: a new virus related to avian sarcoma and leukosis viruses. J Virol 75:2002–2009. doi: 10.1128/JVI.75.4.2002-2009.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Federspiel MJ, Hughes SH. 1997. Retroviral gene delivery. Methods Cell Biol 52:167–177. [PubMed] [Google Scholar]
- 43.Himly M, Foster DN, Bottoli I, Iacovoni JS, Vogt PK. 1998. The DF-1 chicken fibroblast cell line: transformation induced by diverse oncogenes and cell death resulting from infection by avian leukosis viruses. Virology 248:295–304. doi: 10.1006/viro.1998.9290. [DOI] [PubMed] [Google Scholar]
- 44.Moscovici C, Moscovici MG, Jimenez H, Lai MM, Hayman MJ, Vogt PK. 1977. Continuous tissue culture cell lines derived from chemically induced tumors of Japanese quail. Cell 11:95–103. doi: 10.1016/0092-8674(77)90320-8. [DOI] [PubMed] [Google Scholar]
- 45.Reinišová M, Šenigl F, Yin X, Plachý J, Geryk J, Elleder D, Svoboda J, Federspiel MJ, Hejnar J. 2008. A single-amino-acid substitution in the TvbS1 receptor results in decreased susceptibility to infection by avian sarcoma and leukosis virus subgroups B and D and resistance to infection by subgroup E in vitro and in vivo. J Virol 82:2097–2105. doi: 10.1128/JVI.02206-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis, version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 48.Pond SLK, Frost SDW. 2005. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics 21:2531–2533. doi: 10.1093/bioinformatics/bti320. [DOI] [PubMed] [Google Scholar]
- 49.Yang ZH, Wong WSW, Nielsen R. 2005. Bayes empirical Bayes inference of amino acid sites under positive selection. Mol Biol Evol 22:1107–1118. doi: 10.1093/molbev/msi097. [DOI] [PubMed] [Google Scholar]
- 50.Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]