ABSTRACT
Porcine reproductive and respiratory syndrome (PRRS) has become an economically critical factor in swine industry since its worldwide spread in the 1990s. Infection by its causative agent, PRRS virus (PRRSV), was proven to be mediated by an indispensable receptor, porcine CD163 (pCD163), and the fifth scavenger receptor cysteine-rich domain (SRCR5) is essential for virus infection. However, the structural details and specific residues of pCD163 SRCR5 involved in infection have not been defined yet. In this study, we prepared recombinant pCD163 SRCR5 in Drosophila melanogaster Schneider 2 (S2) cells and determined its crystal structure at a high resolution of 2.0 Å. This structure includes a markedly long loop region and shows a special electrostatic potential, and these are significantly different from those of other members of the scavenger receptor cysteine-rich superfamily (SRCR-SF). Subsequently, we carried out structure-based mutational studies to identify that the arginine residue at position 561 (Arg561) in the long loop region is important for PRRSV infection. Further, we showed Arg561 probably takes effect on the binding of pCD163 to PRRSV during virus invasion. Altogether the current work provides the first view of the CD163 SRCR domain, expands our knowledge of the invasion mechanism of PRRSV, and supports a molecular basis for prevention and control of the virus.
IMPORTANCE PRRS has caused huge economic losses to pig farming. The syndrome is caused by PRRSV, and PRRSV infection has been shown to be mediated by host cell surface receptors. One of them, pCD163, is especially indispensable, and its SRCR5 domain has been further demonstrated to play a significant role in virus infection. However, its structural details and the residues involved in infection are unknown. In this study, we determined the crystal structure of pCD163 SRCR5 and then carried out site-directed mutational studies based on the crystal structure to elucidate which residue is important. Our work not only provides structural information on the CD163 SRCR domain for the first time but also indicates the molecular mechanism of PRRSV infection and lays a foundation for future applications in prevention and control of PRRS.
KEYWORDS: CD163, Drosophila S2 cells, PRRSV, SRCR5, crystal structure
INTRODUCTION
Porcine reproductive and respiratory syndrome (PRRS) was first reported in 1987 in the United States (1) and has become a critical economic factor in the swine industry since its worldwide spread in the 1990s (2, 3). PRRS is characterized by reproductive failures in the late-term gestation of sows and respiratory symptoms in pigs of all ages (4–6). Its causative agent, the PRRS virus (PRRSV), was initially isolated in Europe and known as the Lelystad virus, and later it was identified in the United States with the designation VR-2332 (7–9). In 2006, highly pathogenic variants of PRRSV (HP-PRRSV) emerged in China, leading to a devastating fall in pig production throughout the country (10, 11). PRRSV is an enveloped virus containing a single-stranded positive-sense RNA genome of approximately 15 kb (12, 13) and is classified into the order Nidovirales, family Arteriviridae, genus Arterivirus, along with equine arteritis virus (EAV), lactate dehydrogenase-elevating virus of mice (LDV), and simian hemorrhagic fever virus (SHFV) (14–16).
Swine are the only known host, and the differentiated monocytes, particularly porcine alveolar macrophages (PAMs), are the primary target cells for PRRSV (17). In addition, the African green monkey kidney epithelial cell line MA-104 and its derivative, MARC-145, are susceptible to virus infection in vitro (18). PRRSV infection has been further proven to be mediated by the host cell surface receptors (19–21). To date, six putative receptors associated with PRRSV infectivity have been proposed (21), including heparin sulfate (HS) (22), sialoadhesin (Sn/cluster of differentiation 169; CD169) (23), CD163 (24), CD151 (25), vimentin (26), and DC-SIGN (dendritic cell-specific intercellular adhesion molecule-3-grabbing nonintegrin; CD209) (27). An earlier study demonstrated that CD163 confers susceptibility to PRRSV in various cell lines (24). A recent study using gene-edited pigs via the CRISPR-Cas9 system shows that pigs lacking functional porcine CD163 (pCD163) are resistant to PRRSV infection, confirming that it is an indispensable receptor (28).
CD163 is an endocytic receptor in monocytes/macrophages and belongs to class B of the scavenger receptor cysteine-rich (SRCR) superfamily (SRCR-SF) (29, 30). It is structurally a type I membrane protein, consisting of a large extracellular region, a single transmembrane domain, and a short cytoplasmic tail (Fig. 1A). In its extracellular region, there are nine SRCR domains (SRCR1 to -9), separated by two proline-serine-threonine (PST)-rich motifs at a location between SRCR6 and SRCR7 and the end of extracellular region, respectively (31). The fifth SRCR domain (SRCR5) in pCD163 is shown to be essential for PRRSV infection, while the first four N-terminal SRCR domains (SRCR1 to 4) and the cytoplasmic tail are not (32). The remaining SRCR domains (SRCR6 to 9) also may contribute to PRRSV infectivity (32). However, the structural details of pCD163, especially the crucial domain SRCR5, and its specific residues involved in PRRSV infection are not clear yet, hindering our deep understanding of virus pathogenesis.
FIG 1.
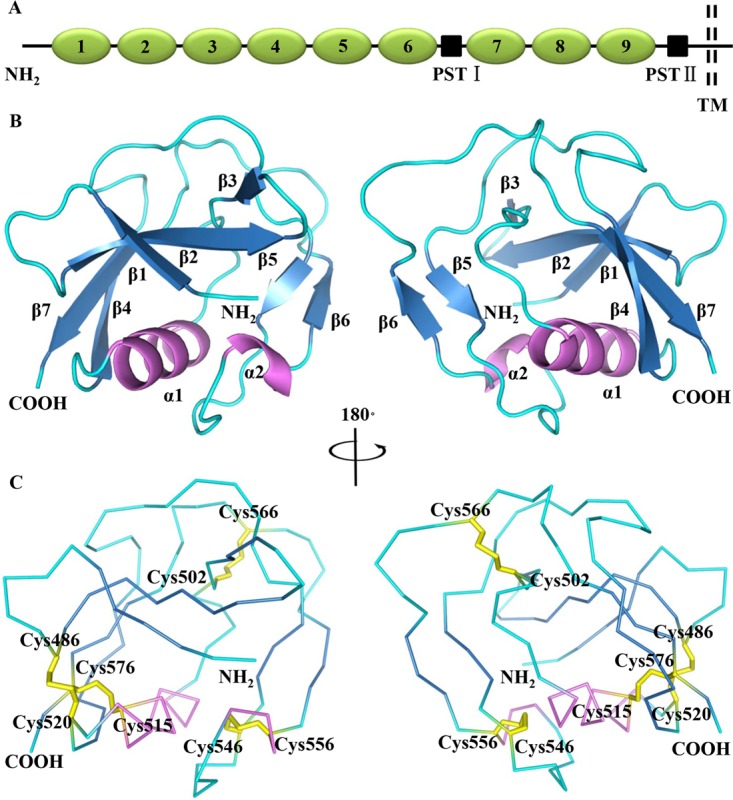
Crystal structure of the fifth scavenger receptor cysteine-rich domain (SRCR5) from porcine CD163 (pCD163). (A) Schematic diagram of pCD163 architecture. pCD163 consists of nine SRCR domains (1 to 9), two proline-serine-threonine (PST)-rich motifs (I and II), a transmembrane (TM) domain, and a cytoplasmic tail. (B) Cartoon diagrams of pCD163 SRCR5 represented in the 180° rotation. The β-strands from β1 to β7 are colored in blue, two helices (α1 and α2) are in violet, and the loop regions are in cyan. The N terminus and the C terminus are also labeled. (C) Ribbon diagrams of pCD163 SRCR5 showing the disulfide bonds represented in the 180° rotation. The disulfide bonds are colored in yellow and the bridged cysteines are labeled.
Here, we expressed recombinant pCD163 SRCR5 in Drosophila melanogaster Schneider 2 (S2) cells. After purification and crystallization, we determined its structure at a high resolution. We then performed structure-based site-directed mutagenesis to elucidate a charged residue important for PRRSV infection. Further, we showed how this residue influenced infection during virus invasion.
RESULTS
Crystal structure of pCD163 SRCR5.
The purified pCD163 SRCR5 exists as an apparent monomer based on its elution volume. Its crystal structure was determined and refined to 2.0 Å in a C2221 space group (Table 1). pCD163 SRCR5 (Pro477-Ser577) adopts a compact heart shape of approximate dimensions of 37 by 35 by 30 Å. As shown in Fig. 1B, the target protein starts with three curved antiparallel β-strands: a long β-strand (β2; Ser487-His494) lies between two short β-strands (β1, Pro477-Val480; β3, Thr497-Gly499), followed by a single α-helix (α1; Leu508-Leu518) and a long β-strand (β4; Thr522-Leu527). The structure extends with a markedly long loop region containing two short β-strands (β5, Glu542-Phe544; β6, Val558-Pro560) and one short 310-helix (α2; Leu553-Leu555) located between the β-strands β5 and β6. It finally ends with a C-terminal long β-strand, β7 (Gly573-Ser577). Altogether, β-strands 1 to 4 and 7 form a twisted five-stranded antiparallel β-sheet, and the β-strands 5 and 6 form a second antiparallel one. The β-strands 1 to 4 and 7 wrap around one side of the α1-helix, and the long loop region surrounds the other.
TABLE 1.
X-ray data collection and model refinement statistics for the crystal structure of pCD163 SRCR5
| Parameter | Value(s) for pCD163 SRCR5e |
|---|---|
| PDB code | 5JFB |
| X-ray source wavelength (Å) | 0.979 |
| Resolution limits (Å) | 50.0–2.0 (2.07–2.0) |
| Space group | C2221 |
| Temp of expt (K) | 100 |
| Cell constants | a = 29.71 Å, b = 73.93 Å, c = 87.53 Å, α = β = γ = 90° |
| Completeness (%) | 97.1 (95.9) |
| Multiplicity | 10.0 (10.6) |
| Rmergea | 0.107 (0.470) |
| No. of observations | 66,922 |
| No. of unique reflections | 6,682 |
| Refinement data | |
| R | 0.208 |
| Rfree | 0.239 |
| Avg B-factor (Å2) | 23.3 |
| RMSD | |
| Bond lengths (Å2) | 0.008 |
| Angle (°) | 1.28 |
| Ramachandran analysis (%) | 97.0,b 3.0,c 0d |
Rmerge = Σ|Ii − <I>|/ΣIi, where Ii is the intensity of the observation and <I> is the mean intensity of the reflections.
Percentage of residues in most favored regions.
Percentage of residues in additional allowed regions.
Percentage of residues in disallowed regions.
Values of the highest-resolution shell are shown in parentheses.
In pCD163 SRCR5, Cys486 and Cys520 form a disulfide bond linking the N terminus of the strand β2 with the C terminus of the α1-helix, Cys515 in the middle of the α1-helix connects to the C-terminal Cys576 of strand β7, and Cys502 and Cys566 in association with Cys546 and Cys556 bridge into two disulfide linkages to restrict loop flexibility (Fig. 1C). These four disulfide bridges are consistent with the typical disulfide linkage pattern (C1-C4, C2-C7, C3-C8, and C5-C6, where “C” is short for cysteine) in the SRCR-SF, confirming the authenticity of the crystal structure (29).
Surface electrostatic potential of pCD163 SRCR5.
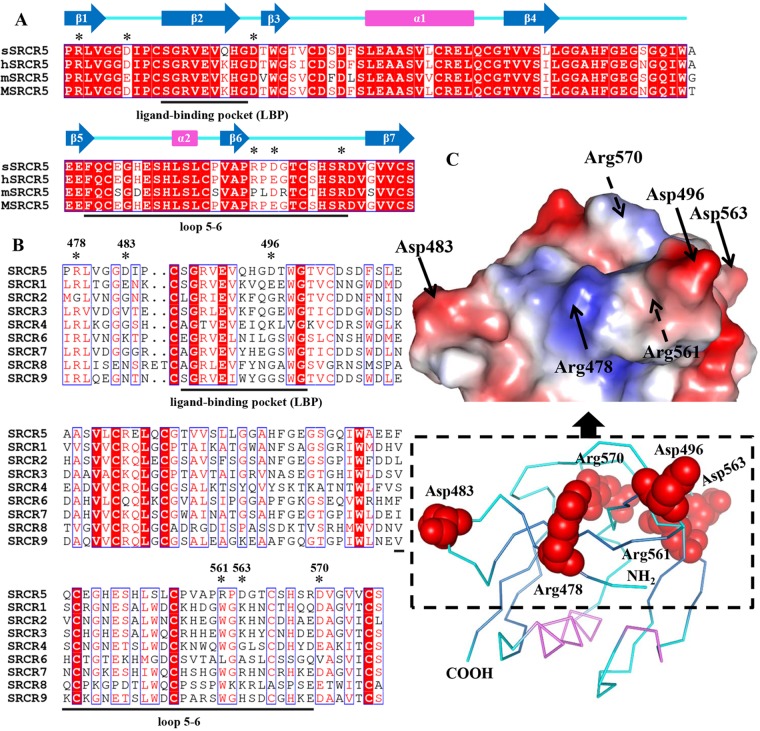
pCD163 SRCR5 includes 14 acidic residues (aspartic acids or glutamic acids) and five basic residues (only arginines). The surface electrostatic potential shows some exterior residues arranged in a special positively and negatively charged distribution. In Fig. 2, basic residues (Arg478 and Arg489, Arg516, and Arg561 and Arg570) are organized into three obvious positively charged clusters (named I, II, and III) situated in the β-strands 1 and 2, the helix α1, and the long loop region (the loop between β-strands 6 and 7), respectively. In contrast, the acidic residues form a continuous negatively charged area (Fig. 2). Interestingly, the basic residues in positively charged clusters and some acidic residues in the negatively charged area jointly constitute two D/E-R-rich regions (D/E-R1 and D/E-R2 in Fig. 2). These two mixed charged regions may form strong electrostatic interactions with their corresponding ligands.
FIG 2.
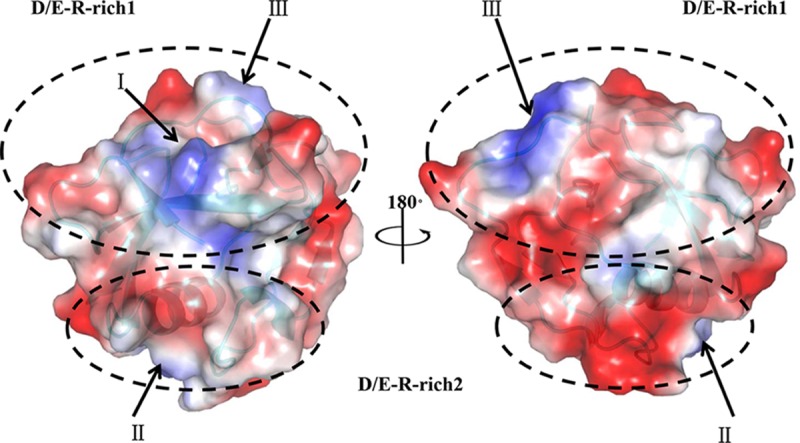
Surface electrostatic potential of pCD163 SRCR5. The images are represented in the 180° rotation to depict basic residues (arginines) in the positively charged clusters (blue) and acidic residues (aspartic acids or glutamic acids) in the negatively charged areas (red). Three positively charged clusters (named I, II, and III) are indicated with arrows. Two D/E-R-rich regions, where D/E represents aspartic acid or glutamic acid and R is short for arginine, are circled in dashed lines. The electrostatic potential is colored from −62 to +62 kiloteslas/charge.
Structural comparison of pCD163 SRCR5 with other members in SRCR-SF.
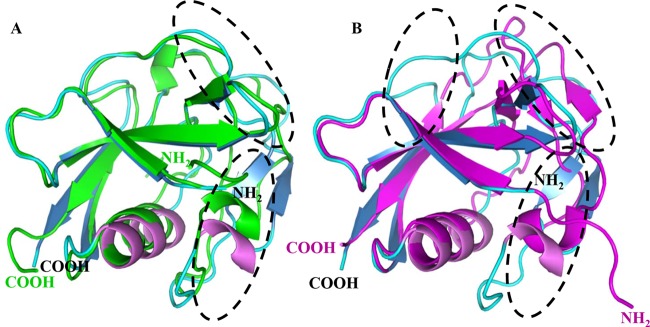
To our knowledge, there are limited structural studies for members of SRCR-SF (33–37). We aligned the structure of pCD163 SRCR5 with that of the Mac-2 binding protein (M2BP) SRCR domain from class A (33) and the third SRCR domain of human CD5 (hCD5 SRCR3) from class B (35), whose crystal structures were first solved into respective classes of SRCR-SF. The primary structural differences between pCD163 SRCR5 and M2BP SRCR domains are the number of cysteines (disulfide bonds) and their structural compositions (33). The crystal structure of pCD163 SRCR5 contains eight cysteines (four disulfide bonds), seven β-strands, and two helices, which are different from its structural template, the M2BP SRCR domain. In the M2BP SRCR domain, there are six cysteines (three disulfide bonds), five β-strands, and three helices (Fig. 3A). A close comparison of pCD163 SRCR5 and M2BP SRCR domains reveals that the root mean square deviation (RMSD) differences are not significant (0.787 Å for 74 matching Cα atoms), suggesting they have similar structural folds. The major differences between them are observed in the long loop region, where the M2BP SRCR domain represents the loop region in an opposed direction. Interestingly, there are more differences between pCD163 SRCR5 and hCD5 SRCR3 in structure. For example, although they both contain four disulfide bonds, seven β-strands, and two helices, pCD163 SRCR5 shows some differences from hCD5 SRCR3 at the N terminus and in the long loop region (only 33 matching Cα atoms) (Fig. 3B) (35). hCD5 SRCR3 has an extended N terminus and displays inverted loop flexibility. All of these differences, especially the long loop region, may contribute to the receptor specificity of pCD163 for PRRSV infection.
FIG 3.
Structural comparison of pCD163 SRCR5 with selected members in SRCR-SF. Crystal structures of pCD163 SRCR5, Mac-2 binding protein (M2BP) SRCR domain (PDB code 1BY2) (A), and the third SRCR domain of human CD5 (hCD5 SRCR3; PDB code 2JA4) (B) were aligned in cartoon diagrams. The pCD163 SRCR5 domain is colored as described for Fig. 1B, the M2BP SRCR domain is in green, and the hCD5 SRCR3 is in magenta. Their N and C termini are labeled, and their differences are circled in dashed lines.
Site-directed mutagenesis of SRCR5 in complete pCD163.
Based on the crystal structure (Fig. 1), we aligned amino acid sequences of CD163 SRCR5 from different species (pig, human, mouse, and monkey; Fig. 4A) as well as the nine SRCR domains of pCD163 (Fig. 4B). As shown in Fig. 4, there are some residues that are conserved in the SRCR5 domain from different species while being variable in nine SRCR domains of pCD163. In this study, we chose charged Arg478, Asp483, Asp496, Arg561, Asp563, and Arg570 (Fig. 4C). We then mutated each residue to the neutral alanine (R478A, D483A, D496A, R561A, D563A, and R570A) and expressed each single-site-mutated pCD163 in PK-15 cells.
FIG 4.
Structure-based sequence alignment of the CD163 SRCR5 from different species and the nine SRCR domains of pCD163. (A) Sequence alignment of the CD163 SRCR5 from different species. The amino acid sequences of CD163 SRCR5 from pig (p; UniProt entry Q2VL90), human (h; UniProt entry Q86VB7), mouse (m; UniProt entry Q2VLH6), and monkey (M; UniProt entry Q2VLG4) are aligned. The amino acids within the so-called loop5-6 region and ligand-binding pocket (LBP) are underlined. Conserved charged residues in these two regions are colored red, and the mutated residues are marked by asterisks. The secondary structural elements of pCD163 SRCR5 are indicated above the aligned sequences. The blue arrows represent the β-strands from β1 to β7, and the violet rectangles represent two helices (α1 and α2). (B) Sequence alignment of the nine SRCR domains of pCD163 (UniProt entry Q2VL90). The amino acids within the so-called loop5-6 region and LBP are underlined. Conserved charged residues in these two regions are colored red, and the mutated residues are marked by asterisks. (C) Site-directed mutagenesis of pCD163 SRCR5. The ribbon diagram and the surface electrostatic potential map of pCD163 SRCR5 are shown with the mutated residues. The mutated residues are labeled as red spheres, and the N and C termini are labeled in the ribbon diagram. The mutated residues are also indicated with arrows in the surface electrostatic potential map, where the solid arrows indicate visible residues and the dashed arrows indicate ones not visible from the front view. The electrostatic potential is colored from −62 to +62 kiloteslas/charge.
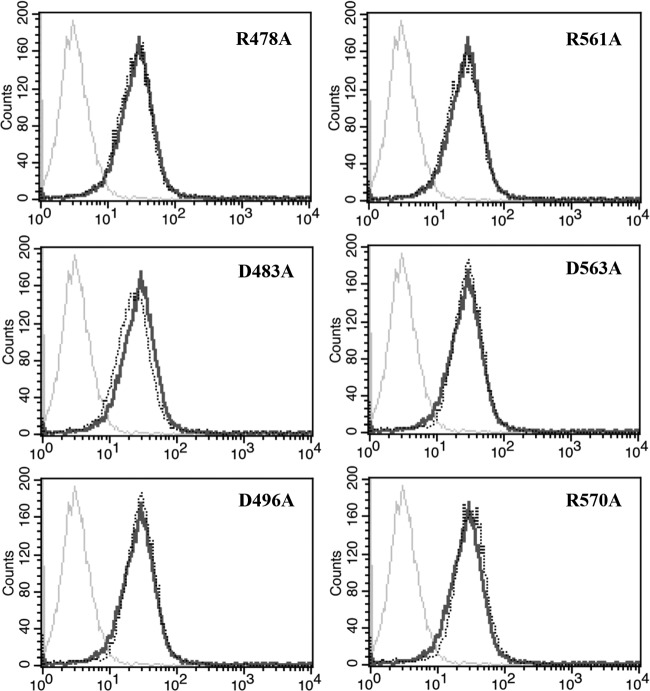
As shown in Fig. 5, PK-15 cells did not express pCD163 at all. After transfection, wild-type pCD163 was expressed at the highest level at 24 h and expressed stably up to 48 h. Thus, we compared all mutated receptors to wild-type pCD163 at 24 h posttransfection. In Fig. 6, all single-site-mutated pCD163 receptors were expressed at almost the same level as the wild-type one, ruling out the influence of the receptor expression level in our subsequent experiments.
FIG 5.
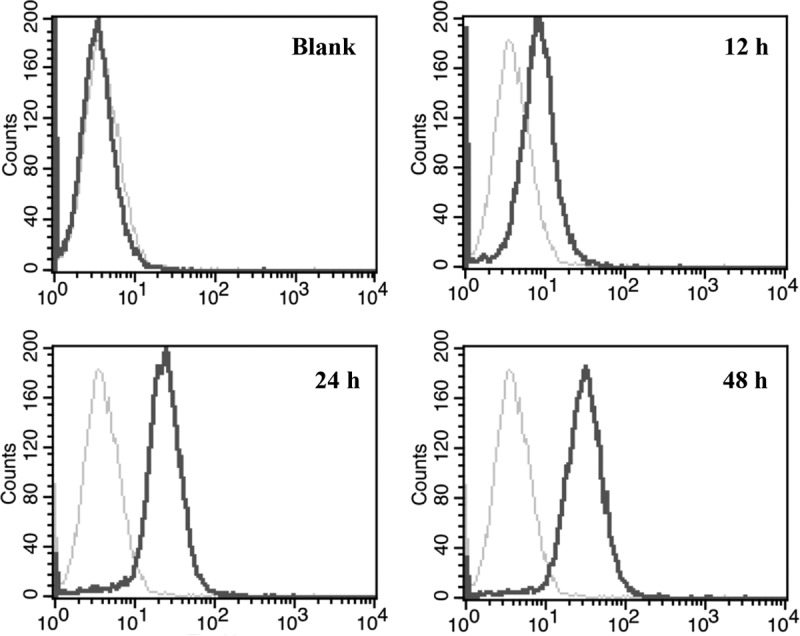
Flow cytometry analysis of wild-type pCD163 expression. To analyze wild-type pCD163 at the indicated time points (12 h, 24 h, and 48 h) posttransfection, the transfected PK-15 cells were incubated with a commercial mouse anti-pCD163 monoclonal antibody (MCA2311GA; AbD Serotec) or isotype control antibody and then incubated with Cy3-conjugated goat anti-mouse IgG (H+L) (Abbkine). Gray lines stand for the isotype antibody controls, and black lines stand for the wild-type receptor stained with anti-pCD163 antibody. The blank PK-15 cells were assayed in parallel as a negative control.
FIG 6.
Flow cytometry analysis of wild-type and mutant pCD163 expression. Expression of wild-type pCD163 and mutant pCD163 at 24 h posttransfection was analyzed. Transfected PK-15 cells were incubated with a commercial mouse anti-pCD163 monoclonal antibody (MCA2311GA; AbD Serotec) or isotype control antibody and then incubated with Cy3-conjugated goat anti-mouse IgG (H+L) (Abbkine). Gray lines stand for the isotype antibody controls, black lines for wild-type receptor, and black dotted lines for mutated receptor stained with anti-pCD163 antibody.
Identification of the specific residues in SRCR5 of pCD163 for PRRSV infection.
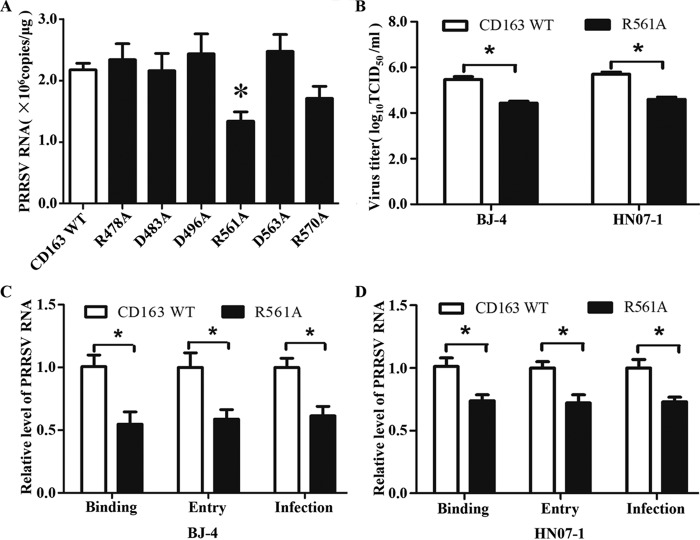
We next detected the effect of mutated residues on PRRSV infection to identify the specific residues. In Fig. 7A, the infected cells bearing a mutated receptor at Asp483 showed viral RNA production almost identical to that of the cells with the wild-type one, suggesting that this residue is not involved in PRRSV infection. Although there was fluctuation, the infected cells with a mutated receptor at Arg478, Asp496, Asp563, or Arg570 showed no significant difference in viral RNA production compared to the cells with the wild-type one (P > 0.05). In contrast, the infected cells with site-directed mutagenesis at Arg561 showed a significant reduction in viral RNA production (almost 50% compared to the wild-type one; P < 0.05). All of these results demonstrate Arg561 is an important residue for PRRSV infection.
FIG 7.
Identification of the specific residues in SRCR5 of pCD163 for PRRSV infection. (A) Real-time PCR analysis of PRRSV infection. The wild type or each mutant pCD163 was transfected into PK-15 cells. After 24 h, the transfected PK-15 cells were inoculated with PRRSV strain BJ-4 at a multiplicity of infection (MOI) of 1 for 12 h. Total RNA from infected PK-15 cells was then extracted, and quantitation of viral RNA was carried out. Data represent means ± SEM from three independent experiments. An asterisk indicates significantly reduced viral RNA production in the mutant pCD163 experimental group compared to the wild-type one (P < 0.05). (B) PRRSV titration assay with mutation at Arg561 in pCD163. Wild-type or R561A mutant pCD163 was transfected into PK-15 cells. After 24 h, the transfected PK-15 cells were inoculated with PRRSV strain BJ-4 or HN07-1 at an MOI of 1 for 48 h. The virus yields were measured by TCID50 assay in MARC-145 cells. Data represent means ± SEM. An asterisk indicates significantly reduced viral titer in the mutant pCD163 experimental group compared to the wild-type one (P < 0.05). (C and D) The binding and entry assays with mutated pCD163 (R561A) for PRRSV BJ-4 and HN07-1, respectively. Wild-type or mutant pCD163 was transfected into PK-15 cells. After 24 h, the transfected PK-15 cells were inoculated with PRRSV strain BJ-4 or HN07-1 at an MOI of 1, and relative quantitation of viral RNA was carried out. Data represent means ± SEM from three independent experiments. An asterisk indicates significantly reduced viral RNA production in the mutant pCD163 experimental group compared to the wild one (P < 0.05).
Analysis of Arg561 for PRRSV infection.
To further demonstrate the importance of Arg561 for PRRSV infection, we carried out PRRSV titration assay with a typical North American (VR2332)-like PRRSV strain, BJ-4, and an HP-PRRSV strain, HN07-1. As seen in Fig. 7B, the mutated receptor of R561A showed a 10-fold decrease for both BJ-4 and HN07-1 production (by almost 1.0 log10; P < 0.05) compared to that of the wild-type one.
We next performed the PRRSV invasion assay to explore how Arg561 influenced virus infection according to previous studies (38, 39). For PRRSV binding assay (Fig. 7C, left), the cells infected by BJ-4 bearing the mutated receptor at Arg561 showed significantly less viral RNA production than those bearing the wild-type one (almost 50% compared to wild-type pCD163; P < 0.05). More importantly, it showed nearly the same reduction ratio as that in PRRSV infection (Fig. 7C, right). For the PRRSV BJ-4 entry assay (Fig. 7C, middle), the mutated receptor at Arg561 showed reduction almost identical to that of PRRSV binding and infection. For HN07-1, the PRRSV binding and entry assays showed similar results (Fig. 7D). Taken together, these results demonstrate Arg561 is important for PRRSV invasion and probably takes effect on the binding of pCD163 to PRRSV during virus infection.
DISCUSSION
Earlier studies showed that pCD163 cooperated with another putative receptor, porcine sialoadhesin (Sn/CD169) (23), to facilitate the PRRSV infection, where the former contributed to the uncoating of PRRSV while the latter promoted the attachment and internalization (invasion) of the virus (40–48). However, one study showed that Sn gene knockout pigs were infected by PRRSV as usual (49), and a recent study using gene-edited pigs demonstrates the indispensability of pCD163 (28). Therefore, pCD163 is a core receptor for PRRSV infection and requires more in-depth research. Despite various studies, there is still a lack of structural details of pCD163, particularly its crucial SRCR5 domain and the specific residues in virus infection. As a result, we carried out our current work with pCD163 SRCR5. Since the low pH has been proven to be required for the entry of PRRSV into the target cells (19, 50), our crystal structure under acidic conditions may represent the functional form of pCD163 SRCR5. Using structure-based mutational research, we then elucidated that Arg561 is an important residue for PRRSV infection and probably takes effect on pCD163 binding to the virus during invasion. To our knowledge, this is the first time these results have been reported in the literature.
Specifically, previous studies demonstrated that the minor envelope glycoproteins GP2 and GP4 of PRRSV interacted with pCD163 (51, 52). The glycosylation of the GP2 and GP4 proteins was also shown to be required for efficient PRRSV interaction with the receptor (53, 54). However, later studies demonstrated that only PRRSV GP4 protein interacted with pCD163 (55), and the glycosylation of GP4 might not play a vital role in this interaction (55, 56). Therefore, PRRSV binding to pCD163 may depend on the amino acid-mediated protein-protein contact. According to previous reports, two regions of pCD163 SRCR5 were speculated to be involved in pCD163-PRRSV interaction (57). One was so-called loop5-6 (58), corresponding to the region from Phe544 to Arg570 in the long loop region of pCD163 SRCR5. A second one was ligand-binding pocket (LBP) (31), and it equals the region covering the strands β2 and β3 in SRCR5 of pCD163. Since CD163 from different species can confer susceptibility to PRRSV (24) and SRCR5 is a crucial domain in the receptor (32), we displayed these two regions in alignment of CD163 SRCR5 from different species (pig, human, mouse, and monkey) and the nine SRCR domains of pCD163 based on our crystal structure (Fig. 4). There are actually some residues arousing our interest. As charged residues may form strong interactions and pCD163 SRCR5 shows a special electrostatic potential (Fig. 2), we chose Arg561, Asp563, and Arg570 from the loop5-6 region, Asp496 in the LBP, and Arg478 and Asp483 in other regions (Fig. 4). We mutated these residues to the neutral alanine and expressed each single-site-mutated pCD163 receptor in PK-15 cells. This cell line was chosen because native PK-15 cells are nonpermissive to PRRSV, while after transfection with pCD163 they support the virus infection and thus are ideal cells for the assay (59). Thereafter we demonstrated that Arg561 is an important residue for PRRSV infection and probably takes effect on pCD163 binding to PRRSV during virus invasion. It was possible that the R561A mutation affects the overall fold, which could affect infection. However, we think that this is unlikely for several reasons. Arg561 is located at a loop region and is flexible, as shown by its high averaged B factor of 20.4 versus 15.8 for the whole. In addition, Arg561 has no interaction with adjacent residues. Moreover, another mutation at Asp563, which is around Arg561 and lies in this flexible loop region, has no effect on PRRSV infection. We have generated the structure with mutation R561A based on the homology modeling method and aligned it with the wild-type one and find no great difference between them (data not shown). Therefore, we believe the R561A mutant is not likely to affect the overall fold. We speculate that Arg561 is involved in a site for binding to PRRSV through electrostatic interaction, and mutation of Arg561 to alanine may disrupt this interaction. We have tried to provide structural data for the complex of pCD163 SRCR5 and PRRSV glycoprotein in hopes of elucidating the actual determinants of the virus-receptor interaction. Unfortunately, we have not obtained their complexes yet. Nonetheless, our work may still shed some light on the interaction between pCD163 and PRRSV.
Indeed, there are some issues left unsolved in the current work. The first one is that only six charged residues in SRCR5 of pCD163 were tested, and PRRSV RNA production or viral titers still remain high with mutation at Arg561. Hence, Arg561 may well be one of several residues contributing to PRRSV infection, and whether remaining charged residues and other neutral residues take effect needs to be determined. A second one is that we supported the structural details of the domain SRCR5, just one out of nine SRCR domains in pCD163. Since the SRCR6 to 9 domains also contribute to pCD163 binding to PRRSV (32, 60), the mechanism of how domains SRCR6 to 9 or SRCR5 to 9 interact with PRRSV demands further exploration. Third, since they share highly conserved amino acid sequences, CD163 SRCR5 from different species may possess a similar structure, and thus the host specificity for PRRSV could not be explained just from the receptor structure. Many other complicated issues in vivo, such as immune surveillance and escape, perhaps are involved in host specificity and need to be addressed in the future. In fact, structural studies on SRCR5 to 9 domains together with site-directed mutagenesis of more residues in pCD163 are now being processed in our laboratory. Trials to obtain complexes of pCD163 and PRRSV glycoprotein(s) are being carried out as well.
In addition to mediating PRRSV infection, CD163 is a well-documented multifunctional receptor (31). In humans, leaked hemoglobin is internalized by CD163 in the case of heme oxidative stress under pathological conditions (58, 61). CD163 also serves as an adhesion receptor for erythroblasts (62) and a receptor for tumor necrosis factor-like weak inducer of apoptosis (TWEAK) (63, 64). Moreover, it functions as a signaling sensor for bacteria (65) and receptors for other viruses, such as African swine fever virus (ASFV) (66) and SHFV (67). These various functions have been shown to be fulfilled by different SRCR domains of CD163 (31). However, there are few structural studies to illuminate them.
We speculate that the lack of structural studies on CD163 arises from the difficulty in preparing the large target protein, which contains many disulfide bonds and glycosylation sites. As a result, we utilized a Drosophila expression system to generate recombinant pCD163 SRCR5 as a secreted protein. We chose this eukaryotic expression system to ensure the proper folding of the disulfide bonds in pCD163 SRCR5, and Drosophila S2 cells are easy to culture in suspension without serum and CO2. In addition, these cells resolve a number of problems in prokaryotic expression systems or mammalian cell expression systems with proper posttranslational modifications and high expression levels (68). The yield of the target protein was up to 20 mg/liter culture medium in the current study, and our crystal structure actually provides the first structural view of the CD163 SRCR domain, as demonstrated by the correct formation of disulfide bonds. As a consequence, it may provide a structural template for CD163 to elucidate its various functions and support structural information for other members in SRCR-SF.
In conclusion, we have provided the crystal structure of pCD163 SRCR5 and an important residue involved in PRRSV infection in our study. The results we obtained actually deepen our understanding of the invasion mechanism of PRRSV, and findings with the specific residue Arg561 support data for the development of drugs and vaccines against PRRSV from the receptor perspective by interrupting PRRSV infection. Furthermore, this work may support genome-edited implications for pCD163 modification to select pigs nonpermissive to PRRSV and substantially reduce the economic losses it causes in the future.
MATERIALS AND METHODS
Materials, cell lines, and viruses.
All chemicals were purchased from Sigma-Aldrich Co. Ltd. (St. Louis, MO, USA) or Sinopharm Chemical Reagent Co. Ltd. (Shanghai, China) unless otherwise stated.
The cell lines used in the current study were Drosophila melanogaster Schneider 2 (S2) cells, porcine kidney PK-15 cells, and MARC-145 cells. The Drosophila S2 cells were kept in Schneider's insect medium supplemented with 10% heat-inactivated fetal bovine serum (FBS; Gibco) and antibiotics (100 U/ml penicillin, 100 μg/ml streptomycin; Gibco) at 28°C in a humidified incubator (69). The PK-15 cells and MARC-145 cells (kept in our laboratory) were maintained routinely in Gibco Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% heat-inactivated FBS and antibiotics at 37°C in a humidified incubator with a 5% CO2 atmosphere.
A typical North American (VR2332)-like PRRSV strain BJ-4 (GenBank accession no. AF331831, isolated in 1996 in China and kindly provided by Hanchun Yang of China Agricultural University, China) and HP-PRRSV strain HN07-1 (isolated during an HP-PRRSV outbreak in Henan province of China in 2007 by our laboratory) were used in this study and prepared according to our laboratory's previous study (70).
Construction of pCD163 SRCR5 expression vector.
The cDNA encoding the pCD163 SRCR5 domain (residues Pro477 to Ser577; the numbering is according to UniProt entry Q2VL90) was amplified from the full-length pCD163 cDNA (GenBank accession no. JX292263) by sticky-end PCR using the primers listed in Table 2. The amplified PCR product was isolated and inserted into the expression vector pMT/BiP/V5-HisA of the Drosophila expression system (Invitrogen) between the BglII and MluI sites. The construct was transformed into competent Escherichia coli strain DH5α and was then screened on a Luria-Bertani (LB) plate (0.5% yeast extract, 1% tryptone, 1% NaCl, 1.5% agar) containing 100 μg/ml ampicillin to select positive colonies. Following selection by colony PCR, the recombinant pCD163 SRCR5 expression vector was verified by sequencing at Shanghai Sangon Biotech Co. Ltd. (Shanghai, China).
TABLE 2.
Primers for PCR, site-directed mutagenesis, and real-time PCR in this study
| Primer name and function | Sequencea |
|---|---|
| Primers for PCR | |
| pCD163-SRCR5, forward-1 | GATCTCCCAGGCTGGTTGGAGG |
| pCD163-SRCR5, forward-2 | TCCCAGGCTGGTTGGAGG |
| pCD163-SRCR5, reverse | CGACGCGTTGAGCAGACTACGCCGACG |
| Site-directed mutagenesis | |
| pCD163-R478A, forward | GCTCAGCCCACAGGAAACCCgccCTGGTTGGAGGGGACATTC |
| pCD163-R478A, reverse | GAATGTCCCCTCCAACCAGggcGGGTTTCCTGTGGGCTGAGC |
| pCD163-D483A, forward | CCCAGGCTGGTTGGAGGGgccATTCCCTGCTCTGGTCGTG |
| pCD163-D483A, reverse | CCCAGGCTGGTTGGAGGggcCATTCCCTGCTCTGGTCGTG |
| pCD163-D496A, forward | GTTGAAGTACAACATGGAgccACGTGGGGCACCGTCTGTG |
| pCD163-D496A, reverse | CACAGACGGTGCCCCACGTggcTCCATGTTGTACTTCAAC |
| pCD163-R561A, forward | CACTCTGCCCAGTAGCACCCgccCCTGACGGGACATGTAGC |
| pCD163-R561A, reverse | GCTACATGTCCCGTCAGGggcGGGTGCTACTGGGCAGAGTG |
| pCD163-D563A, forward | CCCAGTAGCACCCCGCCCTgccGGGACATGTAGCCACAGCAG |
| pCD163-D563A, reverse | CTGCTGTGGCTACATGTCCCggcAGGGCGGGGTGCTACTGGG |
| pCD163-R570A, forward | CGGGACATGTAGCCACAGCgccGACGTCGGCGTAGTCTGCTC |
| pCD163-R570A, reverse | GAGCAGACTACGCCGACGTCggcGCTGTGGCTACATGTCCCG |
| Quantitative real-time PCR | |
| PRRSV-ORF7, forward | AAACCAGTCCAGAGGCAAGG |
| PRRSV-ORF7, reverse | GCAAACTAAACTCCACAGTGTAA |
| GAPDH, forward | CCTTCCGTGTCCCTACTGCCAAC |
| GAPDH, reverse | GACGCCTGCTTCACCACCTTCT |
The boldface letters indicate restriction sites, and the lowercase letters indicate mutated sites.
Expression of recombinant pCD163 SRCR5.
Drosophila S2 cells were transfected with recombinant pCD163 SRCR5 expression vector and the pCoBlast vector by Cellfectin II reagent according to the manufacturer's instructions (Invitrogen). Stably transfected S2 cells were selected in Schneider's insect medium supplemented with 10% FBS, antibiotics, and 25 μg/ml blasticidin S (Invitrogen). The stably transfected pCD163 SRCR5-S2 cells were then scaled up in Sf-900II serum-free medium (Invitrogen) and induced by the addition of a 0.75 mM final concentration of CuSO4 when the cell density reached 4.0 × 106 cells/ml. The culture supernatant was harvested by centrifugation at 200 × g at 4°C for 10 min and then adjusted to pH 7.4, followed by a second centrifugation at 12,000 × g at 4°C for 30 min.
Purification of pCD163 SRCR5.
After filtration, the supernatant containing the target protein was applied to a GE Ni Sepharose excel column preequilibrated with 20 mM Tris-HCl, pH 7.4, 150 mM NaCl. The target protein was then eluted with 20 mM Tris-HCl, pH 7.4, 150 mM NaCl, and 200 mM imidazole. Subsequently, the elution solution containing the target protein was purified by gel filtration chromatography using a GE Superdex 200 10/300 GL prepacked column on the Bio-Rad BioLogic system with 20 mM Tris-HCl, pH 7.4, 150 mM NaCl as the elution buffer. The fraction containing the target protein was collected, dialyzed, and concentrated using a Millipore ultracentrifugation tube (Merck) to 6 mg/ml in 20 mM Tris-HCl, pH 7.4, 20 mM NaCl.
Crystallization, data collection, and structural determination of pCD163 SRCR5.
Crystallization of pCD163 SRCR5 was carried out at room temperature (RT; 25°C) by the sitting-drop vapor diffusion method with an equal volume of the target protein at 6 mg/ml and various crystallization reagents from the crystallization screening kits (Hampton). The primary crystals were acquired with a reservoir solution containing 25% (wt/vol) polyethylene glycol (PEG) 4000, 200 mM (NH4)2SO4 in 100 mM sodium acetate at pH 4.6. Optimization trials were continued to obtain single crystals for data collection.
The crystals were flash-frozen in liquid nitrogen using a cryoprotection solution with 20% glycerol in the crystallization solution. X-ray data sets of the crystals were collected at a wavelength of 0.979 Å and a temperature of 100 K on the beamline BL17U1 at the Shanghai Synchrotron Radiation Facility (SSRF).
Diffraction data sets were processed using the automated data processing pipeline Xia2 with options that ran XDS (71). The initial phase for pCD163 SRCR5 was determined with the molecular replacement (MR) method using the program Molrep in CCP4 suites (72) and the coordinates of the Mac-2 binding protein (M2BP) SRCR domain (PDB code 1BY2) as a search model (33). The structure of pCD163 SRCR5 was refined by the CCP4 program package (72) and manually adjusted iteratively until convergence of the refinement with the molecular graphics program COOT (73). Solvent molecules were added using an Fo-Fc Fourier difference map at 2.5 σ in the final refinement step. Statistics from the data collection and the final model refinement for the structure are summarized in Table 1. The structural figures were generated and aligned with PyMOL (74).
Site-directed mutagenesis of SRCR5 in pCD163.
A construct with complete wild-type pCD163 cDNA integrated into the PiggyBac transposon system (kindly provided by Enmin Zhou, Northwest Agriculture & Forestry University, China) (59) was utilized as a template to generate each single-site mutant encoding pCD163 R478A, D483A, D496A, R561A, D563A, or R570A (the numbering is according to UniProt entry Q2VL90). The primers designed for mutation are listed in Table 2. All mutation constructs were verified by Shanghai Sangon Biotech Co. Ltd.
Cell transfection with wild-type or mutant pCD163.
PK-15 cells were seeded at a density of 4.0 × 105 cells/ml culture medium and incubated overnight. Transfection of PK-15 cells with the same amount of wild-type or mutant pCD163 was then performed using Lipofectamine LTX reagent with Plus reagent according to the manufacturer's instructions (Invitrogen). The expression levels of wild-type pCD163 for different periods (12 h, 24 h, and 48 h) and those of wild-type and each mutant pCD163 at 24 h posttransfection were monitored by flow cytometry. Blank PK-15 cells were also assayed for pCD163 expression as a negative control.
Flow cytometry analyses.
The transfected PK-15 cells were suspended in 1% bovine serum albumin–phosphate-buffered saline-Tween 20 (BSA-PBST) buffer. After centrifugation at 200 × g at 4°C for 5 min, the cells were incubated for 30 min on ice with a commercial mouse anti-pCD163 monoclonal antibody (MCA2311GA; AbD Serotec) or isotype control antibody in 1% BSA–PBST buffer. After washing, the cells were incubated for 30 min on ice with Cy3-conjugated goat anti-mouse IgG (H+L) (Abbkine) in 1% BSA–PBST buffer. After washing, the cells were resuspended with 0.5% paraformaldehyde in BSA–PBST buffer and analyzed. Data on 2.0 × 104 cells were acquired using a FACSCalibur (BD Bioscience) and analyzed by CellQuest Pro software.
PRRSV infection assay.
The transfected PK-15 cells were inoculated with PRRSV strain BJ-4 at a multiplicity of infection (MOI) of 1 at 37°C for 3 h. The viruses not entering into the cells were washed away with sterile culture medium. PPRSV infection was then analyzed by quantitative real-time PCR at 12 h postinoculation (38). Briefly, total RNA from infected PK-15 cells was extracted with TRIzol reagent (Ambion). Viral RNA was quantitated with primers designed for PRRSV ORF7 (listed in Table 2). A plasmid containing PRRSV ORF7 was used as the template to generate a standard curve, and the actual viral RNA copies were calculated based on it. Three replicates of wild-type or each mutant pCD163 were run, and each experiment was independently repeated three times.
PRRSV titration assay.
The transfected PK-15 cells were inoculated with PRRSV strain BJ-4 or HN07-1 at an MOI of 1. The cells were incubated at 37°C for 3 h, and the viruses not entering into the cells were then washed away. At 48 h postinfection, the virus yields were measured by the 50% tissue culture infected dose (TCID50) assay in MARC-145 cells. MARC-145 cells were seeded at a density of 4.0 × 105 cells/ml culture medium in each well of 96-well multiwell plates before inoculation. The cells were then inoculated with PRRSV at a 10-fold serial dilution at 37°C for 1 h. The viruses not entering into the cells were then washed away, and viral titers were determined 5 days postinfection.
PRRSV binding and entry assays.
For PRRSV binding assay (38, 39), the transfected cells were inoculated with PRRSV strain BJ-4 or HN07-1 at an MOI of 1 and incubated at 4°C for 1 h. After the unbound viruses washed away, the level of cell-bound viral RNA (PRRSV ORF7) was normalized with housekeeping glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA and relatively quantified by the 2−ΔΔCT method. For PRRSV entry assay (38, 39), the unbound viruses were washed away and the inoculated cells were then transferred to 37°C for 3 h to allow virus entry. The viruses not entering into the cells were washed, and the entering viral RNA was analyzed by the 2−ΔΔCT method similarly. In parallel, PRRSV infection at 12 h postinoculation was sequentially carried out for comparison. Three replicates of R561A mutant and wild-type pCD163 were run, and each experiment was independently repeated three times.
Statistical analysis.
All experimental data are presented as group means and standard errors of the means (SEM). The experimental data were analyzed using the unpaired, 2-tailed Student t test with Origin software. Differences at the 95% confidence level (P < 0.05) were considered statistically significant.
Accession number(s).
The coordinates of pCD163 SRCR5 were deposited in the Protein Data Bank (PDB code 5JFB).
ACKNOWLEDGMENTS
We thank the staff of the BL17U1 beamline at Shanghai Synchrotron Radiation Facility, Shanghai, People's Republic of China, for assistance during data collection. We thank Norman A. Gregson from University College London for revising the paper and advice.
This work was supported by the Key Project of National Natural Science Funds (31490601), the National Key Basic Research Plan (2014CB542702), and National Natural Science Funds of China (31272546, 31602036, and 31400637). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
We have no conflicts of interest to declare.
REFERENCES
- 1.Keffaber K. 1989. Reproductive failure of unknown etiol-ogy. Am Assoc Swine Pract Newsl 1:1–9. [Google Scholar]
- 2.Neumann EJ, Kliebenstein JB, Johnson CD, Mabry JW, Bush EJ, Seitzinger AH, Green AL, Zimmerman JJ. 2005. Assessment of the economic impact of porcine reproductive and respiratory syndrome on swine production in the United States. J Am Vet Med Assoc 227:385–392. doi: 10.2460/javma.2005.227.385. [DOI] [PubMed] [Google Scholar]
- 3.Lunney JK, Benfield DA, Rowland RR. 2010. Porcine reproductive and respiratory syndrome virus: an update on an emerging and re-emerging viral disease of swine. Virus Res 154:1–6. doi: 10.1016/j.virusres.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Done SH, Paton DJ. 1995. Porcine reproductive and respiratory syndrome: clinical disease, pathology and immunosuppression. Vet Rec 136:32–35. doi: 10.1136/vr.136.2.32. [DOI] [PubMed] [Google Scholar]
- 5.Zimmerman JJ, Yoon KJ, Wills RW, Swenson SL. 1997. General overview of PRRSV: a perspective from the United States. Vet Microbiol 55:187–196. doi: 10.1016/S0378-1135(96)01330-2. [DOI] [PubMed] [Google Scholar]
- 6.Rossow KD. 1998. Porcine reproductive and respiratory syndrome. Vet Pathol 35:1–20. doi: 10.1177/030098589803500101. [DOI] [PubMed] [Google Scholar]
- 7.Wensvoort G, Terpstra C, Pol JM, ter Laak EA, Bloemraad M, de Kluyver EP, Kragten C, van Buiten L, den Besten A, Wagenaar F. 1991. Mystery swine disease in The Netherlands: the isolation of Lelystad virus. Vet Q 13:121–130. doi: 10.1080/01652176.1991.9694296. [DOI] [PubMed] [Google Scholar]
- 8.Collins JE, Benfield DA, Christianson WT, Harris L, Hennings JC, Shaw DP, Goyal SM, McCullough S, Morrison RB, Joo HS, Gorcyca D, Chladek D. 1992. Isolation of swine infertility and respiratory syndrome virus (isolate ATCC VR-2332) in North America and experimental reproduction of the disease in gnotobiotic pigs. J Vet Diagn Investig 4:117–126. doi: 10.1177/104063879200400201. [DOI] [PubMed] [Google Scholar]
- 9.Nelsen CJ, Murtaugh MP, Faaberg KS. 1999. Porcine reproductive and respiratory syndrome virus comparison: divergent evolution on two continents. J Virol 73:270–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tian K, Yu X, Zhao T, Feng Y, Cao Z, Wang C, Hu Y, Chen X, Hu D, Tian X, Liu D, Zhang S, Deng X, Ding Y, Yang L, Zhang Y, Xiao H, Qiao M, Wang B, Hou L, Wang X, Yang X, Kang L, Sun M, Jin P, Wang S, Kitamura Y, Yan J, Gao GF. 2007. Emergence of fatal PRRSV variants: unparalleled outbreaks of atypical PRRS in China and molecular dissection of the unique hallmark. PLoS One 2:e526. doi: 10.1371/journal.pone.0000526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tong GZ, Zhou YJ, Hao XF, Tian ZJ, An TQ, Qiu HJ. 2007. Highly pathogenic porcine reproductive and respiratory syndrome, China. Emerg Infect Dis 13:1434–1436. doi: 10.3201/eid1309.070399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conzelmann KK, Visser N, Van Woensel P, Thiel HJ. 1993. Molecular characterization of porcine reproductive and respiratory syndrome virus, a member of the arterivirus group. Virology 193:329–339. doi: 10.1006/viro.1993.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dokland T. 2010. The structural biology of PRRSV. Virus Res 154:86–97. doi: 10.1016/j.virusres.2010.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Plagemann PG, Moennig V. 1992. Lactate dehydrogenase-elevating virus, equine arteritis virus, and simian hemorrhagic fever virus: a new group of positive-strand RNA viruses. Adv Virus Res 41:99–192. doi: 10.1016/S0065-3527(08)60036-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cavanagh D. 1997. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch Virol 142:629–633. [PubMed] [Google Scholar]
- 16.Snijder EJ, Meulenberg JJ. 1998. The molecular biology of arteriviruses. J Gen Virol 79(Part 5):961–979. doi: 10.1099/0022-1317-79-5-961. [DOI] [PubMed] [Google Scholar]
- 17.Duan X, Nauwynck HJ, Pensaert MB. 1997. Virus quantification and identification of cellular targets in the lungs and lymphoid tissues of pigs at different time intervals after inoculation with porcine reproductive and respiratory syndrome virus (PRRSV). Vet Microbiol 56:9–19. doi: 10.1016/S0378-1135(96)01347-8. [DOI] [PubMed] [Google Scholar]
- 18.Kim HS, Kwang J, Yoon IJ, Joo HS, Frey ML. 1993. Enhanced replication of porcine reproductive and respiratory syndrome (PRRS) virus in a homogeneous subpopulation of MA-104 cell line. Arch Virol 133:477–483. doi: 10.1007/BF01313785. [DOI] [PubMed] [Google Scholar]
- 19.Nauwynck HJ, Duan X, Favoreel HW, Van Oostveldt P, Pensaert MB. 1999. Entry of porcine reproductive and respiratory syndrome virus into porcine alveolar macrophages via receptor-mediated endocytosis. J Gen Virol 80(Part 2):297–305. doi: 10.1099/0022-1317-80-2-297. [DOI] [PubMed] [Google Scholar]
- 20.Van Breedam W, Delputte PL, Van Gorp H, Misinzo G, Vanderheijden N, Duan X, Nauwynck HJ. 2010. Porcine reproductive and respiratory syndrome virus entry into the porcine macrophage. J Gen Virol 91:1659–1667. doi: 10.1099/vir.0.020503-0. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Q, Yoo D. 2015. PRRS virus receptors and their role for pathogenesis. Vet Microbiol 177:229–241. doi: 10.1016/j.vetmic.2015.04.002. [DOI] [PubMed] [Google Scholar]
- 22.Jusa ER, Inaba Y, Kouno M, Hirose O. 1997. Effect of heparin on infection of cells by porcine reproductive and respiratory syndrome virus. Am J Vet Res 58:488–491. [PubMed] [Google Scholar]
- 23.Duan X, Nauwynck HJ, Favoreel HW, Pensaert MB. 1998. Identification of a putative receptor for porcine reproductive and respiratory syndrome virus on porcine alveolar macrophages. J Virol 72:4520–4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Calvert JG, Slade DE, Shields SL, Jolie R, Mannan RM, Ankenbauer RG, Welch SK. 2007. CD163 expression confers susceptibility to porcine reproductive and respiratory syndrome viruses. J Virol 81:7371–7379. doi: 10.1128/JVI.00513-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shanmukhappa K, Kim JK, Kapil S. 2007. Role of CD151, a tetraspanin, in porcine reproductive and respiratory syndrome virus infection. Virol J 4:62. doi: 10.1186/1743-422X-4-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim JK, Fahad AM, Shanmukhappa K, Kapil S. 2006. Defining the cellular target(s) of porcine reproductive and respiratory syndrome virus blocking monoclonal antibody 7G10. J Virol 80:689–696. doi: 10.1128/JVI.80.2.689-696.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang YW, Dryman BA, Li W, Meng XJ. 2009. Porcine DC-SIGN: molecular cloning, gene structure, tissue distribution and binding characteristics. Dev Comp Immunol 33:464–480. doi: 10.1016/j.dci.2008.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whitworth KM, Rowland RR, Ewen CL, Trible BR, Kerrigan MA, Cino-Ozuna AG, Samuel MS, Lightner JE, McLaren DG, Mileham AJ, Wells KD, Prather RS. 2015. Gene-edited pigs are protected from porcine reproductive and respiratory syndrome virus. Nat Biotechnol 34:20–22. doi: 10.1038/nbt.3434. [DOI] [PubMed] [Google Scholar]
- 29.Sarrias MR, Gronlund J, Padilla O, Madsen J, Holmskov U, Lozano F. 2004. The scavenger receptor cysteine-rich (SRCR) domain: an ancient and highly conserved protein module of the innate immune system. Crit Rev Immunol 24:1–37. doi: 10.1615/CritRevImmunol.v24.i1.10. [DOI] [PubMed] [Google Scholar]
- 30.Areschoug T, Gordon S. 2009. Scavenger receptors: role in innate immunity and microbial pathogenesis. Cell Microbiol 11:1160–1169. doi: 10.1111/j.1462-5822.2009.01326.x. [DOI] [PubMed] [Google Scholar]
- 31.Van Gorp H, Delputte PL, Nauwynck HJ. 2010. Scavenger receptor CD163, a Jack-of-all-trades and potential target for cell-directed therapy. Mol Immunol 47:1650–1660. doi: 10.1016/j.molimm.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 32.Van Gorp H, Van Breedam W, Van Doorsselaere J, Delputte PL, Nauwynck HJ. 2010. Identification of the CD163 protein domains involved in infection of the porcine reproductive and respiratory syndrome virus. J Virol 84:3101–3105. doi: 10.1128/JVI.02093-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hohenester E, Sasaki T, Timpl R. 1999. Crystal structure of a scavenger receptor cysteine-rich domain sheds light on an ancient superfamily. Nat Struct Biol 6:228–232. doi: 10.1038/6669. [DOI] [PubMed] [Google Scholar]
- 34.Ojala JR, Pikkarainen T, Tuuttila A, Sandalova T, Tryggvason K. 2007. Crystal structure of the cysteine-rich domain of scavenger receptor MARCO reveals the presence of a basic and an acidic cluster that both contribute to ligand recognition. J Biol Chem 282:16654–16666. doi: 10.1074/jbc.M701750200. [DOI] [PubMed] [Google Scholar]
- 35.Rodamilans B, Munoz IG, Bragado-Nilsson E, Sarrias MR, Padilla O, Blanco FJ, Lozano F, Montoya G. 2007. Crystal structure of the third extracellular domain of CD5 reveals the fold of a group B scavenger cysteine-rich receptor domain. J Biol Chem 282:12669–12677. doi: 10.1074/jbc.M611699200. [DOI] [PubMed] [Google Scholar]
- 36.Garza-Garcia A, Esposito D, Rieping W, Harris R, Briggs C, Brown MH, Driscoll PC. 2008. Three-dimensional solution structure and conformational plasticity of the N-terminal scavenger receptor cysteine-rich domain of human CD5. J Mol Biol 378:129–144. doi: 10.1016/j.jmb.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 37.Chappell PE, Garner LI, Yan J, Metcalfe C, Hatherley D, Johnson S, Robinson CV, Lea SM, Brown MH. 2015. Structures of CD6 and its ligand CD166 give insight into their interaction. Structure 23:1426–1436. doi: 10.1016/j.str.2015.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang Q, Zhang Q, Tang J, Feng WH. 2015. Lipid rafts both in cellular membrane and viral envelope are critical for PRRSV efficient infection. Virology 484:170–180. doi: 10.1016/j.virol.2015.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao J, Xiao S, Xiao Y, Wang X, Zhang C, Zhao Q, Nan Y, Huang B, Liu H, Liu N, Lv J, Du T, Sun Y, Mu Y, Wang G, Syed SF, Zhang G, Hiscox JA, Goodfellow I, Zhou EM. 2016. MYH9 is an essential factor for porcine reproductive and respiratory syndrome virus infection. Sci Rep 6:25120. doi: 10.1038/srep25120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vanderheijden N, Delputte PL, Favoreel HW, Vandekerckhove J, Van Damme J, van Woensel PA, Nauwynck HJ. 2003. Involvement of sialoadhesin in entry of porcine reproductive and respiratory syndrome virus into porcine alveolar macrophages. J Virol 77:8207–8215. doi: 10.1128/JVI.77.15.8207-8215.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Delputte PL, Nauwynck HJ. 2004. Porcine arterivirus infection of alveolar macrophages is mediated by sialic acid on the virus. J Virol 78:8094–8101. doi: 10.1128/JVI.78.15.8094-8101.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Delputte PL, Costers S, Nauwynck HJ. 2005. Analysis of porcine reproductive and respiratory syndrome virus attachment and internalization: distinctive roles for heparan sulphate and sialoadhesin. J Gen Virol 86:1441–1445. doi: 10.1099/vir.0.80675-0. [DOI] [PubMed] [Google Scholar]
- 43.Delputte PL, Van Breedam W, Delrue I, Oetke C, Crocker PR, Nauwynck HJ. 2007. Porcine arterivirus attachment to the macrophage-specific receptor sialoadhesin is dependent on the sialic acid-binding activity of the N-terminal immunoglobulin domain of sialoadhesin. J Virol 81:9546–9550. doi: 10.1128/JVI.00569-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van Gorp H, Van Breedam W, Delputte PL, Nauwynck HJ. 2008. Sialoadhesin and CD163 join forces during entry of the porcine reproductive and respiratory syndrome virus. J Gen Virol 89:2943–2953. doi: 10.1099/vir.0.2008/005009-0. [DOI] [PubMed] [Google Scholar]
- 45.Van Breedam W, Van Gorp H, Zhang JQ, Crocker PR, Delputte PL, Nauwynck HJ. 2010. The M/GP(5) glycoprotein complex of porcine reproductive and respiratory syndrome virus binds the sialoadhesin receptor in a sialic acid-dependent manner. PLoS Pathog 6:e1000730. doi: 10.1371/journal.ppat.1000730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Delrue I, Van Gorp H, Van Doorsselaere J, Delputte PL, Nauwynck HJ. 2010. Susceptible cell lines for the production of porcine reproductive and respiratory syndrome virus by stable transfection of sialoadhesin and CD163. BMC Biotechnol 10:48. doi: 10.1186/1472-6750-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Delputte PL, Van Gorp H, Favoreel HW, Hoebeke I, Delrue I, Dewerchin H, Verdonck F, Verhasselt B, Cox E, Nauwynck HJ. 2011. Porcine sialoadhesin (CD169/Siglec-1) is an endocytic receptor that allows targeted delivery of toxins and antigens to macrophages. PLoS One 6:e16827. doi: 10.1371/journal.pone.0016827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van Breedam W, Verbeeck M, Christiaens I, Van Gorp H, Nauwynck HJ. 2013. Porcine, murine and human sialoadhesin (Sn/Siglec-1/CD169): portals for porcine reproductive and respiratory syndrome virus entry into target cells. J Gen Virol 94:1955–1960. doi: 10.1099/vir.0.053082-0. [DOI] [PubMed] [Google Scholar]
- 49.Prather RS, Rowland RR, Ewen C, Trible B, Kerrigan M, Bawa B, Teson JM, Mao J, Lee K, Samuel MS, Whitworth KM, Murphy CN, Egen T, Green JA. 2013. An intact sialoadhesin (Sn/SIGLEC1/CD169) is not required for attachment/internalization of the porcine reproductive and respiratory syndrome virus. J Virol 87:9538–9546. doi: 10.1128/JVI.00177-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kreutz LC, Ackermann MR. 1996. Porcine reproductive and respiratory syndrome virus enters cells through a low pH-dependent endocytic pathway. Virus Res 42:137–147. doi: 10.1016/0168-1702(96)01313-5. [DOI] [PubMed] [Google Scholar]
- 51.Das PB, Dinh PX, Ansari IH, de Lima M, Osorio FA, Pattnaik AK. 2010. The minor envelope glycoproteins GP2a and GP4 of porcine reproductive and respiratory syndrome virus interact with the receptor CD163. J Virol 84:1731–1740. doi: 10.1128/JVI.01774-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Du Y, Pattnaik AK, Song C, Yoo D, Li G. 2012. Glycosyl-phosphatidylinositol (GPI)-anchored membrane association of the porcine reproductive and respiratory syndrome virus GP4 glycoprotein and its co-localization with CD163 in lipid rafts. Virology 424:18–32. doi: 10.1016/j.virol.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wissink EH, van Wijk HA, Pol JM, Godeke GJ, van Rijn PA, Rottier PJ, Meulenberg JJ. 2003. Identification of porcine alveolar macrophage glycoproteins involved in infection of porcine respiratory and reproductive syndrome virus. Arch Virol 148:177–187. doi: 10.1007/s00705-002-0897-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Das PB, Vu HL, Dinh PX, Cooney JL, Kwon B, Osorio FA, Pattnaik AK. 2011. Glycosylation of minor envelope glycoproteins of porcine reproductive and respiratory syndrome virus in infectious virus recovery, receptor interaction, and immune response. Virology 410:385–394. doi: 10.1016/j.virol.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 55.Wei Z, Tian D, Sun L, Lin T, Gao F, Liu R, Tong G, Yuan S. 2012. Influence of N-linked glycosylation of minor proteins of porcine reproductive and respiratory syndrome virus on infectious virus recovery and receptor interaction. Virology 429:1–11. doi: 10.1016/j.virol.2012.03.010. [DOI] [PubMed] [Google Scholar]
- 56.Li J, Murtaugh MP. 2015. Functional analysis of porcine reproductive and respiratory syndrome virus N-glycans in infection of permissive cells. Virology 477:82–88. doi: 10.1016/j.virol.2015.01.005. [DOI] [PubMed] [Google Scholar]
- 57.Welch SK, Calvert JG. 2010. A brief review of CD163 and its role in PRRSV infection. Virus Res 154:98–103. doi: 10.1016/j.virusres.2010.07.018. [DOI] [PubMed] [Google Scholar]
- 58.Graversen JH, Madsen M, Moestrup SK. 2002. CD163: a signal receptor scavenging haptoglobin-hemoglobin complexes from plasma. Int J Biochem Cell Biol 34:309–314. doi: 10.1016/S1357-2725(01)00144-3. [DOI] [PubMed] [Google Scholar]
- 59.Wang X, Wei R, Li Q, Liu H, Huang B, Gao J, Mu Y, Wang C, Hsu WH, Hiscox JA, Zhou EM. 2013. PK-15 cells transfected with porcine CD163 by PiggyBac transposon system are susceptible to porcine reproductive and respiratory syndrome virus. J Virol Methods 193:383–390. doi: 10.1016/j.jviromet.2013.06.035. [DOI] [PubMed] [Google Scholar]
- 60.Chen Y, Guo R, He S, Zhang X, Xia X, Sun H. 2014. Additive inhibition of porcine reproductive and respiratory syndrome virus infection with the soluble sialoadhesin and CD163 receptors. Virus Res 179:85–92. doi: 10.1016/j.virusres.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 61.Kristiansen M, Graversen JH, Jacobsen C, Sonne O, Hoffman HJ, Law SK, Moestrup SK. 2001. Identification of the haemoglobin scavenger receptor. Nature 409:198–201. doi: 10.1038/35051594. [DOI] [PubMed] [Google Scholar]
- 62.Fabriek BO, Polfliet MM, Vloet RP, van der Schors RC, Ligtenberg AJ, Weaver LK, Geest C, Matsuno K, Moestrup SK, Dijkstra CD, van den Berg TK. 2007. The macrophage CD163 surface glycoprotein is an erythroblast adhesion receptor. Blood 109:5223–5229. doi: 10.1182/blood-2006-08-036467. [DOI] [PubMed] [Google Scholar]
- 63.Bover LC, Cardo-Vila M, Kuniyasu A, Sun J, Rangel R, Takeya M, Aggarwal BB, Arap W, Pasqualini R. 2007. A previously unrecognized protein-protein interaction between TWEAK and CD163: potential biological implications. J Immunol 178:8183–8194. doi: 10.4049/jimmunol.178.12.8183. [DOI] [PubMed] [Google Scholar]
- 64.Moreno JA, Munoz-Garcia B, Martin-Ventura JL, Madrigal-Matute J, Orbe J, Paramo JA, Ortega L, Egido J, Blanco-Colio LM. 2009. The CD163-expressing macrophages recognize and internalize TWEAK: potential consequences in atherosclerosis. Atherosclerosis 207:103–110. doi: 10.1016/j.atherosclerosis.2009.04.033. [DOI] [PubMed] [Google Scholar]
- 65.Fabriek BO, van Bruggen R, Deng DM, Ligtenberg AJ, Nazmi K, Schornagel K, Vloet RP, Dijkstra CD, van den Berg TK. 2009. The macrophage scavenger receptor CD163 functions as an innate immune sensor for bacteria. Blood 113:887–892. doi: 10.1182/blood-2008-07-167064. [DOI] [PubMed] [Google Scholar]
- 66.Sanchez-Torres C, Gomez-Puertas P, Gomez-del-Moral M, Alonso F, Escribano JM, Ezquerra A, Dominguez J. 2003. Expression of porcine CD163 on monocytes/macrophages correlates with permissiveness to African swine fever infection. Arch Virol 148:2307–2323. doi: 10.1007/s00705-003-0188-4. [DOI] [PubMed] [Google Scholar]
- 67.Cai Y, Postnikova EN, Bernbaum JG, Yu SQ, Mazur S, Deiuliis NM, Radoshitzky SR, Lackemeyer MG, McCluskey A, Robinson PJ, Haucke V, Wahl-Jensen V, Bailey AL, Lauck M, Friedrich TC, O'Connor DH, Goldberg TL, Jahrling PB, Kuhn JH. 2015. Simian hemorrhagic fever virus cell entry is dependent on CD163 and uses a clathrin-mediated endocytosis-like pathway. J Virol 89:844–856. doi: 10.1128/JVI.02697-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hou J, Li R, Ma H, Qiao S, Zhang G. 2016. Structural prediction of porcine sialoadhesin V-set Ig-like domain sheds some light on its role in porcine reproductive and respiratory syndrome virus (PRRSV) infection. Front Agric Sci Eng 3:65–71. doi: 10.15302/J-FASE-2016086. [DOI] [Google Scholar]
- 69.Yuan C, Huai Q, Bian CB, Huang M. 2006. The expression, purification and crystallization of monomeric soluble human urokinase receptor. Prog Biochem Biophys 33:277–281. [Google Scholar]
- 70.Qiao S, Feng L, Bao D, Guo J, Wan B, Xiao Z, Yang S, Zhang G. 2011. Porcine reproductive and respiratory syndrome virus and bacterial endotoxin act in synergy to amplify the inflammatory response of infected macrophages. Vet Microbiol 149:213–220. doi: 10.1016/j.vetmic.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 71.Winter G. 2010. xia2: an expert system for macromolecular crystallography data reduction. J Appl Crystallogr 43:186–190. doi: 10.1107/S0021889809045701. [DOI] [Google Scholar]
- 72.CCP4. 1994. The CCP4 suite. Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 73.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 74.DeLano WL. 2002. The PyMOL molecular graphics system. DeLano Scientific, Palo Alto, CA. [Google Scholar]